Abstract
Cisplatin-resistant ovarian cancer occurs in patients with ovarian cancer treated with cisplatin-based chemotherapy, which results in tumor progression during treatment, or recurrence of the tumor within 6 months of the treatment. It is vital that a novel biomarker for diagnosis, or an efficient therapeutic target of cisplatin-resistant ovarian is identified. Long non-coding (lnc)RNAs were determined to serve critical functions in a variety of distinct types of cancer, including ovarian cancer; however, there is limited knowledge regarding the differential expression levels of lncRNAs in cisplatin-resistant and cisplatin-sensitive ovarian cancer. Therefore, in the present study, the expression levels were determined for these cancer types. The lncRNA expression profile in cisplatin-resistant ovarian cancer was analyzed and compared with the results for cisplatin-sensitive ovarian cancer; gene ontology and pathway analysis demonstrated that the dysregulated lncRNAs participated in important biological processes. Subsequently, it was identified that these dysregulated lncRNAs were present in other ovarian cancer tissues and in SKOV3 ovarian cancer cells, as well as its cisplatin-resistant clone, SKOV3/CDDP. In addition, it was revealed that 8 lncRNAs (Enst0000435726, Enst00000585612, Enst00000566734, Enst00000453783, NR_023915, RP11_697E22.2, uc010jub.1 and tcons_00008505) were associated with cisplatin-resistant ovarian cancer. The present study may assist in improving understanding of the initiation and developmental mechanisms underlying cisplatin-resistant ovarian cancer, which could aid future studies in discovering potential biomarkers for diagnosis or therapeutic targets that may be used in clinical treatment.
Keywords: ovarian cancer, long non-coding RNA, cisplatin resistance, therapy target
Introduction
Ovarian cancer is one of the most common female genital tumors, with incidence of ovarian cancer lower than that of cervical and uterine cancer (1). Due to ovarian cancer being highly malignant, having a poor prognosis and the highest mortality rate of all female genital tumors, the five-year survival rate of ovarian cancer is between 20 and 30% in 2013 (2). In addition to these factors, ovarian cancer is difficult to diagnose and treat, as well as there being a rising concern with regard to drug resistance (3). Currently, ovarian cancer is treated with tumor cytoreductive surgery combined with adjuvant platinum-based chemotherapy, including cisplatin (4). The majority of patients with ovarian cancer are sensitive to cisplatin-based chemotherapy initially and have a high rate of remission in the short term; however, abdominal or pelvic recurrence is frequent and drug resistance develops (5,6). Patients with tumors that progress during or recur within 6 months of treatment are considered to be cisplatin-resistant (7).
With progress in the field of life sciences, understanding of the underlying mechanisms of tumor development has grown from the functional gene level to non-coding RNA. Long non-coding RNA (lncRNA), which has >200 nucleotide transcripts, has gained attention in the field of medicine, particularly in oncology, as a result of their abundance, function and mechanism. Various lncRNAs have been demonstrated to be involved in the regulation of tumorigenesis, invasion, metastasis and resistance to cancer treatments (8,9). Although the study of lncRNA is still in the initial stages, important observations have been made; these include the identification of regulator of reprogramming (10) and urothelial cancer-associated 1 (11), which have the ability to regulate the chemosensitivity of hepatocellular carcinoma. In view of the current resistance to lncRNAs previously identified to be dysregulated and associated with chemoresistant ovarian cancer, further exploration of ovarian cancer drug resistance may reveal the mechanisms underlying ovarian cancer resistance, and result in novel methods for the diagnosis and treatment of ovarian cancer.
The aim of the present study was to further investigate dysregulated lncRNAs in cisplatin-resistant ovarian cancer, compared with in cisplatin-sensitive ovarian cancer. This was conducted to assist understanding of the initiation and development mechanisms of ovarian cancer, which could be helpful for discovering potential biomarkers for diagnosis, or novel therapy targets that could be used in clinical treatment.
Materials and methods
Cell lines and reagents
The cisplatin-sensitive SKOV3 ovarian cancer cell line and their cisplatin-resistant clones, SKOV3/CDDP, were obtained from the Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China). Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2 in RPMI-1640 medium with 10% (v/v) fetal bovine serum, 100 U/ml penicillin and 100 U/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All reagents were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany), unless stated otherwise.
Female primary ovarian cancer tissue samples were obtained from the Gynecology Department of Nanjing Maternal and Child Health Hospital (Nanjing, China) from January 2015 to January 2016. In total, there were 6 primary cisplatin-resistant ovarian cancer cases included in the present study, along with 7 age-matched primary cisplatin-sensitive ovarian cancer cases (46 patients with ovarian cancer, 13 patients were treated with cisplatin; 33 patients were treated with paclitaxel combined with doxorubicin). All samples were from epithelial ovarian carcinomas, including 8 serous ovarian carcinomas and 5 mucous type ovarian carcinomas (3 stage I, 4 stage II, 4 stage III, 2 stage IV) (12). Once the tissues were collected, they were washed with 3X RNAlater® (Thermo Fisher Scientific, Inc.) and put into a freezing tube containing 5X RNAlater® solution along with liquid nitrogen, which quick-froze the samples at −70°C. Histopathological diagnoses were all confirmed as ovarian cancer. Informed consent for the use of these samples was obtained from each patient. Ethical approval was obtained from the Nanjing Maternal and Child Health Hospital Ethics Committee.
Total RNA extraction
Tissue samples and cells were lysed in TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and total RNAs extraction was conducted according to the manufacturer's protocol. Quantification and a quality check were performed using Nano-Drop™ and an Agilent 2100 Bio-Analyzer (Agilent Technologies, Inc., Santa Clara, CA, USA), respectively.
lncRNA expression profiling
For lncRNA expression profiling, 3 cisplatin-resistant ovarian cancer samples and 3 cisplatin-sensitive ovarian cancer samples were profiled using an Arraystar lncRNA Microarray v3.0 (Arraystar, Inc., Rockville, MD, USA), as described previously (13). The RNA was purified from 1 mg total RNA following the removal of ribosomal RNA (rRNA) using an mRNA-ONLY™ Eukaryotic mRNA Isolation kit (Epicentre; Illumina, Inc., San Diego, CA, USA). Following this, the full-length of each sample was amplified and transcribed into fluorescent RNA by mRNA-ONLY Eukaryotic mRNA Isolation Kit, Epicentre (Epicentre; Illumina, Inc.) using a random priming method (14) to prevent bias. The labeled RNAs were hybridized onto the Human lncRNA Array v3.0 (Agilent SureHyb; Agilent Technologies, Inc.). Following washing, the arrays were scanned using the Agilent lncRNA Microarray Scanner (Agilent Technologies, Inc.), and the Agilent Feature Extraction software version 11.0.1.1 (Agilent Technologies, Inc.) was used for microarray probe signal data collection. Finally, Agilent GeneSpring GX v12.1 software (Agilent Technologies, Inc.) was employed to normalize the values. lncRNAs and mRNAs for which at least 1 out of 2 groups were flagged in ‘present’ or ‘marginal’ were selected for further data analysis.
lncRNA classification pipeline
In order to elucidate the lncRNA expression pattern in the probe name-centric cisplatin-resistant ovarian cancer gene expression data, clarification of the lncRNAs represented on the Affymetrix microarray (Affymetrix; Thermo Fisher Scientific, Inc.) was conducted via a common lncRNA classification pipeline, using the following strategies: First, the annotations of the microarray data involved the probe name, seqname, gene symbol, gene title, source, chromosome location, sequence and other informative items for the specific probe set; secondly, the seqname was assigned with a GENCODE ID, RefSeq database ID and/or Ensembl gene ID. Seqnames with GENCODE IDs were labeled as ‘Enst.’ Seqnames with Refseq IDs were labeled as ‘NR_’ (non-coding RNA). Seqnames with Ensembl gene IDs were labeled as ‘uc’ (www.genome.ucsc.edu). Thirdly, the seqnames obtained in step 2 were separated by filtering out pseudogenes, rRNAs, microRNAs and other short RNAs; including transfer RNAs, small nuclear RNAs and small nucleolar RNAs (15).
Gene ontology (GO) and pathway analysis
Identification of differentially expressed lncRNAs was conducted via multiple hypothesis testing [false discovery rate (FDR<0.05)], fold-change filtering (absolute fold-change >2.0) and the standard Student's t-test (P<0.05). Identification of significantly enriched biological terms and pathways was completed using GO and pathway analysis of differentially expressed lncRNAs, including antisense lncRNA, intronic lncRNA, enhancer lncRNA, long intergenic noncoding RNAs and other lncRNAs. GO terms and pathway enrichment analysis were based on the Database for Annotation, Visualization and Integrated Discover (DAVID) bioinformatics resource (david.abcc.ncifcrf.gov; version 6.7) and the result of pathway enrichment analysis was confirmed by the Kyoto Encyclopedia of Genes and Genomes (KEGG) online database (www.kegg.jp). Identification of the potential functions of the lncRNAs that were differentially expressed was conducted using functional annotation clustering, using DAVID version 6.7 (david.abcc.ncifcrf.gov) and KEGG (www.kegg.jp), and ranked by enrichment scores.
Validation of differentially expressed lncRNA by reverse-transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA of sample tissues and cells was extracted and reverse transcribed into cDNA with random primers by using a PrimeScript™ Reverse Transcription kit (Takara Bio, Inc., Otsu, Japan), according to the manufacturer's protocol. A standard qPCR was performed to confirm the expression levels of differentially expressed lncRNAs using the Applied Biosystems ViiA 7 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.), following the manufacturer's guidelines. Briefly, the sample mixtures (Table I) were incubated at 95°C for 10 min for an initial denaturation, followed by 40 PCR cycles of incubation at 95°C for 15 sec, 60°C for 30 sec and then 72°C for 30 sec. Each sample was performed in triplicate. The expression levels of lncRNAs were normalized to the internal control, GAPDH, and then quantified using the 2−∆∆Cq method (16). The target genes of the dysregulated lncRNAs were predicted based on the principles of chromosome location of nearby coding genes and of base-pairing (17). The aim of this study was to further explore the dys-regulated lncRNAs in cisplatin resistant ovarian cancer compared to cisplatin sensitive ovarian cancer, which may help the understanding of the initiation and development mechanism of ovarian cancer comprehensively, and probably afford the potential biomarkers for diagnosis or therapy targets for clinical treatment. There were 13 patients who treated with cisplatin in the present study.
Table I.
Primers for qRT-PCR of LncRNAs.
| Seqname | Primers (5′-3′) |
|---|---|
| Enst00000435726 | F: GGAGGTCACTCTCAACACCC |
| R: CAGAGGAGATGAAAGCCATAGA | |
| Enst00000585612 | F: GGAAAGCCTTTAGCCATCGT |
| R: TTCAGGTAGTTGCTTCACATCC | |
| Enst00000566734 | F: AGGACGGTCAGTCATCCTTT |
| R: ATCTTCAGGCACAAAAACCCA | |
| Enst00000453783 | F: GCAGTGCTTGGAGATTGGGA |
| R: TTCATGAGCCCCACACACAA | |
| NR_023915 | F: GCCTACCTGTGGTCTCTTGG |
| R: ACCTCTTTGTGGCCATCACC | |
| RP11_697E22.2 | F: GAAAGAGGGTTTCCGTGCCA |
| R: CGCCACCCTTGGGGTATTT | |
| uc010jub | F: CCAGCAGCCCTCTGGGAA |
| R: AGAAAGGCTGGGCTGAAGTG | |
| tcons_00008505 | F: CTGGGCAACAAGTCCACAGA |
| R: TTAGACCGTCATGGCGGAAG | |
| GAPDH | F: GGTGAAGGTCGGAGTCAACG |
| R: CAAAGTTGTCATGGATGHACC |
F, forward; R, reverse.
Statistical analysis
Values are expressed as the mean ± standard deviation from at least three independent experiments. The differences in lncRNA expression levels were determined by analysis of variance and multiple hypothesis testing followed by the false discovery rate method as a post-hoc test. The sensitivity and specificity were analyzed according to the standard formulas (14). All P-values were two-sided and a value of P<0.05 was considered to indicate a statistically significant difference. Computer-based calculations were conducted using SPSS version 20.0 (IBM Corp., Armonk, NY, USA).
Results
Differential lncRNA expression profiles between cisplatin-resistant ovarian cancer and cisplatin-sensitive ovarian cancer
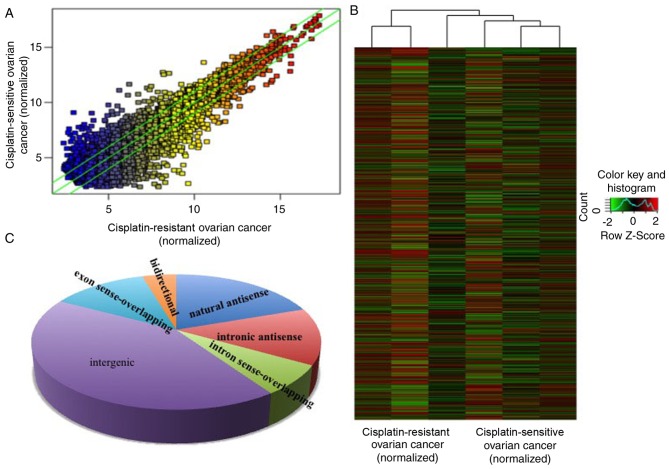
In the present study, the expression levels of lncRNAs in 3 cisplatin-resistant ovarian cancer and 3 age-matched cisplatin-sensitive ovarian cancer samples (enough to screen out the dysregulated lncRNAs) were detected via a high-throughput microarray technique. The patients with tumors that progressed during or recurred within 6 months of the treatment were considered cisplatin-resistant. The results of the microarray revealed that there were 823 upregulated and 765 downregulated lncRNAs in cisplatin-resistant ovarian cancer, compared with cisplatin-sensitive ovarian cancer (Fig. 1A and B) with fold-change filtering (absolute fold-change >2.0), significant difference identified using a Student's t-test (P<0.05) and multiple hypothesis testing (FDR<0.05). According to the nearby coding genes, these differentially expressed lncRNAs included 312 natural antisense, 216 intronic antisense, 114 intron sense-overlapping, 673 intergenic, 201 exon sense-overlapping and 72 bidirectional lncRNAs (Fig. 1C).
Figure 1.
Differential lncRNA expression profiles between tissues of cisplatin-resistant and cisplatin-sensitive ovarian cancer. The lncRNA microarray demonstrated the differences between lncRNA expression in cisplatin-resistant and cisplatin-sensitive ovarian cancer through (A) hot-spot and (B) cluster map analysis. (C) Based on the association of the nearby coding genes, the differentially expressed lncRNAs were classified into certain types, including 312 natural antisense, 216 intronic antisense, 114 intron sense-overlapping, 673 intergenic, 201 exon sense-overlapping and 72 bidirectional lncRNA. lncRNA, long non-coding RNA.
Go and pathway analysis of differentially expressed lncRNAs
To explore the potential functions of the dysregulated lncRNAs in cisplatin-resistant ovarian cancer, the target genes of the lncRNAs were predicted based on the principles of chromosome location of nearby coding genes and of base-pairing (14). Following this, GO analysis was conducted for the lncRNAs and target genes. The GO project (www.geneontology.org) primarily covers 3 areas, including the biological process, molecular function and cellular component, and provides controlled annotations to describe the gene and gene products attributed to any organism (11). The GO-analyzed results indicated that these gene products were primarily located within membrane-bound organelles, extracellular regions, intracellular membrane-bound organelles, cytoplasm, intracellular regions and intracellular, among other locations (Fig. 2A). The genes were predicted to be enriched in the biological processes associated with the cell cycle, namely, regulation of the mitotic cell cycle, cell division, M phase of the mitotic cycle, regulation of the cell cycle process and regulation of the cell cycle, among other processes (Fig. 2B). The molecular functions of these genes included organic cyclic compound binding, protein binding (interaction, selectively and non-covalently, with any protein or protein complex), binding (selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule) and ion binding (Fig. 2C). Furthermore, the pathway analysis demonstrated that these gene products participated in several signaling pathways in humans, including mitogen-activated protein kinase (MAPK) signaling, protein process in the endoplasmic reticulum, Parkinson's disease, endocytosis, ubiquitin mediated proteolysis, p53 signaling pathway, spliceosome, cell cycle, oocyte meiosis and proteasome (Fig. 2D).
Figure 2.
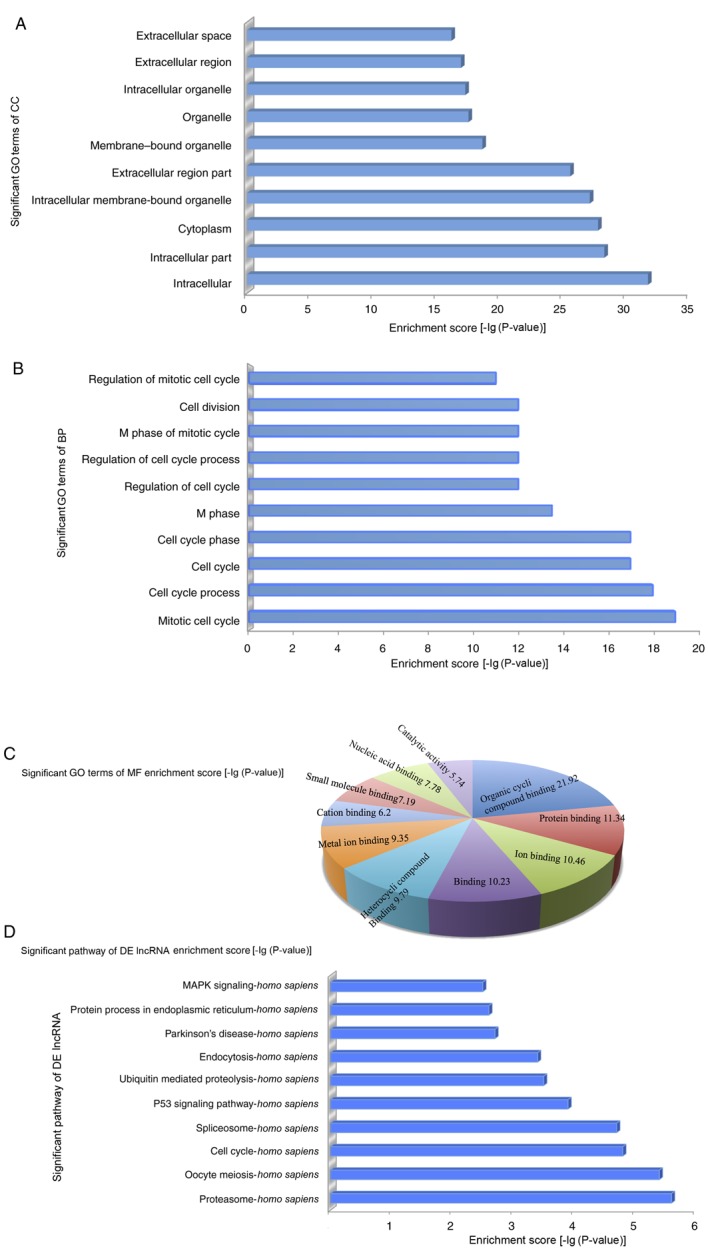
In order to explore the potential functions of dysregulated lncRNAs in cisplatin-resistant ovarian cancer, GO and pathway analysis were performed. (A) The GO analysis data demonstrated that gene products were primarily located on the membrane-bound organelles, extracellular regions, intracellular membrane-bound organelles, cytoplasm, intracellular and intracellular region. (B) Genes were predicted to be enriched in the following biological processes: Regulation of the mitotic cell cycle, cell division, M phase of the mitotic cycle, regulation of the cell cycle process and regulation of the cell cycle. (C) The molecular functions of these genes, including organic cyclic compound binding, protein binding, binding and ion binding. (D) Pathway analysis demonstrated that gene products were involved in several signaling pathways in humans. GO, gene ontology; DE, differently expressed; MF, molecular function; MAPK, mitogen-activated protein kinase; lncRNA, long non-coding RNA.
Discovery of cisplatin-resistant ovarian cancer-associated lncRNAs
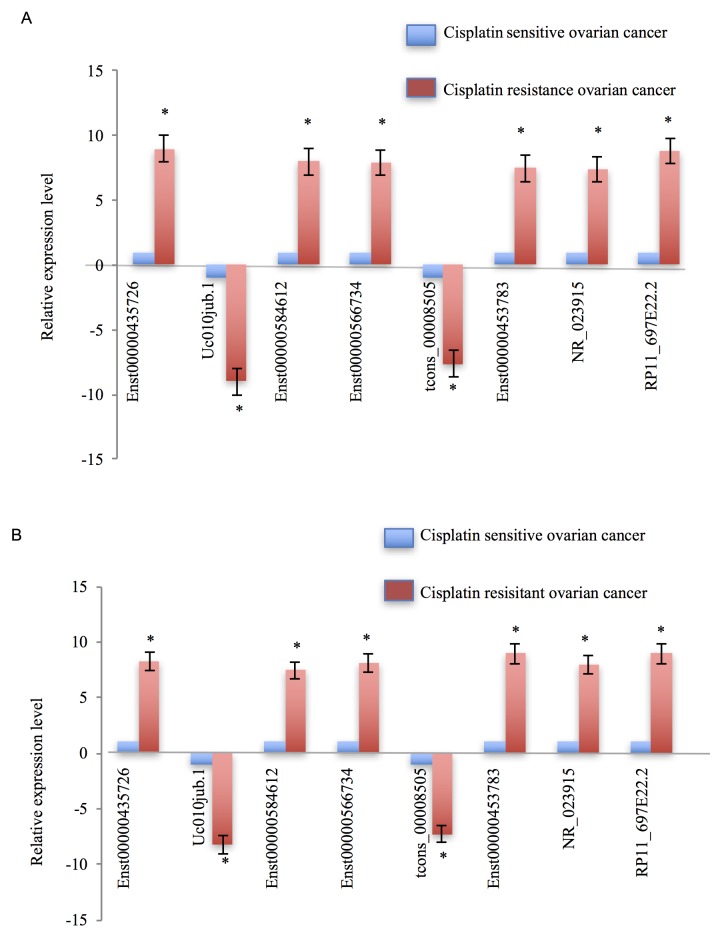
In the present study, the expression levels of those dysregulated lncRNAs were validated not only in a sample pool of 13 patients (46 patients with ovarian cancer; 13 patients were treated with cisplatin; 33 patients were treated with paclitaxel combined with doxorubicin), but also in cisplatin-resistant ovarian cancer cells, SKOV3/CDDP, and cisplatin-sensitive ovarian cancer cells, SKOV3. The differentially expressed lncRNAs were selected by fold-change filtering (absolute fold-change >2.0), Student's t-test (P<0.05) and multiple hypothesis testing (FDR<0.05), and at least 1 out of 2 groups were flagged as ‘present’ or ‘marginal’ following lncRNA expression profiling. Finally, it was identified that 62 lncRNAs exhibited significant differential expression levels in cisplatin-resistant ovarian cancer, compared with in cisplatin-sensitive ovarian cancer controls. Of these 62 dysregulated lncRNAs, 38 lncRNAs were demonstrated to be upregulated and 24 lncRNAs were downregulated. The RT-qPCR results demonstrated that, compared with cisplatin-sensitive ovarian cancer tissues, Enst0000435726, Enst00000585612, Enst00000566734, Enst00000453783, NR_023915 and RP11_697E22.2 were markedly upregulated in cisplatin-resistant ovarian cancer tissues; however, uc010jub.1 and tcons_00008505 were notably downregulated (Fig. 3A). The expression patterns of these eight dysregulated lncRNAs in cisplatin-resistant ovarian cancer cells, compared with the cisplatin-sensitive ovarian cancer cells, appeared concordant with the results from the tissue samples (Fig. 3B).
Figure 3.
Validation of cisplatin-resistant ovarian cancer-associated lncRNAs. (A) First the expression levels of dysregulated lncRNAs were validated in cisplatin-resistant and cisplatin-sensitive ovarian cancer tissues, and 8 lncRNAs were dysregulated in cisplatin-resistant ovarian cancer samples compared with cisplatin-sensitive ovarian cancer samples. (B) Subsequently, the expression levels of these 8 lncRNAs were assessed separately in cisplatin-resistant and cisplatin-sensitive ovarian cancer cells. Tissue and cell data demonstrated consistent results. Each experiment was repeated in triplicate. A positive value denotes the dysregulated lncRNAs that are upregulated, and a negative value means dysregulated lncRNAs that are downregulated. Error bars demonstrate that the differences between cisplatin-resistant ovarian cancer and cisplatin-sensitive ovarian cancer were statistically significant. *P<0.05 vs. cisplatin-sensitive ovarian cancer.
Discussion
Mortality rates associated with ovarian cancer are the highest out of the female reproductive system tumors, demonstrating a serious threat to physical and mental health in females (4–6). Currently, the primary clinical treatment for ovarian cancer is a combination of platinum-based chemotherapy and cytoreductive surgery; however, nearly 70% of patients with ovarian cancer relapse and become multidrug resistant within 6 months (3,4). Improving the survival rate of patients with ovarian cancer is a global concern (18). The mechanism of ovarian cancer resistance involves a variety of genes and a number of different signaling pathways, including the following aspects: Affecting the effective concentration of intracellular changes, including the multidrug resistance gene; expression of the multidrug resistance protein; expression of the lung resistance associated protein (19,20) and affecting drug targets, including β-tubulin expression changes, the cytoskeleton protein gene and the abnormal expression of compartment of uncoupling receptor and ligand (21); DNA damage repair abnormalities, including DNA mismatch repair gene, topoisomerase gene and other changes in the levels of expression or interaction abnormalities (22,23); apoptosis-associated genes, including TP53, survivin, caspase, B cell lymphoma 2, and other gene regulation abnormalities associated with resistance to ovarian cancer (24,25). Although, targeted drugs have been developed in response to a number of the aforementioned mechanisms (26,27), they do not induce a fundamental change in the resistance to ovarian cancer.
Since the human-genome project, lncRNAs have gained attention due to their regulation of histone acetylation, gene methylation, post-transcription translation and other biological processes (28–30). Recently, numerous lncRNAs have been demonstrated to serve critical functions in regulating the physiological behavior of malignant cancer types, including breast, ovarian, gastric and lung cancer, among others. Additionally, lncRNAs have been demonstrated to regulate cancer cell viability, apoptosis, invasion and metastasis (31–37). Although dysregulated lncRNAs between cisplatin-resistant and cisplatin-sensitive ovarian cancer tissues have been identified (38,39), there remains limited knowledge regarding the differentially expressed lncRNAs. The aim of the present study was to improve the understanding of lncRNA expression levels in cisplatin-resistant ovarian cancer.
The results of the microarray assay in the present study revealed dysregulated lncRNAs between cisplatin-resistant and cisplatin-sensitive ovarian cancer tissues, including 312 natural antisense and 673 intergenic (40,41). The data indicate that the resistance behavior of cisplatin-resistant ovarian cancer is potentially associated with these differentially expressed lncRNAs. In order to predict the potential function of these dysregulated lncRNAs, GO analysis was conducted. It was determined that lncRNA regulates several biological processes, including regulation of the mitotic cell cycle, cell division and M phase of the mitotic cycle, which are closely associated with the drug resistance (42) of cancer. The potential functions were classified into 10 categories through analysis of the target gene pool, namely those involving protein binding, binding, heterocyclic compound binding, cation binding, catalytic activity, ion binding, small molecule binding, nucleic acid binding, metal ion binding and organic cyclic compound binding. Notably, it was demonstrated that the dysregulated lncRNAs exhibited binding activity; therefore, these dysregulated lncRNAs may serve important functions in biological processes through regulating the cell cytoskeleton. Furthermore, pathway analysis indicated that these dysregulated lncRNAs mainly participated in signaling pathways in humans, including MAPK signaling, protein process in endoplasmic reticulum, Parkinson's disease, endocytosis, ubiquitin mediated proteolysis, p53 signaling pathway, spliceosome, cell cycle, oocyte meiosis, proteasome and ubiquitin-mediated proteolysis, which have been well studied in the initiation and development of ovarian cancer (32,34,42). The association between oocyte development and cisplatin-resistant ovarian cancer occurrence were investigated. These differentially expressed lncRNAs partially indicated that the function in cisplatin-resistant ovarian cancer corresponded with cisplatin-sensitive ovarian cancer tissues, and that these lncRNAs may be potential biomarkers for diagnosis, or therapeutic targets for cisplatin-resistant ovarian cancer therapy.
The expression levels of these dysregulated lncRNAs were confirmed in a 13-sample pool, in order to avoid heterogeneity of cisplatin-resistant ovarian cancer and individual differences. The differentially expressed lncRNAs were selected as aforementioned. were identified. The expression levels of all 8 dysregulated lncRNAs were confirmed separately in SKOV3 and SKOV3/CDDP cells, and the results were consistent with the results from the tissue samples.
RP11-697E22.2 is an lncRNA that targets the gene hepatocyte nuclear factor 1β. As with all of the 6 lncRNAs, which were observed to be upregulated in the present study, it is an intergenic lncRNA (43). Recently, intergenic and antisense lncRNAs have been demonstrated to regulate cell behavior in a variety of different types of cancer (44–46). The complex mechanisms underlying the function and large quantity of lncRNAs available have resulted in lncRNAs being a popular area of study. These dysregulated lncRNAs identified in the present study are associated with cisplatin-resistant ovarian cancer and may be potential novel biomarkers for diagnosis, or affordable potential targets for the individual therapy of patients with cisplatin-resistant ovarian cancer in the future.
Although the differentially expressed lncRNAs between cisplatin-resistant ovarian cancer and age-matched cisplatin-sensitive ovarian cancer tissues were explored, these lncRNAs may not be associated with ovarian cancer resistance. The data demonstrated the differences in the lncRNA expression profile between cisplatin-resistant and paired cisplatin-sensitive ovarian cancer, and the eight aforementioned lncRNAs may be potential targets for individual therapy. The dysregulated lncRNAs in ovarian cell lines were validated and the results were in accordance with the results from the tissues. The aim of the present study was to identify the differentially expressed lncRNAs between cisplatin-resistant and cisplatin-sensitive ovarian cancer. Investigations into lncRNA target genes are potential avenues of further study. Additionally, validation was performed only in SKOV3 cells, and was a limitation of the present study. Considering the multiple mechanisms underlying drug resistance in ovarian cancer chemotherapy, tissues collected from the other 33 patients may assist in validating the dysregulated lncRNAs in an increased number of ovarian cancer tissues and cells, including paclitaxel-resistant tissues and cells, in the future. It is necessary to validate these results in larger cohorts and additional cell lines, and further studies should aim to investigate the underlying mechanism.
Acknowledgements
The present study was supported by the Science and Technology Development Fund of Shanghai Pudong New Area (grant no. PKJ2016-Y32). The authors wish to thank Kang Chen Bio-tech Ltd., (Shanghai, China) for consultation in the present study.
Funding
The present study was supported by the Science and Technology Development Fund of Shanghai Pudong New Area (grant no. PKJ2016-Y32).
Availability of data and materials
All data and materials are availabile.
Author's contributions
QL was responsible for collecting and analyzing data and writing the article. JZha was responsible for the collection of tissue samples. JZho, BLY and PPL were responsible for the cell experiments. LC and LJ were responsible for the tissue experiments. HL was responsible for analyzing the data and amending the article.
Ethics approval and consent to participate
Informed consent for the use of these samples was obtained from each patient. Ethical approval was obtained from the Nanjing Maternal and Child Health Hospital Ethics Committee.
Consent for publication
Written informed consent for publication was obtained.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Coleman RL, Monk BJ, Sood AK, Herzog TJ. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat Rev Clin Oncol. 2013;10:211–224. doi: 10.1038/nrclinonc.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mo L, Pospichalova V, Huang Z, Murphy SK, Payne S, Wang F, Kennedy M, Cianciolo GJ, Bryja V, Pizzo SV, Bachelder RE. Ascites increases expression/function of multidrug resistance proteins in ovarian cancer cells. PLoS One. 2015;10:e0131579. doi: 10.1371/journal.pone.0131579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khrunin AV, Khokhrin DV, Moisseev AA, Gorbunova VA, Limborska SA. Pharmacogenomic assessment of cisplatin-based chemotherapy outcomes in ovarian cancer. Pharmacogenomics. 2014;15:329–337. doi: 10.2217/pgs.13.237. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Jiang K, Qiu X, Li M, Hao Q, Wei L, Zhang W, Chen B, Xin X. Overexpression of CXCR4 is significantly associated with cisplatin-based chemotherapy resistance and can be a prognostic factor in epithelial ovarian cancer. BMB Rep. 2014;47:33–38. doi: 10.5483/BMBRep.2014.47.1.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khrunin A, Ivanova F, Moisseev A, Khokhrin D, Sleptsova Y, Gorbunova V, Limborska S. Pharmacogenomics of cisplatin-based chemotherapy in ovarian cancer patients of different ethnic origins. Pharmacogenomics. 2012;13:171–178. doi: 10.2217/pgs.11.140. [DOI] [PubMed] [Google Scholar]
- 7.Xu S, Fu GB, Tao Z, OuYang J, Kong F, Jiang BH, Wan X, Chen K. MiR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget. 2015;6:26457–26471. doi: 10.18632/oncotarget.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang J, Ahmad A, Sarkar FH. The role of MicroRNAs in breast cancer migration, invasion and metastasis. Int J Mol Sci. 2012;13:13414–13437. doi: 10.3390/ijms131013414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014;281:1750–1758. doi: 10.1111/febs.12737. [DOI] [PubMed] [Google Scholar]
- 12.Brun JL, Feyler A, Chêne G, Saurel J, Brun G, Hocké C. Long-term results and prognostic factor in patients with epithelial ovarian cancer. Gynecol Oncol. 2000;78:21–27. doi: 10.1006/gyno.2000.5805. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Fu Z, Ji C, Gu P, Xu P, Yu N, Kan Y, Wu X, Shen R, Shen Y. Systematic gene microarray analysis of the lncRNA expression profiles in human uterine cervix carcinoma. Biomed Pharmacother. 2015;72:83–90. doi: 10.1016/j.biopha.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Lv M, Xu P, Wu Y, Huang L, Li W, Lv S, Wu X, Zeng X, Shen R, Jia X, et al. LncRNAs as new biomarkers to differentiate triple negative breast cancer from non-triple negative breast cancer. Oncotarget. 2016;7:13047–13059. doi: 10.18632/oncotarget.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, Lui WM, Wong ST, Leung GK. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Spurlock CF, III, Tossberg JT, Guo Y, Collier SP, Crooke PS, III, Aune TM. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat Commun. 2015;6:6932. doi: 10.1038/ncomms7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 19.Odening KE, Li W, Rutz R, Laufs S, Fruehauf S, Fishelson Z, Kirschfink M. Enhanced complement resistance in drug-selected P-glycoprotein expressing multi-drug-resistant ovarian carcinoma cells. Clin Exp Immunol. 2009;155:239–248. doi: 10.1111/j.1365-2249.2008.03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Connor R, O'Leary M, Ballot J, Collins CD, Kinsella P, Mager DE, Arnold RD, O'Driscoll L, Larkin A, Kennedy S, et al. A phase I clinical and pharmacokinetic study of the multi-drug resistance protein-1 (MRP-1) inhibitor sulindac, in combination with epirubicin in patients with advanced cancer. Cancer Chemother Pharmacol. 2007;59:79–87. doi: 10.1007/s00280-006-0240-7. [DOI] [PubMed] [Google Scholar]
- 21.Obata H, Yahata T, Quan J, Sekine M, Tanaka K. Association between single nucleotide polymorphisms of drug resistance-associated genes and response to chemotherapy in advanced ovarian cancer. Anticancer Res. 2006;26:2227–2232. [PubMed] [Google Scholar]
- 22.Scartozzi M, De Nictolis M, Galizia E, Carassai P, Bianchi F, Berardi R, Gesuita R, Piga A, Cellerino R, Porfiri E. Loss of hMLH1 expression correlates with improved survival in stage III–IV ovarian cancer patients. Eur J Cancer. 2003;39:1144–1149. doi: 10.1016/S0959-8049(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 23.Muenyi CS, States VA, Masters JH, Fan TW, Helm CW, States JC. Sodium arsenite and hyperthermia modulate cisplatin-DNA damage responses and enhance platinum accumulation in murine metastatic ovarian cancer xenograft after hyperthermic intraperitoneal chemotherapy (HIPEC) J Ovarian Res. 2011;4:9. doi: 10.1186/1757-2215-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metzinger DS, Taylor DD, Gercel-Taylor C. Induction of p53 and drug resistance following treatment with cisplatin or paclitaxel in ovarian cancer cell lines. Cancer Lett. 2006;236:302–308. doi: 10.1016/j.canlet.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Raspollini MR, Amunni G, Villanucci A, Castiglione F, Rossi Degl'Innocenti D, Baroni G, Paglierani M, Taddei GL. HER-2/neu and bcl-2 in ovarian carcinoma: Clinicopathologic, immunhistochemical and molecular study in patients with shorter and longer survival. Appl Immunohistochem Mol Morphol. 2006;14:181–186. doi: 10.1097/01.pai.0000155192.94214.f9. [DOI] [PubMed] [Google Scholar]
- 26.Dao MD, Alwan LM, Gray HJ, Tamimi HK, Goff BA, Liao JB. Recurrence patterns after extended treatment with bevacizumab for ovarian, fallopian tube, and primary peritoneal cancers. Gynecol Oncol. 2013;130:295–299. doi: 10.1016/j.ygyno.2013.04.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhillon S. Bevacizumab combination therapy: A review of its use in patients with epithelial ovarian, fallopian tube, or primary peritoneal cancer. BioDrugs. 2013;27:375–392. doi: 10.1007/s40259-013-0043-4. [DOI] [PubMed] [Google Scholar]
- 28.Chen YA, Aravin AA. Non-coding RNAs in transcriptional regulation: The review for current molecular biology reports. Curr Mol Biol Rep. 2015;1:10–18. doi: 10.1007/s40610-015-0002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latos PA, Pauler FM, Koerner MV, Şenergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST, Chu LW, Zhang Z, Zhao H, Zheng SL, Isaacs WB, Xu J. Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis. 2011;32:1655–1659. doi: 10.1093/carcin/bgr187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arase M, Horiguchi K, Ehata S, Morikawa M, Tsutsumi S, Aburatani H, Miyazono K, Koinuma D. Transforming growth factor-β-induced lncRNA-Smad7 inhibits apoptosis of mouse breast cancer JygMC(A) cells. Cancer Sci. 2014;105:974–982. doi: 10.1111/cas.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY, Li PF. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 35.Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al. lncrna-dependent mechanisms of androgen receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K, Liu CY, Zhou LY, Wang JX, Wang M, Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 37.Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen X, Xie B, Ma Z, Yu W, Wang W, Xu D, Yan X, Chen B, Yu L, Li J, et al. Identification of novel long non-coding RNAs in triple-negative breast cancer. Oncotarget. 2015;6:21730–21739. doi: 10.18632/oncotarget.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C, Li Z, Yang Y, Xiang T, Song W, Liu S. Microarray expression profiling of dysregulated long non-coding RNAs in triple-negative breast cancer. Cancer Biol Ther. 2015;16:856–865. doi: 10.1080/15384047.2015.1040957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 41.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Chen Z, Mishra AK, Silva A, Ren W, Pan Z, Wang JH. Chemotherapy-induced differential cell cycle arrest in B cell lymphomas affects their sensitivity to Wee1 inhibition. Haematologica: Haematol. 2017;2017:175992. doi: 10.3324/haematol.2017.175992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hajarnis SS, Patel V, Aboudehen K, Attanasio M, Cobo-Stark P, Pontoglio M, Igarashi P. Transcription factor hepatocyte nuclear factor-1β (HNF-1β) regulates MicroRNA-200 expression through a long Noncoding RNA. J Biol Chem. 2015;290:24793–24805. doi: 10.1074/jbc.M115.670646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui W, Qian Y, Zhou X, Lin Y, Jiang J, Chen J, Zhao Z, Shen B. Discovery and characterization of long intergenic non-coding RNAs (lincRNA) module biomarkers in prostate cancer: An integrative analysis of RNA-Seq data. BMC Genomics. 2015;16(Suppl 7):S3. doi: 10.1186/1471-2164-16-S7-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, Chen H, Hong J, Zou W, Chen Y, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890–6902. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6:9160–9172. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials are availabile.