Abstract
The proprotein convertase subtilisin/kexin type-9 (PCSK9) plays a central role in cardiovascular disease (CVD) by degrading hepatic low-density lipoprotein receptor (LDLR). As such, loss-of-function (LOF) PCSK9 variants that fail to exit the endoplasmic reticulum (ER) increase hepatic LDLR levels and lower the risk of developing CVD. The retention of misfolded protein in the ER can cause ER stress and activate the unfolded protein response (UPR). In this study, we investigated whether a variety of LOF PCSK9 variants that are retained in the ER can cause ER stress and hepatic cytotoxicity. Although overexpression of these PCSK9 variants caused an accumulation in the ER of hepatocytes, UPR activation or apoptosis was not observed. Furthermore, ER retention of endogenous PCSK9 via splice switching also failed to induce the UPR. Consistent with these in vitro studies, overexpression of PCSK9 in the livers of mice had no impact on UPR activation. To elucidate the cellular mechanism to explain these surprising findings, we observed that the 94-kDa glucose-regulated protein (GRP94) sequesters PCSK9 away from the 78-kDa glucose-regulated protein (GRP78), the major activator of the UPR. As a result, GRP94 knockdown increased the stability of GRP78–PCSK9 complex and resulted in UPR activation following overexpression of ER-retained PCSK9 variants relative to WT secreted controls. Given that overexpression of these LOF PCSK9 variants does not cause UPR activation under normal homeostatic conditions, therapeutic strategies aimed at blocking the autocatalytic cleavage of PCSK9 in the ER represent a viable strategy for reducing circulating PCSK9.
Keywords: proprotein convertase subtilisin/kexin type 9 (PCSK9), endoplasmic reticulum stress (ER stress), cardiovascular disease, hepatocyte, GRP78
Introduction
The discovery of PCSK92 has provided a novel therapeutic target for the management of CVD (1, 2). PCSK9 is mainly expressed and secreted by liver hepatocytes where it degrades the LDLR and promotes elevated circulating LDL levels (3, 4), a well-known risk factor of cardiovascular dysfunction (5, 6). Following its characterization, genetic screens revealed that gain-of-function (GOF) mutations in the PCSK9 gene represented a third locus associated with autosomal-dominant hypercholesterolemia (7). In addition to the discovery of the first GOF variant, PCSK9S127R, further investigations led to the identification of a wide range of LOF PCSK9 variants (8, 9). In contrast to GOF mutations, which enhance PCSK9-mediated LDLR degradation and promote increased circulating LDL levels, LOF mutations increase hepatic LDLR expression and reduce circulating LDL levels (10). The Atherosclerosis Risk in Communities (ARIC) study reported that nonsense (C679X/Y142X) and missense (R46L) mutations in PCSK9 were associated with an 88 and 47% reduction in the risk of developing coronary heart disease, respectively (11). Additional LOF variants such as PCSK9G236S, PCSK9N354I, and PCSK9Q152H were subsequently identified and extensively studied in vitro (10, 12). The phenotype associated with such LOF mutations occurs due to the inability of the ER-resident nascent pro-PCSK9 to undergo autocatalytic cleavage and maturation in the ER, thereby leading to its retention. This was found to drive LDLR expression and accounts for the low circulating LDL levels found in patients harboring such mutations (10, 12).
There is now ample evidence supporting the notion that the retention of PCSK9 in the ER leads to a significant reduction of circulating LDL levels. In addition, we have recently demonstrated that ER stress blocks the secretion of PCSK9 in cultured hepatocytes and mice, and this was associated with increased hepatic LDLR expression and reduced circulating LDL levels (13). Furthermore, the retention of PCSK9 in the ER via GRP94 promotes elevated hepatic LDLR levels (14).
Despite the potential benefits of this strategy for reducing circulating LDL levels, ER storage diseases (ERSDs) are a common end point of ER protein retention and contribute to a wide range of human disorders (15, 16). The accumulation of retained/misfolded proteins in the ER leads to ER stress and triggers the UPR (17–19). A well-described form of ERSD occurs as a result of ER arginine vasopressin (VP) retention arising from mutations such as VPG14R and VPG17V. These naturally occurring mutations play a central role in the development of familial neurohypophyseal diabetes insipidus in a manner dependent on ER stress and subsequent UPR activation (20, 21). The UPR signaling cascade consists of three major transducers located at the ER membrane that are activated upon dissociation of GRP78 from their intra-ER domains. These “arms of the UPR” include (a) inositol-requiring protein-1α (IRE1α), which splices and activates X-box–binding protein-1 (XBP1), (b) activating transcription factor-6 (ATF6), and (c) protein kinase RNA (PKR)–like ER kinase (PERK), which promotes the expression of the proapoptotic CCAAT/enhancer–binding protein homologous protein (CHOP). Collectively, this cascade of UPR mediators acts to increase the folding capacity of the ER while reducing its burden by blocking global synthesis and nascent polypeptide influx (18).
Despite the well-documented phenomenon of ERSD, we now report that the ER retention of LOF PCSK9 variants does not cause UPR activation or apoptosis. This hypothesis was tested using a number of LOF PCSK9 variants, including the naturally occurring PCSK9Q152H, in HuH7, HK-2, and N2a cell lines. Furthermore, ER stress and apoptosis were examined in cells treated with a splice-switching oligomer (SSO) to induce the retention of an endogenously expressed PCSK9 splice variant (PCSK9SV) lacking its catalytic domain, which is required for autocatalytic cleavage (22). Finally, coimmunoprecipitation experiments show that GRP94 masks ER-resident pro-PCSK9 from GRP78, which is widely known to act as the major sensor of UPR activation (23, 24). To substantiate this observation, ER PCSK9 retention was shown to cause ER stress/UPR activation following knockdown of GRP94.
Results
Retention of PCSK9 variants in the ER does not induce the UPR
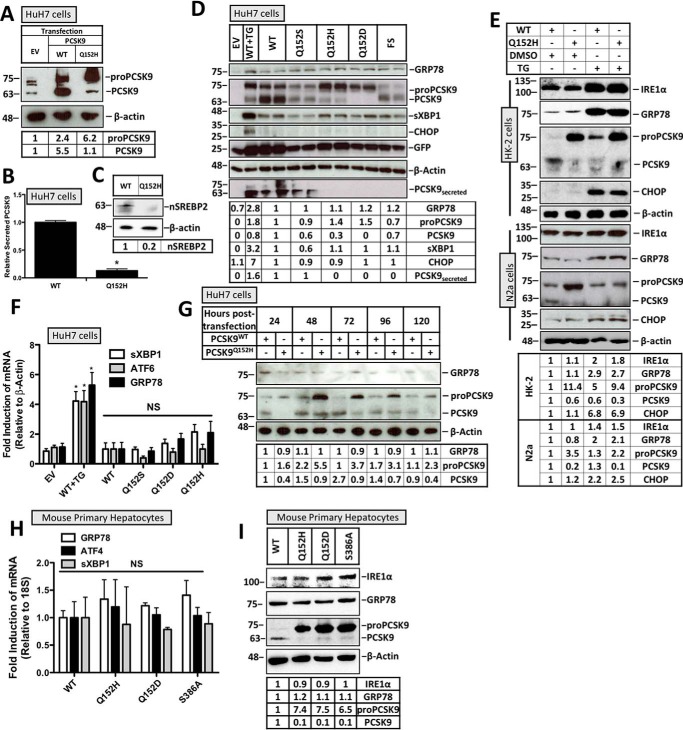
Cultured HuH7 hepatocytes, known to express PCSK9, were transiently transfected with either wildtype PCSK9 (PCSK9WT) or PCSK9Q152H. Endogenous PCSK9 expression was then compared with the cytomegalovirus (CMV)-driven V5-labeled PCSK9 constructs by immunoblotting using an anti-PCSK9 antibody (Fig. 1A). Cells transfected with PCSK9Q152H exhibited 6.2-fold greater levels of ER-resident pro-PCSK9 than that of endogenously expressed PCSK9 found in empty vector (EV)–transfected controls and ∼3-fold greater levels than PCSK9WT-transfected controls. To confirm the cellular retention of PCSK9Q152H, ELISAs were performed on the media from the PCSK9-transfected cells and demonstrated an 87% reduction in the secreted form of the protein (Fig. 1B). Activation of the ER stress–inducible transcription factor SREBP2 (25, 26), a regulator of PCSK9 expression (27), was also examined in PCSK9Q152H-transfected HuH7 cells and found to be 5-fold down-regulated compared with PCSK9WT-transfected control (Fig. 1C). To investigate the effect of PCSK9 variants on UPR activation, HuH7 cells cotransfected with the ER activated indicator (ERAI) plasmid in addition to plasmids encoding a variety of PCSK9 variants were examined via immunoblotting of GRP78 and CHOP. The ERAI plasmid encodes an ER stress–inducible FLAG-XBP1 and was used as an additional tool to characterize UPR activation in our model (13, 28). A group of cells were also treated with the ER stress–inducing agent thapsigargin (TG; 100 nm) to serve as a benchmark of UPR activation. PCSK9 variants examined in this experiment included PCSK9WT and PCSK9Q152S, which undergo autocatalytic cleavage and secretion, as well as PCSK9Q152H, PCSK9Q152D (29), and a C-terminal PCSK9 frameshift variant (PCSK9FS),3 all of which fail to exit the ER. Despite the retention of these PCSK9 variants in the ER, UPR activation was not observed as determined by the absence of GRP78, activated spliced XBP1 (sXBP1), and CHOP induction at the protein level (Fig. 1D). As expected, cells exposed to TG showed increased protein expression of these UPR markers. Furthermore, to investigate whether cells expressing variant PCSK9Q152H exhibited increased susceptibility to UPR activation, transfected cells were also treated with TG. Consistent with data from HuH7 cells, immunoblots of HK-2 and N2a cell lysates confirm that retention of PCSK9Q152H fails to induce UPR activation (Fig. 1E). Furthermore, the level of UPR induction by TG treatment was similar in cells transfected with PCSK9WT and PCSK9Q152H, demonstrating that ER retention of PCSK9 did not enhance the ability of TG to activate the UPR. Additionally, real-time PCR experiments demonstrated that mRNA levels of sXBP1, ATF6, and GRP78 were not significantly different (Fig. 1F). The temporal effect of ER PCSK9 retention on UPR activation was also examined via immunoblotting for GRP78 (Fig. 1G). These data demonstrate that the expression level of PCSK9Q152H in the ER was greatest 48 h post-transfection. Despite this observation, GRP78 expression was not induced by PCSK9Q152H at any of the time points examined. To lend support to our observations made in HuH7, HK-2, and N2a immortalized cell lines, freshly isolated primary mouse hepatocytes transfected with PCSK9 variants PCSK9Q152H, PCSK9Q152D, and PCSK9S386A were also examined for UPR activation. In addition to the aforementioned variants, which fail to undergo autocatalytic cleavage due to mutations at the site of cleavage, PCSK9S386A is an ER-retained variant that fails to undergo autocatalytic cleavage due to a point mutation in the catalytic domain (30). Consistent with our findings in immortalized cell lines, real-time PCR and immunoblot data reveal that the UPR is no more active in cells expressing ER-retained PCSK9 variants than in those expressing the WT secreted form of the protein (Fig. 1, H and I).
Figure 1.
Retention of PCSK9 variants in the ER does not induce the UPR. HuH7 cells were seeded in DMEM and transfected with either EV control, WT, Q152H, Q152S, Q152H, Q152D, or FS V5-labeled PCSK9 variants for 48 h. Cells were also treated with TG (100 nm) for 24 h to serve as a positive control for UPR activation. In addition to V5-PCSK9 transfection, HuH7 cells were also cotransfected with the ERAI plasmid encoding FLAG-XBP1. A, CMV-driven PCSK9 expression was compared with endogenous PCSK9 expression in HuH7 cells. B, an ELISA was run on the media from HuH7 cells transfected with either PCSK9WT or PCSK9Q152H. C, SREBP2 activation, in the form of nuclear SREBP2 (nSREBP2), was also examined in PCSK9Q152H-transfected cells and compared with controls via immunoblotting. D and F, immunoblot and real-time PCR analyses of these cells were performed for ER stress markers ATF6, GRP78, sXBP1, and CHOP. Immunoblot analysis was also completed on V5-PCSK9 in whole cell lysate and medium to examine PCSK9 maturation and secretion, respectively. GFP, originating from the bicistronic pIRES2-EGFP V5-PCSK9 plasmids, served as a transfection control. E, HK-2 and N2a cells transfected with either PCSK9WT or PCSK9Q152H were also examined for UPR activation in the presence or absence of TG. G, a time course experiment was carried out in HuH7 cells transfected with either PCSK9WT or PCSK9Q152H to identify the time point yielding the greatest level of PCSK9 retention. H and I, mouse primary hepatocytes were also transfected with PCSK9Q152H, PCSK9Q152D, and PCSK9S386A and examined for ER stress marker expression of GRP78, ATF4, sXBP1, and IRE1α via real-time PCR and immunoblotting. Differences between treatments were assessed with unpaired Student's t tests. All values are represented as means with error bars representing S.D. B, *, p < 0.05 versus WT. F, *, p < 0.05 versus WT. F and H, nonsignificant (NS).
Retention of VP variants induces the UPR in hepatocytes
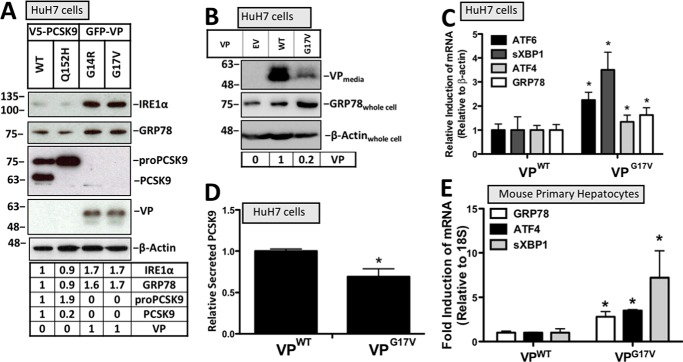
To assess whether the accumulation of misfolded proteins does indeed cause UPR activation in hepatocytes, HuH7 cells transfected with either PCSK9WT or its ER-retention variant PCSK9Q152H were compared with cells transfected with two VP variants, VPG14R and VPG17V, known to induce ER stress in N2a cells (20, 21). Consistent with previous reports, our data demonstrate that ER retention of VP leads to UPR activation as determined by immunoblotting for GRP78 and IRE1α (Fig. 2A). The secretion status of VP in these transfected cells was also examined via immunoblotting the media for VP. A greater level of VP secretion occurred in cells expressing VPWT compared with cells expressing VPG17V, which is consistent with increased ER retention of VPG17V (Fig. 2B). Furthermore, increased intracellular expression of GRP78 was observed in cells expressing VPG17V compared with those expressing VPWT control. Similar to the immunoblot data in Fig. 2, A and B, a significant increase in mRNA levels of sXBP1, ATF4, ATF6, and GRP78 was observed in HuH7 cells transfected with the variant VPG17V (Fig. 2C). Given that chemical ER stress blocks the secretion of PCSK9 from hepatocytes (13), ELISA was used to determine whether PCSK9 secretion status was affected in cells expressing VPG17V. Consistent with the observed VPG17V-induced UPR activation, these cells also secreted significantly less PCSK9 (Fig. 2D). In addition to HuH7 cells, the induction of UPR markers GRP78, ATF4, and sXBP1 was also observed in mouse primary hepatocytes transfected with VPG17V relative to those transfected with VPWT control (Fig. 2E).
Figure 2.
Retention of vasopressin variants induces the UPR. A and C, the relative expression of UPR markers IRE1α, GRP78, ATF6, sXBP1, and ATF4 were examined using immunoblotting and real-time PCR. B, the status of cellular VP retention was examined via immunoblotting of the media from cells transfected with VPWT or VPG17V. D, given that ER stress blocks PCSK9 secretion (13), ELISAs were also used to determine whether VPG17V reduced the content of PCSK9 in the media from HuH7 cells. E, mouse primary hepatocytes were also transfected with VPWT and VPG17V and examined for UPR marker expression of GRP78, ATF4, and sXBP1 using real-time PCR. Differences between treatments were assessed with unpaired Student's t tests. All values are represented as means, and error bars represent S.D. C and E, *, p < 0.05 versus VPWT.
ER retention of a PCSK9 splice variant via RNAi does not cause UPR activation
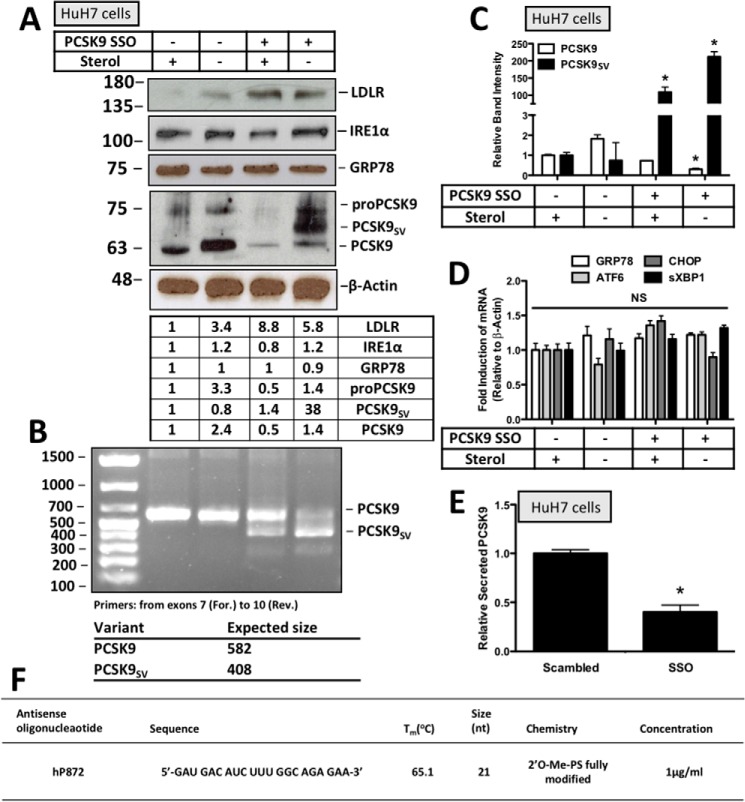
To confirm and extend our overexpression studies using exogenously expressed PCSK9 variants, UPR activation was examined in response to the retention of endogenously expressed PCSK9SV in HuH7 cells. To achieve this, HuH7 cells were transfected with an SSO that promotes the expression of a PCSK9SV lacking exon 8 that encodes the catalytic domain. This form of PCSK9SV fails to undergo autocatalytic cleavage and secretion as demonstrated previously (22). Our data demonstrate that sterol deprivation, which induces PCSK9 expression (13), was necessary to attain the SSO-induced formation of ER-resident PCSK9SV (Fig. 3A). Consistent with our overexpression studies using the CMV-driven PCSK9 plasmids, increasing the ER content of endogenously expressed PCSK9SV also did not correlate with increased expression of the ER stress markers IRE1α and GRP78. Effective alternative splicing of PCSK9 in the presence of the SSO was confirmed via real-time PCR using primers that span exon 8. Given the size of this exon, the PCR product of full-length PCSK9 had an expected size of 582 bp, whereas that of the PCSK9SV lacking exon 8 was 408 bp (Fig. 3B). Quantification of these data shows that the SSO significantly reduced the abundance of PCSK9 mRNA (*, p < 0.05) while increasing the abundance of PCSK9SV mRNA (*, p < 0.05) in the presence and absence of sterol (Fig. 3C). Consistent with the immunoblot data, mRNA levels of ER stress markers were not affected by the SSO (Fig. 3D). To confirm the cellular retention status of PCSK9 resulting from SSO treatment, secreted PCSK9 was examined via ELISA and found to be significantly reduced (*, p < 0.05) (Fig. 3E). The PCSK9 SSO used in this study, also known as hP872, was identical to that reported by Rocha et al. (22) (Fig. 3F).
Figure 3.
ER retention of a PCSK9 splice variant via RNAi does not cause UPR activation. HuH7 cells were seeded in DMEM with and without sterol and transfected with either scrambled siRNA or PCSK9 SSO (1 μm). The SSO used in these studies promotes the expression of a PCSK9SV lacking exon 8 that fails to undergo autocatalytic cleavage and exit from the ER. A, immunoblot analysis of these cells was used to examine PCSK9 expression and the abundance of ER stress markers GRP78 and IRE1α. The status of PCSK9 secretion was also examined indirectly via LDLR immunoblotting. B, to confirm the formation of a PCSK9SV, real-time PCR analysis was carried out for PCSK9 using a forward (For.) primer upstream of exon 8 and a reverse (Rev.) primer downstream of exon 8. The expected sizes of the PCR products from this reaction were 582 and 408 bp for full-length PCSK9 and PCSK9SV, respectively. C, the relative band intensities of PCSK9 and its splice variant were quantified using ImageJ. D, mRNA analysis of ER stress markers GRP78, ATF6, CHOP, and sXBP1 using real-time PCR. E, retention was further confirmed using a PCSK9 ELISA to measure PCSK9 content in the media from these cells. F, sequence and properties of PCSK9 SSO “hP872” as described previously (22). PS, phosphorothioate. Differences between treatments were assessed with unpaired Student's t tests. All values are represented as means, and error bars represent S.D. C, *, p < 0.05 versus the respective sterol-treated controls. E, *, p < 0.05 versus scrambled. NS, nonsignificant.
ER PCSK9 retention does not induce apoptosis
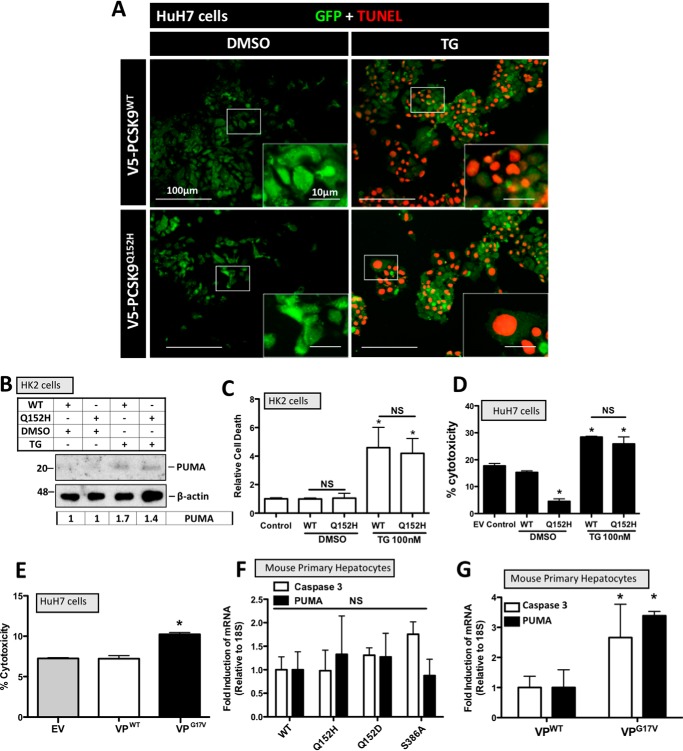
It is well-established that chronic or severe ER stress can trigger apoptotic cell death (31, 32). For this reason, we examined the effect of ER PCSK9 retention on apoptosis as well as selective mediators of apoptosis. HuH7 cells transfected with PCSK9WT and PCSK9Q152H, in the presence or absence of TG (100 nm), were examined for apoptosis-induced DNA damage via TUNEL staining (Fig. 4A). No difference in TUNEL staining was observed between cells expressing PCSK9WT and the PCSK9Q152H retention variant. The abundance of the proapoptotic mediator PUMA was also examined in PCSK9-transfected HK-2 cells, and no significant difference was observed in the presence or absence of TG (100 nm; Fig. 4B). Lastly, cell viability was also assessed in PCSK9-transfected HK-2 and HuH7 cells. Although TG treatment led to a significant increase in the cytotoxicity of HK-2 and HuH7 cells (*, p < 0.05 and †, p < 0.05, respectively), ER retention of PCSK9 did not alter cell viability in either cell line (Fig. 4, C and D). In contrast to PCSK9Q152H, the expression of VPG17V significantly increased cell death compared with VPWT control (Fig. 4E). To confirm our findings, apoptosis marker expression was also examined in mouse primary hepatocytes. Similar to our findings in HuH7 and HK-2 cells, real-time PCR experiments demonstrate that the retention of VPG17V, but not PCSK9Q152H, PCSK9Q152D, or PCSK9S386A, caused an induction of proapoptotic mediators caspase-3 and PUMA relative to WT secreted controls (Fig. 4, F and G).
Figure 4.
ER PCSK9 retention does not induce apoptosis. HuH7 and HK-2 cells were seeded in DMEM and transfected with either PCSK9WT or PCSK9Q152H for 48 h. Cells were also treated with TG (100 nm) or DMSO vehicle control for 24 h. A, HuH7 cells were fixed in 4% paraformaldehyde and stained for apoptosis-induced DNA damage using a 594-labeled TUNEL stain (red). Following TUNEL staining, cells were also stained for GFP to identify cells transfected with the bicistronic pIRES2-EGFP V5-PCSK9 plasmids (green). B, transfected HK-2 cells were also examined for the apoptosis marker PUMA via immunoblot analysis. C and D, cell viability assays were carried out on transfected HK-2 and HuH7 cells using trypan blue stain and lactate dehydrogenase assays, respectively. E, a lactate dehydrogenase assay was also carried out on cells transfected with VPG17V, a mutant known to induce ER stress as a result of ER retention (21). F, freshly isolated mouse primary hepatocytes were transfected with PCSK9 variants PCSK9Q152H, PCSK9Q152D, and PCSK9S386A and examined for the apoptosis markers caspase-3 and PUMA via real-time PCR. G, apoptosis marker expression of caspase-3 and PUMA was also examined in VP-transfected mouse primary hepatocyte using real-time PCR. Differences between treatments were assessed with unpaired Student's t tests. All values are represented as means, and error bars represent S.D. C, *, p < 0.05 versus DMSO-treated WT. D, *, p < 0.05 versus DMSO-treated WT. F and G, *, p < 0.05 versus VPWT. C, D, and E, nonsignificant (NS).
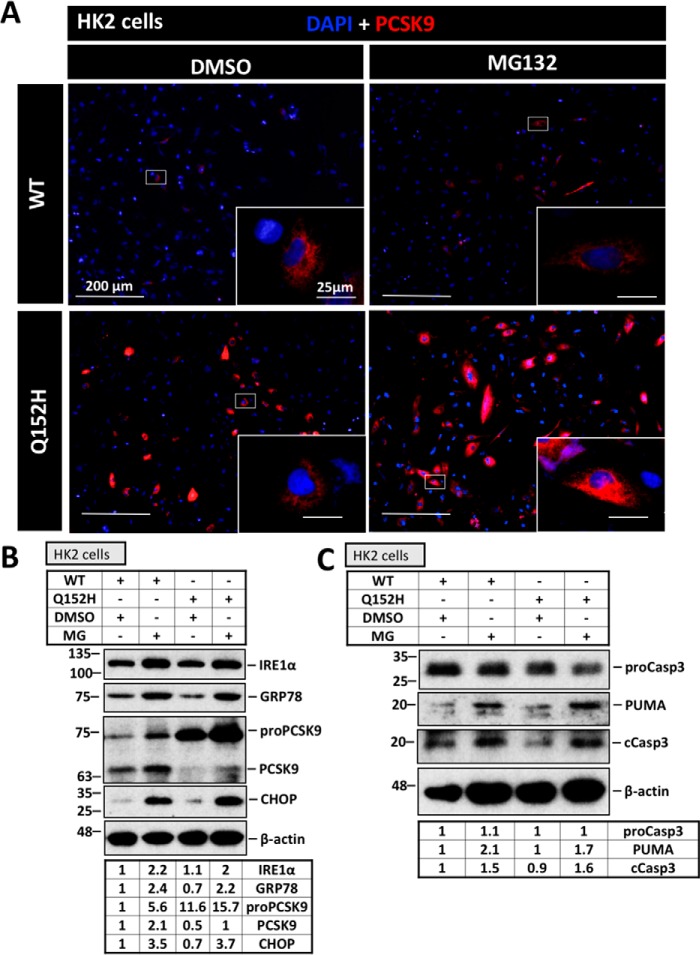
Proteasomal inhibition increases ER PCSK9 content
ER-associated degradation, a process mediated by the proteasome, plays a major role in the removal of misfolded/accumulated proteins from the ER (33). For this reason, we investigated whether the proteasome is responsible for the removal of retained PCSK9 and elicits a protective state against UPR activation in cells transfected with PCSK9Q152H. Moreover, use of the proteasome inhibitor MG132 in combination with CMV-mediated overexpression served as a tool to further enhance exogenous ER-PCSK9 content as a means of causing ER stress. Immunofluorescence staining of V5 was used to visualize the perinuclear localization and accumulation of PCSK9 (Fig. 5A). Representative images show that a greater intensity of staining occurred in PCSK9Q152H-transfected cells than in PCSK9WT-transfected controls. Furthermore, PCSK9Q152H-transfected cells treated with MG132 exhibited more ER-resident PCSK9 than the respective untreated controls, suggesting that the proteasome plays a role in the degradation of PCSK9Q152H. Despite a marked increase in the expression of IRE1α, GRP78, and CHOP in response to MG132, no difference was observed between PCSK9WT and PCSK9Q152H in the presence or absence of MG132 (Fig. 5B). Consistently, MG132 treatment induced the expression of proapoptotic markers cleaved caspase-3 and PUMA, but no difference was observed as a result of PCSK9Q152H expression with respect to its WT counterpart (Fig. 5C).
Figure 5.
Proteasomal inhibition increases ER PCSK9 content. To examine the influence of the proteasome and test whether further increasing the ER content of PCSK9Q152H leads to ER stress, HK-2 cells transfected with V5-labeled PCSK9 were treated with the proteasomal inhibitor MG132 (MG; 1 μm) for 24 h. A, to visualize the extent of ER PCSK9 retention, cells were fixed in 4% paraformaldehyde and stained using a V5 primary antibody and a 594-labeled secondary antibody (red). B, ER stress markers IRE1α, GRP78, and CHOP as well as ER PCSK9 content and maturation were examined via immunoblot analysis. C, apoptosis markers PUMA and cleaved caspase-3 (cCasp3) were also examined using immunoblotting.
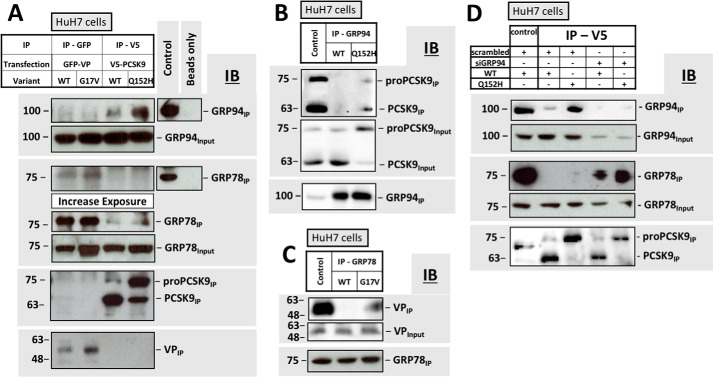
GRP94 masks ER-resident PCSK9 from GRP78
Although GRP78 interacts with a diversity of client proteins (34), recent studies have demonstrated that endogenously expressed PCSK9 interacts with GRP78 to a significantly greater extent following knockdown of GRP94 (14). Given the role of GRP78 as a major trigger of UPR activation, we tested whether GRP94 could mask ER-resident PCSK9 or pro-PCSK9 from detection by GRP78. Furthermore, the interaction between PCSK9Q152H and GRP78 was compared with the interaction between GRP78 and VPG17V. These coimmunoprecipitation experiments demonstrate that the VP retention variant interacts with GRP78 but not GRP94 (Fig. 6A). In contrast, PCSK9 was pulled down by GRP94 and GRP78 with the latter requiring a longer exposure of the immunoblots. Lastly, immunoprecipitation of the ER-retention variants VPG17V and PCSK9Q152H pulled down more GRP78 and GRP94 (respectively) than their secreted WT counterparts. In a reciprocal manner, the interaction between GRP94 and PCSK9 was confirmed via immunoprecipitation of GRP94 (Fig. 6B), and the interaction of GRP78 with VP was confirmed via immunoprecipitation of GRP78 (Fig. 6C). Consistent with Fig. 6A, these immunoblots also demonstrate that GRP78 and GRP94 form stronger interactions with the ER-retention variants of VP and PCSK9 than their WT counterparts. Lastly, coimmunoprecipitation experiments of PCSK9-transfected cells were performed in the presence and absence of small interfering RNA (siRNA) targeted against GRP94 (siGRP94; Fig. 6D). As reported previously (14), we observed a significantly greater level of interaction between PCSK9 and GRP78 under conditions of GRP94 knockdown. Furthermore, we also observed more GRP78 in complex with PCSK9Q152H than with PCSK9WT under such conditions.
Figure 6.
GRP94 masks ER-resident PCSK9 from GRP78. A, coimmunoprecipitation experiments were carried out on cells transfected with VP and its ER-retention variant (VPG17V) and compared with that of cells transfected with PCSK9 and its ER-retention variant (PCSK9Q152H). VP was immunoprecipitated using an anti-GFP capture antibody, whereas PCSK9 was captured using an anti-V5 antibody; effective capture was confirmed by immunoblotting (IB) IPs with anti-GFP and anti-PCSK9 antibodies, respectively. B and C, interactions characterized in A were confirmed by IP of endogenous GRP94 and GRP78 in cells transfected with V5-PCSK9 and GFP-VP, respectively. D, binding of GRP78 to PCSK9WT and PCSK9Q152H was also examined in the presence of siGRP94. B, C, and D, whole cell lysates were used as positive controls for antibody staining.
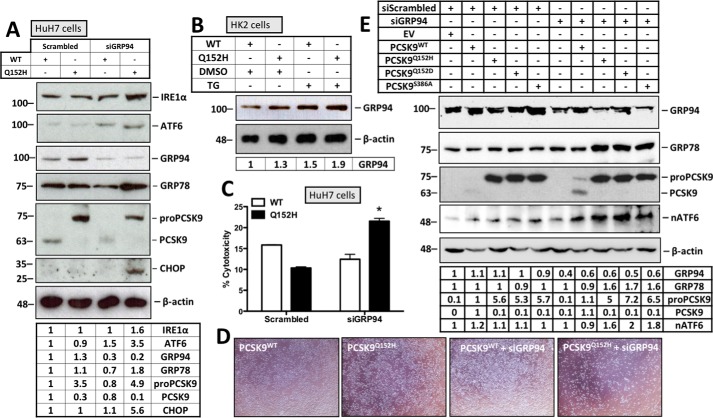
ER retention of PCSK9 variants leads to UPR activation in the absence of GRP94
Given that we observed a greater level of GRP78 association with the PCSK9Q152H variant in the absence of GRP94, our next aim was to determine whether this specific condition would lead to UPR activation. As such, UPR activation was assessed in HuH7 cells cotransfected with either PCSK9WT or PCSK9Q152H in combination with scrambled siRNA or siGRP94. Cells expressing PCSK9Q152H, in the presence of siGRP94, exhibited increased expression of ER stress markers from all three arms of the UPR (ATF6, IRE1α, and CHOP) (Fig. 7A). To further substantiate the relationship between PCSK9 and GRP94, we observed a modest but consistent increase in GRP94 expression in cells transfected with PCSK9Q152H compared with those transfected with the PCSK9WT counterpart (Fig. 7, A and B). Furthermore, cells transfected with both siGRP94 and PCSK9Q152H showed a significant increase in cytotoxicity compared with control groups (Fig. 7, C and D; *, p < 0.05). The effect of GRP94 on UPR activation in response to ER PCSK9 retention was also assessed using PCSK9 variants PCSK9Q152D and PCSK9S386A. Consistent with our findings in Fig. 7A, we observed that cotransfection of HuH7 cells with siGRP94 and ER-retained PCSK9 variants led to increased expression of UPR markers GRP78 and nuclear ATF6 relative to those transfected with siGRP94 and PCSK9WT (Fig. 7E).
Figure 7.
ER retention of the PCSK9Q152H variant leads to UPR activation in the absence of GRP94. A, HuH7 cells were cotransfected with PCSK9WT or PCSK9Q152H and siGRP94. Immunoblot analysis was carried out to examine the expression of ER stress markers IRE1α, ATF6, GRP94, GRP78, and CHOP. B, given that PCSK9 interacts with GRP94, the relative cellular abundance of this chaperone was also examined in HK2 cells transfected with PCSK9WT and PCSK9Q152H. C, cytotoxicity of these cells was examined using a lactate dehydrogenase cell viability assay. D, the relative number of live cells post-transfection was visually apparent via light microscopy. E, in addition to PCSK9Q152H, the ability of other ER-retention PCSK9 variants PCSK9Q152D and PCSK9S386A to cause UPR activation in conditions of reduced GRP94 expression was examined using immunoblotting. Differences between treatments were assessed with unpaired Student's t tests. All values are represented as means, and error bars represent S.D. C, *, p < 0.05 versus scrambled WT control. nATF6, nuclear ATF6.
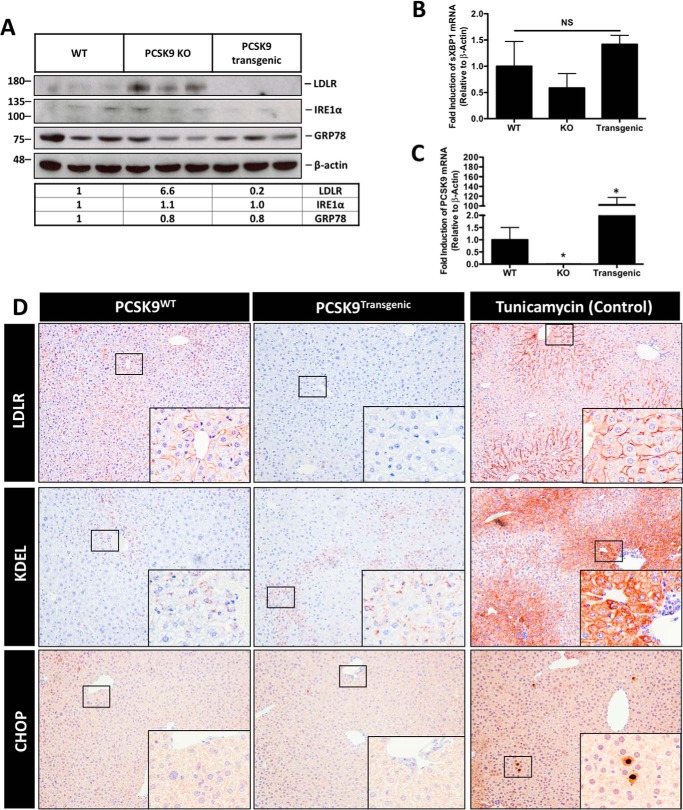
Marked overexpression of mouse PCSK9 does not induce the UPR in murine liver
Based on our cell culture studies, we examined whether transgenic mice overexpressing PCSK9 (PCSK9transgenic), which increases the ER content of nascent PCSK9, would exhibit signs of ER stress. The livers from these mice were examined for UPR activation via immunoblotting for IRE1α and GRP78 (Fig. 8A) and real-time PCR of sXBP1 (Fig. 8B), which yielded no substantial difference between PCSK9WT and PCSK9transgenic mice. Furthermore, PCSK9 knockout (KO) mice showed no induction of the UPR compared with the other groups. LDLR immunoblotting and real-time PCR of PCSK9 served as controls for PCSK9 expression, yielding a significant increase in hepatic LDLR expression and a 100-fold increase in PCSK9 mRNA in PCSK9transgenic mice compared with WT mice (Fig. 8, A and C). Lastly, ER stress was also examined via immunohistochemical staining of the livers for ER stress markers KDEL and CHOP in PCSK9WT and PCSK9transgenic mice as well as a control group of WT mice treated with the ER stress–inducing agent tunicamycin (500 μg/kg for 24 h). Consistent with immunoblot and real-time PCR data, no difference in KDEL or CHOP expression was observed between PCSK9WT and PCSK9transgenic mice (Fig. 8D).
Figure 8.
Marked overexpression of mouse PCSK9 does not induce the UPR in murine liver. To examine whether elevated levels of nascent ER-resident PCSK9 promote UPR activation in vivo, we tested the hypothesis in a transgenic murine model known to express ∼40–100-fold greater levels of PCSK9 than WT controls. A and B, immunoblot and real-time PCR analyses were carried out in the livers of these mice, PCSK9 KO mice and WT controls, to characterize ER stress marker expression of IRE1α, GRP78, and sXBP1. C, a control real-time PCR was also done to confirm the overexpression of PCSK9 in our model. D, formalin-fixed paraffin-embedded liver sections from these mice in addition to mice treated with the ER stress–inducing agent tunicamycin (500 μg/kg; single injection for 24 h) were also stained for ER stress markers KDEL and CHOP. LDLR staining served as a control for the status of PCSK9 expression. Differences between treatments were assessed with unpaired Student's t tests. All values are represented as means, and error bars represent S.D. B, nonsignificant (NS). C, *, p < 0.05 versus WT.
Discussion
A number of ERSDs have been characterized to date (15, 16). Such diseases arise as a result of genetic mutations that lead to the synthesis of a misfolded de novo protein product, which fails to exit the ER (Fig. 9). Frequently, the disease phenotype occurs due to the absence of the mature functional protein at its native site of action. Another well-established mode of action for pathology in ERSDs is chronic activation of the UPR in response to ER stress. A well-characterized ERSD occurring in liver results from a number of established mutations in α1-antitrypsin Z that induce ER stress, liver cirrhosis, and hepatocellular carcinoma (35, 36). Additional examples affecting other tissues include arginine vasopressin mutations (G14R and G17V) in familial neurohypophyseal diabetes insipidus (20, 21, 37), cystic fibrosis transmembrane conductance regulator ΔF508 in cystic fibrosis (38), thyroglobulin mutations in congenital goiter and hyperthyroidism (39), and mutations in the Notch receptor that lead to cerebral autosomal-dominant arteriopathy and leukoencephalopathy (40).
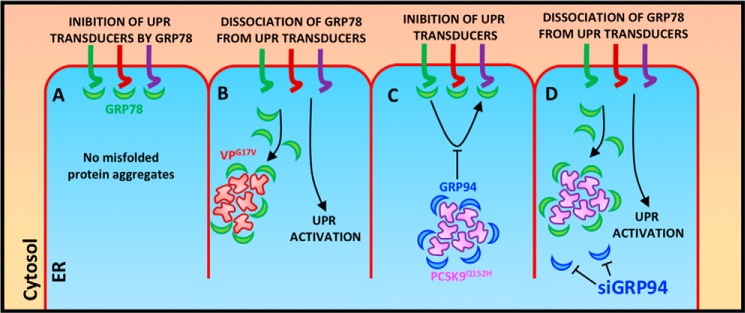
Figure 9.
Model for the masking of ER-resident PCSK9 from UPR sensor GRP78. A, in normal homeostatic conditions, GRP78 is recruited to ER transducers ATF6, IRE1α, and PERK and acts to block UPR activation. B, in the presence of ER-retained protein such as VPG17V, GRP78 dissociates from UPR transducers and interacts with accumulated misfolded proteins to prevent further aggregation. This process liberates ER transducers and leads to UPR activation. C, in contrast to VPG17V, GRP78 is not recruited to ER-retained PCSK9Q152H due to the interaction taking place with GRP94. D, removal of GRP94 from PCSK9 aggregates, however, restores the function of GRP78 as a sensor of UPR activation in response to ER PCSK9 accumulation.
We confirm that overexpression of a mutant form of VP that is retained in the ER resulted in UPR activation and increased cytotoxicity. In contrast to these findings and with striking consistency, overexpression of LOF PCSK9 variants that accumulate in the ER did not exhibit UPR activation or apoptosis. These findings were confirmed using three cell lines (HuH7, HK-2, and N2a) originating from tissues known to express PCSK9 (liver, kidney, and brain, respectively) and with a number of PCSK9 variants (PCSK9Q152H, PCSK9Q152D, and PCSK9FS) known to be retained in the ER. Despite the absence of UPR signaling, a distinct expansion of the ER was observed in HuH7 and HK-2 cells expressing PCSK9Q152H relative to those expressing PCSK9WT controls.4 This phenomenon, which is an initial response to ER stress (41, 42), appears to occur in a manner independent of the UPR. Although the key players in this process are not yet validated, one possibility is that ER stress–inducible genes like SREBP2 promote membrane biogenesis by inducing de novo synthesis and uptake of lipid (43, 44). However, given that ER PCSK9 retention blocks SREBP2 activation (Fig. 1C), increased de novo synthesis is likely not the culprit. PCSK9Q152H expression, however, is known to result in enhanced cell surface LDLR expression (10) and likely allows the cells to acquire the ER expansion–enabling lipid from LDLR-mediated uptake of LDL.
Given that few reports have documented an absence of UPR activation resulting from ER protein retention, the remainder of this discussion will focus on the key differences between PCSK9 and other mutants known to cause ER stress when retained. An initial foundational observation stems from an epidemiological line of evidence: LOF PCSK9 mutations, including PCSK9Q152H, are naturally occurring and confer a substantial reduction in the risk of developing CVD (11, 45, 46). As of yet, the absence/inhibition of circulating PCSK9 has not been associated with deleterious effects other than mild rashes at the site of mAb injection (47). Taken together, these factors imply a positive evolutionary selection pressure for LOF PCSK9 mutations. In support of this notion, the frequency of LOF PCSK9 mutations is relatively high, especially in Africans (48). Indeed, it was recently shown that the PCSK9 mutation C679X, encoding a form of PCSK9 that fails to be secreted (8), has a prevalence of 3.7% in African women (49).
It is also worth considering the implications associated with the observation that nascent PCSK9WT forms pro-PCSK9 oligomers while in the ER (1). Given that ER protein oligomers and/or aggregates are a principal cause of UPR activation and that ER-resident pro-PCSK9 oligomers are naturally occurring, it is therefore evident that cells expressing PCSK9 have adapted a mechanism to prevent constitutive UPR activation and cell death.
Furthermore, in contrast to a number of cell surface/secretory proteins that transit the ER, PCSK9 folding and maturation can occur in a manner independent of ER chaperones. In support of this concept, our previous studies (13) as well as those of others (45) have demonstrated that PCSK9 undergoes maturation in the presence of agents that induce ER Ca2+ depletion regardless of the obligate Ca2+ dependence of ER chaperones. In addition, endogenously expressed PCSK9 does not stably interact with endogenously expressed ER luminal GRP78 (14), which is interesting given the well-established broad client base of this chaperone (50). These findings, in addition to those highlighted in this report, suggest that ER-resident pro-PCSK9 may be undetected and unaffected by major ER chaperones like GRP78. GRP78 is a ubiquitously expressed chaperone involved in the folding of polypeptide chains, nascent chain translocation, resolving the accumulation/removal of misfolded protein in the ER, and modulating the activation of ER stress transducers (51, 52). GRP78 owes its importance in biological systems to its promiscuity, which relies on a client-binding domain known to seek 7–11-residue peptides forming β strands with alternating hydrophobic residues (53, 54). It is estimated that one GRP78-binding site would arise every 36 amino acids within a randomly generated peptide chain (54).
In addition to demonstrating that the interaction between PCSK9 and GRP78 was not as strong as that of PCSK9 and GRP94, our coimmunoprecipitation data also indicate that GRP78 forms a more abundant complex with the retained VPG17V variant than with the VPWT control. These data suggest that ER-retained mutant VPG17V sequesters a greater amount of ER-resident GRP78 (Fig. 6A), likely as a result of increased ER abundance relative to secreted VPWT control. In turn, increased VPG17V-mediated GRP78 sequestration hindered the capacity of this chaperone to act as a blocker of UPR activation and ultimately led to the observed ER stress response (Fig. 2A). Although a similar effect was observed with PCSK9Q152H interacting with GRP94 (versus PCSK9WT) (Fig. 6A), this chaperone is not known to play a direct role as a UPR sensor or mediator of ATF6, IRE1α, and PERK activation (55). Rather, it was recently shown that PCSK9 interacts with GRP78 to a greater extent in the absence of GRP94, likely preventing early binding of PCSK9 to the LDLR in the ER (14). Our data also suggest that GRP94 masks ER-resident pro-PCSK9 oligomers from detection by GRP78. To confirm and extend these findings, we demonstrated that overexpression of the PCSK9Q152H variant induced the UPR following knockdown of GRP94 (Fig. 7A). Furthermore, the onset of ER stress in cells transfected with siGRP94 and PCSK9Q152H was sufficient to cause a significant increase in cytotoxicity 3 days post-transfection as compared with controls (Fig. 7, C and D).
We recently demonstrated that ER PCSK9 retention is associated with a significant reduction of circulating LDL cholesterol (13). We now also confirm that it fails to induce ER stress despite the known link among misfolded protein accumulation, UPR activation, cell death, and disease (18). Our findings demonstrate that PCSK9Q152H likely fails to cause ER stress as a result of a combination of efficient proteasomal degradation (Fig. 5A) and masking by GRP94 (Figs. 6 and 7). In support of these findings, mice expressing 100-fold greater levels of PCSK9 mRNA also failed to exhibit UPR activation (Fig. 8). We are now actively assessing whether the overexpression of LOF variants of PCSK9, including PCSK9Q152H, in the livers of PCSK9 KO mice cause ER stress/UPR activation and/or apoptotic cell death. Taken together, our results indicate that the ER retention of mutant forms of PCSK9 unable to undergo autocatalytic cleavage does not trigger detrimental ER stress, UPR activation, and cytotoxicity. This has important implications for the design of novel therapeutics targeting PCSK9. Specifically, our findings suggest that the development of small molecule inhibitors of PCSK9 autocatalytic cleavage may serve as an effective alternative to the costly biologic PCSK9 inhibitors that are currently available.
Experimental procedures
Cell culture and transfections
HuH7, HK-2, and N2a cells were routinely grown in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich), 100 IU/ml penicillin, and 100 μg/ml streptomycin (both from Gibco, Thermo Fisher Scientific) at 37 °C with 5% CO2. To examine the effect of ER PCSK9 retention on UPR activation, cells were seeded in 6-well plates to a confluence of 60% and transfected with plasmids encoding PCSK9 variants 24 h later. Cells were transfected in penicillin/streptomycin–free medium with Xtremegene transfection reagent (Xtremegene HP, catalogue number 6366236001, Roche Applied Science) at a 3:1 ratio with plasmid DNA (3 μl of transfection reagent/1 μg of DNA/1 ml of medium). To block GRP94 expression, siRNA targeted against GRP94 was purchased from Dharmacon (catalogue number M-006417-02-0005). RNAiMAX (catalogue number 13778030) transfection reagent was used for siRNA transfections; siRNA transfection mixtures contained 100 nm siRNA and 3 μl of RNAiMAX/ml of complete medium. Prior to treatment of cells, transfection mixtures were incubated in Opti-MEM (100 μl of Opti-MEM per treatment; Thermo Fisher Scientific) for 30 min at room temperature prior to transfection according to the manufacturer's instructions. Transfection mixture-containing medium was removed from cells after 24-h incubation and replaced with either fresh medium or medium containing the ER stress–inducing agent TG (100 nm; Sigma-Aldrich) for an additional 24 h prior to cell lysis. The plasmids used in these studies included the bicistronic pIRES2-EGFP plasmids encoding V5-PCSK9WT and its variants PCSK9Q152S, PCSK9Q152H, PCSK9Q152D, and PCSK9FS. VPWT and its variants VPG14R and VPG17V were cloned into pEGFP-N1 as described previously (21), and PCSK9S386A was cloned into a pcDNA3.1 plasmid.
Immunoblotting
Cells were washed in phosphate-buffered saline (PBS) and resuspended in lysis buffer containing SDS and protease inhibitor (catalogue number 4693159001, Roche Applied Science). Total cell protein was normalized using a protein assay (catalogue number 5000121, Bio-Rad). Protein samples were resolved using standard Western blotting procedures on 7 or 10% acrylamide gels and subsequently transferred to nitrocellulose membranes using the Bio-Rad Mini Trans-Blot system (catalogue number 1703930). Membranes were then blocked in 5% skim milk in TBS for 1 h and then incubated in primary antibody overnight for 16 h at 4 °C. The primary antibodies used in this study include the following: anti-ATF6 (catalogue number 70B1413.1, Novus Biologicals), anti-CHOP (catalogue number SC-793, Santa Cruz Biotechnology), anti-FLAG (catalogue number F3165, Sigma-Aldrich), anti-GRP78 (catalogue number 610979, BD Biosciences), anti-IRE1α (catalogue number 3294, Cell Signaling Technology), anti-PCSK9 (catalogue number NB300-959, Novus Biologicals), anti-V5 (catalogue number sc-83849, Santa Cruz Biotechnology), anti-GFP (catalogue number sc-8334, Santa Cruz Biotechnology), anti-GFP (catalogue number NB600-308, Novus Biologicals), and anti-β-actin (catalogue number ab8227, Sigma-Aldrich). Membranes were visualized using EZ-ECL chemiluminescent reagent (catalogue number 20-500-500, Froggabio), and relative band intensities were quantified using ImageLab software (Bio-Rad). Band intensities represent the mean of three replicates adjusted to membranes reprobed for β-actin.
Coimmunoprecipitations
Cells grown in 10-cm dishes were collected in ice-cold nondenaturing IP buffer containing 20 mm Tris HCl, 137 mm NaCl, 1% Nonidet P-40, 2 mm EDTA, protease inhibitor, and phosphatase inhibitor (PhoSTOP, Roche Applied Science). Cells were further lysed by passing through an insulin syringe 20 times. Following lysis, samples were centrifuged at 4 °C for 30 min at 20,000 × g. Supernatants were transferred to new sample tubes, and cell debris–containing tubes were discarded. One milligram of protein from each sample was subsequently incubated with 2 μg of capture antibody targeted against V5 (catalogue number sc-83849, Santa Cruz Biotechnology) or GFP (catalogue number sc-8334, Santa Cruz Biotechnology) for V5-PCSK9– and GFP-VP– transfected cells, respectively, on a rotating platform for 24 h at 4 °C. Following this period, samples were exposed to 100 μl of Protein G magnetic Surebeads (catalogue number 1614023, Bio-Rad) for an additional 2 h on the rotating platform at 4 °C. The beads were then magnetized, and the remaining sample was placed in tubes labeled “input” to serve as controls. The magnetic bead slurry was subjected to four consecutive washes using nondenaturing IP buffer. Protein complexes bound to the beads were collected by boiling the slurry with 100 μl of 4× SDS-PAGE sample/loading buffer. Successful pulldown was confirmed by reprobing immunoblots with anti-PCSK9 antibody (catalogue number NB300-959) and anti-GFP antibody (catalogue number NB600-308).
RNA isolation and real-time PCR
RNA extraction was performed using RNeasy Mini kits (catalogue number 74104, Qiagen) as described previously (7). Total RNA was quantified, normalized, and reverse transcribed into cDNA using a Superscipt Vilo cDNA synthesis kit (catalogue number 11754050, Thermo Fisher Scientific). Real-time PCR was completed using Fast SYBR Green (catalogue number 4385610, Thermo Fisher Scientific). The relative abundance of PCSK9 and PCSK9SV mRNAs from experiments involving SSO was examined via quantitative PCR and agarose gel electrophoresis as described previously (14). Quantification of the relative band intensities was done using ImageLab. DNA was quantified and normalized prior to electrophoresis using a NanoDropTM spectrophotometer.
Immunofluorescence microscopy and TUNEL assay
HuH7 and HK-2 cells were seeded in 4-well chamber slides (catalogue number 177399, Thermo Fisher Scientific) to a confluence of 50% and cultured for 24 h. Cells were then incubated in transfection mixture for 24 h and treated with MG132 (1 μm; catalogue number M8699, Sigma-Aldrich) for an additional 24 h. Following treatments, cells were washed in cold PBS, fixed in 4% paraformaldehyde, and permeabilized in 0.2% Triton X-100 (Sigma-Aldrich). Prior to the 1-h incubation with primary antibodies, cells were blocked in PBS-Tween (PBS-T) containing 5% bovine serum albumin (BSA; catalogue number 05470, Sigma-Aldrich). Primary antibodies used for immunofluorescence staining included the following: anti-V5 (catalogue number sc-83849) and anti-GFP (catalogue number NB600-308). All primary antibodies were diluted 1:100 in PBS-T containing 1% BSA. Following primary antibody incubation, cells were incubated in fluorescently labeled secondary antibodies diluted 1:200 in 1% BSA in PBS-T. These included donkey anti-goat 594 (catalogue number A11058, Thermo Fischer Scientific) and goat anti-rabbit 488 (catalogue number R37116, Thermo Fisher Scientific). Fluorescently labeled cells were then counterstained with DAPI (catalogue number D1306, Thermo Fisher Scientific). TUNEL assays were carried out according to the manufacturer's instructions using a Trevigen kit (catalogue number 4812-30-K, Trevigen).
PCSK9 ELISA
Secreted PCSK9 from cell culture models was measured using the human PCSK9 Quantikinine ELISA kit from R&D Systems (catalogue number DCP900). Briefly, FBS-free medium from transfected cells was collected and centrifuged at 1000 × g for 10 min to remove floating cells/debris. ELISAs were then carried out according to the manufacturer's instructions.
Cell death assays
Lactate dehydrogenase assays were completed according to the manufacturer's instructions (catalogue number 04744926001, Roche Applied Science). A novel cell death assay, based on trypan blue staining of cell debris, was also used to quantify cell death. Briefly, HK-2 cells were transfected with either EV control, PCSK9WT, or PCSK9Q152H in the presence or absence of TG (100 nm). The medium, which contained all cell debris, was collected 24 h later and incubated with 10% trypan blue solution (v/v) (catalogue number 15250061, Thermo Fisher Scientific) for 5 min at room temperature. Samples were then centrifuged at 10,000 × g to isolate the cell debris pellet. The pellet was then washed with isopropanol and subsequently incubated in Hank's balanced salt solution for 30 min at 37 °C to extract trypan blue from the debris. Optical density of trypan blue–containing Hank's balanced salt solution was then measured using a SpectraMax Plus 384 spectrophotometer at 488 nm (Molecular Devices).
Immunohistochemistry
Liver tissues were collected and fixed in formalin, embedded in paraffin, and sectioned at a thickness of 4 μm. KDEL staining was carried out using a 1:40 dilution of the primary antibody with no epitope retrieval (catalogue number ADI-SPA-827, Enzo Life Sciences). CHOP staining was carried out with heat-induced epitope retrieval using a 1:40 dilution of the primary antibody (catalogue number sc-575, Santa Cruz Biotechnology). LDLR staining was carried out using a 1:20 dilution of the primary antibody with no epitope retrieval (catalogue number AF2255, R&D Systems).
Animal studies
12-week-old PCSK9 KO mice or transgenic mice overexpressing mouse PCSK9 (3) and age-matched controls on a C57bl/6 background were housed in a 12-h light/dark cycle and fed normal chow and water ad libitum. Control and PCSK9 KO mice were randomly divided into four groups (n = 5 per group) and treated with either PBS vehicle control or the ER stress–inducing agent tunicamycin (500 μg/kg) for 24 h prior to sacrifice. The McMaster University Animal Research Ethics Board approved all procedures.
Primary hepatocyte isolation
A two-step hepatic perfusion of EGTA (500 m in HEPES buffer, Sigma-Aldrich) and collagenase (0.05% in HEPES buffer, Sigma-Aldrich) was used in 12-week-old male C57bl/6 mice to isolate primary hepatocytes. Following harvest, cells were washed, separated via centrifugation and cell strainers, and plated at a confluence of 1 × 106 cells/well in William's E medium (Gibco, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin.
Statistical analysis
All experiments done during these studies were carried out using a minimum of three replicates. Statistical analysis for differences between groups was performed using two-tailed unpaired Student's t test. Comparisons between plasma PCSK9 concentrations from the same animals before and after treatment were completed using paired two-tailed Student's t test. Statistical tests were completed using Prism software (GraphPad Software, San Diego, CA). Differences between groups were considered significant at p < 0.05, and all values are expressed as mean ± S.D.
Author contributions
P. L., K. P., N. G. S., and R. C. A. conceptualization; P. L., K. P., D. C., N. G. S., and R. C. A. resources; P. L., K. P., and R. C. A. data curation; P. L. and K. P. software; P. L., K. P., J. H. B., and R. C. A. formal analysis; P. L., B. T., and R. C. A. supervision; P. L. and R. C. A. funding acquisition; P. L., K. P., and R. C. A. validation; P. L., K. P., A. A. A.-H., J. H. B., Š. L., N. H., G. G., D. C., and R. C. A. investigation; P. L., K. P., N. G. S., and R. C. A. visualization; P. L., K. P., D. C., N. G. S., and R. C. A. methodology; P. L., K. P., and R. C. A. writing-original draft; P. L., K. P., N. G. S., and R. C. A. project administration; P. L., K. P., A. A. A.-H., J. H. B., Š. L., N. H., G. G., S. A. I., D. C., B. T., N. G. S., and R. C. A. writing-review and editing.
Acknowledgments
We thank Dr. Simon Jackson, Amgen, for constructive insights during these studies as well as Dr. Masayuki Miura, University of Tokyo, for the ERAI plasmid. We would also like to thank St. Joseph's Healthcare Hamilton for the support during the course of these studies.
This work was supported in part by Heart and Stroke Foundation of Ontario Grant T-6146, Heart and Stroke Foundation of Canada Grants G-13-0003064 and G-15-0009389, Canadian Institutes of Health Research (CIHR) Grant 74477 (to R. C. A.); Leducq Foundation Grant 13 CVD 03, CIHR Foundation Grant 148363, and Canada Research Chair 216684 (to N. G. S.); National Institutes of Health Grant DK58111 (to D. R. C.); and St. Joseph's Healthcare Hamilton and CIHR Team Grant in Thromboembolism FRN-79846. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
N. G. Seidah, unpublished data.
P. Lebeau and R. C. Austin, unpublished data.
- PCSK9
- proprotein convertase subtilisin/kexin type-9
- ATF
- activating transcription factor
- CHOP
- CCAAT/enhancer–binding protein homologous protein
- CVD
- cardiovascular disease
- ER
- endoplasmic reticulum
- ERAI
- ER activated indicator
- ERSD
- endoplasmic reticulum storage disease
- EV
- empty vector
- GOF
- gain-of-function
- GRP78
- 78-kDa glucose-regulated protein
- GRP94
- 94-kDa glucose-regulated protein
- IRE1α
- inositol-requiring enzyme-1α
- LDLR
- low-density lipoprotein receptor
- LOF
- loss-of-function
- PERK
- protein kinase RNA–like endoplasmic reticulum kinase
- SSO
- splice-switching oligomer
- TG
- thapsigargin
- UPR
- unfolded protein response
- VP
- vasopressin
- XBP1
- X-box–binding protein-1
- PCSK9SV
- PCSK9 splice variant
- sXBP1
- spliced XBP1
- PUMA
- p53-up-regulated modulator of apoptosis
- siGRP94
- siRNA targeted against GRP94
- IP
- immunoprecipitation
- PCSK9FS
- PCSK9 frameshift variant.
References
- 1. Seidah N. G., Benjannet S., Wickham L., Marcinkiewicz J., Jasmin S. B., Stifani S., Basak A., Prat A., and Chretien M. (2003) The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. U.S.A. 100, 928–933 10.1073/pnas.0335507100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Seidah N. G., Abifadel M., Prost S., Boileau C., and Prat A. (2017) The proprotein convertases in hypercholesterolemia and cardiovascular diseases: emphasis on proprotein convertase subtilisin/kexin 9. Pharmacol. Rev. 69, 33–52 10.1124/pr.116.012989 [DOI] [PubMed] [Google Scholar]
- 3. Zaid A., Roubtsova A., Essalmani R., Marcinkiewicz J., Chamberland A., Hamelin J., Tremblay M., Jacques H., Jin W., Davignon J., Seidah N. G., and Prat A. (2008) Proprotein convertase subtilisin/kexin type 9 (PCSK9): hepatocyte-specific low-density lipoprotein receptor degradation and critical role in mouse liver regeneration. Hepatology 48, 646–654 10.1002/hep.22354 [DOI] [PubMed] [Google Scholar]
- 4. Maxwell K. N., and Breslow J. L. (2004) Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc. Natl. Acad. Sci. U.S.A. 101, 7100–7105 10.1073/pnas.0402133101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daniels T. F., Killinger K. M., Michal J. J., Wright R. W. Jr., and Jiang Z. (2009) Lipoproteins, cholesterol homeostasis and cardiac health. Int. J. Biol. Sci. 5, 474–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hooper A. J., and Burnett J. R. (2013) Anti-PCSK9 therapies for the treatment of hypercholesterolemia. Expert Opin. Biol. Ther. 13, 429–435 10.1517/14712598.2012.748743 [DOI] [PubMed] [Google Scholar]
- 7. Abifadel M., Varret M., Rabès J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., Derré A., Villéger L., Farnier M., Beucler I., Bruckert E., et al. (2003) Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34, 154–156 10.1038/ng1161 [DOI] [PubMed] [Google Scholar]
- 8. Benjannet S., Hamelin J., Chrétien M., and Seidah N. G. (2012) Loss- and gain-of-function PCSK9 variants: cleavage specificity, dominant negative effects, and low density lipoprotein receptor (LDLR) degradation. J. Biol. Chem. 287, 33745–33755 10.1074/jbc.M112.399725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu N. Q., and Li J. J. (2014) PCSK9 gene mutations and low-density lipoprotein cholesterol. Clin. Chim. Acta 431, 148–153 10.1016/j.cca.2014.01.043 [DOI] [PubMed] [Google Scholar]
- 10. Mayne J., Dewpura T., Raymond A., Bernier L., Cousins M., Ooi T. C., Davignon J., Seidah N. G., Mbikay M., and Chrétien M. (2011) Novel loss-of-function PCSK9 variant is associated with low plasma LDL cholesterol in a French-Canadian family and with impaired processing and secretion in cell culture. Clin. Chem. 57, 1415–1423 10.1373/clinchem.2011.165191 [DOI] [PubMed] [Google Scholar]
- 11. Cohen J., Pertsemlidis A., Kotowski I. K., Graham R., Garcia C. K., and Hobbs H. H. (2005) Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 37, 161–165 10.1038/ng1509 [DOI] [PubMed] [Google Scholar]
- 12. Cameron J., Holla O. L., Laerdahl J. K., Kulseth M. A., Ranheim T., Rognes T., Berge K. E., and Leren T. P. (2008) Characterization of novel mutations in the catalytic domain of the PCSK9 gene. J. Intern. Med. 263, 420–431 10.1111/j.1365-2796.2007.01915.x [DOI] [PubMed] [Google Scholar]
- 13. Lebeau P., Al-Hashimi A., Sood S., Lhoták Š., Yu P., Gyulay G., Paré G., Chen S. R., Trigatti B., Prat A., Seidah N. G., and Austin R. C. (2017) Endoplasmic reticulum stress and Ca2+ depletion differentially modulate the sterol regulatory protein PCSK9 to control lipid metabolism. J. Biol. Chem. 292, 1510–1523 10.1074/jbc.M116.744235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poirier S., Mamarbachi M., Chen W. T., Lee A. S., and Mayer G. (2015) GRP94 regulates circulating cholesterol levels through blockade of PCSK9-induced LDLR degradation. Cell Rep. 13, 2064–2071 10.1016/j.celrep.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 15. Callea F., Brisigotti M., Fabbretti G., Bonino F., and Desmet V. J. (1992) Hepatic endoplasmic reticulum storage diseases. Liver 12, 357–362 [DOI] [PubMed] [Google Scholar]
- 16. Rutishauser J., and Spiess M. (2002) Endoplasmic reticulum storage diseases. Swiss Med. Wkly. 132, 211–222 [DOI] [PubMed] [Google Scholar]
- 17. Carlson J. (1990) Endoplasmic reticulum storage disease. Histopathology 16, 309–312 10.1111/j.1365-2559.1990.tb01124.x [DOI] [PubMed] [Google Scholar]
- 18. Austin R. C. (2009) The unfolded protein response in health and disease. Antioxid. Redox Signal. 11, 2279–2287 10.1089/ars.2009.2686 [DOI] [PubMed] [Google Scholar]
- 19. Wu F. L., Liu W. Y., Van Poucke S., Braddock M., Jin W. M., Xiao J., Li X. K., and Zheng M. H. (2016) Targeting endoplasmic reticulum stress in liver disease. Expert Rev. Gastroenterol. Hepatol. 10, 1041–1052 10.1080/17474124.2016.1179575 [DOI] [PubMed] [Google Scholar]
- 20. Birk J., Friberg M. A., Prescianotto-Baschong C., Spiess M., and Rutishauser J. (2009) Dominant pro-vasopressin mutants that cause diabetes insipidus form disulfide-linked fibrillar aggregates in the endoplasmic reticulum. J. Cell Sci. 122, 3994–4002 10.1242/jcs.051136 [DOI] [PubMed] [Google Scholar]
- 21. Yan Z., Hoffmann A., Kaiser E. K., Grunwald W. C. Jr., and Cool D. R. (2011) Misfolding of mutated vasopressin causes ER-retention and activation of ER-stress markers in Neuro-2a cells. Open Neuroendocrinol. J. 4, 136–146 10.2174/1876528901104010136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rocha C. S., Wiklander O. P., Larsson L., Moreno P. M., Parini P., Lundin K. E., and Smith C. I. (2015) RNA therapeutics inactivate PCSK9 by inducing a unique intracellular retention form. J. Mol. Cell. Cardiol. 82, 186–193 10.1016/j.yjmcc.2015.03.009 [DOI] [PubMed] [Google Scholar]
- 23. Pagny S., Lerouge P., Faye L., and Gomord V. (1999) Signals and mechanisms for protein retention in the endoplasmic reticulum. J. Exp. Bot. 50, 157–164 10.1093/jxb/50.331.157 [DOI] [Google Scholar]
- 24. Gardner B. M., Pincus D., Gotthardt K., Gallagher C. M., and Walter P. (2013) Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb. Perspect. Biol. 5, a013169 10.1101/cshperspect.a013169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pai J. T., Brown M. S., and Goldstein J. L. (1996) Purification and cDNA cloning of a second apoptosis-related cysteine protease that cleaves and activates sterol regulatory element binding proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 5437–5442 10.1073/pnas.93.11.5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lhoták S., Sood S., Brimble E., Carlisle R. E., Colgan S. M., Mazzetti A., Dickhout J. G., Ingram A. J., and Austin R. C. (2012) ER stress contributes to renal proximal tubule injury by increasing SREBP-2-mediated lipid accumulation and apoptotic cell death. Am. J. Physiol. Renal Physiol. 303, F266–F278 10.1152/ajprenal.00482.2011 [DOI] [PubMed] [Google Scholar]
- 27. Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., and Goldstein J. L. (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U.S.A. 100, 12027–12032 10.1073/pnas.1534923100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iwawaki T., Akai R., Kohno K., and Miura M. (2004) A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat. Med. 10, 98–102 10.1038/nm970 [DOI] [PubMed] [Google Scholar]
- 29. Abifadel M., Guerin M., Benjannet S., Rabès J. P., Le Goff W., Julia Z., Hamelin J., Carreau V., Varret M., Bruckert E., Tosolini L., Meilhac O., Couvert P., Bonnefont-Rousselot D., Chapman J., et al. (2012) Identification and characterization of new gain-of-function mutations in the PCSK9 gene responsible for autosomal dominant hypercholesterolemia. Atherosclerosis 223, 394–400 10.1016/j.atherosclerosis.2012.04.006 [DOI] [PubMed] [Google Scholar]
- 30. Cameron J., Holla Ø. L., Ranheim T., Kulseth M. A., Berge K. E., and Leren T. P. (2006) Effect of mutations in the PCSK9 gene on the cell surface LDL receptors. Hum. Mol. Genet. 15, 1551–1558 10.1093/hmg/ddl077 [DOI] [PubMed] [Google Scholar]
- 31. Li J., Lee B., and Lee A. S. (2006) Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J. Biol. Chem. 281, 7260–7270 10.1074/jbc.M509868200 [DOI] [PubMed] [Google Scholar]
- 32. Sano R., and Reed J. C. (2013) ER stress-induced cell death mechanisms. Biochim. Biophys. Acta 1833, 3460–3470 10.1016/j.bbamcr.2013.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruggiano A., Foresti O., and Carvalho P. (2014) Quality control: ER-associated degradation: protein quality control and beyond. J. Cell Biol. 204, 869–879 10.1083/jcb.201312042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee A. S. (2005) The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods 35, 373–381 10.1016/j.ymeth.2004.10.010 [DOI] [PubMed] [Google Scholar]
- 35. Teckman J. H., and Perlmutter D. H. (2000) Retention of mutant α1-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am. J. Physiol. Gastrointest. Liver Physiol. 279, G961–974 10.1152/ajpgi.2000.279.5.G961 [DOI] [PubMed] [Google Scholar]
- 36. Lin L., Schmidt B., Teckman J., and Perlmutter D. H. (2001) A naturally occurring nonpolymerogenic mutant of α1-antitrypsin characterized by prolonged retention in the endoplasmic reticulum. J. Biol. Chem. 276, 33893–33898 10.1074/jbc.M105226200 [DOI] [PubMed] [Google Scholar]
- 37. Beuret N., Rutishauser J., Bider M. D., and Spiess M. (1999) Mechanism of endoplasmic reticulum retention of mutant vasopressin precursor caused by a signal peptide truncation associated with diabetes insipidus. J. Biol. Chem. 274, 18965–18972 10.1074/jbc.274.27.18965 [DOI] [PubMed] [Google Scholar]
- 38. Gilbert A., Jadot M., Leontieva E., Wattiaux-De Coninck S., and Wattiaux R. (1998) ΔF508 CFTR localizes in the endoplasmic reticulum-Golgi intermediate compartment in cystic fibrosis cells. Exp. Cell Res. 242, 144–152 10.1006/excr.1998.4101 [DOI] [PubMed] [Google Scholar]
- 39. Kim P. S., Lee J., Jongsamak P., Menon S., Li B., Hossain S. A., Bae J. H., Panijpan B., and Arvan P. (2008) Defective protein folding and intracellular retention of thyroglobulin-R19K mutant as a cause of human congenital goiter. Mol. Endocrinol. 22, 477–484 10.1210/me.2007-0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takahashi K., Adachi K., Yoshizaki K., Kunimoto S., Kalaria R. N., and Watanabe A. (2010) Mutations in NOTCH3 cause the formation and retention of aggregates in the endoplasmic reticulum, leading to impaired cell proliferation. Hum. Mol. Genet. 19, 79–89 10.1093/hmg/ddp468 [DOI] [PubMed] [Google Scholar]
- 41. Schuck S., Prinz W. A., Thorn K. S., Voss C., and Walter P. (2009) Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 187, 525–536 10.1083/jcb.200907074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwarz D. S., and Blower M. D. (2016) The endoplasmic reticulum: structure, function and response to cellular signaling. Cell. Mol. Life Sci. 73, 79–94 10.1007/s00018-015-2052-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nohturfft A., and Zhang S. C. (2009) Coordination of lipid metabolism in membrane biogenesis. Annu. Rev. Cell Dev. Biol. 25, 539–566 10.1146/annurev.cellbio.24.110707.175344 [DOI] [PubMed] [Google Scholar]
- 44. Madison B. B. (2016) Srebp2: a master regulator of sterol and fatty acid synthesis. J. Lipid Res. 57, 333–335 10.1194/jlr.C066712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Benjannet S., Rhainds D., Essalmani R., Mayne J., Wickham L., Jin W., Asselin M. C., Hamelin J., Varret M., Allard D., Trillard M., Abifadel M., Tebon A., Attie A. D., Rader D. J., et al. (2004) NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 279, 48865–48875 10.1074/jbc.M409699200 [DOI] [PubMed] [Google Scholar]
- 46. Cohen J. C., Boerwinkle E., Mosley T. H. Jr., and Hobbs H. H. (2006) Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354, 1264–1272 10.1056/NEJMoa054013 [DOI] [PubMed] [Google Scholar]
- 47. Sabatine M. S., Giugliano R. P., Keech A. C., Honarpour N., Wiviott S. D., Murphy S. A., Kuder J. F., Wang H., Liu T., Wasserman S. M., Sever P. S., Pedersen T. R., and FOURIER Steering Committee and Investigators (2017) Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 376, 1713–1722 10.1056/NEJMoa1615664 [DOI] [PubMed] [Google Scholar]
- 48. Sirois F., Gbeha E., Sanni A., Chretien M., Labuda D., and Mbikay M. (2008) Ethnic differences in the frequency of the cardioprotective C679X PCSK9 mutation in a West African population. Genet. Test. 12, 377–380 10.1089/gte.2008.0013 [DOI] [PubMed] [Google Scholar]
- 49. Hooper A. J., Marais A. D., Tanyanyiwa D. M., and Burnett J. R. (2007) The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis 193, 445–448 10.1016/j.atherosclerosis.2006.08.039 [DOI] [PubMed] [Google Scholar]
- 50. Chen W. T., Tseng C. C., Pfaffenbach K., Kanel G., Luo B., Stiles B. L., and Lee A. S. (2014) Liver-specific knockout of GRP94 in mice disrupts cell adhesion, activates liver progenitor cells, and accelerates liver tumorigenesis. Hepatology 59, 947–957 10.1002/hep.26711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gidalevitz T., Stevens F., and Argon Y. (2013) Orchestration of secretory protein folding by ER chaperones. Biochim. Biophys. Acta 1833, 2410–2424 10.1016/j.bbamcr.2013.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luo S., Mao C., Lee B., and Lee A. S. (2006) GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol. Cell. Biol. 26, 5688–5697 10.1128/MCB.00779-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Flynn G. C., Pohl J., Flocco M. T., and Rothman J. E. (1991) Peptide-binding specificity of the molecular chaperone BiP. Nature 353, 726–730 10.1038/353726a0 [DOI] [PubMed] [Google Scholar]
- 54. Blond-Elguindi S., Cwirla S. E., Dower W. J., Lipshutz R. J., Sprang S. R., Sambrook J. F., and Gething M. J. (1993) Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–728 10.1016/0092-8674(93)90492-9 [DOI] [PubMed] [Google Scholar]
- 55. Marzec M., Eletto D., and Argon Y. (2012) GRP94: an HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim. Biophys. Acta 1823, 774–787 10.1016/j.bbamcr.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]