Abstract
Human epidermal growth factor receptor 1 or 2 (HER1/2), and fibroblast growth factor receptor 1 (FGFR1) signaling serve critical roles in the progression of breast cancer; however, cross-talk between HER1/2 and FGFR1 signaling has not been extensively studied. In the present study, the copy number variation status of FGFR1 and HER1/2, and the clinical implications and prognostic relevance of this, were evaluated in invasive ductal breast cancer (IDC) tissue samples. Quantitative polymerase chain reaction and fluorescence in situ hybridization were used to assess gene copy number variation in IDC samples, and the clinical characteristics and survival curves of patients with IDC were analyzed. The amplification of FGFR1 was identified in 16.0% of the samples (12 of 75), of HER1 in 26.7% (20 of 75), of HER2 in 37.3% (28 of 75), and of FGFR1 and HER1/2 simultaneously in 8.0% (6 of 75). FGFR1 and HER1/2 co-amplification were significantly correlated with distant metastasis (P=0.035), recurrence (P=0.026) and decreased disease-free survival time (P=0.042). This was the case for patients undergoing endocrine therapy (P=0.002) and chemotherapy (P=0.044). Taken together, the results indicate that patients with FGFR1 and HER1/2 co-amplification may exhibit a less favorable prognosis compared with patients with either FGFR1, HER1/2 amplification or without amplification.
Keywords: FGFR1, HER1, HER2, breast cancer, prognosis, copy number variation
Introduction
Based on the 2014 World Health Organization report, breast cancer has the second highest incidence of mortality for females in China (1). Breast cancer is a heterogeneous disease; numerous frequent gene copy number variations (CNVs) have been identified, including gene amplification of fibroblast growth factor receptor 1 (FGFR1) (2), human epidermal growth factor receptor 1 (HER1), human epidermal growth factor 2 (HER2), GATA3, PIK3CA, MAP3K1, TBX3, RUNX1, CBFB, AFF2, PIK3R1, PTPN22, PTPRD, NF1, SF3B1 and CCND3 (3). Preliminary data from the next-generation genome sequencing of primary breast cancer has confirmed that CNVs may occur in a large selection of genes (3), and indicate that these variations may lead to different clinical consequences.
HER1 (also known as EGFR or ErbB1) and HER2 (also known as ErbB2) belong to the ErbB family of signaling proteins, which comprises four members: HER1, HER2, ErbB3 and ErbB4. ErbB receptors are often amplified, mutated and/or overexpressed in breast cancer (4,5). Between 15 and 20% of newly diagnosed invasive breast carcinomas overexpress HER2 or exhibit HER2 gene amplification (6). The frequency of HER1 overexpression in breast cancer is variable, reportedly ranging from 7 to 43% (7–13).
Activation of the ErbB family of receptor tyrosine kinases via their cognate epidermal growth factor-like peptide ligands constitutes a major event in the signaling pathways that control the proliferation, survival, angiogenesis and metastasis of breast cancer cells (14). Therefore, ErbB family member receptors are attractive potential therapeutic targets in breast cancer. At present, numerous tyrosine kinase inhibitors that target ErbBs have been successfully developed and approved to treat cancer patients. Trastuzumab (also known as Herceptin), a humanized monoclonal antibody against the extracellular portion of the HER2 protein, is in widespread clinical use (15). Notably, interactions between HER2 and other ErbB receptors, including HER1 and ErbB3, have been suggested as a possible mechanism for the resistance to trastuzumab. Once activated by the binding of its specific ligands, HER1 is the preferred heterodimerization partner for HER2; HER1/HER2 heterodimers are more stable than HER1 homodimers, and binding of HER1 with HER2 can potentiate and amplify the growth signals from HER1 activation (16–18).
FGFR1 has also been investigated and may be amplified in 8–15% of all cases of breast cancer (19–21). FGFR1 is a member of the FGFR family, which exhibit a highly conserved structure between members and throughout evolution. FGFRs are receptors for fibroblast growth factors; the interaction between fibroblast growth factors and FGFRs is associated with the regulation of cell proliferation, survival, migration and differentiation during development and adult life. The mutation and amplification of FGFRs causes the aberrant activation of downstream pathways, promoting cell cycle progression and mesenchymal transformation while inhibiting apoptosis. Amplification of the FGFR1 gene (at 8p11-12) is the most common alteration to FGFR1 (22–24). FGFR1 amplification may also drive resistance to endocrine therapy (25).
FGFR and EGFR signaling may mediate the downstream phosphoinositide-3-kinase/Akt serine/threonine kinase (PI3K/AKT) pathway. In this pathway, activated EGFR binds GRB2-associated binding protein 1 together with growth factor receptor-bound protein 2 to recruit PI3K (26). In addition, FGFRs commonly mediate the PI3K/AKT pathway via FGFR substrate 2a and other adaptor molecules (27). Major pathways downstream of activated EGFRs/FGFRs, besides PI3K/AKT, include extracellular signal-regulated kinase/mitogen-activated protein kinase 1 (ERK/MAPK) and protein kinase C/phospholipase C (PKC/PLC) pathways. All of these pathways serve an important role in cell proliferation, migration, differentiation and the inhibition of apoptosis.
In the present study, the gene amplification statuses of HER1, HER2 and FGFR1 were evaluated in 75 cases of invasive ductal breast cancer (IDC). Quantitative polymerase chain reaction (qPCR) and fluorescence in situ hybridization (FISH) were used to assess the gene CNV. A statistical analysis revealed an association between CNVs and the clinical prognosis.
Materials and methods
Patients and tissue samples
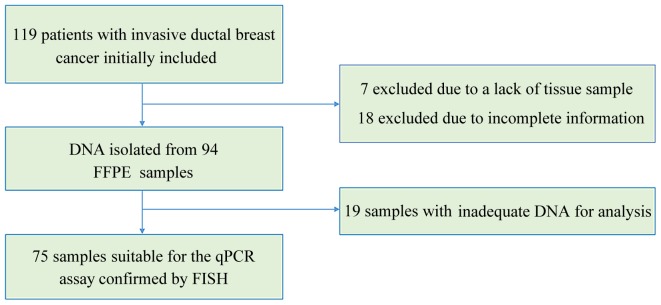
The records of the Department of Pathology of West China Hospital (Chengdu, China) were retrospectively examined and 119 records of IDC cases were initially included in the study (Fig. 1). Of the 119 cases, 7 were excluded as there was no tissue specimen available and 18 were excluded due to incomplete information. Of the remaining 94 formalin-fixed paraffin-embedded (FFPE) IDC tissue samples, 75 were suitable for qPCR detection. The samples qualified for qPCR if IDC was >70% of the FFPE sample and if they were acquired prior to systemic treatment. Data regarding clinical characteristics were gathered while preserving patient anonymity. The expression status of ER and PR were obtained from clinical pathology reports. The tumors were regarded as estrogen or progesterone receptor-positive if ≥1% of tumor cells were stained positively, according to previously reported criteria (28). The study was approved by the Ethics Committee of West China Hospital (no. 2013-191) and written informed consent was obtained from all patients.
Figure 1.
Flowchart showing the inclusion criteria of the study. FFPE, formalin-fixed paraffin-embedded; qPCR, quantitative polymerase chain reaction; FISH, fluorescence in situ hybridization
DNA isolation and qPCR
The tumor areas of each IDC sample were identified on slides stained with hematoxylin and eosin, and matched with corresponding FFPE tumor tissues. DNA was extracted from 4-µm-thick FFPE tissue sections using the QIAamp DNA FFPE Tissue Kit (Qiagen GmbH, Hilden, Germany). DNA was quantified with the Nanodrop2000 (Thermo Fisher Scientific, Inc., Wilmington, DE, USA) and samples with A260/280 absorbance ratios <1.70, or ≥1.95, were excluded. qPCR was performed with the Bio-Rad CFX96 system with SsoFast Evagreen Supermix (both Bio-Rad, Hercules, CA, USA) and quantified using the 2−ΔΔCq method (29). A total of 100 ng DNA was used per reaction and each reaction was performed at 98°C for 2 min, 98°C for 5 sec and 60°C for 10 sec, for 40 cycles. Primer sequences are included in Table I. Transferrin receptor and GAPDH were used as reference genes. Additionally, 50 normal samples were used as a control group, which were also obtained from Department of Pathology of West China Hospital and reported to possess no tumor cells in the initial pathologist reports. The relative amplification levels of FGFR1, HER1 and HER2 were normalized to the mean of the reference genes in order to calculate the relative CNV, compared with the control samples, using the 2−ΔΔCq method. A gene was considered to be amplified where the fold-change compared to the control group was ≥2.
Table I.
Primers used for the quantitative polymerase chain reaction detection of TFRC, GAPDH, FGFR1, HER1 and HER2.
| Gene | GenBank no. | Oligo type | Oligo sequence | Target size (bp) |
|---|---|---|---|---|
| TFRC | NC_000003.12 | Forward | 5′-ACTTCCTCTCTCCCTACGTATC-3′ | 105 |
| Reverse | 5′-GCAGTTTCAAGTTCTCCAGTAAAG-3′ | |||
| GAPDH | NG_007073.2 | Forward | 5′-CCTCAAGATCATCAGCAATGCCTC-3′ | 100 |
| Reverse | 5′-GTGGTCATGAGTCCTTCCACGATA-3′ | |||
| FGFR1 | NC_000008.11 | Forward | 5′-AGGCTGTGCTGTTGCACCTA-3′ | 128 |
| Reverse | 5′-ATCCGGGGCAGTTGCTAGTC-3′ | |||
| HER1 | NG_007726.3 | Forward | 5′-CGGGACGTTTCGTTCTTCGG-3′ | 130 |
| Reverse | 5′-GAAAGTTGGGAGCGGTTCGG-3′ | |||
| HER2 | NG_007503.1 | Forward | 5′-ATGAGCTACCTGGAGGATGT-3′ | 103 |
| Reverse | 5′-CCAGCCCGAAGTCTGTAATTT-3′ |
TFRC, transferrin receptor; FGFR1, fibroblast growth factor receptor 1; HER1, human epidermal growth factor receptor 1; HER2, human epidermal growth factor receptor 2.
FISH assays
FISH was performed on the tumor tissue samples with the following commercially available locus-specific and chromosome enumeration probes: HER1 (EGFR Spectrum Orange) with centromere 7 (CEP 7 Spectrum Green); HER2 (EGFR Spectrum Orange) with centromere 17 (CEP 17 Spectrum Green; all from LBP Medicine Science & Technology, Co., Ltd., Guangzhou, China); and FGFR1 (Orange) with centromere 8 (CEP 8 Spectrum Green; Empire Genomics, Buffalo, NY, USA). De-paraffinized 4-mm tumor sections were heated in antigen retrieval solution (sodium citrate, pH 6.0) in the microwave for 16 min, then in pepsin solution (LBP Medicine Science & Technology, Co., Ltd.) for 20 min at 43°C. The slides were dehydrated with 70, 85 and 100% ethanol. The tissue sections with probes for EGFR/CEP 7 or HER2/CEP 17 were denatured in a ThermoBrite hybridization chamber (IRIS International, Inc., Norwood, MA, USA) at 85°C for 5 min, followed by 20 h hybridization at 43°C. The tissue sections with probes for FGFR1/CEP 8 were denatured at 85°C for 5 min, followed by 20 h hybridization at 40°C. Following hybridization, washes were performed according to the supplier protocols. Slides were counterstained with 0.2 µmol/l 4′,6-diamidino-2-phenylindole in an anti-fade solution, and viewed with a fluorescence microscope. From each slide, a total of 60 tumor cells were evaluated; the gene and centromere copy number, and average predominant gene as defined by the mean ratio of the gene copy number vs. centromere copy number of each slide were estimated. A oncogene-to-centromere signal ratio ≥2 was considered to indicate amplification (30,31).
Statistical analysis
Statistical significance was assessed using SPSS software version 22.0 for Windows (IBM SPSS, Armonk, NY, USA). P<0.05 was considered to indicated a statistically significant difference. The association between clinical characteristics and gene CNV were analyzed using χ2 tests. Kaplan-Meier survival curves were plotted, and the significance of differences between survival curves was determined using the log-rank test.
Results
CNV status of FGFR1, HER1 and HER2
The CNV status of FGFR1, HER1 and HER2 were detected by qPCR and confirmed by FISH analysis for 75 patients with primary IDC (Table II; Fig. 2). It was demonstrated that 16.0% of the samples exhibited FGFR1 amplification (12 of 75), 26.7% HER1 amplification (20 of 75), 37.3% HER2 amplification (28 of 75), 50.7% HER1/2 amplification (38 of 75), 8.0% FGFR1 and HER1/2 co-amplification (6 of 75) and 42.7% samples exhibited no amplification (32 of 75). All samples detected by qPCR were positively confirmed by FISH analysis (100%).
Table II.
Clinicopathological characteristics of invasive ductal breast cancer with FGFR1 amplification, HER1/2 amplification, FGFR1 and HER1/2 co-amplification or no amplification.
| FGFR1 amplification, n (%) | HER1/2 amplificationa, n (%) | FGFR1 and HER1/2 co-amplification, n (%) | FGFR1 or HER1/2 amplification, n (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | + | − | P-value | + | − | P-value | + | − | P-value | + | − | P-value |
| Total | 12 (16.0) | 63 (84.0) | 38 (50.7) | 37 (49.3) | 6 (8.0) | 69 (92.0) | – | 43 (57.3) | 32 (42.7) | |||
| Age, years | 0.223 | 0.688 | 0.411 | 0.304 | ||||||||
| ≤50 | 6 (50.0) | 43 (68.3) | 24 (63.2) | 25 (67.6) | 3 (50.0) | 46 (66.7) | 17 (39.5) | 9 (28.1) | ||||
| >50 | 6 (50.0) | 20 (31.7) | 14 (36.8) | 12 (32.4) | 3 (50.0) | 23 (33.3) | 26 (60.5) | 23 (71.9) | ||||
| Tumor size | 0.312 | 0.174 | 0.495 | 0.289 | ||||||||
| T0-T2 | 12 (100.0) | 58 (92.1) | 34 (89.5) | 36 (97.3) | 6 (100.0) | 64 (92.8) | 39 (90.7) | 31 (96.9) | ||||
| T3-T4 | 0 (0.0) | 5 (7.9) | 4 (10.5) | 1 (2.7) | 0 (0.0) | 5 (7.2) | 4 (9.3) | 1 (3.1) | ||||
| Nodal status | 0.324 | 0.569 | 0.274 | 0.486 | ||||||||
| N0 | 5 (41.7) | 36 (57.1) | 22 (53.7) | 19 (46.3) | 2 (33.3) | 39 (56.5) | 25 (58.1) | 16 (50.0) | ||||
| N1-N3 | 7 (58.3) | 27 (42.9) | 16 (47.1) | 18 (52.9) | 4 (66.7) | 30 (43.5) | 18 (41.9) | 16 (50.0) | ||||
| Menopausal status | 0.084 | 0.289 | 0.244 | 0.147 | ||||||||
| Menopausal | 8 (66.7) | 25 (39.7) | 19 (50.0) | 14 (37.8) | 4 (66.7) | 29 (42.0) | 22 (51.2) | 11 (34.4) | ||||
| Premenopausal | 4 (33.3) | 38 (60.3) | 19 (50.0) | 23 (62.2) | 2 (33.3) | 40 (58.0) | 21 (48.8) | 21 (65.6) | ||||
| Clinical stage | 0.588 | 0.188 | 0.714 | 0.485 | ||||||||
| I–II | 10 (83.3) | 48 (76.2) | 27 (71.1) | 31 (83.8) | 5 (83.3) | 53 (76.8) | 32 (74.4) | 26 (81.3) | ||||
| III–IV | 2 (16.7) | 15 (23.8) | 11 (28.9) | 6 (16.2) | 1 (16.7) | 16 (23.2) | 11 (25.6) | 6 (18.7) | ||||
| ER Status | 0.642 | 0.007b | 0.438 | 0.003b | ||||||||
| ER+ | 9 (75.0) | 43 (68.3) | 21 (55.3) | 31 (83.8) | 5 (83.3) | 47 (68.1) | 19 (44.2) | 4 (12.5) | ||||
| ER- | 3 (25.0) | 20 (31.7) | 17 (44.7) | 6 (16.2) | 1 (16.7) | 22 (31.9) | 24 (55.8) | 28 (87.5) | ||||
| PR Status | 0.223 | 0.063 | 0.086 | 0.045b | ||||||||
| PR+ | 6 (50.0) | 43 (68.3) | 21 (44.7) | 28 (75.7) | 2 (33.3) | 47 (68.1) | 19 (44.2) | 7 (21.9) | ||||
| PR- | 6 (50.0) | 20 (31.7) | 17 (55.3) | 9 (24.3) | 4 (66.7) | 22 (31.9) | 24 (55.8) | 25 (78.1) | ||||
| HER2 | 0.538 | <0.001b | 0.769 | 0.001b | ||||||||
| 0-1+ | 8 (66.7) | 31 (49.2) | 15 (39.5) | 24 (64.9) | 3 (50.0) | 36 (52.2) | 20 (46.5) | 19 (59.4) | ||||
| 2+ | 2 (16.7) | 17 (27.0) | 6 (15.8) | 13 (35.1) | 1 (16.7) | 18 (26.1) | 7 (16.3) | 12 (37.5) | ||||
| 3+ | 2 (16.7) | 15 (23.8) | 17 (44.7) | 0 (0.0) | 2 (33.3) | 15 (21.7) | 16 (37.2) | 1 (3.1) | ||||
| Recurrence | 0.184 | 0.157 | 0.026b | 0.216 | ||||||||
| Yes | 1 (8.3) | 1 (98.4) | 2 (5.3) | 0 (0.0) | 1 (16.7) | 1 (1.4) | 41 (95.3) | 32 (100.0) | ||||
| No | 11 (91.7) | 62 (1.6) | 36 (94.7) | 37 (100.0) | 5 (83.3) | 68 (98.6) | 2 (4.7) | 0 (0.0) | ||||
| Distant metastasis | 0.341 | 0.051 | 0.035b | 0.249 | ||||||||
| Yes | 2 (16.7) | 5 (7.9) | 6 (15.8) | 1 (2.7) | 2 (33.3) | 5 (7.2) | 24 (55.8) | 16 (50.0) | ||||
| No | 10 (83.3) | 58 (92.1) | 32 (84.2) | 36 (97.3) | 4 (66.7) | 64 (92.8) | 19 (44.2) | 16 (50.0) | ||||
Including 20 HER1+ patients, 28 HER2+ patients, and 10 HER1+ and HER2+ patients.
Statistically significant (P<0.05). FGFR1, fibroblast growth factor receptor 1; HER1, human epidermal growth factor receptor 1; HER2, human epidermal growth factor receptor 2; ER, estrogen receptor; PR, progesterone receptor.
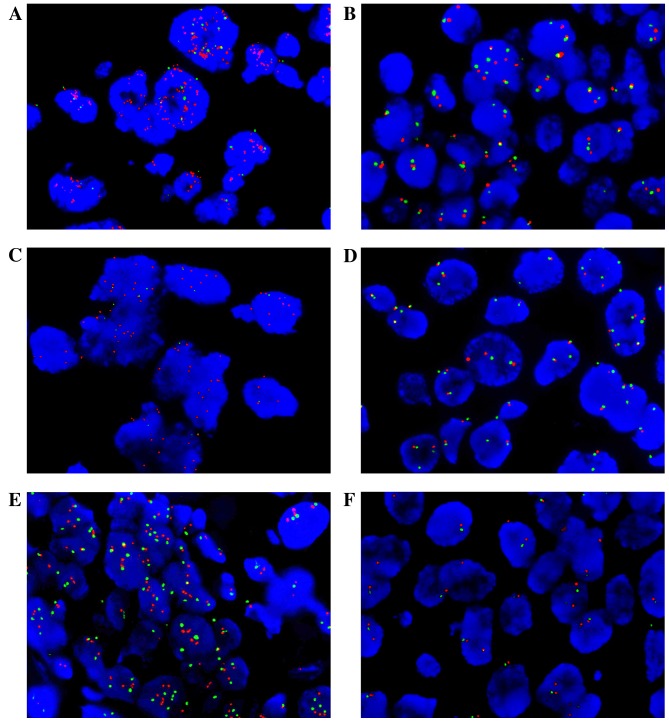
Figure 2.
Fluorescence in situ hybridization assays confirmed the quantitative polymerase chain reaction-detected FGFR1, HER1 and HER2 copy number variation in invasive ductal breast cancer tumor tissue samples. The detected signal for gene probes is displayed as red and for centromere probes as green. (A) FGFR1 amplification and (B) non-amplification. (C) HER1 amplification and (D) non-amplification. (E) HER2 amplification and (F) non-amplification. Magnification, ×100. FGFR, fibroblast growth factor receptor; HER1, human epidermal growth factor receptor 1; HER2, human epidermal growth factor 2.
Baseline clinical characteristics
A total of 75 patients with primary IDC were included (Table II). The median age at diagnosis of IDC was 48.4 years (range, 29–72 years). Patients were grouped according to the amplification status of FGFR1 or HER1/2. The amplification of HER1/2 was significantly associated with estrogen receptor (P=0.007) and HER2 (P<0.001) expression status. The frequency of local recurrence (P=0.026) and distant metastasis (P=0.035) were significantly higher in the subgroup with FGFR1 and HER1/2 co-amplification. A total of 5 patients developed a distant metastasis, of which 1 patient had metastasis at the time of diagnosis, and 2 patients had distant metastases and local recurrence simultaneously. There were 5 cases that resulted in mortality in the entire group; 2 were directly associated with breast cancer, whereas the other cases were uncertain.
The mean disease-free survival time was 25.7 months (range, 0–41 months; Table III); the mean overall survival time was 26.4 months (range, 12–41 months). Tumor size was correlated with disease-free survival time (P=0.006). Nodal status was significantly associated with disease-free (P=0.021) and overall (P=0.009) survival times. Additionally, the clinical stage of the cancer was significantly associated with overall survival time (P=0.042).
Table III.
Analysis of clinicopathological characteristics with disease-free and overall survival.
| Disease-free survival | Overall survival | ||||
|---|---|---|---|---|---|
| Parameter | n (%) | Log-rank | P-value | Log-rank | P-value |
| Age, years | 0.111 | 0.739 | 1.675 | 0.196 | |
| ≤50 | 49 (65.3) | ||||
| >50 | 26 (34.7) | ||||
| Tumor size | 7.672 | 0.006a | 1.967 | 0.160 | |
| T0-T2 | 70 (93.3) | ||||
| T3-T4 | 5 (6.7) | ||||
| Nodal status | 5.353 | 0.021a | 6.738 | 0.009a | |
| N0 | 41 (54.7) | ||||
| N1-N3 | 34 (45.3) | ||||
| Menopausal status | 0.008 | 0.927 | 0.614 | 0.433 | |
| Menopausal | 33 (44.0) | ||||
| Premenopausal | 42 (56.0) | ||||
| Clinical stage | 1.900 | 0.168 | 4.151 | 0.042a | |
| I–II | 58 (77.3) | ||||
| III–IV | 17 (22.7) | ||||
| ER status | 0.906 | 0.341 | 0.221 | 0.638 | |
| ER+ | 52 (69.3) | ||||
| ER- | 23 (30.7) | ||||
| PR status | 1.666 | 0.197 | 1.436 | 0.231 | |
| PR+ | 49 (65.3) | ||||
| PR- | 26 (34.7) | ||||
| HER2 | 1.827 | 0.401 | 0.883 | 0.643 | |
| 0-1+ | 39 (52.0) | ||||
| 2+ | 19 (25.3) | ||||
| 3+ | 17 (22.7) | ||||
| FGFR1 amplification | 0.786 | 0.375 | 0.934 | 0.334 | |
| + | 12 (16.0) | ||||
| − | 63 (84.0) | ||||
| HER1/2 amplification | 3.628 | 0.057 | 1.908 | 0.167 | |
| + | 38 (50.7) | ||||
| − | 37 (49.3) | ||||
| FGFR1 and HER1/2 co-amplification | 4.136 | 0.042a | 0.394 | 0.530 | |
| + | 6 (8.0) | ||||
| − | 69 (92.0) | ||||
Statistically significant (P<0.05). ER, estrogen receptor; PR, progesterone receptor; FGFR1, fibroblast growth factor receptor 1; HER1, human epidermal growth factor receptor 1; HER2, human epidermal growth factor receptor 2.
Prognostic significance of FGFR1 and HER1/2 co-amplification
FGFR1 and HER1/2 co-amplification was significantly associated with local recurrence and distant metastasis, as mentioned previously (Table II). To further investigate the association between FGFR1 and HER1/2 co-amplification and prognosis, FGFR1 and HER1/2 co-amplification status was assessed with a Kaplan-Meier analysis (Fig. 3). Based on this analysis, FGFR1 and HER1/2 co-amplification was significantly associated with reduced disease-free survival time (P=0.042; Fig. 3A). This was true for patients receiving chemotherapy (P=0.044) or endocrine therapy (P=0.002; Table IV). However, FGFR1 amplification (Fig. 3B) and HER1/2 amplification (Fig. 3C) individually exhibited no significant correlation with disease-free (P=0.375 and P=0.057, respectively) or overall (P=0.334 and P=0.167, respectively) survival time (Table III).
Figure 3.
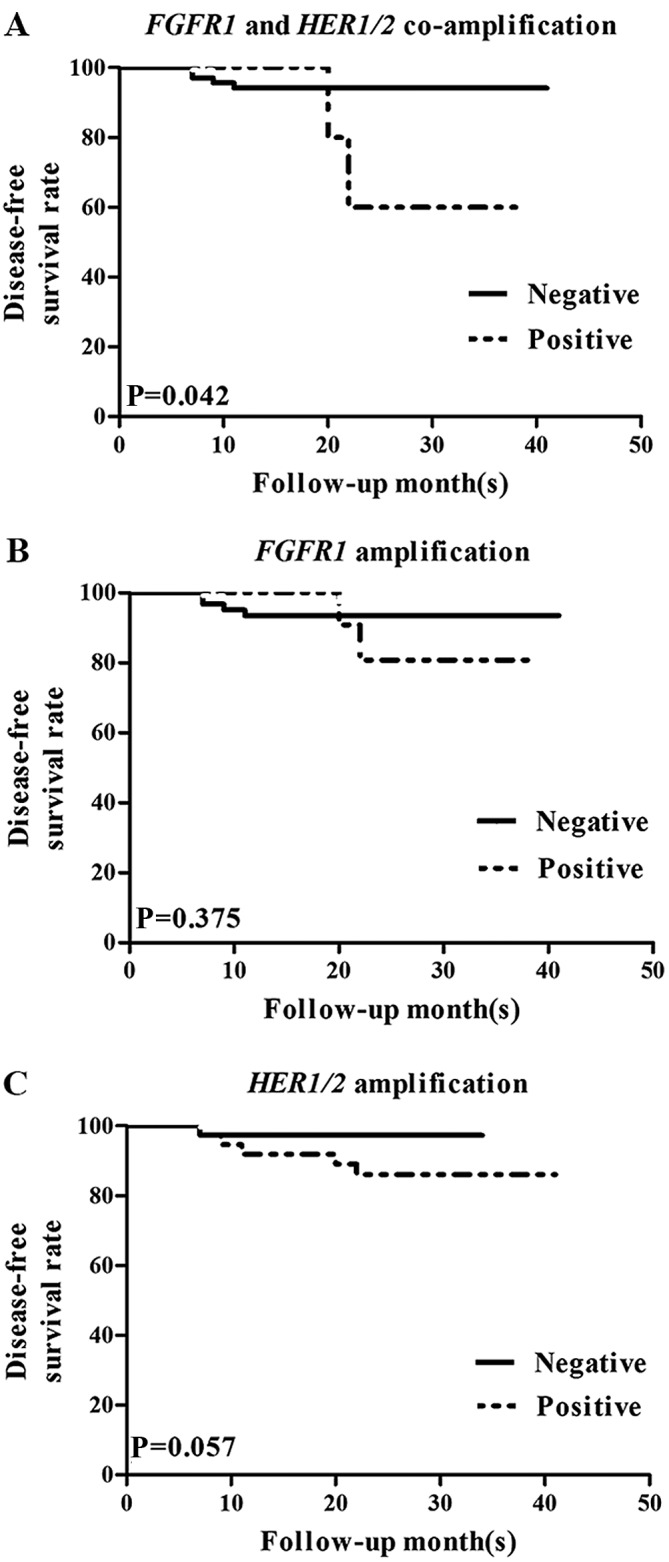
Kaplan-Meier survival analysis of disease-free survival time in association with FGFR1 and HER1/2 amplification in invasive ductal breast cancer. (A) The co-amplification of FGFR1 and HER1/2 was significantly associated with decreased disease-free survival time, whereas (B) FGFR1 and (C) HER1/2 amplification were not individually correlated with disease-free survival time. FGFR, fibroblast growth factor receptor; HER1/2, human epidermal growth factor receptor 1 or 2.
Table IV.
Association between treatment response and FGFR1 and HER1/HER2 co-amplification.
| FGFR1 and HER1/2 co-amplification, n (%) | Disease-free survival time | ||||
|---|---|---|---|---|---|
| Treatment | n | Positive | Negative | Log-rank | P-value |
| Chemotherapy | 74 | 6 (8.1) | 68 (91.9) | 4.038 | 0.044a |
| Endocrine therapy | 50 | 6 (12.0) | 44 (88.0) | 9.730 | 0.002a |
Statistically significant (P<0.05). FGFR1, fibroblast growth factor receptor 1; HER1/2, human epidermal growth factor receptor 1 or 2.
Discussion
The present retrospective study aimed to investigate the association between the CNV of FGFR1, HER1 and HER2, and the prognosis of patients with IDC. In the present study, FGFR1 was amplified in 15.6% of samples, and the amplification rates of HER1 and HER2 were 26.7 and 37.3%, respectively. In other studies, the amplification rates of FGFR1, HER1 and HER2 have been reported as ~10, ~15 and ~20%, respectively (6,32,33). These differences may be due to differences in the methods for detection, classification standards for amplification, or sample sizes between the studies.
In the present study, the group of patients with co-amplification of FGFR1 and HER1/2 was significantly more likely to experience recurrence and distant metastasis. Co-amplification also influenced disease-free survival time, with the co-amplification experiencing less favorable outcomes. No association between prognosis and amplification of FGFR1 or HER1/2 alone was identified. The data of the present study contrasted from previous indications that the amplification of FGFR1 is associated with poor survival time (20), as FGFR1 amplification did not affect disease-free or overall survival time in the current study. The difference may be due to the constitution of the sample; different types of breast cancer or a different number of patients may have caused the drift. The amplification of FGFR1 was, however, associated with menopausal status; the majority of patients with FGFR1 amplification were menopausal, in contrast to the patients without FGFR1 amplification. HER1/2 amplification status was associated with the ER and HER2 protein expression statuses. This result was consistent with the view that HER2 amplification is highly associated with its protein overexpression (6).
In addition, the response to therapy in the FGFR1 and HER1/2 co-amplification group was investigated. The data included patients who underwent chemotherapy and endocrine therapy. The 6 patients who harbored FGFR1 and HER1/2 co-amplification had poor outcomes following chemotherapy or endocrine therapy. Insensitivity to therapy may have caused the group of patients to have unfavorable outcomes. Accordingly, further study is required to investigate whether, and how, FGFR1 and HER1/2 co-amplification can influence tumor resistance to drug therapy.
The amplification of HER1 is not as common in breast cancer as HER2 amplification, and the effect of HER1 amplification on patients with breast cancer remains unclear (34,35). By contrast, HER2 is already a therapeutic target, and the effect of HER2 amplification for patients with breast cancer has been confirmed (14,36). Previously the amplification of FGFR1 has been regarded as an independent prognostic factor and a potential therapeutic target in breast cancer, and it may be associated with resistance in endocrine therapy (25). FGFR1 amplification is rarely exhibited in HER2-amplified tumors (25); the genes are mutually exclusive methods for activating similar downstream pathways, including PI3K/AKT, ERK/MAPK and PKC/PLC (29). Further investigation is required to study how the co-activation of FGFR and HER1/2 could affect downstream pathways.
In conclusion, the expression levels of FGFR1, HER1, HER2 were detected using qPCR and FISH. A specific group of patients with co-amplification of FGFR1 and HER1/2 was identified to be associated with worse prognosis, and correlated with resistance to endocrine therapy and chemotherapy. This finding suggests that the gene statuses of FGFR1, HER1 and HER2 can be predictive of clinical outcome, and that different therapeutic strategies should be implemented for these patients in order to improve the prognosis.
Competing interests
The authors declare that they have no competing interests.
Glossary
Abbreviations
- HER1/2
human epidermal growth factor receptor 1 or 2
- FGFR
fibroblast growth factor receptor
- IDC
invasive ductal breast cancer
- CNV
copy number variation
- AKT
Akt serine/threonine kinase
- PI3K
phosphoinositide-3-kinase
- ERK
extracellular signal-regulated kinase
- MAPK
mitogen-activated protein kinase 1
- PKC
protein kinase C
- PLC
phospholipase C
- qPCR
quantitative polymerase chain reaction
- FISH
fluorescence in situ hybridization
- FFPE
formalin-fixed paraffin-embedded
References
- 1.Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27:2–12. doi: 10.1186/s40880-015-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rougé C, Lasorsa L, Letessier A, Ginestier C, Monville F M, et al. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res. 2005;3:655–667. doi: 10.1158/1541-7786.MCR-05-0128. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network: Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Normanno N, Bianco C, Strizzi L, Mancino M, Maiello MR, De Luca A, Caponigro F, Salomon DS. The ErbB receptors and their ligands in cancer: An overview. Curr Drug Targets. 2005;6:243–257. doi: 10.2174/1389450053765879. [DOI] [PubMed] [Google Scholar]
- 5.Troyer KL, Lee DC. Regulation of mouse mammary gland development and tumorigenesis by the ERBB signaling network. J Mammary Gland Biol Neoplasia. 2001;6:7–21. doi: 10.1023/A:1009560330359. [DOI] [PubMed] [Google Scholar]
- 6.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 7.DiGiovanna MP, Stern DF, Edgerton SM, Whalen SG, Moore D, II, Thor AD. Relationship of epidermal growth factor receptor expression to ErbB-2 signaling activity and prognosis in breast cancer patients. J Clin Oncol. 2005;23:1152–1160. doi: 10.1200/JCO.2005.09.055. [DOI] [PubMed] [Google Scholar]
- 8.Wrba F, Reiner A, Ritzinger E, Holzner JH, Reiner G. Expression of epidermal growth factor receptors (EGFR) on breast carcinomas in relation to growth fractions, estrogen receptor status and morphological criteria. An immunohistochemical study. Pathol Res Pract. 1988;183:25–29. doi: 10.1016/S0344-0338(88)80153-5. [DOI] [PubMed] [Google Scholar]
- 9.Suo Z, Risberg B, Kalsson MG, Willman K, Tierens A, Skovlund E, Nesland JM. EGFR family expression in breast carcinomas. c-erbB-2 and c-erbB-4 receptors have different effects on survival. J Pathol. 2002;196:17–25. doi: 10.1002/path.1003. [DOI] [PubMed] [Google Scholar]
- 10.Nieto Y, Nawaz F, Jones RB, Shpall EJ, Nawaz S. Prognostic significance of overexpression and phosphorylation of epidermal growth factor receptor (EGFR) and the presence of truncated EGFRvIII in locoregionally advanced breast cancer. J Clin Oncol. 2007;25:4405–4413. doi: 10.1200/JCO.2006.09.8822. [DOI] [PubMed] [Google Scholar]
- 11.Koletsa T, Kotoula V, Karayannopoulou G, Nenopoulou E, Karkavelas G, Papadimitriou CS, Kostopoulos I. EGFR expression and activation are common in HER2 positive and triple-negative breast tumours. Histol Histopathol. 2010;25:1171–1179. doi: 10.14670/HH-25.1171. [DOI] [PubMed] [Google Scholar]
- 12.Hwangbo W, Lee JH, Ahn S, Kim S, Park KH, Kim CH, Kim I. EGFR gene amplification and protein expression in invasive ductal carcinoma of the breast. Korean J Pathol. 2013;42:107–115. doi: 10.4132/KoreanJPathol.2013.47.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhargava R, Gerald WL, Li AR, Pan Q, Lal P, Ladanyi M, Chen B. EGFR gene amplification in breast cancer: Correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod Pathol. 2005;18:1027–1033. doi: 10.1038/modpathol.3800438. [DOI] [PubMed] [Google Scholar]
- 14.Normanno N, Bianco C, De Luca A, Maiello MR, Salomon DS. Target-based agents against ErbB receptors and their ligands: A novel approach to cancer treatment. Endocr Relat Cancer. 2003;10:1–21. doi: 10.1677/erc.0.0100001. [DOI] [PubMed] [Google Scholar]
- 15.de Azambuja E, Procter MJ, van Veldhuisen DJ, Agbor-Tarh D, Metzger-Filho O, Steinseifer J, Untch M, Smith IE, Gianni L, Baselga J, et al. Trastuzumab-associated cardiac events at 8 years of median follow-up in the Herceptin Adjuvant trial (BIG 1–01) J Clin Oncol. 2014;32:2159–2165. doi: 10.1200/JCO.2013.53.9288. [DOI] [PubMed] [Google Scholar]
- 16.Graus-Porta D, Beerli RR, Daly JM, Hynes NE. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Worthylake R, Opresko LK, Wiley HS. ErbB-2 amplification inhibits down-regulation and induces constitutive activation of both ErbB-2 and epidermal growth factor receptors. J Biol Chem. 1999;274:8865–8874. doi: 10.1074/jbc.274.13.8865. [DOI] [PubMed] [Google Scholar]
- 18.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 19.Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15:441–451. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- 20.Elsheikh Elbauomy S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, Ellis IO, Reis-Filho JS. FGFR1 amplification in breast carcinomas: A chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9:R23. doi: 10.1186/bcr1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reis-Filho JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, Grigoriadis A, Sarrio D, Savage K, Dexter T, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–6662. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- 22.Penault-Llorca F, Bertucci F, Adélaïde J, Parc P, Coulier F, Jacquemier J, Birnbaum D, deLapeyrière O. Expression of FGF and FGF receptor genes in human breast cancer. Int J Cancer. 1995;61:170–176. doi: 10.1002/ijc.2910610205. [DOI] [PubMed] [Google Scholar]
- 23.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi OP, Ethier SP. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer. 1999;81:1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, Natrajan R, Marchio C, Iorns E, Mackay A, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baird K, Davis S, Antonescu CR, Harper UL, Walker RL, Chen Y, Glatfelter AA, Duray PH, Meltzer PS. Gene expression profiling of human sarcomas: Insights into sarcoma biology. Cancer Res. 2005;65:9226–9235. doi: 10.1158/0008-5472.CAN-05-1699. [DOI] [PubMed] [Google Scholar]
- 27.Tenhagen M, van Diest PJ, Ivanova IA, van der Wall E, van der Groep P. Fibroblast growth factor receptors in breast cancer: Expression, downstream effects, and possible drug targets. Endocr Relat Cancer. 2012;19:R115–R129. doi: 10.1530/ERC-12-0060. [DOI] [PubMed] [Google Scholar]
- 28.Hammond ME, Hayes DF, Wolff AC, Mangu PB, Temin S. American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Oncol Pract. 2010;6:196–197. doi: 10.1200/JOP.777003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Russell PA, Yu Y, Young RJ, Conron M, Wainer Z, Alam N, Solomon B, Wright GM. Prevalence, morphology, and natural history of FGFR1-amplified lung cancer, including squamous cell carcinoma, detected by FISH and SISH. Mod Pathol. 2014;27:1621–1631. doi: 10.1038/modpathol.2014.71. [DOI] [PubMed] [Google Scholar]
- 31.Cihoric N, Savic S, Schneider S, Ackermann I, Bichsel-Naef M, Schmid RA, Lardinois D, Gugger M, Bubendorf L, Zlobec I, Tapia C. Prognostic role of FGFR1 amplification in early-stage non-small cell lung cancer. Br J Cancer. 2014;110:2914–2922. doi: 10.1038/bjc.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, Rodriguez C, Theillet C. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: Definition of phenotypic groups. Cancer Res. 1997;57:4360–4367. [PubMed] [Google Scholar]
- 33.Kersting C, Tidow N, Schmidt H, Liedtke C, Neumann J, Boecker W, van Diest PJ, Brandt B, Buerger H. Gene dosage PCR and fluorescence in situ hybridization reveal low frequency of egfr amplifications despite protein overexpression in invasive breast carcinoma. Lab Invest. 2004;84:582–587. doi: 10.1038/labinvest.3700077. [DOI] [PubMed] [Google Scholar]
- 34.Bhargava R, Gerald WL, Li AR, Pan Q, Lal P, Ladanyi M, Chen B. EGFR gene amplification in breast cancer: Correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod Pathol. 2005;18:1027–1033. doi: 10.1038/modpathol.3800438. [DOI] [PubMed] [Google Scholar]
- 35.Iqbal N, Iqbal N. Human epidermal growth factor receptor 2 (HER2) in cancers: Overexpression and therapeutic implications. Mol Biol Int. 2014;2014:852748. doi: 10.1155/2014/852748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]