Abstract
Post-translational modification of serine/threonine residues in nucleocytoplasmic proteins with GlcNAc (O-GlcNAcylation) is an essential regulatory mechanism in many cellular processes. In Drosophila, null mutants of the Polycomb gene O-GlcNAc transferase (OGT; also known as super sex combs (sxc)) display homeotic phenotypes. To dissect the requirement for O-GlcNAc signaling in Drosophila development, we used CRISPR/Cas9 gene editing to generate rationally designed sxc catalytically hypomorphic or null point mutants. Of the fertile males derived from embryos injected with the CRISPR/Cas9 reagents, 25% produced progeny carrying precise point mutations with no detectable off-target effects. One of these mutants, the catalytically inactive sxcK872M, was recessive lethal, whereas a second mutant, the hypomorphic sxcH537A, was homozygous viable. We observed that reduced total protein O-GlcNAcylation in the sxcH537A mutant is associated with a wing vein phenotype and temperature-dependent lethality. Genetic interaction between sxcH537A and a null allele of Drosophila host cell factor (dHcf), encoding an extensively O-GlcNAcylated transcriptional coactivator, resulted in abnormal scutellar bristle numbers. A similar phenotype was also observed in sxcH537A flies lacking a copy of skuld (skd), a Mediator complex gene known to affect scutellar bristle formation. Interestingly, this phenotype was independent of OGT Polycomb function or dHcf downstream targets. In conclusion, the generation of the endogenous OGT hypomorphic mutant sxcH537A enabled us to identify pleiotropic effects of globally reduced protein O-GlcNAc during Drosophila development. The mutants generated and phenotypes observed in this study provide a platform for discovery of OGT substrates that are critical for Drosophila development.
Keywords: O-linked N-acetylglucosamine (O-GlcNAc), O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT), Drosophila genetics, CRISPR/Cas, polycomb, post-translational modification (PTM), Drosophila development, Host cell factor, hypomorphic sxc mutant, Mediator complex
Introduction
Nucleocytoplasmic post-translational modification of protein serine/threonine residues with GlcNAc, otherwise known as O-GlcNAcylation, is a key regulator of several cellular signaling events (1). O-GlcNAc transfer is mediated by O-GlcNAc transferase (OGT),3 whereas O-GlcNAcase (OGA) removes the modification from proteins. The OGT donor substrate UDP-GlcNAc is one of the critical regulators of O-GlcNAcylation and is a product of the hexosamine biosynthetic pathway (2). Change in flux through the hexosamine biosynthetic pathway downstream of glucose availability leads to altered UDP-GlcNAc levels and consequently impinges upon levels of nucleocytoplasmic protein O-GlcNAcylation (3). Thus, O-GlcNAc signaling is an important transducer of cellular glucose levels, modulating the function of the O-GlcNAcylated substrates by multiple mechanisms, including changes in enzyme activity (4), protein stability (5, 6), oligomerization (7), and solubility (8). Protein O-GlcNAcylation has also been demonstrated to occur co-translationally and was shown to increase the stability of nascent protein chains (9). Modulation of protein function by O-GlcNAcylation ultimately leads to altered transcriptional profiles (10, 11). Increasing evidence associates deregulation of O-GlcNAc signaling with disease states such as cancer, diabetes, and neurodegeneration (12). Point mutations in OGT that segregate with X-linked intellectual disability have recently been described (13, 14).
Loss or knockdown of OGT in metazoa leads to lethality at various stages of development (15–18). Mouse embryonic stem cells are not viable in the absence of ogt, and tissue-specific ogt knockout leads to a range of phenotypes in nervous and immune systems (15, 19, 20). Reduction in OGT levels in Xenopus and zebrafish leads to severe growth defects (17, 18). In Drosophila, OGT (also known as sxc (super sex combs), henceforth referred to only as sxc) mutants die as pharate adults (21). sxc is a Polycomb group (PcG) gene that contributes to control of HOX gene expression and specification of segmental identity (16). The Drosophila embryonic O-GlcNAcome is dynamic, with increased numbers of proteins becoming O-GlcNAc-modified with developmental time (22). Polyhomeotic (Ph), a core component of the PRC1, has been identified as a key O-GlcNAc substrate (8). Reduced O-GlcNAcylation of a Ser/Thr-rich stretch in Ph leads to its aggregation and is associated with misexpression of downstream HOX genes (8). Interestingly, lethality of sxc mutants can be rescued by transgenic overexpression of catalytically defective Drosophila OGT (DmOGT) point mutants (23). When one of the catalytically compromised DmOGT mutants, DmOGTH537A, was used to rescue pupal lethality of sxc nulls, the efficiency of the rescue was about 80% relative to the rescue with DmOGTWT. The in vitro catalytic activity of DmOGTH537A is about 6% of that of DmOGTWT (23). Another point mutant, DmOGTK872M, in which the catalytic lysine residue is mutated, lacks any detectable activity in vitro and does not rescue pupal lethality of sxc mutants. These observations imply that a minimal level of protein O-GlcNAcylation is sufficient to support a complete life cycle in Drosophila. In addition, it also implies that the functionality of the most critical O-GlcNAc substrates in addition to Ph is still retained to a large extent in sxc null flies rescued by the DmOGTH537A mutant.
The recent emergence of CRISPR/Cas9 gene-editing technology allows the generation of flies with precise point mutations in sxc to begin to link phenotypes to mechanisms. Bacteria utilize CRISPR/Cas9 as a defense system against viral pathogens (24). Harnessing the endonuclease activity of Cas9 targeted to a specific genomic target by providing a single guide RNA, dsDNA breaks (DSBs) can be introduced. Repair of these DSBs by homologous recombination can be exploited to create precise point mutants. Since the first report exploiting the CRISPR/Cas9 technique to engineer targeted DSB mutants, this gene-editing strategy has been used to generate null mutants in numerous organisms (25, 26). Generation of animals with precise point mutations has been achieved in zebrafish (27) and mice (28). In Drosophila, CRISPR/Cas9 technology has been used to produce protein nulls (29), to create defined deletions (30), to tag proteins (31), to insert FRT/attP sites in endogenous loci (31), to activate transcription in vivo (32), to decipher functional implications of miRNA-miRNA response element interaction (33), and also to create a mutagenic chain reaction aimed at generating autocatalytic mutations to produce homozygous loss-of-function mutations (34). More recently, point mutants have also been generated by several groups (35–37).
Human host cell factor 1 (Hcf1) has been reported previously as an O-GlcNAc protein (38). A transcriptional regulator, Hcf1 is required as a host cell factor for human herpes simplex virus infection (39). Hcf1 is a large protein that is proteolytically processed by OGT into N-terminal Hcf1N and C-terminal Hcf1C products that regulate different phases of the cell cycle (40). Apart from O-GlcNAcylating Hcf1, mammalian OGT is also essential for this proteolytic processing of Hcf (41). Intriguingly, whereas Drosophila Hcf (dHcf) is also extensively O-GlcNAcylated (22), its proteolytic processing is instead performed by a separate protease, Taspase I (8, 22, 42). O-GlcNAcylation of Hcf has been proposed to prevent its aggregation (8). dHCf is a multifunctional protein, underlined by virtue of genetic interaction of a null allele, dHcfHR1, with components of the PcG, Trithorax (TrxG), and Enhancer of Trithorax and Polycomb (ETP) group (43). Because dHcf is not a proteolytic substrate of OGT in Drosophila, this is an attractive system to dissect the role of dHcf O-GlcNAcylation. Flies null for dHcf display pleiotropic phenotypes that are enhanced or suppressed in various PcG, TrxG, and ETP mutant backgrounds (43). Several phenotypes of the dHcfHR1 mutant are enhanced by an allele of an ETP gene skuld (skd) (43). skd encodes the Drosophila orthologue of human MED13, a component of the Mediator complex, which is a conduit connecting transcription factor signals to RNA polymerase II transcriptional machinery (44, 45).
The effect of reduced as opposed to complete loss of protein O-GlcNAc at the organismal level has not been previously investigated. Here, we investigated the genetic interaction between sxc/OGTH537A and dHcfHR1, a dHcf null allele (43). Using hypomorphic sxcH537A homozygotes, we demonstrate that O-GlcNAc signaling is required for wing vein formation and tolerance to increased temperature. In addition, variation in scutellar bristle numbers is enhanced in sxcH537A mutants simultaneously lacking dHcf or having reduced skd function. In summary, these results outline the requirement of O-GlcNAc signaling in several pathways in Drosophila.
Results
Highly efficient gene editing with CRISPR/Cas9 generates precise sxc mutants
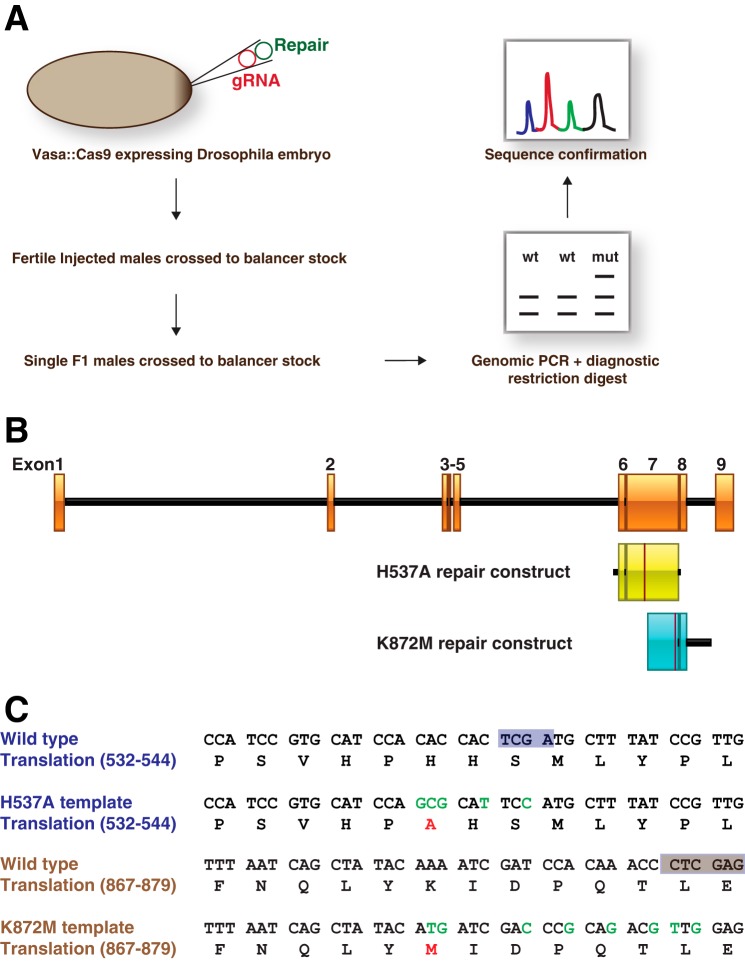
Given that sxc is a maternal effect gene and resides at a locus that is not amenable to producing germ line clones lacking the maternal copy using the FRT-flipase system, current approaches to eliminate the maternal copy have relied on using the UAS-GAL4 system (8, 46). To enable reliable and physiological phenotypic characterization of the requirement of the O-GlcNAc modification for Drosophila development, we embarked on producing a precise hypomorphic OGT point mutant, sxcH537A and a catalytically dead mutant, sxcK872M utilizing the CRISPR/Cas9 gene-editing technology in combination with homologous recombination (Fig. 1A, Table S1). Single guide RNA (sgRNA) was designed using the Zhang laboratory web tool (crispr.mit.edu).4 To facilitate homologous repair–based gene editing, repair constructs carrying the desired OGT hypomorphic (H537A) or catalytically dead (K872M) mutations were cloned into a pGEX6P1 plasmid (Fig. 1B). The homologous arms on either side of the mutations were about 1 kb long, with the repair cassette targeting exon 7 of the OGT genomic region for both of the mutations (Fig. 1B). In addition to the necessary mutations changing the codon to Ala in place of His at position 537 or Met in place of Lys at position 872, silent mutations were introduced in wobble positions of adjacent codons (Fig. 1C). This strategy was employed to decrease the chances of the repaired DNA being subjected to further Cas9 nuclease cleavage and also to enable a robust screening assay exploiting the elimination of TaqI (H537A) or XhoI (K872M) restriction enzyme sites (Fig. 1C).
Figure 1.
Strategy to generate sxcH537A mutants using the CRISPR/Cas9 gene-editing technology. A, experimental outline of the CRISPR/Cas9 homologous recombination scheme adopted to generate sxc mutant flies. gRNA and the respective homologous repair plasmids were injected into vasa::Cas9 embryos (Bloomington stock 51323). F1 males derived from injected embryos were allowed to mate with balancer chromosome stocks, sacrificed, and genotyped using restriction fragment length polymorphism assay to determine the presence of a genetic lesion. Genomic DNA from flies that were resistant to restriction digestion was sequenced to confirm the nature of the lesion. B, sxc genomic region with exons depicted as orange boxes and introns as black lines. The extent of the genomic DNA supplied for homologous repair carrying either the H537A or the K872M mutations is shown in the yellow and blue boxes, respectively. The red line highlighted within each of these boxes marks the site of the introduced mutations in the repair constructs. C, genomic DNA sequence of the repair region carrying the mutation in the WT and mutant scenarios are shown. Below the DNA sequence is the translated protein. The changes that were made in the mutant DNA construct are highlighted in green, and the expected change in protein translation is marked in red. The TaqI and XhoI restriction sites are marked with light purple or brown boxes, respectively. Successful incorporation of the mutant sequence or an indel will lead to the loss of the restriction sites.
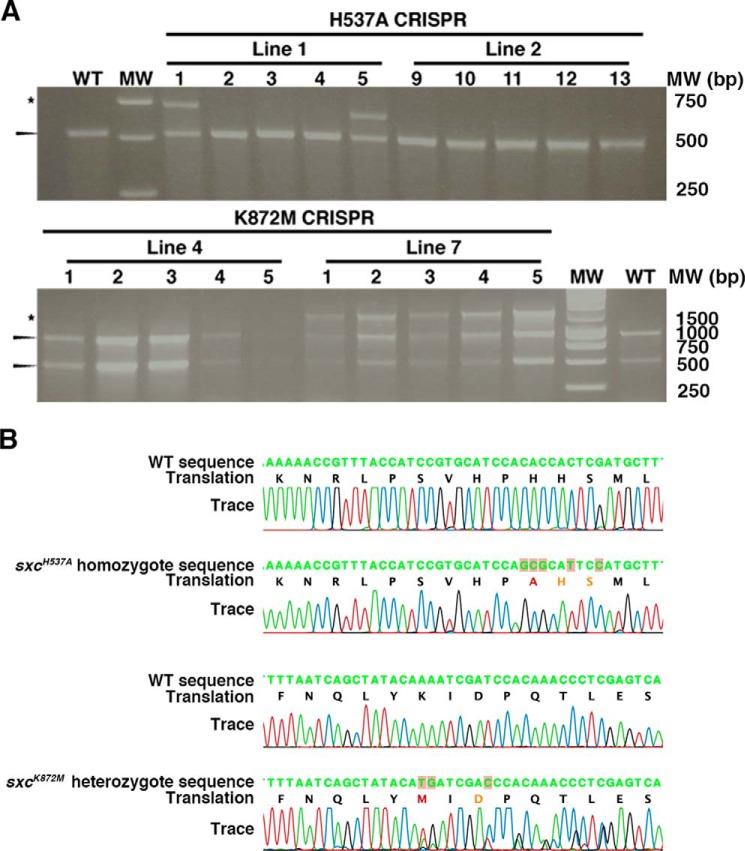
Both the sgRNA and the repair plasmids were injected into the vasa::Cas9 fly line (47). Injected adult males were mated with balancer chromosome stock to eliminate the X chromosome carrying the Cas9 transgene and to balance the putative mutant chromosome. F1 males resulting from this cross were allowed to mate before sacrificing and isolating whole genomic DNA. Isolated genomic DNA was subjected to PCR followed by restriction analyses with TaqI (H537A) or XhoI (K872M). At least five individual F1 males from each of the 23 (H537A) and 8 (K872M) fertile parental lines were assessed in this manner (Table 1). A representative gel demonstrating the restriction assay from two different parental lines for each mutation is shown in Fig. 2A. Two lines were positive with the XhoI restriction assay while screening for the K872M mutation. Sequencing the PCR product confirmed that at least one F1 male from each of these two parental lines was positive for the precise K872M mutation. Thus, the efficiency of generating the K872M precise point mutation was 25%. Neither of the sxcK872M lines produce homozygotes or complement the well-characterized sxc null alleles, sxc1 or sxc6 (48). The sxcK872M is therefore a recessive lethal allele. Thus, the successful generation of such an allele using the CRISPR/Cas9 technique implies that loss of OGT catalysis can be tolerated during male germ cell development.
Table 1.
Efficiency of generating sxcH537A mutants using a CRISPR/Cas9 approach
| Mutant | Parental lines tested, PCR + restriction digestion | Precise mutations | Indels | Efficiency of precise mutation % |
|---|---|---|---|---|
| H537A | 23 | 6 | 4 | 26 |
| K872M | 8 | 2 | 2 | 25 |
Figure 2.
Confirmation of sxcH537A and sxcK872M mutant lines derived by the CRISPR/Cas9 technique. A, representative gels demonstrating the loss of TaqI (above) or XhoI (below) restriction sites in potential sxcH537A or sxcK872M mutants, respectively. Genomic DNA from F1 males was extracted and subjected to PCR amplification followed by restriction digest with TaqI or XhoI. Shown are restriction digests of genomic DNA from five F1 males, each derived from two injected parents. The arrowheads mark the digested band, whereas the asterisk marks the band resistant to TaqI (above) or XhoI (below). B, sequencing chromatograms of WT (top), the putative sxcH537A homozygote line 1.5 genomic DNA (second), WT (third), and the putative sxcK872M heterozygote line 7.11 (bottom). These data confirm the incorporation of a desired mutation that would lead to the His-537 to Ala mutation in addition to the two silent mutations that were introduced into the wobble positions in the adjacent codons. For the Lys-872 to Met mutants, the presence of multiple peaks in the chromatogram demonstrates the heterozygosity of the locus.
Screening for the H537A mutation revealed a total of six lines that were positive in the TaqI restriction assay. Sequencing showed that at least one F1 male from each of these six parental lines was positive for the precise H537A mutation, establishing the rate of generating a precise mutation at 26%. In addition, four of the six lines also carried insertions/deletions leading to sxc null. From the parental line 1, one of the lines that triggered the TaqI assay (line 1.1) was assessed by genomic sequencing and was found to have a 63-bp insertion resulting in a frameshift that would only code for an OGT truncation (residues 1–537). Line 1.1 did not complement either the sxc1 or sxc6 alleles and was found to be recessive lethal. On the other hand, sequencing of line 1.5 heterozygotes confirmed that it was a precise H537A mutation, henceforth referred to as sxcH537A. sxcH537A homozygotes could be derived, and their mutant status was further confirmed by sequencing (Fig. 2B). The codon specifying the His-537 to Ala mutation and the additional wobble mutations were also present in the homozygous sxcH537A mutants. Furthermore, upon sequencing the entire region of the ∼2-kb homologous recombination genomic boundaries, we did not observe nonspecific mutation(s) that might have been introduced during the gene-editing process. A key concern with the use of any gene-editing approach is the possibility of off-target mutagenesis. All of the potential off-targets predicted by the web tool used for gRNA selection were sequenced in the sxcH537A (Table S2) and sxcK872M (Table S3) mutants and confirmed to be WT. Thus, we have achieved highly efficient gene editing with CRISPR/Cas9 to generate sxc hypomorphic mutants in an otherwise endogenous background that will help interrogate the function of O-GlcNAc in development.
Reduced O-GlcNAcylation is associated with wing vein phenotype and developmental lethality
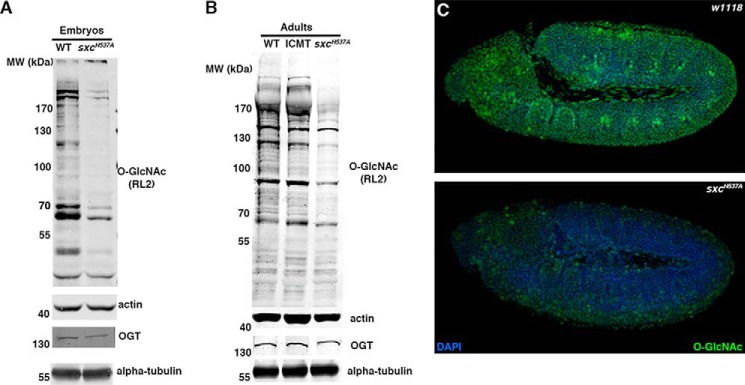
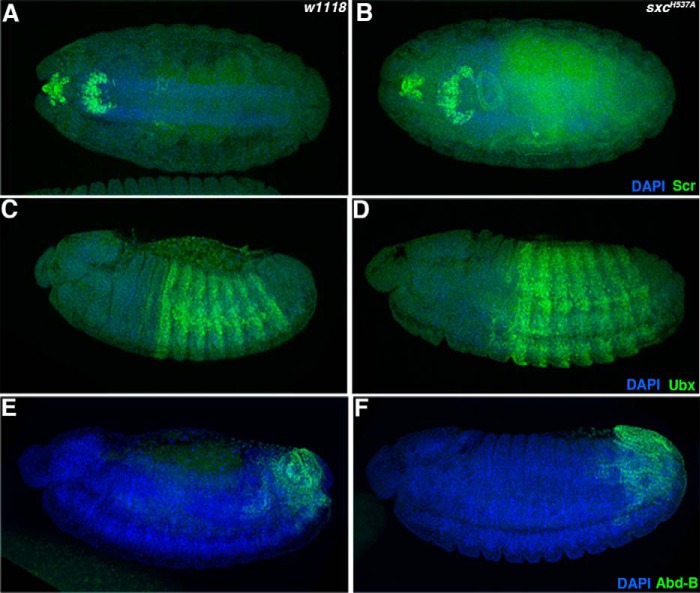
We probed the levels of global O-GlcNAc and OGT in the sxcH537A mutant embryos (Fig. 3A) and adults (Fig. 3B). Immunoblots with a commercial O-GlcNAc antibody (RL2) revealed a large reduction in protein O-GlcNAcylation in F2 embryos that lack WT maternal and zygotic contribution and in adults (Fig. 3, A and B). However, OGT protein levels are comparable between WT and sxcH537A mutant embryos or adults (Fig. 3, A and B). Immunostaining sxcH537A homozygous embryos using RL2 antibody revealed a global reduction in O-GlcNAc levels as compared with the WT embryos (Fig. 3C). However, the reduced O-GlcNAc levels in sxcH537A embryos do not lead to a change in the expression domains of Hox proteins, Scr, Ubx, and Abd-B, as compared with the WT (Fig. 4).
Figure 3.
The hypomorphic sxcH537A mutants have reduced O-GlcNAc levels. A, O-GlcNAc levels are severely reduced in sxcH537A embryos. Either WT or sxcH537A homozygous embryos were collected, dechorionated, lysed, and subjected to SDS-PAGE and immunoblotted with anti-O-GlcNAc (RL2) or anti-OGT antibodies. The blots were normalized with either anti-actin or anti-α-tubulin antibodies, respectively. B, O-GlcNAc levels are severely reduced in sxcH537A adults. WT, balancer (IF/CyO; MKRS/TM6 (ICMT)), and sxcH537A homozygous adults were lysed, and the lysates were used for immunoblotting with anti-O-GlcNAc (RL2) or anti-OGT antibodies. The blots were normalized with either anti-actin or anti-α-tubulin antibodies, respectively. C, WT (w1118; top) or sxcH537A (bottom) homozygous embryos were immunostained with anti-O-GlcNAc antibody (RL2). Shown are stage 9–11 embryos of each of the genotypes.
Figure 4.
Reduced O-GlcNAc levels in sxcH537A mutants does not affect Hox gene expression pattern. Stage 13–14 WT (w1118; A, C, and E) or sxcH537A (B, D, and F) embryos were immunostained with anti-Scr (A and B), anti-Ubx (C and D), or anti-Abd-B (E and F) antibodies. The expression domains of all of these Hox genes tested remained unchanged. All of the embryos are aligned along the anterior-posterior axis with the anterior to the left. Embryos are depicted in either dorsal (A and B) or lateral (C–F) views.
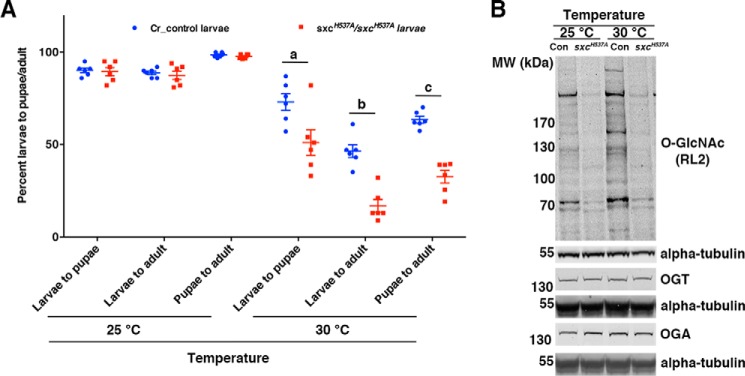
To assess whether reduced O-GlcNAc levels in the sxcH537A mutants resulted in defects during larval/pupal development, Cr control (generation outlined under “Experimental procedures”) or sxcH537A mutant L1 larvae were transferred onto fresh food vials, and the numbers of pupae formed as well as adults eclosed were evaluated. When the larvae were collected from embryos grown at 25 °C, there was no difference in the percentage of larvae developing to pupae or adults between Cr control and sxcH537A mutants (Fig. 5A). Given that increased temperature affects the viability of sxc null flies (46), pupae formation and adult eclosion was also assessed at 30 °C. Larval to pupal or adult development was significantly affected in sxcH537A mutants as compared with Cr control flies at 30 °C (Fig. 5A). Whereas 73 and 46% Cr control larvae develop into pupae and adults, respectively, only 51 and 17% of sxcH537A mutant larvae develop to pupae and adults (Fig. 5A). Pupal to adult development was 63 and 33% in Cr control and sxcH537A mutants, respectively (Fig. 5A). The increased lethality of sxcH537A homozygotes was associated with the inability to increase total O-GlcNAc levels at 30 °C as compared with the Cr control (Fig. 5B), which appears to be independent of OGT or OGA protein levels (Fig. 5B). In summary, it appears that the ability to increase O-GlcNAc levels with an increase in temperature during Drosophila development is protective to the organism. We next went on to investigate whether global reduction in O-GlcNAc levels in the sxcH537A affects dHcf function.
Figure 5.
Reduced O-GlcNAc levels in sxcH537A mutants leads to increased larval/pupal lethality at higher temperature. A, lethality at higher temperature is increased in sxcH537A homozygotes. Either Cr control or sxcH537A F1 larvae (25 per vial, 100 larvae per experiment; n = 6) were transferred to fresh food vials at 25 or 30 °C, and the numbers of pupae formed and adults eclosed were counted. Development to pupae/adults from larvae or to adulthood from pupae was significantly reduced in sxcH537A homozygotes compared with Cr control flies (a and b, p < 0.001; c, p < 0.05; t test with Holm–Sidak correction). B, O-GlcNAc levels remain unaltered at higher temperature in sxcH537A embryos. Age-matched stage 16 Cr control or sxcH537A embryos were collected at either 25 or 30 °C, dechorionated, lysed, and subjected to SDS-PAGE and immunoblotted with anti-O-GlcNAc (RL2), anti-OGT, or anti-OGA antibodies. The blots were normalized with either rabbit anti-α-tubulin (O-GlcNAc blot), mouse anti-α-tubulin (OGT and OGA blots), or antibodies. This blot is representative of three biological replicates.
Hypomorphic OGT phenotype is enhanced on reducing levels of transcriptional modulators
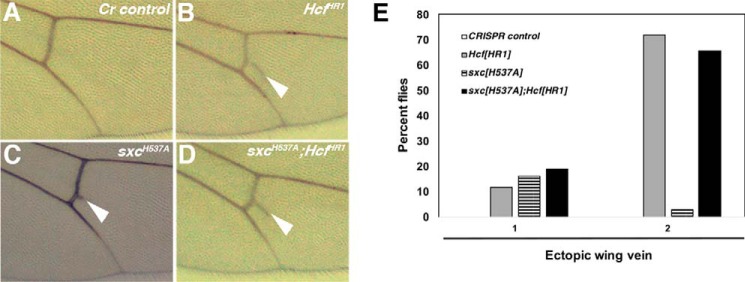
One of the striking phenotypes observed in 22% of sxcH537A adults was an ectopic wing vein emerging from the posterior cross-vein (Fig. 6, A and B). Homozygotes for dHcf null allele, dHcfHR1, display a similar phenotype (43). We therefore assessed the genetic interaction between the dHcfHR1 null allele and the sxcH537A hypomorph, given that dHcf is a well-characterized O-GlcNAcylated protein in humans (41) and Drosophila (22). A previous report has characterized the genetic interaction between skd1 (a hypomorphic recessive lethal skd allele) and the dHcfHR1 allele resulting in enhancement of the ectopic wing vein phenotype, along with extra scutellar bristle and genitalia rotation phenotypes (43). There was no enhancement of the ectopic wing vein phenotype in sxcH537A; dHcfHR1 double homozygotes (Fig. 6, D and E) compared with dHcfHR1 homozygotes (Fig. 6, C and E). Moreover, the genitalia rotation phenotype was not observed in any of the genotypes tested.
Figure 6.
Ectopic wing vein phenotype of dHcfHR1 mutants is not enhanced in sxcH537A mutants. A, image of the wing of an adult fly from the Cr control stock. There is no ectopic wing vein material seen arising from the posterior cross-vein in any of the control fly wings. Also marked are the longitudinal veins (L4 and L5). B, in HcfHR1 homozygotes, ectopic wing vein material is seen deposited in most flies, marked by the white arrowhead. C, in sxcH537A homozygotes, this phenotype is not as penetrant. D, the number of sxcH537A; HcfHR1 double homozygous flies having ectopic wing vein phenotype is comparable with penetrance seen in HcfHR1 homozygotes. E, the number of adult flies having ectopic wing vein deposition arising from the posterior cross-vein from Cr control (white bar), HcfHR1 homozygotes (gray bar), sxcH537A homozygotes (hatched bar), and sxcH537A; HcfHR1 double homozygotes (black bar) were counted. The graph represents the percentage of flies from each of the above genotypes having the ectopic wing vein in either one or both of the wings. None of the Cr control flies have ectopic wing veins, whereas quite a high percentage of HcfHR1 homozygotes display this phenotype. The proportion of sxcH537A; HcfHR1 double homozygotes have similar levels of the ectopic wing vein phenotype.
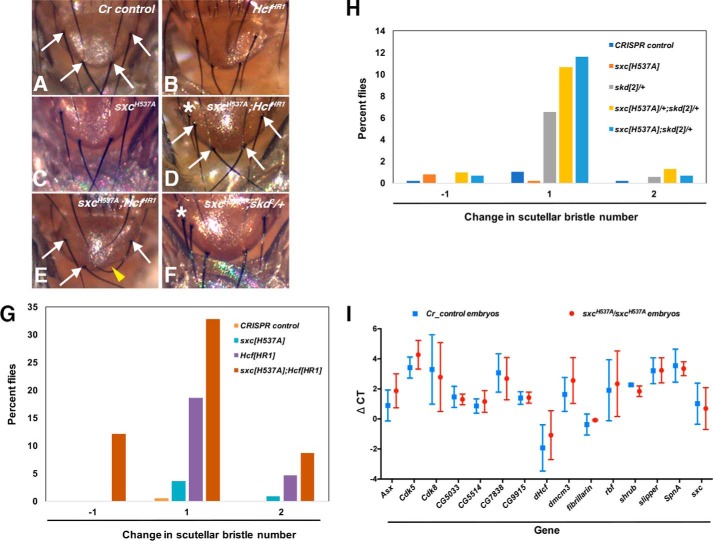
There are four scutellar bristles in most Drosophila species (49). In a previous study, skd1 heterozygotes were found to have normal bristle numbers, whereas about a third of skd1 heterozygotes in a dHcfHR1 background possessed extra scutellar bristles (43). In our experiments, all of the Cr control flies had the normal component of four scutellar bristles (Fig. 7, A, G, and H). On examining sxcH537A homozygotes (n = 111), about 5% of the flies were found to have either one or two extra scutellar bristles (Fig. 7, C and G). In dHcfHR1 homozygotes, the percentage of flies with extra scutellar bristles was 18% (Fig. 7, B and G). Interestingly, 41% of sxcH537A; dHcfHR1 double homozygotes (n = 58) had one or two extra scutellar bristles, whereas 12% were missing a scutellar bristle (Fig. 7, D, E, and G). The defect in flies scored for a missing bristle was the complete loss of the mechanosensory organ as opposed to accidental bristle damage (Fig. 7E). These data therefore demonstrate an interaction between the sxcH537A and dHcfHR1 alleles, specifically in the determination and/or function of the sensory organ precursor (SOP) cells essential for bristle formation. Furthermore, we also investigated whether the deregulation of scutellar bristle number is affected by PcG (Polycomb; Pc) and TrxG (brahma; brm) genes (Table 2). On reducing one copy of Pc (Pc1, an amorphic recessive lethal allele) in either sxcH537A/+ or sxcH537A background, a normal number of scutellar bristles was observed, indicating no genetic interaction with respect to this phenotype (Table 2). However, the super sex combs phenotype (sex combs in the second and third pairs of thoracic legs) observed in Pc1/+ flies (21% of all males scored) was enhanced in an sxcH537A/+ (56%) or sxcH537A (66%) background, revealing a role of the catalytic activity of sxc in Polycomb function (Table 3). Cr control or sxcH537A flies did not exhibit the super sex combs phenotype (Table 3). On performing a genetic interaction between sxcH537A and brm2 alleles, only a small percentage of sxcH537A/+; brm2/+ (5%) or sxcH537A; brm2/+ (4.8%) flies were found to have the scutellar bristle phenotype (Table 2).
Figure 7.
sxcH537A extra scutellar bristle phenotype is enhanced in Hcf null background. A, Cr control; B, HcfHR1 homozygotes; C, sxcH537A homozygotes; D, sxcH537A; HcfHR1 double homozygotes; E, sxcH537A; HcfHR1 double homozygotes. F, sxcH537A; skd2/+ flies were treated with Flynap, and scutellar images were captured. The white arrows mark the four scutellar bristles. Homozygous HcfHR1 or sxcH537A homozygotes predominantly possess four scutellar bristles. However, in sxcH537A; HcfHR1 double homozygotes, over half of the flies have either extra (D) or missing (E) scutellar bristle(s). Flies of the genotype sxcH537A; skd2/+ also have slightly increased extra scutellar bristle phenotype (F). The extra scutellar bristle is marked with an asterisk in D and F. The yellow arrowhead marks the missing scutellar bristle in E. G, the number of scutellar bristles in adult flies from Cr control (orange bars, n = 199), HcfHR1 (blue bars, n = 43), sxcH537A (purple bars, n = 111), and sxcH537A; HcfHR1 double homozygotes (brown bars, n = 58) were counted. The graph represents the percentage of flies from each of the above genotypes having either one less (−1) or one (1) or two (2) more than the four scutellar bristles mostly observed in control flies. All of the control (Cr control) flies have only four scutellar bristles with minor deviation toward an extra one or two scutellar bristles in HcfHR1 or sxcH537A homozygotes. However, a significant proportion of sxcH537A; HcfHR1 double homozygotes have varying scutellar bristle numbers. H, the number of scutellar bristles in adult flies from Cr control (dark blue, n = 492), sxcH537A (orange bars, n = 507), skd2/+ (gray bars, n = 184), sxcH537A/+; skd2/+ (yellow bars, n = 309), and sxcH537A; skd2/+ (light blue bars, n = 146) flies were counted. The graph represents the percentage of flies from each of the above genotypes having either one less (−1) or one (1) or two (2) more than the four scutellar bristles mostly observed in control flies. There is a modest increase in the percentage of sxcH537A/+; skd2/+ or sxcH537A; skd2/+ flies having extra scutellar bristles as compared with skd2/+ flies. I, quantitative real-time PCR was performed to detect the transcripts potentially downstream of dHcf apart from sxc and dHcf transcripts. The graph represents the ΔCT values of the respective transcripts in either Cr control (blue squares) or sxcH537A (red circles) stage 7–11 embryos. The experiments were repeated three times, and no significant difference was observed in the levels of any of the transcripts assessed (t test with Holm–Sidak correction).
Table 2.
Scutellar bristle phenotype of sxcH537A mutants is not affected by reduction of Polycomb function
Flies of the respective genotypes were scored for the number of scutellar bristles. The percentage of flies exhibiting either less or more than the normal scutellar bristle number of four are listed.
| Genotype | Number of flies scored | Percentage of flies with decreased scutellar bristle number | Percentage of flies with increased scutellar bristle number |
|---|---|---|---|
| Cr control | 388 | 0.3 | 1 |
| sxcH537A | 302 | 1 | 0.3 |
| Pc1/+ | 247 | 0.4 | 0 |
| sxcH537A/+; Pc1/+ | 197 | 0 | 0.5 |
| sxcH537A; Pc1/+ | 150 | 2 | 0 |
| brm2/+ | 99 | 0 | 0 |
| sxcH537A/+; brm2/+ | 302 | 0.3 | 5 |
| sxcH537A; brm2/+ | 104 | 0 | 4.8 |
Table 3.
Super sex combs phenotype of Pc1 is enhanced in sxcH537A background
Males of the respective genotypes were scored for the presence of sex combs on second and third thoracic legs. Percentage of flies exhibiting the super sex combs phenotype are listed.
| Genotype | Number of males scored | Percentage of males, super sex combs phenotype |
|---|---|---|
| Cr control | 203 | 0 |
| sxcH537A | 153 | 0 |
| Pc1/+ | 124 | 21 |
| sxcH537A/+; Pc1/+ | 115 | 56 |
| sxcH537A; Pc1/+ | 80 | 66 |
To investigate whether reduced O-GlcNAc levels in the sxcH537A homozygotes also impinge upon skd function or vice versa, interaction between sxcH537A and a hypomorphic recessive lethal skd allele, skd2 (the skd1 stock is not publicly available), was assessed. About 7% of the skd2 heterozygotes (n = 184) displayed extra scutellar bristles. Slightly higher abnormal scutellar bristle numbers were observed in both sxcH537A/+; skd2/+ double heterozygotes (13%, n = 309; Fig. 7H) and sxcH537A; skd2/+ flies (13%, n = 146; Fig. 7, F and H), indicating a genetic interaction between the sxcH537A and skd2 alleles, albeit to a lesser extent than that observed between the sxcH537A and dHcfHR1 alleles. Adults of the genotype sxcH537A; skd2/+; dHcfHR1 could not be derived, implying that loss of OGT and dHcf activity in skd heterozygotes leads to developmental lethality.
In light of the genetic interaction between sxcH537A and dHcfHR1 alleles, we investigated whether dHcf function is affected in sxcH537A mutants. Knockdown of dHcf in S2 cells was previously reported to lead to transcriptional up-regulation of fibrillarin and CG5033 (50). There is also evidence that dHcf interacts with Drosophila elongation factors dE2F1 and dE2F2 (51). Data from human cell lines implicate a role for HCF1 in transcriptional control of E2F-bound genes (52). Transcription of several genes (Table S4), including ASXL, CDK5, and CDK8, is deregulated on HCF1 knockdown (52). We investigated the changes in transcript levels of dHcf/HCF1 downstream targets derived from both of these studies (50, 52) in Cr control and sxcH537A embryos. The transcript levels of all of the dHcf/HCF1 downstream targets investigated remained unchanged when compared with those in Cr control embryos (Fig. 7I). In summary, these data implicate sxc, dHcf, and skd in a common pathway that is responsible for scutellar bristle determination. Nevertheless, the molecular details of how reduced O-GlcNAc levels in the sxcH537A mutants contribute to this phenotype remain to be investigated.
Discussion
Using CRISPR/Cas9 technology, we have been able to produce an important tool in the form of a hypomorphic sxc mutant. This is particularly useful, given that sxc is a maternal effect gene and that its genomic locus impedes production of mutants that lack maternal as well as zygotic gene products using the Flipase/FRT system (53). Previous studies have circumvented this hurdle using various transgenic approaches (8, 23, 46). However, nonendogenous, constitutive expression of transgenic OGT can lead to artifacts. In addition, our previous observation that minimal OGT glycosyltransferase activity is sufficient to sustain Drosophila development through multiple generations was an added impetus to produce catalytically deficient OGT mutants in an otherwise endogenous background (23). The sxcH537A mutant provides a platform to investigate the role of OGT catalytic activity in Drosophila development. Utilizing a restriction assay to screen for potential mutants, we have harnessed the CRISPR/Cas9 gene-editing technology to create precise sxc point mutations. We were able to produce two precise sxc point mutations, sxcH537A and sxcK872M, at an efficiency of 25%, starting from fertile injected males for each of the mutations.
Phenotypic analysis of the sxcK872M mutant that codes for a catalytically dead mutant could not be pursued because this mutation is recessive lethal. This observation is supported by the fact that the previously published sxc1 or sxc6 mutant alleles cannot be complemented by the sxcK872M allele. In addition, we were able to derive null alleles from the H537A gRNA injections that also did not complement sxc1 or sxc6 lethality. These results establish the specificity of the gRNAs used in our CRISPR/Cas9 approach to create the sxc point mutations. Specificity of the mutagenesis was also highlighted by the significant reduction of O-GlcNAc levels in sxcH537A homozygotes. The lack of derepression of Hox genes in sxcH537A F2 embryos reiterates our earlier finding that a minimal level of O-GlcNAcylation is sufficient to support Drosophila development (23). The data obtained in the current work eliminate the potential artifacts of overexpression and the possibility that WT and mutant forms of OGT form heteromeric complexes. In this scenario of significantly reduced global O-GlcNAc that does not lead to Hox gene derepression, it will be interesting to investigate the dynamics of Ph O-GlcNAcylation and consequently its aggregation/loss of function (8). This is relevant because the loss of Ph function leads to derepression of Hox genes in embryos and larval imaginal discs (16, 54).
The reduced levels of protein O-GlcNAcylation in sxcH537A homozygotes are associated with larval and pupal lethality at elevated temperatures. It has previously been reported that elevated temperature leads to lethality during embryogenesis in maternal or zygotic sxc mutants (46). The endogenous sxcH537A mutant has enabled us to identify the specific requirement of catalytic activity of OGT as opposed to the OGT interactome, at post-embryonic stages of development. It opens up the possibility that the O-GlcNAc modification, akin to glycosylation in the secretory pathway, is essential for stabilizing misfolded proteins at higher temperatures. Heat stress in mammalian cells is associated with increased cellular O-GlcNAc levels. Reducing OGT catalytic activity by genetic or chemical means renders the cells more susceptible to thermal stress (55, 56). Heat-stressed OGT−/− mouse embryonic fibroblasts have reduced levels of specific heat-shock proteins (57). Downstream of OGT/O-GlcNAc cycling, the levels of these heat-shock proteins are proposed to be regulated by GSK3β-dependent phosphorylation of heat-shock factor 1 (57). Several proteins with diverse functions were demonstrated to be hyper-O-GlcNAc–modified and up-regulated on heat stress in monkey fibroblasts (58). Heat stress–induced heat-shock protein 70 has been described to bind to O-GlcNAcylated proteins, preventing their misfolding (59). Increased hsp70 levels on heat stress are probably downstream of O-GlcNAcylated Sp1 (60). Nevertheless, the mechanistic details of how O-GlcNAc–dependent thermoprotection occurs in Drosophila require further analyses.
Scutellar bristles arise from progenitors in the larval wing imaginal disc epithelium known as SOPs. Clusters of cells that express proneural genes of the achaete-scute (ac-sc) complex are subjected to selection by Notch-Delta signaling–mediated lateral inhibition. This process leads to specification of SOPs (61–64). Once specified, the SOPs go on to differentiate into mechanosensory organs via a complex, orchestrated pathway (65). A GATA-1 family transcription factor, Pannier (Pnr), is an activator of ac-sc, specifically required for the specification of the dorsocentral bristles that are nonscutellar mechanosensory organs (66). The extra bristle phenotype of the pnrD1 allele is enhanced by the Pc1 allele, implying PcG-mediated control of SOP determination (66). However, we do not observe an interaction between Pc1 and sxcH537A with respect to bristle numbers in the scutellum. Moreover, the phenotype observed in sxcH537A; HcfHR1 double homozygotes is one wherein there is increased variation in the number of scutellar bristles, with some flies also having a reduced number of bristles. These observations therefore imply that the specification of scutellar SOPs in sxcH537A flies is not via the influence of OGT catalytic activity on PcG function.
The extra scutellar bristle phenotype is enhanced significantly in sxcH537A; HcfHR1 double homozygotes when compared with either sxcH537A or HcfHR1 homozygotes. This phenotype is also enhanced in HcfHR1 homozygotes that have a single copy of the skd1 allele (43). However, we observe a weaker genetic interaction between sxcH537A and skd2 alleles as compared with the interaction between sxcH537A and HcfHR1. This implies that the pathways potentially affected by reduced O-GlcNAc levels in sxcH537A flies are able to tolerate the presence of a hypomorphic copy of skd more effectively than an HcfHR1 null background. None of the other phenotypes described for the skd1/+; HcfHR1 flies were recapitulated in either sxcH537A; HcfHR1 or sxcH537A; skd1/+ animals, indicating specific roles for O-GlcNAc in dHcf and/or Mediator complex function. Nevertheless, reduction in O-GlcNAc levels is not tolerated in animals both having reduced skd levels and lacking dHcf. Interestingly, point mutations in human OGT and MED12, another Mediator component, co-segregate in individuals affected with X-linked intellectual disability (XLID) (13, 14). Mutations have also been identified in human HCF1 that are associated with X-linked mental retardation (67, 68). Moreover, rare variants of both MED12 and HCF1 were shared only by the affected siblings in a family affected by a severe form of XLID (69). We observe a common pathway being affected when orthologs of XLID genes are used in genetic interaction experiments. Therefore, the scutellar bristle number phenotype is potentially a readout in Drosophila to genetically dissect the contribution of OGT/O-GlcNAc function in XLID.
In conclusion, we have demonstrated successful generation of catalytically hypomorphic sxc mutants using a simple, transferable assay to screen for mutagenesis by CRISPR/Cas9 gene editing. Analysis of the sxcH537A thus obtained has helped uncover several phenotypes that are a result of a reduction in protein O-GlcNAcylation. Either the reduced O-GlcNAcylation of dHcf or conversely decreased dHcf function impinging upon OGT function(s) affects normal scutellar bristle numbers. Moreover, Drosophila embryos possess a dynamic O-GlcNAcome that could contribute to phenotypes described in this study and others that remain to be discovered (22, 70, 71). Apart from other applications, the hypomorphic sxcH537A mutant is a tool that can be used to investigate the role of dHcf O-GlcNAc, potentially developed as a model to investigate the role of OGT in XLID and investigate O-GlcNAc occupancy in the Ph Ser/Thr-rich stretch. Moreover, investigating the O-GlcNAcome in sxcH537A mutants would help in narrowing down key transducers of O-GlcNAc signaling in Drosophila development. This analysis will be particularly informative in eliminating the functionally inconsequential O-GlcNAcylation events and establish the role of O-GlcNAc signaling in Drosophila development.
Experimental procedures
Drosophila genetics, scutellar imaging, and immunostaining
The following stocks from the Bloomington Drosophila Stock Centre were used: w1118 WT, vasa::Cas9 (BL51323), HcfHR1, skd2/TM6, brm2/TM6, and Pc1/TM1. CRISPR/Cas9 injections were performed at the University of Cambridge fly facility into embryos from the vasa::Cas9 line (Bloomington stock: 51323). Microinjections were carried out with a mixture of 100 ng/μl gRNA plasmid with 300 ng/μl repair construct mix. Injected founder male flies were crossed with IF/CyO; MKRS/TM6 balancer stock. At least 10 male F1 sxc*/CyO potential germ line mutants were crossed again with IF/CyO; MKRS/TM6 virgins. This ensured the outcrossing of the vasa::Cas9-carrying X chromosome. The F1 males were then snap-frozen for genotyping as outlined below. Stocks of either sxcH537A/CyO or sxcK872M/CyO were established from F2 progeny of sequence-confirmed mutants. Furthermore, the genotype of sxcH537A homozygotes derived from the sxcH537A/CyO stock was confirmed. In addition, all of the predicted off-target sites were PCR-amplified and checked for the presence of any lesions compared with the genomic DNA from the BL51323 line. None of the predicted off-target sites were found to have mutations. To perform Western blots with whole flies, either WT or the sxcH537A flies were snap-frozen and processed as outlined below. The control flies (Cr control) were derived by crossing the flies from the stock used for microinjection (Bloomington Stock: BL51323) using a similar crossing scheme as that used to derive the sxcH537A homozygotes. This ensured maintenance of the genetic background and the loss of the vasa::Cas9-carrying X chromosome.
The number of scutellar bristles was assessed in the genotypes Cr control, sxcH537A, HcfHR1, skd2/+, sxcH537A; HcfHR1, sxcH537A; skd2/+, sxcH537A/+; skd2/+, brm2/+, sxcH537A/+; brm2/+, sxcH537A; brm2/+, Pc1/+, sxcH537A/+; Pc1/+, and sxcH537A; Pc1/+, using a Motic SMZ microscope. Images from representative flies treated with FlyNap (Carolina Biological Sciences) were acquired using a Leica E24 HD dissection microscope. The presence of sex combs on second and third thoracic legs was scored for the genotypes Cr control, sxcH537A, Pc1/+, sxcH537A/+; Pc1/+, and sxcH537A; Pc1/+, using a Motic SMZ microscope.
Fixing and immunostaining of embryos was performed as described previously (72). The following antibodies were used: mouse anti-O-GlcNAc (1:250, RL2, Abcam) and mouse antibodies from the Developmental Studies Hybridoma Bank (anti-Scr (1:50), anti-Abd-B (1:50), and anti-Ubx (1:50)) with the respective fluorescent secondary antibodies (Invitrogen). Microscopic images were obtained with Leica SP8 confocal microscope and processed using Volocity (Improvision) software.
Cloning and restriction fragment length polymorphism assay to detect mutants
gRNA sites were chosen using the website crispr.mit.edu,4 and annealing oligonucleotides were designed with the appropriate overhangs and cloned into the BpiI-cut pCFD3-dU63gRNA vector (Table S1). Inserts were confirmed by DNA sequencing.
Repair templates were generated by PCR of either a 2160-bp (H537A) or a 2063-bp (K872M) region of the Drosophila genome from S2 cell genomic DNA using GoTaq G2 polymerase (Promega). The PCR product was cloned into pGEX6P1 plasmid. The insert sequence was confirmed by DNA sequencing. The desired mutation, in addition to the silent mutations, was introduced by site-directed mutagenesis following the Stratagene QuikChange mutagenesis kit but using KOD Hot Start polymerase (Novagen) and subsequently confirmed by DNA sequencing.
To assess and confirm generation of CRISPR/Cas9 gene editing, candidate homo-/heterozygous flies were frozen in Eppendorf tubes and homogenized in 50 μl of squishing buffer (10 mm Tris-HCl, pH 8, 1 mm EDTA, 25 mm NaCl, and 200 μg/ml freshly added Proteinase K (Roche Applied Science)). The homogenate was incubated at 37 °C for 30 min, followed by inactivation of Proteinase K at 95 °C for 3 min, and centrifuged. 1 μl of supernatant was used per 25-μl PCR. 5 μl of PCRs was digested with TaqI (H537A PCR) or XhoI (K872M PCR), followed by electrophoresis of the digested products. Samples that were resistant to TaqI and XhoI digestion were sequenced. A second PCR was performed on potential heterozygous CRISPR mutants with primer pairs out with the repair construct to confirm that the observed sequencing result was not due to random integration of the repair plasmid. The second PCR product was also sequenced. To determine any potential mutagenesis at any of the predicted off-target sites, PCRs were performed with the requisite primers (Table S2), followed by sequencing.
Eclosion rate experiments
For eclosion rate experiments, Cr control or sxcH537A homozygote flies were transferred to apple juice agar plates thinly smeared with yeast paste at either 25 or 30 °C. After an overnight collection, 25 F1 larvae were transferred to fresh food vials. Four such vials were set up per biological replicate (n = 6; a total of 600 F1 larvae were thus scored for each genotype). The number of pupae formed was assessed by counting the number of pupal cases per food vial. In addition, the number of adult flies eclosing from each vial was also recorded. We report the percentage of F1 larvae forming pupae/adults and the number of pupae giving rise to adults. t tests were performed for statistical analyses.
To harvest embryos for Western blotting, embryos were collected for 1 h and further aged (to stage 16) for either 13.5 or 11 h at 25 or 30 °C, respectively, before dechorionating and snap-freezing the embryos. The frozen embryos were subjected to Western blot analysis as outlined below.
Western blotting
To prepare total embryo lysates, embryos were collected on apple juice agar plates at 25 °C overnight (0–16 h). The embryos thus collected were dechorionated with bleach and snap-frozen in dry ice. The frozen embryos were homogenized in lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Triton X-100, 1 μm GlcNAcstatin C, 5 mm sodium fluoride, 2 mm sodium orthovanadate, 1 mm benzamidine, 0.2 mm phenylmethylsulfonyl fluoride, 5 μm leupeptin, and 1 mm DTT). For Western blots, five anesthetized adult flies were frozen on dry ice. The frozen flies were homogenized in 50 μl of lysis buffer, followed by the addition of an equal volume of 3× SDS Laemmli buffer. Lysates were then heated for 5 min at 95 °C and centrifuged at 16,000 × g for 10 min, and supernatants were collected. Protein concentrations were estimated using the 660-nm protein assay (Thermo Scientific). 30 μg of the crude lysate was subjected to SDS-PAGE and transferred onto nitrocellulose membrane before immunoblotting with RL2 (1:1000; Abcam), rabbit anti-OGT (H-300, 1:1000; Santa Cruz Biotechnology, Inc.), rabbit anti-OGA (1:1000; Sigma), mouse anti-α-tubulin (1:10,000; DSHB), and/or rabbit anti-actin (1:5000; Sigma) and the respective IR dye conjugated secondary antibodies (LI-COR or Life Technologies; 1:10,000).
Quantitative real-time PCR
Quantitative real-time PCR was performed with Cr control and sxcH537A homozygous embryos. Cr control and sxcH537A were transferred to apple juice agar plates thinly smeared with yeast paste at 25 °C. Fresh plates were used to collect embryos for 2 h. The plates were then changed, and the embryos were allowed to age for 3 h. RNA isolation (Qiagen RNAeasy Plus kit), quantification (Nanodrop), and cDNA generation (Bio-Rad Iscript cDNA synthesis kit) were then performed as per the manufacturer's instructions. cDNA equivalent to 100 pg of input total RNA was subjected to quantitative real-time PCR (Quanta Biosciences) in a Bio-Rad CFX Connect system. Primers used for dHcf downstream targets (Table S4) were from either published literature (50) or an online tool for Drosophila primers (73). The reported threshold cycle (CT) values were used to compute ΔCT values as described (74). Three biological replicates were used to determine the ΔCT values, and t tests with the Holm–Sidak method to correct for multiple comparisons were used for statistical analysis.
Author contributions
D. M., A. T. F., and D. M. F. v. A. conceived the study. D. M. performed the Drosophila experiments and phenotypic analyses. A. T. F. performed molecular biology. D. M., A. T. F., and D. M. F. v. A. interpreted the data and wrote the manuscript.
Acknowledgments
Fly stocks were obtained from the Drosophila Stock Center (Bloomington, IN). Microinjections were performed at the University of Cambridge Department of Genetics Fly Facility. We thank the University Imaging Facility, Dundee, which is supported by Wellcome Trust Technology Platform Award 097945/B/11/Z and MRC Next Generation Optical Microscopy Award MR/K015869/1.
This work was supported by Wellcome Trust Investigator Award 110061 (to D.M.F.v.A.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S4.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- OGT
- O-GlcNAc transferase
- OGA
- O-GlcNAcase
- DSB
- dsDNA break
- gRNA
- guide RNA
- sgRNA
- single guide RNA
- bp
- base pair(s)
- kb
- kilobase pair(s)
- SOP
- sensory organ precursor
- XLID
- X-linked intellectual disability.
References
- 1. Hart G. W. (2014) Three decades of research on O-GlcNAcylation: a major nutrient sensor that regulates signaling, transcription and cellular metabolism. Front. Endocrinol. 5, 183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hart G. W., Slawson C., Ramirez-Correa G., and Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 10.1146/annurev-biochem-060608-102511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buse M. G., Robinson K. A., Marshall B. A., Hresko R. C., and Mueckler M. M. (2002) Enhanced O-GlcNAc protein modification is associated with insulin resistance in GLUT1-overexpressing muscles. Am. J. Physiol. Endocrinol. Metab. 283, E241–E250 10.1152/ajpendo.00060.2002 [DOI] [PubMed] [Google Scholar]
- 4. Erickson J. R., Pereira L., Wang L., Han G., Ferguson A., Dao K., Copeland R. J., Despa F., Hart G. W., Ripplinger C. M., and Bers D. M. (2013) Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature 502, 372–376 10.1038/nature12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Olivier-Van Stichelen S., and Hanover J. A. (2014) X-inactivation normalizes O-GlcNAc transferase levels and generates an O-GlcNAc-depleted Barr body. Front. Genet. 5, 256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chu C. S., Lo P. W., Yeh Y. H., Hsu P. H., Peng S. H., Teng Y. C., Kang M. L., Wong C. H., and Juan L. J. (2014) O-GlcNAcylation regulates EZH2 protein stability and function. Proc. Natl. Acad. Sci. U.S.A. 111, 1355–1360 10.1073/pnas.1323226111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yi W., Clark P. M., Mason D. E., Keenan M. C., Hill C., Goddard W. A. 3rd, Peters E. C., Driggers E. M., and Hsieh-Wilson L. C. (2012) Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337, 975–980 10.1126/science.1222278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gambetta M. C., and Müller J. (2014) O-GlcNAcylation prevents aggregation of the Polycomb group repressor Polyhomeotic. Dev. Cell 31, 629–639 10.1016/j.devcel.2014.10.020 [DOI] [PubMed] [Google Scholar]
- 9. Zhu Y., Liu T. W., Cecioni S., Eskandari R., Zandberg W. F., and Vocadlo D. J. (2015) O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat. Chem. Biol. 11, 319–325 10.1038/nchembio.1774 [DOI] [PubMed] [Google Scholar]
- 10. Vella P., Scelfo A., Jammula S., Chiacchiera F., Williams K., Cuomo A., Roberto A., Christensen J., Bonaldi T., Helin K., and Pasini D. (2013) Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell 49, 645–656 10.1016/j.molcel.2012.12.019 [DOI] [PubMed] [Google Scholar]
- 11. Jang H., Kim T. W., Yoon S., Choi S. Y., Kang T. W., Kim S. Y., Kwon Y. W., Cho E. J., and Youn H. D. (2012) O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell 11, 62–74 10.1016/j.stem.2012.03.001 [DOI] [PubMed] [Google Scholar]
- 12. Bond M. R., and Hanover J. A. (2015) A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol. 208, 869–880 10.1083/jcb.201501101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bouazzi H., Lesca G., Trujillo C., Alwasiyah M. K., and Munnich A. (2015) Nonsyndromic X-linked intellectual deficiency in three brothers with a novel MED12 missense mutation [c.5922G>T (p.Glu1974His)]. Clin. Case Rep. 3, 604–609 10.1002/ccr3.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Niranjan T. S., Skinner C., May M., Turner T., Rose R., Stevenson R., Schwartz C. E., and Wang T. (2015) Affected kindred analysis of human X chromosome exomes to identify novel X-linked intellectual disability genes. PLoS One 10, e0116454 10.1371/journal.pone.0116454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shafi R., Iyer S. P., Ellies L. G., O'Donnell N., Marek K. W., Chui D., Hart G. W., and Marth J. D. (2000) The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 10.1073/pnas.100471497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gambetta M. C., Oktaba K., and Müller J. (2009) Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325, 93–96 10.1126/science.1169727 [DOI] [PubMed] [Google Scholar]
- 17. Kenwrick S., Amaya E., and Papalopulu N. (2004) Pilot morpholino screen in Xenopus tropicalis identifies a novel gene involved in head development. Dev. Dyn. 229, 289–299 10.1002/dvdy.10440 [DOI] [PubMed] [Google Scholar]
- 18. Webster D. M., Teo C. F., Sun Y., Wloga D., Gay S., Klonowski K. D., Wells L., and Dougan S. T. (2009) O-GlcNAc modifications regulate cell survival and epiboly during zebrafish development. BMC Dev. Biol. 9, 28 10.1186/1471-213X-9-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O'Donnell N., Zachara N. E., Hart G. W., and Marth J. D. (2004) Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell Biol. 24, 1680–1690 10.1128/MCB.24.4.1680-1690.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Swamy M., Pathak S., Grzes K. M., Damerow S., Sinclair L. V., van Aalten D. M., and Cantrell D. A. (2016) Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat. Immunol. 17, 712–720 10.1038/ni.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ingham P. W. (1984) A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell 37, 815–823 10.1016/0092-8674(84)90416-1 [DOI] [PubMed] [Google Scholar]
- 22. Mariappa D., Selvan N., Borodkin V., Alonso J., Ferenbach A. T., Shepherd C., Navratilova I. H., and vanAalten D. M. F. (2015) A mutant O-GlcNAcase as a probe to reveal global dynamics of protein O-GlcNAcylation during Drosophila embryonic development. Biochem. J. 470, 255–262 10.1042/BJ20150610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mariappa D., Zheng X., Schimpl M., Raimi O., Ferenbach A. T., Müller H. A., and van Aalten D. M. (2015) Dual functionality of O-GlcNAc transferase is required for Drosophila development. Open Biol. 5, 150234 10.1098/rsob.150234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A., and Horvath P. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 [DOI] [PubMed] [Google Scholar]
- 25. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., and Charpentier E. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Doudna J. A., and Charpentier E. (2014) Genome editing: the new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- 27. Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Kaini P., Sander J. D., Joung J. K., Peterson R. T., and Yeh J. R. (2013) Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One 8, e68708 10.1371/journal.pone.0068708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., Zhang F., and Jaenisch R. (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bassett A. R., Tibbit C., Ponting C. P., and Liu J. L. (2013) Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4, 220–228 10.1016/j.celrep.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., Harrison M. M., Wildonger J., and O'Connor-Giles K. M. (2013) Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035 10.1534/genetics.113.152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baena-Lopez L. A., Alexandre C., Mitchell A., Pasakarnis L., and Vincent J. P. (2013) Accelerated homologous recombination and subsequent genome modification in Drosophila. Development 140, 4818–4825 10.1242/dev.100933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin S., Ewen-Campen B., Ni X., Housden B. E., and Perrimon N. (2015) In vivo transcriptional activation using CRISPR/Cas9 in Drosophila. Genetics 201, 433–442 10.1534/genetics.115.181065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bassett A. R., Azzam G., Wheatley L., Tibbit C., Rajakumar T., McGowan S., Stanger N., Ewels P. A., Taylor S., Ponting C. P., Liu J. L., Sauka-Spengler T., and Fulga T. A. (2014) Understanding functional miRNA-target interactions in vivo by site-specific genome engineering. Nat. Commun. 5, 4640 10.1038/ncomms5640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gantz V. M., and Bier E. (2015) Genome editing: the mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348, 442–444 10.1126/science.aaa5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zimmer C. T., Garrood W. T., Puinean A. M., Eckel-Zimmer M., Williamson M. S., Davies T. G., and Bass C. (2016) A CRISPR/Cas9 mediated point mutation in the α6 subunit of the nicotinic acetylcholine receptor confers resistance to spinosad in Drosophila melanogaster. Insect. Biochem. Mol. Biol. 73, 62–69 10.1016/j.ibmb.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mao C. X., Wen X., Jin S., and Zhang Y. Q. (2017) Increased acetylation of microtubules rescues human tau-induced microtubule defects and neuromuscular junction abnormalities in Drosophila. Dis. Model Mech. 10, 1245–1252 10.1242/dmm.028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Douris V., Papapostolou K. M., Ilias A., Roditakis E., Kounadi S., Riga M., Nauen R., and Vontas J. (2017) Investigation of the contribution of RyR target-site mutations in diamide resistance by CRISPR/Cas9 genome modification in Drosophila. Insect. Biochem. Mol. Biol. 87, 127–135 10.1016/j.ibmb.2017.06.013 [DOI] [PubMed] [Google Scholar]
- 38. Ruan H. B., Han X., Li M. D., Singh J. P., Qian K., Azarhoush S., Zhao L., Bennett A. M., Samuel V. T., Wu J., Yates J. R. 3rd, Yang X. (2012) O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 16, 226–237 10.1016/j.cmet.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kristie T. M., Liang Y., and Vogel J. L. (2010) Control of α-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim. Biophys. Acta 1799, 257–265 10.1016/j.bbagrm.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Julien E., and Herr W. (2003) Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 22, 2360–2369 10.1093/emboj/cdg242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Capotosti F., Guernier S., Lammers F., Waridel P., Cai Y., Jin J., Conaway J. W., Conaway R. C., and Herr W. (2011) O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 144, 376–388 [DOI] [PubMed] [Google Scholar]
- 42. Capotosti F., Hsieh J. J., and Herr W. (2007) Species selectivity of mixed-lineage leukemia/trithorax and HCF proteolytic maturation pathways. Mol. Cell Biol. 27, 7063–7072 10.1128/MCB.00769-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rodriguez-Jato S., Busturia A., and Herr W. (2011) Drosophila melanogaster dHCF interacts with both PcG and TrxG epigenetic regulators. PLoS One 6, e27479 10.1371/journal.pone.0027479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Treisman J. (2001) Drosophila homologues of the transcriptional coactivation complex subunits TRAP240 and TRAP230 are required for identical processes in eye-antennal disc development. Development 128, 603–615 [DOI] [PubMed] [Google Scholar]
- 45. Yin J. W., and Wang G. (2014) The Mediator complex: a master coordinator of transcription and cell lineage development. Development 141, 977–987 10.1242/dev.098392 [DOI] [PubMed] [Google Scholar]
- 46. Radermacher P. T., Myachina F., Bosshardt F., Pandey R., Mariappa D., Müller H. A., and Lehner C. F. (2014) O-GlcNAc reports ambient temperature and confers heat resistance on ectotherm development. Proc. Natl. Acad. Sci. U.S.A. 111, 5592–5597 10.1073/pnas.1322396111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M., and O'Connor-Giles K. M. (2014) Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sinclair D. A., Syrzycka M., Macauley M. S., Rastgardani T., Komljenovic I., Vocadlo D. J., Brock H. W., and Honda B. M. (2009) Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. U.S.A. 106, 13427–13432 10.1073/pnas.0904638106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sheldon B. L., and Milton M. K. (1972) Studies on the scutellar bristles of Drosophila melanogaster. II. Long-term selection for high bristle number in the Oregon RC strain and correlated responses in abdominal chaetae. Genetics 71, 567–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Furrer M., Balbi M., Albarca-Aguilera M., Gallant M., Herr W., and Gallant P. (2010) Drosophila Myc interacts with host cell factor (dHCF) to activate transcription and control growth. J. Biol. Chem. 285, 39623–39636 10.1074/jbc.M110.140467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tyagi S., Chabes A. L., Wysocka J., and Herr W. (2007) E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol. Cell 27, 107–119 10.1016/j.molcel.2007.05.030 [DOI] [PubMed] [Google Scholar]
- 52. Parker J. B., Yin H., Vinckevicius A., and Chakravarti D. (2014) Host cell factor-1 recruitment to E2F-bound and cell-cycle-control genes is mediated by THAP11 and ZNF143. Cell Rep. 9, 967–982 10.1016/j.celrep.2014.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chou T. B., and Perrimon N. (1996) The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144, 1673–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dura J. M., and Ingham P. (1988) Tissue- and stage-specific control of homeotic and segmentation gene expression in Drosophila embryos by the polyhomeotic gene. Development 103, 733–741 [DOI] [PubMed] [Google Scholar]
- 55. Sohn K. C., Lee K. Y., Park J. E., and Do S. I. (2004) OGT functions as a catalytic chaperone under heat stress response: a unique defense role of OGT in hyperthermia. Biochem. Biophys. Res. Commun. 322, 1045–1051 10.1016/j.bbrc.2004.08.023 [DOI] [PubMed] [Google Scholar]
- 56. Zachara N. E., O'Donnell N., Cheung W. D., Mercer J. J., Marth J. D., and Hart G. W. (2004) Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress: a survival response of mammalian cells. J. Biol. Chem. 279, 30133–30142 10.1074/jbc.M403773200 [DOI] [PubMed] [Google Scholar]
- 57. Kazemi Z., Chang H., Haserodt S., McKen C., and Zachara N. E. (2010) O-linked β-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-3β-dependent manner. J. Biol. Chem. 285, 39096–39107 10.1074/jbc.M110.131102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zachara N. E., Molina H., Wong K. Y., Pandey A., and Hart G. W. (2011) The dynamic stress-induced “O-GlcNAc-ome” highlights functions for O-GlcNAc in regulating DNA damage/repair and other cellular pathways. Amino Acids 40, 793–808 10.1007/s00726-010-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guinez C., Mir A. M., Leroy Y., Cacan R., Michalski J. C., and Lefebvre T. (2007) Hsp70-GlcNAc-binding activity is released by stress, proteasome inhibition, and protein misfolding. Biochem. Biophys. Res. Commun. 361, 414–420 10.1016/j.bbrc.2007.07.020 [DOI] [PubMed] [Google Scholar]
- 60. Lim K. H., and Chang H. I. (2006) O-linked N-acetylglucosamine suppresses thermal aggregation of Sp1. FEBS Lett. 580, 4645–4652 10.1016/j.febslet.2006.07.040 [DOI] [PubMed] [Google Scholar]
- 61. Hartenstein V., and Posakony J. W. (1989) Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development 107, 389–405 [DOI] [PubMed] [Google Scholar]
- 62. Brennan K., Tateson R., Lieber T., Couso J. P., Zecchini V., and Arias A. M. (1999) The abruptex mutations of notch disrupt the establishment of proneural clusters in Drosophila. Dev. Biol. 216, 230–242 10.1006/dbio.1999.9501 [DOI] [PubMed] [Google Scholar]
- 63. Romani S., Campuzano S., Macagno E. R., and Modolell J. (1989) Expression of achaete and scute genes in Drosophila imaginal discs and their function in sensory organ development. Genes Dev. 3, 997–1007 10.1101/gad.3.7.997 [DOI] [PubMed] [Google Scholar]
- 64. Culí J., and Modolell J. (1998) Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 12, 2036–2047 10.1101/gad.12.13.2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. García-Bellido A., and de Celis J. F. (2009) The complex tale of the achaete-scute complex: a paradigmatic case in the analysis of gene organization and function during development. Genetics 182, 631–639 10.1534/genetics.109.104083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Biryukova I., and Heitzler P. (2008) Drosophila C-terminal binding protein, dCtBP is required for sensory organ prepattern and sharpens proneural transcriptional activity of the GATA factor Pnr. Dev. Biol. 323, 64–75 10.1016/j.ydbio.2008.08.014 [DOI] [PubMed] [Google Scholar]
- 67. Huang L., Jolly L. A., Willis-Owen S., Gardner A., Kumar R., Douglas E., Shoubridge C., Wieczorek D., Tzschach A., Cohen M., Hackett A., Field M., Froyen G., Hu H., Haas S. A., et al. (2012) A noncoding, regulatory mutation implicates HCFC1 in nonsyndromic intellectual disability. Am. J. Hum. Genet. 91, 694–702 10.1016/j.ajhg.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yu H. C., Sloan J. L., Scharer G., Brebner A., Quintana A. M., Achilly N. P., Manoli I., Coughlin C. R. 2nd, Geiger E. A., Schneck U., Watkins D., Suormala T., Van Hove J. L., Fowler B., Baumgartner M. R., et al. (2013) An X-linked cobalamin disorder caused by mutations in transcriptional coregulator HCFC1. Am. J. Hum. Genet. 93, 506–514 10.1016/j.ajhg.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Prescott T. E., Kulseth M. A., Heimdal K. R., Stadheim B., Hopp E., Gambin T., Coban Akdemir Z. H., Jhangiani S. N., Muzny D. M., Gibbs R. A., Lupski J. R., and Stray-Pedersen A. (2016) Two male sibs with severe micrognathia and a missense variant in MED12. Eur. J. Med. Genet. 59, 367–372 10.1016/j.ejmg.2016.06.001 [DOI] [PubMed] [Google Scholar]
- 70. Selvan N., Williamson R., Mariappa D., Campbell D. G., Gourlay R., Ferenbach A. T., Aristotelous T., Hopkins-Navratilova I., Trost M., and van Aalten D. M. F. (2017) A mutant O-GlcNAcase enriches Drosophila developmental regulators. Nat. Chem. Biol. 13, 882–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu T. W., Myschyshyn M., Sinclair D. A., Cecioni S., Beja K., Honda B. M., Morin R. D., and Vocadlo D. J. (2017) Genome-wide chemical mapping of O-GlcNAcylated proteins in Drosophila melanogaster. Nat. Chem. Biol. 13, 161–167 10.1038/nchembio.2247 [DOI] [PubMed] [Google Scholar]
- 72. Müller H. A. (2008) Immunolabeling of embryos. Methods Mol. Biol. 420, 207–218 10.1007/978-1-59745-583-1_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hu Y., Sopko R., Foos M., Kelley C., Flockhart I., Ammeux N., Wang X., Perkins L., Perrimon N., and Mohr S. E. (2013) FlyPrimerBank: an online database for Drosophila melanogaster gene expression analysis and knockdown evaluation of RNAi reagents. G3 3, 1607–1616 10.1534/g3.113.007021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]