Abstract
As a kind of nanometric lipidic vesicles, exosomes have been presumed to play a leading role in the regulation of tumor microenvironment through exosomes-mediated transfer of proteins and genetic materials. Tumor-derived exosomes are recognized as a critical determinant of the tumor progression. Intriguingly, some current observations have identified that exosomes are essential for several intercellular exchanges of proteins, messenger RNAs, noncoding RNAs (including long noncoding RNAs and microRNAs) as well as to the process of cancer metastasis and drug resistance. Herein, we review the role of exosomes and their molecular cargos in cancer invasion and metastasis, summarize how they interact with antitumor agents, and highlight their translational implications.
Keywords: Exosomes, contents, metastasis, drug resistance, cancer
Introduction
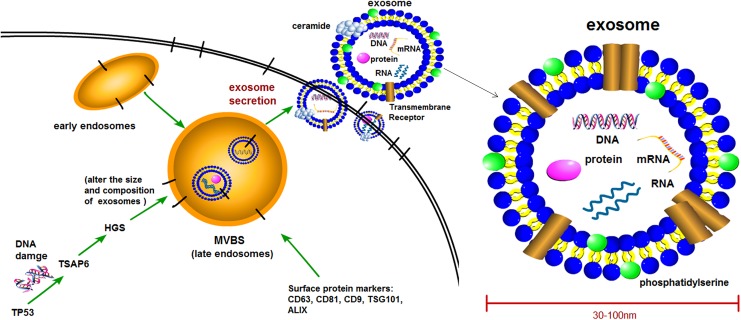
Exosomes are spherical bilayered membrane vesicles with an average diameter of 30 to 100 nm and “saucer-shape” morphology.1 They are the only type of extracellular vesicles formed from endosomal compartment invaginations, which are called multivesicular bodies (MVBs) and are released in the endosomal network.2–4 After the fusion of MVBs with plasma membrane, the internal contents are released into the extracellular space as the form of “exosomes” (Figure 1). Exosomes can be found in a range of fluids, including blood, plasma, saliva, urine, synovial fluid, amniotic fluid, malignant ascite, and pleural effusions.5–9 The biogenesis mechanisms of exosomes have not been fully elucidated, but they are secreted from nearly all cell types including normal cells and diseased cells.10–12 In particular, tumor cells possess more exosomes-releasing properties when compared to normal cells.13 Exosomes from different cell phenotypes and body fluids contain various bioactive molecules such as proteins (including oncoproteins, tumor suppressor proteins, and transcriptional regulators), lipids (including phosphatidylcholines, phosphatidylethanolamines, and phosphatidylserines),14–16 DNA (including single-strand DNA, genomic DNA, XX DNA, and retrotransposon elements),17–20 and RNAs (including messenger RNAs [mRNAs], long noncoding RNAs [lncRNAs], microRNAs [miRNAs], and other non-coding RNAs [ncRNAs]).21–23 Circulating exosomes are complex molecular assemblies of these bioactive molecules, and these biomolecules permit horizontal transfer of oncogenic traits from the primary tumor to recipient target cells located in distant organs. Pioneering works call this phenomenon “genometastasis”24 as an implication of the intercellular trafficking of oncogenic macromolecules via exosomes to explain cancer metastasis.25 Since Abdouh et al 26 confirmed the validity of the genometastatic theory in human cells for the first time, recent studies provide more evidences to support this idea, and experimental data demonstrate the role of circulating factors carried by exosomes in conferring mesenchymal–epithelial transition (MET),27 mediating oncogenic transformation28 and contributing to cancer metastasis.
Figure 1.
Generation and the structure of exosome. Exosomes are vesicles with a phospholipid bilayer membrane; they are considered to be secreted from intracellular multivesicular bodies (MVBs or late endosomes) into extracellular space. The exosomes contain a range of proteins, RNAs, mRNAs, and DNA molecular cargoes, with surface protein markers including CD9, CD63, CD81, TSG101, and ALIX, and there are equal or asymmetrical distributions of phospholipids between the 2 leaflets of exosome membrane. mRNAs indicates messenger RNAs; TBS, Tris-buffered saline; TSG101, tumor-susceptibility gene-101; ALG-2-interacting protein X (ALIX).
Although it is possible that horizontal transformation may require preexisting alterations in recipient cells,29 Abdouh et al 30 also reported that human cells carrying a single oncosuppressor mutation would show an increased uptake of exosomes when exposed to blood-circulating cancer factors and were capable of integrating cancer factors at metastatic sites. This further demonstrates that oncogenic factors transferred via circulating cancer exosomes induce malignant transformation of target cells even at distant distance.
The constituent information about the exosomal molecular cargos can be obtained in public access databases, such as ExoCarta31 and EVPedia.32 Under the stimulation of external extreme conditions such as oxidative stress and serum starvation, most cells can produce exosomes containing altered RNA contents.33,34
In addition, mutated oncogenes may also alter exosome secretion.1,35,36 Tumor cells release exosomes into the circulation, leading to significant increase of exosomes in patients with cancer compared to the healthy controls,13 and the abnormal exosomes may be associated with decreased overall survival.37 Tumor-secreted exosomes can even influence organotropism. It is proved that tumor-derived exosomal molecules guide exosomes to specific organs and promote organ-specific metastasis because exosomes from different cancer recapitulate the organ specificity of their cell of origin and prepare premetastatic niches as well.38,39
There are several approaches for the isolation of exosomes: Ultracentrifugation1,40–42 and commercial kits (ExoQuick kit, System Biosciences, USA; Total Exosome Isolation Reagent, Invitrogen, USA; Qiagen miRNeasy Kit, Qiagen, Germany) are usually used. Generally, traditional ultracentrifugation-based approach can provide cleaner exosomes, but this method is labor intensive and time consuming and in need of expensive laboratory equipment and larger sample volumes. For the commercial kit approach, the acquired exosomes contain additional molecular complexes that are unnecessary. Therefore, an improved isolation technology is required to obtain exosomes effectively.39
In this article, we review the role of exosomes and its molecular cargos in tumor invasion and metastasis, summarize how they interact with antitumor drugs, and show the potential of clinical application.
Exosomes in Tumor Metastasis and Drug Resistance
Exosomes were considered “garbage bag” of unnecessary cellular materials when they were first discovered.11,43 However, many important biological functions of exosomes have been convincingly demonstrated in the recent years. They are involved in intercellular communication, immune system modulation, cell growth enrichment, energy pathways maintenance, and propagation of viruses via releasing intracellular cargoes (eg, proteins, lipids, DNA, and RNAs including tumor-derived ncRNAs).40,44–46 One type of the exosomes, tumor-derived exosomes, have been found to apparently facilitate tumor growth and metastasis.47 Rather than simple cellular debris, tumor-derived exosomes act as extracellular organelles with roles in remodeling tumor microenvironment by delivering messages for cell–cell communication.48–51 It is a well-built means of intercellular exchange via exosomes, and especially, the exosomes transport the nucleic acids and proteins from tumor cells to neighboring cells in tumor microenvironments.52
First, tumor-derived exosomes induce proinflammatory phenomena. They interact with their proteins and miRNAs to result in an inflammatory behavior, and this is essential for recruiting inflammatory CCR6+CD4+ Th17+ into specific tumor sites53 to contribute to proliferation, angiogenesis, and metastasis of malignant cells in tumor microenvironment.54,55 For instance, when taken up by resident cells, tumor-derived exosomal integrins (ITG) increase proinflammatory S100 gene expression.38 Second, tumor-derived exosomes mediate vascular leakiness as a key feature of premetastatic niche formation.56 Similar to cytokines, tumor-derived exosomes can recruit bone marrow-derived cells to premetastatic tumor tissue, thus contributing to creating a permissive microenvironment for tumor metastases.57 In this way, they could remodel the extracellular matrix (ECM) to support tumor growth and a prometastatic phenotype.58,59 Third, tumor-derived exosomes are involved in immune response. They suppress the activation of effector T-cells, trigger the apoptosis of activated T-cells, and help metastatic cells to escape tumor immune surveillance.60,61 Fourth, tumor-derived exosomes promote tumor progression via regulating drug resistance: (1) tumor cells can utilize the exosome secretion to extrude anticancer drugs; (2) exosomal molecular cargoes can compete with anticancer drugs, especially antibody-based drugs, to bind with oncogenic targets, thus lowering their therapeutic effect62,63; and (3) exosomes contribute to the lateral transfer of drug resistance from drug-resistant cells to drug-sensitive cells.64
Exosomes and Tumor Metastasis
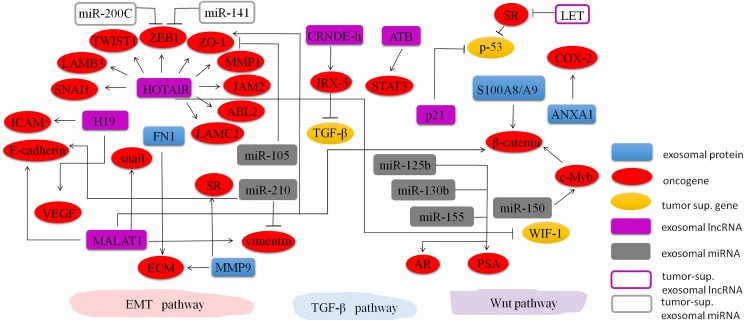
Exosomes can mediate important cancer-related pathways to enhance tumor invasion and metastasis through different manners, including controlling angiogenesis,65–69 modulating stromal cells,70–72 remodeling the ECM,73 transferring malignant traits,30 and establishing the premetastatic niche.74–77 Exosomes containing ncRNAs and proteins play significant roles in these pathways (Table 1 and Figure 2). However, the biological function of exosomal ncRNAs and proteins in tumor invasion and metastasis remains unclear.
Table 1.
Functions of Exosomal Noncoding RNAs and Proteins in Tumor Metastasis and Invasion.
| Tumor Types | Oncogene/Suppressor | Functions in Tumors | Reference | |
|---|---|---|---|---|
| Exosomal proteins | ||||
| EDIL-3 | Urinary bladder cancer | Oncogene | High-grade muscle invasive | Beckham et al 78 Street et al 79 |
| FN1 | Colorectal cancer | Oncogene | Tumor progression and metastasis; ECM degradation; EMT | Akakura et al 80 |
| MMP9 | Colorectal cancer | Oncogene | ECM degradation; EMT by TGF-β/Smad signaling pathway | Lampropoulos et al 81 |
| S100A8 and S100A9 | Colorectal cancer | Oncogene | Involves in distal metastasis by Wnt/β-catenin pathway | Ichikawa et al 82 |
| H-ras | Intestinal epithelial cells | Oncogene | Exhibit EV uptake and transforming effects | Lee et al 29 |
| ITGα6, ITGβ4, and ITGβ1 | Breast cancer | Oncogene | Lung metastasis | Hoshino et al 38 |
| ITGαv and ITGβ5 | Pancreatic cancer | Oncogene | Liver metastasis | Hoshino et al 38 |
| ITGβ3 | Breast cancer | Oncogene | Brain metastasis | Hoshino et al 38 |
| Exosomal miRNA | ||||
| let-7 miRNA family | Gastric cancer | Oncogene | Promote metastasis | Ohshima et al 83 |
| miR-31 | Breast cancer | Tumor suppressor | Opposing local invasion and metastatic colonization | Ragusa et al 41 |
| miR-105 | Breast cancer | Oncogene | Promote metastasis | Zhou et al 84 |
| miR-130 | Breast cancer | Oncogene | Enhances cell proliferation and migration | Ragusa et al 41 Kruger et al 85 |
| miR-328 | Breast cancer | Oncogene | Promote metastasis | Kruger et al 85 |
| miR-150 | Monocytic | Oncogene | Cell migration | Zhang et al 86 |
| miR-210 | Colorectal cancer | Oncogene | Promote EMT, metastasis | Bigagli et al 87 |
| miR-200c, miR-141 | Colorectal cancer | Tumor suppressor | Cell proliferation, invasion, migration, EMT | Tanaka et al 88 |
| miR-125b, miR-130b, miR-155 | Prostate cancer | Oncogene | Triggering neoplastic transformation and MET | Abd Elmageed et al 27 |
| Exosomal lncRNA | ||||
| MALAT1 | Liver cancer | Oncogene | Regulate alternate splicing, cancer metastasis, and tumor recurrence | Tripathi et al 89 Yang et al 90 |
| ATB | Liver cancer | Oncogene | Mediate TGF-β signaling pathway, promote the invasion–metastasis cascade | Yuan et al 91 |
| CRNDE-h | Colorectal cancer | Oncogene | Reduce the sensitivity to the cytostatic effect of TGF-β | Liu et al 92 |
| lncRNA-TUC339 | Hepatic cancer | Oncogene | Promote the growth and spread | Kogure et al 93 |
| lncRNA-H19 | Liver cancer | Oncogene | EMT, hepatic metastases | Conigliaro et al 94 |
| lncRNA-HOTAIR | Urothelial bladder cancer | Oncogene | Tumor migration and invasion | Berrondo et al 95 Yan et al 96 |
| lncRNA-p21 | Multiple cancers | Oncogene | Affect global gene expression | Gezer et al 97 Dimitrova et al 98 |
Abbreviations: EDIL-3, epidermal growth factor-like repeats and discoidin I-like domain 3; FN1, fibronectin-1; ECM, extracellular matrix; EMT, epithelial–mesenchymal transition; MMP9, matrix metalloproteinase-9; TGF-β, transforming growth factor β; ITG, integrins; miRNA, microRNA; MET, mesenchymal–epithelial transition; lncRNA, long noncoding RNA; MALAT1, metastasis-associated lung adenocarcinoma transcript-1; CRNDE-h, colorectal neoplasia differentially expressed-h.
Figure 2.
Regulating network of exosomal proteins and ncRNAs in tumors. Molecular signaling of exosomal nc-RNAs (ie, exosomal miRNAs [exo-miRNAs] and exosomal lncRNAs [exo-lncRNAs]) and proteins in most frequent cancer-related pathways involved in cancer metastasis (ie, EMT, TGF-β, and Wnt). These bioactive molecules carried in exosomes take effects in metastasis promotion or repression and interact with others. EMT pathway is a process by which epithelial cells lose their cell–cell adhesion and acquire migratory property to become mesenchymal stem cells; TGF-β pathway is involved in cellular processes including cell growth, differentiation, and invasion; and Wnt pathway regulates crucial aspects of cell fate determination and cell migration during embryonic development. ncRNAs indicates noncoding RNAs; miRNA, microRNA; lncRNAs, long noncoding RNAs; EMT, epithelial–mesenchymal transition; TGF-β, transforming growth factor β.
Association between exosomal proteins and tumor metastasis
Exosomes contain numerous proteins, which may be associated with the growth and survival of metastatic tumor cells. Chen et al 40 revealed some upregulated exosomal proteins from patients with colorectal cancer (CRC) are involved in the modulation of pretumorigenic microenvironment for metastasis and invasiveness. These exosomal proteins, such as fibronectin-1, mediate cytoskeletal organization and actin dynamics to enable cell adhesion and motility.80 Matrix metalloproteinase-9 (MMP9), a protein found to be overexpressed in exosomes of patients with CRC, plays a key role in ECM degradation via the activation of transforming growth factor β (TGF-β)/Smad signaling pathway.81 Additionally, some exosomal protein markers could be used as predictive markers for organ-specific metastasis. For example, epidermal growth factor (EGF)-like repeats and discoidin I-like domain 3 protein from urinary exosomes of patients with bladder cancer can be used for predicting muscle metastasis.78,79 Furthermore, surface proteins of exosomes are also involved in metastatic tropism. Hoshino et al revealed a close correlation between exosomal ITG and metastatic tropism.38 They found that exosomal ITG could regulate local microenvironments within future metastatic organs. They also reveal that the expression of exosomal ITGα6 and its partners ITGβ4 and ITGβ1 was associated with lung metastasis; ITGαv and ITGβ5 were linked to liver metastasis, while ITGβ3 underlied organotropism to the brain.38 Recent studies show that the metastatic tumor cells-derived exosomes are enriched with proteins, such as S100A8 and S100A9.99 Exosomal S100A8/A9 proteins can be upregulated from tumor cells with high activity of Wnt/β-catenin pathway, and these proteins are able to recruit leukocytes and probably indirectly involved in the formation of premetastatic niches in distal metastatic organs.82
Association between exosomal miRNAs and tumor metastasis
Micro-RNAs, a large class of ncRNAs, play pivotal roles in various cancers through the repression of downstream cancer-associated mRNAs.100 Since miRNAs are detected in serum from patients with diffuse large B-cell lymphoma first,101 different exosomal miRNAs have been reported in distinct types of malignancies, including tongue cancer,102 breast cancer,103 lung cancer,104 ovarian cancer,105 prostate cancer,106 CRC,107,108 and gastric cancer.83 These findings support that these miRNAs are potential biomarkers for cancer diagnosis and prognosis.109 There are abundant miRNAs in the exosomes. Notably, the expression and distribution exosomal miRNAs are often dysregulated in cancerous tissue. Cancer-derived exosomes transfer miRNAs into target cell cytoplasm and then result in downregulation of expression of specific target genes, thus affecting cell function. This process is possibly modulated by important signaling pathways in recipient cells, such as Wnt, Ras, TGF-β, and p53 signaling pathways.110,111 Intriguingly, packaging of miRNAs into exosomes is not stochastic, miRNAs are preferentially selected into CD63-positive exosomes via a ceramide-dependent pathway, and some oncogenic signaling, such as Kirsten rat sarcoma viral oncogene homolog, may result in selective packaging of miRNA into exosomes, thus generating different cancer-specific exosome profiles.112,113
Among those exosomal miRNAs mentioned earlier, some miRNAs are closely associated with tumor invasion and metastasis. For example, exosomal miR-31 from breast cancer cells influences metastasis by suppressing local invasion and metastatic colonization, and it is an onco-miR whose overexpression enhances cancer cell proliferation and migration.41 Similarly, miR-130a and miR-328 are observed to be upregulated in exosomes derived from metastatic MDA-MB-231 cells.85 Moreover, the let-7 miRNA family, including let-7a, let-7b, let-7c, let-7d, let-7e, and let-7i, is packaged into exosomes from metastatic AZ-P7a gastric cancer cell to maintain their oncogenesis and invasiveness.83 Thus, this miRNA family has a vital role in the delivery of oncogenic signals to promote metastasis. Le et al 114 clearly demonstrated that exosomes containing miR-200 could transfer the metastatic capability between metastatic and nonmetastatic cancer cells. Bigagli and her group also found that exosomes containing miR-210 might be considered as epithelial–mesenchymal transition (EMT) promoting signal that guides metastatic cells to free new sites of dissemination with low level of vimentin and high level of E-cadherin.87 Zhang et al 86 showed that exosomes from the human monocytic cells human acute monocytic leukemia cells (THP-1) with a high level of miR-150 could deliver miR-150 into the human endothelial cells human microvascular endothelial cells (HMEC-1) and then affect MYB proto-oncogene (c-Myb) expression and cell migration. Likewise, exosomes derived from MDA-MB-231 cells and Michigan Cancer Foundation-10 (MCF-10) cells harbor plentiful miR-105, which was responsible for promoting metastasis by suppressing the expression of tight junction protein zonula occludens 1 (ZO-1).84 Abd Elmageed et al 27 revealed a new role for prostate cancer (PC) cell-derived exosomes in triggering neoplastic transformation and MET through trafficking of oncogenic factors (including onco-miRNAs miR-125b, miR-130b, and miR-155) into adipose-derived stem cells of patients with PC. In contrast, some exosomal miRNAs are negatively correlated with invasion ability. Shota et al 88 observed an increased expression of exosomal miR-200c and miR-141 in SW620 and SW620/OxR CRC cells after decitabine treatment, accompanied by the acquisition of epithelial cell-like characteristics. So exosomal miR-200c and miR-141 may be an indicator for MET and potentially associated with metastasis inhibition.
Association between exosomal lncRNAs and tumor metastasis
The lncRNAs from exosomes may serve as signaling molecules in various biological processes mediating intercellular communication. High levels of exosomal lncRNAs not only provide a novel diagnostic method for early prediction but also show predictive potential for tumor invasion and metastasis.115,116 Notably, quantitative comparisons prove that exosomes lack mRNAs but contain abundant lncRNAs.117,118 A subsequent attempt points out that lncRNAs occupy 3.36% of the total exosomal RNA content.119 Liu et al 92 also indicated that exosomal colorectal neoplasia differentially expressed-h (CRNDE-h) levels were associated with distant metastasis and lymph node metastasis, accompanied by high expression of iroquois homeobox protein-5 in the metastatic sites.120 Besides, this exosomal lncRNA decreased the sensitivity to the cytostatic effect of TGF-β and granted a growth performance to tumor cells in TGF-β/SMADS pathway.120 Kogure and colleagues97 suggested that the exosome-mediated transfer of lncRNA-TUC339 could improve the growth of cancer cells and facilitate the spread of hepatocellular carcinoma (HCC). Through the secretion of exosomes containing lncRNA-H19, CD90+ liver cancer cells can modulate endothelial–mesenchymal phenotype and involve in hepatic metastases by inducing an increase in vascular EGF and intercellular adhesion molecule-1 transcripts,121 thus influencing tumor microenvironment in a prometastatic way.122 Through the release of exosomes containing metastasis-associated lung adenocarcinoma transcript-1 (MALAT1), liver cancer cells can modify RNA alternate splicing by adjusting the levels of phosphorylated to dephosphorylated serine/arginine-rich (SR) proteins.89 This process is connected with metastasis and recurrence of liver cancer as well as a diverse range of cancers.90 It has also been suggested that MALAT1 could alter the protein expression of E-cadherin, ZO-1, β-catenin, vimentin, and snail, which are involved in EMT.93
Through the release of exosomes containing lncRNA-ATB, the TGF-β signaling pathway can be modulated. In detail, lncRNA-ATB binds to interleukin 11 and triggers signal transducer and activator of transcription 3 (STAT3) signaling pathway to promote the invasion–metastasis cascade.91 Analogously, exosomal lncRNA-p21 has been reported to regulate mRNA translation and to suppress the p53 to affect global gene expression by binding to the heterogeneous nuclear ribonucleoprotein K (hnRNP-K) complex.94,117,123 Besides, exosomal lncRNA-HOTAIR in urothelial bladder cancer cells can regulate the expression of EMT genes, including SNAI1, LAMB3, TWIST1, MMP1, ZEB1, ZO-1, JAM2, ABL2, and LAMC2, thus inducing tumor migration and invasion.124 Yan et al 125 also showed that HOTAIR was involved in migration and invasion through inhibiting the canonical Wnt pathway antagonist protein WIF-1 in vitro.
However, some other exosomal lncRNAs show a function to inhibit tumor metastasis. The lncRNA-LET binds to and destabilizes nuclear factor of activated T-cells 90 kDa (NF90),126 thus suppressing cell growth and metastasis in HCC.127 With these observations, exosomal lncRNAs may be useful for diagnosis and prevention of metastasis. Exosomal miRNAs have been extensively studied in various cancers,98 whereas exosomal lncRNAs are being pursued for similar goals. Although the specific association of exosomes with cancer carcinogenesis and metastasis remains unclear, their stable property makes them to be widely used in cancer research. It is reported that exosomes remain stable even at a temperature range from 4°C to 42°C.95
Exosomes and Drug Resistance
Exosomes-mediated drug resistance
Drug resistance is a major challenge for cancer therapy. Several studies reveal that exosomes are involved in the modulation of chemosensitivity by transferring the resistant phenotype to recipient cells.64 Transport of ncRNAs, including miRNAs and lncRNAs, mediated by exosomes, is believed to be an effective mechanism for acquiring drug resistance in cancer cells (Table 2). For example, in ovarian cancer, exosomal transfer of miR-433 can promote paclitaxel resistance through the induction of cellular senescence.126 In HCC, exosomal transfer of lncRNA-ROR can increase TGF-β-dependent chemoresistance.127,98 In breast cancer, exosomal transfer of lncRNA urothelial cancer associated-1 (UCA1) can increase tamoxifen resistance in estrogen receptor-positive MCF-7 cells through mTOR signaling pathway.96 Exosomal transfer of lncRNA UCA1 can also increase chemoresistance of bladder cancer cells via activating the Wnt signaling pathway.128 Exosomes expressing full-length human epidermal growth factor 2 molecules enable them to bind and sequester Trastuzumab, thus lowering the therapeutic effectiveness of Trastuzumab in breast cancer.62 In renal cancer, exosome-transmitted lncRNA Activated in RCC with Sunitinib Resistance (lncARSR) can disseminate Sunitinib resistance by acting as a competing endogenous RNA for miR-34 and miR-449, thus facilitating expression of receptor tyrosine kinase (AXL) and c-MET.129 Exosomes are speculated as a novel target for cancer therapy because they can promote angiogenic phenotype and cell-to-cell adhesion in target cells.130
Table 2.
Functions of Exosomal ncRNAs in Tumor Drug Resistance.
| Tumor Types | Function in Drug Resistance | Drug | Reference | |
|---|---|---|---|---|
| Exosomal miRNA | ||||
| miR-433 | Ovarian cancer | Promote | Paclitaxel | Weiner-Gorze et al 126 |
| Exosomal lncRNA | ||||
| lncRNA-ROR | Hepatocellular cancer | Promote | Chemotherapeutic drugs | Takahashi et al 127 Nawaz et al 98 |
| lncRNA UCA1 | Breast cancer | Promote | Tamoxifen | Xu et al 96 |
| lncRNA UCA1 | Bladder cancer | Promote | Chemotherapeutic drugs | Fan et al 128 |
| HER2 | Breast cancer | Promote | Trastuzumab | Ciravolo et al 62 |
| lncARSR | Renal cancer | Promote | Sunitinib | Qu et al 129 |
Abbreviations: ncRNAs, noncoding RNAs; miRNA, microRNA; lncRNA, long noncoding RNAs; UCA1, urothelial cancer associated-1; HER2, human epidermal growth factor-2; lncARSR.
A study suggests that the acid microenvironment of the tumor–host interface may stimulate the output of exosomes,131 and such exosomes could release several contents that transport a resistance phenotype to sensitive tumor cells by changing cell cycle control and stimulating antiapoptosis programs.132 It has also been proposed that EMT inducers can increase resistance to chemotherapy.133 Bigagli et al 114 proved the chemosensitivity of metastatic cells undergoing EMT was diminished compared to adherent HCT-8 colon cancer cells, since exosomes secreted from HCT-8 cells might impact the EMT program and made the metastatic cells spontaneously insensitive to chemotherapeutic strategies. There is also a deduction that vesicles such as exosomes are able to move between cells using specific structures called nanotubes.133 However, it is not clear which of these theories may be correct.
Therapeutic potential of exosomes in drug resistance of cancer
Treatment with locked nucleic acids targeting exosomal lncRNA and miRNAs can restore drug sensitivity. Through the untiring efforts of researchers, the lncRNAs and miRNAs are considered as putative targets for cancer therapy via exosome-mediated mechanism, such as lncRNA-H19 and lnc-ARSR9.121,134 Exosomal ncRNAs-based therapeutic protocols, such as antisense oligonucleotides, have been proved to be capable of restraining pathological lncRNAs. Therefore, these exosomal ncRNAs are inspiring and appealing for cancer therapy. In addition, there is a speculation that exosomes can deliver drugs to selective targets as a novel platform in a drug delivery system. In particular, exosomes are capable of crossing the blood–brain barrier without inducing an immune response.135 However, a set of troubles have been suffered during the clinical development of exosomes-based therapeutic technologies for cancers, including their indefinite functions in the complex networks, the difficulty in accurate quantification of exosomal lncRNAs and miRNAs, the hard transport of several lncRNAs and miRNAs antagonists or mimics as well as the unclear clinical pharmacokinetics and drug toxicity.39 Therefore, more studies are needed to verify the potential of applying exosomes and their contents in cancer therapy. Exosomes-based drugs are expected to enter clinical trials, and their great potential may be revealed.
Prospects and Challenges
From the above, we conclude that tumor-derived exosomes not only discard cellular waste but also trigger signaling pathways and drug resistance in target cells and facilitate the growth of metastatic cells. It is encouraging that nanomaterials or microbodies, such as exosomes-mediated delivery system, provide more possibility. Exosomes containing MAGE family member A3 (MAGE-3) peptides show minimal toxic effects after treatment in patients with stages III/IV melanoma.135 However, small RNA-based drugs, such as miRNAs and circRNAs, face different kinds of challenges due to the lack of targeted delivery strategies in cancer-targeted therapy.136
Switching to a wider scientific horizon, exosomes are still on initial exploration at present, and only a few molecular mechanisms about exosomes in cancer progression are discovered. In addition, the strategies and methods used for investigating exosomes are still limited and challenging. The identification methods of exosomes are diverse; for instance, the size of pelleted particles can be measured using dynamic light scattering such as Brookhaven instruments BI-9000 digital correlator (Brookhaven instruments, USA), Zetasizer Nano ZS (Malvern, UK), LM10 NanoSight (Malvern, UK), or BI200-SM goniometer (Brookhaven instruments, USA), configured with a solid-state laser tuned at 532 nm.122,124 The morphology of exosomes is suitable to be examined under a transmission electron microscope (TEM) at 80 keV, with a diameter of approximately 30 to 100 nm under the TEM ultimately. The typical electron micrographs can be captured by a megapixel digital camera, such as Erlangshen 11 digital camera (Gatan, USA).124 Exosomes can also be identified by Western blot analysis with exosomal surface protein markers antibodies such as anti-CD9, anti-CD63, anti-CD81, anti-CD82, and so on.137 However, from the reported studies, we can see that the identification of exosomes is not coherent and comprehensive, which cannot avoid the occurrence of false-positive or false-negative results.
Currently, the large-scale profiles of miRNAs or lncRNAs in exosomes are usually obtained using microarrays of next-generation sequencing followed by the extraction of exosomes, which can be customized and time reduced.138 Besides, quantitative polymerase chain reaction (PCR), digital PCR, and NanoString nCounter Gene Expression Assay (NanoString nCounter system, USA) can be used for detection of miRNAs or lncRNAs, with features of expensive and uneffective but high sensitivity.139 Exosome protein can be isolated by optimized lysis buffer140 and quantitated by a microBCA Protein Assay kit (Pierce, USA). When the focus is on the number of total exosomal proteins in patients with cancer, a quantitative proteomic analysis can be used.141
In conclusion, more details regarding the functional contents of exosomes involved in cancer progression is highly desired, and exploring their biological mechanism will improve the clinical applicability of exosomes in cancer diagnosis and therapy.
Abbreviations
- CRC
colorectal cancer
- CRNDE-h
colorectal neoplasia differentially expressed-h
- ECM
extracellular matrix
- EMT
epithelial–mesenchymal transition
- HCC
hepatocellular carcinoma
- ITG
integrins
- lncRNAs
long noncoding RNAs
- MET
mesenchymal–epithelial transition
- miRNAs
microRNAs
- mRNAs
messenger RNAs
- MVBs
multivesicular bodies
- ncRNAs
noncoding RNAs
- PCR
polymerase chain reaction
- SR
serine/arginine-rich
- TEM
transmission electron microscope
- TGF-β
transforming growth factor β
- UCA1
urothelial cancer associated-1
Footnotes
Authors’ Note: Chengcheng Zhang, Qing Ji, and Yue Yang contributed equally to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by International Cooperation Key Project of National Natural Science Foundation of China (81520108031) and National Natural Science Foundation of China (81573749 and 81473478), Key Project of Shanghai Municipal Science and Technology Commission (16401970500), Program of Shanghai Academic Research Leader (16XD1403600), Program for Shanghai Outstanding Medical Academic Leader, Natural Science Foundation of Shanghai, China (16ZR1437700), and Research Project for Practice Development of National TCM Clinical Research Bases (JDZX2015067).
References
- 1. Sun Y, Zheng W, Guo Z, et al. A novel TP53 pathway influences the HGS-mediated exosome formation in colorectal cancer. Sci Rep. 2016;6:28083 doi:10.1038/srep28083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hurley JH, Odorizzi G. Get on the exosome bus with ALIX. Nat Cell Biol. 2012;14(7):654–655. doi:10.1038/ncb2530 [DOI] [PubMed] [Google Scholar]
- 3. Urbanelli L, Magini A, Buratta S, et al. Signaling pathways in exosomes biogenesis, secretion and fate. Genes. 2013;4(2):152–170. doi:10.3390/genes4020152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yanez-Mo M, Siljander PR, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066 doi:10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gonzalez-Begne M, Lu B, Han X, et al. Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J Proteome Res. 2009;8(3):1304–1314. doi:10.1021/pr800658c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garcia JM, Garcia V, Pena C, et al. Extracellular plasma RNA from colon cancer patients is confined in a vesicle-like structure and is mRNA-enriched. RNA. 2008;14(7):1424–1432. doi:10.1261/rna.755908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keller S, Rupp C, Stoeck A, et al. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72(9):1095–1102. doi:10.1038/sj.ki.5002486 [DOI] [PubMed] [Google Scholar]
- 8. Dear JW, Street JM, Bailey MA. Urinary exosomes: a reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics. 2013;13(10-11):1572–1580. doi:10.1002/pmic.201200285 [DOI] [PubMed] [Google Scholar]
- 9. Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7(3):e30679 doi:10.1371/journal.pone.0030679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang C, Robbins PD. The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol. 2011;2011:842849 doi:10.1155/2011/842849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi:10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83(11):1484–1494. doi:10.1016/j.bcp.2011.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi:10.1016/j.ygyno.2008.04.033 [DOI] [PubMed] [Google Scholar]
- 14. Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89(2):205–212. doi:10.1016/j.biochi.2006.10.014 [DOI] [PubMed] [Google Scholar]
- 15. Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. Biochim Biophys Acta. 2014;1841(1):108–120. doi:10.1016/j.bbalip.2013.10.004 [DOI] [PubMed] [Google Scholar]
- 16. Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289(7):3869–3875. doi:10.1074/jbc.C113.532267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180 doi:10.1038/ncomms1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cai J, Han Y, Ren H, et al. Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J Mol Cell Biol. 2013;5(4):227–238. doi:10.1093/jmcb/mjt011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burger D, Schock S, Thompson CS, Montezano AC, Hakim AM, Touyz RM. Microparticles: biomarkers and beyond. Clin Sci (Lond). 2013;124(7):423–441. doi:10.1042/cs20120309 [DOI] [PubMed] [Google Scholar]
- 20. Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24(6):766–769. doi:10.1038/cr.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282 doi:10.1038/ncomms1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Record M, Subra C, Silvente-Poirot S, Poirot M. Exosomes as intercellular signalosomes and pharmacological effectors. Biochem Pharmacol. 2011;81(10):1171–1182. doi:10.1016/j.bcp.2011.02.011 [DOI] [PubMed] [Google Scholar]
- 23. Raimondo F, Morosi L, Chinello C, Magni F, Pitto M. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics. 2011;11(4):709–720. doi:10.1002/pmic.201000422 [DOI] [PubMed] [Google Scholar]
- 24. Garcia-Olmo D, Garcia-Olmo DC, Ontanon J, Martinez E, Vallejo M. Tumor DNA circulating in the plasma might play a role in metastasis. The hypothesis of the genometastasis. Histol Histopathol. 1999;14(4):1159–1164. [DOI] [PubMed] [Google Scholar]
- 25. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi:10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- 26. Abdouh M, Zhou S, Arena V, et al. Transfer of malignant trait to immortalized human cells following exposure to human cancer serum. J Exp Clin Cancer Res. 2014;33:86 doi:10.1186/s13046-014-0086-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abd Elmageed ZY, Yang Y, Thomas R, et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells. 2014;32(4):983–997. doi:10.1002/stem.1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hamam D, Abdouh M, Gao ZH, Arena V, Arena M, Arena GO. Transfer of malignant trait to BRCA1 deficient human fibroblasts following exposure to serum of cancer patients. J Exp Clin Cancer Res. 2016;35:80 doi:10.1186/s13046-016-0360-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee TH, Chennakrishnaiah S, Meehan B, et al. Barriers to horizontal cell transformation by extracellular vesicles containing oncogenic H-ras. Oncotarget. 2016;7(32):51991–52002. doi:10.18632/oncotarget.10627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abdouh M, Hamam D, Gao ZH, Arena V, Arena M, Arena GO. Exosomes isolated from cancer patients’ sera transfer malignant traits and confer the same phenotype of primary tumors to oncosuppressor-mutated cells. J Exp Clin Cancer Res. 2017;36(1):113 doi:10.1186/s13046-017-0587-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40(Database issue):D1241–D1244. doi:10.1093/nar/gkr828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim DK, Kang B, Kim OY, et al. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles. 2013;2 doi:10.3402/jev.v2i0.20384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi:10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 34. Eldh M, Ekstrom K, Valadi H, et al. Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS One. 2010;5(12):e15353 doi:10.1371/journal.pone.0015353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Manohar S, Harlow M, Nguyen H, Li J, Hankins GR, Park M. Chromatin modifying protein 1A (Chmp1A) of the endosomal sorting complex required for transport (ESCRT)-III family activates ataxia telangiectasia mutated (ATM) for PanC-1 cell growth inhibition. Cell Cycle. 2011;10(15):2529–2539. doi:10.4161/cc.10.15.15926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hupalowska A, Miaczynska M. The new faces of endocytosis in signaling. Traffic. 2012;13(1):9–18. doi:10.1111/j.1600-0854.2011.01249.x [DOI] [PubMed] [Google Scholar]
- 37. Silva J, Garcia V, Rodriguez M, et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012;51(4):409–418. [DOI] [PubMed] [Google Scholar]
- 38. Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi:10.1038/nature15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15(1):39 doi:10.1186/s12943-016-0524-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen Y, Xie Y, Xu L, et al. Protein content and functional characteristics of serum-purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int J Cancer. 2017;140(4):900–913. doi:10.1002/ijc.30496 [DOI] [PubMed] [Google Scholar]
- 41. Ragusa M, Statello L, Maugeri M, et al. Highly skewed distribution of miRNAs and proteins between colorectal cancer cells and their exosomes following Cetuximab treatment: biomolecular, genetic and translational implications. Oncoscience. 2014;1(2):132–157. doi:10.18632/oncoscience.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ahadi A, Khoury S, Losseva M, Tran N. A comparative analysis of lncRNAs in prostate cancer exosomes and their parental cell lines. Genomics Data. 2016;9:7–9. doi:10.1016/j.gdata.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262(19):9412–9420. [PubMed] [Google Scholar]
- 44. Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73(10):1907–1920. doi:10.1016/j.jprot.2010.06.006 [DOI] [PubMed] [Google Scholar]
- 45. Nolte-’t Hoen EN, Buschow SI, Anderton SM, Stoorvogel W, Wauben MH. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood. 2009;113(9):1977–1981. doi:10.1182/blood-2008-08-174094 [DOI] [PubMed] [Google Scholar]
- 46. Fevrier B, Vilette D, Archer F, et al. Cells release prions in association with exosomes. Proc Natl Acad Sci U S A. 2004;101(26):9683–9688. doi:10.1073/pnas.0308413101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Filipazzi P, Burdek M, Villa A, Rivoltini L, Huber V. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin Cancer Biol. 2012;22(4):342–349. doi:10.1016/j.semcancer.2012.02.005 [DOI] [PubMed] [Google Scholar]
- 48. Clayton A, Mason MD. Exosomes in tumour immunity. Curr Oncol. 2009;16(3):46–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Corrado C, Raimondo S, Saieva L, Flugy AM, De Leo G, Alessandro R. Exosome-mediated crosstalk between chronic myelogenous leukemia cells and human bone marrow stromal cells triggers an interleukin 8-dependent survival of leukemia cells. Cancer Lett. 2014;348(1-2):71–76. doi:10.1016/j.canlet.2014.03.009 [DOI] [PubMed] [Google Scholar]
- 50. Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl). 2013;91(4):431–437. doi:10.1007/s00109-013-1020-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hong BS, Cho JH, Kim H, et al. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics. 2009;10:556 doi:10.1186/1471-2164-10-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, Pegtel DM. Exosomes: fit to deliver small RNA. Commun Integr Biol. 2010;3(5):447–450. doi:10.4161/cib.3.5.12339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Deng ZB, Zhuang X, Ju S, et al. Exosome-like nanoparticles from intestinal mucosal cells carry prostaglandin E2 and suppress activation of liver NKT cells. J Immunol. 2013;190(7):3579–3589. doi:10.4049/jimmunol.1203170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30(7):1073–1081. doi:10.1093/carcin/bgp127 [DOI] [PubMed] [Google Scholar]
- 55. Bondar T, Medzhitov R. The origins of tumor-promoting inflammation. Cancer Cell. 2013;24(2):143–144. doi:10.1016/j.ccr.2013.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17(6):816–826. doi:10.1038/ncb3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi:10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015;219:278–294. doi:10.1016/j.jconrel.2015.06.029 [DOI] [PubMed] [Google Scholar]
- 59. Vella LJ. The emerging role of exosomes in epithelial–mesenchymal-transition in cancer. Front Oncol. 2014;4:361 doi:10.3389/fonc.2014.00361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Andreola G, Rivoltini L, Castelli C, et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med. 2002;195(10):1303–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huber V, Fais S, Iero M, et al. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: role in immune escape. Gastroenterology. 2005;128(7):1796–1804. [DOI] [PubMed] [Google Scholar]
- 62. Ciravolo V, Huber V, Ghedini GC, et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol. 2012;227(2):658–667. doi:10.1002/jcp.22773 [DOI] [PubMed] [Google Scholar]
- 63. Safaei R, Larson BJ, Cheng TC, et al. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther. 2005;4(10):1595–1604. doi:10.1158/1535-7163.mct-05-0102 [DOI] [PubMed] [Google Scholar]
- 64. Sousa D, Lima RT, Vasconcelos MH. Intercellular transfer of cancer drug resistance traits by extracellular vesicles. Trends Mol Med. 2015;21(10):595–608. doi:10.1016/j.molmed.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 65. Kucharzewska P, Christianson HC, Welch JE, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci U S A. 2013;110(18):7312–7317. doi:10.1073/pnas.1220998110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ekstrom EJ, Bergenfelz C, von Bulow V, et al. WNT5A induces release of exosomes containing pro-angiogenic and immunosuppressive factors from malignant melanoma cells. Mol Cancer. 2014;13:88 doi:10.1186/1476-4598-13-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tadokoro H, Umezu T, Ohyashiki K, Hirano T, Ohyashiki JH. Exosomes derived from hypoxic leukemia cells enhance tube formation in endothelial cells. J Biol Chem. 2013;288(48):34343–34351. doi:10.1074/jbc.M113.480822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ji H, Greening DW, Barnes TW, et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13(10-11):1672–1686. doi:10.1002/pmic.201200562 [DOI] [PubMed] [Google Scholar]
- 69. Lokody I. Genetics: exosomally derived miR-105 destroys tight junctions. Nat Rev Cancer. 2014;14(6):386–387. doi:10.1038/nrc3747 [DOI] [PubMed] [Google Scholar]
- 70. Rana S, Malinowska K, Zoller M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia. 2013;15(3):281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Luga V, Zhang L, Viloria-Petit AM, et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151(7):1542–1556. doi:10.1016/j.cell.2012.11.024 [DOI] [PubMed] [Google Scholar]
- 72. Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70(23):9621–9630. doi:10.1158/0008-5472.can-10-1722 [DOI] [PubMed] [Google Scholar]
- 73. Roma-Rodrigues C, Fernandes AR, Baptista PV. Exosome in tumour microenvironment: overview of the crosstalk between normal and cancer cells. BioMed Res Int. 2014;2014:179486 doi:10.1155/2014/179486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71(11):3792–3801. doi:10.1158/0008-5472.can-10-4455 [DOI] [PubMed] [Google Scholar]
- 75. Boelens MC, Wu TJ, Nabet BY, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159(3):499–513. doi:10.1016/j.cell.2014.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bobrie A, Krumeich S, Reyal F, et al. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72(19):4920–4930. doi:10.1158/0008-5472.can-12-0925 [DOI] [PubMed] [Google Scholar]
- 77. Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28(5):1551–1558. doi:10.3892/or.2012.1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Beckham CJ, Olsen J, Yin PN, et al. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J Urol. 2014;192(2):583–592. doi:10.1016/j.juro.2014.02.035 [DOI] [PubMed] [Google Scholar]
- 79. Street JM, Yuen PS, Star RA. Bioactive exosomes: possibilities for diagnosis and management of bladder cancer. J Urol. 2014;192(2):297–298. doi:10.1016/j.juro.2014.05.050 [DOI] [PubMed] [Google Scholar]
- 80. Akakura N, Hoogland C, Takada YK, et al. The COOH-terminal globular domain of fibrinogen gamma chain suppresses angiogenesis and tumor growth. Cancer Res. 2006;66(19):9691–9697. doi:10.1158/0008-5472.can-06-1686 [DOI] [PubMed] [Google Scholar]
- 81. Lampropoulos P, Zizi-Sermpetzoglou A, Rizos S, Kostakis A, Nikiteas N, Papavassiliou AG. TGF-beta signalling in colon carcinogenesis. Cancer Lett. 2012;314(1):1–7. doi:10.1016/j.canlet.2011.09.041 [DOI] [PubMed] [Google Scholar]
- 82. Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol Cancer Res. 2011;9(2):133–148. doi:10.1158/1541-7786.mcr-10-0394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ohshima K, Inoue K, Fujiwara A, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5(10):e13247 doi:10.1371/journal.pone.0013247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhou W, Fong MY, Min Y, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–515. doi:10.1016/j.ccr.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kruger S, Abd Elmageed ZY, Hawke DH, et al. Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer. 2014;14:44 doi:10.1186/1471-2407-14-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39(1):133–144. doi:10.1016/j.molcel.2010.06.010 [DOI] [PubMed] [Google Scholar]
- 87. Bigagli E, Luceri C, Guasti D, Cinci L. Exosomes secreted from human colon cancer cells influence the adhesion of neighboring metastatic cells: role of microRNA-210. Cancer Biol Ther. 2016:1–8. doi:10.1080/15384047.2016.1219815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tanaka S, Hosokawa M, Ueda K, Iwakawa S. Effects of decitabine on invasion and exosomal expression of miR-200c and miR-141 in oxaliplatin-resistant colorectal cancer cells. Biol Pharm Bull. 2015;38(9):1272–1279. doi:10.1248/bpb.b15-00129 [DOI] [PubMed] [Google Scholar]
- 89. Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi:10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yang X, Xie X, Xiao YF, et al. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360(2):119–124. doi:10.1016/j.canlet.2015.02.035 [DOI] [PubMed] [Google Scholar]
- 91. Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi:10.1016/j.ccr.2014.03.010 [DOI] [PubMed] [Google Scholar]
- 92. Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7(51):85551–85563. doi:10.18632/oncotarget.13465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sun R, Qin C, Jiang B, et al. Down-regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial–mesenchymal transition. Mol BioSyst. 2016;12(3):952–962. doi:10.1039/c5mb00685f [DOI] [PubMed] [Google Scholar]
- 94. Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi:10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Malik ZA, Kott KS, Poe AJ, et al. Cardiac myocyte exosomes: stability, HSP60, and proteomics. Am J Physiol Heart Circ Physiol. 2013;304(7):H954–H965. doi:10.1152/ajpheart.00835.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xu CG, Yang MF, Ren YQ, Wu CH, Wang LQ. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. 2016;20(20):4362–4368. [PubMed] [Google Scholar]
- 97. Kogure T, Yan IK, Lin WL, Patel T. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer. 2013;4(7-8):261–272. doi:10.1177/1947601913499020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Nawaz M, Fatima F, Nazarenko I, et al. Extracellular vesicles in ovarian cancer: applications to tumor biology, immunotherapy and biomarker discovery. Expert Rev Proteomics. 2016;13(4):395–409. doi:10.1586/14789450.2016.1165613 [DOI] [PubMed] [Google Scholar]
- 99. Coghlin C, Murray GI. Biomarkers of colorectal cancer: recent advances and future challenges. Proteomics Clin Appl. 2015;9(1-2):64–71. doi:10.1002/prca.201400082 [DOI] [PubMed] [Google Scholar]
- 100. Romero-Cordoba SL, Salido-Guadarrama I, Rodriguez-Dorantes M, Hidalgo-Miranda A. . miRNA biogenesis: biological impact in the development of cancer. Cancer Biol Ther. 2014;15(11):1444–1455. doi:10.4161/15384047.2014.955442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–675. doi:10.1111/j.1365-2141.2008.07077.x [DOI] [PubMed] [Google Scholar]
- 102. Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14(9):2588–2592. doi:10.1158/1078-0432.ccr-07-0666 [DOI] [PubMed] [Google Scholar]
- 103. King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421 doi:10.1186/1471-2407-12-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Aushev VN, Zborovskaya IB, Laktionov KK, et al. Comparisons of microRNA patterns in plasma before and after tumor removal reveal new biomarkers of lung squamous cell carcinoma. PLoS One. 2013;8(10):e78649 doi:10.1371/journal.pone.0078649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kobayashi M, Salomon C, Tapia J, Illanes SE, Mitchell MD, Rice GE. Ovarian cancer cell invasiveness is associated with discordant exosomal sequestration of Let-7 miRNA and miR-200. J Transl Med. 2014;12:4 doi:10.1186/1479-5876-12-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Westermann AM, Schmidt D, Holdenrieder S, et al. Serum microRNAs as biomarkers in patients undergoing prostate biopsy: results from a prospective multi-center study. Anticancer Res. 2014;34(2):665–669. [PubMed] [Google Scholar]
- 107. Yang Y, Gu X, Zhou M, Xiang J, Chen Z. Serum microRNAs: a new diagnostic method for colorectal cancer. Biomed Rep. 2013;1(4):495–498. doi:10.3892/br.2013.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kalimutho M, Del Vecchio Blanco G, Di Cecilia S, et al. Differential expression of miR-144* as a novel fecal-based diagnostic marker for colorectal cancer. J Gastroenterol. 2011;46(12):1391–1402. doi:10.1007/s00535-011-0456-0 [DOI] [PubMed] [Google Scholar]
- 109. Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6(3):267–283. doi:10.1586/epr.09.17 [DOI] [PubMed] [Google Scholar]
- 110. Ji H, Chen M, Greening DW, et al. Deep sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA-enrichment signatures. PloS One. 2014;9(10):e110314 doi:10.1371/journal.pone.0110314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tovar-Camargo OA, Toden S, Goel A. Exosomal microRNA biomarkers: emerging frontiers in colorectal and other human cancers. Expert Rev Mol Diagn. 2016;16(5):553–567. doi:10.1586/14737159.2016.1156535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Cha DJ, Franklin JL, Dou Y, et al. KRAS-dependent sorting of miRNA to exosomes. Elife. 2015;4:e07197 doi:10.7554/eLife.07197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285(23):17442–17452. doi:10.1074/jbc.M110.107821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Le MT, Hamar P, Guo C, et al. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. 2014;124(12):5109–5128. doi:10.1172/jci75695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Pandey GK, Mitra S, Subhash S, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26(5):722–737. doi:10.1016/j.ccell.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 116. Veltri WR. Non-coding RNAs as biomarkers for metastatic prostate cancer. The Lancet Oncol. 2014;15(13):1412–1413. doi:10.1016/s1470-2045(14)71124-6 [DOI] [PubMed] [Google Scholar]
- 117. Gezer U, Ozgur E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38(9):1076–1079. doi:10.1002/cbin.10301 [DOI] [PubMed] [Google Scholar]
- 118. Batagov AO, Kuznetsov VA, Kurochkin IV. Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genomics. 2011;12(suppl 3):S18 doi:10.1186/1471-2164-12-s3-s18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Huang X, Yuan T, Tschannen M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319 doi:10.1186/1471-2164-14-319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Liu T, Zhang X, Yang YM, Du LT, Wang CX. Increased expression of the long noncoding RNA CRNDE-h indicates a poor prognosis in colorectal cancer, and is positively correlated with IRX5 mRNA expression. Onco Targets Ther. 2016;9:1437–1448. doi:10.2147/ott.s98268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Fellig Y, Ariel I, Ohana P, et al. H19 expression in hepatic metastases from a range of human carcinomas. J Clin Pathol. 2005;58(10):1064–1068. doi:10.1136/jcp.2004.023648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Conigliaro A, Costa V, Lo Dico A, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155 doi:10.1186/s12943-015-0426-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Dimitrova N, Zamudio JR, Jong RM, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54(5):777–790. doi:10.1016/j.molcel.2014.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Berrondo C, Flax J, Kucherov V, et al. Expression of the long non-coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS One. 2016;11(1):e0147236 doi:10.1371/journal.pone.0147236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Yan TH, Lu SW, Huang YQ, et al. Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumour Biol. 2014;35(10):10249–10257. doi:10.1007/s13277-014-2344-8 [DOI] [PubMed] [Google Scholar]
- 126. Weiner-Gorzel K, Dempsey E, Milewska M, et al. Overexpression of the microRNA miR-433 promotes resistance to paclitaxel through the induction of cellular senescence in ovarian cancer cells. Cancer Med. 2015;4(5):745–758. doi:10.1002/cam4.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi:10.1016/j.fob.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Fan Y, Shen B, Tan M, et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014;281(7):1750–1758. doi:10.1111/febs.12737 [DOI] [PubMed] [Google Scholar]
- 129. Qu L, Ding J, Chen C, et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer cell. 2016;29(5):653–668. doi: 10.1016/j.ccell.2016.03.004 [DOI] [PubMed] [Google Scholar]
- 130. Tominaga N, Yoshioka Y, Ochiya T. A novel platform for cancer therapy using extracellular vesicles. Adv Drug Deliv Rev. 2015;95:50–55. doi:10.1016/j.addr.2015.10.002 [DOI] [PubMed] [Google Scholar]
- 131. Taraboletti G, D’Ascenzo S, Giusti I, et al. Bioavailability of VEGF in tumor-shed vesicles depends on vesicle burst induced by acidic pH. Neoplasia. 2006;8(2):96–103. doi:10.1593/neo.05583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Chen WX, Liu XM, Lv MM, et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One. 2014;9(4):e95240 doi:10.1371/journal.pone.0095240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Hoshino H, Miyoshi N, Nagai K, et al. Epithelial–mesenchymal transition with expression of SNAI1-induced chemoresistance in colorectal cancer. Biochem Biophys Res Commun. 2009;390(3):1061–1065. doi:10.1016/j.bbrc.2009.10.117 [DOI] [PubMed] [Google Scholar]
- 134. Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26(2):155–165. doi:10.1038/modpathol.2012.160 [DOI] [PubMed] [Google Scholar]
- 135. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. doi:10.1038/nbt.1807 [DOI] [PubMed] [Google Scholar]
- 136. Gong Z, Yang J, Li J, et al. Novel insights into the role of microRNA in lung cancer resistance to treatment and targeted therapy. Curr Cancer Drug Targets. 2014;14(3):241–258. [DOI] [PubMed] [Google Scholar]
- 137. Wang J, Zhou Y, Lu J, et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol. 2014;31(9):148 doi:10.1007/s12032-014-0148-8 [DOI] [PubMed] [Google Scholar]
- 138. Huang YK, Yu JC. Circulating microRNAs and long non-coding RNAs in gastric cancer diagnosis: an update and review. World J Gastroenterol. 2015;21(34):9863–9886. doi:10.3748/wjg.v21.i34.9863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Mohankumar S, Patel T. Extracellular vesicle long noncoding RNA as potential biomarkers of liver cancer. Brief Funct Genomics. 2016;15(3):249–256. doi:10.1093/bfgp/elv058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Alhamdani MS, Schroder C, Hoheisel JD. Analysis conditions for proteomic profiling of mammalian tissue and cell extracts with antibody microarrays. Proteomics. 2010;10(17):3203–3207. doi:10.1002/pmic.201000170 [DOI] [PubMed] [Google Scholar]
- 141. Lim JW, Mathias RA, Kapp EA, et al. Restoration of full-length APC protein in SW480 colon cancer cells induces exosome-mediated secretion of DKK-4. Electrophoresis. 2012;33(12):1873–1880. doi:10.1002/elps.201100687 [DOI] [PubMed] [Google Scholar]