Abstract
Objective
Receptors and ion channels of smooth muscle cells (SMCs) and endothelial cells (ECs) are integral to the regulation of vessel diameter and tissue blood flow. Physiological roles of ion channels and receptors in skeletal muscle and mesenteric arteries have been identified however their gene expression profiles are undefined. We tested the hypothesis that expression profiles for ion channels and receptors governing vascular reactivity vary with cell type, vascular bed and age.
Methods
Mesenteric and superior epigastric arteries were dissected from Old (24–26 months) and Young (3–6 months) C57BL/6J mice. ECs and SMCs were collected for analysis with custom qRT-PCR arrays to determine expression profiles of 80 ion channel and receptor genes. Bioinformatics analyses were applied to gain insight into functional interactions.
Results
We identified 68 differences in gene expression with respect to cell type, vessel type and age. Heat maps illustrate differential expression and distance matrices predict patterns of co-expression. Gene networks based upon protein-protein interaction datasets and KEGG pathways illustrate biological processes affected by specific differences in gene expression.
Conclusion
Differences in gene expression profiles are most pronounced between microvascular ECs and SMCs with subtle variations between vascular beds and age groups.
Keywords: bioinformatics, gene expression, network construction, vascular endothelium, vascular smooth muscle
INTRODUCTION
Ion channels and receptors mediate vasomotor control in resistance arteries by regulating membrane potential (Vm), intracellular [Ca2+] and complementary signaling events in both vascular smooth muscle cells (SMCs) and endothelial cells (ECs)1,2. The present study was designed to investigate representative categories of ion channels and receptors to determine how their gene expression profiles vary with cell type, vascular bed and biological age. Mesenteric and superior epigastric arteries were chosen for two reasons. First, these vessels have been the subject of physiological studies in young and old mice 3–6, thus analyses of gene expression profiles provide a novel context to identify directions for future research in aging. Second, inclusion of vessels from functionally distinct areas of the body enables comparison of expression profiles in tissues that differ in embryonic origin and function. The list of genes chosen for this study is not exhaustive for those involved in vasomotor control, however emphasis was placed on including genes families that are commonly studied (e.g. adrenergic and cholinergic receptors) as well as those whose role in vascular function are less clear (e.g. purinergic receptors). We questioned how gene expression in microvascular resistance arteries (diameters: 150–200 μm) may differ with cell type and vascular bed as well as how these relationships may be affected by aging. Each family of receptors and ion channels comprising the present study is introduced by its common protein name and gene symbol; these are listed collectively in Supplementary Table 1.
Calcium Channels
Calcium channels are critical for providing Ca2+ that activates SMC contraction 7 and are integral to endothelium-dependent SMC relaxation 8–10. Calcium channels mediate Ca2+ entry from the extracellular fluid, Ca2+ release from intracellular stores, and couple Ca2+ entry with store depletion. In the plasma membrane of SMCs, VGCCs open in response to depolarization, enabling Ca2+ influx. L-type VGCCs, including CaV 1.2 (Cacna1c) and 1.3 (Cacna1d), are a primary source of Ca2+ influx in vascular SMCs. In addition, P/Q- and N-type type VGCCs (CaV 2.1;Cacna1a and 2.2; Cacna1b) are also activated by depolarization and can be distinguished based on inhibition by ω-agitoxin IVA or ω-conotoxin GVIA, respectively 11–13. T-Type VGCCs (CaV 3.1;Cacna1g and 3.2;Cacna1h) activate at more negative Vm and produce transient Ca2+ currents 7,11. In ECs, multiple isoforms of plasma membrane TRP channel isoforms (discussed below) are primary sources of Ca2+ influx eliciting vasodilation through both NO production and EDH14,15.
Calcium channels in the endoplasmic reticulum (ER) of both ECs and SMCs include ryanodine receptors (Ryr1-3) and inositol 1,4,5-trisphosphate receptors (Itpr1-3). When activated, these channels release Ca2+ from internal stores. Activation of a cluster of RyRs (i.e. Ca2+ sparks) leads to SMC relaxation through activation of nearby BKCa channels while their collective activation elicits SMC contraction via a global increase in [Ca2+]i 1. In contrast to RyR, both local 16 and widespread activation of SMC IP3Rs lead to SMC contraction 17. In a reciprocal manner, activation of Ca2+ channels in the ER of ECs promotes SMC relaxation. Thus, local (Ca2+ pulsars) 18 and widespread (Ca2+ wave) activation 19 of IP3Rs in ECs leads to endothelium-dependent hyperpolarization (EDH) and the release of vasodilator autacoids (e.g., NO and prostacyclin). Three isoforms of RyRs 20 and IP3Rs participate in Ca2+ release,21 with distribution profiles that vary across vascular beds.
Store-operated Ca2+ entry in SMCs and ECs is mediated largely by STIMs (Stim1-2), which act as Ca2+ sensors on the ER stores. Upon store depletion, STIM1 translocates to the plasma membrane and interacts with a CRAC channel to form Orai1 (Orai1) tetramers, thereby allowing Ca2+ influx and refilling of ER Ca2+ stores 22. The TRP channels (discussed below), particularly TRPC channels, may also participate in this process through interacting with Stim1 and Orai 23.
Potassium Channels
Calcium-activated K+ channels are activated in response to an increase in [Ca2+]i 24,25, with K+ efflux leading to hyperpolarization. Subtypes of KCa channels are distinguished by their conductance into 3 categories: large (BKCa; Kcnma1, Kcnmb1), intermediate (IKCa; Kcnn4) and small (SKCa; Kcnn1, Kcnn2, Kcnn3). The BKCa channels are particularly important in SMCs, where they are central to negative feedback regulation of myogenic tone. For example, local activation of BKCa channels by Ca2+ sparks arising from nearby RyR clusters evokes STOCs 26 that oppose vasoconstriction. Membrane depolarization and/or global increases in [Ca2+]i, often from L-type VGCCs, leads to widespread BKCa activation that opposes SMC contraction 1,17. In contrast to BKCa, the voltage-insensitive IKCa and SKCa channels effect vasodilation primarily through their expression in the plasma membrane of ECs 27. In response to a rise in [Ca2+]i, activation of IKCa and SKCa channels evokes EDH and, through electrical coupling to SMCs via gap junctions, promotes vasodilation through closure of L-type VGCCs 9,28.
Inwardly rectifying K+ (Kir) channels exert their effects through conduction of strong inward current when Vm falls below the equilibrium potential for K+ and small outward currents when Vm rises above EK 29. Strong inward rectifiers (Kir 2.1; Kcnj2 and Kir 2.2; Kcnj12) are prominent subtypes in vascular SMCs, where their activation contributes to the maintenance of resting Vm 30. A unique property of Kir channels is their ability to amplify hyperpolarization through a negative slope conductance, however this role varies between vascular beds 31. Recent findings implicate a key role for Kir2.1 in capillary ECs of the brain to cerebral blood flow in response to an elevation of [K+]o 32. When coupled with sulfonylurea receptors, Kir6.1 (Kcnj8) and Kir6.2 (Kcnj11) form KATP channels, whose activation (e.g., by ADP) leads to membrane hyperpolarization and vasodilation 30,33. As with Kir channels, the role of KATP channels in regulating vasomotor tone and tissue blood flow varies between vascular beds 34,35.
Transient Receptor Potential (TRP) Channels
The TRP channels enable transmembrane flux of cations with varying selectivity and consist of several major families including canonical (TRPC; Trpc1-6), vanilloid (TRPV; Trpv1), melastatin (TRPM; Trpm4)), and ankyrin (TRPA; Trpa1) channels 15,36. TRP channels are highly expressed in both SMC and ECs, are activated by a wide range of physiological stimuli and are involved in the regulation of Vm, store-operated Ca2+ entry (SOCE) 37, receptor-operated Ca2+ entry (ROCE) 38, Ca2+ signaling 39,40 and myoendothelial signaling 41. Activation of TRP channels in SMCs typically leads to vasoconstriction, while activation TRP channels in ECs produces vasodilation as well as changes in permeability and angiogenesis. Recent reviews explores the specific physiological roles for each TRP family member and isoform in vascular SMCs1,15 and ECs15.
Neurotransmitter and Neuropeptide Receptors
Perivascular nerves regulate blood flow through the release of neurotransmitters, which modulate a multitude of downstream signaling events via binding to their receptors on membranes of SMCs, ECs and/or adjacent nerves 42. Activation of sympathetic nerves causes release of NE and NPY, resulting in vasoconstriction. Norepinephrine binds to adrenoreceptors (ARs) on both SMCs and ECs. The αARs predominate in most vascular beds and mediate vasoconstriction via Gq- (α1AR; Adra1a, Adra1b, Adra1c) and Gi- (α2AR; Adra2a, Adra2b, Adra2c) mediated increases in SMC [Ca2+]i 43. In turn, βARs (Adrb1, Adrb2, Adrb3) promote vasodilation through Gs-mediated decreases in SMC [Ca2+]i and release of NO from ECs 44. Neuropeptide Y effects vasoconstriction through activation of SMC Y1 (Npy1r) and Y2 (Npy2r) receptors, which produce Gα-mediated increases in SMC [Ca2+]i 45 and potentiate the effects of αAR agonists 46.
Many conduit and resistance arteries are also innervated by perivascular sensory nerves, which elicit vasodilation and oppose sympathetic vasoconstriction through the release of CGRP and SP. CGRP-mediated vasodilation requires assembly of its receptor components on SMCs and ECs, including RAMP1 (Ramp1), CALCRL (Calcrl) and RCP (Cgrprcp)47. Upon binding of CGRP, relaxation of SMCs occurs primarily through cAMP-mediated activation of K+ channels 48, while EC-dependent dilations can occur via cAMP and PKA-mediated increases in NO 49. Substance P effects vasodilation through binding to NK1 (Tacr1) receptors on ECs, which mediate increases in [Ca2+]i and NO production 45.
Purinergic Receptors
Perivascular sympathetic and sensory nerves each release ATP as a co-transmitter that, along with circulating purines, can bind to purinergic receptors on both SMCs and ECs 50. Arteries express two major classes of P2 receptors on SMCs, ECs and nerves which form homodimers and heterodimers of different subtypes and can produce either vasoconstriction or vasodilation depending on the specific isoform, cell type and vascular bed 42. P2X receptors (subtypes P2X1-7; P2rx1-7) are ATP-gated cation channels that mediate Ca2+ and/or Na+ influx and membrane depolarization which, in SMCs, activates of L-type VGCCs leading to increased [Ca2+]i. This signaling pathway is mediated via P2X1 receptors in SMCs from many vascular beds 51; few studies address the role of other P2X isoforms due to limited specific pharmacological agents. P2X receptors are also expressed to a lesser extent on ECs, with activation contributing to vasodilation via EDH along with production of NO and prostacyclin 52.
In contrast to the ionotropic P2X receptors, P2Y receptors (P2ry1,2,4,6,11,12,13,14) are metabotropic, eliciting their effects on SMCs and ECs through activation of G-proteins following ATP, ADP, or UTP binding in accord with receptor subtype 53. Analogous to the effects of P2X receptor activation, P2Y activation on ECs leads to vasodilation via increased [Ca2+]i, NO production, and EDH, with the relative contributions of NO vs. EDH varying with vessel size and vascular bed 54. Multiple P2Y subtypes also have been reported in SMCs, with most (but not all) studies demonstrating that their activation leads to vasoconstriction via increased [Ca2+]i 55,56. Overall, purinergic signaling is more complex and heterogeneous when compared to other receptor classes in the vasculature. For example, beyond their roles in vasomotor control, purinergic receptors mediate perivascular nerve activity, cell proliferation, cell migration and angiogenesis. A recent review 57 provides an in-depth analysis of purinergic receptor expression, pharmacology and function in the vasculature.
Peroxisome Proliferator-Activated Receptors
Peroxisome proliferator-activated receptors are nuclear receptors that modulate transcription following hormone activation, heterodimerization with retinoid X receptors and binding to PPAR response elements on DNA58. Included in this family are two subtypes studied here- PPARα (Ppara) and PPARγ (Pparg). Each subtype appears to be protective against cardiovascular disease through their roles in nutrient metabolism, angiogenesis, proliferation, cell migration, inflammation and oxidative stress 59,60. Although their roles in vasomotor control are not well defined, PPARs may facilitate normal endothelial function through NO- and NFκB-dependent signaling 61. For example, aortae of PPARα−/− mice exhibit impaired NO-signaling and increased oxidative stress 62 In SMCs, disruption of PPARγ via dominant negative mutation leads to hypercontractile responses to pharmacological agonists, likely through a RhoA-dependent mechanism 63 while inhibition of PPARγ increases myogenic constriction in mesenteric arteries 64, possibly via increased AngII signaling 65.
Angiotensin II Receptors
Angiotensin II receptors are G-protein coupled receptors of the peptide hormone angiotensin II, an integral mediator of the renin-angiotensin system as well as cardiovascular function in health and disease66. The main subtypes of these receptors are Type 1 (Agtr1a, Agtr1b) and Type 2 (Agtr2) receptors (AT1 and AT2, respectively). AT1a and AT1b receptors are responsible for the majority of physiological effects of angiotensin II and both are expressed in SMCs and ECs of resistance arteries 66. Activation of AT1a and AT1b receptors initiates multiple signaling cascades, ultimately contributing to vasoconstriction, cell proliferation, cell hypertrophy, superoxide production, endothelin release, lipid peroxidation, adhesion molecule expression and vascular matrix expansion (See 67 for in-depth review). Recent studies using cell-specific deletion of Agtr1a in either SMCs or ECs 68 highlight the critical role of these receptors in the pathogenesis of hypertension and aneurisms. AT2 receptors oppose the effects of AT1 activation by participating in vasodilation, apoptosis, growth inhibition, NO production and collagen synthesis. However, the roles of AT2 receptors in the vasculature require further study as expression profiles and vasomotor effects demonstrated in rodent vessels have not been replicated in human vessels 67.
Rationale and significance of bioinformatics analysis
While qRT-PCR data analysis and calculation of the ΔΔCT and 2−ΔΔCT values are standard approaches for identifying differential gene expression between experimental groups, there is a gap in understanding both common and unique changes happening between the cell type, vessel type and age groups in this study. Bioinformatics data analysis techniques allow mining of gene expression data and combine this information with publicly available knowledge from protein-protein interaction (PPI) datasets. Further insight into the biological processes affected in each comparison is provided by pathway mapping. These approaches are applied routinely for drawing inferences from other techniques used to evaluate gene expression (e.g., microarrays, RNAseq) and protein expression (e.g., mass spectrometry) but have rarely been used for qPCR studies.
Earlier studies have looked at the expression of single or subsets of respective genes in particular blood vessels to evaluate differences between vascular beds, cell type and to evaluate designated experimental conditions, however differences in techniques and experimental design across studies confound a comprehensive picture of ion channel and receptor expression in the resistance vasculature. The purpose of this study was twofold: First, to test the hypothesis that expression of 80 representative genes of ion channels and receptors associated with the regulation of tissue blood flow varies with: (1) cell type: SMC vs. EC, (2) vascular bed: MA vs. SEA, and (3) biological age: 3–6 months vs. 24–26 months. Second, to apply bioinformatics analyses to investigate relationships among the expression of these representative genes. Pathway and gene network analyses were thereby applied to explore how the present data may be applied to predict downstream function and signaling pathways.
MATERIALS AND METHODS
Animal Care
All procedures were approved by the University of Missouri Animal Care and Use Committee and were performed in accord with the National Research Council’s Guide for the Care and Use of Laboratory Animals. Experiments were performed using Young (3–6 months, n=5) and Old (24–26 months, n=5) C57Bl/6 male mice obtained from the National Institute on Aging Colony at Charles River (Wilmington, MA). Mice were housed at the University of Missouri (MU) animal facility for 1–4 weeks prior to use on a 12: 12 hour light: dark cycle maintained at ~23°C with fresh tap water and standard chow available ad libitum. Mice were anaesthetized with pentobarbital sodium (60 mg/kg) via intraperitoneal injection, the abdomen was shaved and, following confirmation of deep anesthesia, the chest cavity was opened, the heart was removed to ensure death by exsanguination and tissues were harvested for obtaining blood vessels.
Tissue Collection
Entire abdominal muscles were removed and SEAs were dissected bilaterally to free them from surrounding tissue as described 69. The intact mesentery and small intestine were excised and first- and second- order MAs were dissected as described 4. Several vessel segments were collected separately for each mouse then digested at 34°C (SEA: 30 min; MA: 20 min) in a buffer (pH 7.4) composed of (in mM): 137 NaCl, 5 KCl, 1 MgCl2, 10 HEPES, 10 glucose, 2 CaCl2, 0.1% bovine serum albumin (US Biochemical 10856, Cleveland, OH, USA), 0.62 mg/mL papain (P-4762; Sigma Chemical Co., St. Louis, MO, USA), 1.5 mg/mL collagenase type H (C-8051; Sigma), and 1 mg/mL dithioerythritol (D-8255; Sigma). Respective samples were transferred to a Petri dish on the stage of a microscope (GFL, Zeiss, Thornwood, NY, USA) and gently triturated through a heat-polished micropipette (internal diameter, 80–120 μm according to vessel diameters) secured in a micromanipulator. Cells were sampled while visualizing dispersed vessels at 200X magnification to ensure pure, cell-type specific samples. The endothelium was discerned as intact “tubes” 69 surrounded by dispersed SMCs that typically adopted a “C” shape on the bottom of the dish. For each vessel type, ~5,000 ECs and ~500 SMCs were collected per mouse by aspiration; a fresh micropipette was used for each cell type. Micropipettes were pulled (P-97; Sutter Instruments, Novato, CA, USA) from borosilicate glass capillary tubes (#1B100-4; World Precision Instruments, Sarasota, FL, USA) and heat-polished with a custom-built microforge.
RNA Extraction, cDNA Synthesis and qRT-PCR Array
Isolated cells were placed immediately into 100 μL of lysis buffer from the RNAqueous Micro Kit with DNase from Ambion Inc. (Austin, TX, USA) and total RNA was extracted per manufacturer’s guidelines in a final volume of 18 μL elution buffer. RNA samples were stored at −80 °C prior to their use in PCR arrays.
For positive controls, samples of intact tissues (brain, heart, small intestine and spleen) from a Young mouse were processed as follows: 50 mg tissue was flash frozen, pulverized and added to 1 mL TRI Reagent (T9424; Sigma) then incubated for 5 min at room temperature (RT). The supernatant containing RNA was then treated with chloroform (C2432; Sigma, 0.2 mL/mL TRI Reagent). Aqueous phase was separated by centrifugation at 12,000 × g for 15 min and RNA precipitated with isopropanol (I6504; Sigma, 0.5 mL/mL TRI Reagent) followed by centrifugation at 12,000 × g for 10 min. The formed pellet was washed with 1 mL of 75% EtOH made in diethyl pyrocarbonate-treated H2O, centrifuged at 10,000 × g for 5 min and air-dried. RNA was dissolved in RNase-free H2O by incubating at 58°C for 10 min. Samples were stored at −80°C until reverse transcribed.
Each RNA sample was evaluated using the Nanodrop 2000 and Qubit RNA HS Assay (ThermoFisher, Waltham, MA, USA) to determine the RNA concentration (2–6 ng/μL for ECs and SMCs; >1000 ng/μL for control tissues) and to verify its integrity 70 (OD260/OD280nm absorption ratio >1.8; OD260/OD230nm absorption ratio >1.5). RNA samples were diluted to 1 ng/μL prior to reverse transcription with the High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol. Resulting cDNA samples were stored at −20°C until used for qRT-PCR arrays.
The qRT-PCR was performed using Custom Taqman PreAmplification primer pools and custom Fast Taqman Array Plates created to our specifications by Applied Biosystems (Foster City, CA, USA). Each array was comprised of 40 target genes of interest, 5 endogenous control genes (18S, Dna2 Gapdh, Gusb, Actb) 71 and 2 cell type specific genes (Nos3 for EC and Acta2 for SMC) (Supplementary Table 1). Five biological replicates (each containing a single cell type collected from a separate mouse) were run from each of 8 experimental groups (Young SEA EC, Young SEA SMC, Young MA EC, Young MA SMC, Old SEA EC, Old SEA SMC, Old MA EC, Old MA SMC) according to the manufacturer‘s protocol. The run for each array began with a preamplification reaction (7.5ng cDNA in a 30 μL reaction) followed by PCR amplification using Applied Biosystems Taqman Fast Advanced Master Mix (10 μL reactions) on the Applied Biosystems Step One Plus Real Time PCR System using thermal cycling conditions stated by the manufacturer (95°C for 20 s; 95°C for 3 s, 60°C for 30 s; 40 cycles). A no RT reaction was also performed using the Gapdh primer to confirm lack of contamination from genomic DNA, along with an internal plate control. All assays were inventoried products from Applied Biosystems (Supplementary Table 1), validated to meet their quality control specifications. Control tissues (above) verified assays not previously run to ensure their reactivity and efficiency (data not shown). In total, 44 assay plates were used to complete the study.
Data Analysis for qRT-PCR Array
The qRT-PCR array datasets were analyzed with the ΔΔCT method 72. Using Expression Suite Software V1.0.3 (Applied Biosystems) 73, CT values for all samples were calculated, where CT is defined as the fractional cycle number at which fluorescence exceeded the fixed 0.2 threshold. CT values were exported to Microsoft Excel and (in accord with manufacturer’s guidelines), for samples where a CT value was reported as “Undetermined” (i.e., never reached threshold), a value of 40 (the total number of cycles) was substituted to allow statistical analysis. Expression Suite determined 18S as the ideal endogenous control gene based upon stable expression across cell types, vessel types and ages. The distribution of average CT values across the groups ranged from 10 to ≤ 40 (Supplementary Table 2, Supplementary Figures 1–3). To address outliers in the biological replicates within groups, we performed a normalization technique prior to further analysis as follows: If all other replicates within a group for a specific gene had a CT =40, the outlier replicates were replaced with 40. If the statistical outlier had a CT < 40, the value was replaced with the mean of the remaining replicates within the group. Δ CT, ΔΔCT and fold change (Supplementary Tables 2–5) were calculated for each experimental group, using the following formulae:
-
(i)
-
(ii)
-
(iiii)
-
(iv)
The formula 2−ΔΔCT was applied to the analysis of relative expression (i.e., fold change) 72. For each gene in the arrays, fold-change comparisons were made based on cell type (SMC vs EC), vessel type (MA vs SEA), and age (Old vs Young) across each experimental group. An unpaired, equal variance t-test was used to compare average CT values from the five biological replicates within each experimental group to determine significant differences (P ≤ 0.05) and thereby identify genes that were differentially expressed based upon cell type, vessel type and age group. P-values were then adjusted to account for multiple comparisons using the Benjamini-Hochberg procedure74 with P-values remaining < 0.05 considered significant. All adjusted P-values are listed alongside fold-change values in Supplementary Tables 3–5. Variance box plots (Supplementary Figures 1–3) show the variance of CT measurements for biological replicates across all genes used to calculate the differential expression levels of the genes for each comparison. Each “Box” represents the range of values for the expression level of all genes in one replicate, along with the standard deviation. The plots were created using R software standard libraries, with ggplot2 and reshape libraries used to calculate the variance and create the boxplots.
TIGR Multiexperiment Viewer (T-MeV; version 4.9) 75 was used to perform hierarchical clustering, heat map visualization, coexpression analysis and biological theme discovery. Sorting gene expression data using hierarchical clustering and visualizing changes in expression with heat maps (Figure 2) facilitates identifying groups of genes that have higher or lower expression within a defined comparison. Generally, genes responding in the same manner tend to be involved in related functions and associated pathways. To achieve a color gradient in heat maps, significant increases in gene expression were scaled according to the following scheme using 2−ΔΔCT values for fold change: 1 −10 = 1; 10–50 = 2; 50–100 = 3; 100–1000 = 4; and >1000 = 5, creating five possible shades of red. To increase the numerical spread for genes with reduced expression, values (v) were scaled as:
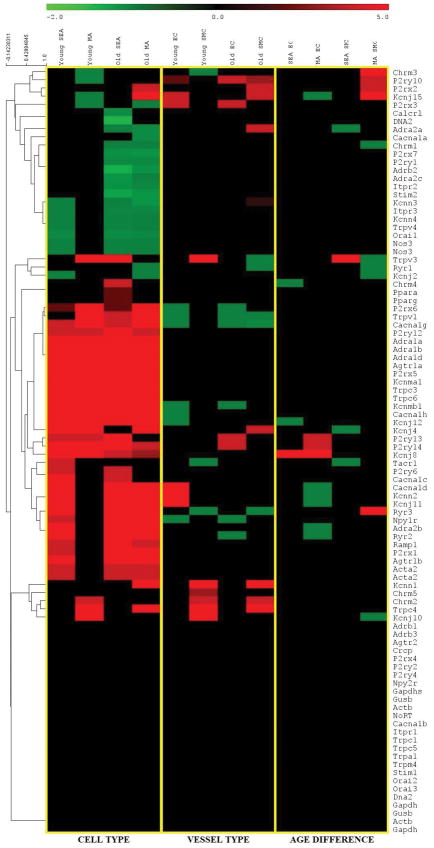
Figure 2. Heat map representation and hierarchical clustering of gene expression changes.
Data represent differential gene expression profiles (2−ΔΔCT i.e., fold-changes) for genes listed along ordinate. Yellow vertical lines designate 3 levels of analyses for respective column headings: Left: Cell type (SMC vs. EC) across vessel type and age group. Center: Vessel type (MA vs. SEA) across cell type and age group; Right: Age (Young vs. Old) across cell type and vessel type. Control genes (Nos3, Acta2) are listed twice based upon their use in both arrays compiled for all genes depicted. Red indicates increased expression and green indicates decreased expression; color intensity indicates the magnitude of difference in expression according to horizontal scale at top. Black depicts genes with no significant difference in expression. Values were hierarchically clustered based on average linkage analysis as depicted in the dendrogram on the left of the heatmap.
-
(v)
to create a gradient of values between 0 and −2. Across all comparisons of cell type, vessel type and age, a hierarchical analysis was applied to cluster differentially expressed genes.
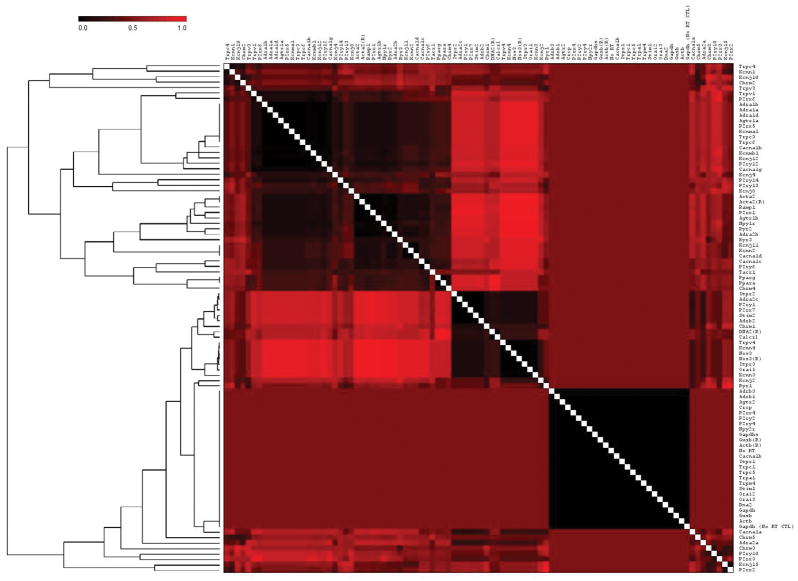
Gene coexpression analysis using matrix visualization (Figure 3) helps to identify genes having a similar expression pattern with other genes in terms of the Pearson correlation coefficient (PCC). T-MeV was used to construct the gene-gene coexpression matrix, which identifies similarities and differences between two genes based on their expression profiles across all samples tested. Perfect similarity between gene expression (i.e., coexpression) is depicted by PCC=1 value (bright red) while dissimilar gene expression patterns are shown by PCC=0 (black), with intermediate values falling throughout the red-to-black gradient. The clustering of genes shown in the dendrogram of Figure 3 is based on the Euclidian distance matrix and represents clusters of genes that tend to be coexpressed together.
Figure 3. Coexpression of genes.
The clustered matrix was obtained using the Pearson distance between expression profiles of genes across all comparisons of cell type, vessel type, and biological age. A PCC of 0 (black) indicates no interaction; a PCC of 1 (bright red) indicates 100% interaction; shading indicates level of interaction according to scale at top left. Diagonal white line indicates each gene compared to self. Relationships between groups of genes are represented by the dendrogram on the left of the figure. Its’ hierarchical clustering is based on the Euclidian distance matrix depicting the level of dissimilarity between the PCC of clusters of genes. The dendrogram illustrates the organization of these clusters with relation to other clusters within the gene list. Genes clustered more closely within the dendrogram (i.e., shorter brackets) have a higher degree of similarity in their coexpression profiles.
Gene network analysis (Figure 4) was performed by visualizing genes in Cytoscape, with functions extended by the addition of plugins 76. Genes are represented and grouped as nodes according to expression levels. Cytoscape utilizes a node list file that defines the parameters for the nodes in the network, in this case, the expression levels of each gene. These data are combined with an edge list file that defines the properties of the nodes using known and predicted protein information mined from PPI databases. The mouse protein-protein network of the STRING database 77,78 is built from highly curated experimental interactions obtained from public databases such as The Protein Data Bank 79 and text-mining based on published literature. This PPI network was used as a framework to generate a network whereby protein names are nodes and the distances between nodes are defined by the confidence score of the interaction between respective proteins. In the present study, expression changes (or lack thereof) of each gene are integrated into this analysis to elucidate links within a network according to their expression patterns (Figure 4).
Figure 4. Protein-Protein Interaction Networks.
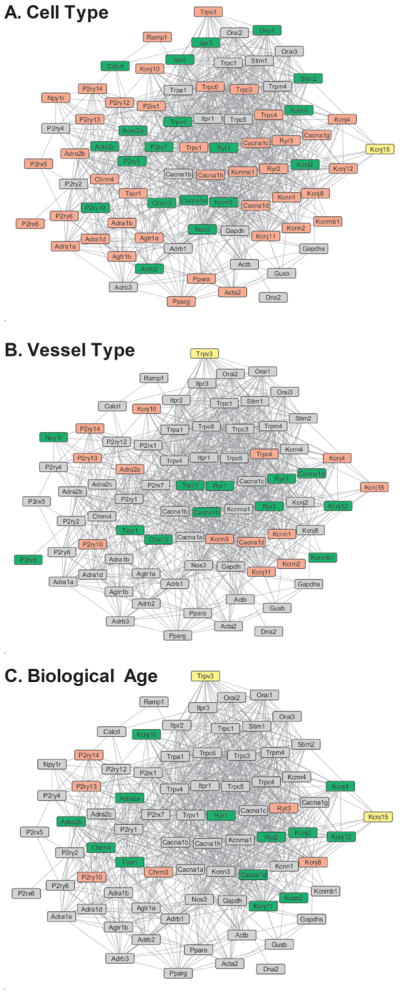
Images are networks depicting predicted PPIs based on STRING database mining for all measured genes based on Cell Type (SMC vs.EC), Vessel Type (MA vs. SEA) and Biological Age (Young vs. Old). The PPI network data forms the backbone and connections of the network, with lines indicating likely interactions between the proteins coded by each gene. The color coding of the nodes represents the change (of lack thereof) in expression level of the gene. Genes in red have increased expression, green have decreased expression, yellow have variable expression changes, and gray have no differential expression within each comparison group.
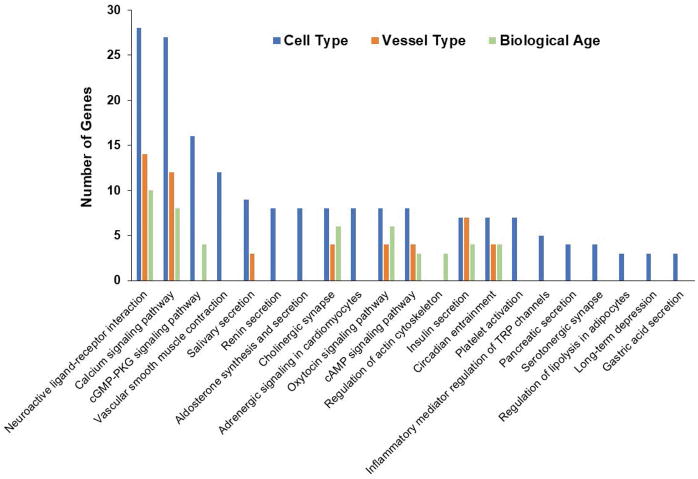
Differentially expressed genes were also linked to common physiological functions using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. A rankings-based analysis of the 80 target genes (Figure 5) determined KEGG pathways that include ≥ 5 differentially expressed genes from the PCR arrays. These correlations provide insight into pathways and associated networks contributing to functional differences between experimental groups; e.g., cell type, vessel type and the effects of aging as studied here.
Figure 5. Number of Differentially expressed genes mapped to KEGG pathways.
The chart depicts the number of differentially expressed genes from each comparison that were found to be part of a metabolic pathway as classified in the KEGG database. The chart has been truncated at 5 genes per pathway, and therefore does not list any pathways mapped to < 5 differentially expressed genes.
RESULTS
Gene expression: CT, ΔCT and fold-change values
Each of the 2 custom arrays included 40 genes of interest + 7 control genes + no RT control (Supplementary Table 1). Average CT values for each EC and SMC sample are listed in Supplementary Table 2, with fold-changes in expression listed in Supplementary Tables 3–5. Across comparisons of cell type, vascular bed and age a total of 68 genes had significant (P < 0.05) differences in expression as shown in a Venn diagram (Figure 1; listed in Supplementary Table 6). Within these 68 differentially-expressed genes, cell type (SMC vs. EC) comparisons included 66 significant differences across vessel type and age. Vessel type comparisons (SEA vs. MA) had 26 significant differences across cell type and biological age, while age had 23 significant differences across vessel type and cell type (Figure 1 and Supplementary Tables 3–6). Many differentially-expressed genes overlap between groups.
Figure 1. Differential gene expression based on cell type, vessel type and age.
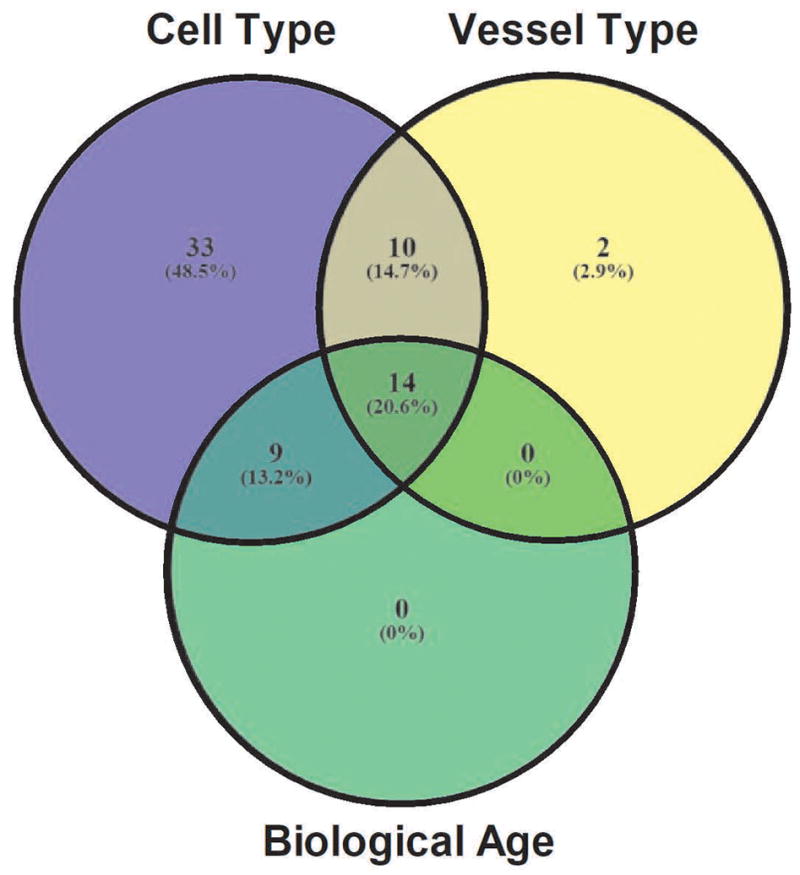
Venn diagram depicts number of differentially expressed genes using 2−ΔΔCT (fold-change) values and P-values at a 0.05 threshold (Avg CT P < 0.05): SMC vs. EC (purple), MA vs. SEA (yellow) and Young vs. Old (green). Intersections represent differentially expressed genes that are shared between comparisons.
With significant differences in gene expression for each of our 3 group comparisons, data were normalized to the control gene 18s (equations i and ii in Methods), yielding the ΔCT values for each gene listed in Supplementary Table 2. The ΔCTs were then used to generate ΔΔCT and 2−ΔΔCT values for each gene across all combinations of cell type, vessel type and age; adjusted P-values are given with respective fold-changes (Supplementary Tables 3–5). These data provide a physiological index of differences in gene expression for bioinformatics and network analysis.
Heat maps and distance matrices
Heat maps and coexpression analysis were generated with T-MeV based upon the program’s default parameters for all genes of interest. In the heat map with hierarchical clustering (Figure 2), the gene differential expression scale was prepared to depict a color gradient ranging from −2 to +5 to best visualize unchanged genes and the differentially expressed genes, which spanned a range of fold-change values > 106 (Supplementary Tables 3–5). In the heat map, black represents genes with no significant change in expression (i.e., average ΔCT P-value > 0.05) for the listed gene. The green gradient represents genes with significantly lower expression, while the red gradient represents genes with significantly increased expression. The average linkage analysis method 80 was used in hierarchical clustering analysis (Figure 2). The greatest number of differences in gene expression occurred with cell type, with fewer differences between vessel types while biological age had the smallest affect.
A gene distance matrix (Figure 3) was generated to gain insight into potential coexpression between each of the target and control genes in the study. This matrix illustrates (1) coexpression level of gene pairs within the matrix and (2) hierarchical clustering of genes with similar coexpression patterns for groups ion channel and receptor genes across cell type, vessel type, and age. The hierarchical clustering highlights several large groups of genes with similar coexpression patterns across all genes tested, observable both in the dendogram and visually as large blocks with similar color patterns in the matrix (Figure 3).
Network and functional analysis: Cytoscape and KEGG pathway analysis
To better understand the potential for physical and functional interactions involving the genes and resultant proteins in this study, the gene list was analyzed using publicly available mouse protein-protein interactions data from the STRING database 77,78. The STRING database contains PPI datasets for multiple species, each curated on the basis of several metrics that culminate in a single confidence score that defines the level of physical and/or functional interaction between a pair of designated proteins. A constrained interaction list containing only the target receptors and ion channels in this study was used to build the backbone of the network in Cytoscape 76, which was overlaid by our values for differentially expressed genes. The Allegro plugin 81 in Cytoscape was used to sort and build the network based on confidences score between proteins. Nodes in the network depict genes and were colored to match changes in gene expression (Figure 4). Red depicts genes with increased expression, green depicts genes with decreased expression and yellow indicates “ambiguous” genes having both increased and decreased expression within the 4 sub-comparisons in the same category (i.e., different colors within the 4 columns of the heatmaps for cell type, for vessel type and/or age group). Gray indicates genes with no differential expression. These networks highlight both the interconnected nature of differentially expressed genes and the unique patterns by which they vary based on cell type, vessel type and biological age.
Following analysis of gene/protein interactions using Cytoscape, the KEGG pathway resource further characterize the specific pathways affected by the up- and down-regulated genes characterized in the present study. Figure 5 depicts a summary of the number of differentially expressed genes mapped to metabolic pathways as classified in the KEGG database. The highest mapped pathways are calcium signaling, neuroactive ligand-receptor interaction and cGMP-PKG signaling. This outcome is consistent with selecting target genes based upon their association with vasomotor control.
DISCUSSION
This study was designed to test the hypothesis that the expression of ion channels and receptors associated with vasomotor control vary with cell type (SMC vs. EC), vessel type (SEA vs. MA) and biological age (Old vs. Young) using the mouse as a model system. Finding the greatest differences in gene expression between SMCs and ECs was expected given the distinct roles between the intima and media in vascular function. The major differences between cell types encompassed multiple ion channel and receptor families including those whose roles in vascular function are well-characterized and those whose roles have yet to be defined. While fewer between vessel types than between cell types, differences centered on calcium signaling in both SMCs and ECs. Differences in transcript levels need not translate to stoichiometric differences in protein expression, but even subtle changes in transcription may contribute to regional heterogeneity in vascular function. Remarkably, age was the variable having the fewest differences in gene expression but even here the most apparent differences were in channels and receptors related to calcium signaling. Relatively few differences between age groups for either cell type or vessel type is also consistent with healthy aging exerting only subtle effects on vascular function.
Differential gene expression based on cell type
Across all genes tested, 16 had higher or lower expression (as depicted in the heat maps) across all 4 cell type comparisons, indicating that many differences between ECs and SMCs persist across vascular beds and ages. All 16 of these genes had increased expression in SMCs vs. ECs (Figure 2, Supplementary Table 3) and many are known to exert vasomotor effects via SMCs including: adrenergic receptors (Adra1a, and Adra1d)43, angiotensin II receptors (Agtr1a) 66,82, T-type Ca2+ channels (Cacna1g and Cacna1h) 83, BKCa channels (Kcnma1 and Kcnmb1) 10, NPY receptors (Npy1r) 42, and TRP C3 (Trpc3) and C6 (Trpc6) channels 15. Several genes with increased expression in SMCs have controversial or unclear roles in vasomotor function. For example, 4 purinergic receptor genes (P2rx5, P2rx6, P2ry12 and P2ry14) had greater expression in SMCs of both young and old SEAs and MAs however evidence for their functional significance is limited. Previous studies have typically failed to detect P2rx5 84 or P2rx6 85 in either SMCs or ECs from MAs or found P2rx5 expression at the limit of detection 85,86. Expression of P2rx6 has been reported for pig aortic SMCs 87 and for cultured human vascular ECs 88. However, because cell culture alters the expression of ion channels and receptors in vascular SMCs and ECs 2,89, it can be difficult to compare such findings to native cells with respect to “constitutive” gene expression.
Interpreting expression and/or functional data regarding P2X receptors is complicated by the ability of these channels to form heteromultimers. For example, detection of expression and/or currents though P2X5 receptors may result from a P2X1/5 heteromultimer 90. P2ry12 and P2ry14 gene expression and protein function have indicated SMC-specific expression, with a role in vasoconstriction defined in multiple studies and vascular beds 57,91,92. The presence of P2Y12 receptors on ECs remains unclear. Functional studies in thoracic aorta suggest no role in endothelium-dependent relaxation of SMCs 93, but the expression of P2ry12 has been demonstrated in ECs of the blood-brain barrier 94. Our study suggests that due to robust expression of P2ry12 on SMCs and negligible expression on ECs, these receptors on SMCs are likely of greater physiological significance in vasomotor control than on ECs. Cell-specific differences in purinergic receptor expression point to their potential as future targets for pharmacological treatments within the resistance vasculature.
When comparing gene expression based on cell type, a remarkable trend was the relative consistency in increased- or decreased levels across vessel type and biological age. However, there were a subset of 22 genes for which only 3 of 4 subgroups changed in the same direction (increased or decreased), with Young MA being the outlier 19 times (Figure 2, Supplementary Table 3); data were internally consistent between biological replicates (see Supplementary Table 2 of raw CT data). The relatively lower difference in gene expression between SMCs and ECs in MAs from young vessels suggests that it may be more difficult to target one cell layer vs. the other in physiological experiments as well as clinical applications. Combined with other differences between vessel types, these data may question the use of MAs from young mice as a “typical” resistance artery and highlight the importance of evaluating function in multiple vascular beds.
Differential gene expression based on vascular bed
In contrast to differences in gene expression based on cell type, there were few differences based on vessel type. In fact, none of the genes included in this study had significantly increased or decreased expression across Young or Old ECs or SMCs from MA vs. SEA. Further separating the data based on cell type (which accounts for the most differences in gene expression), SMCs expressed 4 genes that were differentially expressed in SMCs from MA vs. SEA (Figure 2, Supplementary Table 4). One subtype of the sarcoplasmic reticulum-localized ryanodine receptor (Ryr3) were expressed less in the SMCs of MA vs. SEA, and one subtype each of small-conductance Ca2+-activated K+ channels (Kcnn1), muscarinic receptors (Chrm2) and TRPC channels (Trpc4) had higher expression in SMCs from MA vs. SEA. Given the importance of RyRs and SK channels to the regulation of Ca2+ signaling and vascular tone 10,95, large differences in the expression of these genes in SEAs vs. MAs suggests that differences in SMC Ca2+ signaling pathways contribute to functional heterogeneity in respective (mesenteric and skeletal muscle) vascular beds.
In ECs, 6 genes were expressed differently in MAs vs. SEAs, with 3 (P2ry10, P2rx3 and Trpv1) at higher levels in MAs. The functional roles of P2Y10 (a “purinergic” receptor reclassified as an orphan receptor) and P2X3 proteins are not established in the vasculature. Finding increased levels of P2rx3 in MA vs. SEA ECs is consistent with a recent study that failed to detect its expression in skeletal muscle femoral arteries 96 while pointing to this receptor as a potential cause of regional heterogeneity in vascular function. Our results for Trpv1 expression are consistent with a study showing decreased Trpv1 mRNA expression in rat mesenteric vs. skeletal muscle arteries 97. Expression of 2 genes was lower in ECs from MA vs. SEA: P2rx6 and Npy1r. Negligible expression of P2rx6 in MAs is consistent with earlier findings 85 and underscores different roles of purinergic receptor subtypes across vascular beds. The variable expression of NPY1 receptors in ECs from MA vs. SEA is novel, particularly in light of the belief that this receptor’s vasomotor effects are typically mediated via expression on SMCs 8,98. The potent vascular effects of NPY and the paucity of studies exploring its NPY1 receptor-mediated endothelial effects suggest that this difference in expression based on vascular bed warrants further study.
Differential gene expression based on age
Age-wise comparisons yielded the lowest proportion of differentially expressed genes (Figures 1, 2). Similar to MA vs SEA comparisons, there were no genes with significantly increased or decreased expression across all 4 subgroups tested. In contrast, subdividing the samples by cell type highlights only 1 difference in expression in Old vs. Young samples. Endothelial cells had increased Kcnj8 (Kir 6.1) expression in Old vs. Young. Kir 6.1 subunits are a critical component of KATP channels, which are impaired in some arteries during aging 99, suggesting that an increase in Kir 6.1 may alter the function of KATP channels with advanced age. All 22 other significant differences in gene expression in Old vs. Young were observed in just 1 of the 4 sample groups tested. Because no differences in gene expression were related age alone (Figure 1), broadly interpreting the physiological significance of these 22 differences remains difficult. However, these unique patterns of gene expression may contribute to heterogeneity in age-related changes between vascular beds.
Gene coexpression and molecular interaction networks
Construction of the coexpression matrix provides insight in gene expression trends across samples of both cell types, vessel types and ages (Figure 3). The lack of many distinct patterns of coexpression (blocks of similar color in the matrix) highlights the large degree of heterogeneity in gene coexpression patterns within the vasculature. Examination of the matrix demonstrates many strong associations in expression patterns between individual and/or families of ion channels and receptors, some of which serve as a “positive control” for the coexpression analysis. For example, Ramp1 and Calcrl, the genes for two critical protein components of a functional CGRP receptor, are shown to be highly coexpressed with each other, as predicted (note bright red color in Ramp1-Calcrl, Figure 3). Other expected observations help validate the hierarchical clustering within the matrix, including the tight clustering of controls repeated in both arrays (Acta2, Nos3, Gapdh, Actb, Gusb). The genes corresponding to many closely-related proteins also were grouped together, including L-type VGCCs (Cacna1c, Cacna1d), α1-ARs (Adra1a, Adra1b, Adra1c), PPARs (Ppara, Pparg). In contrast, many gene families included in this study clustered heterogeneously, with some members tightly associated and others with greater Euclidean distances. For example, of the TRPC channel genes, some clustered closely (Trpc3 and TrpC6, Trpc1 and TrpC5), while Trpc4 clustered separately from other groups. Understanding patterns of coexpression both within and between gene families provides insight into which specific targets may have unique physiological functions in the vasculature.
It is also important to note that the genes shown to demonstrate moderate-to-high coexpression across all samples are not necessarily the same genes with differential expression based on cell type, vascular bed or aging. Similarly, coexpression of two genes does not necessarily indicate a high chance of protein-protein interaction (PPI), as shown in the molecular interaction networks (Figure 4A–C). This point is illustrated by comparing Npy1r and Calcrl in our analyses. Across all samples tested, Npy1r and Calcrl exhibited high coexpression (i.e., Pearson coefficient >0.75 and bright red color in Figure 3). However, these genes showed differential expression in opposite directions for both cell type (Figure 4A), only 1 differentially expressed in vessel type (Figure 4B) and no change in expression of either gene with aging (Figure 4C). In addition, examination of the protein-protein interactions of these genes using the STRING database and illustrated with Cytoscape show no demonstrated or predicted interactions between these proteins (Figure 5). Interestingly, the interaction networks highlighted that both Npy1r and Calcrl likely have protein interaction with P2ry14, which had increased expression in SMC vs. EC, MA vs. SEA and Young vs. Old samples (Figure 5). These 3 genes/proteins participate in the KEGG pathway “neuroactive ligand-receptor interaction,” which was one of the most significant physiological pathways identified in this study based on the number of differentially expressed genes in each comparison (Figure 5). Clearly, neither gene expression nor network analysis alone will guarantee findings of altered protein expression or function. Only functional studies comparing cell type, vessel type and aging can ultimately determine the physiological significance of the interactions suggested by the present findings. Nevertheless, our gene expression measurements coupled with bioinformatics analysis tools provide a reference from which to identify the best receptor and ion channel candidates to include in future studies of signaling events in resistance arteries.
Summary
Our findings illustrate that transcript levels for ion channels and receptors differed to the greatest extent with cell type while relatively few were altered significantly by advanced age. That the pattern of differentially expressed genes was not uniform across cell type or vessel type highlights the heterogeneous effect aging has in different vascular beds. Additional studies are needed to extend these results with respect to protein expression and functional significance in the vasculature. In total, the gene expression measurements, coexpression data, and network associations identified here illustrate the application of bioinformatics to the analysis of qPCR gene expression data sets as they pertain to the vasculature.
Perspectives
In mouse superior epigastric vs. mesenteric arteries, 80 ion channel and receptor genes display the greatest differential expression between cell type (smooth muscle vs. endothelium), followed by vessel type and biological age (~4 vs. ~24 mo). Bioinformatic analysis provides insight into gene coexpression patterns, likely protein-protein interactions and signaling pathways. These findings highlight changes in gene expression with respect to their interactions and physiological functions and may serve to direct future studies of altered signaling within the vascular wall.
Supplementary Material
Acknowledgments
GRANTS:
This research was supported by National Institutes of Health grant R37HL041026 to SSS.
EMB was supported by K99HL129196
Anjana Ramnath contributed to initial components of informatics data analysis.
ABBREVIATIONS
- [Ca2+]i
intracellular calcium concentration
- Ang II
angiotensin II
- AR
adrenergic receptor
- BKCa
large conductance calcium-activated potassium channel
- CALCRL
calcitonin receptor-like receptor
- cAMP
cyclic adenosine monophosphate
- CGRP
calcitonin gene-related peptide
- CRAC
calcium release activated channel
- CT
critical threshold
- EC
endothelial cell
- EDH
endothelium-dependent hyperpolarization
- ER
endoplasmic reticulum
- EK
Equilibrium (Nernst) potential for potassium
- IKCa
intermediate conductance calcium-activated potassium channel
- IP3R
inositol (1,4,5)-trisphosphate receptors
- KATP
ATP-sensitive potassium channel
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- Kir
inwardly-rectifying potassium channels
- MA
mesenteric arteries
- NE
norepinephrine
- NO
nitric oxide
- NPY
neuropeptide Y
- PCC
Pearson correlation coefficient
- PKA
protein kinase A
- PPAR
peroxisome proliferator-activated receptor
- PPI
protein-protein interaction
- RAMP1
receptor activity modifying protein 1
- ROCE
receptor-operated calcium entry
- RyR
ryanodine receptor
- SEA
superior epigastric artery
- SKCa
small conductance calcium-activated potassium channel
- SMC
smooth muscle cell
- SOCE
store-operated calcium entry
- SP
substance P
- STIM
stromal interaction molecule
- STOC
spontaneous transient outward current
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins
- TRP
transient receptor potential
- VGCC
voltage-gated calcium channels
- Vm
membrane potential
Footnotes
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
AUTHOR CONTRIBUTIONS
Author contributions: E.M.B., R.L.S. and S.S.S. conception and design of research; E.M.B. and R.L.S. performed experiments; E.M.B., S.S., R.L.S., T.J. and S.S.S. analyzed data; E.M.B., S.S., R.L.S., T.J. and S.S.S. interpreted results of experiments; E.M.B. and S.S. prepared figures; E.M.B. drafted manuscript; E.M.B., S.S., R.L.S., T.J. and S.S.S. edited and revised manuscript; E.M.B., S.S., R.L.S., T.J. and S.S.S. approved final version of manuscript.
References
- 1.Tykocki NR, Boerman EM, Jackson WF. Smooth Muscle Ion Channels and Regulation of Vascular Tone in Resistance Arteries and Arterioles. Comprehensive Physiology. 2017;7:485–581. doi: 10.1002/cphy.c160011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nilius B, Droogmans G. Ion channels and their functional role in vascular endothelium. Physiol Rev. 2001;81:1415–1459. doi: 10.1152/physrev.2001.81.4.1415. [DOI] [PubMed] [Google Scholar]
- 3.Boerman E, Segal S. Aging Impairs Spontaneous Ca2+ Signaling and Regulation by CGRP in Smooth Muscle Cells of Mouse Mesenteric Arteries. FASEBJ. 2016;30:945.943–945.943. [Google Scholar]
- 4.Boerman EM, Segal SS. Depressed perivascular sensory innervation of mouse mesenteric arteries with advanced age. J Physiol. 2016;594:2323–2338. doi: 10.1113/JP270710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Socha MJ, Boerman EM, Behringer EJ, Shaw RL, Domeier TL, Segal SS. Advanced age protects microvascular endothelium from aberrant Ca2+ influx and cell death induced by hydrogen peroxide. J Physiol. 2015:2155–2169. doi: 10.1113/JP270169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behringer EJ, Shaw RL, Westcott EB, Socha MJ, Segal SS. Aging impairs electrical conduction along endothelium of resistance arteries through enhanced Ca2+-activated K+ channel activation. Arterioscler Thromb Vasc Biol. 2013;33:1892–1901. doi: 10.1161/ATVBAHA.113.301514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HR, Appel S, Vetterkind S, Gangopadhyay SS, Morgan KG. Smooth muscle signalling pathways in health and disease. J Cell Mol Med. 2008;12:2165–2180. doi: 10.1111/j.1582-4934.2008.00552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feletou M. The endothelium. San Rafael, Calif: Morgan & Claypool Life Sciences; 2011. [PubMed] [Google Scholar]
- 9.Garland CJ, Hiley CR, Dora KA. EDHF: spreading the influence of the endothelium. Br J Pharmacol. 2011;164:839–852. doi: 10.1111/j.1476-5381.2010.01148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ledoux J, Werner M, Brayden J, Nelson M. Calcium-activated potassium channels and the regulation of vascular tone. Physiology (Bethesda) 2006;21:69–78. doi: 10.1152/physiol.00040.2005. [DOI] [PubMed] [Google Scholar]
- 11.Catterall WA. Structure and regulation of voltage-gated Ca2+ channels. Annual review of cell and developmental biology. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- 12.Mintz IM, Venema VJ, Swiderek KM, Lee TD, Bean BP, Adams ME. P-type calcium channels blocked by the spider toxin omega-Aga-IVA. Nature. 1992;355:827–829. doi: 10.1038/355827a0. [DOI] [PubMed] [Google Scholar]
- 13.Whorlow SL, Angus JA, Wright CE. Selectivity of omega-conotoxin GVIA for N-type calcium channels in rat isolated small mesenteric arteries. Clin Exp Pharmacol Physiol. 1996;23:16–21. [PubMed] [Google Scholar]
- 14.Yao X, Garland CJ. Recent developments in vascular endothelial cell transient receptor potential channels. Circ Res. 2005;97:853–863. doi: 10.1161/01.RES.0000187473.85419.3e. [DOI] [PubMed] [Google Scholar]
- 15.Earley S, Brayden JE. Transient receptor potential channels in the vasculature. Physiol Rev. 2015;95:645–690. doi: 10.1152/physrev.00026.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzales AL, Amberg GC, Earley S. Ca2+ release from the sarcoplasmic reticulum is required for sustained TRPM4 activity in cerebral artery smooth muscle cells. American journal of physiology Cell physiology. 2010;299:C279–288. doi: 10.1152/ajpcell.00550.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amberg GC, Navedo MF. Calcium dynamics in vascular smooth muscle. Microcirculation. 2013;20:281–289. doi: 10.1111/micc.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ledoux J, Taylor MS, Bonev AD, Hannah RM, Solodushko V, Shui B, Tallini Y, Kotlikoff MI, Nelson MT. Functional architecture of inositol 1,4,5-trisphosphate signaling in restricted spaces of myoendothelial projections. Proc Natl Acad Sci U S A. 2008;105:9627–9632. doi: 10.1073/pnas.0801963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Félétou M. The Endothelium: Part 1: Multiple Functions of the Endothelial Cells—Focus on Endothelium-Derived Vasoactive Mediators. San Rafael, CA: Morgan & Claypool Life Sciences; 2011. Calcium Signaling in Vascular Cells and Cell-to-Cell Communications; pp. 19–39. [PubMed] [Google Scholar]
- 20.Fill M, Copello JA. Ryanodine receptor calcium release channels. Physiol Rev. 2002;82:893–922. doi: 10.1152/physrev.00013.2002. [DOI] [PubMed] [Google Scholar]
- 21.Foskett J, White C, Cheung K, Mak D. Inositol trisphosphate receptor Ca2+ release channels. Physiol Rev. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trebak M. STIM/Orai signalling complexes in vascular smooth muscle. J Physiol. 2012;590:4201–4208. doi: 10.1113/jphysiol.2012.233353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Deng X, Hewavitharana T, Soboloff J, Gill DL. Stim, ORAI and TRPC channels in the control of calcium entry signals in smooth muscle. Clin Exp Pharmacol Physiol. 2008;35:1127–1133. doi: 10.1111/j.1440-1681.2008.05018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson MT, Quayle JM. Physiological roles and properties of potassium channels in arterial smooth muscle. Am J Physiol. 1995;268:C799–822. doi: 10.1152/ajpcell.1995.268.4.C799. [DOI] [PubMed] [Google Scholar]
- 25.Jackson W. Ion channels and vascular tone. Hypertension. 2000;35:173–178. doi: 10.1161/01.hyp.35.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson MT, Cheng H, Rubart M, Santana LF, Bonev AD, Knot HJ, Lederer WJ. Relaxation of arterial smooth muscle by calcium sparks. Science. 1995;270:633–637. doi: 10.1126/science.270.5236.633. [DOI] [PubMed] [Google Scholar]
- 27.Busse R, Edwards G, Félétou M, Fleming I, Vanhoutte PM, Weston AH. EDHF: bringing the concepts together. Trends Pharmacol Sci. 2002;23:374–380. doi: 10.1016/s0165-6147(02)02050-3. [DOI] [PubMed] [Google Scholar]
- 28.Bagher P, Segal SS. Regulation of blood flow in the microcirculation: role of conducted vasodilation. Acta Physiol (Oxf) 2011;202:271–284. doi: 10.1111/j.1748-1716.2010.02244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quayle JM, McCarron JG, Brayden JE, Nelson MT. Inward rectifier K+ currents in smooth muscle cells from rat resistance-sized cerebral arteries. Am J Physiol. 1993;265:C1363–1370. doi: 10.1152/ajpcell.1993.265.5.C1363. [DOI] [PubMed] [Google Scholar]
- 30.Quayle JM, Nelson MT, Standen NB. ATP-sensitive and inwardly rectifying potassium channels in smooth muscle. Physiol Rev. 1997;77:1165–1232. doi: 10.1152/physrev.1997.77.4.1165. [DOI] [PubMed] [Google Scholar]
- 31.Smith PD, Brett SE, Luykenaar KD, Sandow SL, Marrelli SP, Vigmond EJ, Welsh DG. KIR channels function as electrical amplifiers in rat vascular smooth muscle. J Physiol. 2008;586:1147–1160. doi: 10.1113/jphysiol.2007.145474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longden TA, Dabertrand F, Koide M, Gonzales AL, Tykocki NR, Brayden JE, Hill-Eubanks D, Nelson MT. Capillary K(+)-sensing initiates retrograde hyperpolarization to increase local cerebral blood flow. Nat Neurosci. 2017;20:717–726. doi: 10.1038/nn.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Standen N. KATP Channels in Vascular Smooth Muscle:Structure, Regulation and Functional Roles. J Clin Basic Cardiol. 2003;6:7–14. [Google Scholar]
- 34.Duncker DJ, Oei HH, Hu F, Stubenitsky R, Verdouw PD. Role of KATP channels in regulation of systemic, pulmonary, and coronary vasomotor tone in exercising swine. Am J Physiol Heart Circ Physiol. 2001;280:H22–33. doi: 10.1152/ajpheart.2001.280.1.H22. [DOI] [PubMed] [Google Scholar]
- 35.Faraci FM, Heistad DD. Regulation of the cerebral circulation: role of endothelium and potassium channels. Physiol Rev. 1998;78:53–97. doi: 10.1152/physrev.1998.78.1.53. [DOI] [PubMed] [Google Scholar]
- 36.Clapham DE, Runnels LW, Strübing C. The TRP ion channel family. Nat Rev Neurosci. 2001;2:387–396. doi: 10.1038/35077544. [DOI] [PubMed] [Google Scholar]
- 37.Lindsey SH, Tribe RM, Songu-Mize E. Cyclic stretch decreases TRPC4 protein and capacitative calcium entry in rat vascular smooth muscle cells. Life Sci. 2008;83:29–34. doi: 10.1016/j.lfs.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 38.Guibert C, Beech DJ. Positive and negative coupling of the endothelin ETA receptor to Ca2+-permeable channels in rabbit cerebral cortex arterioles. J Physiol. 1999;514(Pt 3):843–856. doi: 10.1111/j.1469-7793.1999.843ad.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonkusare SK, Bonev AD, Ledoux J, Liedtke W, Kotlikoff MI, Heppner TJ, Hill-Eubanks DC, Nelson MT. Elementary Ca2+ signals through endothelial TRPV4 channels regulate vascular function. Science. 2012;336:597–601. doi: 10.1126/science.1216283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan MN, Gonzales AL, Pires PW, Bruhl A, Leo MD, Li W, Oulidi A, Boop FA, Feng Y, Jaggar JH, Welsh DG, Earley S. Localized TRPA1 channel Ca2+ signals stimulated by reactive oxygen species promote cerebral artery dilation. Sci Signal. 2015;8:ra2. doi: 10.1126/scisignal.2005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian X, Francis M, Solodushko V, Earley S, Taylor MS. Recruitment of dynamic endothelial Ca2+ signals by the TRPA1 channel activator AITC in rat cerebral arteries. Microcirculation. 2013;20:138–148. doi: 10.1111/micc.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westcott EB, Segal SS. Perivascular innervation: A multiplicity of roles in vasomotor control and myoendothelial signaling. Microcirculation. 2013;20:217–238. doi: 10.1111/micc.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guimarães S, Moura D. Vascular adrenoceptors: an update. Pharmacol Rev. 2001;53:319–356. [PubMed] [Google Scholar]
- 44.Queen LR, Ferro A. B-adrenergic receptors and nitric oxide generation in the cardiovascular system. Cell Mol Life Sci. 2006;63:1070–1083. doi: 10.1007/s00018-005-5451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brain SD, Cox HM. Neuropeptides and their receptors: innovative science providing novel therapeutic targets. Br J Pharmacol. 2006;147(Suppl 1):S202–211. doi: 10.1038/sj.bjp.0706461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wahlestedt C, Edvinsson L, Ekblad E, Håkanson R. Neuropeptide Y potentiates noradrenaline-evoked vasoconstriction: mode of action. J Pharmacol Exp Ther. 1985;234:735–741. [PubMed] [Google Scholar]
- 47.Russell FA, King R, Smillie SJ, Kodji X, Brain SD. Calcitonin gene-related peptide: physiology and pathophysiology. Physiol Rev. 2014;94:1099–1142. doi: 10.1152/physrev.00034.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelson MT, Huang Y, Brayden JE, Hescheler J, Standen NB. Arterial dilations in response to calcitonin gene-related peptide involve activation of K+ channels. Nature. 1990;344:770–773. doi: 10.1038/344770a0. [DOI] [PubMed] [Google Scholar]
- 49.Gray DW, Marshall I. Human α-calcitonin gene-related peptide stimulates adenylate cyclase and guanylate cyclase and relaxes rat thoracic aorta by releasing nitric oxide. Br J Pharmacol. 1992;107:691–696. doi: 10.1111/j.1476-5381.1992.tb14508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erlinge D, Burnstock G. P2 receptors in cardiovascular regulation and disease. Purinergic Signal. 2008;4:1–20. doi: 10.1007/s11302-007-9078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lamont C, Vial C, Evans RJ, Wier WG. P2X1 receptors mediate sympathetic postjunctional Ca2+ transients in mesenteric small arteries. Am J Physiol Heart Circ Physiol. 2006;291:H3106–3113. doi: 10.1152/ajpheart.00466.2006. [DOI] [PubMed] [Google Scholar]
- 52.Harrington LS, Mitchell JA. P2X1 receptors and the endothelium. Mem Inst Oswaldo Cruz. 2005;100(Suppl 1):111–112. doi: 10.1590/s0074-02762005000900019. [DOI] [PubMed] [Google Scholar]
- 53.Ralevic V. Purines as neurotransmitters and neuromodulators in blood vessels. Curr Vasc Pharmacol. 2009;7:3–14. doi: 10.2174/157016109787354123. [DOI] [PubMed] [Google Scholar]
- 54.You J, Johnson TD, Marrelli SP, Bryan RM. Functional heterogeneity of endothelial P2 purinoceptors in the cerebrovascular tree of the rat. Am J Physiol. 1999;277:H893–900. doi: 10.1152/ajpheart.1999.277.3.H893. [DOI] [PubMed] [Google Scholar]
- 55.Govindan S, Taylor CW. P2Y receptor subtypes evoke different Ca(2+) signals in cultured aortic smooth muscle cells. Purinergic Signal. 2012;8:763–777. doi: 10.1007/s11302-012-9323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bény JL. Characterization of purine receptors in mouse thoracic aorta. J Cardiovasc Pharmacol. 2004;44:171–177. doi: 10.1097/00005344-200408000-00005. [DOI] [PubMed] [Google Scholar]
- 57.Burnstock G, Ralevic V. Purinergic signaling and blood vessels in health and disease. Pharmacol Rev. 2014;66:102–192. doi: 10.1124/pr.113.008029. [DOI] [PubMed] [Google Scholar]
- 58.Duan SZ, Usher MG, Mortensen RM. PPARs: the vasculature, inflammation and hypertension. Curr Opin Nephrol Hypertens. 2009;18:128–133. doi: 10.1097/MNH.0b013e328325803b. [DOI] [PubMed] [Google Scholar]
- 59.Kvandová M, Majzúnová M, Dovinová I. The role of PPARγ in cardiovascular diseases. Physiol Res. 2016;65:S343–S363. doi: 10.33549/physiolres.933439. [DOI] [PubMed] [Google Scholar]
- 60.Oyekan A. PPARs and their effects on the cardiovascular system. Clin Exp Hypertens. 2011;33:287–293. doi: 10.3109/10641963.2010.531845. [DOI] [PubMed] [Google Scholar]
- 61.Kleinhenz JM, Kleinhenz DJ, You S, Ritzenthaler JD, Hansen JM, Archer DR, Sutliff RL, Hart CM. Disruption of endothelial peroxisome proliferator-activated receptor-gamma reduces vascular nitric oxide production. Am J Physiol Heart Circ Physiol. 2009;297:H1647–1654. doi: 10.1152/ajpheart.00148.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silswal N, Parelkar N, Andresen J, Wacker MJ. Restoration of Endothelial Function in Pparα (−/−) Mice by Tempol. PPAR Res. 2015;2015:728494. doi: 10.1155/2015/728494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pelham CJ, Ketsawatsomkron P, Groh S, Grobe JL, de Lange WJ, Ibeawuchi SR, Keen HL, Weatherford ET, Faraci FM, Sigmund CD. Cullin-3 regulates vascular smooth muscle function and arterial blood pressure via PPARγ and RhoA/Rho-kinase. Cell Metab. 2012;16:462–472. doi: 10.1016/j.cmet.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ketsawatsomkron P, Lorca RA, Keen HL, Weatherford ET, Liu X, Pelham CJ, Grobe JL, Faraci FM, England SK, Sigmund CD. PPARγ regulates resistance vessel tone through a mechanism involving RGS5-mediated control of protein kinase C and BKCa channel activity. Circ Res. 2012;111:1446–1458. doi: 10.1161/CIRCRESAHA.112.271577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carrillo-Sepulveda MA, Keen HL, Davis DR, Grobe JL, Sigmund CD. Role of vascular smooth muscle PPARγ in regulating AT1 receptor signaling and angiotensin II-dependent hypertension. PLoS One. 2014;9:e103786. doi: 10.1371/journal.pone.0103786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mehta PK, Griendling KK. Angiotensin II cell signaling: physiological and pathological effects in the cardiovascular system. Am J Physiol Cell Physiol. 2007;292:C82–97. doi: 10.1152/ajpcell.00287.2006. [DOI] [PubMed] [Google Scholar]
- 67.Berry C, Touyz R, Dominiczak AF, Webb RC, Johns DG. Angiotensin receptors: signaling, vascular pathophysiology, and interactions with ceramide. Am J Physiol Heart Circ Physiol. 2001;281:H2337–2365. doi: 10.1152/ajpheart.2001.281.6.H2337. [DOI] [PubMed] [Google Scholar]
- 68.Chen D, Coffman TM. AT1 Angiotensin receptors-vascular and renal epithelial pathways for blood pressure regulation. Curr Opin Pharmacol. 2015;21:122–126. doi: 10.1016/j.coph.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 69.Socha MJ, Segal SS. Isolation of microvascular endothelial tubes from mouse resistance arteries. J Vis Exp. 2013:e50759. doi: 10.3791/50759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 71.Zampieri M, Ciccarone F, Guastafierro T, Bacalini MG, Calabrese R, Moreno-Villanueva M, Reale A, Chevanne M, Bürkle A, Caiafa P. Validation of suitable internal control genes for expression studies in aging. Mech Ageing Dev. 2010;131:89–95. doi: 10.1016/j.mad.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 72.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔ CT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 73.Expression Suite Software V1.0.3 [Online] Applied Biosystems. http://www.thermofisher.com/us/en/home/technical-resources/software-downloads/expressionsuite-software.html.
- 74.Fleming I, Busse R. Molecular mechanisms involved in the regulation of the endothelial nitric oxide synthase. Am J Physiol Regul Integr Comp Physiol. 2003;284:R1–12. doi: 10.1152/ajpregu.00323.2002. [DOI] [PubMed] [Google Scholar]
- 75.Howe EA, Sinha R, Schlauch D, Quackenbush J. RNA-Seq analysis in MeV. Bioinformatics. 2011;27:3209–3210. doi: 10.1093/bioinformatics/btr490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sokal R, Michener C. A statistical method for evaluating systematic relationships. University of Kansas Science Bulletin. 1958;38:1409–1438. [Google Scholar]
- 81.Yoon JS, Jung W-H. A GPU-accelerated bioinformatics application for large-scale protein interaction networks. Asia Pacific Bioinformatics Conference; Incheon. South Korea. 2011. [Google Scholar]
- 82.Hughes AD. Molecular and cellular mechanisms of action of angiotensin II (AT1) receptors in vascular smooth muscle. J Hum Hypertens. 1998;12:275–281. doi: 10.1038/sj.jhh.1000635. [DOI] [PubMed] [Google Scholar]
- 83.Kuo IY, Wölfle SE, Hill CE. T-type calcium channels and vascular function: the new kid on the block? J Physiol. 2011;589:783–795. doi: 10.1113/jphysiol.2010.199497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lewis CJ, Evans RJ. Lack of run-down of smooth muscle P2X receptor currents recorded with the amphotericin permeabilized patch technique, physiological and pharmacological characterization of the properties of mesenteric artery P2X receptor ion channels. Br J Pharmacol. 2000;131:1659–1666. doi: 10.1038/sj.bjp.0703744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gitterman DP, Evans RJ. Properties of P2X and P2Y receptors are dependent on artery diameter in the rat mesenteric bed. Br J Pharmacol. 2000;131:1561–1568. doi: 10.1038/sj.bjp.0703760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Phillips JK, Hill CE. Neuroreceptor mRNA expression in the rat mesenteric artery develops independently of innervation. Int J Dev Neurosci. 1999;17:377–386. doi: 10.1016/s0736-5748(99)00032-5. [DOI] [PubMed] [Google Scholar]
- 87.Chin TY, Chueh SH. Distinct Ca2+ signalling mechanisms induced by ATP and sphingosylphosphorylcholine in porcine aortic smooth muscle cells. Br J Pharmacol. 2000;129:1365–1374. doi: 10.1038/sj.bjp.0703190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Glass R, Loesch A, Bodin P, Burnstock G. P2X4 and P2X6 receptors associate with VE-cadherin in human endothelial cells. Cell Mol Life Sci. 2002;59:870–881. doi: 10.1007/s00018-002-8474-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beech DJ. Ion channel switching and activation in smooth-muscle cells of occlusive vascular diseases. Biochem Soc Trans. 2007;35:890–894. doi: 10.1042/BST0350890. [DOI] [PubMed] [Google Scholar]
- 90.Lambrecht G. Agonists and antagonists acting at P2X receptors: selectivity profiles and functional implications. Naunyn Schmiedebergs Arch Pharmacol. 2000;362:340–350. doi: 10.1007/s002100000312. [DOI] [PubMed] [Google Scholar]
- 91.Meister J, Le Duc D, Ricken A, Burkhardt R, Thiery J, Pfannkuche H, Polte T, Grosse J, Schöneberg T, Schulz A. The G protein-coupled receptor P2Y14 influences insulin release and smooth muscle function in mice. J Biol Chem. 2014;289:23353–23366. doi: 10.1074/jbc.M114.580803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haanes KA, Edvinsson L. Characterization of the contractile P2Y14 receptor in mouse coronary and cerebral arteries. FEBS Lett. 2014;588:2936–2943. doi: 10.1016/j.febslet.2014.05.044. [DOI] [PubMed] [Google Scholar]
- 93.Dol-Gleizes F, Marés AM, Savi P, Herbert JM. Relaxant effect of 2-methyl-thio-adenosine diphosphate on rat thoracic aorta: effect of clopidogrel. Eur J Pharmacol. 1999;367:247–253. doi: 10.1016/s0014-2999(98)00985-6. [DOI] [PubMed] [Google Scholar]
- 94.Ceruti S, Vigano F, Colombo L, Deli M, Rosa P, Sperlágh B, Abbracchio M, Kittel A. Investigation of ATP-metabolizing enzymes and purinergic receptors in a new in vitro blood-brain barrier model system. Purinergic Signal. 2010;6:359–366. [Google Scholar]
- 95.Gollasch M, Lohn M, Furstenau M, Nelson MT, Luft FC, Haller H. Ca2+ channels, Ca2+ sparks, and regulation of arterial smooth muscle function. Z Kardiol. 2000;89(Suppl 2):15–19. doi: 10.1007/s003920070095. [DOI] [PubMed] [Google Scholar]
- 96.Lewis CJ, Evans RJ. P2X receptor immunoreactivity in different arteries from the femoral, pulmonary, cerebral, coronary and renal circulations. J Vasc Res. 2001;38:332–340. doi: 10.1159/000051064. [DOI] [PubMed] [Google Scholar]
- 97.Tóth A, Czikora A, Pásztor ET, Dienes B, Bai P, Csernoch L, Rutkai I, Csató V, Mányiné IS, Pórszász R, Edes I, Papp Z, Boczán J. Vanilloid receptor-1 (TRPV1) expression and function in the vasculature of the rat. J Histochem Cytochem. 2014;62:129–144. doi: 10.1369/0022155413513589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hodges GJ, Jackson DN, Mattar L, Johnson JM, Shoemaker JK. Neuropeptide Y and neurovascular control in skeletal muscle and skin. Am J Physiol Regul Integr Comp Physiol. 2009;297:R546–555. doi: 10.1152/ajpregu.00157.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang HQ, Subbotina E, Ramasamy R, Coetzee WA. Cardiovascular KATP channels and advanced aging. Pathobiol Aging Age Relat Dis. 2016;6:32517. doi: 10.3402/pba.v6.32517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.