Abstract
Robust fluorescence-based biosensors are emerging as critical tools for high-throughput strain improvement in synthetic biology. Many biosensors are developed in model organisms where sophisticated synthetic biology tools are also well established. However, industrial biochemical production often employs microbes with phenotypes that are advantageous for a target process, and biosensors may fail to directly transition outside the host in which they are developed. In particular, losses in sensitivity and dynamic range of sensing often occur, limiting the application of a biosensor across hosts. Here we demonstrate the optimization of an Escherichia coli-based biosensor in a robust microbial strain for the catabolism of aromatic compounds, Pseudomonas putida KT2440, through a generalizable approach of modulating interactions at the protein-DNA interface in the promoter and the protein-protein dimer interface. The high-throughput biosensor optimization approach demonstrated here is readily applicable towards other allosteric regulators.
Keywords: Whole cell biosensor, Aromatic catabolism, Transcription factor, PcaU, Shikimate
Graphical abstract
Highlights
-
•
A biosensor optimized for a robust, industrially useful P. putida strain.
-
•
Modulation of protein-DNA and protein-protein interactions pursued.
-
•
Offers a generalized optimization protocol for transcription factor-based sensors.
-
•
Intracellular metabolite production and detection made possible in P. putida.
-
•
Functional biosensor in P. putida will allow high throughput strain evolution.
1. Introduction
The application of sensor-reporter elements (Dietrich et al., 2010) in whole cell biosensing of small molecules has gained substantial popularity as an approach for functional improvement of enzymes (Harrington et al., 2017, Tang et al., 2013) and metabolic pathways (Rogers et al., 2016, Tang and Cirino, 2011, Yang et al., 2017), as well as to enhance the efficiency of production strains (Binder et al., 2013, Lehning et al., 2017, Mahr et al., 2015). In sensor-reporter systems, the output signal from the reporter is proportional to the product titer (Rogers and Church, 2016), which in turn correlates with the conversion efficiency of the enzyme, pathway, or strain. To this end, we previously reported an engineered sensor derived from a protocatechuate (PCA) inducible transcription factor, PcaU from Acinetobacter baylyi ADP1 (Jha et al., 2014). PcaU is an IclR-type transcription factor and an activator of the operon involved in catabolism of PCA (Kok et al., 1998). Structurally conserved, tetrameric IclR transcription factors remain bound to the operator both in the presence and absence of the effector molecule and in the presence of effector molecule, the sigma factor is expected to be recruited (Molina-Henares et al., 2006). The optimized sensor regulates the expression of green fluorescent protein (GFP) as a reporter in Escherichia coli. Using the PcaU-GFP sensor-reporter system, we demonstrated that even a modest dynamic range was sufficient to couple whole cell sensing to the powerful screening efficiency of flow cytometry and select a rare ‘strong’ phenotype (enzymatic activity) from a large pool of ‘weaker’ phenotype (Jha et al., 2014).
For the development of robust industrial strains, model organisms such as E. coli that have well developed synthetic biology tools, including multiple biosensors for molecules of interest, may not be ideal for a given target application. Instead, the use of alternate microbial strains that exhibit beneficial, difficult to transfer phenotypes (e.g., high flux through target pathways, thermal or pH tolerance, etc.) will often enable more efficient and cost-effective process design. However, the deployment of biosensor-based screening in a new host organism may not directly transfer, and any loss of sensitivity and dynamic range can reduce the utility of the biosensor. Pseudomonas putida KT2440 is one such strain that has increasingly been investigated as a potential microbial cell factory for producing target chemicals due to its high toxicity tolerance and high natural flux in aromatic-catabolic pathways (Martínez-García and de Lorenzo, 2017, Nikel et al., 2016). Given the ability of biosensor-based technologies to make rapid, often non-intuitive changes to microbial production of target compounds and the difficulty in transitioning those to alternate hosts, there remains a key gap in the literature on how to engineer biosensor technologies originally developed in model organisms to strains that may be more useful for industrial applications, such as P. putida.
PCA, the molecule of interest for which a biosensor was previously developed in E. coli, is an important central intermediate in aromatic catabolism (Fuchs et al., 2011) and siderophore biosynthesis (Fox et al., 2008) as well as a key hub molecule in the conversion of sugars to cis,cis-muconic acid and other shikimate-derived products (Frost and Draths, 1995). Most studies reported to date have targeted conversion of PCA-derived muconic acid to adipic and terephthalic acid (Lu et al., 2016, Niu et al., 2002, Vardon et al., 2015) and other polymer precursors (Rorrer et al., 2016, Suastegui et al., 2016). By engineering these pathways, PCA has been successfully produced in a diverse set of host organisms ranging from E. coli (Draths and Frost, 1995, Koppisch et al., 2012), to P. putida (Johnson et al., 2016), and to yeast (Curran et al., 2013, Suastegui et al., 2016, Weber et al., 2012). Given the ability of P. putida to convert aromatic carbon to value-added chemicals with high flux (Linger et al., 2014, Salvachúa et al., 2015, Vardon et al., 2015) and an interest in establishing this robust microbe as an industrially relevant production strain (Nikel et al., 2016), transitioning the PCA sensor-reporter system in P. putida was pursued in this study, wherein we demonstrate a generalizable method in promoter and protein engineering to regain sensitivity and dynamic range while transitioning transcription factor-based biosensors between microbes.
2. Material and methods
2.1. Construction of P. putida strains with gene knockouts
Pseudomonas putida KT2440 (ATCC# 47054) knockout variant CJ072 was generated by deleting the genes pcaH and pcaG, which encode the two-subunit protocatechuate 3,4-dioxygenase, from the P. putida KT2440 genome using plasmid pCJ011 and the antibiotic/sucrose method of selection/counter-selection of gene replacement (Blomfield et al., 1991, Marx, 2008) as previously described (Johnson and Beckham, 2015). Similarly, pobA, encoding the 4-hydroxybenzoate hydroxylase, and a portion of pobR, which encodes the transcription factor that regulates expression of pobA, were deleted from the P. putida KT2440 genome to generate strain CJ182 using plasmid pCJ041. pCJ041 was constructed by amplifying a 1 kb 5′ targeting region with primer pair oCJ292 (5′-AGTGAGCGCAACGCAATTAATGTGAGTTAGCGAACTTTAGTAAAGGCTGGGCTTTCAGTTCATC-3′) and oCJ293 (5′-GCGGCCGCGGGCTGCGAGCTACGGG-3′) and a 1 kb 3′ targeting region with primer pair oCJ296 (5′-CCTGACCCGTAGCTCGCAGCCCGCGGCCGCGTGTGGATCAGCCGCCGTC-3’) and oCJ297 (5′-CCCTGAGTGCTTGCGGCAGCGTGAAGCTAGGCCCGCTTCGGTAAGGTCG-3’) from P. putida KT2440 gDNA and these were assembled using NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs) according to the manufacturer’s instructions into pK18mobsacB (ATCC# 87097) (Kvitko and Collmer, 2011, Schäfer et al., 1994) amplified linearly with primers oCJ288 (5′-CTAGCTTCACGCTGCCGCAAG-3′) and oCJ289 (5′-CTAACTCACATTAATTGCGTTGCGCTCACTG-3′). These PCR amplifications were performed using Q5® Hot Start High-Fidelity 2X Master Mix (New England Biolabs). The assemblies were transformed into NEB® 5-alpha F'Iq competent Escherichia coli (New England Biolabs) according to the manufacturer's instructions. Clones were screened by diagnostic restriction digest and those that were correctly assembled were confirmed using Sanger sequencing performed by GENEWIZ, Inc. Deletion of pobAR from the P. putida KT2440 genome using pCJ041 was confirmed by amplification of a 2094 bp product in diagnostic colony PCR using MyTaq™ HS Red Mix (Bioline) with primers oCJ298 (5′-ACCTTTCATCTGCGGACC-3′) and oCJ299 (5′-ATCTGTGGCACCCACTTG-3′) rather than the 3942 bp product generated when pobA and pobR are present.
2.2. Plasmid construction
The sequences of the primers, genes for sensor and reporter and the vector backbone used in this study are provided in Supplementary material (Section 3). Plasmid constructs were generated using broad host range vector, pBTL-2 (Addgene plasmid# 22806) (Lynch and Gill, 2006) as a backbone. The vector backbone was PCR amplified using pBTL-2_Fwd/pBTL-2_Rev primers (Fragment 1). The genetic sequences encoding the transcription factor based sensor, PcaU, along with the regulatory region from the E. coli-specific sensor-reporter system was PCR amplified from a previously constructed plasmid (Jha et al., 2014) using pBTL-2_pcaU_overlap_Fwd/PcaUpromo_sfGFP_overlap_Rev primers (Fragment 2). The genetic sequence of the reporter, superfolder GFP (sfGFP) (Pédelacq et al., 2006), was generated as a PCR fragment using sfGFP_Fwd/pBTL-2_sfGFP_overlap_Rev primers (Fragment 3). Fragments 1, 2 and 3 were assembled using NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs) to create a circular plasmid (Supplementary Fig. S1), such that sensor-reporter cassette with intergenic regulatory region was bound by bidirectional soxR and tonB transcription terminators in pBTL-2. The construct was named pBTL-2_pcaU_1 (pPcaU1).
2.3. Construction of a diversified promoter library
The oligonucleotides used for diversification of the PcaU promoter are provided in the Supplementary material (Section 3). Using pPcaU1 as a template for PCR and wobble containing oligonucleotides PcaU_promoter_diversify_1 with equimolar mix of PcaU_promoter_diversify_3a and PcaU_promoter_diversify_3b, DNA fragments were made containing complete randomization of three positions each on the -35 and -10 promoter regions and randomization of one position in the proposed transcription initiation (TI) site. Hence, seven positions with complete randomization gave a library diversity of 47. A deletion between the promoter and the operator was incorporated by the equimolar mix of two oligonucleotides (PcaU_promoter_diversify_3a and PcaU_promoter_diversify_3b) that differed by a single deletion of a base and overlapped with the region between the operator and the promoter. The fragments were assembled using overlap extension PCR (Horton et al., 1990) to generate a promoter library with a theoretical diversity of approximately 33,000.
2.4. Homology modeling and determining dimer interface of the PcaU transcription factor
The inducer binding domain (IBD) of PcaU (residues 103-278) was modeled as a symmetric homodimer with a C2 symmetry as described earlier (Jha et al., 2015). The crystal structure of a putative transcriptional regulator from Rhodococcus sp RHA1 (PDB code 2IA2) showing 36% sequence homology with the PcaU was used as a template for symmetric homology modeling using the Fold-and-Dock protocol in Rosetta (Das et al., 2009).
2.5. Construction of a PcaU dimer interface library
To perform saturation mutagenesis at two positions (residues 147 and 148) at the putative dimer interface as predicted by symmetric homology modeling, DNA fragments were created using primers pBTL-2_pcaU_overlap_Fwd and PcaU_TD_randomize_Fwd with ‘NNK’ codons corresponding to the two selected positions and primers PcaU_TD_randomize_Rev/pBTL-2_sfGFP_overlap_Rev for rest of the genetic sequence for the sensor and the sfGFP reporter including the intergenic region in the sensor plasmid construct. The sensor plasmid with optimized promoter sequence (pPCAU1.1) was used as a template for PCR. The two DNA fragments along with the pBTL-2 backbone amplified using pBTL-2_Fwd/pBTL-2_Rev primers were assembled using NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs).
2.6. Transformation of plasmid constructs in P. putida strains
P. putida KT2440 strains (native, CJ072, or CJ182) were grown in 10 ml of standard lysogeny broth (LB) rich medium, overnight at 30 °C and 225–250 rpm in a shaker, centrifuged, and washed twice with 300 mM sucrose using one-fifth the volume of the original culture and finally resuspended in 300 mM sucrose and glycerol at 20% final concentration at a volume one-twentieth the volume of the original culture. For each transformation reaction, approximately 35 μl of competent cells was mixed with approximately 20 ng of plasmid DNA and electroporated in 1 mm cuvettes at a voltage of 1.6 kV, 25 μF capacitance and 200 ohm resistance using an electroporater (BioRad). Typical time constants of 4.7–5 ms were observed for a successful electroporation. The cells were immediately diluted in 1 ml of rich media [LB or Super Optimal Broth with Catabolite repression (SOC)] and recovered at 30 °C and 225–250 rpm in a shaker for 1–2 h. The recovered cells were centrifuged at 5000 rpm for 3 mins using a table-top centrifuge and 900 μl of supernatant was discarded. The pellet was resuspended in the remaining supernatant and plated on an LB agar plate supplemented with 50 μg/ml of kanamycin (Kan50). Usually very high transformation efficiency was observed resulting in a lawn of transformed colonies on 85 mm plates. A small scoop from the lawn was grown and stored as glycerol stocks (for monoclonals) or all the cells in the lawn were scraped, diluted to an optical density at 600 nm (OD600) of ~3 in LB media (supplemented with Kan50 and 20% glycerol) and stored as glycerol stocks (for libraries). The glycerol stocks were stored in a −80 °C freezer.
2.7. Culture growth, induction and flow cytometry
A small scoop from the glycerol stocks was inoculated in 2 ml LB with Kan50 and grown overnight at 30 °C and 225–250 rpm in a shaker. This ‘seed’ culture was diluted 100-fold in a fresh growth media and grown for 4–5 h until an OD600 of ~0.6. Approximately 300 μl culture was dispensed in wells in a deepwell 96-well block for growth at different induction conditions. “Uninduced” (UI) cultures were deprived from any exogenous supplementation of an inducer, while “Induced” cultures were supplemented with appropriate concentration of small aromatic molecules under study. Appropriate volumes of stocks of PCA and 4-hydroxybenzoate (dissolved in equimolar sodium hydroxide) and sodium benzoate and catechol (dissolved in water) were added to each well for dose-response evaluation of the sensors. The deepwell block was incubated at 30 °C and 1000 rpm in a plate shaker for 16–18 h. Each culture (2 μl) was diluted 100-fold in PBS (phosphate buffer saline) with 1% sucrose and analyzed using LSR II (BD Biosciences), Accuri (BD Biosciences) or FACSAria III (BD Biosciences) at an excitation and emission wavelengths of 488 nm and 530 nm respectively. The average fluorescence intensity of the dense population in gated FSC/SSC (forward and side scatter determining the size and granularity of cells) scatter plot was noted.
Alternatively, in particular for the libraries, cultures were directly inoculated from the glycerol stocks, grown in 50 ml falcon tubes for 6–7 h to an OD600 of ~0.6 and then distributed in smaller culture tubes and induced with appropriate concentrations of PCA. The culture tubes were incubated at 30 °C and 225–250 rpm in a shaker for 16–18 h before diluted 100-fold in PBS with 1% sucrose and analyzed or used for fluorescence-activated cell sorting (FACS) in FACSAria III (BD Biosciences). Post FACS rounds, the cells were grown for ~20 h to saturation and then aliquoted and stored as glycerol stocks in a −80 °C freezer.
3. Results and discussion
3.1. P. putida KT2440 strains used in the study
Two variants of P. putida KT2440 were utilized in this study. CJ072 had genes encoding the PCA-3,4-dioxygenease (pcaHG) deleted which made it incapable of metabolizing PCA via the native β-ketoadipate pathway, thereby permitting a build-up of PCA in the cell upon endogenous exposure to PCA. CJ182 had genes encoding 4-hydroxybenzoate hydroxylase (pobA) and the regulator (pobR) deleted. This strain is expected to be incapable of metabolizing 4HB and converting it to PCA. These two mutant strains along with the native strain were crucial for experimental evaluation of the PCA biosensor since they constituted a series of strains with variable PCA accumulation efficiency. While native P. putida KT2440 strain catabolized PCA to the downstream molecules in the β-ketoadipate pathway (Harwood and Parales, 1996), CJ072 accumulated any PCA formed or failed to metabolize PCA that was exogenously supplemented. Finally, CJ182 that had mutations upstream of PCA in the β-ketoadipate pathway, was not expected to make PCA from 4HB, but if PCA provided exogenously, it was capable of catabolizing PCA to the downstream molecules in the β-ketoadipate pathway.
3.2. PCA biosensor design using E. coli adapted sensor-promoter-reporter sequence
An E. coli-adapted PCA biosensor (Jha et al., 2014) consisted of genes encoding the PcaU transcription factor (TF) from Acinetobacter baylyi ADP1 (codon-optmized for E. coli) and the sfGFP reporter. The pcaU-sfgfp promoter region in the plasmid contained the native pcaU-pcaI intergenic region (from A. baylyi ADP1) but included an E. coli-compatible ribosome binding site (RBS). The TF-promoter-reporter cassette from this plasmid construct was assembeled into a broad host range vector, pBTL-2. The new plasmid construct, pPcaU1 (for pBTL-2_pcaU_1) (Supplementary Fig. S1) was transformed into P. putida KT2440 strains (native and CJ072) and grown to mid-log phase at 30 °C before induction with PCA up to a final concentration of 10 mM and further grown overnight at 30 °C. Cell fluorescence measurements using flow cytometry (ex/em 488/530) did not show any signal increase upon induction with PCA (Supplementary Fig. S2A).
3.3. Strategy for evolution of PCA sensor in P. putida
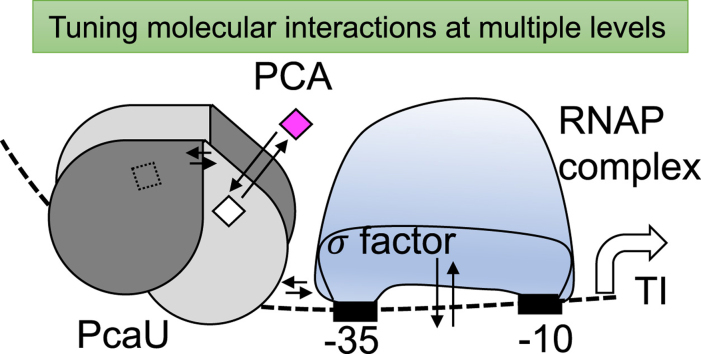
The PcaU transcription activator, which belongs to an IclR family of transcription factors (Molina-Henares et al., 2006) and is a tetramer (a dimer of dimers), is expected to undergo multiple key interactions with the DNA operator, PCA ligand, and sigma factor of the RNA polymerase for recruitment and transcription. We speculated that manipulating any of these key interactions, along with alteration of the transcription initiation site and RBS, would be capable of increasing the sensitivity and dynamic range of the sensor. Our strategy for evolution of the biosensor was to modulate some of these key interactions for enhancing the response of the biosensor.
3.3.1. Evolution of pcaU-sfgfp intergenic region for PCA response in P. putida
In our earlier work (Jha et al., 2014), we demonstrated that a transcriptional regulator and the promoter sequence from one host bacterium may require tuning to function in a different host strain. To enable an Acinetobacter pcaU-pcaI intergenic region to operate in E. coli as a pcaU-sfgfp regulatory region, the inclusion of an E. coli-compatible RBS (AAGGAG) was sufficient to achieve an appreciable dynamic range (Jha et al., 2014). Further optimization of promoter regions (-35/-10 sites), the transcription initiation site (TI), and the distance between the promoter and operator also resulted in enhancement of the dynamic range by 50%. With the aim of optimizing the PCA biosensor for P. putida in a similar way, we created a library from the randomization of three positions each in the -35 and -10 sites (Fig. 1A) and a base in the TI, incorporating a deletion between the PcaU operator and promoter site with a 50% probability. This library was screened in the P. putida strain CJ072 which is expected to be incapable of metabolizing exogenously provided PCA.
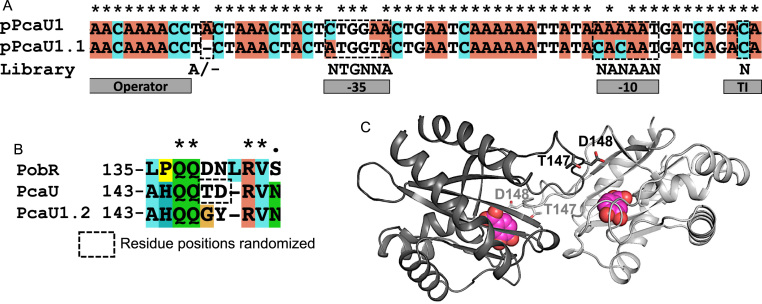
Fig. 1.
Libraries for promoter and protein evolution inP. putida. (A) Alignment of the evolved PcaU promoter (pPcaU1.1) with an E. coli adapted promoter (pPcaU1). Positions randomized to create the library are shown below the alignment. The PcaU operator is only partially shown. (B) Alignment of homologous PobR and PcaU sequences in the region with high sensitivity to mutagenesis in PobR. The D139N mutation in a PobR variant exhibited an enhanced contrast ratio (Jha et al., 2016), and randomization was carried out on PcaU at analogous positions. The corresponding region in PcaU1.2 is aligned to show TD→GY mutation. (C) Homology model of PcaU inducer binding domain as a dimer and docked with PCA (shown in spheres), showing spatial orientation of T147 and D148 (shown in sticks).
Using a FACSAria III flow cytometer (BD Biosciences), four rounds of FACS were performed. After two rounds of sorting of the top 5% of induced population (grown in 10 mM PCA) based on fluorescence intensity, one round of negative selection (the bottom 50% ‘dark’ population of culture grown in the absence of PCA, also referred as uninduced or UI culture) was performed to eliminate constitutively active variants. The fourth round of FACS to select the top 1% fluorescent population grown in 10 mM PCA and subsequent plating and evaluation of monoclonal isolates resulted in the identification of a variant that showed a distinct response when dosed with 1 mM PCA (Supplementary Fig. S2B). The isolated variant showed mutations in both -35 and -10 sites along with the intended deletion between the promoter and the operator (Fig. 1A). The new version of the sensor plasmid was dubbed pPcaU1.1 (for pBTL-2_pcaU_1.1).
3.3.2. Evolution of the PcaU dimer interface for enhanced response switch
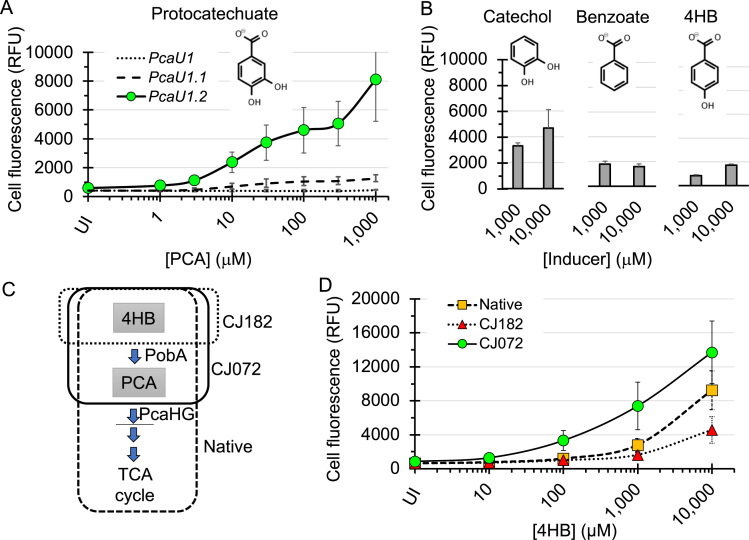
Since pPcaU1.1 showed a very low contrast ratio [(Induced fluorescence signal)/(Uninduced fluorescence signal)< 3], further enhancement of both sensitivity and amplitude in a dose-response curve was necessary. To achieve this, the knowledge gained from mutagenesis studies in an expected PobR dimer interface was utilized for PcaU (Jha et al., 2016). In the previous work, the mutation D139N in PobR resulted in a gain in response amplitude of the transcription factor. The PcaU sequence alignment with PobR showed 55% sequence identity and presented T147 of PcaU as a corresponding amino acid to D139 in PobR (Fig. 1B). Symmetric homology modeling using Rosetta (Das et al., 2009) of PcaU carried out in accordance with our previous work (Jha et al., 2015) also placed T147 at the homodimer interface of the PcaU inducer binding domain and was thus chosen for alteration (Fig. 1C). Since D148 in PcaU is also at the dimer interface and could be a better representation for D139 in PobR, it was also selected for mutagenesis. Complete randomization of the PcaU sequence positions 147 and 148 using ‘NNK’ codons that code for all twenty amino acids and a stop codon, yielded a library with a theoretical diversity of > 400. The plasmid library containing the optimized promoter from pPcaU1.1, was transformed in strain CJ072, grown in the absence of any inducer (uninduced), and was subjected to FACS where the bottom 40% of the ‘dark’ population was selected to eliminate high background and constitutive variants. Subsequently, the sorted population was grown in 0.01–10 mM PCA, and the top 1% of the population based on fluorescence intensity was collected from the 0.01 mM PCA induction, with the aim of selecting a sensitive variant of the sensor. The second round of FACS yielded several monoclonal isolates that showed comparable dynamic range with the E. coli PCA biosensor (Jha et al., 2014). The most sensitive and responsive variant with T147G/D148Y mutations was named pPcaU1.2 (for pBTL-2_pcaU_1.2), and was able to detect exogenously supplemented PCA at concentrations below 0.003 mM and showed a contrast ratio of over twelvefold (Fig. 2A, Supplementary Fig. S2C). Other variants in the second round of sorting, for example T147S/D148F or T147P/D148F, did not show any response at sub-millimolar concentrations of PCA but showed a steep gain in response beyond 3 mM exogenous PCA concentration (Supplementary Fig. S3).
Fig. 2.
Whole cell biosensing of protocatechuate inP. putida. (A) Response plot of different generations of the PcaU-based sensor in P. putida when exogenously dosed with PCA. UI (for uninduced cultures) refers to microbial cells grown in the absence of exogenously supplemented PCA. (B) Specificity evaluation of the evolved sensor, pPcaU1.2, against similar aromatic molecules. Error bars in (A) and (B) are standard deviations from more than three independent experiments performed by multiple researchers. (C) Schematic showing the ability of 4-hydroxybenzoate to be metabolized to PCA and beyond depending on the genotype of P. putida strains, namely CJ182 (no conversion to PCA, dotted box), native P. putida KT2440 (conversion to PCA, which is then converted to downstream metabolites, dashed box) and CJ072 (accumulates PCA, solid box). (D) Intracellular accumulation of PCA and sensor response in P. putida strains. Error bars are standard deviations from two independent experiments performed by different researchers on different days. UI (for uninduced cultures) refers to microbial cells grown in the absence of exogenously supplemented 4HB. RFU: Relative Fluorescence Units.
3.4. Dose-response and inducer specificity test of evolved biosensor in P. putida
Inducer specificity in a biosensor is key to its function and utility. Accordingly, the evolved PCA biosensor was tested against aromatic molecules with similar functional groups, namely benzoate, 4-hydroxybenzoate (4HB), and catechol. Since the tested molecules were relatively smaller than PCA, any rejection of the molecule from the binding pocket due to steric hindrance was unlikely. Cross reactivity between PCA and 4HB in their native transcription factors, PcaU and PobR respectively, was previously shown to be relevant only for PcaU, which revealed visible activation with 4HB while PobR was completely unresponsive to PCA (Jha et al., 2015). Specificity evaluation of pPcaU1.2 was carried out in CJ072 and another knockout strain, CJ182, which is incapable of metabolizing 4HB. Low but measurable activation of the sensor was observed with benzoate and 4HB (Fig. 2B), while catechol showed a signal that was an appreciable fraction of PCA response (Fig. 2B). It is important to note that the intracellular concentration of each molecule in a host organism could differ at the same exogenous concentration of each molecule, and this cannot be evaluated in the current experimental approach. Additionally, as mentioned earlier, wild type P. putida KT2440 is capable of catabolizing a variety of aromatic molecules including PCA, 4HB, benzoate, and catechol, which would result in the underestimation of the concentration dependent sensor activation. In this work, we created P. putida knockout strains CJ182 and CJ072, which are incapable of utilizing 4HB and PCA, respectively, but other molecules (catechol and benzoate) tested in this work would still undergo conversion into downstream products of β-ketoadipate pathway in these strains resulting in reduced actual concentrations of those molecules in the extracellular and intracellular environments.
3.5. Intracellular PCA production and sensing
In addition, the pPcaU1.2 sensor plasmid was tested in a dynamic PCA production environment with different strains of P. putida. Depending on the genotype of the strain, 4HB would either accumulate (CJ182),be converted to PCA as an end product (CJ072), or be further metabolized via the β-ketoadipate pathway (native P. putida KT2440) (Fig. 2C). The response to 4HB feeding in different strains harboring the sensor was consistent with the expected PCA level in the cell. The sensor response was highest in CJ072 (due to PCA accumulation), followed by native (in which PCA is metabolized via the β-ketoadipate pathway) with the lowest response observed in CJ182 (where 4HB is not converted into PCA) (Fig. 2D). The low response observed in CJ182 was the result of 4HB cross-reactivity (Fig. 2B).
4. Conclusions
Our work demonstrates the optimization of a biosensor for transfer from one organism to an alternate host organism. The PcaU-based sensor was optimized in P. putida KT2440 using FACS, adding P. putida to a small list of microorganisms where FACS has been applied in conjunction with a biosensor (Schallmey et al., 2014). The optimized sensor responds to both PCA and catechol (with an appreciable contrast ratio), which are the key intermediates in the carbon flow via protocatechuate and catechol branches of β-ketoadipate pathway (Harwood and Parales, 1996). With only a few examples of established sensor-reporter systems in P. putida (Garmendia et al., 2008), and limited efforts to establish sensor-reporter systems in host organisms beyond E. coli and S. cerevisiae and in non-model organisms (DeLorenzo et al., 2017), this work demonstrates intracellular production and detection of a key metabolic intermediate in P. putida. Furthermore, the promoter and protein evolution approach to altering the dynamics of protein-DNA and protein-protein interactions that resulted in modulated response of the allosteric regulator is widely applicable to designing suitable biosensors with applications in optimization and regulation of biosynthetic pathways (Dahl et al., 2013, Raman et al., 2014).
Acknowledgements
The work was supported by the U.S. Department of Energy (DOE) Energy Efficiency and Renewable Energy (EERE) Bioenergy Technologies Office (BETO) from the Agile BioFoundry to T.D. (under Contract NL0032182) and G.T.B. and the DOE Science Undergraduate Laboratory Internships (to J.B.). Preliminary work was supported by the Defense Threat Reduction Agency (CBCALL12-LS-6-0622 to C.E.M.S.) and computational resources provided by LANL institutional computing (W13_SynBio). C.W.J., P.K. and G.T.B. also thank DOE EERE BETO for funding under Contract DE-AC36-08GO28308 with the National Renewable Energy Laboratory. The U.S. Government retains and the publisher, by accepting the article for publication, acknowledges that the U.S. Government retains a nonexclusive, paid up, irrevocable, worldwide license to publish or reproduce the published form of this work, or allow others to do so, for U.S. Government purposes.
Acknowledgments
Notes
The authors declare competing financial interest. Whole cell biosensors are subject of patent applications by Los Alamos National Laboratory.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2018.03.001.
Contributor Information
Ramesh K. Jha, Email: rjha@lanl.gov.
Taraka Dale, Email: tdale@lanl.gov.
Appendix A. Supplementary material
Supplementary material
References
- Binder S., Siedler S., Marienhagen J., Bott M., Eggeling L. Recombineering in Corynebacterium glutamicum combined with optical nanosensors: a general strategy for fast producer strain generation. Nucleic Acids Res. 2013;41:6360–6369. doi: 10.1093/nar/gkt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomfield I.C., Vaughn V., Rest R.F., Eisenstein B.I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- Curran K.A., Leavitt J.M., Karim A.S., Alper H.S. Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab. Eng. 2013;15:55–66. doi: 10.1016/j.ymben.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Dahl R.H., Zhang F., Alonso-Gutierrez J., Baidoo E., Batth T.S., Redding-Johanson A.M., Petzold C.J., Mukhopadhyay A., Lee T.S., Adams P.D., Keasling J.D. Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 2013;31:1039–1046. doi: 10.1038/nbt.2689. [DOI] [PubMed] [Google Scholar]
- Das R., André I., Shen Y., Wu Y., Lemak A., Bansal S., Arrowsmith C.H., Szyperski T., Baker D. Simultaneous prediction of protein folding and docking at high resolution. Proc. Natl. Acad. Sci. 2009;106:18978–18983. doi: 10.1073/pnas.0904407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorenzo D.M., Henson W.R., Moon T.S. Development of chemical and metabolite sensors for Rhodococcus opacus PD630. ACS Synth. Biol. 2017 doi: 10.1021/acssynbio.7b00192. [DOI] [PubMed] [Google Scholar]
- Dietrich J.A., McKee A.E., Keasling J.D. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu. Rev. Biochem. 2010;79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- Draths K.M., Frost J.W. Environmentally compatible synthesis of catechol from D-glucose. J. Am. Chem. Soc. 1995;117:2395–2400. [Google Scholar]
- Fox D.T., Hotta K., Kim C.-Y., Koppisch A.T. The missing link in petrobactin biosynthesis: asbF encodes a (−)-3-Dehydroshikimate dehydratase. Biochemistry. 2008;47:12251–12253. doi: 10.1021/bi801876q. [DOI] [PubMed] [Google Scholar]
- Frost J.W., Draths K.M. Biocatalytic syntheses of aromatics from D-glucose: renewable microbial sources of aromatic compounds. Annu. Rev. Microbiol. 1995;49:557–579. doi: 10.1146/annurev.mi.49.100195.003013. [DOI] [PubMed] [Google Scholar]
- Fuchs G., Boll M., Heider J. Microbial degradation of aromatic compounds — from one strategy to four. Nat. Rev. Microbiol. 2011;9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- Garmendia J., De Las Heras A., Galvão T.C., De Lorenzo V. Tracing explosives in soil with transcriptional regulators of Pseudomonas putida evolved for responding to nitrotoluenes. Microb. Biotechnol. 2008;1:236–246. doi: 10.1111/j.1751-7915.2008.00027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L.B., Jha R.K., Kern T.L., Schmidt E.N., Canales G.M., Finney K.B., Koppisch A.T., Strauss C.E.M., Fox D.T. Rapid Thermostabilization of Bacillus thuringiensis Serovar Konkukian 97–27 Dehydroshikimate dehydratase through a structure-based enzyme design and whole cell activity assay. ACS Synth. Biol. 2017;6:120–129. doi: 10.1021/acssynbio.6b00159. [DOI] [PubMed] [Google Scholar]
- Harwood C.S., Parales R.E. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- Horton R.M., Cai Z.L., Ho S.N., Pease L.R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Jha R.K., Chakraborti S., Kern T.L., Fox D.T., Strauss C.E.M. Rosetta comparative modeling for library design: engineering alternative inducer specificity in a transcription factor. Proteins Struct. Funct. Bioinforma. 2015;83:1327–1340. doi: 10.1002/prot.24828. [DOI] [PubMed] [Google Scholar]
- Jha R.K., Kern T.L., Fox D.T., Strauss C.E.M. Engineering an Acinetobacter regulon for biosensing and high-throughput enzyme screening in E. coli via flow cytometry. Nucleic Acids Res. 2014;42:8150–8160. doi: 10.1093/nar/gku444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha R.K., Kern T.L., Kim Y., Tesar C., Jedrzejczak R., Joachimiak A., Strauss C.E.M. A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor. Nucleic Acids Res. 2016:gkw687. doi: 10.1093/nar/gkw687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.W., Beckham G.T. Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Metab. Eng. 2015;28:240–247. doi: 10.1016/j.ymben.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Johnson C.W., Salvachúa D., Khanna P., Smith H., Peterson D.J., Beckham G.T. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 2016;3:111–119. doi: 10.1016/j.meteno.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok R.G., D’Argenio D.A., Ornston L.N. Mutation analysis of PobR and PcaU, closely related transcriptional activators in acinetobacter. J. Bacteriol. 1998;180:5058–5069. doi: 10.1128/jb.180.19.5058-5069.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppisch, A.T., Fox, D.T., Hotta, K., Welsh, J.D., 2012. Production of industrially relevant compounds in prokaryotic organisms. US20120196339 A1.
- Kvitko, B.H., Collmer, A., 2011. Construction of Pseudomonas syringae pv. tomato DC3000 Mutant and Polymutant Strains, In: Plant Immunity, Methods in Molecular Biology. Humana Press, pp. 109–128. 〈https://dx.doi.org/10.1007/978-1-61737-998-7_10〉. [DOI] [PubMed]
- Lehning C.E., Siedler S., Ellabaan M.M.H., Sommer M.O.A. Assessing glycolytic flux alterations resulting from genetic perturbations in E. coli using a biosensor. Metab. Eng. 2017;42:194–202. doi: 10.1016/j.ymben.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger J.G., Vardon D.R., Guarnieri M.T., Karp E.M., Hunsinger G.B., Franden M.A., Johnson C.W., Chupka G., Strathmann T.J., Pienkos P.T., Beckham G.T. Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. 2014;111:12013–12018. doi: 10.1073/pnas.1410657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Lu F., Chen J., Yu W., Huang Q., Zhang J., Xu J. Production of diethyl terephthalate from biomass-derived muconic acid. Angew. Chem. 2016;128:257–261. doi: 10.1002/anie.201509149. [DOI] [PubMed] [Google Scholar]
- Lynch M.D., Gill R.T. Broad host range vectors for stable genomic library construction. Biotechnol. Bioeng. 2006;94:151–158. doi: 10.1002/bit.20836. [DOI] [PubMed] [Google Scholar]
- Mahr R., Gätgens C., Gätgens J., Polen T., Kalinowski J., Frunzke J. Biosensor-driven adaptive laboratory evolution of l -valine production in Corynebacterium glutamicum. Metab. Eng. 2015;32:184–194. doi: 10.1016/j.ymben.2015.09.017. [DOI] [PubMed] [Google Scholar]
- Martínez-García E., de Lorenzo V. Molecular tools and emerging strategies for deep genetic/genomic refactoring of pseudomonas. Curr. Opin. Biotechnol. 2017;47:120–132. doi: 10.1016/j.copbio.2017.06.013. [DOI] [PubMed] [Google Scholar]
- Marx C.J. Development of a broad-host-range sacB-based vector for unmarked allelic exchange. BMC Res. Notes. 2008;1:1. doi: 10.1186/1756-0500-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Henares A.J., Krell T., Eugenia Guazzaroni M., Segura A., Ramos J.L. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol. Rev. 2006;30:157–186. doi: 10.1111/j.1574-6976.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- Nikel P.I., Chavarría M., Danchin A., de Lorenzo V. From dirt to industrial applications: Pseudomonas putida as a synthetic Biology chassis for hosting harsh biochemical reactions. Curr. Opin. Chem. Biol., Synth. Biol. * Synth. Biomol. 2016;34:20–29. doi: 10.1016/j.cbpa.2016.05.011. [DOI] [PubMed] [Google Scholar]
- Niu W., Draths K.M., Frost J.W. Benzene-Free synthesis of adipic acid. Biotechnol. Prog. 2002;18:201–211. doi: 10.1021/bp010179x. [DOI] [PubMed] [Google Scholar]
- Pédelacq J.-D., Cabantous S., Tran T., Terwilliger T.C., Waldo G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Raman S., Rogers J.K., Taylor N.D., Church G.M. Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. 2014;111:17803–17808. doi: 10.1073/pnas.1409523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J.K., Church G.M. Genetically encoded sensors enable real-time observation of metabolite production. Proc. Natl. Acad. Sci. 2016;113:2388–2393. doi: 10.1073/pnas.1600375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J.K., Taylor N.D., Church G.M. Biosensor-based engineering of biosynthetic pathways. Curr. Opin. Biotechnol. 2016;42:84–91. doi: 10.1016/j.copbio.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Rorrer N.A., Dorgan J.R., Vardon D.R., Martinez C.R., Yang Y., Beckham G.T. Renewable unsaturated polyesters from muconic acid. ACS Sustain. Chem. Eng. 2016;4:6867–6876. [Google Scholar]
- Salvachúa D., Karp E.M., Nimlos C.T., Vardon D.R., Beckham G.T. Towards lignin consolidated bioprocessing: simultaneous lignin depolymerization and product generation by bacteria. Green. Chem. 2015;17:4951–4967. [Google Scholar]
- Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Schallmey M., Frunzke J., Eggeling L., Marienhagen J. Looking for the pick of the bunch: high-throughput screening of producing microorganisms with biosensors. Curr. Opin. Biotechnol. 2014;26:148–154. doi: 10.1016/j.copbio.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Suastegui M., Matthiesen J.E., Carraher J.M., Hernandez N., Rodriguez Quiroz N., Okerlund A., Cochran E.W., Shao Z., Tessonnier J.-P. Combining metabolic engineering and electrocatalysis: application to the production of Polyamides from sugar. Angew. Chem. Int. Ed. 2016;55:2368–2373. doi: 10.1002/anie.201509653. [DOI] [PubMed] [Google Scholar]
- Tang S.-Y., Cirino P.C. Design and application of a mevalonate-responsive regulatory protein. Angew. Chem. Int. Ed. 2011;50:1084–1086. doi: 10.1002/anie.201006083. [DOI] [PubMed] [Google Scholar]
- Tang S.-Y., Qian S., Akinterinwa O., Frei C.S., Gredell J.A., Cirino P.C. Screening for enhanced triacetic acid lactone production by recombinant Escherichia coli expressing a designed Triacetic acid lactone reporter. J. Am. Chem. Soc. 2013;135:10099–10103. doi: 10.1021/ja402654z. [DOI] [PubMed] [Google Scholar]
- Vardon D.R., Franden M.A., Johnson C.W., Karp E.M., Guarnieri M.T., Linger J.G., Salm M.J., Strathmann T.J., Beckham G.T. Adipic acid production from lignin. Energy Environ. Sci. 2015;8:617–628. [Google Scholar]
- Weber C., Brückner C., Weinreb S., Lehr C., Essl C., Boles E. Biosynthesis of cis,cis-muconic acid and its aromatic precursors, catechol and protocatechuic acid, from renewable feedstocks by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012;78:8421–8430. doi: 10.1128/AEM.01983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Wang J., Pang Q., Zhang F., Wang J., Wang Q., Qi Q. Pathway optimization and key enzyme evolution of N -acetylneuraminate biosynthesis using an in vivo aptazyme-based biosensor. Metab. Eng. 2017;43:21–28. doi: 10.1016/j.ymben.2017.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material