Abstract
The development of colorectal cancer (CRC) involves genetic and epigenetic modifications, and aberrant DNA methylation within gene promoters is a primary mediator of epigenetic inheritance in CRC. The present study evaluated whether promoter methylation of four CRC candidate genes [protocadherin γ subfamily A12 (PCDH-γ-A12), solute carrier family 19 A 1 (SLC19A1), cAMP responsive element binding protein (CREB) and cylindromatosis (CYLD) contributed to the risk and metastasis of CRC by screening a total of 42 CRC and 42 adjacent normal tissue samples. DNA methylation was measured by methylation-specific polymerase chain reaction (MSP). Polymerase chain reaction (PCR) products were bisulfite converted and validated by sequencing. The χ2 test was employed to assess the association between promoter methylation and a series of clinicopathological characteristics. The promoters of PCDH-γ-A12 and SLC19A1 were observed to be more frequently methylated in CRC tissues than normal tissues. In addition, significantly higher methylation of the PCDH-γ-A12 and SLC19A1 promoters was also observed in CRC tissues with lymph metastasis compared with those without lymph metastasis. In addition, no association was observed between CREB and CYLD methylation and the occurrence and metastasis of CRC. These results suggest that the hypermethylation of the PCDH-γ-A12 and SLC19A1 promoters may contribute to the occurrence and metastasis of CRC in the Han Chinese population.
Keywords: colorectal cancer, methylation-specific polymerase chain reaction, metastasis, promoter, protocadherin γ subfamily A12, solute carrier family 19 A 1
Introduction
Colorectal cancer (CRC) is one of the most common digestive malignancies, and it arises through well-defined sequential multi-step carcinogenesis that transforms normal glandular epithelium into invasive adenocarcinomas (1,2). The development of CRC involves genetic and epigenetic modifications. Aberrant DNA methylation within gene promoters is a primary mediator of epigenetic inheritance in CRC (3,4).
DNA methylation typically occurs in CpG islands and it refers to the enzymatic addition of a methyl group to the 5′ position of cytosine by DNA methyltransferases to produce 5-methyl cytosine. Methylation of CpG islands in the gene promoter region may induce chromatin conformational modifications that inhibit access of transcriptional machinery, altering gene expression levels (5,6). Therefore, promoter methylation is commonly associated with gene silencing and promoter demethylation with gene expression (7,8).
There are complex changes of DNA methylation in a number of carcinomas, and particularly in CRC (9,10). Numerous genes are aberrantly methylated in CRC patients, including adenomatous polyposis coli (APC), WNT5A, mutL homolog 1 (MLH1), cyclin-dependent kinase inhibitor 2A (CDKN2A) and Ras association domain-containing protein 1 (11–15). Aberrant DNA methylation of gene promoters in CRC is involved in its occurrence, progression, diagnosis, staging, prognosis and response to chemotherapy (16).
The protocadherin gamma subfamily A12 (PCDH-γ-A12) gene encodes a cell surface adhesion protein that serves essential roles in cell-cell and cell-matrix interactions and tumor metastasis (17,18). The solute carrier family 19 A 1 (SLC19A1) gene encodes a membrane protein that is involved in the regulation of intracellular concentrations of folate (19). SLC19A1 gene mutation is associated with the risk of CRC (20). The cAMP responsive element binding protein (CREB) gene encodes a transcription factor that induces the transcription of genes in response to hormonal stimulation of the cAMP pathway (21,22). P300/CREB binding protein genes promote cancer progression in colon cancer cell lines with microsatellite instability (23). Cylindromatosis (CYLD) encodes a cytoplasmic protein with three cytoskeletal-associated protein-glycine-conserved domains, and it regulates cell proliferation, apoptosis, cell movement and cell differentiation (24–27). CYLD is downregulated or lost in colon carcinoma cell lines compared with primary human colonic epithelial cells. The functional relevant loss of CYLD expression may contribute to tumor development and progression, and it may provide a new target for therapeutic strategies (28).
Promoter methylation of the PCDH-γ-A12, SLC19A1, CREB and CYLD genes has been demonstrated to regulate their gene expression levels, and hypermethylation of these promoters has been observed in acute lymphoblastic leukemia (29), breast cancer (30,31) and malignant melanoma (32). However, hypermethylation of the PCDH-γ-A12, SLC19A1, CREB and CYLD promoters has not been investigated in CRC. In light of the previous findings, the aim of the present study was to investigate whether PCDH-γ-A12, SLC19A1, CREB and CYLD gene promoter methylation contributed to the risk of CRC.
Materials and methods
Tissue sample collection
In this study, CRC patients who had not received radiotherapy, chemotherapy, targeted therapy or dendritic cell/cytokine-induced killer therapy prior to surgery were recruited between June 2012 and April 2013 (Table I). CRC samples, normal adjacent tissue samples and matched metastatic lymph node samples were collected at the time of surgery from 42 primary sporadic CRC patients at the Department of Gastrointestinal Surgery in the Affiliated Hospital of Ningbo University, China. Tissues were immediately preserved in liquid nitrogen at −80°C following removal from the body and stored at −80°C until use. Normal adjacent tissues were collected from at least 5 cm away from the edge of the tumor, and there were no obvious tumor cells, as evaluated by a pathologist. Tumor stage was determined according to Dukes' staging system, and cellular differentiation was graded according to Broders' grading system. Informed consent was given by all subjects. The Human Research Ethics Committee of Ningbo University approved all aspects of the study.
Table I.
Clinical profiles of the colorectal cancer patients.
| Characteristics | Subgroup | Patients, n |
|---|---|---|
| Gender | Male | 28 |
| Female | 14 | |
| Age (years) | ≤60 | 16 |
| >60 | 26 | |
| TNM stage | 1, 2 | 21 |
| 3, 4 | 21 | |
| Lymph metastasis | Yes | 21 |
| No | 21 | |
| Distant metastasis | Yes | 8 |
| No | 34 | |
| CEA | ≥5.0 ng/ml | 15 |
| <5.0 ng/ml | 27 | |
| CA19-9 | ≥37 U/ml | 9 |
| <37 U/ml | 33 | |
| Tumor location | Colon | 26 |
| Rectum | 16 | |
| Differentiation | Poor | 10 |
| Moderate | 32 | |
| Good | 0 | |
| Tumor size | <5 cm | 28 |
| ≥5 cm | 14 | |
| Histological | Adenocarcinoma | 40 |
| classification | Mucinous adenocarcinoma | 2 |
| Undifferentiated carcinoma | 0 |
TNM, tumor-node-metastasis; CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19-9.
DNA isolation and bisulfite modification
Genomic DNA was isolated using a QIAamp DNA mini kit (Qiagen GmbH, Hilden, Germany). The concentration and quality of genomic DNA were determined using the NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The concentration of DNA was above 30 ng/µl and the purity of DNA was at the A260/A280 ratio of 1.7–1.9. DNA was bisulfite-treated with the EZ DNA Methylation-Gold kit (Zymo Research, Orange, CA, USA). Following the completion of bisulfite modification, all unmethylated cytosines in CpG islands were converted to uracil, while methylated cytosines remained unchanged.
Methylation-specific polymerase chain reaction (MSP) and bisulfite sequencing
The methylated and unmethylated primers (Table II) were designed using the Primer Premier 6.0 program (Premier Biosoft International, Palo Alto, CA, USA). MSP was performed in a total volume of 20 µl containing 2 µl bisulfite modified DNA, 1 µmol each of forward and reverse primers, 10 µl Premix Taq (Takara Biotechnology Co., Ltd., Dalian, China) and 7 µl double-distilled water with the following cycling parameters: 10 min of denaturation at 95°C followed by 55 cycles of 30 sec at 95°C, 45 sec at 72°C and a final extension for 10 min at 72°C. Polymerase chain reaction (PCR) products were then loaded and electrophoresed on 2% agarose gels, stained with ethidium bromide, and visualized under UV illumination. In order to confirm the result of methylation- and unmethylation-specific PCR, PCR products randomly obtained from the group were sequenced bidirectionally by Invitrogen (Thermo Fisher Scientific, Inc.) with the same primers used for MSP.
Table II.
List of all primers used.
| Gene | Subgroup | Sense (5′-3′) | Antisense (5′-3′) | Size (bp) |
|---|---|---|---|---|
| PCDH-γ-A12 | M | ATTAAGGTGGTGGCGGTGGAT | GACGCCGACGCTCCTATCAA | 449 |
| U | AAGGTGGTGGTGGTGGATAG | ACCAACACTCCTATCAAAC | 443 | |
| SLC19A1 | M | TTGTTGTAGCGGTGTTGGAAGG | TCCGCCGCAACCTACGAAT | 361 |
| U | TTTGTTGTAGTGGTGTTGGAAG | TTCCACCACAACCTACAAAT | 363 | |
| CREB | M | CGGCGGTTAAGAGTAGAGTTA | GCGTCACTCACCAACACT | 492 |
| U | TGGTGGTTAAGAGTAGAGTTA | TCACTCACCAACACTCCAC | 489 | |
| CYLD | M | AGTTGGTGGTAGCGTAGCG | CATTCACTAACCTCGAACGA | 495 |
| U | TGGTGGTAGTGTAGTGTTT | TCACTAACCTCAAACAACA | 489 |
PCDH-γ-A12, protocadherin γ subfamily A12; SLC19A1, solute carrier family 19 A 1; CREB, cAMP responsive element binding protein; CYLD, cylindromatosis; M, methylated; U, unmethylated.
Statistics
Statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS) statistical software package (version 16.0; SPSS, Inc., Chicago, IL, USA), and the results were obtained using GraphPad Prism version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). All analyses were two-sided, and P<0.05 was considered to indicate a statistically significant difference.
Results
Methylation rates of promoters in CRC vs
normal tissues
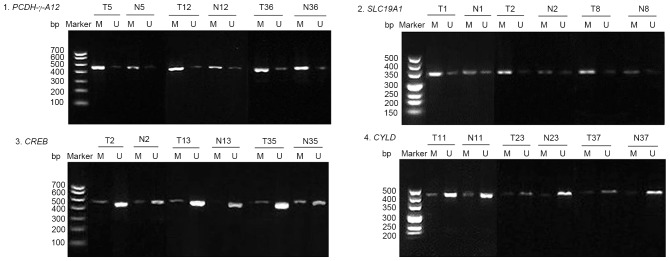
A total of 42 pairs of CRC and adjacent normal tissues were examined, and representative results of the agarose gel electrophoresis were selected (Fig. 1). The results revealed that the methylation rates of PCDH-γ-A12, SLC19A1, CREB and CYLD promoters in CRC were 83.33% (35/42), 78.57% (33/42), 26.19% (11/42) and 14.29% (6/42), while the methylation rates of these promoters in normal tissues were 57.14% (24/42), 45.24% (19/42), 11.90% (5/42) and 11.90% (5/42). PCDH-γ-A12 and SLC19A1 gene promoters were more frequently methylated in CRC tissues than in normal tissues (83.33% vs. 57.14%, P=0.009 and 78.57% vs. 42.54%, P=0.002), while there was no significant difference in methylation rates of CREB and CYLD gene promoters between CRC tissues and normal tissues (26.19% vs. 11.90%, P=0.095 and 14.29% vs. 11.90%, P=0.746; Table III).
Figure 1.
Representative results for methylation status of protocadherin γ subfamily A12 (PCDH-γ-A12), solute carrier family 19 A 1 (SLC19A1), cAMP responsive element binding protein (CREB) and cylindromatosis (CYLD) genes in colorectal cancer tissues (T) and adjacent normal tissues (N). M, methylated; U, unmethylated.
Table III.
Methylation status of PCDH-γ-A12, SLC19A1, CREB and CYLD genes in colorectal cancer and normal tissues.
| Gene | Group | Total | M | U | M% | χ2 | P-value |
|---|---|---|---|---|---|---|---|
| PCDH-γ-A12 | Cases | 42 | 35 | 7 | 83.33 | 6.891 | 0.009 |
| Controls | 42 | 24 | 18 | 57.14 | |||
| SLC19A1 | Cases | 42 | 33 | 9 | 78.57 | 9.894 | 0.002 |
| Controls | 42 | 19 | 23 | 45.24 | |||
| CREB | Cases | 42 | 11 | 31 | 26.19 | 2.779 | 0.095 |
| Controls | 42 | 5 | 37 | 11.90 | |||
| CYLD | Cases | 42 | 6 | 36 | 14.29 | 0.105 | 0.746 |
| Controls | 42 | 5 | 37 | 11.90 |
PCDH-γ-A12, protocadherin γ subfamily A12; SLC19A1, solute carrier family 19 A 1; CREB, cAMP responsive element binding protein; CYLD, cylindromatosis; M, methylated; U, unmethylated.
Methylation rates of promoters in lymph vs. non-lymph metastasis CRC tissues
In addition, the methylation rates of PCDH-γ-A12, SLC19A1, CREB and CYLD promoters in lymph metastasis CRC tissues were 100.00% (21/21), 95.24% (20/21), 33.33% (7/21) and 19.05% (4/21), while the methylation rates of these promoters in non-lymph metastasis CRC tissues were 66.67% (14/21), 61.90% (13/21), 19.05% (4/21) and 9.52% (2/21). PCDH-γ-A12 and SLC19A1 gene promoters were more frequently methylated in lymph metastasis CRC tissues than non-lymph metastasis CRC tissues (100.00% vs. 66.67%, P=0.013% and 95.24% vs. 61.90%, P=0.024), while there was no significant difference in the methylation rate of CREB and CYLD gene promoters between lymph metastasis CRC tissues and non-lymph metastasis CRC tissues (33.33% vs. 19.05%, P=0.292 and 19.05% vs. 9.52%, P=0.659; Table IV).
Table IV.
Methylation status of PCDH-γ-A12, SLC19A1, CREB and CYLD genes in lymph metastasis and non-lymph metastasis colorectal cancer tissues.
| Gene | Subgroup | Total | M | U | M% | χ2 | P-value |
|---|---|---|---|---|---|---|---|
| PCDH-γ-A12 | Cases | 21 | 21 | 0 | 100.00 | 6.171 | 0.013 |
| Controls | 21 | 14 | 7 | 66.67 | |||
| SLC19A1 | Cases | 21 | 20 | 1 | 95.24 | 5.091 | 0.024 |
| Controls | 21 | 13 | 8 | 61.90 | |||
| CREB | Cases | 21 | 7 | 14 | 33.33 | 1.109 | 0.292 |
| Controls | 21 | 4 | 17 | 19.05 | |||
| CYLD | Cases | 21 | 4 | 17 | 19.05 | 0.194 | 0.659 |
| Controls | 21 | 2 | 19 | 9.52 |
PCDH-γ-A12, protocadherin γ subfamily A12; SLC19A1, solute carrier family 19 A 1; CREB, cAMP responsive element binding protein; CYLD, cylindromatosis; M, methylated; U, unmethylated.
Bisulphite sequencing of PCDH-γ-A12, SLC19A1, CREB and CYLD genes
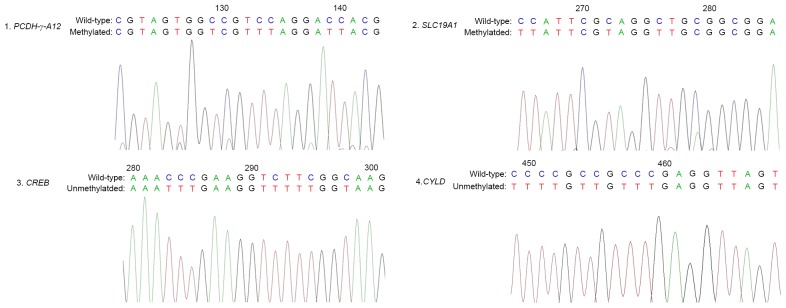
In order to confirm the results of the PCR-based methylation analysis describe above, high-resolution bisulfite genomic sequencing was performed in the stochastic samples derived from the methylation PCR experiments. In agreement with the MSP results, CpG dinucleotides of the PCDH-γ-A12 and SLC19A1 promoters in the samples demonstrated extensive hypermethylation, whereas the CREB and CYLD promoters were unmethylated at these CpG dinucleotides (Fig. 2).
Figure 2.
Bisulphite sequencing of protocadherin γ subfamily A12 (PCDH-γ-A12), solute carrier family 19 A 1 (SLC19A1), cAMP responsive element binding protein (CREB) and cylindromatosis (CYLD) genes.
Correlation between methylation status of promoters and clinicopathological factors
The correlation between the methylation status of the PCDH-γ-A12, SLC19A1, CREB and CYLD gene promoters and the clinicopathological characteristics of CRC is shown in Table V. There was no significant difference in clinicopathological factors, including sex, age, tumor-node-metastasis stage, lymph node status, metastasis status, tumor location, differentiation status, tumor size and histological grade. There was also no correlation between the methylation status of the PCDH-γ-A12, SLC19A1, CREB and CYLD gene promoters and the serum levels of carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9).
Table V.
Association between PCDH-γ-A12, SLC19A1, CREB and CYLD methylation in CRC serum and clinicopathological features.
| PCDH-γ-A12 | SLC19A1 | CREB | CYLD | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Subgroup | Patient, n | M | U | P-value | M | U | P-value | M | U | P-value | M | U | P-value |
| Gender | Male | 28 | 25 | 3 | 0.306 | 21 | 7 | 0.690 | 7 | 21 | 1 | 4 | 24 | 1.000 |
| Female | 14 | 10 | 4 | 12 | 2 | 4 | 10 | 2 | 12 | |||||
| Age (years) | ≤60 | 16 | 14 | 2 | 0.887 | 10 | 6 | 0.109 | 4 | 12 | 1 | 3 | 13 | 0.846 |
| >60 | 26 | 21 | 5 | 23 | 3 | 7 | 19 | 3 | 23 | |||||
| TNM stage | 1, 2 | 21 | 18 | 3 | 1.000 | 17 | 4 | 1.000 | 6 | 15 | 0.547 | 2 | 19 | 0.659 |
| 3, 4 | 21 | 17 | 4 | 16 | 5 | 5 | 16 | 4 | 17 | |||||
| Lymph metastasis | Yes | 21 | 21 | 0 | 0.013 | 20 | 1 | 0.024 | 7 | 14 | 0.292 | 4 | 17 | 0.756 |
| No | 21 | 14 | 7 | 13 | 8 | 4 | 17 | 2 | 19 | |||||
| Distant metastasis | Yes | 8 | 7 | 1 | 1.000 | 8 | 0 | 0.245 | 4 | 4 | 0.209 | 3 | 5 | 0.128 |
| No | 34 | 28 | 6 | 25 | 9 | 7 | 27 | 3 | 31 | |||||
| CEA | ≥5.0 ng/ml | 15 | 12 | 3 | 1.000 | 10 | 5 | 0.313 | 3 | 12 | 0.754 | 3 | 12 | 0.742 |
| <5.0 ng/ml | 27 | 23 | 4 | 23 | 4 | 8 | 19 | 3 | 24 | |||||
| CA19-9 | ≥37 U/ml | 9 | 7 | 2 | 1.000 | 6 | 3 | 0.600 | 3 | 6 | 0.903 | 2 | 7 | 0.818 |
| <37 U/ml | 33 | 28 | 5 | 27 | 6 | 8 | 25 | 4 | 29 | |||||
| Tumor location | Colon | 26 | 21 | 5 | 0.887 | 23 | 3 | 0.109 | 8 | 18 | 0.618 | 3 | 23 | 0.846 |
| Rectum | 16 | 14 | 2 | 10 | 6 | 3 | 13 | 3 | 13 | |||||
| Differentiation | Poor | 10 | 9 | 1 | 0.871 | 9 | 1 | 0.570 | 4 | 6 | 0.468 | 2 | 8 | 0.941 |
| Moderate | 32 | 26 | 6 | 24 | 8 | 7 | 25 | 4 | 28 | |||||
| Good | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Tumor size | <5 cm | 28 | 23 | 5 | 1.000 | 22 | 6 | 1.000 | 7 | 21 | 1 | 3 | 25 | 0.640 |
| ≥5 cm | 14 | 12 | 2 | 11 | 3 | 4 | 10 | 3 | 11 | |||||
| Histological classification | Adenocarcinoma | 40 | 33 | 7 | 1.000 | 31 | 9 | 1.000 | 10 | 30 | 1 | 4 | 36 | 0.558 |
| Mucinous adenocarcinoma | 2 | 2 | 0 | 2 | 0 | 1 | 1 | 1 | 1 | |||||
| Undifferentiated carcinoma | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
PCDH-γ-A12, protocadherin γ subfamily A12; SLC19A1, solute carrier family 19 A 1; CREB, cAMP responsive element binding protein; CYLD, cylindromatosis; M, methylated; U, unmethylated; TNM, tumor-node-metastasis; CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19-9.
Discussion
Cancer develops through a multi-step process which results from the progressive accumulation of genetic and epigenetic alterations (33). Epigenetic modifications, which have a fundamental role in the regulation of gene expression, involve DNA methylation, specific histone modifications and non-coding RNA interventions (34,35). As one of the main epigenetic modifications, DNA methylation of promoters often downregulates gene transcription, while DNA demethylation of promoters activates gene expression. DNA methylation-mediated tumor suppressor gene silencing may contribute to tumor progression (7,36). Aberrant DNA methylation of gene promoters has become a promising biomarker for the early diagnosis of diseases (37–41).
In the colon, aberrant DNA methylation arises extremely early, initially in normal-appearing mucosa, and it may be part of the age-associated field defects observed in sporadic CRC (42). Hypermethylation in CpG islands has been demonstrated to be a novel mechanism of tumor suppressor gene silencing (7,8). A number of genes have now been demonstrated to be hypermethylated in colorectal tumors, including APC (11), MLH1 (43) and O6-methylguanine DNA methyltransferase (44). For example, the inactivation of the cyclin-dependent kinase inhibitor P16/CDKN2A/INK4a by methylation leads to the disruption of cell-cycle regulation and potentially provides a growth advantage to affected cells (45).
PCDH-γ-A12 is a member of the protocadherin γ gene cluster, which includes 22 genes divided into 3 subfamilies (subfamily A, B and C) (46). The exon of PCDH-γ-A12 encodes the extracellular region, which includes six cadherin ectodomains and a transmembrane region. These cadherin-like cell adhesion proteins most likely serve a critical role in the establishment and function of specific cell-cell connections in the brain and cancer (18). The hypermethylation of PCDH-γ-A12 induces the downregulation of PCDH-γ-A12 gene transcription by rendering the chromatin structure inaccessible to the transcription machinery in a variety of tumors including bladder cancer, breast cancer, acute lymphoblastic leukemia and non-small cell lung cancer (17,29,47,48). The present study in CRC provides new evidence for the contribution of PCDH-γ-A12 promoter hypermethylation to the occurrence and metastasis of CRC.
SLC19A1 encodes a membrane protein that is a transporter of folate, and is involved in the regulation of intracellular concentrations of folate. SLC19A1 is also a major transporter of antifolate drugs used for certain types of cancer chemotherapy, including methotrexate (MTX) (30). The expression of SLC19A1 is downregulated following exposure to MTX in breast cancer, and a reverse correlation was identified between the promoter methylation and mRNA levels of SLC19A1. A variant of the SLC19A1 gene is associated with metastatic colorectal cancer (20). The present study in CRC adds new evidence for the contribution of SLC19A1 promoter hypermethylation to the occurrence and metastasis of CRC.
Certain studies have focused on the correlation between colorectal cancer clinical features and the methylation of certain genes, including p15, APC and E-cadherin, suggesting that the inactivation of certain tumor suppressor genes through aberrant promoter methylation of CpG islands may serve a role in the development of colorectal cancer (49,50). Multiple methylation pathways may be involved in the tumorigenesis of CRC and associated with the aggressiveness of clinical disease (37). In the present study, the correlation between the methylation of PCDH-γ-A12, SLC19A1, CREB and CYLD and colorectal cancer clinical features was examined. However, no significant correlation was identified between PCDH-γ-A12, SLC19A1, CREB and CYLD methylation and the clinical features, which may be due to the lack of power in the samples used.
CEA is a member of a family of cell surface glycoproteins that are excessively produced in the majority of human colorectal carcinomas (51). CEA measurement is mainly used as a tumor marker to monitor colorectal carcinoma treatment, to identify recurrences following surgical resection and to localize cancer spread through measurement of biological fluids (52,53). CA19-9 is a useful tumor-associated antigen for the serological detection of colorectal carcinomas, and may be used to monitor patients with advanced colorectal carcinomas (54). One aim of the present study was to observe whether the status of PCDH-γ-A12, SLC19A1, CREB and CYLD promoter methylation had a correlation with the serum level of CEA and CA19-9. However, no significant correlation was observed between PCDH-γ-A12, SLC19A1, CREB and CYLD promoter methylation and the serum level of CEA and CA19-9. This may imply that aberrant methylation of PCDH-γ-A12, SLC19A1, CREB and CYLD combined with conventional tumor markers could serve as complementary markers in the diagnosis of CRC. However, further study is necessary to confirm this hypothesis.
In conclusion, PCDH-γ-A12 and SLC19A1 promoters, but not CREB and CYLD promoters, are hypermethylated and contribute to the occurrence and metastasis of colorectal cancer. These findings may provide a new direction in the detection and treatment of CRC. Future research is required to determine the detailed mechanisms of how the PCDH-γ-A12 and SLC19A1 genes contribute to the risk of CRC.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the Natural Science Foundation of Zhejiang Province (LY15H160015 and LS14H26001), the K. C. Wong Magna Fund in Ningbo University, the Science and Technology Innovation Team of Ningbo (2011B82014), the Specialized Research Fund for the Social Development of Hangzhou (20160533B21) and the Scientific Innovation Fund of the Affiliated Hospital of Hangzhou Normal University.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Author's contributions
MY and SD designed the research. CZ, JL, TH and CC conducted the experiments. CZ, QH and HJ analyzed the data. The manuscript was drafted by CZ and SD. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Human Research Ethics Committee of Ningbo University and written informed consent was obtained from all participants.
Consent for publication
All participants provided written informed consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers (Basel) 2013;5:676–713. doi: 10.3390/cancers5020676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk CM, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 7.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 8.Zhang CJ, Zhou JX, Liu J, Ma ZY, Zhang SW, Dou K, Huang HW, Cai T, Liu R, Zhu JK, et al. The splicing machinery promotes RNA-directed DNA methylation and transcriptional silencing in Arabidopsis. EMBO J. 2013;32:1128–1140. doi: 10.1038/emboj.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwitalla S, Ziegler PK, Horst D, Becker V, Kerle I, Begus-Nahrmann Y, Lechel A, Rudolph KL, Langer R, Slotta-Huspenina J, et al. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell. 2013;23:93–106. doi: 10.1016/j.ccr.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 10.Amatu A, Sartore-Bianchi A, Moutinho C, Belotti A, Bencardino K, Chirico G, Cassingena A, Rusconi F, Esposito A, Nichelatti M, et al. Promoter CpG island hypermethylation of the DNA repair enzyme MGMT predicts clinical response to dacarbazine in a phase II study for metastatic colorectal cancer. Clin Cancer Res. 2013;19:2265–2272. doi: 10.1158/1078-0432.CCR-12-3518. [DOI] [PubMed] [Google Scholar]
- 11.Gay LJ, Mitrou PN, Keen J, Bowman R, Naguib A, Cooke J, Kuhnle GG, Burns PA, Luben R, Lentjes M, et al. Dietary, lifestyle and clinicopathological factors associated with APC mutations and promoter methylation in colorectal cancers from the EPIC-Norfolk study. J Pathol. 2012;228:405–415. doi: 10.1002/path.4085. [DOI] [PubMed] [Google Scholar]
- 12.Rawson JB, Mrkonjic M, Daftary D, Dicks E, Buchanan DD, Younghusband HB, Parfrey PS, Young JP, Pollett A, Green RC, et al. Promoter methylation of Wnt5a is associated with microsatellite instability and BRAF V600E mutation in two large populations of colorectal cancer patients. Br J Cancer. 2011;104:1906–1912. doi: 10.1038/bjc.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakagawa H, Nuovo GJ, Zervos EE, Martin EW, Jr, Salovaara R, Aaltonen LA, de la Chapelle A. Age-related hypermethylation of the 5′ region of MLH1 in normal colonic mucosa is associated with microsatellite-unstable colorectal cancer development. Cancer Res. 2001;61:6991–6995. [PubMed] [Google Scholar]
- 14.Shima K, Nosho K, Baba Y, Cantor M, Meyerhardt JA, Giovannucci EL, Fuchs CS, Ogino S. Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: Cohort study and literature review. Int J Cancer. 2011;128:1080–1094. doi: 10.1002/ijc.25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliveira C, Velho S, Domingo E, Preto A, Hofstra RM, Hamelin R, Yamamoto H, Seruca R, Schwartz S., Jr Concomitant RASSF1A hypermethylation and KRAS/BRAF mutations occur preferentially in MSI sporadic colorectal cancer. Oncogene. 2005;24:7630–7634. doi: 10.1038/sj.onc.1208906. [DOI] [PubMed] [Google Scholar]
- 16.Coppedè F. Epigenetic biomarkers of colorectal cancer: Focus on DNA methylation. Cancer Lett. 2014;342:238–247. doi: 10.1016/j.canlet.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y, Lemon W, Liu PY, Yi Y, Morrison C, Yang P, Sun Z, Szoke J, Gerald WL, Watson M, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morishita H, Yagi T. Protocadherin family: Diversity, structure, and function. Curr Opin Cell Biol. 2007;19:584–592. doi: 10.1016/j.ceb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Stanislawska-Sachadyn A, Mitchell LE, Woodside JV, Buckley PT, Kealey C, Young IS, Scott JM, Murray L, Boreham CA, McNulty H, et al. The reduced folate carrier (SLC19A1) c.80G>A polymorphism is associated with red cell folate concentrations among women. Ann Hum Genet. 2009;73:484–491. doi: 10.1111/j.1469-1809.2009.00529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang L, Zhang T, Xie C, Liao X, Yu Q, Feng J, Ma H, Dai J, Li M, Chen J, et al. SLCO1B1 and SLC19A1 gene variants and irinotecan-induced rapid response and survival: A prospective multicenter pharmacogenetics study of metastatic colorectal cancer. PLoS One. 2013;8:e77223. doi: 10.1371/journal.pone.0077223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaywitz AJ, Greenberg ME. CREB: A stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 22.Altarejos JY, Montminy M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ionov Y, Matsui S, Cowell JK. A role for p300/CREB binding protein genes in promoting cancer progression in colon cancer cell lines with microsatellite instability. Proc Natl Acad Sci USA. 2004;101:1273–1278. doi: 10.1073/pnas.0307276101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickstrom SA, Masoumi KC, Khochbin S, Fassler R, Massoumi R. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. 2010;29:131–144. doi: 10.1038/emboj.2009.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Donnell MA, Perez-Jimenez E, Oberst A, Ng A, Massoumi R, Xavier R, Green DR, Ting AT. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat Cell Biol. 2011;13:1437–1442. doi: 10.1038/ncb2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Sun L, Huo L, Liu M, Li D, Zhou J. CYLD regulates angiogenesis by mediating vascular endothelial cell migration. Blood. 2010;115:4130–4137. doi: 10.1182/blood-2009-10-248526. [DOI] [PubMed] [Google Scholar]
- 27.Alameda JP, Fernandez-Acenero MJ, Moreno-Maldonado R, Navarro M, Quintana R, Page A, Ramirez A, Bravo A, Casanova ML. CYLD regulates keratinocyte differentiation and skin cancer progression in humans. Cell Death Dis. 2011;2:e208. doi: 10.1038/cddis.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellerbrand C, Bumes E, Bataille F, Dietmaier W, Massoumi R, Bosserhoff AK. Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis. 2007;28:21–27. doi: 10.1093/carcin/bgl081. [DOI] [PubMed] [Google Scholar]
- 29.Taylor KH, Pena-Hernandez KE, Davis JW, Arthur GL, Duff DJ, Shi H, Rahmatpanah FB, Sjahputera O, Caldwell CW. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Res. 2007;67:2617–2625. doi: 10.1158/0008-5472.CAN-06-3993. [DOI] [PubMed] [Google Scholar]
- 30.Yang R, Li WW, Hoang BH, Kim H, Banerjee D, Kheradpour A, Healey JH, Meyers PA, Bertino JR, Gorlick R. Quantitative correlation between promoter methylation and messenger RNA levels of the reduced folate carrier. BMC Cancer. 2008;8:124. doi: 10.1186/1471-2407-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demura M, Bulun SE. CpG dinucleotide methylation of the CYP19 I.3/II promoter modulates cAMP-stimulated aromatase activity. Mol Cell Endocrinol. 2008;283:127–132. doi: 10.1016/j.mce.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Massoumi R, Kuphal S, Hellerbrand C, Haas B, Wild P, Spruss T, Pfeifer A, Fassler R, Bosserhoff AK. Down-regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J Exp Med. 2009;206:221–232. doi: 10.1084/jem.20082044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiba T, Marusawa H, Ushijima T. Inflammation-associated cancer development in digestive organs: Mechanisms and roles for genetic and epigenetic modulation. Gastroenterology. 2012;143:550–563. doi: 10.1053/j.gastro.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Mossman D, Scott RJ. Long term transcriptional reactivation of epigenetically silenced genes in colorectal cancer cells requires DNA hypomethylation and histone acetylation. PLoS One. 2011;6:e23127. doi: 10.1371/journal.pone.0023127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31:1609–1622. doi: 10.1038/onc.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097–5102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- 37.Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, Park J, Kim DH. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res. 2009;15:6185–6191. doi: 10.1158/1078-0432.CCR-09-0111. [DOI] [PubMed] [Google Scholar]
- 38.Richards KL, Zhang B, Sun M, Dong W, Churchill J, Bachinski LL, Wilson CD, Baggerly KA, Yin G, Hayes DN, et al. Methylation of the candidate biomarker TCF21 is very frequent across a spectrum of early-stage nonsmall cell lung cancers. Cancer. 2011;117:606–617. doi: 10.1002/cncr.25472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang D, Hong Q, Shen Y, Xu Y, Zhu H, Li Y, Xu C, Ouyang G, Duan S. The diagnostic value of DNA methylation in leukemia: A systematic review and meta-analysis. PLoS One. 2014;9:e96822. doi: 10.1371/journal.pone.0096822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang D, Shen Y, Dai D, Xu Y, Xu C, Zhu H, Huang T, Duan S. Meta-analyses of methylation markers for prostate cancer. Tumour Biol. 2014;35:10449–10455. doi: 10.1007/s13277-014-2300-7. [DOI] [PubMed] [Google Scholar]
- 41.Sui X, Wang D, Geng S, Zhou G, He C, Hu X. Methylated promoters of genes encoding protocadherins as a new cancer biomarker family. Mol Biol Rep. 2012;39:1105–1111. doi: 10.1007/s11033-011-0837-8. [DOI] [PubMed] [Google Scholar]
- 42.Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29–39. doi: 10.1023/A:1025806911782. [DOI] [PubMed] [Google Scholar]
- 43.Malhotra P, Anwar M, Kochhar R, Ahmad S, Vaiphei K, Mahmood S. Promoter methylation and immunohistochemical expression of hMLH1 and hMSH2 in sporadic colorectal cancer: A study from India. Tumour Biol. 2014;35:3679–3687. doi: 10.1007/s13277-013-1487-3. [DOI] [PubMed] [Google Scholar]
- 44.Pietrantonio F, Perrone F, de Braud F, Castano A, Maggi C, Bossi I, Gevorgyan A, Biondani P, Pacifici M, Busico A, et al. Activity of temozolomide in patients with advanced chemorefractory colorectal cancer and MGMT promoter methylation. Ann Oncol. 2014;25:404–408. doi: 10.1093/annonc/mdt547. [DOI] [PubMed] [Google Scholar]
- 45.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 46.Yagi T. Clustered protocadherin family. Dev Growth Differ 50 Suppl. 2008;1:S131–S140. doi: 10.1111/j.1440-169X.2008.00991.x. [DOI] [PubMed] [Google Scholar]
- 47.Reinert T, Modin C, Castano FM, Lamy P, Wojdacz TK, Hansen LL, Wiuf C, Borre M, Dyrskjot L, Orntoft TF. Comprehensive genome methylation analysis in bladder cancer: Identification and validation of novel methylated genes and application of these as urinary tumor markers. Clin Cancer Res. 2011;17:5582–5592. doi: 10.1158/1078-0432.CCR-10-2659. [DOI] [PubMed] [Google Scholar]
- 48.Tommasi S, Karm DL, Wu X, Yen Y, Pfeifer GP. Methylation of homeobox genes is a frequent and early epigenetic event in breast cancer. Breast Cancer Res. 2009;11:R14. doi: 10.1186/bcr2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin SY, Yeh KT, Chen WT, Chen HC, Chen ST, Chiou HY, Chang JG. Promoter CpG methylation of tumor suppressor genes in colorectal cancer and its relationship to clinical features. Oncol Rep. 2004;11:341–348. [PubMed] [Google Scholar]
- 50.Lind GE, Thorstensen L, Lovig T, Meling GI, Hamelin R, Rognum TO, Esteller M, Lothe RA. A CpG island hypermethylation profile of primary colorectal carcinomas and colon cancer cell lines. Mol Cancer. 2004;3:28. doi: 10.1186/1476-4598-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benchimol S, Fuks A, Jothy S, Beauchemin N, Shirota K, Stanners CP. Carcinoembryonic antigen, a human tumor marker, functions as an intercellular adhesion molecule. Cell. 1989;57:327–334. doi: 10.1016/0092-8674(89)90970-7. [DOI] [PubMed] [Google Scholar]
- 52.Ding W, Wang J, Wang F, Wang G, Wu Q, Ju S, Cong H, Wang H. Serum sAPRIL: A potential tumor-associated biomarker to colorectal cancer. Clin Biochem. 2013;46:1590–1594. doi: 10.1016/j.clinbiochem.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 53.Baek JY, Yeo HY, Chang HJ, Kim KH, Kim SY, Park JW, Park SC, Choi HS, Kim DY, Oh JH. Serpin B5 is a CEA-interacting biomarker for colorectal cancer. Int J Cancer. 2014;134:1595–1604. doi: 10.1002/ijc.28494. [DOI] [PubMed] [Google Scholar]
- 54.Petrioli R, Licchetta A, Roviello G, Pascucci A, Francini E, Bargagli G, Conca R, Miano ST, Marzocca G, Francini G. CEA and CA19.9 as early predictors of progression in advanced/metastatic colorectal cancer patients receiving oxaliplatin-based chemotherapy and bevacizumab. Cancer Invest. 2012;30:65–71. doi: 10.3109/07357907.2011.629380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.