Abstract
Chlamydomonas reinhardtii adapts to the stress of CO2-limiting conditions through the induction of a set of genes including CAH1, which encodes a periplasmic carbonic anhydrase. CAH1 is up-regulated under low-CO2 conditions (air containing 0.04% [v/v] CO2) in the presence of light, whereas it is down-regulated under high-CO2 conditions (5% [v/v] CO2) or in the dark. In an effort to identify cis-elements involved in the transcriptional regulation of CAH1, a series of 5′-nested deletions of the region upstream of CAH1 were fused to a promoterless arylsulfatase reporter gene (ARS). The upstream region from −651 to +41 relative to the transcription start site was sufficient to regulate the expression of ARS with kinetics similar to those of endogenous CAH1. Deletion of the region between −651 and −294 resulted in a significant decrease in the level of arylsulfatase activity expressed under low-CO2 conditions. The 543-bp upstream region from −651 to −109, without any promoter elements, CAAT-box, or TATA-box, could confer CO2 and light responsiveness on the β2-tubulin minimal promoter. This 543-bp region was divided into two parts: a 358-bp silencer region from −651 to −294, which represses the minimal promoter activity under high-CO2 conditions, and a 185-bp enhancer region from −293 to −109, which activates the promoter under low-CO2 conditions in the presence of light.
When microalgae are exposed to the stress of CO2-limiting conditions, they express a set of genes involved in a carbon-concentrating mechanism (CCM) that functions to take up inorganic carbon from the external environment into the cells and to elevate the CO2 level around Rubisco (Aizawa and Miyachi, 1986; Badger and Price, 1994). This adaptation to the stress of low-CO2 conditions suggests the existence of a sensory mechanism by which cells perceive changes in the levels of environmental CO2 and a pathway by which the signal indicative of the shortage of CO2 is transduced into the induction of specific genes.
In Chlamydomonas reinhardtii, several genes regulated in response to changes in CO2 concentration have been isolated, including Ala:α-ketoglutarate aminotransferase (Chen et al., 1996), mitochondrial CA (Eriksson et al., 1996), and chloroplast envelope protein LIP-36 (Chen et al., 1997). CAH1, encoding a periplasmic CA, is one of the most well characterized of these genes (Dionisio-Sese et al., 1990; Fujiwara et al., 1990; Fukuzawa et al., 1990). CAH1 is not transcribed when cells are maintained under high-CO2 conditions, whereas transcripts of this gene accumulate at a significant level within 1 h after transfer to low-CO2 conditions. However, this induction does not occur when cells are cultured in the dark or in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea, indicating that photosynthetic electron flow is required. Interestingly, CAH2 is regulated in a manner opposite to that of CAH1. It is transcribed preferentially under high-CO2 conditions, even in the dark, and light has a negative effect on the expression of CAH2 (Fujiwara et al., 1990).
Ronen-Tarazi et al. (1995) have described an analysis of a low-CO2-inducible promoter in a cyanobacterium, Synechococcus sp. PCC7942, produced by fusing a 380-bp fragment derived from the region upstream of cmpA with a promoterless chloramphenicol acetyltransferase reporter gene. However, no detailed analysis of the CO2-responsive elements in this strain has been reported.
In eukaryotes, an analysis of CO2-responsive promoters of β-CA1 and β-CA2, which encode mitochondrial CA isozymes in C. reinhardtii, has been reported (Villand et al., 1997). Both of these genes are up-regulated under low-CO2 conditions and they share an identical 194-bp sequence in their 5′-upstream region. This 194-bp region was shown to be enough to drive a promoterless ARS reporter gene. However, it is not known whether this CO2-dependent gene regulation is mediated by activation under low-CO2 conditions or by repression under high-CO2 conditions.
In this paper, we present direct evidence, from experiments using a series of chimeric fusion constructs containing CAH1 upstream regions, the β2-tubulin minimal promoter and an ARS reporter gene, indicating that the CO2-responsive transcriptional regulation of CAH1 is mediated by both enhancer and silencer regions in its 5′-upstream region.
MATERIALS AND METHODS
Cells and Culture Conditions
Chlamydomonas reinhardtii strain 5D (nit1-305, cw-15) (Tam and Lefebvre, 1993) was maintained in TAP medium (Gorman and Levine, 1965) under continuous illumination (150 μmol m−2 s−1) at 28°C. For the high-CO2 conditions, cells were cultured in modified HSM medium (Sueoka, 1960), supplemented with 20 mm MOPS (pH 7.2) and 0.4 mm MgSO4 (HSM+S) under aeration with air enriched with 5% (v/v) CO2. For low-CO2 conditions, cultures were bubbled with ordinary air containing 0.04% (v/v) CO2.
Chimeric Constructs
DNA fragments of various lengths containing portions of the 5′-upstream region of CAH1 were amplified by PCR. The oligonucleotide primer p-Cla1 (5′-GCATCGGTGTTCA AGTGGGTTGCAGGTA-3′), which is complementary to the CAH1 upstream sequence from position +20 to +41 relative to the transcription initiation site, was used in combination with p-Xho2 (5′-TTCTCGAGGTTCTCCACCTTGCCAGCGCAC-3′), p-Xho3 (5′-TTCTCGAGTCAGCTTCTCTCCCGCCAGCAT-3′), p-Xho4 (5′-ATCTCGAGAGATTTTCACCG GTTGGAAGGA-3′), p-Xho5 (5′-AACTCGAGGTATGACATGGGTGCCGGAACT-3′) and p-Xho6 (5′-CCCTCGAG GCTGCAGACTGTGCGCATGCAG-3′), to amplify upstream regions between −818 and +41, −651 and +41, −293 and +41, −200 and +41, and −151 and +41, respectively. These PCR products were blunt-ended, phosphorylated, XhoI- digested, and inserted into the SalI-EcoRV restriction sites of the plasmid pJD54, which contains a promoterless ARS gene (Davies et al., 1992). These chimeric constructs were named pCAO2, pCAO3, pCAO4, pCAO5, and pCAO6, respectively.
To generate constructs containing the β2-tubulin minimal promoter (pCT series), two additional primers were synthesized. CAup-Kpn4 (5′-ATGGTACCTTAAAACCAGAAGCT GCATTTC-3′) hybridizes to the region just upstream of the CAAT box (between −109 and −130) and CAup-Kpn7 (5′-ATCAGCTGACAACGCTGCCAACGTGGTGGC-3′) corresponds to the region between −294 and −315. Primer sets p-Xho3 and CAup-Kpn4, p-Xho3 and CAup-Kpn7, and p-Xho4 and CAup-Kpn4 were used for PCR. The amplified fragments were cloned into the blunt-ended KpnI site of the plasmid pJD100 (Davies and Grossman, 1994), and the resulting chimeric constructs were named pCT34, pCT37, and pCT44, respectively.
Transformation
The host Chlamydomonas strain 5D (nit1-305, cw-15) was co-transformed with the chimeric constructs and plasmid pMN24, containing the entire nitrate reductase gene (Fernandez et al., 1989), by the glass beads method with slight modifications (Kindle, 1990). Cells cultured to a concentration of 1 to 2 × 106 cells mL−1 were collected by centrifugation, and resuspended in TAP(NO3) (TAP medium in which NH4Cl was replaced by KNO3) at a concentration of 2 × 108 cells mL−1. Ten micrograms of pMN24 and 50 μg of a chimeric construct were added to 5 mL of the cell suspension as supercoiled DNA, and vortexed with glass beads for 30 s. The glass beads were allowed to settle, and the supernatant was diluted with TAP(NO3) medium and centrifuged. The pelleted cells were resuspended in TAP(NO3) and spread onto TAP(NO3) agar plates. After 1 week, the nit+ colonies that appeared on the plates were used for further analysis.
Screening of ARS-Expressing Transformants
The cells from nit+ colonies were grown in liquid TAP(NO3) medium in 96-well microtiter plates. These cultures were used to inoculate HSM+S medium containing 0.3 mm X-SO4 (Sigma, St. Louis). The cells were cultured under high-CO2 conditions for 1 d, and then transferred to low-CO2 conditions. Transformants expressing arylsulfatase were identified as those showing a blue color. As a more sensitive assay, 0.8 mm N-SO4 (Sigma) was used as a substrate. To the culture, an equal volume of a solution containing 4% (w/v) SDS and 0.4 m Na acetate (pH 4.8) and 0.2 volume of 10 mg mL−1 tetrazotized-o-dianisidine (Sigma) were added to visualize the arylsulfatase activity.
Quantification of Arylsulfatase Activity
Cell cultures were centrifuged and the supernatants were assayed for arylsulfatase activity by adding 50 μL of 100 mm imidazole, 25 μL of 1 m Tris-HCl (pH 10.0), and 5 μL of 80 mm N-SO4 to 420 μL of supernatant (Ohresser et al., 1997). The mixture was incubated at 37°C, and the reaction was stopped by adding 500 μL of a solution containing 4% (w/v) SDS and 0.4 m Na acetate (pH 4.8). The absorbance was measured at 540 nm immediately after addition of 100 μL of 10 mg mL−1 tetrazotized-o-dianisidine, and the value was normalized by dividing by the chlorophyll content of the culture.
Northern-Blot Analysis
Total RNA was isolated as described by Chomczynski et al. (1987). Ten micrograms of total RNA was electrophoresed in a denaturing agarose gel and blotted onto a nylon membrane (Hybond-N+, Amersham). Two radiolabeled probes were used for hybridization, a 32P-terminally labeled 40-mer oligonucleotide (p-CA1-5′; 5′-GGTGTTCAAGTGGGTTGCAGGTAATGACTCAACGCAGGGT-3′), which hybridizes to the 5′ untranslated region of CAH1, and 0.8- and 1.2-kb BamHI fragments corresponding to the ARS coding region, which were excised from plasmid pJD27 containing ARS cDNA (de Hostos et al., 1989).
RESULTS
A 692-bp Region between −651 and +41 Sufficient for CO2- and Light-Responsive Gene Regulation
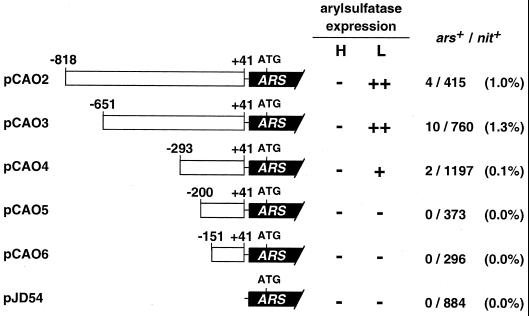
In an effort to identify the regions involved in CO2-responsive transcriptional regulation of CAH1, a series of 5′-nested deletions produced by PCR were fused to an ARS reporter gene (Davies et al., 1992). Five chimeric constructs were generated, named pCAO2, pCAO3, pCAO4, pCAO5, and pCAO6, that contain portions of the upstream region up to positions −818, −651, −293, −200, and −151, respectively (Fig. 1). These constructs were introduced into the host Chlamydomonas strain 5D (nit1-305, cw-15) together with pMN24 DNA (Fernandez et al., 1989), and nit+ colonies exhibiting arylsulfatase activity under low-CO2 conditions were selected. When cells were transformed with pCAO2 or pCAO3, 1.0% and 1.3% of the total nit+ colonies exhibited arylsulfatase activity. However, when cells were transformed with pCAO4 only 0.1% of the nit+ colonies expressed arylsulfatase activity, and the levels of activity were much lower than those obtained with pCAO2 or pCAO3. When cells were transformed with pCAO5 or pCAO6, no transformants expressing arylsulfatase activity were obtained among the nit+ colonies tested (Fig. 1).
Figure 1.
Schematic drawings of the chimeric constructs. A series of 5′-nested deletions of the CAH1 upstream region, represented by white boxes, were fused to the promoterless ARS reporter gene, represented by black bars. Numbering on the white boxes indicates positions relative to the transcription start site. Strain 5D was co-transformed with these chimeric constructs and pMN24, and nit+ transformants were analyzed for arylsulfatase expression under high- (H) and low-CO2 conditions (L). High (++), low (+), or no (−) arylsulfatase activity is indicated. The number of arylsulfatase-expressing colonies among the nit+ transformants is indicated at the far right as ars+/nit+. The nucleotide sequence of the 5′-upstream region of CAH1 will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases with the accession no. AB026126.
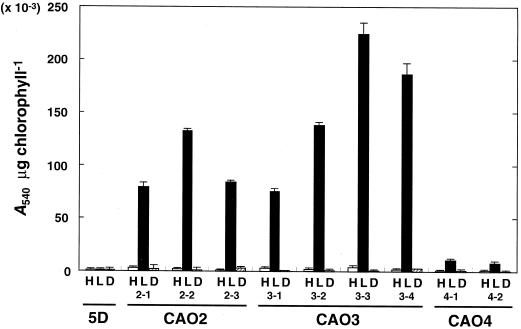
Among the arylsulfatase-expressing clones, transformants containing the appropriate CAH1 upstream regions, which had been generated by PCR, were selected by Southern-blot analysis (data not shown). The arylsulfatase activity expressed by the individual transformants was quantified under high-CO2 conditions in the light, under low-CO2 conditions in the light, and under low-CO2 conditions in the dark (Fig. 2). After 8 h of incubation under these conditions, arylsulfatase activity was measured by a 12-h enzyme reaction as described in “Materials and Methods.” No activity was detected in the host strain 5D because the endogenous ARS gene is repressed by excess SO42− in the culture medium (de Hostos et al., 1988). The strains CAO2 and CAO3, harboring pCAO2 and pCAO3, respectively, exhibited arylsulfatase activity only under low-CO2 conditions in the light, indicating that the 692-bp region between −651 and +41 was sufficient for CO2-responsive regulation. Strain CAO4 harboring pCAO4 also exhibited arylsulfatase activity under low-CO2 conditions in the light, but the level of activity was much lower than that of CAO2 or CAO3. This result suggests that the 358-bp region between −651 and −294 may function as an enhancer element under low-CO2 conditions.
Figure 2.
Quantification of arylsulfatase activity in individual transformants. Cells cultured under high-CO2 conditions were maintained under high-CO2 in the light (H), transferred to low-CO2 in the light (L), or to low-CO2 in the dark (D). After an 8-h incubation under these conditions, arylsulfatase activity was measured by a 12-h enzyme reaction, as described in “Materials and Methods.” 5D is the host strain used for DNA transformation. CAO2, CAO3, and CAO4 represent transformants harboring pCAO2, pCAO3, and pCAO4, respectively. The results are the average of three determinations, with sd represented by the bars above the graph.
The expression of CAH1 is not induced in the dark, even if cells are shifted from high- to low-CO2 conditions (Fujiwara et al., 1990; Fukuzawa et al., 1990). The light-requirement for arylsulfatase induction in the transformants was also tested as shown in Figure 2. Light-dependent expression of arylsulfatase activity was observed in all of the tested transformants containing the chimeric constructs pCAO2, pCAO3, and pCAO4.
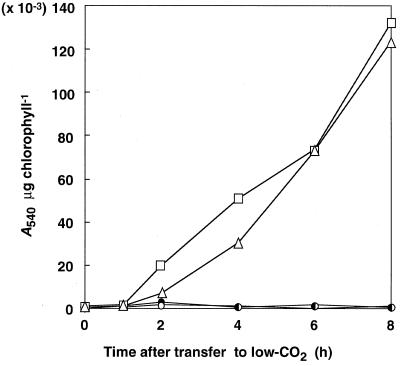
The arylsulfatase activity in strains CAO2-1 and CAO3-1 was measured at various times after transfer from high- to low-CO2 conditions by 3-h enzyme reactions as described in “Materials and Methods” (Fig. 3). Under high-CO2 conditions, the activity in both strains remained undetectable as in the host strain 5D (data not shown). Activity was detected 2 h after transfer to low-CO2 conditions and the levels increased for up to 8 h. This induction of arylsulfatase activity showed a good correlation with the induction of periplasmic CA, which accumulates within 2 h after transfer to low-CO2 conditions and continues to increase for up to 8 h (Dionisio-Sese et al., 1990; Rawat and Moroney, 1995). These results suggest that the 692-bp region between −651 and +41 is sufficient to regulate the expression of the ARS reporter gene with kinetics similar to those of endogenous CAH1.
Figure 3.
Time course of changes in arylsulfatase activity in transformants harboring the chimeric constructs. Cells grown under high-CO2 conditions were transferred to low-CO2 conditions or maintained under high-CO2 conditions. The arylsulfatase activity was measured by 3-h enzyme reactions as described in “Materials and Methods.” □, CAO2–1, low-CO2; ▵, CAO3–1, low-CO2; ●, 5D, high-CO2; ○, 5D, low-CO2.
Accumulation of CAH1/ARS Chimeric mRNA in Transgenic Chlamydomonas Cells
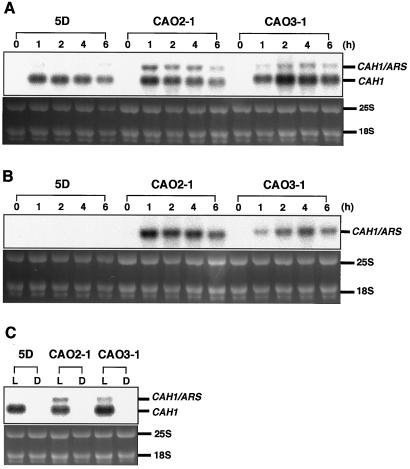
To evaluate the expression of the ARS reporter gene at the mRNA level, 10 μg of total RNA from each of the transgenic Chlamydomonas strains CAO2-1 and CAO3-1, isolated 0, 1, 2, 4, and 6 h after transfer from high- to low-CO2 conditions, was electrophoresed and hybridized with a 32P-labeled oligonucleotide probe, p-CA1-5′, which is specific for the 5′-untranslated region of CAH1 (Fig. 4A). This probe hybridized with both transcripts from the endogenous CAH1 and those from the introduced chimeric constructs containing the 5′-flank of CAH1 and the ARS coding region. In all strains tested, 2.0-kb CAH1 mRNA was detected 1 h after transfer to low-CO2 conditions, and the amount reached a maximum 4 h after the transfer. In CAO2-1 and CAO3-1, 2.5-kb transcripts from the chimeric construct (CAH1/ARS) were expressed, showing kinetics similar to that of endogenous CAH1. To distinguish the CAH1/ARS chimeric transcripts from the premature CAH1 mRNA observed in lanes containing RNA from the host strain 5D, the membrane was reprobed with 32P-labeled ARS cDNA (Fig. 4B). The chimeric transcripts accumulated only in the transgenic Chlamydomonas cells grown under low-CO2 conditions, and no signal was detected in the case of the host strain 5D under either high- or low-CO2 conditions. These results indicate that CO2-responsive expression of arylsulfatase encoded by pCAO2 and pCAO3 was regulated at the mRNA level with kinetics similar to endogenous CAH1.
Figure 4.
Northern-blot analyses of CAH1/ARS chimeric transcripts in the transformants CAO2-1 and CAO3-1. A and B, Cells grown under high-CO2 conditions were transferred to low-CO2 conditions and total RNA was isolated 0, 1, 2, 4, and 6 h after the change in CO2 level. Ten micrograms of RNA from each of the transformants was electrophoresed in a denaturing agarose gel, blotted onto a membrane, and hybridized with 32P-labeled p-CA1-5′ (A) or ARS cDNA (B). CAH1/ARS represents the 2.5-kb transcripts from the chimeric constructs. C, CAO2-1 and CAO3-1 were grown under high-CO2 conditions in the light and subsequently transferred to low-CO2 conditions in the light (L) or dark (D). Total RNA was isolated 4 h after transfer to each of these conditions. The oligonucleotide probe p-CA1-5′ was used for hybridization.
To examine the effect of light on induction of the CAH1/ARS chimeric genes in CAO2-1 and CAO3-1, total RNA samples were isolated from cells grown under low-CO2 conditions in the dark or in the light for 4 h, and hybridization was performed using 32P-labeled p-CA1-5′ as a probe (Fig. 4C). Chimeric transcripts or transcripts from the endogenous CAH1 did not accumulate in the dark, indicating that the CAH1/ARS chimeric genes in pCAO2 and pCAO3 are regulated by light at the mRNA level.
A 543-bp Region between −651 and −109 Conferring CO2 Responsiveness on the β2-Tubulin Minimal Promoter
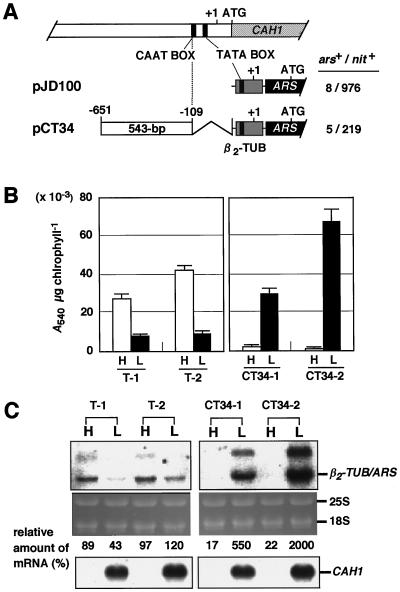
The region between −651 and +41 contains the transcriptional start site and putative promoter elements, TATA- and CAAT boxes. To test whether CO2-responsive regulatory cis-elements are located in the region upstream of the promoter region, the 543-bp region between -651 and -109, which does not contain these promoter elements, was inserted into the plasmid pJD100 (Davies and Grossman, 1994). In pJD100, the ARS reporter gene is driven by a constitutive β2-tubulin minimal promoter. The resulting plasmid, pCT34 (Fig. 5A) was introduced into Chlamydomonas cells and transformants harboring only one copy of the intact chimeric construct were grown under high- and low-CO2 conditions for 4 h. Arylsulfatase activity and mRNA levels were measured (Fig. 5, B and C). Transformants harboring the chimeric construct pJD100 or pCT34 were named T strains and CT34 strains, respectively. It was reported that pJD100 drives low-level constitutive arylsulfatase expression when transformants are grown heterotrophically in TAP medium (Davies and Grossman, 1994). However, under photoautotrophic conditions (Fig. 5B), the T strains exhibited slightly higher levels of arylsulfatase activity under high-CO2 conditions than low-CO2 conditions. Two representative graphs of the activity displayed by six T strains tested are shown in Figure 5B. In C. reinhardtii, tubulin genes are up-regulated during cell division for the formation of the mitotic spindle apparatus and for the assembly of new flagella (Ares and Howell, 1982), and expression of pJD100 is reported to increase slightly during cell division (Davies and Grossman, 1994). It is supposed that higher levels of expression of pJD100 were observed in T strains under high-CO2 conditions because the cells divided more frequently under high- than under low-CO2 conditions.
Figure 5.
A, Schematic drawings of the chimeric constructs. The 543-bp region from −651 to −109 (white box), which does not include sequences downstream from the CAAT box, was fused to a β2-tubulin minimal promoter (shaded box)-driven ARS reporter gene (pJD100), and the resulting chimeric construct was named pCT34. B, Quantification of arylsulfatase activity in individual transformants. The activity in at least five independent transformants was measured, and the results for two representative transformants are shown. Cells grown under high-CO2 conditions were maintained under high-CO2 (H) or transferred to low-CO2 conditions (L), and the arylsulfatase activity was measured 4 h after the change in CO2 level. Bars above the graph indicate the sd for the average of three determinations. C, Northern-blot analysis of the β2-TUB/ARS chimeric transcripts in transformants T-1, T-2, CT34-1, and CT34-2. Total RNA was isolated from cells adapted to high- (H) or low-CO2 conditions (L) for 4 h. Ten micrograms of RNA was electrophoresed and hybridized with 32P-labeled ARS cDNA (upper autoradiographs) or p-CA1-5′ (lower autoradiographs). β2-TUB/ARS represents the transcript from the chimeric constructs. Values of the relative amount of mRNA represent the percentage of the amount of the chimeric transcript over the average amount in three independent T strains (T-1, T-2, and T-3; not shown).
On the other hand, transformants CT34-1 and CT34-2 expressed arylsulfatase activity with the same kinetics as endogenous CAH1 (Fig. 5B). Under high-CO2 conditions, they exhibited reduced activity compared with T-1 and T-2. Northern-blot analysis revealed that the reduction in arylsulfatase activity was due to a decrease in the levels of chimeric transcripts expressed from the β2-tubulin-ARS hybrid gene (β2-TUB/ARS) (Fig. 5C). These results indicate that the 543-bp region functions as a transcriptional silencer under high-CO2 conditions.
Under low-CO2 conditions, these transformants exhibited 3- to 7-fold higher levels of activity and accumulated 6- to 20-fold higher levels of the β2-TUB/ARS mRNA compared with T strains harboring the β2-tubulin minimal promoter-driven ARS. These results indicate that the 543-bp region also functions as a transcriptional enhancer under low-CO2 conditions. An additional band observed above the β2-TUB/ARS mRNA (Fig. 5C) may be premature mRNA containing ARS introns.
The 543-bp Region Is Divided into a Silencer Region and an Enhancer Region
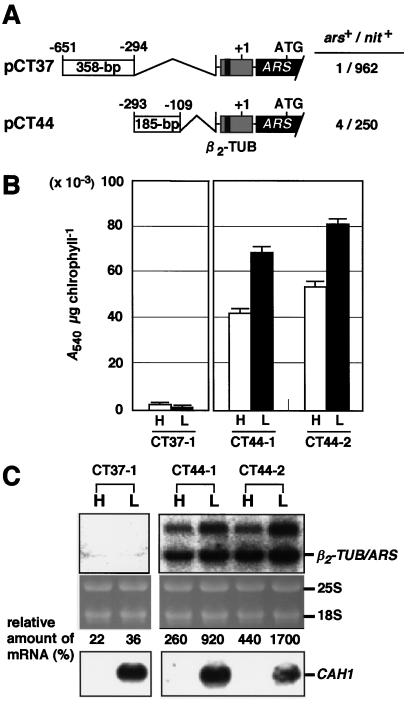
To identify which parts of the 543-bp region function as a silencer or an enhancer, the 543-bp fragment was divided into two parts: a 358-bp region from −651 to −294, and a 185-bp region from −293 to −109. The two fragments were then inserted into pJD100 to generate pCT37 and pCT44 (Fig. 6A). Transformants harboring the chimeric construct pCT37 or pCT44 were named CT37 and CT44, respectively.
Figure 6.
A, Schematic drawings of the chimeric constructs. The CAH1 5′-upstream region between −651 and −109 was divided into two fragments at position −293 and each fragment was fused to the ARS reporter gene driven by the β2-tubulin minimal promoter. The chimeric construct pCT37 contains the 385-bp region from −651 to −294, and pCT44 contains the 185-bp region from −293 to −109, respectively. B, Quantification of arylsulfatase activity in individual transformants. The arylsulfatase activity under high- (H) and low-CO2 conditions (L) was quantified in transformants containing pCT37 (CT37-1) or pCT44 (CT44-1 and CT44-2). Results for two representative CT44 strains among four tested are shown. Bars above the graph indicate the sd for the average of three determinations. C, Northern-blot analysis of β2-TUB/ARS chimeric transcripts in transformants CT37, CT44-1, and CT44-2. Ten micrograms of total RNA from cells exposed to high- (H) or low-CO2 conditions (L) for 4 h was electrophoresed in each lane. Radiolabeled ARS cDNA (upper autoradiographs) or p-CA1-5′ (lower autoradiographs) were used as probes. Values of the relative amount of mRNA represent the percentage of the amount of the chimeric transcripts in CT37 and CT44 over the average amount in three independent T strains.
It was expected that the 358-bp region functioned as an low-CO2-dependent enhancer, because deletion of this region from pCAO3 resulted in a decrease in the level of arylsulfatase activity under low-CO2 conditions (Figs. 1 and 2). Unexpectedly, however, in the presence of this region, the basal activity of the β2-tubulin minimal promoter was repressed under both high- and low-CO2 conditions (CT37-1 in Fig. 6B). Northern-blot analysis revealed that the amount of β2-TUB/ARS mRNA in CT37-1 was less than one-third of that in T strains (Fig. 6C). These results suggest that the 358-bp region functions as a transcriptional silencer independent of the external CO2 level.
In the transformants CT44-1 and CT44-2 harboring pCT44 (Fig. 6A), which does not contain the 358-bp region, arylsulfatase expression was no longer repressed under high-CO2 conditions. The arylsulfatase activity levels in two representatives of four strains tested are shown in Figure 6B (CT44-1 and CT44-2). This result strongly supports our speculation that the region from -651 to -294 functions as a silencer. Furthermore, under low-CO2 conditions, the CT44 strains (CT44-1 and CT44-2) exhibited 8- to 10-fold higher arylsulfatase activity (Fig. 6B) and accumulated 9- to 17-fold higher levels of β2-TUB/ARS mRNA (Fig. 6C) than those in T-1 and T-2 cells containing pJD100 (Fig. 5, B and C), indicating that the 185-bp region functions as a transcriptional enhancer under low-CO2 conditions. These results indicate that both the 358-bp silencer region from −651 to −294 and the 185-bp enhancer region from −293 to −109 are sufficient to regulate the expression of CAH1 in response to the external CO2 level.
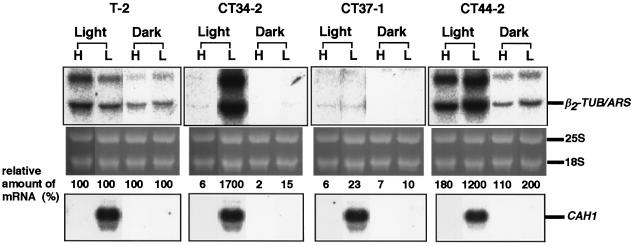
The light responsiveness of these chimeric constructs was also assayed by northern-blot analysis using total RNA samples isolated from cells incubated under high- or low-CO2 conditions in the light or in the dark. In CT34-2, the chimeric transcript accumulated under low-CO2 conditions only in the light as in the case of the endogenous CAH1 (Fig. 7). This result suggests that the 543-bp region is sufficient for light-dependent transcriptional regulation.
Figure 7.
Light-responsive expression of the ARS reporter gene in transgenic C. reinhardtii cells. Cells were incubated under high- (H) or low-CO2 conditions (L) either in the light or in the dark for 4 h. Ten micrograms of RNA from these cells was electrophoresed and blotted onto a membrane, and then hybridized with ARS cDNA (upper autoradiographs) or p-CA1-5′ (lower autoradiographs). Values of the relative amount of mRNA represent the percentage of the amount of the chimeric transcripts in CT34-2, CT37-1 and CT44-2 over the amount in the transformant T-2.
In the presence of the 185-bp region, 2-fold higher levels of β2-TUB/ARS mRNA were expressed even in the dark when the cells were exposed to low-CO2 conditions (CT44-2 in Fig. 7) compared with T-2 in the dark. When CT44-2 cells were cultured under low-CO2 conditions in the light, a 12-fold higher level of β2-TUB/ARS mRNA was detected compared with that in T-2 cells. These findings suggest that the 185-bp region is sufficient to confer light-dependent responsiveness on the β2-tubulin minimal promoter.
To clarify whether the repression under high-CO2 conditions is dependent on light, expression of the chimeric constructs was examined under high-CO2 conditions both in the light and in the dark (Fig. 7). In the presence of the 358-bp region (CT34-2 and CT37-1 in Fig. 7), β2-TUB/ARS mRNA was scarcely detected under high-CO2 conditions regardless of illumination. In CT44-2 harboring pCT44, which lacks the 358-bp region, β2-TUB/ARS mRNA accumulated under high-CO2 conditions both in the light and in the dark. These results indicate that the silencer element represses transcription in a manner independent of exposure to light. Furthermore, when CT37-1 cells were cultured in the dark, the amount of β2-TUB/ARS mRNA that accumulated under either high- or low-CO2 conditions was less than one-tenth of that observed in T-2 cells. This result suggests that the silencer region represses transcription even under low-CO2 conditions in the absence of light.
DISCUSSION
In this study, we fused a series of 5′-nested deletions of the region upstream of CAH1 to an ARS reporter gene and identified regions essential for CO2-responsive gene regulation (Fig. 1). As shown in Figure 2, quantification of arylsulfatase activity in transgenic C. reinhardtii revealed that the 692-bp region between −651 and +41 relative to the transcription start site is sufficient for full induction of the arylsulfatase activity under low-CO2 conditions. Deletion of a 358-bp region (−651 to −294) resulted in a great reduction of the arylsulfatase activity but did not completely abolish CO2-responsiveness (Fig. 2). Therefore, we expected that the 358-bp region might contain an enhancer element functioning under low-CO2 conditions. However, this region could not enhance transcription of the β2-tubulin minimal promoter, and instead was found to repress it (Fig. 6; CT37-1). One possible explanation for this result is that a silencer element located in this region works more dominantly than the enhancer element. No transformants expressing arylsulfatase activity were obtained when cells were transformed with pCAO5 or pCAO6 (Fig. 1). Although there is no evidence that these chimeric constructs were successfully incorporated into the C. reinhardtii genome, it is possible that they lack sequence elements essential to the arylsulfatase expression under low-CO2 conditions, assuming that transformation frequency does not vary among different chimeric constructs.
The 543-bp upstream region between −651 to −109 without any promoter elements, CAAT-box, or TATA-box was sufficient to confer CO2 and light responsiveness on the β2-tubulin minimal promoter (Figs. 5 and 7). This 543-bp region was divided into two parts, a 358-bp silencer region from −651 to −294 and a 185-bp enhancer region from −293 to −109 (Fig. 6). The enhancer region activated the β2-tubulin minimal promoter-driven ARS transcription in a light-dependent manner under low-CO2 conditions (Fig. 7; CT44-2), while the silencer region repressed transcription in a manner independent of light illumination under high-CO2 conditions and independent of the CO2 level in the dark (Fig. 7; CT37-1). It is supposed that, under low-CO2 conditions in the light, only the enhancer is able to achieve a sufficiently high level of gene activation to overcome the silencer effect and CAH1 mRNA accumulates.
Previously, photosynthetic red light has been shown to be essential to activate the CAH1 expression at the mRNA level (Dionisio-Sese et al., 1990). In addition to the photosynthesis-dependent processes, a blue-light-stimulated mechanism is thought to be involved in CAH1 transcript regulation. Also, the CAH1 expression is shown to depend on the phase of the cell cycle (Marcus et al., 1986) and circadian rhythm (Rawat and Moroney, 1995; Fujiwara et al., 1996). It is possible that these regulations might be mediated by the enhancer and silencer regions.
One question arises from comparison of the arylsulfatase expression patterns in cells harboring pCAO4 or pCT44. Why does deletion of the 358-bp region between −651 and −294 from pCT34 result in high-level arylsulfatase expression, as seen in cells harboring pCT44 under high-CO2 conditions, whereas deletion of the same region from pCAO3 results in low-level expression, as seen in cells harboring pCAO4 under low-CO2 conditions? A simple explanation is that region −109 to +41, which is present in pCAO4 but absent from pCT44, functions as a silencer under high-CO2 conditions (CAO4 in Fig. 2). Consistent with this hypothesis, cells harboring pCT44, which contains neither the silencer region from −651 to −294 nor the other possible silencer region from −109 to +41, show no repression of arylsulfatase expression under high-CO2 conditions, whereas cells harboring pCAO4, which contains the silencer region from −109 to +41, show normal repression of arylsulfatase expression.
The 543-bp region that is sufficient for CO2-responsive transcriptional regulation was compared with upstream regions of other low-CO2-inducible genes, such as those that encode mitochondrial β-CA isozymes (Villand et al., 1997) and chloroplast envelope protein LIP-36 (Chen et al., 1997). A conserved CGCGCC sequence, which extends from −319 to −313 in the region upstream of CAH, was found in all of the genes. Additionally, two 12-mer sequences, GGGTTGAANTCCC (−553 to −541 in CAH1) and AACCCCNGNTGCA (−157 to −145), were also found in the upstream regions of β-CA genes. It has been reported that the expression of β-CA1 and β-CA2 is regulated in a manner similar to that of CAH1, not only being responsive to the external CO2 concentration, but also light, acetate, and circadian rhythms (Eriksson et al., 1998). Interestingly, another conserved sequence, AGCGGCTCGC (−168 to −159 in CAH1), was found in the region upstream of CAH2, which is regulated by CO2 and light in a manner opposite to that of CAH1 (Fujiwara et al., 1990). Perhaps these sequence motifs function as CO2- and/or light-responsive regulatory elements in C. reinhardtii.
Two sequence motifs that function in the promoters of higher plants were also detected. The first is a G-box-like sequence motif, CACGTTG, found at −310 to −304. The G-box is a ubiquitous, cis-acting element of plant genes to which bZIP proteins called G-box factors bind (Menkens et al., 1995). It is known that the G-box plays a role in the response of diverse promoters to factors such as light, anaerobiosis, and hormones including abscisic acid, ethylene, and auxin. The second is the sequence ATTTTCAC that is identical to a part of the ATCATTTTCACT light-responsive cis-element box III (Green et al., 1987). This motif lies within the enhancer region (−290 to −283). It is possible that this sequence is involved in the light-dependent transcriptional activation of CAH1.
Our results demonstrate that CAH1 is regulated by enhancer and silencer elements in response to the external CO2 level and light (Figs. 6 and 7). The presence of the enhancer element in the region upstream of CAH1 and the fact that a mutant that does not induce CAH1 under low-CO2 conditions has been isolated (Fukuzawa et al., 1998) strongly suggest that regulatory mechanisms for transcriptional activation are functioning in C. reinhardtii. Considering the existence of Chlorella ellipsoidea mutants in which the CCM is not repressed under high-CO2 conditions (Matsuda and Colman, 1996), negative regulatory mechanisms that repress CCM under CO2-abundant conditions are operating in photosynthetic eukaryotes. In a cyanobacterium, Synechococcus sp. PCC 7942, positive and negative regulatory elements have also been shown in the promoter region of cmpA, which encodes a 42-kD low-CO2-inducible protein (Ronen-Tarazi et al., 1995). These findings strongly suggest that CO2 sensing and signaling mechanisms that control photosynthetic properties are commonly functioning in aquatic photosynthetic organisms.
ACKNOWLEDGMENTS
We thank Dr. John P. Davies for providing pJD54, pJD100, and pJD27, and for helpful suggestions. We also thank Dr. Paul A. Lefebvre for providing strain 5D and plasmid pMN24 and for technical advice.
Footnotes
This work was supported by the Japanese Ministry of Education, Science and Culture (grant nos. 09660357 and 10170219) and by the Japan Society for the Promotion of Science (grant no. JSPS–RFTF97R16001).
LITERATURE CITED
- Aizawa K, Miyachi S. Carbonic anhydrase and CO2 concentrating mechanisms in microalgae and cyanobacteria. FEMS Microbiol Rev. 1986;39:215–233. [Google Scholar]
- Ares M, Howell SH. Cell cycle stage-specific accumulation of mRNAs encoding tubulin and other polypeptides in Chlamydomonas. Proc Natl Acad Sci USA. 1982;79:5577–5581. doi: 10.1073/pnas.79.18.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Price GD. The role of carbonic anhydrase in photosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:369–392. [Google Scholar]
- Chen Z-Y, Burow MD, Mason CB, Moroney JV. A low-CO2-inducible gene encoding an alanine: α-ketoglutarate aminotransferase in Chlamydomonas reinhardtii. Plant Physiol. 1996;112:677–684. doi: 10.1104/pp.112.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z-Y, Lavigne LL, Mason CB, Moroney JV. Cloning and over expression of two cDNAs encoding the low-CO2-inducible chloroplast envelope protein LIP-36 from Chlamydomonas reinhardtii. Plant Physiol. 1997;114:265–273. doi: 10.1104/pp.114.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Davies JP, Grossman AR. Sequences controlling transcription of the Chlamydomonas reinhardtii β2-tubulin gene after deflagellation and during the cell cycle. Mol Cell Biol. 1994;14:5165–5174. doi: 10.1128/mcb.14.8.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Weeks DP, Grossman AR. Expression of the arylsulfatase gene from the β2-tubulin promoter in Chlamydomonas reinhardtii. Nucleic Acids Res. 1992;20:2959–2965. doi: 10.1093/nar/20.12.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos EL, Schilling J, Grossman AR. Structure and expression of the gene encoding the periplasmic arylsulfatase of Chlamydomonas reinhardtii. Mol Gen Genet. 1989;218:229–239. doi: 10.1007/BF00331273. [DOI] [PubMed] [Google Scholar]
- de Hostos EL, Togasaski RK, Grossman A. Purification and biosynthesis of a derepressible periplasmic arylsulfatase from Chlamydomonas reinhardtii. J Cell Biol. 1988;106:29–37. doi: 10.1083/jcb.106.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisio-Sese ML, Fukuzawa H, Miyachi S. Light-induced carbonic anhydrase expression in Chlamydomonas reinhardtii. Plant Physiol. 1990;94:1103–1110. doi: 10.1104/pp.94.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Karlsson J, Ramazanov Z, Gardeström P, Samuelsson G. Discovery of an algal mitochondrial carbonic anhydrase: molecular cloning and characterization of a low-CO2-induced polypeptide in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1996;93:12031–12034. doi: 10.1073/pnas.93.21.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Villand P, Gardeström P, Samuelsson G. Induction and regulation of expression of a low-CO2-induced mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 1998;116:637–641. doi: 10.1104/pp.116.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez E, Schnell R, Ranum LPW, Hussey SC, Silflow CD, Lefebvre PA. Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1989;86:6449–6453. doi: 10.1073/pnas.86.17.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Fukuzawa H, Tachiki A, Miyachi S. Structure and differential expression of two genes encoding carbonic anhydrase in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:9779–9783. doi: 10.1073/pnas.87.24.9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Ishida N, Tsuzuki M. Circadian expression of the carbonic anhydrase gene, Cah1, in Chlamydomonas reinhardtii. Plant Mol Biol. 1996;32:745–749. doi: 10.1007/BF00020215. [DOI] [PubMed] [Google Scholar]
- Fukuzawa H, Fujiwara S, Yamamoto Y, Dionisio-Sese ML, Miyachi S. cDNA cloning, sequence, and expression of carbonic anhydrase in Chlamydomonas reinhardtii: regulation by environmental CO2 concentration. Proc Natl Acad Sci USA. 1990;87:4383–4387. doi: 10.1073/pnas.87.11.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa H, Ishizaki K, Miura K, Matsueda S, Inoue T, Kucho K, Ohyama K. Isolation and characterization of high-CO2 requiring mutants from Chlamydomonas reinhardtii by gene tagging. Can J Bot. 1998;76:1–6. [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ, Kay SA, Chua N-H. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J. 1987;6:2543–2549. doi: 10.1002/j.1460-2075.1987.tb02542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus Y, Schuster G, Michaels A, Kaplan A. Adaptation to CO2 level and changes in the phosphorylation of thylakoid proteins during the cell cycle of Chlamydomonas reinhardtii. Plant Physiol. 1986;80:604–607. doi: 10.1104/pp.80.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Colman B. A new screening method for algal photosynthetic mutants. Plant Physiol. 1996;110:1283–1291. doi: 10.1104/pp.110.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR. The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci. 1995;20:506–510. doi: 10.1016/s0968-0004(00)89118-5. [DOI] [PubMed] [Google Scholar]
- Ohresser M, Matagne RF, Loppes R. Expression of the arylsulfatase reporter gene under the control of the nit1 promoter in Chlamydomonas reinhardtii. Curr Genet. 1997;31:264–271. doi: 10.1007/s002940050204. [DOI] [PubMed] [Google Scholar]
- Rawat M, Moroney JV. The regulation of carbonic anhydrase and ribulose-1,5-bisphosphate carboxylase/oxygenase activase by light and CO2 in Chlamydomonas reinhardtii. Plant Physiol. 1995;109:937–944. doi: 10.1104/pp.109.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen-Tarazi M, Schwarz R, Bouevitch A, Lieman-Hurwitz J, Erez J, Kaplan A. Response of photosynthetic microorganisms to changing ambient concentration of CO2. In: Joint I, editor. Molecular Ecology of Aquatic Microbes. NATO ASI Series. G38. Berlin: Springer-Verlag; 1995. pp. 323–334. [Google Scholar]
- Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1960;46:83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam L-W, Lefebvre PA. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics. 1993;135:375–384. doi: 10.1093/genetics/135.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villand P, Eriksson M, Samuelsson G. Carbon dioxide and light regulation of promoters controlling the expression of mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Biochem J. 1997;327:51–57. doi: 10.1042/bj3270051. [DOI] [PMC free article] [PubMed] [Google Scholar]