Abstract
RNA polymerase II (Pol II) is the central enzyme that transcribes eukaryotic protein-coding genes to produce mRNA. The mushroom toxin α-amanitin binds Pol II and inhibits transcription at the step of RNA chain elongation. Pol II from yeast binds α-amanitin with micromolar affinity, whereas metazoan Pol II enzymes exhibit nanomolar affinities. Here, we present the high-resolution cryo-EM structure of α-amanitin bound to and inhibited by its natural target, the mammalian Pol II elongation complex. The structure revealed that the toxin is located in a pocket previously identified in yeast Pol II but forms additional contacts with metazoan-specific residues, which explains why its affinity to mammalian Pol II is ∼3000 times higher than for yeast Pol II. Our work provides the structural basis for the inhibition of mammalian Pol II by the natural toxin α-amanitin and highlights that cryo-EM is well suited to studying interactions of a small molecule with its macromolecular target.
Keywords: transcription, RNA polymerase II, RNA polymerase, cryo-electron microscopy, structural biology
Introduction
The toxin α-amanitin occurs in poisonous amanita mushrooms and inhibits Pol II,2 the enzyme that transcribes protein-coding genes in eukaryotes to produce mRNA (1). The toxin α-amanitin is a modified peptide that comprises eight amino acids forming two ring systems (1). It contains the noncanonical amino acid residues dihydroxy isoleucine (Ile(OH)2), hydroxyl proline (Hyp), and hydroxyl tryptophan (Trp(OH)), which contribute to its toxicity and its affinity for the Pol II enzyme (2).
Previous structural work used Pol II from the yeast Saccharomyces cerevisiae to reveal that α-amanitin binds in a pocket of the enzyme formed by the polymerase bridge helix, an element of the active center, and the RPB1 funnel domain helices α21 and α23, and loop α23-α24 (3). The subsequent structure of a yeast Pol II elongation complex with bound DNA template and RNA transcript further showed that α-amanitin contacts two elements of the polymerase active center: the bridge helix and the trigger loop (4). The toxin in particular traps the trigger loop, which is a mobile element that undergoes folding for catalyzing extension of the RNA chain and is also important for the translocation of nucleic acids to the next DNA template position after catalysis (4).
Different organisms vary strongly in their sensitivity to α-amanitin (5), and their Pol II enzymes bind α-amanitin with different affinities (2). Whereas Pol II from yeasts such as S. cerevisiae binds α-amanitin with micromolar affinity (2), metazoan enzymes show much higher affinity, with mammalian Pol II binding with nanomolar affinity (6). The reasons for this dramatic variation in eukaryotic species remain unknown. Here we provide the cryo-EM structure of α-amanitin bound to its natural target, the mammalian Pol II elongation complex, and describe contacts of the toxin with Pol II that explain why its affinity is much higher for the mammalian enzyme.
Results
To investigate the binding affinity variation of α-amanitin in eukaryotic species, we determined the cryo-EM structure of α-amanitin bound to its natural target, the mammalian Pol II elongation complex (EC). We purified Pol II from pig thymus (“Experimental procedures”) and added human Gdown1 (hGdown1) (Fig. 1A), which is often associated with Pol II in metazoan cells (7). The EC was formed with a DNA-RNA scaffold that was highly similar to a previously used one (8). The EC was active in RNA synthesis and was inhibited after α-amanitin addition (Fig. 1B). The EC sample was cross-linked with BS3, incubated with α-amanitin, and immediately applied to EM grids before flash freezing. Cryo-EM analysis revealed a homogeneous distribution of particles that could be classified easily (supporting Figs. S1 and S2D). 134,512 particle images were extracted and used for 3D reconstruction, resulting in a cryo-EM density map at a nominal resolution of 3.4 Å (supporting Fig. S1).
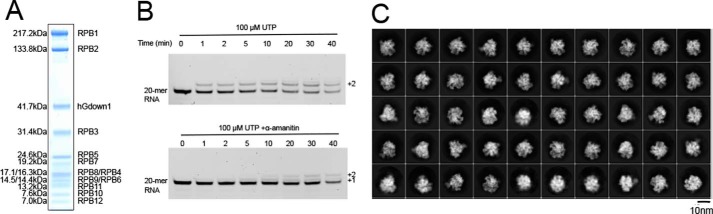
Figure 1.
Pig (S. scrofa) Pol II purification, in vitro RNA extension assay, and exemplary 2D classes of the data set. A, SDS-PAGE analysis of the Pol II–hGdown1 complex. B, the reconstituted Pol II–hGdown EC is active in RNA extension and inhibited by α-amanitin. In the absence of α-amanitin (upper panel), two uridine residues were incorporated into the RNA of the scaffold upon incubation with 100 mm UTP, as expected from the presence of two templating adenine bases downstream. In the presence of α-amanitin (lower panel), nucleotide addition is slowed down, and addition of only one uridine residue was observed, as expected from impaired Pol II translocation. C, representative 2D classes generated from the cryo-EM data set.
To obtain an atomic model of the mammalian Pol II EC-α-amanitin complex, we placed the previously refined bovine Pol II structure (8) into the density and adjusted it locally. There was no density for hGdown1, which apparently dissociated from the complex. The region around the Pol II active center, including α-amanitin and its binding pocket, was well resolved, with an estimated local resolution of ∼3.0 Å (supporting Fig. S2, A and B). There were no other significant additional densities observed.
We could build an atomic model for α-amanitin and define its chemical interactions with Pol II (Figs. 2 and 3 and Table 1). The structure was completed by classification and refinement focused on the flexible Pol II stalk subcomplex RPB4–RPB7, upstream DNA, and the mobile trigger loop, followed by manual adjustments and real-space refinement (supporting Fig. S1; see also “Experimental procedures”).
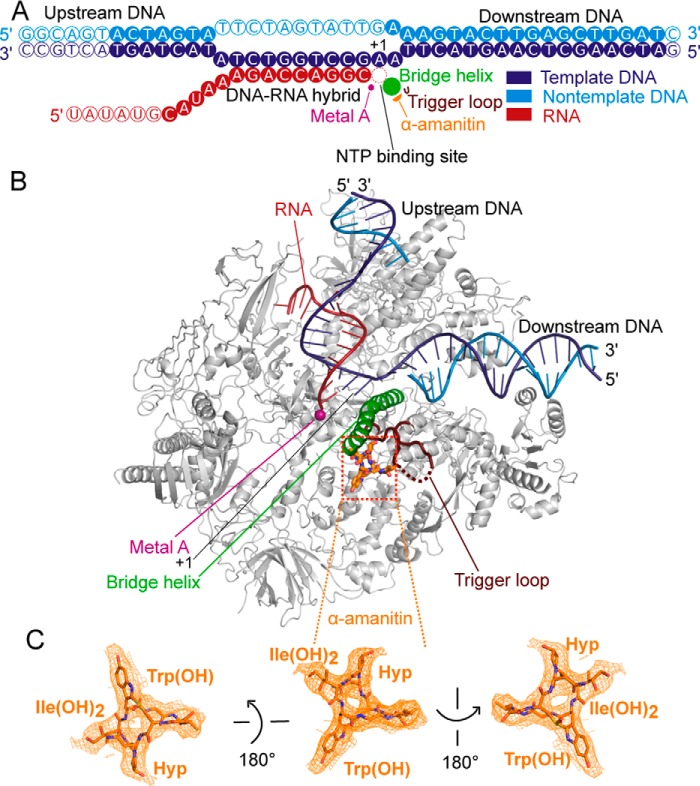
Figure 2.
Cryo-EM structure of mammalian Pol II EC bound by α-amanitin. A, nucleic acid scaffold is depicted schematically. Filled and unfilled circles represent modeled and not modeled nucleotides, respectively. The nucleotide-binding site (red dashed circle), bridge helix (green), the catalytic metal ion A (pink), trigger loop (brown), and α-amanitin (orange) are indicated. The color code is used throughout. B, overview of the structure. Pol II is shown as a silver ribbon model, and other elements are colored as in A. C, electron density for α-amanitin (orange mesh) in three different views. Important contact moieties with Pol II are indicated. Nitrogen, oxygen, and sulfur atoms are blue, red, and yellow, respectively.
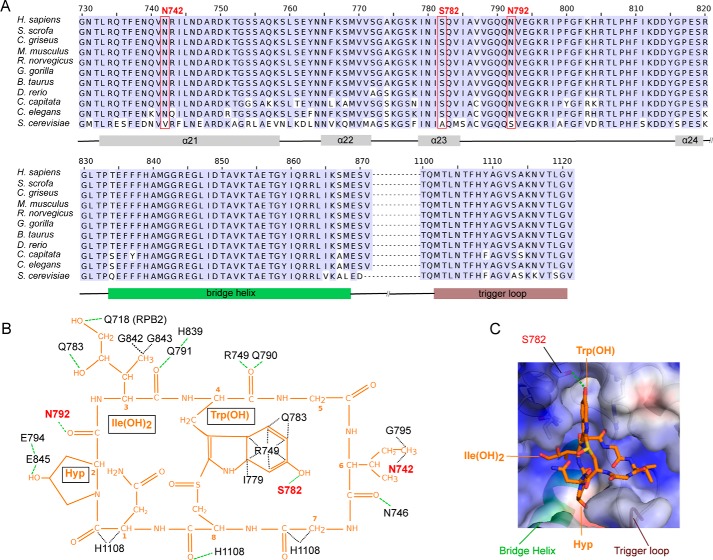
Figure 3.
Interactions of mammalian Pol II with α-amanitin. A, sequence alignment of residues forming the α-amanitin-binding pocket in RPB1 between various metazoan species and the yeast S. cerevisiae (bottom row). The red boxes indicate amino acid residues that form metazoan-specific interactions with α-amanitin. Helices α21 to α24, bridge helix, and trigger loop are indicated at the bottom of the sequence alignment. B, schematic overview of Pol II–amanitin interactions. The chemical structure of α-amanitin is shown in orange. RPB1 residues conserved over eukaryotes are labeled in black, whereas metazoan-specific amanitin-interacting residues are labeled in red. The green dashed lines indicate hydrogen bonds, whereas black dashed lines show other interactions. C, surface representation of the amanitin-binding Pol II pocket. Positively and negatively charged surfaces are in blue and red, respectively. The bridge helix, trigger loop, and RPB1 residue Ser782 are indicated.
Table 1.
Hydrogen bonds between α-amanitin and S. scrofa Pol II
| α-Amanitin residue | α-Amanitin atom | Pol II residue (RPB1) | Pol II atom | Length | Present in yeast Pol II EC–α-amanitin complex |
|---|---|---|---|---|---|
| Å | |||||
| 2 | OD(D) | Glu845 | OE1(A) | 3.1 | Yes |
| 2 | O(A) | Asn792 | N(D) | 3.6 | Yes |
| 2 | O(A) | Asn792 | ND2(D) | 2.8 | No |
| 3 | OD(A) | Gln718 (RPB2) | NE2(D) | 2.8 | Yes |
| 3 | OG(A) | Gln783 | NE2(D) | 3.5 | Yes |
| 3 | O(A) | Gln791 | NE2(D) | 3.6 | Yes |
| 4 | O(A) | Arg749 | NE | 3.6 | Yes |
| 4 | O(A) | Gln790 | N(D) | 2.5 | Yes |
| 4 | OH2 | Ser782 | OG | 2.5 | No |
| 6 | O(A) | Asn746 | ND2(D) | 3.7 | Yes |
| 8 | O(A) | His1108 | NE2(D) | 3.6 | Yes |
The structure of the Pol II EC is highly similar to the previously determined structure of the bovine counterpart (8). Pig Pol II differs from bovine Pol II in only five residues: RPB1 Glu1968, RPB5 Glu32 and Asp46, RPB6 Ser126, and RPB9 Phe11. The EC adopts the post-translocation state with a straight bridge helix, different from the slightly bent bridge helix observed in the yeast Pol II-α-amanitin crystal structure (4), which is thought to reflect a translocation intermediate. The trigger loop adopts a conformation that most closely resembles the “wedged” conformation previously observed in the yeast EC bound by α-amanitin (4). However, residue Leu1104 (Leu1081 in yeast), which forms a wedge behind the bridge helix in the yeast structure (4), protrudes ∼2 Å less in between the bridge helix and the polymerase cleft module, essentially not forming a wedge anymore, and consistent with the observed straight bridge helix. We refer to this slightly altered trigger loop conformation as “unwedged” because it is likely that it is adopted after the wedged conformation and before the addition of the next nucleotide.
The position and binding pocket of α-amanitin is as observed in the yeast EC (4) (Figs. 2B and 3B and Table 1). Most contacts between α-amanitin and yeast Pol II observed in the EC are conserved in the mammalian complex, as expected by the high conservation of residues involved in binding the toxin (Fig. 3, A and B). Conserved contacts are also formed by His1108 (yeast His1085) in the trigger loop of Pol II.
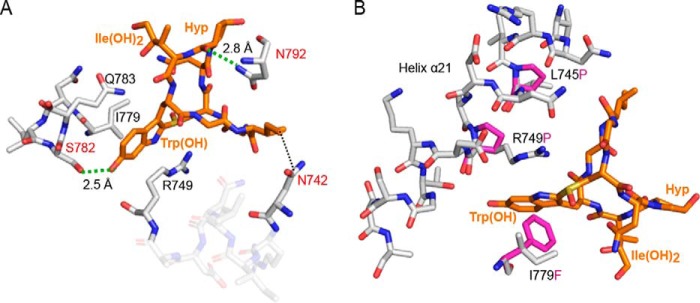
Three differences in α-amanitin-Pol II contacts are observed. First, the side chain of RPB1 residue Ser782, conserved over mammals and other metazoan species, forms an additional hydrogen bond with the hydroxyl group in the indole ring of the tryptophan in α-amanitin (Figs. 3, A–C, and 4A). Ser782 lines the bottom of a cage, formed by the universally conserved Pol II residues Arg749, Ile779, and Gln783, for the indole ring of α-amanitin (Figs. 3C and 4A). The yeast residue corresponding to mammalian Ser782 is Ala759 and cannot form this hydrogen bond. Second, Asn792 forms an additional hydrogen bond with its side chain to the backbone carbonyl group of 4,5-dihydroxyisoleucine in α-amanitin (Figs. 3B and 4A). This contact is not present in the yeast Pol II–amanitin complex, because the yeast counterpart of mammalian Asn792 is Ser769, and the observed hydrogen bond is thus not possible. There is a third residue in the amanitin-binding pocket that differs, Asn742 (Fig. 3, A and B), which corresponds to Val719 in yeast, but this is unlikely to contribute strongly to the difference in affinity because in both structures these residues form van der Waals contacts with the side chain of isoleucine in α-amanitin.
Figure 4.
Extra hydrogen bonds in mammalian and binding pocket mutation analysis. A, two metazoan-specific hydrogen bonds are indicated with green dashed lines, and the corresponding bond lengths are indicated between α-amanitin and mammalian RPB1. B, modeling of site-specific mutations in the α-amanitin–binding pocket that confer resistance to α-amanitin in Mus musculus. The Pol II model is shown with gray sticks, whereas the mutated amino acids are shown with magenta sticks.
Thus, compared with the yeast structure, two additional hydrogen bonds are formed between α-amanitin and the mammalian EC. It is known that two additional hydrogen bonds can give rise to enthalpy changes that account for changes in dissociation constants by 3 orders of magnitude (9, 10). We therefore suggest that the two additional hydrogen bonds account for the much higher affinity of mammalian Pol II for the toxin. This interpretation is supported by known biochemical data obtained with amanitin derivatives that lack certain functional groups (1, 11). In particular, alkylation of the hydroxyl group in the indole ring is predicted to prevent hydrogen bond formation and is known to decrease toxicity and inhibitory potential of amanitin (1).
The structure also suggests the molecular basis for α-amanitin resistance arising from mutations in the binding pocket in Pol II enzymes from mice (12) and Drosophila (13). Modeling shows that mutation I779F in mouse RPB1 leads to a steric clash that likely prevents α-amanitin from binding (Fig. 4B). The additional mouse mutations L745P and R749P likely destabilize helix α21, which forms part of the binding pocket (Fig. 4B). The Drosophila melanogaster Rpb1 mutations N792D and N793D (13) are predicted to disrupt hydrogen bonds between Pol II and amanitin, thereby decreasing affinity.
Discussion
More than one century after the discovery of α-amanitin (14), we now provide an atomic model of its structure in complex with its natural target, the mammalian Pol II EC. This work provides the structural basis of mammalian Pol II inhibition by α-amanitin. Whereas insights into the mechanism of transcription inhibition by α-amanitin were already derived from structures of the yeast Pol II (3) and the yeast EC (4), our current work additionally provides a molecular explanation for the long-standing observation that α-amanitin has a much higher affinity for mammalian Pol II, compared with the yeast enzyme. Most notably, we observe two additional, well defined hydrogen bonds that are possible in mammalian Pol II enzymes, but not in yeast Pol II, explaining the tighter binding of the toxin to the former.
Together with recent studies (15, 16), our work also shows that cryo-EM can now be used to study the detailed interactions of small molecules with proteins, as required for drug design. We note that such applications of cryo-EM still often require that the target molecule or complex has a critical size. In the future, further developments of cryo-EM will, however, likely remove this limitation such that the inhibition of target molecules and complexes of lower molecular weight by small molecules can also be studied.
Experimental procedures
Purification of Sus scrofa RNA polymerase II and recombinant hGdown1
S. scrofa Pol II was purified essentially as described for the bovine Pol II preparation (8), except that pig thymus instead of bovine thymus was used. Briefly, thymus was homogenized, and the supernatant was filtered. After polyethyleneimine precipitation, Pol II was purified with a MacroPrepQ column, followed by ammonium sulfate precipitation and an affinity column with 8WG16 (αRPB1 CTD) antibody-coupled Sepharose, a UNO-Q anion exchange column, and finally a Sephacryl S-300 HiLoad sizing column. The typical yield was 2–4 mg from ∼500 g of thymus.
Recombinant expression and purification of hGdown1 was performed as described previously (8). After elution from the UnoQ column (Bio-Rad), Pol II was combined with a 3-fold molar excess of hGdown1 and incubated for 2–3 h before applying the complex to a HiPrep 16/60 Sephacryl S-300 HR column (GE Healthcare). Fractions containing the Pol II–hGdown1 complex were collected and concentrated to a concentration of 2–3 mg/ml using an Amicon Ultra-15 centrifugal filter unit (100-kDa molecular mass cut-off) (Merck). Sample aliquots were snap frozen in liquid nitrogen and stored at −80 °C prior to use.
Elongation complex preparation
The DNA scaffold used for the EC is the same as the one used for the bovine RNA polymerase II with the transcription elongation factor 5,6-dichloro-1-β-d-ribofuranosyl benzimidazole-induced sensitivity factor complex (17) (template DNA, 5′-GATCAAGCTCAAGTACTTAAGCCTGGTCTATACTAGTACTGCC-3′; and nontemplate DNA 5′-GGCAGTACTAGTATTCTAGTATTGAAAGTACTTGAGCTTGATC-3′) but slightly different with the one for the bovine EC structure (8). A 20-nt RNA (RNA 5′-UAUAUGCAUAAAGACCAGGC-3′) was used for the formation of a 9-nt DNA–RNA hybrid and 11 nt of exiting RNA. The scaffold was annealed (18), and a 1.5-fold molar excess of scaffold was added to the Pol II–hGdown complex. The sample was incubated on ice for 10 min and subsequently incubated for an additional 15 min at 20 °C while shaking at 550 rpm. The complex was cross-linked with 3 mm BS3 (Thermo Scientific) on ice for 30 min. The cross-linking reaction was quenched with 50 mm ammonium bicarbonate and applied to a Superdex 200 increase 10/300 GL column (GE Healthcare) pre-equilibrated with Pol II buffer (5 mm HEPES, pH 7.25 at 25 °C, 150 mm NaCl, 10 μm ZnSO4, 10 mm DTT). The peak containing the complex was pooled and concentrated to a concentration of 473 μm. A 1.5-fold molar excess of α-amanitin was added to the elongation complex. The sample was incubated on ice for 20 min.
EM
4 μl of the protein solution was applied to glow-discharged Quantifoil R2/2 gold grids (Quantifoil) and plunged into liquid ethane after blotting with a FEI Vitrobot Mark IV (FEI, Hillsboro, OR). The data were acquired on a FEI Titan Krios, operated at 300 keV, and equipped with a Gatan K2 Summit direct electron detector and a Quantum GIF. Micrographs were collected automatically with the software package EPU (FEI) at a nominal magnification of 130K (1.07 Å per pixel) in counting mode. The dose rate was 3.8 e−/pixel/s. Three images were acquired per foil hole. Each micrograph was collected with a total dose of 35 electrons per square angstrom over a 10-s exposure, fractionated into 40 frames (0.25 s each). Defocus values ranged from 1 to 3 μm. Micrograph frames were aligned and corrected with MotionCor2 (19). Unless otherwise noted, data processing was performed using RELION 2.1 (20). Contrast transfer function parameters were estimated using Gctf (21). Initial 2D classes were calculated from 2,909 manually selected particles from 37 micrographs. The initial 2D classes were used as templates for autopicking. After manual inspection of all 2,049 micrographs, a total of 207,410 particles were obtained. Two rounds of 2D classification were performed, and bad particles were removed. The resulting data set of 134,512 particles was used for further refinement and focused classification refinement in 3D. The Bos taurus Pol II structure (8) (EMDataBank accession code EMD-3219) was low-pass filtered to 40 Å as an initial model for 3D refinement. Initial 3D refinement followed by movie processing and particle polishing yielded a reconstruction at an overall resolution of 3.4 Å (gold-standard Fourier shell correlation criterion 0.143, RELION 2.1). Focused 3D classification without image alignment was performed on the α-amanitin–binding pocket, the Pol II stalk (RPB4–RPB7), and upstream DNA, followed by global 3D refinement.
Model building and refinement
Model building was based on the previously published bovine Pol II structure (8) (Protein Data Bank accession code 5FLM). The model was manually fitted in COOT (22). The α-amanitin molecule was taken from a S. cerevisiae α-amanitin–bound Pol II structure (4) (Protein Data Bank accession code 2VUM). The α-amanitin molecule was rigid body fitted into the density. The structure was refined in real space with special restraints to the nucleic acids and α-amanitin using PHENIX (23).
Transcription assay
Template DNA and RNA were mixed at a molar ratio of 1:1 and annealed as described (18). The annealed DNA–RNA was mixed with Pol II–hGdown complex at a molar ratio of 1:2 and incubated at 28 °C for 10 min. Nontemplate DNA was added and incubated at 28 °C for an additional 10 min. The elongation complex was mixed with α-amanitin or buffer (control) at the same molar ratio used for the complex formation. The sample was subsequently incubated on ice for 20 min. 100 μm UTP was added to both control and experimental reactions. The reaction was incubated in transcription buffer (20 mm Na-HEPES, pH 7.5, 60 mm (NH4)2SO4, 8 mm MgSO4, 10 μm ZnSO4, 10 mm DTT, 10% (v/v) glycerol) at 28 °C, and samples were taken at the indicated time points. The reaction was stopped by adding stop buffer (50 mm EDTA, 6.4 m urea, 1-fold TBE (Sigma–Aldrich)) to the reaction. The product RNA was separated using a 20% denaturing urea polyacrylamide gel (300 V) and visualized using a GE Typhoon FLA 9500 (GE Healthcare).
Author contributions
X. L., L. F., and C. W. data curation; X. L., L. F., and P. C. formal analysis; X. L. and L. F. validation; X. L. and L. F. investigation; X. L. and L. F. visualization; X. L., L. F., and C. W. methodology; X. L. and P. C. writing-original draft; X. L., L. F., and P. C. writing-review and editing; L. F., C. W., and P. C. supervision; P. C. conceptualization; P. C. resources; P. C. project administration.
Supplementary Material
Acknowledgments
We thank Carrie Bernecky, Christian Dienemann, Christoph Engel, Björn Schwalb, Seychelle M. Vos, and Dimitry Tegunov for help.
This work was supported by Deutsche Forschungsgemeinschaft Grants SFB860 and SPP1935 (to P. C.), European Research Council Advanced Investigator Grant TRANSREGULON Grant Agreement 693023 (to P. C.), and funds from the Volkswagen Foundation (to P. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1 and S2.
The atomic coordinates and structure factors (code 6EXV) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The electron density reconstruction and final model were deposited with the EMDataBank under accession code EMDB-3981.
- Pol II
- RNA polymerase II
- EC
- elongation complex
- hGdown1
- human Gdown1
- nt
- nucleotide(s).
References
- 1. Wieland T. (1986) Peptides of Poisonous Amanita Mushrooms, Springer Series in Molecular Biology, Springer-Verlag, New York Inc., New York [Google Scholar]
- 2. Wieland T., and Faulstich H. (1991) Fifty years of amanitin. Experientia 47, 1186–1193 10.1007/BF01918382 [DOI] [PubMed] [Google Scholar]
- 3. Bushnell D. A., Cramer P., and Kornberg R. D. (2002) Structural basis of transcription: α-amanitin–RNA polymerase II cocrystal at 2.8 Å resolution. Proc. Natl. Acad. Sci. U.S.A. 99, 1218–1222 10.1073/pnas.251664698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brueckner F., and Cramer P. (2008) Structural basis of transcription inhibition by α-amanitin and implications for RNA polymerase II translocation. Nat. Struct. Mol. Biol. 15, 811–818 10.1038/nsmb.1458 [DOI] [PubMed] [Google Scholar]
- 5. Cochet-Meilhac M., Nuret P., Courvalin J. C., and Chambon P. (1974) Animal DNA-dependent RNA polymerases: 12. determination of the cellular number of RNA polymerase B molecules. Biochim. Biophys. Acta 353, 185–192 10.1016/0005-2787(74)90183-X [DOI] [PubMed] [Google Scholar]
- 6. Bensaude O. (2011) Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Which compound to choose? How to evaluate its activity? Transcription 2, 103–108 10.4161/trns.2.3.16172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu X., Malik S., Negroiu C. C., Hubbard K., Velalar C. N., Hampton B., Grosu D., Catalano J., Roeder R. G., and Gnatt A. (2006) A mediator-responsive form of metazoan RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 103, 9506–9511 10.1073/pnas.0603702103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bernecky C., Herzog F., Baumeister W., Plitzko J. M., and Cramer P. (2016) Structure of transcribing mammalian RNA polymerase II. Nature 529, 551–554 10.1038/nature16482 [DOI] [PubMed] [Google Scholar]
- 9. Hubbard R. E., and Kamran Haider M. (2010) Hydrogen bonds in proteins: role and strength. In Encyclopedia of Life Sciences, John Wiley & Sons, Inc., New York: 10.1002/9780470015902.a0003011.pub2 [DOI] [Google Scholar]
- 10. Klebe G. (2015) Applying thermodynamic profiling in lead finding and optimization. Nat. Rev. Drug Discov. 14, 95–110 10.1038/nrd4486 [DOI] [PubMed] [Google Scholar]
- 11. Baumann K., Zanotti G., and Faulstich H. (1994) A β-turn in α-amanitin is the most important structural feature for binding to RNA polymerase II and three monoclonal antibodies. Protein Sci. 3, 750–756 10.1002/pro.5560030504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartolomei M. S., and Corden J. L. (1995) Clustered α-amanitin resistance mutations in mouse. Mol. Gen. Genet. 246, 778–782 10.1007/BF00290727 [DOI] [PubMed] [Google Scholar]
- 13. Bartolomei M. S., and Corden J. L. (1987) Localization of an α-amanitin resistance mutation in the gene encoding the largest subunit of mouse RNA polymerase II. Mol. Cell. Biol. 7, 586–594 10.1128/MCB.7.2.586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schlesinger H., and Ford W. W. (1907) On the chemical properties of amanita-toxin. J. Biol. Chem. 3, 279–283 [Google Scholar]
- 15. Wong W., Bai X.-C., Sleebs B. E., Triglia T., Brown A., Thompson J. K., Jackson K. E., Hanssen E., Marapana D. S., Fernandez I. S., Ralph S. A., Cowman A. F., Scheres S. H. W., and Baum J. (2017) Mefloquine targets the Plasmodium falciparum 80S ribosome to inhibit protein synthesis. Nat. Microbiol. 2, 17031 10.1038/nmicrobiol.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao Y., Cao E., Julius D., and Cheng Y. (2016) TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 10.1038/nature17964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bernecky C., Plitzko J. M., and Cramer P. (2017) Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp. Nat. Struct. Mol. Biol. 24, 809–815 10.1038/nsmb.3465 [DOI] [PubMed] [Google Scholar]
- 18. Brueckner F., Hennecke U., Carell T., and Cramer P. (2007) CPD damage recognition by transcribing RNA polymerase II. Science 315, 859–862 10.1126/science.1135400 [DOI] [PubMed] [Google Scholar]
- 19. Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., and Agard D. A. (2017) MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernandez-Leiro R., and Scheres S. H. W. (2017) A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 73, 496–502 10.1107/S2059798316019276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang K. (2016) Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 10.1016/j.jsb.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Torices R., and Muñoz-Pajares A. J. (2015) PHENIX: An R package to estimate a size-controlled phenotypic integration index. Appl. Plant Sci. 3, 10.3732/apps.1400104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.