Abstract
The low-density lipoprotein receptor (LDLR) and related receptors are important for the transport of diverse biomolecules across cell membranes and barriers. Their functions are especially relevant for cholesterol homeostasis and diseases, including neurodegenerative and kidney disorders. Members of the LDLR-related protein family share LDLR class A (LA) repeats providing binding properties for lipoproteins and other biomolecules. We previously demonstrated that short linker regions between these LA repeats contain conserved O-glycan sites. Moreover, we found that O-glycan modifications at these sites are selectively controlled by the GalNAc-transferase isoform, GalNAc-T11. However, the effects of GalNAc-T11–mediated O-glycosylation on LDLR and related receptor localization and function are unknown. Here, we characterized O-glycosylation of LDLR-related proteins and identified conserved O-glycosylation sites in the LA linker regions of VLDLR, LRP1, and LRP2 (Megalin) from both cell lines and rat organs. Using a panel of gene-edited isogenic cell line models, we demonstrate that GalNAc-T11–mediated LDLR and VLDLR O-glycosylation is not required for transport and cell-surface expression and stability of these receptors but markedly enhances LDL and VLDL binding and uptake. Direct ELISA-based binding assays with truncated LDLR constructs revealed that O-glycosylation increased affinity for LDL by ∼5-fold. The molecular basis for this observation is currently unknown, but these findings open up new avenues for exploring the roles of LDLR-related proteins in disease.
Keywords: glycosylation, glycosyltransferase, lipid metabolism, lipoprotein receptor, low-density lipoprotein (LDL), GALNT, O-glycosylation, lipophorin receptor
Introduction
The low-density lipoprotein receptor (LDLR)3 and related receptors are membrane-bound cell surface receptors with important endocytic functions for lipoproteins and a variety of diverse extracellular ligands (1). The founding member LDLR is important for maintaining cholesterol homeostasis, and deleterious mutations in LDLR lead to decreased LDL catabolism and elevated levels of plasma LDL-cholesterol (2). The LDLR superfamily includes the VLDL receptor (VLDLR), LDLR-related protein 1 (LRP1), LDLR-related protein 1B (LRP1B), LDLR-related protein 2 (LRP2 or megalin), and LDLR-related protein 8 (LRP8 or ApoER2), as well as more distantly related receptors such as the sortilin-related receptor (1). These members also have important roles in cardiovascular diseases, as well as neurodegenerative and proteinuric renal diseases (3, 4). The ectodomains of LDLR-related proteins share characteristic structural features including LDLR-type A repeats (LA) (complement-like cysteine-rich ligand binding repeats), epidermal growth factor-like repeats, and Tyr–Trp–Thr–Asp β-propeller domains (1).
The LA modules consist of ∼40 amino acids each with three disulfide bridges (Cys1–3, Cys2–5, and Cys4–6). They are found as clusters of seven repeats in LDLR, eight in VLDLR, and multiple clusters in LRP1 and LRP2, and they constitute the dominant ligand-binding region of these receptors (see Fig. 1) (5). The functional structure of the LA module requires a calcium ion coordinated by conserved acidic residues found between Cys4 and Cys6 in the LA sequence, and binding to different ligands appears to require different subsets of LA modules (6–9). The LA modules are bound by the ER-resident receptor-associated protein (RAP) early in the secretory pathway, and this interaction, believed to prevent premature intracellular binding to ligands, is lost at lower pH such as in later Golgi compartments (5, 9, 10). The LA modules are interspaced by a short linker sequence mostly formed by four residues ending in Thr with the sequence motif XXC6XXXTC1XX, although some linkers are longer. Recently we demonstrated that the evolutionary conserved Thr residues in these linkers of all LDLR-related proteins carry O-glycans (11). Moreover, we found that only one of the many polypeptide GalNAc-transferase (GalNAc-T) isoenzymes, GalNAc-T11, which initiates O-glycosylation of proteins, was responsible for introducing O-glycans at the Thr in the XXC6XXXTC1XX motif of linkers in LDLR and presumably other receptors with this motif.
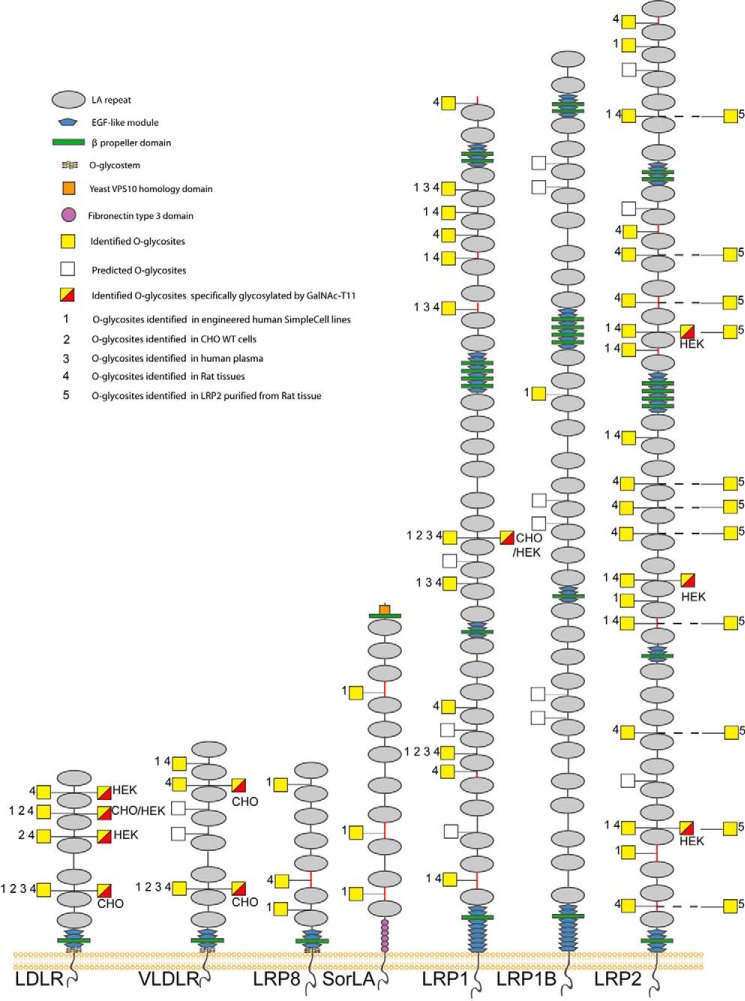
Figure 1.
Graphic depiction of O-glycosites in LA linkers of human LDLR-related proteins. The depiction summarizes all identified O-glycosites (filled yellow squares) and predicted O-glycosites (open squares) in LA linker regions of the LDLR family proteins. LA linkers with the relaxed sequence motif XXC6XnTC1XX (n = 3–5) are indicated as black lines, and other linkers are indicated with red lines. Glycosites previously identified in engineered human SimpleCell lines (labeled 1) (15–17), in CHO WT cells (labeled 2) (18), in human plasma (labeled 3) (19), and in rat liver, kidney, and brain tissues (labeled 4) and purified rat LRP2 (labeled 5) in the present study are shown. Detailed information about O-glycosites is presented in Data Set S1, O-glycopeptides identified in the present study are described in Data Set S2, and (glyco)peptides identified in purified LRP2 are described in Data Set S4. Glycosites specifically regulated by GalNAc-T11 (yellow and red squares) were identified by comparative analysis of isolated shed LDLR expressed in HEK293 SC cells with and without KO of GALNT11 as previously reported (11) or by differential dimethyl-labeling O-glycoproteomics (17) of CHO and HEK293 SC cell lines (indicated) with and without KO of GALNT11 in unpublished studies. Note that some LA modules are not conserved between rat and human (Table S1).
Both N- and O-glycosylation of LDLR have been reported previously (7, 12). O-Glycosylation in the stem region is important for cell-surface expression and stability of this receptor as demonstrated with the CHO ldlD cell line deficient in the UDP-Glc/GlcNAc C4-epimerase (12). Previous studies have also suggested that O-glycosylation of LDLR in the N-terminal domain may be important for LDL binding and uptake; however, the nature and positions of these O-glycans were not identified (13, 14). In the present study, we explored the functional role of the identified O-glycans in the short linker regions between LA modules (11). We took advantage of the finding that these O-glycans are specifically generated by GalNAc-T11, whereas the O-glycans important for stability of LDLR on the cell membrane are directed by multiple GalNAc-Ts (11). Using genetically engineered human HepG2 and HEK293 cells, as well as Chinese hamster ovary (CHO) cells, we demonstrate that the O-glycans in the LA ligand-binding region of LDLR, as well as VLDLR, are important for high-affinity lipoprotein binding and uptake and that the sialic acids carried by these O-glycans are essential for this. These findings have important implications for our understanding of the function and regulation of the LDLR-related proteins and their roles in diseases.
Results
O-Glycosites in the linker regions of class A repeats of LDLR-related proteins
We have previously identified a total of 17 O-glycosylation sites in evolutionary conserved linker regions in between LDLR LA repeats within the sequence motif XXC6XXXTC1XX of most of the LDLR-related proteins in a large number of engineered human SimpleCells (SCs) (Fig. 1, labeled 1) (15–17). SCs are genetically engineering cell lines with knockout of the COSMC gene, which is an essential chaperone for the core1 synthase, C1GALT1, that elongates the initial O-GalNAc glycans in most cell types. Thus, the O-glycan structures in SCs have the rather homogeneous GalNAc or in some cells NeuAcα2,6GalNAc structures facilitating sensitive identification by MS using lectin enrichment of glycopeptides in cell digests (15). The identified O-glycosites in LDLR-related proteins include two of the four XXC6XXXTC1XX motifs in LDLR, two of five in VLDLR, two of two in LRP8, five of eight in LRP1, one of six in LRP1B, and seven of thirteen in LRP2. Additional O-glycosites in linker regions with slightly different sequences were also identified (Fig. 1, linkers shown in red). Analysis of LDLR in HEK293 and CHO cells suggested that recombinant shed LDLR had partial occupancy of O-glycans in the LA regions, whereas O-glycosylation of the juxtamembrane region was more complete (11).
The SimpleCell glycoproteomics strategy involves genetic truncation of O-glycosylation for efficient enrichment and sensitive MS sequencing, and here we first sought to confirm O-glycosylation of the linker regions in LDLR-related proteins in WT cells and rat organs. We previously did confirm some of the LDLR, VLDLR, and LRP1 sites in WT CHO cells (18), as well as in rat liver and human plasma (19) (Fig. 1, labeled 2 and 3, and Data Set S1). We extended these studies to include comprehensive analysis of LDLR-related proteins in rat liver, brain, and kidney (Fig. 1, labeled 4, and Data Set S2), and we were able to confirm four of the four predicted glycosites in the XXC6XXXTC1XX linker regions of LDLR, three of five in VLDLR, seven of eight in LRP1, and eight of ten in LRP2 (rat LRP2 has three fewer predicted glycosites than human between LA1–2, 23–24, and 30–31).
To further confirm these O-glycans, we analyzed LRP2 purified from rat kidney. The neuraminidase-treated and trypsin-digested LRP2 yielded coverage of ∼57% of the ectodomain of LRP2 (Data Set S4). We identified 11 O-glycosites with six found in the 10 XXC6XXXTC1XX motifs (Fig. 1 and Data Set S4). In most cases the corresponding peptides without O-glycans were also identified, which may suggest incomplete stoichiometry in agreement with our previous studies of secreted LDLR, although the purified LRP2 may include intracellular immature glycoforms (11).
Trafficking and cell-surface expression of LDLR is not dependent on GalNAc-T11
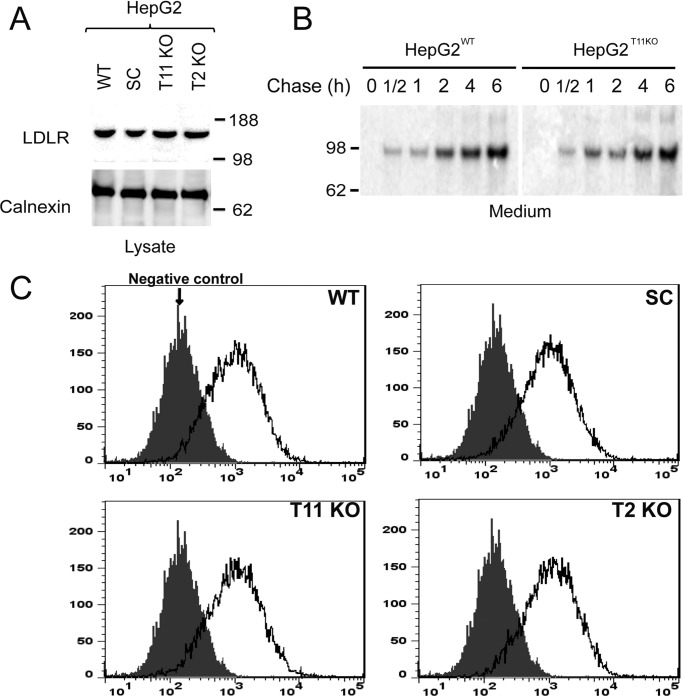
It is well established that LDLR requires O-glycosylation in the stem region for stability at the cell surface (12), so we first established that surface expression of LDLR in human liver HepG2 cells was unaffected by the loss of GalNAc-T11–mediated O-glycosylation of the linker regions. Knockout of GALNT11 or the COSMC chaperone required for the core 1 synthase C1GALT1 to truncate O-glycans to the initial GalNAc residue in HepG2 cells did not affect expression, trafficking, or shedding of the endogenous LDLR, as evaluated by SDS-PAGE Western blotting analysis of total cell lysates (Fig. 2A) and 35S-labeled pulse-chase experiments (Fig. 2B). Flow-cytometric analysis of nonpermeabilized HepG2 cells further confirmed that the cell-surface expression of endogenous LDLR was unaffected by the loss of GalNAc-T11–mediated O-glycosylation or capacity for elongation of O-glycans (Fig. 2C).
Figure 2.
Expression and shedding of LDLR is similar in HepG2 WT and mutant cells. A, SDS-PAGE Western blotting analysis of total cell extracts of HepG2 WT, SC, GALNT11 KO, and GALNT2 KO cell lines stained with anti-LDLR antibody (upper panel) and anti-calnexin (lower panel). The only immunoreactive band corresponds to the expected migration of the mature form of LDLR (molecular mass, 160 kDa). A representative experiment of at least three is shown. B, 35S-pulse-chase analysis of shed LDLR from HepG2 WT and GALNT11 KO cells using immunoprecipitation with a polyclonal anti-LDLR antibody from culture media. The signals were exposed in a PhosphorImager, and the only immunoreactive band corresponds to the expected migration of shed LDLR (molecular mass, 97 kDa). The cells were pulsed for 30 min and chased as indicated. A representative experiment of two is shown. C, flow cytometry analysis of LDLR surface expression on nonpermeabilized HepG2 WT and mutant cells as indicated. A representative experiment of at least three is shown.
Interestingly, the general truncation of O-glycans to the unsialylated GalNAc residue by COSMC knockout in HepG2 cells (20) did not appear to affect cell surface stability or shedding of the LDLR (Fig. 2C). This is, however, at least partly in agreement with previous studies of LDLR shedding in the CHO ldlD cell line, where complete loss of O-glycosylation results in rapid shedding and degradation of LDLR, whereas allowing only GalNAc glycosylation by supplementing growth medium with GalNAc alone at least partly retained cell-surface expression (20).
LDL ligand binding and uptake by LDLR is affected by GalNAc-T11–mediated O-glycosylation
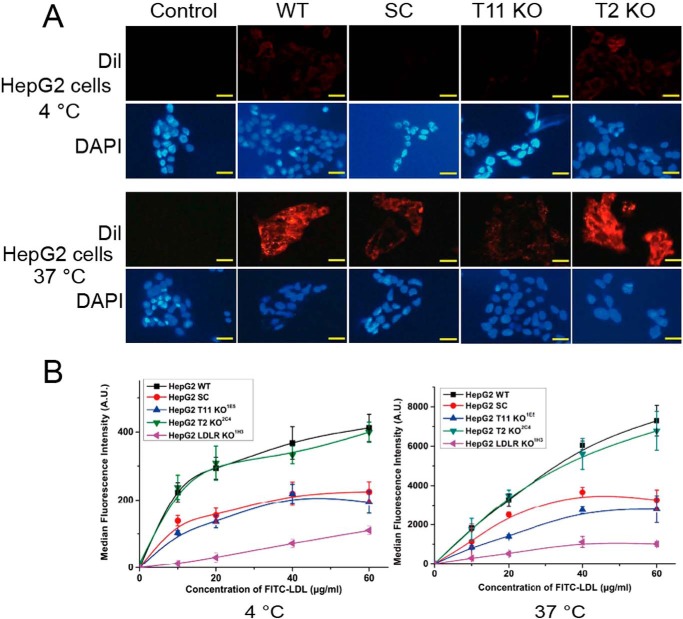
Fresh fluorescently labeled DiI-LDL was used in binding and uptake studies with isogenic HepG2 cells with isogenic cells with knockout of COSMC (SC), GALNT11, or GALNT2 (Fig. 3A). Initial assays were performed by exposing cells to 20 μg/ml DiI-LDL for 4 h at 4 °C for binding and 37 °C for uptake, followed by washing, air drying, fixation, and analysis by direct fluorescent microscopy. Both binding and uptake of DiI-LDL were substantially reduced in HepG2 cells with knockout of COSMC and GALNT11, whereas knockout of the isoenzyme control GALNT2 had no effect (Fig. 3A). Interestingly, knockout of COSMC or GALNT11 produced the same reduction, suggesting that the elongated O-glycans with sialic acids are required for the function.
Figure 3.
LDL binding and uptake is affected by GalNAc-T11 O-glycosylation of LDLR in HepG2 cells. A, representative fluorescence microscopy images of HepG2 cells after incubation with 20 μg/ml DiI-LDL at 4 °C (upper panel) and 37 °C (lower panel) for 4 h. DiI-LDL– (red) and 4′,6′-diamino-2-phenylindole–stained (DAPI) nuclei (blue) are shown. The scale bar indicates 10 μm. HepG2 cells were grown for 24 h on gelatinized coverslips in LPDS medium prior to treatment. At least three different experiments were performed. B, quantification of LDL binding (left panel) and internalization (right panel) of HepG2 WT and mutant cell lines cultured in 24 wells with different concentrations of FITC-LDL at 4 or 37 °C for 4 h. The values represent the mean of triplicate determinations (n = 3); error bars represent ±S.D.
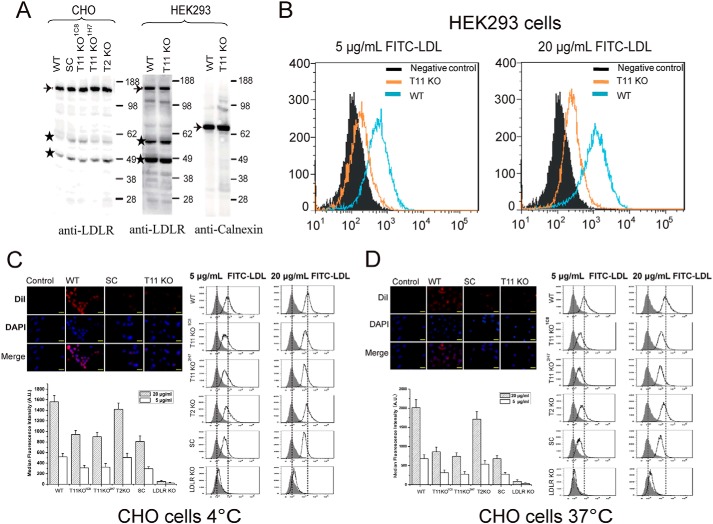
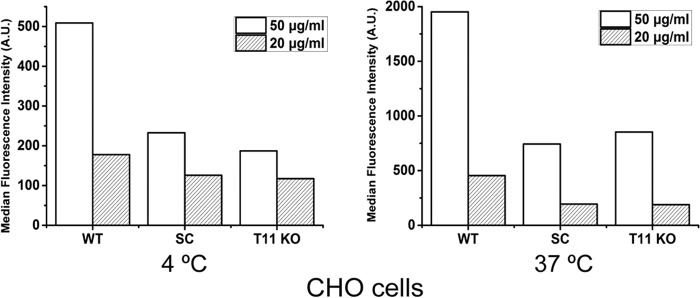
We next performed a dose study (10–60 μg/ml) using FITC-LDL and including HepG2 cells with knockout of LDLR (Fig. 3B and Fig. S2). A strong dose-dependent reduction of LDL binding and uptake was found in HepG2 cells without GANT11 and COSMC, which was intermediary to the reduction found upon knockout of the LDLR (Fig. 3B). Interestingly, both binding and uptake of FITC-LDL appeared to saturate at 40 μg/ml in HepG2 cells with deficiency in O-glycosylation, as well as in LDLR, whereas saturation was not evident at 60 μg/ml with WT cells (Fig. 3B). We confirmed that these observations were not cell type–specific by testing isogenic HEK293 and CHO cells with similar knockouts (Fig. 4). Interestingly, knockout of Ldlr in CHO cells produced a particularly pronounced reduction of binding and uptake of LDL compared with knockout of Galnt11 or Cosmc (Fig. 4, C and D). Because truncation of O-glycans by knockout of COSMC produced similar effects as knockout of GALNT11, we tested the effect of sialic acid capping alone by pretreatment of cells with neuraminidase to remove sialic acids on WT CHO and HepG2 cells and found similar reduction in binding and uptake of FITC-LDL (Fig. 5). In summary, these data indicate that sialic acid residues on O-glycans in the LA linker regions directed by GalNAc-T11 enhance the capacity of LDLR for binding and uptake of LDL.
Figure 4.
Analysis of the expression and function of LDLR on CHO and HEK293 WT and mutant cell clones. A, SDS-PAGE Western blotting analysis of LDLR expressed in CHO (left panel) and HEK293 (right panel) cells using anti-LDLR antibody (Ab30532, Abcam). Analysis of calnexin expression in HEK293 cells was evaluated with anti-calnexin (ADI-SPA-860) specific for the human protein. Equal amounts of cell lysate protein based on BCA were loaded, and the blots show a representative example from at least three repeat experiments. Bands labeled with stars were considered unspecific. B, FACS analysis of uptake of two concentrations of FITC-LDL by HEK293 WT and GALNT11 KO cells at 37 °C for 4 h. The experiment was performed twice with similar results, and representative results are shown. C, FACS analysis of binding of FITC-LDL by CHO WT, SC, and Galnt11 KO cells 4 °C for 4 h. D, FACS analysis of uptake of FITC-LDL by CHO WT, SC, and Galnt11 KO cells at 37 °C for 4 h. The experiments in C and D were performed in triplicate (n = 3), and the data shown represent averages with error bars.
Figure 5.
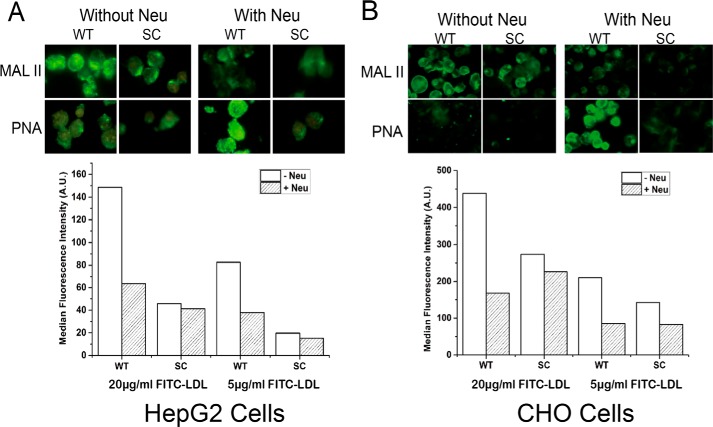
Analysis of effect of neuraminidase pretreatment on LDL uptake. HepG2 WT cells (A) and CHO WT cells (B) were pretreated with and without neuraminidase and subjected to lectin immunocytology (upper panels) and FITC-LDL binding assays (lower panels). PNA and Maackia amurensis lectin II (MAL II) lectins were used to demonstrate the loss of sialic acids by the neuraminidase treatment. FACS analysis of FITC-LDL binding was performed with HepG2 and CHO WT and SC cells, and pretreatment with neuraminidase affected binding to WT cells but not to SC cells, which is in agreement with the established O-glycosylation capacity of these cells where WT produces sialylated core 1 O-glycans, and SC produces the nonsialylated GalNAc O-glycans. The experiment was performed twice with similar results.
Recombinant LDLR ectodomains require O-glycans for efficient inhibition of LDL binding and uptake by WT CHO cells
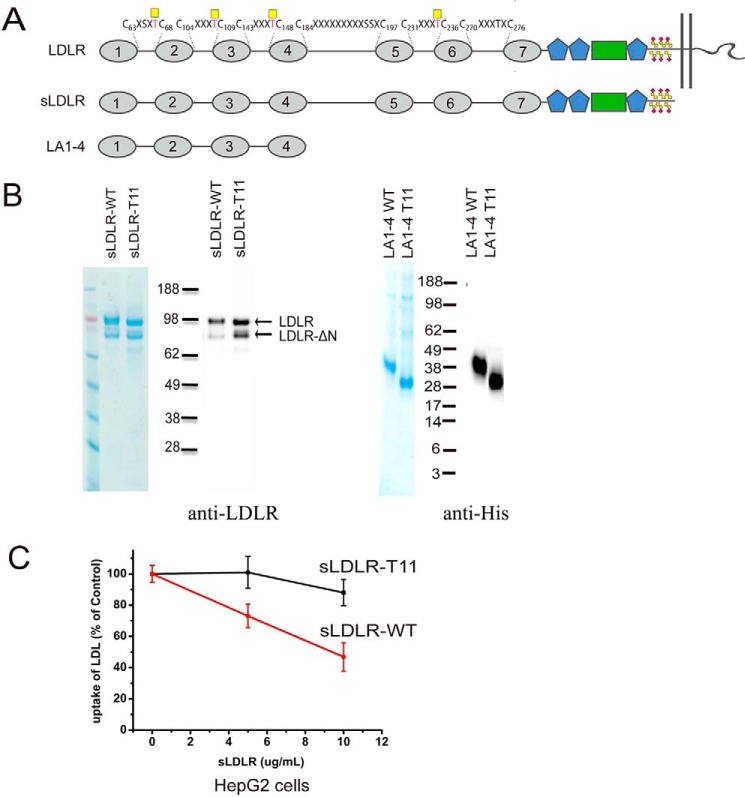
We expressed truncated secreted constructs of human LDLR in CHO cells with and without knockout of Galnt11 and isolated them to apparent homogeneity (Fig. 6, A and B). The full secreted ectodomains of LDLR, sLDLR, expressed in WT CHO cells exhibited substantially better inhibition of FITC-LDL uptake by HepG2 cells compared with the same construct expressed in CHO cells without Galnt11 (Fig. 6C and Fig. S3). Moreover, direct binding assays with LDL and sLDLR, as well as a further truncated construct LA1–4, showed that the constructs expressed in CHO without Galnt11 exhibited an ∼5-fold lower affinity compared with the same constructs expressed in WT CHO cells (Table 1), emphasizing that loss of O-glycosylation by GalNAc-T11 results in an LDLR that binds LDL with much lower affinity.
Figure 6.
Analysis of truncated LDLR constructs and inhibition of LDL uptake. A, graphic depiction of truncated LDLR constructs sLDLR and LA1–4 used. B, SDS-PAGE Coomassie and Western blotting analyses of purified sLDLR and LA1–4 using anti-LDLR (Abcam, Ab30532) or anti-His. sLDLR and LA1–4 were expressed in CHO cells and purified using Jacalin lectin chromatography or nickel–nitrilotriacetic acid, respectively, and further purified by ion exchange with a yield of ∼30 μg from 100 ml of growth medium. The presence of two molecular weight forms of sLDLR is likely due to N-terminal truncation as reported previously (57). Note minor differences in migration of sLDLR and LA1–4 derived from cells with and without Galnt11, presumably because of a loss of O-glycans at the three glycosites in the linker region (Thr67, Thr108, and Thr147). Loading was based on BCA quantification of purified proteins, but the yield of sLDLR isolated from cells with knockout of Galnt11 appeared higher based on the Coomassie staining and WB. C, quantification of FITC-LDL uptake by HepG2 WT cells in the presence of sLDLR with and without O-glycans introduced by GalNAc-T11. 20 μg/ml FITC-LDL was preincubated with different concentrations of sLDLR as indicated (based on the BCA quantification) and subsequently incubated with HepG2 WT cells for 1 h at 37 °C. The values represent the means of triplicate determinations (n = 3); error bars represent ± S.D.
Table 1.
ELISA-based binding analysis of the interaction between LDLR fragments and LDL
| EC50 (means ± S.D.) | N | |
|---|---|---|
| nm | ||
| sLDLR WT | 4.52 ± 1.25a | 4 |
| sLDLR T11KO | 30.61 ± 5.06 | 4 |
| LA 1–4 WT | 16.76 ± 2.4 | 3 |
| LA 1–4 T11KO | 73.98 ± 15.44 | 3 |
VLDLR also require O-glycans for VLDL binding and uptake
VLDLR contains eight LA modules with five of the linker regions predicted to carry O-glycans, although we have only identified three of these yet with our shotgun glycoproteomics strategy (Fig. 1). However, we have found in preliminary studies that the two identified sites are dependent on GALNT11 in CHO and HEK293 cells. We therefore tested binding and uptake of FITC-VLDL by isogenic CHO cells with and without knockout of Cosmc (SC) or Galnt11 (1C8 clone) and found that loss of either gene resulted in substantial loss of binding and uptake of VLDL, similar to what was found for LDL (Fig. 7). We were unable to confirm cell-surface expression of VLDLR in mutant cells because of a lack of antibodies, and we predict that expression similar to LDLR is not affected by loss of GalNAc-T11–mediated glycosylation (Fig. 2).
Figure 7.
Analysis of FITC-VLDL binding and uptake in CHO WT and mutant cell lines. CHO WT, SC, and Galnt11 KO (1C8 clone) were incubated with two different concentrations of FITC-VLDL (20 or 50 μg/ml) at 4 °C for binding (left panel) or 37 °C for internalization (right panel) for 4 h. The FACS analysis and quantification was performed as described in the legend to Fig. 4. The experiment was performed twice with similar results, and a representative data set is presented.
Conservation of GalNAc-T11–directed O-glycans in LA linkers in Drosophila
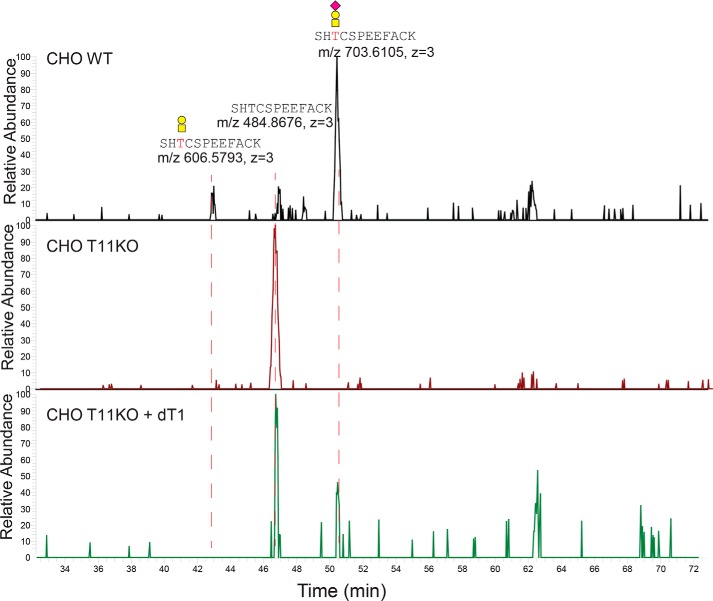
We previously demonstrated that the Drosophila ortholog of GalNAc-T11, dGalNAc-T1 (dT1), or l(2)35Aa, is essential for viability (21, 22). Drosophila members of the LDLR-related protein family consist of the lipophorin receptors (Lpr), which share a similar organization of domains including seven or eight LA modules (23). Lprs are essential for the efficient uptake of neutral lipids in Drosophila and include Lpr1 and Lpr2, of which Lpr2-E is the major receptor involved in the uptake by nurse cells and oocytes (24). We expressed a C-terminal HA-tagged full coding construct of Lpr2-E in WT CHO cells with and without KO of Galnt11, as well as the combination of KO of Galnt11 and co-expression of dGalNAc-T1 (Fig. S4). The naturally shed ectodomain of Lpr2-E without the HA tag was enriched by Jacalin chromatography of glycoproteins with sialylated ST (NeuAcα2–3Galβ1–3GalNAcα1-O-Ser/Thr) O-glycans followed by trypsin digestion and direct LC-MS/MS analysis without prior neuraminidase treatment. We obtained an overall sequence coverage for Lpr2-E of 45% with the sample from CHO WT and CHO Galnt11 KO and 35% from CHO Galnt11 KO with the co-expression of dGalNAc-T1. The linker region (LA2–3) with the consensus motif C6RSHTC1 was identified as glycopeptide only in CHO cells with expression of human GalNAc-T11 (WT) or co-expression of Drosophila dGalNAc-T1, whereas in CHO Galnt11 KO, we only identified the corresponding unglycosylated peptide (Fig. 8 and Data Set S3). These results suggest that the mammalian and Drosophila orthologs share the unique capabilities in glycosylating LDLR-related protein linker regions.
Figure 8.
Extracted ion chromatographs (XICs) of the identified glycopeptides and peptides at the LA2–3 linker of Lpr2-E. LC–MS/MS data of the tryptic digest of the lectin enriched O-glycoproteins from Lpr2-E expressing CHO WT (top panel), CHO Galnt11 KO (middle panel), and CHO Galnt11KO with stable expression of dGalNAc-T1 (bottom panel) were extracted with the theoretical m/z of the identified linker region glycopeptides and peptides. XICs of different ionic forms of the identified peptide sequence with or without one of the predicted glycans (HexNAc, HexNAcNeuAc, HexHexNAc, and HexHexNAcNeuAc) were used for search. The identified glycoforms are annotated, and XIC peaks are labeled with the accurate m/z of the (glyco)peptides and charge states.
Discussion
Protein O-glycosylation of the GalNAc-type serves diverse and highly specific roles in fine-tuning protein functions (25), and here we provide conclusive evidence for a novel role in modulating the binding properties of the large family of LDLR-related proteins. Other types of protein O-glycosylation including O-Fuc, O-Glc, and O-GlcNAc modulate the binding properties and signaling of the Notch receptors, and these types of glycosylation serve important roles in development and diseases (26). GalNAc-type O-glycosylation is unique and orchestrated by a large family of isoenzymes that enables differential and perhaps dynamic regulation of single glycosites. The specific modification of the LA linkers in LDLR-related proteins by the GalNAc-T11 isoform presented here is perhaps the most exclusive function of a GalNAc-T isoenzyme identified to date. We show that GalNAc-T11 directed O-glycosylation of the LA linkers in the ligand-binding domains of LDLR and VLDLR markedly affect the binding affinity and uptake of their respective lipoprotein targets, and it is likely that this is a general function for all LDLR-related proteins.
Our results are in agreement with and extend early studies suggesting the existence of O-glycans in addition to the well characterized glycans in the juxtamembrane region of LDLR (27), and they may provide an explanation for the otherwise puzzling finding that a Monensin-resistant CHO line, MonR-31, with apparent loss of these glycans showed reduced binding and uptake of LDL (13, 14, 28). The MonR-31 cell line is unfortunately no longer available, but our results suggest that the lectin selection used to establish the cell line resulted in deletion of the GALNT11 gene. Studies over decades with lectin-resistant CHO mutants demonstrate that the O-glycans in the juxtamembrane region of LDLR are essential for stability of the receptor at the cell surface (12), but importantly O-glycosylation in the juxtamembrane “mucin-like” domain with high density of O-glycans is redundantly achieved by multiple GalNAc-T isoforms and presumably not regulated (11). In contrast, the single O-glycosites in the LA modules are selectively controlled by the GalNAc-T11 isoform and therefore amenable to differential and perhaps dynamic site-specific regulation in cells and organs.
It is increasingly becoming apparent that site-specific regulation of selected O-glycosites by the large family of GalNAc-T isoenzymes are involved in fine-tuning protein function with important roles in health and diseases (25). The most illustrative example to date is the unique role of the GalNAc-T3 isoform in phosphate homeostasis, where O-glycosylation co-regulates proprotein processing of FGF23 in a complex interplay with proprotein convertases and the Golgi protein kinase Fam20C, within the short sequence element 176RHTR↓S, where O-glycosylation of Thr178 directed by GalNAc-T3 inhibits processing and phosphorylation of Ser181 by Fam20C inhibits O-glycosylation (29, 30). Congenital deficiency in GALNT3 cause hyperphosphatemia and ectopic ossifications, whereas deficiency in Fam20C causes osteosclerotic dysplasia and hypophosphatemia (31, 32), and GALNT3 is a GWAS candidate for low mineral bone density (33). Many of the other 20 GALNT genes have been identified as candidate genes for dispositions to common diseases (25, 34), and we were recently able to provide extensive validation for the role of GALNT2 in dyslipidemia in man, primates, and rodents and identified several site-specifically regulated O-glycoprotein targets (19). The substrate specificities and functions of GalNAc-Ts have mainly been studied by in vitro enzyme assays using short peptides as acceptors in the past (35), and this approach has provided important insights into specific functions of many individual isoenzymes including GalNAc-T11. However, although GalNAc-T11 does function with a number of short peptide substrates (21, 22, 35), it only functions in vitro with the LA linker regions in the context of the entire folded LA-modules (11). This may suggest that GalNAc-T11 is unique among the GalNAc-T isoforms in recognizing folded domains as substrates. Other glycosyltransferases initiating O-Fuc, O-Glc, and O-GlcNAc recognize small folded domains such as epidermal growth factor-like and thrombospondin type 1 repeats (26, 36), and these domains share a design with three conserved disulfide bridges with the LA modules (37, 38). However, in all cases the acceptor sites are located in the folded domain, whereas the substrate site for GalNAc-T11 is located in the short linker between the folded domains. We only discovered the unique function of GalNAc-T11 through O-glycoproteomics with glycoengineered cell lines (11). Availability of isogenic cell lines with and without individual GALNT genes are important discovery platforms for the isoform-specific functions as shown here and in combination with our quantitative differential glycoproteomics approach enable unbiased discovery of nonredundant biological functions of the large family of GalNAc-T isoenzymes (17). We also used genetic dissection to demonstrate the functional importance of the structure of the O-glycans. Thus, knockout of COSMC in CHO and HepG2 cells, which results in truncation of the normal core1 O-glycans to the simple GalNAc residue, produced the same marked change in the function of LDLR, demonstrating the importance of the O-glycan structure and the terminal sialic acid (Figs. 3–5).
Cellular trafficking of LDLR-related proteins is complex and essential for their functions. These receptors are transported through the ER-Golgi secretory pathway bound to the receptor-associated protein RAP (LRPAP1 or α2-macroglobulin receptor-associated protein) that acts as a chaperone and presumably protect receptors from early binding to ligands until dissociation occurs at the lower pH in later Golgi compartments (39). Studies with Escherichia coli produced LA modules without glycans established that RAP binds to multiple LA modules through electrostatic interaction between three calcium-coordinating acidic residues in each LA and lysine residues (9, 40, 41). In LRP1 the most juxtamembrane cluster of LA modules LA3–6 that contains linker O-glycans (Fig. 1 and Table S1) appears to be the most important (9, 41). In a recent study of the complete LRP1 protein expressed in HEK293 cells, presumably with GalNAc-T11 directed O-glycosylation, strong nanomolar interaction with RAP was demonstrated (42), suggesting that O-glycosylation of the linkers in LA modules may not play a role in the interaction with RAP, which is also in line with our finding that surface expression and shedding of LDLR was unaffected by LA module O-glycosylation (Fig. 2, A and C). The co-crystal structure of RAP with the LA modules 3 and 4 in a complex shows that the acceptor Thr in the LA linker region is facing toward RAP at a distance of ∼10 Å from the closest amino acid of RAP (9), suggesting that O-glycosylation may occur only after release from RAP.
The ligand binding of LDLR-related proteins resembles the interaction with RAP and depends on the conserved acidic residues in the LA modules (9). Many studies of the ligand binding properties of LDLR and VLDLR have been performed with E. coli produced truncated receptor constructs (43–45), which would exclude analysis of the O-glycans found in the LA linkers. Similarly, studies with LDLR expressed in insect cells are likely to exclude these O-glycans or at least completely lack sialic acids (9). However, the O-glycans are not essential for binding, but they induce a marked enhancement in extracellular binding and uptake of LDL and VLDL by LDLR and VLDLR in cell assays. This was clearly shown for LDLR in CHO cells, where deficiency in O-glycans produced intermediary LDL binding and uptake compared with complete LDLR deficiency and further supported by direct binding assays showing detectable but ∼5-fold lower binding affinity of sLDLR to LDL in the absence of O-glycans (Fig. 4, C and D, and Table 1). How O-glycans exert these effects requires structural studies with appropriately glycosylated receptor constructs, but we envision that the sialylated O-glycans may participate directly in the ligand interaction by, for example, adding to the negative charges of the conserved aspartic acids in the LA module or perhaps more likely induce local conformational effects that organize consecutive LA modules in more favorable binding mode.
The implications of our findings for the many diseases and conditions associated with the diverse functions of LDLR-related proteins require further studies. Genetic deficiency in GALNT11 has not been identified so far in man, and it is predicted to be extremely rare if occurring (34), but GALNT11 has been identified as a GWAS candidate gene for chronic kidney decline (46). The GWAS signal for chronic kidney decline resides in intron one of the GALNT11 gene, suggesting that the genetic predisposition is a result of altered gene regulation, similar to what has been established for the role of GALNT2 in dyslipidemia (19, 47). Our study did not directly address the functional role of O-glycans on other LDLR-related proteins including LRP2; however, GalNAc-T11 is highly expressed in proximal tubules of the kidney, and LRP2 is the major endocytic receptor responsible for reabsorbtion of proteins from the glomerular filtrate (21, 48). Given the common structure of the ligand binding LA modules and conservation of LA linker O-glycan sites directed by GalNAc-T11, we propose that the O-glycans on all the LDLR-related proteins serve similar roles in modulating ligand binding. Moreover, it seems plausible that the identification of GALNT11 as a GWAS candidate for chronic kidney disease may relate to altered glycosylation and function of LRP2, and further studies into this are clearly warranted. Given the important roles of LDLR and VLDLR in lipoprotein metabolism and hypercholesterolemic conditions (49) and the dramatic effect GalNAc-T11 directed O-glycosylation has on these receptors, it may be surprising that GALNT11 has not been associated with cholesterol disorders. We are in progress with such studies, but we did provide evidence that the unique substrate specificity of GalNAc-T11 is at least partially conserved by the Drosophila ortholog dGalNAc-T1 also designated l(2)35Aa, which is essential for viability (21, 22). In summary, our study presents conclusive evidence for an important functional role of O-glycans in modulating the ligand binding properties of the large family of LDLR-related proteins, which provides a novel understanding of the function and potentially regulation of these receptors with wide implications for common diseases.
Experimental procedures
O-Glycoproteomic analysis of rat organs
Samples of ∼100 mg of tissue from rat liver, brain, or kidney were homogenized in 300 μl of lysis buffer containing 1% RapiGest (Waters) in 50 mm ammonium bicarbonate using an IKA Ultra Turbax blender at maximum speed for 20 s followed by 30 s of sonication using a Sonic Dismembrator (Fischer Scientific). Homogenates were boiled for 5 min, diluted with 50 mm ammonium bicarbonate to 0.1% RapiGest, reduced with 20 mm DTT in 50 mm ammonium bicarbonate at 60 °C for 45 min, alkylated with 40 mm iodoacetamide in 50 mm ammonium bicarbonate at room temperature for 35 min, followed by addition of 20 mm DTT to terminate alkylation, and digestion with trypsin (50 μg/sample) (Roche) at 37 °C, ON. Digests were treated with concentrated 6 μl TFA at 37 °C for 30 min, cleared by centrifugation, and purified on C18 Sep-Pak (Waters). The digests were desialylated with 1 unit of neuraminidase (N3001, Sigma) in 50 mm sodium citrate (pH 5.0) at 37 °C overnight followed by SepPak purification and lyophilization. The samples were dissolved in lectin-binding buffer (175 mm Tris, pH 7.4) and subjected to Jacalin-agarose (AL1153, Vector Laboratories) lectin weak affinity chromatography (LWAC) performed as described previously with PNA (19), with minor modifications. The glycopeptides were eluted with 3 × 1-column volume 0.7 m galactose in lectin buffer (175 mm Tris, pH 7.4). Lectin weak affinity chromatography fractions were screened by LC-MS for glycopeptide content, and fractions enriched in glycopeptides were pooled and submitted to LC-MS analysis using EASY-nLC 1000 UHPLC (Thermo Scientific) interfaced via NanoSpray Flex ion source to an LTQ-Orbitrap Fusion Pro spectrometer (Thermo Scientific), as previously described (17). Data processing was performed using Proteome Discoverer 1.4 software (Thermo Scientific).
Gene targeting of GALNT11and LDLR in HepG2 cells
A CompoZTM custom zinc finger nuclease targeting construct for GALNT11 with the following binding and cutting (parentheses) sites, 5′-GACCGCTTGGGCTAC(CACAGA)GATGTGCCAGACACAAGG-3′ was obtained from (Merck Millipore) and GFP/Crimson-tagged as previously described (50). Guide RNAs targeting exon2 of LDLR was designed using a gUIDEbookTM, Desktop Genetics-Horizon Discoveries gRNA design algorithm and cloned into dual gRNA and Cas9–2A-GFP expression vectorpX458 (Addgene plasmid no. 48138). Zinc finger nuclease or gRNA plasmids were transfected into HepG2 cells by electroporation using Amaxa Nucleofector (Lonza), and clones were selected using a workflow with bulk sorting by FACS 48 h after transfection, followed by 1–2 weeks in culture, and cloning by limiting dilution in 96-well plates as recently reported (51). Clones with frameshift mutations were identified by Indel Detection by Amplicon Analysis using the following primers (52): GALNT11-F, 5′-AGCTGACCGGCAGCAAAATTGTCGCTGACTAACTTCACTCTTTTG-3′; GALNT11-R, 5′-TCTCAGACCATGTGGCTTCA-3′; LDLR-F, 5′-AGCTGACCGGCAGCAAAATTGGATTCTGGCGTTGAGAGACC-3′; and LDLR-R, 5′-GTGCCTGTAATCCCAGCACT-3′. All selected clones were Sanger sequenced in the target site for confirmation (Fig. S1). The HepG2 COSMC knockout and HepG2 GALNT2 knockout clones were described previously (17). All HepG2 clones were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mmol/liter l-glutamine, and 1% nonessential amino acids. All reagents for cell culture were obtained from Gibco.
Expression and purification of secreted LDLR
A construct (LA1–4) containing residues 1–196 of the coding region of human LDLR was generated by PCR using a plasmid containing the entire coding region and cloned into pcDNA3.1/myc-His (C-terminal tags) (Invitrogen). The following primers were used: LA-1-F, 5′-CCCAAGCTTGCCACCATGGGGCCCTGGGGCTGGAA-3′; and LA-4-R, 5′-CCGCTCGAGGGGGCTACTGTCCCCTTGGA-3′. The plasmid encoding LA1–4 and a bicistronic enhanced GFP (EGFP) plasmid encoding human full-length LDLR with a C-terminal V5 tag (53) were expressed in CHO WT and CHO Galnt11 KO cells (1C8 clone). The sLDLR was purified as follows: after 48 h of culture the medium (300 ml) was collected and dialyzed twice against 50 mm Tris (pH 7.4) at 4 °C, diluted into 2× binding buffer (350 mm Tris, pH 7.4), and applied to a 1-ml packed Jacalin-agarose column (Vector Laboratories). Following washing by 40–50 column volumes (CV) of binding buffer O-glycoproteins were eluted with 0.8 m galactose in binding buffer and detected by Western blotting using anti-LDLR (ab30532, Abcam). The secreted His-tagged LA1–4 construct was purified on nickel affinity chromatography (Invitrogen) by mixing the culture media 3:1 (v/v) with 4× binding buffer (200 mm Tris, pH 8.0, 1.2 m NaCl) and applied to 0.3 ml of packed nickel–nitrilotriacetic acid–agarose (Invitrogen) pre-equilibrated in binding buffer (50 mm Tris, pH 8.0, 300 mm NaCl). The column was washed with binding buffer and eluted with binding buffer containing 250 mm imidazole. Fractions containing LA1–4 were analyzed by SDS-PAGE and pooled. Further purification of sLDLR and LA1–4 were achieved by anion-exchange MonoQ (5/50 GL, GE Healthcare) chromatography on ÄKTA FPLC interfaced by UNICORN 4.12 control software. Enriched pooled fractions were centrifuged at 14,000 rpm at 4 °C for 30 min and loaded on the MonoQ pre-equilibrated in 25 mm Bis-Tris (pH 6.5) with 10 mm NaCl. The column was washed with 15 CV and eluted using a linear gradient of 10 mm-1 m NaCl. Fractions were analyzed by SDS-PAGE Coomassie and Western blotting with anti-LDLR (ab30532, Abcam) or anti-His antibody (A00186, GenScript). Purified proteins were quantified by BCA protein assay kit (Thermo Scientific).
SDS-PAGE Western blotting analysis
NuPAGE Novex Bis-Tris 4–12% gels were used with transfer to nitrocellulose membranes for 90 min (0.45 μm, Bio-Rad) 200 V. The blots were blocked in 5% skimmed milk in TBS-T and washed three times in TBS-T prior to overnight incubation at 4 °C with rabbit polyclonal anti-LDLR antibody (ab30532, Abcam) or rabbit polyclonal anti-calnexin antibody (1:2,000) (Enzo Life Sciences). The blots were developed with ECL (Pierce catalog no. 32106, Thermo Scientific) after incubation with horseradish peroxidase-conjugated secondary antibody (Dako) for 1 h. Calnexin was used to normalize.
Metabolic labeling and immunoprecipitation
HepG2 WT and GALNT11 KO cells were cultured in methionine and cysteine-free RPMI 1640 (Gibco) media containing 2 mmol/liter l-glutamine for 30 min, before replacement of the same medium containing 0.1 mCi/ml Promix ([35S]methionine/[35S]cysteine) (PerkinElmer Life Sciences). For pulse-chase, cells were labeled for 30 min and chased for 0–6 h in complete RPMI 1640 medium containing 5 mm methionine and cysteine. For co-immunoprecipitation the LDL receptor was immunoprecipitated overnight at 4 °C using a polyclonal anti-LDLR antibody (ab30532, Abcam) and protein G–Sepharose (Amersham Biosciences). The immunoprecipitate was analyzed by NuPAGE after incubation in LDS sample buffer (Invitrogen) for 10 min at 95 °C. The bands were quantified using a PhosphorImager (STORM 840, Molecular Dynamics) and ImageQuant software (Molecular Dynamics).
LDLR functional assays
The cells were cultured in 24-well plates and used at ∼60% confluence. CHO cells were cultured with EX CELL® CD CHO fusion medium (Sigma) with 2% glutamine. HepG2 and HEK293 cells were cultured in Dulbecco's modified Eagle's medium containing 10% FBS, and 1 day before FBS was substituted with 10% lipoprotein-deficient serum (LPDS) (Alfa Aesar). The cells were incubated for 4 h at 4 °C for binding and 37 °C for uptake with different concentrations of FITC-LDL (54) or DiI-LDL (Alfa Aesar), followed by washing twice in PBS with 1% BSA, fixation in 4% formaldehyde for 10 min, and washing twice with PBS with 1% BSA. FITC-LDL was analyzed by FACS in a LSR-II flow cytometer (Beckman Coulter) with 10,000 events acquired for data analysis, and the results were expressed as the mean fluorescence of activated gated cells, selected in a forward versus side scatter window. DiI-LDL was analyzed with cells mounted in ProLong Gold antifade reagent (4′,6′-diamino-2-phenylindole) (Invitrogen) using a Zeiss Axioskop 2 plus with an AxioCam MR3. Bit depth and pixel dimensions were 36 bits and 1,388 × 1,040 pixels, respectively. Analysis of FITC-VLDL was performed as described for LDL (54). All measurements have been performed in triplicate. Competition assays with secreted LDLR constructs were performed with 20 μg/ml FITC-LDL or DiI-LDL preincubated with the indicated concentrations of secreted LDLR constructs at 37 °C before addition to cells for 4 h.
Neuraminidase pretreatment
The cells for binding and uptake assays were incubated with Clostridium perfringens neuraminidase (Sigma, N3001) diluted in PBS to a final concentration of 0.1 unit/ml at 37 °C for 4 h, followed by washing twice with in medium containing LPDS. Controls for neuraminidase included staining with biotinylated PNA (B-1073) and biotinylated Maackia amurensis lectin II (B-1265) lectins followed by Alexa Fluor® 488–conjugated streptavidin (Life Technologies, S32354).
LDLR expression
HepG2 cells cultured in medium with LPDS for 24 h, resuspended in PBS with 1% BSA, blocked 30 min with PBS with 10% FBS, washed twice with PBS with 1% BSA, and incubated with rabbit polyclonal anti-LDLR (1:100; 3 mg/liter; Ab30532) for 1 h at room temperature followed by FITC-conjugated swine anti-rabbit IgG (1:100; Dako) and analysis in a LSR-II flow cytometer.
Direct LDL-binding assays
A modified ELISA binding assay with purified sLDLR and LA1–4 coated in 96-well plates in 10 mm Tris-HCl (pH 7.4) and 50 mm NaCl, 2 mm CaCl2 (buffer A) was used (55). The plates were blocked with 5% BSA and incubated with freshly purified human LDL in buffer A for 2 h at room temperature, before washing four times with buffer A and once with buffer A containing 0.1% Tween 20, followed by incubation with goat polyclonal anti-apolipoprotein B (Abcam, UK) for 1 h, peroxidase-conjugated mouse anti-goat IgG (Thermo Scientific) for 1 h, and development with 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) substrate (Sigma–Aldrich) and photometric quantification at 405 nm. All data were corrected for unspecific binding and relativized to maximum, and EC50 values were extracted from binding curves after fitting the data to a five-parameter logistic equation (SigmaPlot 13.0; Systat Software Inc.). In control experiments, mouse monoclonal anti-LDL receptor (PROGEN) and horse anti-mouse horseradish peroxidase-linked secondary antibody (Cell Signaling Technology) were used to detect the amount of LDLR fragments bound to plate and calculate the surface coverage percentage. Experiments were carried out at two surface subsaturating coverage percentages and [L] ≫ [R] conditions, obtaining the same binding values.
Recombinant expression of Drosophila pgant35A and Lpr2-E
The full coding region of Drosophila melanogaster pgant35A was derived from pUAST-pgant35A (56) and subcloned into pcDNA3 (Invitrogen) to generate pcDNA3-pgant35A. The ORF of D. melanogaster lpr2E with a C-terminal 3× HA tag in pUAST-lpr2E was kindly provided by Joaquim Culi (Sevilla, Spain; Ref. 23) and used to generate pcDNA3-lpr2E. The pcDNA3-pgant35A construct was expressed in CHO Galnt11 KO cells (1C8 clone) by electroporation and G418 selection, and stable clones were identified by PCR with primers PCDNAFOR and dT1-KPS (5′-CGATTCGCCAGTGGTAATGCTGGC-3′). The pcDNA3-lpr2E construct was stably expressed in CHO WT, CHO Galnt11 KO, and CHO Galnt11 KO with expression of pgant35A, using electroporation and Zeocin selection. Stable clones were screened by immunocytochemistry and Western blotting analysis using a monoclonal anti-HA antibody (Santa Cruz).
Analysis of recombinant shed Drosophila Lpr2-E
Culture medium from CHO cells stably expressing full coding C-terminal HA-tagged Drosophila Lpr2-E construct was diluted twice in 175 mm Tris (pH 7.5) and loaded twice onto a 0.8-ml Jacalin lectin column, followed by 20× CV washing in the same buffer and 2× CV washing with 50 mm ammonium bicarbonate added. Bound O-glycoproteins were eluted by heating the lectin slurry in 0.1% RapiGest. The eluate was heated for 10 min at 80 °C, reduced with 20 mm DTT at 60 °C for 45 min, alkylated with 40 mm iodoacetamide in 50 mm ammonium bicarbonate at room temperature for 35 min followed by addition of 20 mm DTT to terminate alkylation, and finally digested with trypsin (1 μg/sample) (Roche) at 37 °C overnight. Digests were treated with concentrated TFA at 37 °C for 30 min, cleared by centrifugation, purified on C18 Sep-Pak (Waters), and lyophilized. Each sample was dissolved in 0.1% TFA and submitted to LC-MS and high-energy collision dissociation/electron transfer dissociation–MS/MS.
Author contributions
S. W., Y. M., K. T. S., and H.C. designed and performed experiments, analyzed data and wrote the manuscript. Y. N. contributed to CRISPR/Cas9 design. Z. Y., W. T., C. K. G., E. L. -N., N. B. P., and C. C. performed experiments. A. B. -V., C. M., K. B. U. provided materials and performed direct LDL binding assays. N. G. S., R. N., E. I. C. provided materials and contributed to data interpretation and manuscript preparations. R. H.-G., L. H., E. P. B., and S. Y. V. contributed to the design of experiments.
Supplementary Material
Acknowledgments
We thank Olav M. Andersen for valuable comments and discussions on the manuscript. We thank Joaquim Culi for sharing invaluable reagents.
This work was supported by the Læge Sofus Carl Emil Friis og hustru Olga Doris Friis' Legat, the Kirsten og Freddy Johansen Fonden, the Lundbeck Foundation, the A.P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til Almene Formaal, the Mizutani Foundation, the Novo Nordisk Foundation, the Danish Research Council Sapere Aude Research Talent Grant (to K. T. S.), and the Danish National Research Foundation (DNRF107). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1, Figs. S1–S4, and Data Sets S1–S4.
- LDLR
- low-density lipoprotein receptor
- SC
- SimpleCells
- GalNAc-T
- UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase
- VLDLR
- very low-density lipoprotein receptor
- LRP
- LDLR-related protein
- LA
- LDLR class A repeats
- Jacalin
- jackfruit agglutinin
- CV
- column volume(s)
- GWAS
- genome-wide association studies
- Lpr
- lipophorin receptor
- RAP
- receptor-associated protein
- CHO
- Chinese hamster ovary
- sLDLR
- shed recombinant LDLR
- HA
- hemagglutinin
- PNA
- peanut agglutinin
- LPDS
- lipoprotein-deficient serum
- XIC
- extracted ion chromatograph
- DiI
- 1,19-dioctadecyl-3,3,3,3-tetramethyl-indocarbocyanine perchlorate.
References
- 1. Dieckmann M., Dietrich M. F., and Herz J. (2010) Lipoprotein receptors: an evolutionarily ancient multifunctional receptor family. Biol. Chem. 391, 1341–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jeon H., and Blacklow S. C. (2005) Structure and physiologic function of the low-density lipoprotein receptor. Annu. Rev. Biochem. 74, 535–562 10.1146/annurev.biochem.74.082803.133354 [DOI] [PubMed] [Google Scholar]
- 3. Zlokovic B. V., Deane R., Sagare A. P., Bell R. D., and Winkler E. A. (2010) Low-density lipoprotein receptor-related protein-1: a serial clearance homeostatic mechanism controlling Alzheimer's amyloid β-peptide elimination from the brain. J. Neurochem. 115, 1077–1089 10.1111/j.1471-4159.2010.07002.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nielsen R., Christensen E. I., and Birn H. (2016) Megalin and cubilin in proximal tubule protein reabsorption: from experimental models to human disease. Kidney Int. 89, 58–67 10.1016/j.kint.2015.11.007 [DOI] [PubMed] [Google Scholar]
- 5. Blacklow S. C. (2007) Versatility in ligand recognition by LDL receptor family proteins: advances and frontiers. Curr. Opin Struct. Biol. 17, 419–426 10.1016/j.sbi.2007.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Russell D. W., Brown M. S., and Goldstein J. L. (1989) Different combinations of cysteine-rich repeats mediate binding of low density lipoprotein receptor to two different proteins. J. Biol. Chem. 264, 21682–21688 [PubMed] [Google Scholar]
- 7. Daly N. L., Scanlon M. J., Djordjevic J. T., Kroon P. A., and Smith R. (1995) Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. U.S.A. 92, 6334–6338 10.1073/pnas.92.14.6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rudenko G., Henry L., Henderson K., Ichtchenko K., Brown M. S., Goldstein J. L., and Deisenhofer J. (2002) Structure of the LDL receptor extracellular domain at endosomal pH. Science 298, 2353–2358 10.1126/science.1078124 [DOI] [PubMed] [Google Scholar]
- 9. Fisher C., Beglova N., and Blacklow S. C. (2006) Structure of an LDLR–RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol. Cell 22, 277–283 10.1016/j.molcel.2006.02.021 [DOI] [PubMed] [Google Scholar]
- 10. Beglova N., and Blacklow S. C. (2005) The LDL receptor: how acid pulls the trigger. Trends Biochem. Sci. 30, 309–317 10.1016/j.tibs.2005.03.007 [DOI] [PubMed] [Google Scholar]
- 11. Pedersen N. B., Wang S., Narimatsu Y., Yang Z., Halim A., Schjoldager K. T., Madsen T. D., Seidah N. G., Bennett E. P., Levery S. B., and Clausen H. (2014) Low density lipoprotein receptor class A repeats are O-glycosylated in linker regions. J. Biol. Chem. 289, 17312–17324 10.1074/jbc.M113.545053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kingsley D. M., Kozarsky K. F., Hobbie L., and Krieger M. (1986) Reversible defects in O-linked glycosylation and LDL receptor expression in a UDP-Gal/UDP-GalNAc 4-epimerase–deficient mutant. Cell 44, 749–759 10.1016/0092-8674(86)90841-X [DOI] [PubMed] [Google Scholar]
- 13. Yoshimura A., Yoshida T., Seguchi T., Waki M., Ono M., and Kuwano M. (1987) Low binding capacity and altered O-linked glycosylation of low density lipoprotein receptor in a monensin-resistant mutant of Chinese hamster ovary cells. J. Biol. Chem. 262, 13299–13308 [PubMed] [Google Scholar]
- 14. Seguchi T., Merkle R. K., Ono M., Kuwano M., and Cummings R. D. (1991) The dysfunctional LDL receptor in a monensin-resistant mutant of Chinese hamster ovary cells lacks selected O-linked oligosaccharides. Arch. Biochem. Biophys. 284, 245–256 10.1016/0003-9861(91)90292-Q [DOI] [PubMed] [Google Scholar]
- 15. Steentoft C., Vakhrushev S. Y., Vester-Christensen M. B., Schjoldager K. T., Kong Y., Bennett E. P., Mandel U., Wandall H., Levery S. B., and Clausen H. (2011) Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 10.1038/nmeth.1731 [DOI] [PubMed] [Google Scholar]
- 16. Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., et al. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 10.1038/emboj.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schjoldager K. T., Joshi H. J., Kong Y., Goth C. K., King S. L., Wandall H. H., Bennett E. P., Vakhrushev S. Y., and Clausen H. (2015) Deconstruction of O-glycosylation: GalNAc-T isoforms direct distinct subsets of the O-glycoproteome. EMBO Rep. 16, 1713–1722 10.15252/embr.201540796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang Z., Halim A., Narimatsu Y., Jitendra Joshi H., Steentoft C., Schjoldager K. T., Alder Schulz M., Sealover N. R., Kayser K. J., Paul Bennett E., Levery S. B., Vakhrushev S. Y., and Clausen H. (2014) The GalNAc-type O-glycoproteome of CHO cells characterized by the SimpleCell strategy. Mol. Cell. Proteomics 13, 3224–3235 10.1074/mcp.M114.041541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khetarpal S. A., Schjoldager K. T., Christoffersen C., Raghavan A., Edmondson A. C., Reutter H. M., Ahmed B., Ouazzani R., Peloso G. M., Vitali C., Zhao W., Somasundara A. V., Millar J. S., Park Y., Fernando G., et al. (2016) Loss of function of GALNT2 lowers high-density lipoproteins in humans, nonhuman primates, and rodents. Cell Metab. 24, 234–245 10.1016/j.cmet.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schjoldager K. T., Vakhrushev S. Y., Kong Y., Steentoft C., Nudelman A. S., Pedersen N. B., Wandall H. H., Mandel U., Bennett E. P., Levery S. B., and Clausen H. (2012) Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells. Proc. Natl. Acad. Sci. U.S.A. 109, 9893–9898 10.1073/pnas.1203563109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwientek T., Bennett E. P., Flores C., Thacker J., Hollmann M., Reis C. A., Behrens J., Mandel U., Keck B., Schäfer M. A., Haselmann K., Zubarev R., Roepstorff P., Burchell J. M., Taylor-Papadimitriou J., et al. (2002) Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals: one subfamily composed of l(2)35Aa is essential in Drosophila. J. Biol. Chem. 277, 22623–22638 10.1074/jbc.M202684200 [DOI] [PubMed] [Google Scholar]
- 22. Ten Hagen K. G., and Tran D. T. (2002) A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is essential for viability in Drosophila melanogaster. J. Biol. Chem. 277, 22616–22622 10.1074/jbc.M201807200 [DOI] [PubMed] [Google Scholar]
- 23. Parra-Peralbo E., and Culi J. (2011) Drosophila lipophorin receptors mediate the uptake of neutral lipids in oocytes and imaginal disc cells by an endocytosis-independent mechanism. PLoS Genet. 7, e1001297 10.1371/journal.pgen.1001297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rodenburg K. W., Smolenaars M. M., Van Hoof D., and Van der Horst D. J. (2006) Sequence analysis of the non-recurring C-terminal domains shows that insect lipoprotein receptors constitute a distinct group of LDL receptor family members. Insect Biochem. Mol. Biol. 36, 250–263 10.1016/j.ibmb.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 25. Schjoldager K. T., and Clausen H. (2012) Site-specific protein O-glycosylation modulates proprotein processing: deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim. Biophys. Acta 1820, 2079–2094 10.1016/j.bbagen.2012.09.014 [DOI] [PubMed] [Google Scholar]
- 26. Takeuchi H., and Haltiwanger R. S. (2014) Significance of glycosylation in Notch signaling. Biochem. Biophys. Res. Commun. 453, 235–242 10.1016/j.bbrc.2014.05.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davis C. G., Elhammer A., Russell D. W., Schneider W. J., Kornfeld S., Brown M. S., and Goldstein J. L. (1986) Deletion of clustered O-linked carbohydrates does not impair function of low density lipoprotein receptor in transfected fibroblasts. J. Biol. Chem. 261, 2828–2838 [PubMed] [Google Scholar]
- 28. Shite S., Seguchi T., Yoshida T., Kohno K., Ono M., and Kuwano M. (1988) A new class mutation of low density lipoprotein receptor with altered carbohydrate chains. J. Biol. Chem. 263, 19286–19289 [PubMed] [Google Scholar]
- 29. Kato K., Jeanneau C., Tarp M. A., Benet-Pagès A., Lorenz-Depiereux B., Bennett E. P., Mandel U., Strom T. M., and Clausen H. (2006) Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis: secretion of fibroblast growth factor 23 requires O-glycosylation. J. Biol. Chem. 281, 18370–18377 10.1074/jbc.M602469200 [DOI] [PubMed] [Google Scholar]
- 30. Tagliabracci V. S., Engel J. L., Wiley S. E., Xiao J., Gonzalez D. J., Nidumanda Appaiah H., Koller A., Nizet V., White K. E., and Dixon J. E. (2014) Dynamic regulation of FGF23 by Fam20C phosphorylation, GalNAc-T3 glycosylation, and furin proteolysis. Proc. Natl. Acad. Sci. U.S.A. 111, 5520–5525 10.1073/pnas.1402218111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Topaz O., Shurman D. L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., Khamaysi Z., Behar D., Petronius D., Friedman V., Zelikovic I., Raimer S., Metzker A., Richard G., and Sprecher E. (2004) Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat. Genet. 36, 579–581 10.1038/ng1358 [DOI] [PubMed] [Google Scholar]
- 32. Simpson M. A., Hsu R., Keir L. S., Hao J., Sivapalan G., Ernst L. M., Zackai E. H., Al-Gazali L. I., Hulskamp G., Kingston H. M., Prescott T. E., Ion A., Patton M. A., Murday V., George A., et al. (2007) Mutations in FAM20C are associated with lethal osteosclerotic bone dysplasia (Raine syndrome), highlighting a crucial molecule in bone development. Am. J. Hum. Genet. 81, 906–912 10.1086/522240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duncan E. L., Danoy P., Kemp J. P., Leo P. J., McCloskey E., Nicholson G. C., Eastell R., Prince R. L., Eisman J. A., Jones G., Sambrook P. N., Reid I. R., Dennison E. M., Wark J., Richards J. B., et al. (2011) Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 7, e1001372 10.1371/journal.pgen.1001372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hansen L., Lind-Thomsen A., Joshi H. J., Pedersen N. B., Have C. T., Kong Y., Wang S., Sparso T., Grarup N., Vester-Christensen M. B., Schjoldager K., Freeze H. H., Hansen T., Pedersen O., Henrissat B., et al. (2015) A glycogene mutation map for discovery of diseases of glycosylation. Glycobiology 25, 211–224 10.1093/glycob/cwu104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kong Y., Joshi H. J., Schjoldager K. T., Madsen T. D., Gerken T. A., Vester-Christensen M. B., Wandall H. H., Bennett E. P., Levery S. B., Vakhrushev S. Y., and Clausen H. (2015) Probing polypeptide GalNAc-transferase isoform substrate specificities by in vitro analysis. Glycobiology 25, 55–65 10.1093/glycob/cwu089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moreira Bde S., Sampaio R. F., Furtado S. R., Dias R. C., and Kirkwood R. N. (2016) The relationship between diabetes mellitus, geriatric syndromes, physical function, and gait: a review of the literature. Curr, Diabetes Rev. 12, 240–251 10.2174/1573399811666150615142746 [DOI] [PubMed] [Google Scholar]
- 37. Lira-Navarrete E., Valero-González J., Villanueva R., Martínez-Júlvez M., Tejero T., Merino P., Panjikar S., and Hurtado-Guerrero R. (2011) Structural insights into the mechanism of protein O-fucosylation. PLoS One 6, e25365 10.1371/journal.pone.0025365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Valero-González J., Leonhard-Melief C., Lira-Navarrete E., Jiménez-Osés G., Hernández-Ruiz C., Pallarés M. C., Yruela I., Vasudevan D., Lostao A., Corzana F., Takeuchi H., Haltiwanger R. S., and Hurtado-Guerrero R. (2016) A proactive role of water molecules in acceptor recognition by protein O-fucosyltransferase 2. Nat. Chem. Biol. 12, 240–246 10.1038/nchembio.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bu G. (2001) The roles of receptor-associated protein (RAP) as a molecular chaperone for members of the LDL receptor family. Int. Rev. Cytol. 209, 79–116 10.1016/S0074-7696(01)09011-8 [DOI] [PubMed] [Google Scholar]
- 40. Andersen O. M., Christensen P. A., Christensen L. L., Jacobsen C., Moestrup S. K., Etzerodt M., and Thogersen H. C. (2000) Specific binding of α-macroglobulin to complement-type repeat CR4 of the low-density lipoprotein receptor-related protein. Biochemistry 39, 10627–10633 10.1021/bi000498h [DOI] [PubMed] [Google Scholar]
- 41. Jensen G. A., Andersen O. M., Bonvin A. M., Bjerrum-Bohr I., Etzerodt M., Thøgersen H. C., O'Shea C., Poulsen F. M., and Kragelund B. B. (2006) Binding site structure of one LRP-RAP complex: implications for a common ligand-receptor binding motif. J. Mol. Biol. 362, 700–716 10.1016/j.jmb.2006.07.013 [DOI] [PubMed] [Google Scholar]
- 42. De Nardis C., Lössl P., van den Biggelaar M., Madoori P. K., Leloup N., Mertens K., Heck A. J., and Gros P. (2017) Recombinant expression of the full-length ectodomain of LDL receptor-related protein 1 (LRP1) unravels pH-dependent conformational changes and the stoichiometry of binding with receptor-associated protein (RAP). J. Biol. Chem. 292, 912–924 10.1074/jbc.M116.758862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Simmons T., Newhouse Y. M., Arnold K. S., Innerarity T. L., and Weisgraber K. H. (1997) Human low density lipoprotein receptor fragment: successful refolding of a functionally active ligand-binding domain produced in Escherichia coli. J. Biol. Chem. 272, 25531–25536 10.1074/jbc.272.41.25531 [DOI] [PubMed] [Google Scholar]
- 44. Yasui N., Nogi T., and Takagi J. (2010) Structural basis for specific recognition of reelin by its receptors. Structure 18, 320–331 10.1016/j.str.2010.01.010 [DOI] [PubMed] [Google Scholar]
- 45. Martínez-Oliván J., Arias-Moreno X., Velazquez-Campoy A., Millet O., and Sancho J. (2014) LDL receptor/lipoprotein recognition: endosomal weakening of ApoB and ApoE binding to the convex face of the LR5 repeat. FEBS J. 281, 1534–1546 10.1111/febs.12721 [DOI] [PubMed] [Google Scholar]
- 46. Gorski M., Tin A., Garnaas M., McMahon G. M., Chu A. Y., Tayo B. O., Pattaro C., Teumer A., Chasman D. I., Chalmers J., Hamet P., Tremblay J., Woodward M., Aspelund T., Eiriksdottir G., et al. (2015) Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int. 87, 1017–1029 10.1038/ki.2014.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roman T. S., Marvelle A. F., Fogarty M. P., Vadlamudi S., Gonzalez A. J., Buchkovich M. L., Huyghe J. R., Fuchsberger C., Jackson A. U., Wu Y., Civelek M., Lusis A. J., Gaulton K. J., Sethupathy P., Kangas A. J., et al. (2015) Multiple hepatic regulatory variants at the GALNT2 GWAS locus associated with high-density lipoprotein cholesterol. Am. J. Hum. Genet. 97, 801–815 10.1016/j.ajhg.2015.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Christensen E. I., Verroust P. J., and Nielsen R. (2009) Receptor-mediated endocytosis in renal proximal tubule. Pflugers Arch. 458, 1039–1048 10.1007/s00424-009-0685-8 [DOI] [PubMed] [Google Scholar]
- 49. Brown M. S., and Goldstein J. L. (1976) Receptor-mediated control of cholesterol metabolism. Science 191, 150–154 10.1126/science.174194 [DOI] [PubMed] [Google Scholar]
- 50. Duda K., Lonowski L. A., Kofoed-Nielsen M., Ibarra A., Delay C. M., Kang Q., Yang Z., Pruett-Miller S. M., Bennett E. P., Wandall H. H., Davis G. D., Hansen S. H., and Frödin M. (2014) High-efficiency genome editing via 2A-coupled co-expression of fluorescent proteins and zinc finger nucleases or CRISPR/Cas9 nickase pairs. Nucleic Acids Res. 42, e84 10.1093/nar/gku251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lonowski L. A., Narimatsu Y., Riaz A., Delay C. E., Yang Z., Niola F., Duda K., Ober E. A., Clausen H., Wandall H. H., Hansen S. H., Bennett E. P., and Frödin M. (2017) Genome editing using FACS enrichment of nuclease-expressing cells and indel detection by amplicon analysis. Nat. Protoc. 12, 581–603 10.1038/nprot.2016.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang Z., Steentoft C., Hauge C., Hansen L., Thomsen A. L., Niola F., Vester-Christensen M. B., Frödin M., Clausen H., Wandall H. H., and Bennett E. P. (2015) Fast and sensitive detection of indels induced by precise gene targeting. Nucleic Acids Res. 43, e59 10.1093/nar/gkv126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Canuel M., Sun X., Asselin M. C., Paramithiotis E., Prat A., and Seidah N. G. (2013) Proprotein convertase subtilisin/kexin type 9 (PCSK9) can mediate degradation of the low density lipoprotein receptor-related protein 1 (LRP-1). PLoS One 8, e64145 10.1371/journal.pone.0064145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Etxebarria A., Benito-Vicente A., Alves A. C., Ostolaza H., Bourbon M., and Martin C. (2014) Advantages and versatility of fluorescence-based methodology to characterize the functionality of LDLR and class mutation assignment. PLoS One 9, e112677 10.1371/journal.pone.0112677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang S., Henry L., Ho Y. K., Pownall H. J., and Rudenko G. (2010) Mechanism of LDL binding and release probed by structure-based mutagenesis of the LDL receptor. J. Lipid Res. 51, 297–308 10.1194/jlr.M000422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bennett E. P., Chen Y. W., Schwientek T., Mandel U., Schjoldager K., Cohen S. M., and Clausen H. (2010) Rescue of Drosophila melanogaster l(2)35Aa lethality is only mediated by polypeptide GalNAc-transferase pgant35A, but not by the evolutionary conserved human ortholog GalNAc-transferase-T11. Glycoconj. J. 27, 435–444 10.1007/s10719-010-9290-5 [DOI] [PubMed] [Google Scholar]
- 57. Tveten K., Holla Ø. L., Cameron J., Strøm T. B., Berge K. E., Laerdahl J. K., and Leren T. P. (2012) Interaction between the ligand-binding domain of the LDL receptor and the C-terminal domain of PCSK9 is required for PCSK9 to remain bound to the LDL receptor during endosomal acidification. Hum. Mol. Genet. 21, 1402–1409 10.1093/hmg/ddr578 [DOI] [PubMed] [Google Scholar]
- 58. Brown M. S., and Goldstein J. L. (1974) Familial hypercholesterolemia: defective binding of lipoproteins to cultured fibroblasts associated with impaired regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity. Proc. Natl. Acad. Sci. U.S.A. 71, 788–792 10.1073/pnas.71.3.788 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.