Abstract
Cellular plasticity is now recognized as a fundamental feature of tissue biology. The steady-state differentiation of stem and progenitor cells into mature cells is, in itself, the index form of cellular plasticity in adult organisms. Following injury, when it is critical to quickly regenerate and restore tissue integrity and function, other types of cellular plasticity may be crucial for organismal survival. In these contexts, alterations in the epigenetic landscape of tissues are likely to occur in order to allow normally restricted cell fate transitions. Epigenetic mechanisms, particularly DNA methylation and histone modifications, have been shown to play an important role in regulating such plasticity. Relevant mechanisms have been well studied in the context of the direct reprograming of somatic cells into induced pluripotent stem cells. Indeed, epigenetic regulation of cell fate is part and parcel of normal embryonic development and is a central regulator of cellular diversity. This is normally thought to involve the establishment of divergent chromatin patterns that culminate in cells with distinct and what were previously thought to be irreversible fates. This brief review aims to put some of these new observations in the larger context of regeneration after injury.
Cellular plasticity
In multicellular organisms, individual progenitor cells are thought to undergo progressive cell fate restriction on the path to forming fully mature differentiated cells. This concept was promulgated by Conrad Waddington through his conceptualization of an epigenetic landscape for the embryo [1]. However, his diagram did not directly address the restriction of cell identity in adult tissues [2•]. Seminal experimental work in hematopoiesis reinforced his paradigm. This thinking was naturally extended to solid tissues. However, modern experimental evidence has revealed that cell state is remarkably dynamic, especially after injury in epithelia.
It is likely that some forms of adult cellular plasticity are central for organismal survival following injury, particularly when it is critical to quickly restore tissue integrity and function after the loss of cells [3,4]. Plasticity phenomena were initially described on the basis of careful histologic and marking experiments and can now be categorized into a few baskets based on stringent genetic lineage tracing with cell type specific markers: (1) a mature cell can dedifferentiate and revert into a progenitor cell of the same lineage, (2) a mature cell can transdifferentiate into another mature cell, and (3) a progenitor cell can transdetermine and convert into another type of progenitor cell. With regard to dedifferentiation, it is now known that a fully differentiated secretory cell in the mammalian airway can dedifferentiate into a stem cell following ablation of the original stem cell population [5]. Similar examples of dedifferentiation have been reported in fly testis [6,7], and in the stomach and intestine [8–11]. With regard to transdifferentiation, there is evidence that mature δ-cells of the pancreas and the hepatocytes of the liver can convert into insulin producing β-cells and biliary epithelial cells, respectively [12,13]. In the case of transdetermination, work in the fly imaginal disks revealed that progenitor cells could adopt the behavior of related but distinct progenitors [14–16]. The basis of these forms of plasticity is just beginning to be defined. Some of it is likely based on the nature of pre-existing transcriptional networks. But clearly, in the context of injury and environmental perturbation, there must be a rewiring of the epigenetic landscape in the sense that cells of a particular fate can be redirected into another distinct fate, despite the fact that these paths don’t normally exist in the embryo or in steady state adult tissues. In emerging new data, epigenetics, in the more restricted modern usage of the term (inheritable, non-genetic histone and DNA alteration), is also clearly at play in regulating plasticity after injury.
There are three major classes of epigenetic modifiers that govern gene expression: (1) DNA methylation, (2) histone marks, and (3) non-coding RNAs. Proteins that read, write, and/or erase DNA and histone modifications are well described to play key roles in the regulation of cell identity. When promoters and transcription start sites are methylated, activating transcription factors are prevented from binding these regulatory elements or repressive chromatin remodeling complexes are recruited to these regions and result in the repression of gene expression [17–19]. Histone modifications often result in an alteration of the distance between nucleosomes, and have an impact on chromatin compaction and result in the recruitment of histone-modifying complexes that activate or repress gene expression [20]. Genomic imprinting is a prominent example of epigenetic regulation during development. X-chromosome inactivation is regulated by histone modifications and the action of a non-coding RNA, called Xist [21–23].
Polycomb group (PcG) proteins are important epigenetic regulators that act in synergy during development to deposit repressive histone marks that govern tissue-specific gene expression in adulthood [24,25]. The polycomb repressive complex (PRC)-2 mediates the deposition of H3K27me3 via the catalytically active SET-domain-containing proteins Ezh1 and Ezh2, whereas the other two core PRC2 members, Suz12 and Eed, are required for complex stability [26].
The epigenetic basis of cellular plasticity has been very well studied during the direct reprogramming of somatic cells into induced pluripotent stem cells (iPSCs). In addition to alterations of the transcriptional network, ectopic expression of reprogramming transcription factors generates a chromatin landscape that is highly similar to that of embryonic stem cells (ESCs) [27–30•]. Similarly, open chromatin in ESCs is maintained through the action of chromatin-modifying complexes [31•–33]. The INO80 complex, a SWI/SNF family chromatin remodeler, has been shown to play a role in ESC self-renewal and direct reprogramming. INO80 is recruited to pluripotency loci and mediates the maintenance of an accessible chromatin state [31•]. During reprogramming, chromatin alterations are also caused by the induction of locus-specific DNA demethylation [30•,34–36]. Following reprogramming of the female fibroblast cells into iPSCs, the somatic epigenome is globally reversed into an epigenetic state similar to ES cells. In this case, the previously silenced X chromosome is reactivated, indicating that the epigenetic marks can be erased upon reprogramming [30•]. The newly activated X chromosome undergoes random X inactivation upon subsequent differentiation of iPSCs, suggesting that the newly forming epigenetic state can be re-established, independent of the previous epigenetic landscape [30•].
The molecular epigenetic basis of cellular plasticity in adult tissues
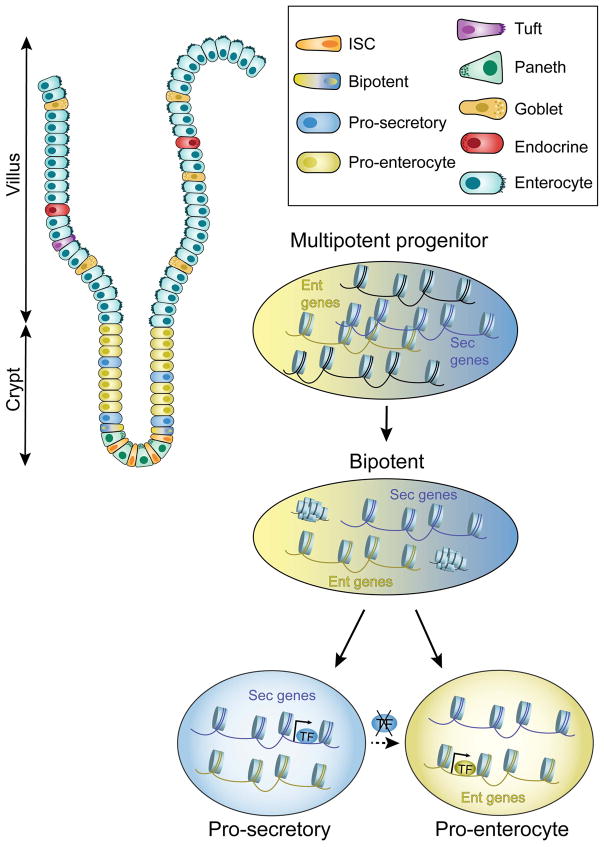
In many ways, the index form of adult cellular plasticity is the steady state differentiation of stem and progenitors cells into mature cells [37]. In the case of the intestine, multipotent stem cells possess a broadly permissive chromatin configuration that presumably allows multiple pathways of differentiation to occur (Figure 1) [38••]. During intestinal stem cell differentiation, Notch-mediated lateral inhibition governs the cell fate choice between a secretory and an enterocyte lineage. Interestingly, both secretory and absorptive progenitors showed comparable levels of activating histone marks, H3K4me2 and H3K27ac. Similarly, DNaseI hypersensitivity suggested open chromatin states that allow for either final cell fate choice in both sets of progenitors. The binding of a secretory-specific transcription factor, ATOH1, in intestinal stem cells promotes secretory progenitor cell differentiation. When Atoh1 is depleted from specified secretory cells, increased enterocyte progenitors are formed (Figure 1) [38••]. This fate acquisition or transdifferentiation is possible because enterocyte-associated chromatin is retained in its open configuration in secretory progenitors. Thus, intestinal progenitors possess broadly open chromatin that allows cell fate switching based on the presence or absence of particular lineage-restricted transcription factors. Presumably, if differentiation was associated with the closing of chromatin linked to alternative lineage-specific genes, plasticity would be restricted.
Figure 1. Intestinal progenitors maintain an accessible chromatin state that underlies cell plasticity.
Although chromatin states become restricted in the course of the differentiation of intestinal stem cells into mature secretory and enterocyte cells, secretory progenitor cells maintain an open chromatin configuration at enterocyte loci that allows the conversion of secretory into enterocyte progenitors (normally regulated by lateral inhibition). Specifically, upon the loss of a secretory transcription factor, the secretory progenitor cell transdifferentiates into an enterocyte progenitor cell. Blue: secretory-associated factors; yellow: enterocyte-associated factors.
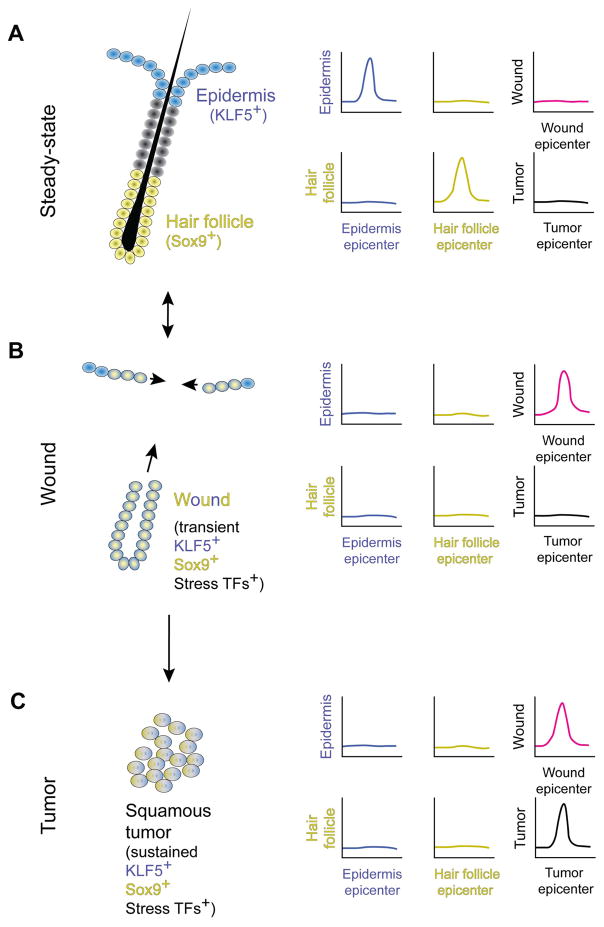
In the steady state epidermis and hair follicle, the respective stem cells express Klf5 and Sox9, and these lineage-associated transcription factors are required for the maintenance of these stem cells. The expression of these genes is regulated by specific epidermal and hair follicle epicenters within super enhancers (Figure 2A) [39••,40]. During wound repair, both Klf5 and Sox9 are expressed simultaneously. And this dual Klf5 and Sox9 expression in “wound stem cells” are necessary for repair. In the instance of wound cells, the transient co-expression of Klf5 and Sox9 is associated with (1) a new wound epicenter, (2) the loss of epidermal and hair follicle epicenters, and (3) the expression of activating stress-associated transcription factors (Figure 2B) [39••]. After wound repair, the steady state expression of Klf5 and Sox9 is restored in epidermal and hair follicle stem cells, respectively. In tumors, wound epicenters do occur, but are also associated with new tumor epicenters as well as a sustained expression of Klf5, Sox9, and stress-associated transcription factors (Figure 2C) [39••]. Therefore, while epigenetic plasticity is critical for proper wound repair, it must be tightly regulated to prevent cancer.
Figure 2. Epicenters located within super enhancers govern normal epithelial cell fate and wound repair requires the dual activation of epidermal and hair follicle gene expression through a wound-specific epicenter.
(A) The epidermis and hair follicle represent two distinct cell fates that are maintained by separate stem cells. The epidermal stem cells express Klf5 and the hair follicle stem cells express Sox9. In each case, their expression is regulated by a specific epicenter within a larger super enhancer. (B) In the case of wounding, stress-induced regulatory elements are activated to transiently allow cell plasticity. A new wound epicenter results in the co-expression of Klf5 and Sox9. This transient co-expression of epidermal and hair follicle genes is required for wound repair. (C) In tumors, the expression of stress-induced and epidermal and hair follicle lineage-specific transcription factors are sustained, resulting in the expression of oncogenes. This induction is engendered through the formation of a new tumor epicenter that occurs alongside a wound epicenter. Blue: epidermis; yellow: hair follicle; magenta: wound; black: tumor.
The epigenetic regulation of cellular plasticity in lungs
In lung, as in other tissues, epigenetic mechanisms regulating cell plasticity are just beginning to be explored. During development, conditional loss of Ezh2 (a SET-domain-containing subunit of the PRC2 complex, responsible for deposition of H3K27me3 [26]) results in defective branching morphogenesis and impaired alveolarization [41,42]. The loss of Ezh2 throughout the embryonic lung endoderm results in the precocious appearance of basal progenitor-like cells, possibly at the expense of secretory cells [41]. Therefore, Ezh2 seems to restrict the basal cell lineage during lung development, and allows proper differentiation of secretory cell population.
Proper saccule and alveoli formation in the developing lung epithelium also relies on the function of a histone deacetylase (Hdac3). Loss of Hdac3 leads to impaired spreading of alveolar epithelial type 1 cells and consequent defective sacculation at E18.5, a stage when type 1 cells expand substantially to line the increasing alveolar surface. Hdac3-mediated deacetylation results in the loss of the expression of miRNA17-92 which is required for the proper regulation of transforming growth factor β (TGF-β) signaling [43]. Overexpression of miRNA17-92 blocks epithelial differentiation, leading to increased number of early progenitors [44–46]. Again, this data points to a role for epigenetic regulation in normal cell fate differentiation during embryogenesis, but its role in the adult and in regeneration following injury is unclear.
DNA methylation has been shown to regulate the promoter activity of mouse surfactant protein b (Sftpb) since its expression is negatively correlated with DNA methylation level at the Sftpb promoter. The unmethylated Sftpb promoter possesses an active chromatin configuration marked by H3K4me3, an active histone modification. Specifically, Brahma-related gene-1 (Brg1), a catalytic subunit of the SWI/SNF chromatin-remodeling complex, is recruited to the Sftpb promoter in cells that express this surfactant protein. In mouse lung epithelial cell lines, Brg1 interacts with Nkx2.1, the cardinal lung lineage-specifying factor, facilitating its binding to the Sftpb promoter which ultimately leads to its increased transcription and surfactant production [47]. Loss of Brg1 in epithelial cells decreases the level of active histone mark H3K4me3 at the Sftpb promoter, leading to decreased surfactant protein expression [47]. Thus, DNA methylation and histone marking both regulate the surfactant protein b expression in cooperation with the lung lineage-specifying transcription factor Nkx2.1.
The Hopx protein, a transcription cofactor and a target of Nkx2.1 and Gata6, is expressed in the developing airway epithelium, in an overlapping pattern with Hdac2, a histone deacetylase. Hopx controls the maturation of alveolar epithelial type 2 cells and the expression of surfactant proteins [48]. Depletion of Hopx results in impaired development of type 2 cells, increased surfactant protein expression and defective alveolar formation [48]. Hopx also interacts with Hdac2, implying a potential epigenetic regulation of type 2 cells. Further studies are required to provide evidence for such regulation in the lung epithelium [46].
Epigenetic alterations have been associated with various lung diseases such as idiopathic pulmonary fibrosis (IPF) [49,50], chronic obstructive pulmonary disease (COPD) [51,52], and lung cancer [53–55]. A more complete description of the epigenetic changes that occur in patients is likely to allow the assessment of causality. Since epigenetic modulators have entered the clinic, it is possible that the rational application of epigenetic modulators can be used to treat disease-associated pathologic plasticity.
Future directions
Very little is currently known about the epigenetic states of specific cell types in the setting of epithelial tissue injury, and in turn very little is known about the epigenetic basis of adult cell plasticity. Once clear epigenetic patterns are established for a variety of normal cell types within a given lineage, either at the population or single cell level, we can begin to assess whether open chromatin configurations, such as in intestinal progenitors, form a paradigm for explaining plasticity. Conversely, epigenetic marks may themselves be altered by injury and this might provide the basis for an altered landscape that permits cell fate transitions not evident in the steady state tissue.
As in ES and iPS cell culture, the status of the chromatin configuration of the primary cells in culture is likely to be a critical determinant of their potency and differentiation capacity. Indeed, despite the robust expansion of human airway basal stem cells in culture, the expanded cells lose some of their functions and differentiation potential [56]. Whether the epigenome of the expanded cells is altered in culture and whether these changes play a causal role in the deterioration of cellular functions remains to be demonstrated. Such knowledge is necessary for the safe and effective use of stem cells for screening purposes, and particularly when they are contemplated as therapeutic agents.
It will also be important to assess the epigenome in highly defined models of plasticity. For example, in the airway epithelium, mature secretory cells are able to dedifferentiate and acquire a stem cell fate when stem cells are ablated. In this context, the most mature secretory cells resist dedifferentiation [5].
Understanding the epigenetic state of secretory cell subpopulations of varying states of maturity is likely to contribute to our understanding of the mechanisms that lead to cell identity “locking”.
Finally, epigenetic therapies are in preclinical and clinical trials for many diseases [49–55]. Most of the current epigenetic modifiers such as DNA methyltransferase inhibitors and Hdac inhibitors globally affect cellular states. Thus, much more specific knowledge of the epigenome of individual cells in individual tissues is necessary. Large-scale single-cell epigenomes are likely to provide deep insights into our understanding of differentiation in various tissues, regeneration, plasticity, and pathology.
Acknowledgments
J.R. is a Howard Hughes Medical Institute Faculty Scholar, a New York Stem Cell Foundation Robertson Investigator, a Maroni Research Scholar at Massachusetts General Hospital, and a member of the Ludwig Institute for Cancer Research of Harvard Medical School. This work was supported by grants from the NIH R01HL116756, R01HL118185. We apologize to the myriad authors whose work we could not include in this brief review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Waddington CH. The strategy of the genes. George Allen Unwin Ltd; 1957. [Google Scholar]
- 2•.Rajagopal J, Stanger BZ. Perspective Plasticity in the Adult : How Should the Waddington Diagram Be Applied to Regenerating Tissues. Dev Cell. 2016;36:133–137. doi: 10.1016/j.devcel.2015.12.021. This review revisits the Waddington diagram in the context of cellular plasticity during adult regeneration. It suggests that the epigenetic landscape in adult tissues must be dynamic to facilitate the cellular plasticity that is required following injury. [DOI] [PubMed] [Google Scholar]
- 3.Blanpain C, Fuchs E. Plasticity of epithelial stem cells in tissue regeneration. Science. 2014;344:1242281–1242281. doi: 10.1126/science.1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tata PR, Rajagopal J. Cellular plasticity: 1712 to the present day. Curr Opin Cell Biol. 2016;43:46–54. doi: 10.1016/j.ceb.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–23. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kai T, Spradling A. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature. 2004:428. doi: 10.1038/nature02436. [DOI] [PubMed] [Google Scholar]
- 7.Brawley C, Matunis E. Regeneration of Male Germline Stem Cells by Spermatogonial Dedifferentiation in Vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- 8.van Es JH, Sato T, van de Wetering M, Luybimova A, Gregorieff A, Zeinstra L, van den Born M, Korving J, Martens AC, van den Oudenaarden, Alexander Clevers H. Dll1 marks early secretory progenitors in gut crypts that can revert to stem cells upon tissue damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, Van Den Born M, Korving J, De Sauvage F, Van Es JH, et al. Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Stange D, Koo B, Huch M, Sibbel G, Basak O, Lyubimova A, Kujala P, Bartfeld S, Koster J, Geahlen J, Peters P, Van Es J, van de Wetering M, Mills J, Clevers H. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tetteh PW, Farin HF, Clevers H. Plasticity within stem cell hierarchies in mammalian epithelia. Trends Cell Biol. 2015;25:100–108. doi: 10.1016/j.tcb.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE, Stanger BZ. Robust cellular reprogramming occurs spontaneously during liver regeneration Robust cellular reprogramming occurs spontaneously during liver regeneration Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chera S, Baronnier D, Ghila L, Cigliola V, Jensen JN, Gu G, Furuyama K, Thorel F, Gribble FM, Reimann F, et al. Diabetes Recovery By Age-Dependent Conversion of Pancreatic δ-Cells Into Insulin Producers. Nature. 2014;514:503–507. doi: 10.1038/nature13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadorn E. Transdetermination in cells. Sci Am. 1968:219. doi: 10.1038/scientificamerican1168-110. [DOI] [PubMed] [Google Scholar]
- 15.Sustar A, Schubiger G. A transient cell cycle shift in Drosophila imaginal disc cells precedes multipotency. Cell. 2005;120:383–393. doi: 10.1016/j.cell.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Johnston LA. Regeneration and Transdetermination: New Tricks from Old Cells. Cell. 2005;120:288–290. doi: 10.1016/j.cell.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 17.Reik W. Epigenetic Reprogramming in Mammalian Development. Science. 2001;293:1089–1094. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 18.Willyard C. A new Twist on epigenetics. Nat News. 2017:542. [Google Scholar]
- 19.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 20.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JT, Jaenisch R. The (epi)genetic control of mammalian X-chromosome inactivation. Curr Opin Genet Dev. 1997;7:274–80. doi: 10.1016/s0959-437x(97)80138-4. [DOI] [PubMed] [Google Scholar]
- 22.Reik W, Lewis A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat Rev Genet. 2005;6:403–410. doi: 10.1038/nrg1602. [DOI] [PubMed] [Google Scholar]
- 23.Mak W, Nesterova TB, Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N. Reactivation of the Paternal X Chromosome in Early Mouse Embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- 24.Geisler SJ, Paro R. Trithorax and Polycomb group-dependent regulation: a tale of opposing activities. Development. 2015;142:2876–2887. doi: 10.1242/dev.120030. [DOI] [PubMed] [Google Scholar]
- 25.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 26.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Adachi K, Schöler HR. Directing reprogramming to pluripotency by transcription factors. Curr Opin Genet Dev. 2012;22:416–422. doi: 10.1016/j.gde.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly Reprogrammed Fibroblasts Show Global Epigenetic Remodeling and Widespread Tissue Contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. This paper demonstrates, during the reprogramming of female fibroblasts into iPSCs, that the global epigenetic state is reversed into an ES-like chromatin landscape. The previously silenced X chromosome is reactivated following reprogramming, and the newly activated X chromosome undergoes random X inactivation upon subsequent differentiation of iPSCs. This paper shows efficient erasure of pre-existing epigenetic imprints following reprogramming, and the reestablishment of appropriate epigenetic marks upon iPSCs differentiation. [DOI] [PubMed] [Google Scholar]
- 31•.Wang L, Du Y, Ward JM, Shimbo T, Lackford B, Zheng X, Maio Y-L, Zhou B, Han L, Fargo DC, et al. INO80 Dependent Promoter Access Facilitates Activation of Pluripotency Genes in Embryonic Stem Cell Self-Renewal, Reprogramming, and Blastocyst Development. Cell Stem Cell. 2014;14:575–591. doi: 10.1016/j.stem.2014.02.013. This paper illustrates a role for the INO80 complex in ESC self-renewal, directed reprogramming, and blastocyst development. INO80 is recruited to pluripotency loci together with reprogramming transcription factors, and mediates the maintenance of open chromatin and the recruitment of additional factors and RNA polymerase II. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gasper-Maia A, Alajem A, Polesso F, Sridharan R, Mason M, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci. 2009;106:5187–5191. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherwood RI, Hashimoto T, O’Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, Gifford DK. Discovery of non-directional and directional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soufi A, Donahue G, Zaret KS. Facilitators and Impediments of the Pluripotency Reprogramming Factors’ Initial Engagement with the Genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magnani L, Eeckhoute J, Lupien M. Pioneer factors: Directing transcriptional regulators within the chromatin environment. Trends Genet. 2011;27:465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Xie W, Ren B. Enhancing Pluripotency and Lineage Specification. Science. 2013;341:245–247. doi: 10.1126/science.1236254. [DOI] [PubMed] [Google Scholar]
- 38••.Kim T-H, Li F, Ferreiro-Neira I, Ho L-L, Luyten A, Nalapareddy K, Long H, Verzi M, Shivdasani RA. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. This paper demonstrates the basis of the cellular plasticity of intestinal progenitor cells. Both secretory and absorptive progenitor cells possess accessible chromatin states that allow both secretory and absorptive cell fates to be established. Specific mature daughter lineages are established following fate-specifying transcription factor binding during the process of lateral inhibition. However, the established secretory cell progenitor can be re-directed towards the absorptive cell fate upon loss of the secretory-lineage associated transcription factor ATOH1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Ge Y, Gomez NC, Adam RC, Nikolova M, Yang H, Verma A, Lu CPJ, Polak L, Yuan S, Elemento O, et al. Stem Cell Lineage Infidelity Drives Wound Repair and Cancer. Cell. 2016;169:636–642e14. doi: 10.1016/j.cell.2017.03.042. This paper demonstrates lineage infidelity and transient activation of both epidermis and hair follicle super enhancers in stem cells during the process of wound closure. In tumors, lineage-associated and wound-associated signals are maintained and additionally tumor enhancers are activated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whyte Wa, Orlando Da, Hnisz D, Abraham BJ, Charles Y, Kagey MH, Rahl PB, Lee TI, Young Ra. Master Transcription Factors and Mediator Establish Super- Enhancers at Key Cell Identity Genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snitow ME, Li S, Morley MP, Rathi K, Lu MM, Kadzik RS, Stewart KM, Morrisey EE. Ezh2 represses the basal cell lineage during lung endoderm development. development. 2015;142:108–117. doi: 10.1242/dev.116947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galvis La, Holik AZ, Short KM, Pasquet J, Lun ATL, Blewitt ME, Smyth IM, Ritchie ME, Asselin-Labat M-L. Repression of Igf1 expression by Ezh2 prevents basal cell differentiation in the developing lung. Development. 2015;142:1458–1469. doi: 10.1242/dev.122077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Frank DB, Morley MP, Zhou S, Wang X, Lu MM, Lazar MA, Morrisey EE. HDAC3-Dependent Epigenetic Pathway Controls Lung Alveolar Epithelial Cell Remodeling and Spreading via miR-17-92 and TGF-β Signaling Regulation. Dev Cell. 2016:36. doi: 10.1016/j.devcel.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted Deletion Reveals Essential and Overlapping Functions of the miR-17-92 Family of miRNA Clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Y, Thomson M, Wang HYF, Hammond SM, Hogan BLM. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morrisey EE, Hogan BLM. Preparing for the First Breath: Genetic and Cellular Mechanisms in Lung Development. Dev cell. 2010;18:997–1003. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Y, Vo T, Millien G, Tagne J-B, Kotton D, Mason RJ, Williams MC, Ramirez MI. Epigenetic mechanisms modulate thyroid transcription factor 1-mediated transcription of the surfactant protein B gene. J Biol Chem. 2010;285:2152–64. doi: 10.1074/jbc.M109.039172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin Z, Gonzales L, Kolla V, Rath N, Zhang Y, Lu MM, Kimura S, Ballard PL, Beers MF, Epstein JA, et al. Hop functions downstream of Nkx2. 1 and GATA6 to mediate HDAC-dependent negative regulation of pulmonary gene expression. Am J Physiol Lung Cell Mol Physiol. 2006;291:L191–9. doi: 10.1152/ajplung.00385.2005. [DOI] [PubMed] [Google Scholar]
- 49.Pandit KV, Milosevic J, Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Transl Res. 2011;157:191–199. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 50.Pang M, Zhuang S. Histone deacetylase: a potential therapeutic target for fibrotic disorders. J Pharmacol Exp Ther. 2010;335:266–72. doi: 10.1124/jpet.110.168385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito K, Ito M, Elliott WM, Cosio B, Caramori G, Kon OM, Barczyk A, Hayashi S, Adcock IM, Hogg JC, et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med. 2005;352:1967–76. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- 52.Qiu W, Baccarelli A, Carey VJ, Boutaoui N, Bacherman H, Klanderman B, Rennard S, Agusti A, Anderson W, Lomas DA, et al. Variable DNA methylation is associated with chronic obstructive pulmonary disease and lung function. Am J Respir Crit Care Med. 2012;185:373–381. doi: 10.1164/rccm.201108-1382OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolden JE, Shi W, Jankowski K, Kan C-Y, Cluse L, Martin BP, MacKenzie KL, Smyth GK, Johnstone RW. HDAC inhibitors induce tumor-cell-selective pro-apoptotic transcriptional responses. Cell Death Dis. 2013;4:e519. doi: 10.1038/cddis.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho AS, Turcan S, Chan TA. Epigenetic therapy: Use of agents targeting deacetylation and methylation in cancer management. Onco Targets Ther. 2013;6:223–232. doi: 10.2147/OTT.S34680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hatzimichael E, Crook T. Cancer epigenetics: new therapies and new challenges. J Drug Deliv. 2013;2013:529312. doi: 10.1155/2013/529312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mou H, Vinarsky V, Tata PR, Brazauskas K, Choi SH, Crooke AK, Zhang B, Solomon GM, Turner B, Bihler H, et al. Dual SMAD Signaling Inhibition Enables Long-Term Expansion of Diverse Epithelial Basal Cells. Cell Stem Cell. 2016;19:217–231. doi: 10.1016/j.stem.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]