Abstract
AIM
To investigate the effect of probiotic supplementation during the development of an experimental model of colitis associated colon cancer (CAC).
METHODS
C57BL/6 mice received an intraperitoneal injection of azoxymethane (10 mg/kg), followed by three cycles of sodium dextran sulphate diluted in water (5% w/v). Probiotic group received daily a mixture of Lactobacillus acidophilus, Lactobacillus rhamnosus and Bifidobacterium bifidum. Microbiota composition was assessed by 16S rRNA Illumina HiSeq sequencing. Colon samples were collected for histological analysis. Tumor cytokines was assessed by Real Time-PCR (Polymerase Chain Reaction); and serum cytokines by Multiplex assay. All tests were two-sided. The level of significance was set at P < 0.05. Graphs were generated and statistical analysis performed using the software GraphPad Prism 5.0. The project was approved by the institutional review board committee.
RESULTS
At day 60 after azoxymethane injection, the mean number of tumours in the probiotic group was 40% lower than that in the control group, and the probiotic group exhibited tumours of smaller size (< 2 mm) (P < 0.05). There was no difference in richness and diversity between groups. However, there was a significant difference in beta diversity in the multidimensional scaling analysis. The abundance of the genera Lactobacillus, Bifidobacterium, Allobaculum, Clostridium XI and Clostridium XVIII increased in the probiotic group (P < 0.05). The microbial change was accompanied by reduced colitis, demonstrated by a 46% reduction in the colon inflammatory index; reduced expression of the serum chemokines RANTES and Eotaxin; decreased p-IKK and TNF-α and increased IL-10 expression in the colon.
CONCLUSION
Our results suggest a potential chemopreventive effect of probiotic on CAC. Probiotic supplementation changes microbiota structure and regulates the inflammatory response, reducing colitis and preventing CAC.
Keywords: Intestinal microbiota, Chemoprevention, Lactobacillus acidophilus, Lactobacillus rhamnosus, Bifidobacterium bifidum
Core tip: Intestinal microbiota has an essential role in carcinogenesis, acting in promotion of inflammation, proliferation and neoplastic progression. Probiotic supplementation is an alternative means of favourably modulating the intestinal microbiota. In this study, we investigate the effect of supplementation with a Lactobacillus acidophilus, Lactobacillus rhamnosus and Bifidobacterium bifidum mixture during the development of an experimental model of colitis-associated colon cancer. Probiotic supplementation on colorectal cancer changed the microbiota and reduced inflammation in the colon, probably by regulating the inflammatory response, and reducing inflammatory cell infiltration by lowering chemokine expression, thus preventing colitis.
INTRODUCTION
Colon cancer involves a complex and heterogeneous mechanism, mostly induced by the accumulation of somatic mutations over time, which are caused by environmental factors, diet, microbial exposure and metabolites and the host immune response[1]. Although the causes of colon cancer are not well established, inflammation has been implicated from initiation to promotion of the disease, even for those tumours that do not have a direct causal relationship to inflammation[2]. Tumour-promoting inflammation is a hallmark of cancer, and there is strong evidence that inflammation plays a critical role in cancer development[3]. The balance between the expression of mediators and immunological modulators, as well as the amount and activation of different types of inflammatory cells in the tumour microenvironment, will determine the tumour growth rate[4].
The intestinal microbiota may act as a link between colon cancer-promoting factors and the stages of carcinogenesis[5]. Alteration in microbial composition and diversity is considered essential for the promotion of inflammation, proliferation and neoplastic progression[6]. Studies evaluating the composition of the microbiota in colorectal cancer (CRC) identified that bacteria such as Bacteroides, Parabacteroides, Alistipes, Akkermansia spp. Porphyromonadaceae, Coriobacteridae, Staphylococcaceae and Methanobacteriales are commonly increased. Others, such as Bifidobacterium, Lactobacillus, Ruminococcus, Faecalibacterium spp., Roseburia and Treponema, are consistently decreased[6]. However, these association studies cannot determine if this diversity is a cause or a consequence of CRC. As a result, methods that can selectively manipulate the microbiota have emerged as a strategy that may aid in the prevention of cancer.
The intestinal microbiota can be modulated by several factors such as environment, radiation, surgery, medicines, aging, diet, lifestyle and host genetic. Not coincidentally, these factors are also related to inflammation and colon cancer risk[7-9]. Another important way of modulating the intestinal microbiota is through the supplementation of bacterial strains (probiotics). Probiotic supplements are monoassociated cultures or a mix of living microorganisms; Lactobacillus rhamnosus, Lactobacillus reuteri, L. acidophilus, Lactobacillus bulgaricus, Bifidobacterium infantis, Bifidobacterium bifidum and Saccharomyces boulardii are commonly used as probiotics[10]. Probiotic strains are usually found in dairy products such as yogurts and cheeses or used as food supplements or drugs.
The beneficial effects of probiotic bacteria were recognized more than 100 years ago by Metchnikoff[11]. Modifications to the microbial community can prevent or treat various gastrointestinal disorders such as inflammatory bowel disease and irritable bowel syndrome[12], as well as systemic diseases such as eczema[13], respiratory infections[14], asthma[15] and diabetes[16]. Mechanistically, probiotics may reduce cancer risk by exerting several effects, including destruction of potential carcinogens, reducing microbial genotoxicity, altering the metabolites produced by the microbiota, producing anti-tumourigenic and anti-mutagenic compounds, competing with pathogenic bacteria, increasing the intestinal barrier, increasing the innate immune response of the host and modulating cell proliferation and anti-apoptotic pathways[17-21].
Although the effects of probiotics have been investigated in in vitro experiments, animal models, and some human gastrointestinal diseases, little is known about the interaction between probiotic supplementation, changes in the intestinal microbiota and neoplastic transformations of the gastrointestinal mucosa[19,22].
Thus, the aim of this work is to investigate the effect of supplementation of a Lactobacillus acidophilus, Lactobacillus rhamnosus and Bifidobacterium bifidum mixture on the intestinal microbiota, inflammation and neoplastic alterations in the gastrointestinal mucosa during the development of an experimental model of colitis associated colon cancer.
MATERIALS AND METHODS
Animals
Eight-week-old, male C57BL/6 mice weighing approximately 25 g were provided by the central laboratory of the State University of Campinas (UNICAMP) (Campinas, SP, Brazil). All experiments were conducted in order to minimize the pain and discomfort of the animals. Animals were maintained in cages with a maximum of 5 animals, with free access to water and food, in a bed of wood shavings, controlled temperature by air-conditioned, in a light-dark cycle of 12 h. Intragastric gavage administration was done carefully, with the animal immobilized, using gavage needle appropriate for mice. All procedures were performed according to the Ethical Principles in Animal Experimentation adopted by the Brazilian Society of Laboratory Animal Science (SBCAL), with the current law n° 11.794 of October 8, 2008 and the decree n° 6.899 of July 15, 2009. The Ethics Committee on Animal Use (CEUA) of UNICAMP approved the project, according to protocol no. 2761-1.
Experimental design
Animals were randomly divided into two experimental groups (control and probiotic), both of which received a standard diet (AIN93-M). The probiotic group received by gavage daily 0.6 billion CFU (colony forming units) each of Lactobacillus acidophilus, Lactobacillus rhamnosus and Bifidobacterium bifidum, diluted in 200 μL of drinking water, while the control group received 200 μL of drinking water daily. Treatment starts one week before colon cancer induction and finished one day before animal sacrifice. Each group started with 15 animals, which were identified and monitored individually throughout the experiment, and their weights were evaluated weekly. Colon cancer induction was done by intraperitoneally injection of 10 mg/kg of Azoxymethane (Sigma-Aldrich®). After 1 wk, 2.5% dextran sulphate sodium (DSS, MW 36-50 kDa) (MP Biomedical, Inc) was supplied in the drinking water for 5 d, followed by 14 d with unsupplemented drinking water. This cycle was repeated two additional times, and the mice were sacrificed 10 d after the last cycle, according Greten et al[23]. Tumour samples were collected for RT-PCR and cytokine analysis. Colon tissues were collected for western blotting. Colon faeces were collected for 16S rRNA sequencing. Blood was collected to obtain serum for analysis of serum cytokines. Tumour and colon samples were collected and frozen immediately in liquid nitrogen and all samples were stored at -80 °C until analysis.
Microbiota analysis by 16S rRNA sequencing
DNA was extracted from faecal material using a QIAamp DNA Stool Mini kit (Qiagen, Hilden, Germany) and 50 ng were used for cDNA library synthesis with the Rapid Library Preparation Kit (Roche Applied Science, Mannheim, Germany), according to the manufacturer’s instructions. The cDNA was analysed with a Bioanalyzer and High Sensitive DNA Kit (Agilent Technologies Inc., Santa Clara, CA, United States) to ensure equimolar use of the samples in PCR. These samples were then sequenced with a 16S Metagenomic Sequencing® Illumina Kit combined with the HiSeq 2500 System (Illumina) sequencer, according to the manufacturer’s instructions. Sequence reads obtained from the V4 region of the 16S gene were analysed according to the UPARSE pipeline[24], using the USEARCH v9.2.64 package. For OTU clustering a threshold of 97% similarity was used through the UPARSE-OTU algorithm. α- and β-diversity analyses were calculated using the R package Phyloseq v.1.19.1[25] and vegan 2.4_2 packets, using the OTU table normalized to the smallest sample size. Taxa with differential abundance between groups were identified using the Kruskal-Wallis test (P < 0.05). In the bar plot are shown those taxa with relative abundance greater than 1%.
Cytokines analysis
Analysis of cytokines in serum and tumour tissue was performed by multiplex immunoassay (Bio Plex Pro Mouse Cytokine 23 Plex Panel - Bio Rad, Code: M60009RDPD) according to the manufacturer’s instructions. Tumor tissue protein were extracted previously with appropriate protein extraction buffer, in TissueLyser equipment for 3 min at 30 rpm. After 20 min of rest, the samples were centrifuged at 11000 rpm for 30 min at 4 °C. The supernatant was collected and used for analysis after protein quantification.
Real-time PCR
Tumour tissue was extracted using the RNeasy Mini Kit (Qiagen®) on the QIAcube (Qiagen®), according to the manufacturer’s recommendations. RNA was quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, United States) at a wavelength of 260 nm. cDNA was synthesized following the recommendations of the cDNA Synthesis Kit (Thermo-Scientific). Real-time PCR reactions were performed using the TaqMan™ system (Applied Biosystems), which consists of a pair of primers and a fluorophore- labelled probe. The cycling conditions used were: 50 °C for 2 min, 95 °C for 10 min and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The relative gene expression values were obtained by analyzing the results in the Applied Biosystems 7500 System SDS Software program. Expression levels of the genes of interest were normalized to that of glyceraldehyde 3-phosphate dehydrogenase (Gapdh; Mm99999915_g1TaqMan®). The genes of interest studied were IL-6 (Mm00446190_m1TaqMan®), IL-1β (Mm00434228_m1 TaqMan®), TNF (Mm00443258_m1 TaqMan®), IL-10 (Mm01288386_m1 TaqMan®), IL-4 (Mm00445259_m1TaqMan®), IL-13 (Mm00434204_m1 TaqMan®) and Tgfβ1 (TaqMan Mm01178820_m1).
Inflammatory index
Colons and tumours were removed for histology. Tissues were processed and fixed on microscopic slides and stained with haematoxylin and eosin. The inflammatory index includes verification of the severity of the areas of epithelial degeneration, focal or multifocal areas, erosions of the epithelium, presence of ulcers, tissue hyperplasia and size of the affected area. Analysis of the inflammatory index was performed according to Cooper et al[26].
Western blotting
Colon tissue was extracted using a protein extraction buffer (1% Triton X-100, 100 mmol/L Tris (pH 7.4), 100 mmol/L sodium pyrophosphate, 100 mmol/L sodium fluoride, 10 mmol/L EDTA, 10 mmol/L sodium vanadate, 2 mmol/L phenylmethanesulfonyl fluoride and 0.1 mg/mL aprotinin), Laemmli sample buffer containing 100 mmol/L DTT was added and the mixture was heated to 100 °C for 5 min (ref). For total extracts, similar-sized aliquots were subjected to 8%-15% SDS-PAGE. The samples were electrophoresed for the separation of the proteins, being labelled with a marker of known molecular weight (Thermo Scientific PageRuler Prestained Protein Ladder). Using a wet transfer apparatus, the resolved proteins were blotted onto nitrocellulose membranes (Bio-Rad). Then, membranes were blocked with 5% milk solution and incubated overnight at 4 °C with specific antibodies. The antibodies used were anti-phospho-IKK Ser180/Ser181 (Santa Cruz Biotechnology, SC-23470-R), anti-IKK alpha (Santa Cruz Biotechnology, SC-7183), anti-TNF (Cell Signaling Technology, Cell-3707), anti-IL10 (Cell Signaling Technology, Cell-12163) and anti-β-Tubulin (Cell Signaling Technology, Cell-2146). Bands of interest were distinguished according to the protein ladder molecular weight. These membranes were exposed to a chemiluminescence solution (SuperSignal West Pico Chemiluminescent Substrate (Pierce)) and band intensities were revealed by optical densitometry of developed autoradiographs or in a ChemiDoc MP (Bio-Rad).
Statistical analysis
Results were expressed as mean ± SD. The primary outcome was number of tumors. Intestinal microbiota abundance and diversity, inflammatory index and cytokines expression were the secondary outcomes. The Mann-Whitney U test was used for comparisons between two groups for continuous variables and Chi-square test was used to compare categorical variables. All tests were two-sided. The level of significance was set at P < 0.05. Graphs were generated and statistical analysis performed using the software GraphPad Prism 5.0. Statistical analysis for microbiota data were described in “Microbiota analysis by 16S rRNA sequencing” section.
RESULTS
Probiotic supplementation reduces tumour incidence in a colitis associated colorectal model
Using a CRC model associated with colitis, we investigated the role of probiotic supplementation in the development of CRC. To this end, mice received an intraperitoneal injection of azoxymethane, followed by three cycles of treatment with sodium dextran sulphate (DSS) diluted in water. Two hundred microliters of water or probiotics (0.6 × 109 L. acidophilus, 0.6 × 109 L. rhamnosus and 0.6 × 109 B. bifidum) was provided daily by gavage. The number of tumours was quantified at day 60 after azoxymethane injection. As shown in Figure 1A, the mean number of tumours was 9.7 (± 5.7) (n = 33) in the control group and 5.8 (± 3.3) (n = 29) in the probiotic group, which represents a 40% reduction (P = 0.001). There was no difference in mean tumour size between the groups [control = 3.5 cm (± 1.4) (n = 32), probiotic = 3.0 cm (± 1.7) (n = 29), P = 0.14]; however, the probiotic group presented more tumours of smaller size (< 2 mm) (P = 0.0002) (Figure 1B). These results are represented in the images in Figure 1C. There was no statistically significant difference between initial and final mean weights [control = 1.5 (± 2.7) (n = 31), probiotic = 0.3 (± 3.7) (n = 31), P = 0.21] (Figure 1D).
Figure 1.
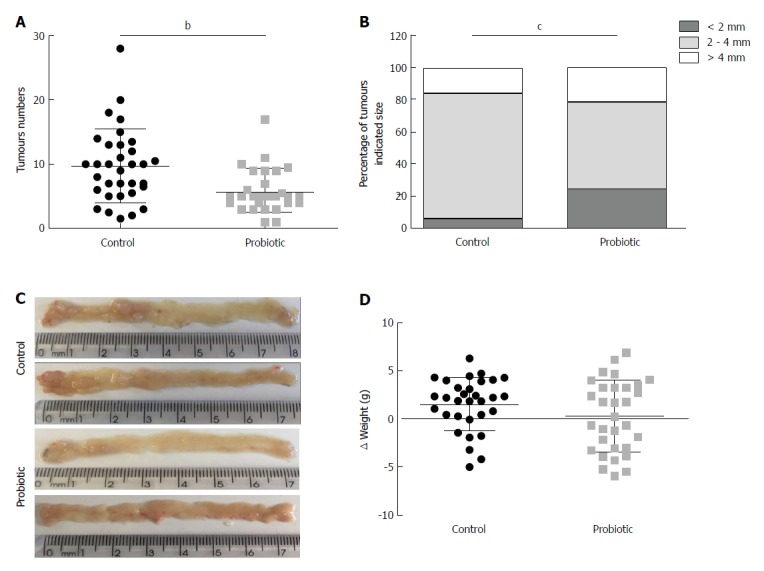
Probiotic supplementation reduces tumour incidence in a colorectal cancer model associated with colitis. Number (A) and size (B) of colon tumours in the control (n = 33) and probiotic (n = 29) groups. C: Representative images of tumours in the colons of the control and probiotic groups at day 60 after injection with azoxymethane. D: Change in body weight during treatment with azoxymethane and DSS in the control (n = 31) and probiotic (n = 31) groups. aP < 0.05; bP < 0.01; cP < 0.001; Mann-whitney U test (A and D), Chi-square test (B). Data are from three independent experiments and are presented as mean and standard deviation.
Probiotic supplementation changes the gut microbiota in the colon
Next, we investigated how probiotic strain supplementation interfered with the abundance and diversity of the intestinal microbiota in the colon and cecum of the probiotic group compared with the control group. Faecal samples from the cecum and colon were collected on day 60 and immediately frozen until the date of DNA extraction. The microbiota profile was characterized by 16S rRNA Illumina HiSeq sequencing. Our results indicate that probiotic supplementation did not change the alpha diversity of the intestinal microbiota. Comparisons of species richness by total number of operational taxonomic units and Chao1 index did not reveal differences between groups; neither were there differences in diversity assessed by Shannon index (Figure 2A-C). However, multidimensional ordering analysis showed a difference in beta diversity between the control and probiotic groups; in this analysis a closer proximity between points indicated a higher similarity between samples (Figure 2D). It is possible to distinguish two fields in the plot, probiotic in the lower right quadrant and control in the upper left quadrant. There was a significant difference between the control and probiotic groups according to Permanova analysis (P < 0.001). Rarefaction curves and the Richness diversity index are shown in the supplemental material (Supplementary Figure 1). Moreover, probiotic supplementation modulated the intestinal microbiota in the colon at the phylum level, generating an increase in bacteria of the phylum Actinobacteria (P < 0.05) (Figure 2E and F). In addition, in the taxonomic analysis we highlight the statistically significant difference found at the genus level of Lactobacillus, Bifidobacterium, Akkermansia, Allobaculum, Clostridium XI and Clostridium XVIII, which were increased in the probiotic group, while Clostridium XIVa was reduced in probiotic group (Figure 3A and B); other genera with a statistically significant difference between groups are shown in the supplemental material (Supplementary Figure 2). Taxonomy plots on the class, order and family levels are shown in the supplemental material (Supplementary Figure 3).
Figure 2.

Probiotic supplementation changes the intestinal microbiota composition. A-C: Microbiota alpha-diversity in the faecal content of the cecum and colon from control and probiotic groups. Differences and comparisons of species richness (A), total number of operational taxonomic units, (B) Chao1 Richness Index, (C) Shannon Diversity Index. Data are expressed as means ± SEM. Control cecum, n = 8; Probiotic cecum, n = 4; Control colon, n = 7; Probiotic colon, n = 10, Kruskal-Wallis test. D: Non-metric multidimensional scaling (nMDS) analysis of colon and cecum faecal content from control and probiotic-treated animals. Each point represents a sample, groups identified by coloration, according to legend. E, F. Relative abundance at the phylum level in the cecum and colon from control and treated with probiotic Groups. E: Each bar represents an individual animal, the colour of each cell indicates the relative abundance of bacterial phyla. Taxa with relative abundance greater than 1% are shown. F: Box-plot represents log relative normalized abundance at the phylum level in the cecum and colon from control and probiotic-treated groups. ‘C’ for Control and ‘P’ for Probiotic. Only the significant P values obtained by the Kruskal-Wallis test are shown as q value.
Figure 3.
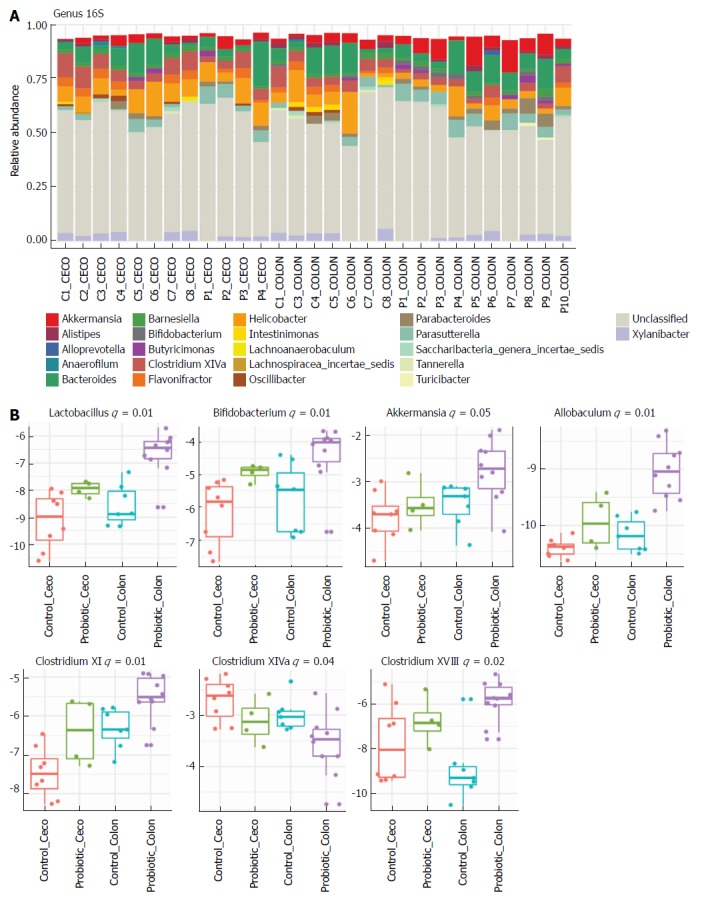
Probiotic supplementation changes the gut microbiota in the colon at the genus level. Relative abundance at the genus level in the cecum and colon from control and probiotic-treated groups at day 60 after colon cancer induction. A: Each bar represents an individual animal, and the colour of each cell indicates the relative abundance of the bacterial genus. Taxa with relative abundance greater than 1% are shown. ‘C’ for Control and ‘P’ for Probiotic. B: Box-plot represents log relative normalized abundance at the genus level in the cecum and colon from control and probiotic-treated groups. Only significant P values (q value) obtained by the Kruskal-Wallis test are shown. Control cecum, n = 8; Probiotic cecum, n = 4; Control Colon, n = 7; Probiotic colon, n = 10. Kruskal-Wallis test.
Probiotic supplementation modulates inflammation in a colitis associated colorectal carcinogenesis model
At day 60, the colonic tissue were extracted and prepared for histological analysis, and tissue inflammation was assessed by determining the inflammatory index. The probiotic group had a lower inflammatory index than the control group [control = 7.9 (± 1.6), n = 9, probiotic = 4.2 (± 1.0), n = 9; P = 0.0005], an approximately 46% reduction (Figure 4A and B). Spleen weight did not differ significantly between treatments [control = 0.18 (± 0.05), n = 21; probiotic = 0.12 (± 0.06), n = 25; P = 0.38] (Figure 4C).
Figure 4.
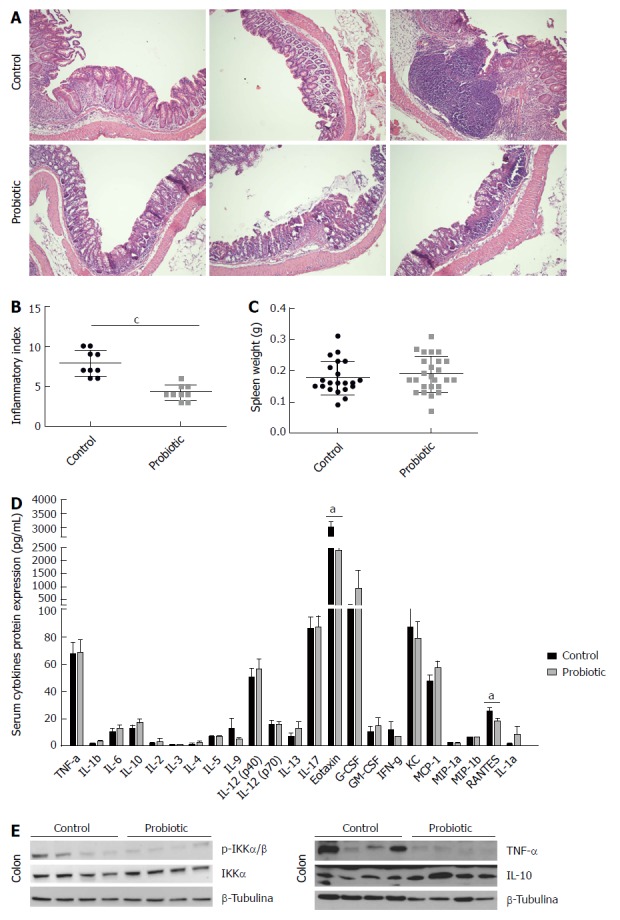
Probiotic supplementation modulates inflammation in colorectal carcinogenesis associated with colitis. Representative images (A) and inflammatory index (B) of mouse colons from control and probiotic groups at day 60 after azoxymethane injection stained with haematoxylin and eosin. cP < 0.001 Mann-Whitney U test, data representative of two independent experiments presented as mean and standard deviation. C: Spleen weight in control group (n = 21) and probiotic (n = 25) mice. Mann-Whitney U test. Data from two independent experiments are presented as mean and standard deviation. D: Concentration of serum cytokines analysed by Bio-Plex Multiplex cytokine assay at day 60 after injection with azoxymethane. aP < 0.05; Mann-Whitney U test. Data from two independent experiments, presented as mean and standard deviation. E: Representative western blot images of two independent experiments showing colon lysates of control and probiotic group mice; p-IKKα/β, IKKα, TNF-α, IL10 and β-tubulin.
In addition, serum cytokines were measured to assess whether probiotic supplementation could modulate systemic inflammation. There was an increase in important chemokines in the control group compared with the probiotic group: RANTES [control = 26.0 (± 8.4), P = 14; probiotic = 18.5 (± 6.9), n = 15; P = 0.03] and Eotaxin [control = 3010 (± 704.4), n = 14, probiotic = 2384 (± 854.4), n = 15; P = 0.02]. Increases in the mean levels of the cytokines IL-10 and IL-13 and a decrease in IFN-γ were observed, but these differences were not statistically significant. Other cytokines assessed did not differ between the control and probiotic groups (Figure 4D).
We further investigated whether intracellular inflammatory pathways in colonic tissue could be modulated by probiotic supplementation. Our results demonstrated that the probiotic group had lower expression of the phosphorylated protein IKK, reduced TNF-α expression and increased IL-10 expression, which indicates less activation of inflammatory pathways (Figure 4E).
Probiotic supplementation does not modulate cytokine expression in the tumour microenvironment
In order to evaluate the expression of cytokines in the tumour environment, we evaluated the expression of cytokines in the tumour tissue by RT-PCR (Figure 5A-G) and by Bio-Plex Multiplex cytokine assay (Figure 5H) at day 60 after injection with azoxymethane. We observed an interesting but non-significant increase in the mRNA expression of the cytokine TGF-β, which was approximately 25% higher in the probiotic group [control = 1.00 (± 0.29) n = 17, probiotic = 1.24 (± 0.47) n = 21, P = 0.06] (Figure 5E). Changes in other cytokines were not statistically significant.
Figure 5.
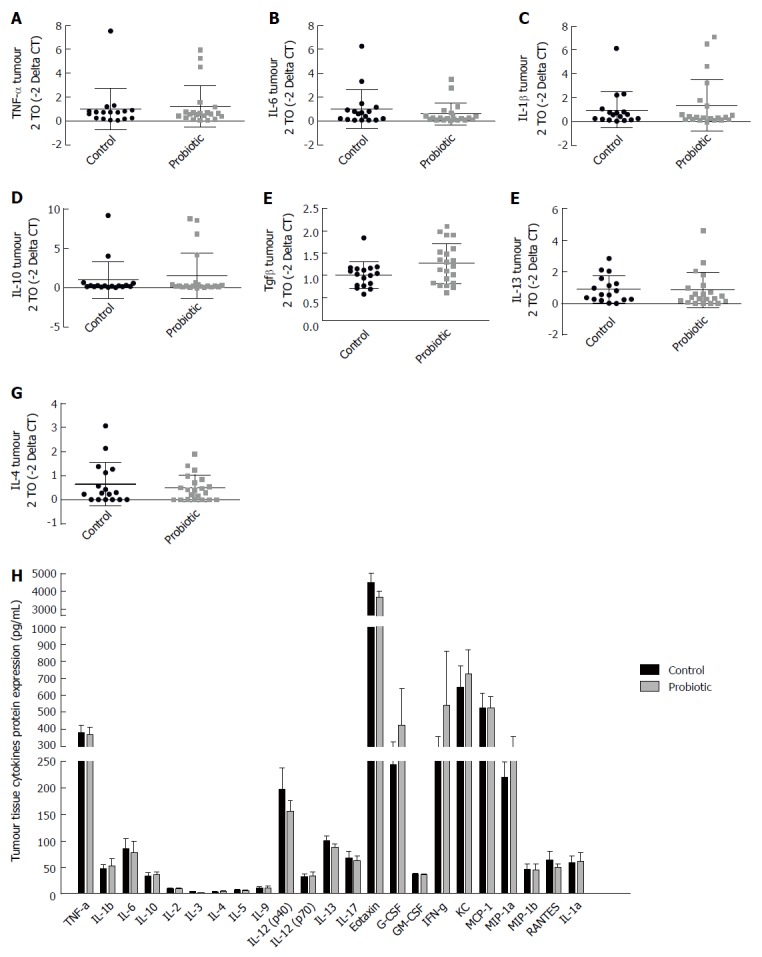
Probiotic supplementation does not modify the tumour microenvironment. Real-time RT-PCR analysis of TNF-α (A), IL-6 (B), IL-1β (C), IL-10 (D), TGF-β1 (E), and IL-13 (F), IL-4(G) in colon tumour tissue of control and probiotic groups; results are presented relative to those of Gapdh. H: Concentration of cytokines in the tumour tissue analysed by Bio-Plex Multiplex cytokine assay at day 60 after injection with azoxymethane. Mann-Whitney U test. Data from two independent experiments, presented as mean and standard deviation.
DISCUSSION
The present study demonstrates that supplementation with Lactobacillus rhamnosus, Lactobacilus acidophilus and Bifidobacterium bifidum reduced the colorectal tumour burden in mice, preventing colitis with a change in microbiota composition, reduction of inflammatory pathways in the colon, and modulation of cytokine and chemokine expression. To the best of our knowledge, no other study has evaluated the impact of the association of these strains in probiotic supplementation on the richness, diversity and abundance of the colon microbiota in colitis-associated cancer (CAC).
Prior reports showed that the isolated treatment with Lactobacillus acidophilus, Lactobacillus rhamnosus or Bifidobacterium bifidum are associated with tumour suppressive effects in colon cancer cell lines and in experimental tumour models[27-30]. Moreover, clinical studies showed that Lactobacillus and Bifidobacterium are frequently reduced in patients with intestinal bowel disease or CRC[31]. The enrichment or depletion of different microbial strains and the change in microbial diversity is considered essential for the promotion of inflammation, proliferation and neoplastic progression[32]. Here, we used the association of Lactobacillus acidophilus, Lactobacillus rhamnosus and Bifidobacterium bifidum to assess if it can favourably alter the microbiota composition[33].
In this study, the alpha diversity (i.e., the number of different taxa or microbial species that could be detected in one sample) was assessed by the Shannon index and richness by the Chao index in the gut microbiota of the colon, and there was no difference between the control and probiotic groups. Otherwise, a significant difference was observed in beta diversity (i.e., the diversity in microbial community between different samples, accessed by the microbial composition abundances) and in the microbial composition at the genus and phylum levels. Based on these facts, it is possible to affirm that probiotic supplementation could change the structure of the microbiota.
The phylum Actinobacteria was increased in CAC supplemented with probiotics. Interestingly, Gao et al[34] using 16S rDNA sequencing observed that at the phylum level the number of Actinobacteria and Firmicutes decreased in the gut of CRC patients. Actinobacteria is a phylum of gram-positive bacteria, and the genus Bifidobacterium is one of its main components[35]; accordingly, probiotic supplementation increased the prevalence of this genus in CAC. Importantly, analysing the mucosa-adherent microbiota, Chen et al[36] identified reduced Bifidobacterium, Faecalibacterium and Blautia in CRC patients, while Fusobacterium, Porphyromonas, Peptostreptococcus and Mogibacterium were enriched. Similarly, colonic mucosa samples of patients with CRC presented a reduced amount of B. longum and B. bifidum compared with those in patients with diverticulitis[31]. These data suggest that probiotic supplementation alters the CAC microbiota to an anti-neoplastic one. Consistent with this hypothesis, probiotic supplementation increased the abundance of Lactobacillus in CAC. In particular, Lactobacillus not only prevents DMH induced colon carcinogenesis in rats[37,38] but also ameliorates inflammation in an experimental model of colon cancer[28,39,40]. Akkermansia muciniphila is another intestinal bacterium which may have potential anti-inflammatory properties in metabolic disorders, and it has been inversely associated with obesity, diabetes, cardiometabolic diseases and low-grade inflammation[41]. In other colitis model, such as interleukin10 knockout mice, supplementation with Lactobacillus plantarum LPOnlly ameliorates colon inflammation by microbiome alteration[42], while a combination of Bifidobacterium longum, Lactobacillus acidophilus, Enterococcus faecalis improved epithelial-barrier function and reduced proinflammatory cytokines secretion[43]. A. muciniphila was reduced in ulcerative colitis patients[44], but was positively associated with CRC patients[45]. As a mucin-degrading commensal bacterium, it can impair intestinal barrier function, promoting colitis[46]. In contrast, other studies found that A. muciniphila increases the density of mucus-producing goblet cells, restoring the mucus layer[47]. In an experimental study, orally administered A. muciniphila extracellular vesicles protected against DSS-induced colitis, reducing proinflammatory cytokine expression, increasing colon length and reducing inflammatory cell infiltration of the colon wall[48]. We found an increase trend in A. muciniphila abundance in the probiotic group, which, in association with other microorganisms, may have prevented colitis in our study.
Furthermore, the abundance of short-chain fatty acid (SCFA)-producing bacteria, like Allobaculum, was increased in the probiotic group. SCFA (acetate, propionate, and butyrate) are produced through the fermentation of non-digestible carbohydrates by intestinal bacteria and play an important role in maintaining intestinal health with anti-inflammatory and antineoplastic properties[49]. Antineoplastic properties of SCFAs are linked to the production of anti-inflammatory cytokines such as IL10 and TGF-β[50]. Likewise, Bifidobacterium and S. thermophilus also stimulate the release of TGF-β[51]. Considering that, we found an increase in the expression of TGF-β mRNA in tumour tissues of the probiotic group. It is possible that TGF-β may be involved in the anti-inflammatory and anti-neoplastic effects of probiotic supplementation, and this deserves further investigation. Another mechanism by which probiotics can prevent inflammation is by modulating signalling pathways, inhibiting the PI3K/Akt and IKK/NF-κB pathway, thereby modulating cytokine and chemokine secretion[22,52]. In our study, probiotic supplementation reduced proinflammatory pathway activity, decreasing IKK activation in colon, suggesting a local effect. Notably, a similar result was observed in a study with ulcerative colitis patients patients where probiotic consumption increased IL-10, and decreased TNF-α and IL-1β, inhibiting NF-κB expression[53].
In accordance with a reduced activity of NF-κB, our data demonstrate that the probiotic-supplemented group presented reduced expression of CCL5/RANTES in serum. Given that CCL5/RANTES may promote tumour growth by stimulating proliferative pathways and angiogenesis and recruiting inflammatory cells[54-56], it is plausible to hypothesize that those chemokines are involved in tumour development, collaborating to reduced tumour burden in the probiotic-supplemented group. On the other hand, probiotic supplementation reduced the expression of eotaxin, a chemokine primarily responsible for eosinophil recruitment during inflammation, which may contribute to preventing the recruitment of inflammatory cells to the colon in the probiotic group. Eosinophils are potent proinflammatory cells, capable to produce and release cytotoxic proteins, cytokines, and metabolites reactive to oxygen, causing severe damage to the tissue. Eosinophils accumulation is common in patients with ulcerative colitis and active inflammatory bowel disease[57,58].
In aggregate these studies indicate that intestinal microbiota modulates carcinogenesis in different steps of carcinogenesis. Interestingly, the probiotic supplementation composition used in this study have its effects more pronounced in tumour initiation and promotion, as we found decreased tumour number and smaller tumour size in probiotic group.
It is plausible that probiotic supplementation can be included in clinical practice, preventing CRC in patients at higher risk of colitis. However, it is necessary to conduct a clinical trial to confirm this hypothesis. In conclusion, our results suggest a potential chemopreventive effect of probiotic supplementation on CRC. Microbiota changed by probiotic supplementation promote intestinal homeostasis and regulate the inflammatory response, reducing inflammatory cell infiltration by lowering chemokine expression, thus preventing CAC (Figure 6).
Figure 6.
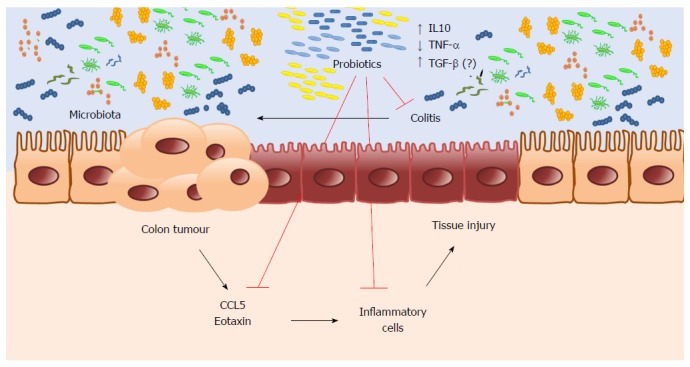
Microbiota modification by probiotic supplementation reduces colitis modulating cytokines expression preventing colon cancer development in mice.
ARTICLE HIGHLIGHTS
Research background
Derangement in intestinal microbial composition impacts in mucosal inflammation, tumour promotion and neoplastic progression. Given that the intestinal microbiota can be modulated by several factors, and probiotic supplementation is an interesting alternative to re-establish intestinal eubiosis. Furthermore, in vitro studies and experiments with animal models demonstrated that several bacteria strains (probiotics) can modulate proliferative, apoptotic and inflammatory pathways; increase the innate immune response; produce anti-tumourigenic and anti-mutagenic compounds and destroy carcinogens; reduce genotoxicity; and increase intestinal barrier. Thus, probiotic modulation of intestinal microbiota has emerged as a potential chemopreventive agent.
Research motivation
Despite the idea that probiotic supplementation could prevent colorectal cancer (CRC), little is known about the supplementation of a mix of bacterial probiotic strains as well as its impact in the intestinal microbiota composition and neoplastic transformations of the intestinal mucosa. Our data can contribute to solve the gaps in the literature of whether this mix of probiotic, dose and time of supplementation used was able to alter the alpha and beta diversity of the intestinal microbiota, and how this treatment impact in colitis, serum cytokines and neoplastic development.
Research objectives
The aim of this work is to investigate the effect of supplementation of a Lactobacillus acidophilus, Lactobacillus rhamnosus and Bifidobacterium bifidum mixture on the intestinal microbiota composition, inflammation and neoplastic alterations in the colon during the development of an experimental model of colitis associated colon cancer (CAC). Overall, this study intents to strengthen data from preclinical studies, encouraging clinical trials to investigate their role in preventing colitis and CRC in humans.
Research methods
We used an experimental model of CAC. C57BL/6 mice received intraperitoneal injection of Azoxymethane, followed by 3 cycles of 2.5% dextran sulphate sodium in drinking water, with an interval of 14 days between cycles. The intervention group received by gavage daily 0.6 billion CFU (colony forming units) each of Lactobacillus acidophilus, Lactobacillus rhamnosus and Bifidobacterium bifidum, diluted in 200 μL of drinking water, while the control group received 200 μL of drinking water daily. Colon tissues were collected for inflammatory index analysis in histological sheets and western blotting to assess inflammatory proteins expression. Cytokines expression in serum and tumour tissue was performed by multiplex immunoassay, and in tumour samples were also used Real Time-PCR. Microbiota analysis was done from colon faeces using 16S rRNA sequencing method.
Research results
Probiotic supplementation reduces tumour incidence in a colitis associated colorectal model, we found decreased tumour number and smaller tumour size in probiotic group. In parallel, probiotic supplementation changes the gut microbiota in the colon. We did not detect any change in alpha diversity of the intestinal microbiota, but a difference in beta diversity and in the microbial composition at the genus and phylum level. In addition, probiotic supplementation reduced 46% the inflammatory index compared to the control group. Overall, these results highlight the potential for use of these probiotics mixture to human colitis to reduce inflammation and prevent colon cancer. Thus, further clinical trials are needed to confirm these preclinical insights.
Research conclusions
We found that supplementation with Lactobacillus acidophilus, Lactobacillus rhamnosus and Bifidobacterium bifidum during colitis associated colorectal carcinogenesis model changed intestinal microbiota, without altering richness and diversity of intestinal microbiota. Lactobacillus and Bifidobacterium increased in probiotic group and may be responsible for chemopreventive effect of probiotic supplementation on CRC. In summary, we suggest that probiotic supplementation could prevent CAC development by changes in microbiota composition which promotes intestinal homeostasis and regulates the inflammatory response, reducing inflammatory cell infiltration by lowering chemokine expression.
Research perspectives
The present study made biological plausible that probiotic supplementation can reduce inflammation and prevent CRC in patients with colitis. Therefore, clinical trials are needed to confirm this hypothesis and increase the therapeutic arsenal against this haunted disease.
ACKNOWLEDGMENTS
We thank Renata Bagarolli, Carla Bueno Silva and Patrícia Villas Boas for contributions to animal gavage; Luiz Janeri (in memorium), Josimo Pinheiro, Dioze Guadagnini and Andrey Santos from State University of Campinas (UNICAMP) for technical assistance and the staff of the Life Sciences Core Facility (LaCTAD) from State University of Campinas (UNICAMP) for Cell Biology and Genomics analysis. We thank the compounding pharmacy “Ao Pharmacêutico”, Campinas – Brazil for the donation of probiotics.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Brazil
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Supported by São Paulo Research Foundation (FAPESP, Brazil, No. 07607-8/2013); and National Research Council (CNPq, No. 150127/2016-2, No. 306821/2010-9).
Institutional animal care and use committee statement: The Ethics Committee on Animal Use (CEUA) of UNICAMP approved the project, according to protocol no. 2761-1.
Conflict-of-interest statement: No potential conflicts of interest were disclosed.
Data sharing statement: No additional data are available.
ARRIVE guidelines statement: The authors have read the ARRIVE Guidelines, and the manuscript was prepared and revised according to the ARRIVE Guidelines.
Peer-review started: March 7, 2018
First decision: April 3, 2018
Article in press: April 23, 2018
P- Reviewer: Chiba T, Drastich P, Kreisel W, Pellicano R S- Editor: Gong ZM L- Editor: A E- Editor: Huang Y
Contributor Information
Maria Carolina S Mendes, Department of Internal Medicine, Faculty of Medical Sciences, State University of Campinas (UNICAMP), Campinas, São Paulo 13083-887, Brazil.
Daiane SM Paulino, Department of Internal Medicine, Faculty of Medical Sciences, State University of Campinas (UNICAMP), Campinas, São Paulo 13083-887, Brazil.
Sandra R Brambilla, Department of Internal Medicine, Faculty of Medical Sciences, State University of Campinas (UNICAMP), Campinas, São Paulo 13083-887, Brazil.
Juliana A Camargo, Department of Internal Medicine, Faculty of Medical Sciences, State University of Campinas (UNICAMP), Campinas, São Paulo 13083-887, Brazil.
Gabriela F Persinoti, Brazilian Bioethanol Science and Technology Laboratory (CTBE), Brazilian Center for Research in Energy and Materials (CNPEM), Campinas, São Paulo 13083-970, Brazil.
José Barreto C Carvalheira, Department of Internal Medicine, Faculty of Medical Sciences, State University of Campinas (UNICAMP), Campinas, São Paulo 13083-887, Brazil. jbcc@g.unicamp.br.
References
- 1.Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22:557–566. doi: 10.3748/wjg.v22.i2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Q, Gao R, Wu W, Qin H. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumour Biol. 2013;34:1285–1300. doi: 10.1007/s13277-013-0684-4. [DOI] [PubMed] [Google Scholar]
- 6.O’Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13:691–706. doi: 10.1038/nrgastro.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tennyson CA, Friedman G. Microecology, obesity, and probiotics. Curr Opin Endocrinol Diabetes Obes. 2008;15:422–427. doi: 10.1097/MED.0b013e328308dbfb. [DOI] [PubMed] [Google Scholar]
- 8.Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. 2016;22:713–722. doi: 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
- 9.Brennan CA, Garrett WS. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu Rev Microbiol. 2016;70:395–411. doi: 10.1146/annurev-micro-102215-095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 11.Mantovani A. From phagocyte diversity and activation to probiotics: back to Metchnikoff. Eur J Immunol. 2008;38:3269–3273. doi: 10.1002/eji.200838918. [DOI] [PubMed] [Google Scholar]
- 12.Didari T, Mozaffari S, Nikfar S, Abdollahi M. Effectiveness of probiotics in irritable bowel syndrome: Updated systematic review with meta-analysis. World J Gastroenterol. 2015;21:3072–3084. doi: 10.3748/wjg.v21.i10.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JY, Kwon JH, Ahn SH, Lee SI, Han YS, Choi YO, Lee SY, Ahn KM, Ji GE. Effect of probiotic mix (Bifidobacterium bifidum, Bifidobacterium lactis, Lactobacillus acidophilus) in the primary prevention of eczema: a double-blind, randomized, placebo-controlled trial. Pediatr Allergy Immunol. 2010;21:e386–e393. doi: 10.1111/j.1399-3038.2009.00958.x. [DOI] [PubMed] [Google Scholar]
- 14.King S, Glanville J, Sanders ME, Fitzgerald A, Varley D. Effectiveness of probiotics on the duration of illness in healthy children and adults who develop common acute respiratory infectious conditions: a systematic review and meta-analysis. Br J Nutr. 2014;112:41–54. doi: 10.1017/S0007114514000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mennini M, Dahdah L, Artesani MC, Fiocchi A, Martelli A. Probiotics in Asthma and Allergy Prevention. Front Pediatr. 2017;5:165. doi: 10.3389/fped.2017.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, Buys NJ. Glucose- and glycaemic factor-lowering effects of probiotics on diabetes: a meta-analysis of randomised placebo-controlled trials. Br J Nutr. 2016;115:1167–1177. doi: 10.1017/S0007114516000076. [DOI] [PubMed] [Google Scholar]
- 17.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bienenstock J, Gibson G, Klaenhammer TR, Walker WA, Neish AS. New insights into probiotic mechanisms: a harvest from functional and metagenomic studies. Gut Microbes. 2013;4:94–100. doi: 10.4161/gmic.23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlando A, Russo F. Intestinal microbiota, probiotics and human gastrointestinal cancers. J Gastrointest Cancer. 2013;44:121–131. doi: 10.1007/s12029-012-9459-1. [DOI] [PubMed] [Google Scholar]
- 20.Bron PA, Kleerebezem M, Brummer RJ, Cani PD, Mercenier A, MacDonald TT, Garcia-Ródenas CL, Wells JM. Can probiotics modulate human disease by impacting intestinal barrier function? Br J Nutr. 2017;117:93–107. doi: 10.1017/S0007114516004037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geier MS, Butler RN, Howarth GS. Probiotics, prebiotics and synbiotics: a role in chemoprevention for colorectal cancer? Cancer Biol Ther. 2006;5:1265–1269. doi: 10.4161/cbt.5.10.3296. [DOI] [PubMed] [Google Scholar]
- 22.Thomas CM, Versalovic J. Probiotics-host communication: Modulation of signaling pathways in the intestine. Gut Microbes. 2010;1:148–163. doi: 10.4161/gmic.1.3.11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 25.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper HS, Murthy SN, Shah RS, Sedergran DJ. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69:238–249. [PubMed] [Google Scholar]
- 27.Urbanska AM, Bhathena J, Martoni C, Prakash S. Estimation of the potential antitumor activity of microencapsulated Lactobacillus acidophilus yogurt formulation in the attenuation of tumorigenesis in Apc(Min/+) mice. Dig Dis Sci. 2009;54:264–273. doi: 10.1007/s10620-008-0363-2. [DOI] [PubMed] [Google Scholar]
- 28.Gamallat Y, Meyiah A, Kuugbee ED, Hago AM, Chiwala G, Awadasseid A, Bamba D, Zhang X, Shang X, Luo F, et al. Lactobacillus rhamnosus induced epithelial cell apoptosis, ameliorates inflammation and prevents colon cancer development in an animal model. Biomed Pharmacother. 2016;83:536–541. doi: 10.1016/j.biopha.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Jacouton E, Chain F, Sokol H, Langella P, Bermúdez-Humarán LG. Probiotic Strain Lactobacillus casei BL23 Prevents Colitis-Associated Colorectal Cancer. Front Immunol. 2017;8:1553. doi: 10.3389/fimmu.2017.01553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuugbee ED, Shang X, Gamallat Y, Bamba D, Awadasseid A, Suliman MA, Zang S, Ma Y, Chiwala G, Xin Y, et al. Structural Change in Microbiota by a Probiotic Cocktail Enhances the Gut Barrier and Reduces Cancer via TLR2 Signaling in a Rat Model of Colon Cancer. Dig Dis Sci. 2016;61:2908–2920. doi: 10.1007/s10620-016-4238-7. [DOI] [PubMed] [Google Scholar]
- 31.Gueimonde M, Ouwehand A, Huhtinen H, Salminen E, Salminen S. Qualitative and quantitative analyses of the bifidobacterial microbiota in the colonic mucosa of patients with colorectal cancer, diverticulitis and inflammatory bowel disease. World J Gastroenterol. 2007;13:3985–3989. doi: 10.3748/wjg.v13.i29.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Yang Y, Huycke MM. Microbiome-driven carcinogenesis in colorectal cancer: Models and mechanisms. Free Radic Biol Med. 2017;105:3–15. doi: 10.1016/j.freeradbiomed.2016.10.504. [DOI] [PubMed] [Google Scholar]
- 33.Thomas AM, Jesus EC, Lopes A, Aguiar S Jr, Begnami MD, Rocha RM, Carpinetti PA, Camargo AA, Hoffmann C, Freitas HC, Silva IT, Nunes DN, Setubal JC, Dias-Neto E. Tissue-Associated Bacterial Alterations in Rectal Carcinoma Patients Revealed by 16S rRNA Community Profiling. Front Cell Infect Microbiol. 2016;6:179. doi: 10.3389/fcimb.2016.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao R, Kong C, Huang L, Li H, Qu X, Liu Z, Lan P, Wang J, Qin H. Mucosa-associated microbiota signature in colorectal cancer. Eur J Clin Microbiol Infect Dis. 2017;36:2073–2083. doi: 10.1007/s10096-017-3026-4. [DOI] [PubMed] [Google Scholar]
- 35.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verma A, Shukla G. Probiotics Lactobacillus rhamnosus GG, Lactobacillus acidophilus suppresses DMH-induced procarcinogenic fecal enzymes and preneoplastic aberrant crypt foci in early colon carcinogenesis in Sprague Dawley rats. Nutr Cancer. 2013;65:84–91. doi: 10.1080/01635581.2013.741746. [DOI] [PubMed] [Google Scholar]
- 38.Verma A, Shukla G. Synbiotic (Lactobacillus rhamnosus + Lactobacillus acidophilus+inulin) attenuates oxidative stress and colonic damage in 1,2 dimethylhydrazine dihydrochloride-induced colon carcinogenesis in Sprague-Dawley rats: a long-term study. Eur J Cancer Prev. 2014;23:550–559. doi: 10.1097/CEJ.0000000000000054. [DOI] [PubMed] [Google Scholar]
- 39.Gosai V, Ambalam P, Raman M, Kothari CR, Kothari RK, Vyas BR, Sheth NR. Protective effect of Lactobacillus rhamnosus 231 against N-Methyl-N’-nitro-N-nitrosoguanidine in animal model. Gut Microbes. 2011;2:319–325. doi: 10.4161/gmic.18755. [DOI] [PubMed] [Google Scholar]
- 40.Zahran WE, Elsonbaty SM, Moawed FSM. Lactobacillus rhamnosus ATCC 7469 exopolysaccharides synergizes with low level ionizing radiation to modulate signaling molecular targets in colorectal carcinogenesis in rats. Biomed Pharmacother. 2017;92:384–393. doi: 10.1016/j.biopha.2017.05.089. [DOI] [PubMed] [Google Scholar]
- 41.Cani PD, de Vos WM. Next-Generation Beneficial Microbes: The Case of Akkermansia muciniphila. Front Microbiol. 2017;8:1765. doi: 10.3389/fmicb.2017.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen H, Xia Y, Zhu S, Yang J, Yao J, Di J, Liang Y, Gao R, Wu W, Yang Y, et al. Lactobacillus plantarum LPOnlly alters the gut flora and attenuates colitis by inducing microbiome alteration in interleukin10 knockout mice. Mol Med Rep. 2017;16:5979–5985. doi: 10.3892/mmr.2017.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi CZ, Chen HQ, Liang Y, Xia Y, Yang YZ, Yang J, Zhang JD, Wang SH, Liu J, Qin HL. Combined probiotic bacteria promotes intestinal epithelial barrier function in interleukin-10-gene-deficient mice. World J Gastroenterol. 2014;20:4636–4647. doi: 10.3748/wjg.v20.i16.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vigsnæs LK, Brynskov J, Steenholdt C, Wilcks A, Licht TR. Gram-negative bacteria account for main differences between faecal microbiota from patients with ulcerative colitis and healthy controls. Benef Microbes. 2012;3:287–297. doi: 10.3920/BM2012.0018. [DOI] [PubMed] [Google Scholar]
- 45.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8:e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, Park SK, Jeon SG, Roh TY, Myung SJ, et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One. 2013;8:e76520. doi: 10.1371/journal.pone.0076520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008;19:587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 50.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 51.Donkor ON, Ravikumar M, Proudfoot O, Day SL, Apostolopoulos V, Paukovics G, Vasiljevic T, Nutt SL, Gill H. Cytokine profile and induction of T helper type 17 and regulatory T cells by human peripheral mononuclear cells after microbial exposure. Clin Exp Immunol. 2012;167:282–295. doi: 10.1111/j.1365-2249.2011.04496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai C, Zheng CQ, Meng FJ, Zhou Z, Sang LX, Jiang M. VSL#3 probiotics exerts the anti-inflammatory activity via PI3k/Akt and NF-κB pathway in rat model of DSS-induced colitis. Mol Cell Biochem. 2013;374:1–11. doi: 10.1007/s11010-012-1488-3. [DOI] [PubMed] [Google Scholar]
- 53.Hakansson A, Molin G. Gut microbiota and inflammation. Nutrients. 2011;3:637–682. doi: 10.3390/nu3060637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allavena P, Germano G, Marchesi F, Mantovani A. Chemokines in cancer related inflammation. Exp Cell Res. 2011;317:664–673. doi: 10.1016/j.yexcr.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 55.Oppermann M. Chemokine receptor CCR5: insights into structure, function, and regulation. Cell Signal. 2004;16:1201–1210. doi: 10.1016/j.cellsig.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 56.Aldinucci D, Colombatti A. The inflammatory chemokine CCL5 and cancer progression. Mediators Inflamm. 2014;2014:292376. doi: 10.1155/2014/292376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matthews AN, Friend DS, Zimmermann N, Sarafi MN, Luster AD, Pearlman E, Wert SE, Rothenberg ME. Eotaxin is required for the baseline level of tissue eosinophils. Proc Natl Acad Sci USA. 1998;95:6273–6278. doi: 10.1073/pnas.95.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zezos P, Patsiaoura K, Nakos A, Mpoumponaris A, Vassiliadis T, Giouleme O, Pitiakoudis M, Kouklakis G, Evgenidis N. Severe eosinophilic infiltration in colonic biopsies predicts patients with ulcerative colitis not responding to medical therapy. Colorectal Dis. 2014;16:O420–O430. doi: 10.1111/codi.12725. [DOI] [PubMed] [Google Scholar]


