Abstract
A Chlamydomonas reinhardtii chloroplast transformant, designated MU7, carrying a chimeric (rbcL promoter: β-glucuronidase [GUS]: psaB 3′ end) gene whose transcripts have been found previously to be unstable in light (half-life of 20 min in light as opposed to a half-life of 5 h in the dark), was used to study the role of electron transport and of the redox state in the degradation of chloroplast transcripts in the light. Blocking photosynthetic electron transport with 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) prevented the light-dependent breakdown of the pool of GUS transcripts in MU7 cells. Diamide, an oxidizing agent, caused a measurable delay in the degradation of GUS transcripts in the light. The addition of dithiothreitol (DTT), a dithiol reductant, to MU7 cells in which GUS transcript levels were stabilized by DCMU induced degradation of GUS transcripts. Similarly, DTT induced a decrease in the levels of GUS transcripts when added to MU7 cells in the dark period of the light/dark cycle, a period in which GUS transcript levels normally increase. The levels of transcripts of endogenous chloroplast genes were affected by DCMU and DTT in the same direction as levels of GUS transcripts. The results suggest a regulatory role of the redox state in the degradation of chloroplast transcripts in C. reinhardtii.
The expression of genes in chloroplasts is regulated by both external factors and endogenous processes (Gruissem and Tonkyn, 1993; Sugita and Sugiura, 1996; Goldschmidt-Clermont, 1998). Control points have been identified at transcriptional (Mullet, 1993) and posttranscriptional levels (Gillham et al., 1994; Rochaix, 1996; Danon, 1997), but, despite recent progress, the molecular mechanisms regulating the individual molecular processes of chloroplast gene expression are in most cases not fully understood.
Light is the most important external signal that influences chloroplast gene expression; it has been found to affect transcription (Klein and Mullet, 1990; Sexton et al., 1990; Salvador et al., 1993b), RNA processing (Deshpande et al., 1997), transcript stability (Salvador et al., 1993a, 1993b), translation (Danon and Mayfield, 1991), protein modification (Allen, 1992; Elich et al., 1993; Danon and Mayfield, 1994b), and turnover of proteins (Ohad et al., 1990) at the molecular level. In most studies, the effect of light on the basic molecular processes in chloroplasts appears to be mediated by photosynthetic electron transport (e.g. Mohamed and Jansson, 1991; Deshpande et al., 1997). Non-cyclic electron transport in chloroplasts produces reducing power and a proton gradient across the thylakoid membrane, the energy of which is preserved mostly in the ATP pool.
How reducing power and/or ATP concentration is linked to the regulation of the molecular machinery in chloroplasts is not clear, but a number of studies suggest that protein redox carriers act as signal transducers between electron transport and the various molecular processes (Levings and Siedow, 1995). In the best-studied example—the light-dependent translation of transcripts of the chloroplast psbA gene in Chlamydomonas reinhardtii—a redox signal appeared to be carried by a series of thiol proteins from the chloroplast electron transport chain to a multiprotein complex that activates translation by binding specifically to psbA transcripts (Yohn et al., 1998b). Proteins of this regulatory redox chain have been isolated and partially characterized (Kim and Mayfield, 1997; Yohn et al., 1998a). It has been suggested (Levings and Siedow, 1995) that similar signal transduction pathways might be involved in other light-modulated molecular processes in chloroplasts.
The effect of light on the stability of chloroplast transcripts has been studied in some detail in the unicellular green alga C. reinhardtii (Salvador et al., 1993a, 1993b). C. reinhardtii cells growing in 12-h light/12-h dark cycles were found to degrade chloroplast transcripts up to five times faster in the light period than in the dark period of the 24-h cycles (Salvador et al., 1993b). Sequences in the 5′ untranslated regions of mRNA were found to be crucial for breakdown of C. reinhardtii chloroplast transcripts (Kuchka et al., 1989; Rochaix et al., 1989; Nickelsen et al., 1994), in particular for accelerated transcript degradation in the light (Salvador et al., 1993a). Putative protein factors have been identified that are thought to stabilize chloroplast transcripts by binding to sequence elements in their 5′ untranslated regions (Nickelsen et al., 1994). These findings suggest that interactions of regulatory proteins with sequence elements of the 5′ untranslated regions of mRNAs are involved in light/dark control of transcript longevity in the C. reinhardtii chloroplast. The molecular mechanism that links the light/dark signal and a possible binding of regulatory proteins to RNA is not known, but a signal pathway similar to the redox pathway found in the C. reinhardtii chloroplast for light regulation of translation of psbA transcripts (Danon, 1997) appears feasible.
To begin to dissect the molecular machinery that links the light/dark signal to the regulation of degradation of chloroplast transcripts, we inhibited chloroplast electron transport in a transgenic cell line of C. reinhardtii and monitored levels of transcripts of a chimeric β-glucuronidase (GUS) reporter gene that have been shown previously to be very unstable in light. The results show that photosynthetic non-cyclic electron transport is required for light-dependent RNA decay in the chloroplasts of C. reinhardtii. Altering the redox state of C. reinhardtii cells had a notable effect on chloroplast transcript levels, suggesting that non-cyclic electron transport is signaled to the RNA degrading machinery via redox carriers.
MATERIALS AND METHODS
Growth of Algae
Chloroplast transformants of Chlamydomonas reinhardtii were grown on minimal high-salt medium (Sueoka, 1960) on agar plates (1.5%, w/v) or in liquid cultures. High-density cultures were grown in glass tubes in a temperature-controlled water bath (32°C) in 12-h dark/12-h light cycles (light intensity approximately 500 W/m2). Cells were mixed by bubbling with air supplemented with 2% (w/v) CO2. Cell numbers were kept at approximately 1 × 106 cells/mL by daily dilutions of the cultures with fresh high-salt medium. The light-sensitive mutant CC-373 (ac-uc-2-21), obtained from the Chlamydomonas Genetics Center at Duke University, was maintained in low light on agar plates on minimal medium supplemented with 2.5 g/L potassium acetate and used for transformation as described previously (Blowers et al., 1990). In some experiments 3-(3,4-dichlorophenyl)1,1-dimethylurea (DCMU), diazenedicarboxylic acid bis(N,N-dimethylamide) (diamide), β-mercaptoethanol, and/or 1,3 dithiothreitol (DTT) at various concentrations were added to the cultures. Cell density was monitored by counting in a hemocytometer.
Chloroplast Transformation
To introduce chimeric rbcL:GUS gene constructs into the chloroplasts of C. reinhardtii, agar-plated cells of the atpB-deficient mutant CC-373 were bombarded with DNA-coated gold particles essentially as described previously (Boynton et al., 1988; Blowers et al., 1989). Transgenic cell lines were selected and maintained on agar plates in high-light conditions and grown in liquid high-salt medium for analyses of gene expression.
DNA- and RNA-Blot Analyses
DNA was extracted from C. reinhardtii transformants as described previously (Blowers et al., 1990). The presence of the chimeric rbcL:GUS gene in the chloroplast genome was verified by slot-blot analyses using the radiolabeled coding region of the GUS gene as a probe (Blowers et al., 1990). Total RNA from C. reinhardtii was isolated and analyzed by RNA gel (northern) blots as described previously (Salvador et al., 1993a). Radiolabeled double-stranded probes (described by Salvador et al., 1993a) were used to detect transcripts of the atpB, rbcL, psaB, and GUS genes. Hybridization signals on autoradiograms were quantified using image analysis software (Digital Science 1D, Kodak, Rochester, NY).
In Vivo Measurement of Transcription Rates of Chloroplast Genes
Rates of chloroplast gene transcription were measured by determining the incorporation of [32P]orthophosphate over a 10-min time interval into newly synthesized RNA in phosphate-starved cells, as described previously (Salvador et al., 1993a).
Plasmids
Construction of plasmid pMU7, containing 290 bp upstream of the start site of rbcL gene transcription and the complete 5′ untranslated region of the rbcL gene fused to the coding region of the Escherichia coli uidA gene, was described previously (Salvador et al., 1993b). Plasmid pMU31 contains 66 bp upstream of the start site of rbcL gene transcription, the complete 5′ untranslated region, and 243 bp of rbcL gene coding sequences fused to the coding region of the bacterial uidA gene. The rbcL sequences were amplified by PCR using the oligonucleotides 5′-TCTATGCTCGACTGATAAGACAAGTACATAAATTTGCTAGTTTACAT-3′ and 5′-GTAACAACAAGCTTGGTAACG-3′ as 5′ and 3′ primers, respectively. The agarose-purified PCR fragment was cut with XhoI/HindIII, subcloned in pBluescript SK+ (Stratagene, La Jolla, CA) that had been cut with the same enzymes, and finally inserted into the transformation vector pCrc32 (Blowers et al., 1993) upstream of the GUS gene coding region using the XhoI and SmaI sites for cloning.
Cell Viability Assays
To assess the viability of C. reinhardtii cells in the presence of relatively high concentrations (1–20 mm) of DTT under the conditions described in the experiments, cell cultures treated for 1 h with 20 mm DTT were diluted 1,000-fold with fresh high-salt medium and plated in 100-μL aliquots on high-salt agar plates. Numbers of colonies on the plates were counted after approximately 2 weeks of growth in high-light conditions and compared with the number of colonies on control plates. The capacity of C. reinhardtii cells for photosynthetic oxygen evolution and respiration after 1 h of incubation with 20 mm DTT in the dark was measured with a Clark-type oxygen electrode (Hansatech, King's Lynn, UK).
RESULTS
Photosynthetic Electron Transport Is Required for Light-Dependent Degradation of Chloroplast Transcripts
In C. reinhardtii cells growing in 12-h light/12-h dark cycles, transcripts of chloroplast genes are degraded faster in the light period than in the dark period of the 24-h light/dark cycle (Salvador et al., 1993b). To find out whether light-driven electron transport plays a role in light/dark regulation of transcript degradation in the C. reinhardtii chloroplast, we determined the effect of DCMU, a specific inhibitor of non-cyclic photosynthetic electron transport that binds tightly to the Qb site of photosystem II, on the abundance of chloroplast transcripts in light (Fig. 1). A C. reinhardtii chloroplast transformant designated MU7, which carries a chimeric (rbcL promoter:GUS:psaB 3′ end) reporter gene construct whose transcripts have been found to be approximately 15 times less stable in light (t1/2 = 20 min) than in darkness (t1/2 = 5 h) (Salvador et al., 1993a), was used in these analyses. We determined previously by measuring β-glucuronidase activity that the chimeric GUS gene is translated in MU7 (U. Klein and M.L. Salvador, unpublished data).
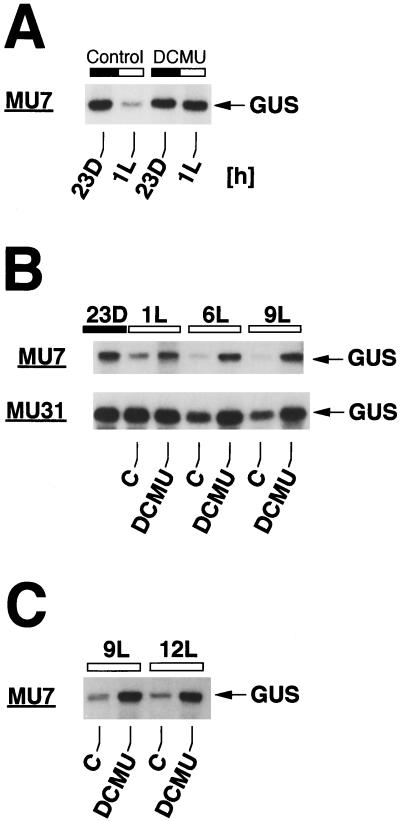
Figure 1.
DCMU prevents degradation of GUS transcripts in the light in C. reinhardtii chloroplast transformants. Algae were grown in 12-h dark/12-h light periods, and total RNA was isolated at the time points indicated. RNA was separated in a 1.3% (w/v) agarose/formaldehyde gel, transferred to a nylon membrane (Zetaprobe, Bio-Rad, Hercules, CA), and hybridized to a 32P-labeled GUS probe. The membrane was exposed at −80°C to x-ray film (XAR-5, Kodak) with an intensifying screen. DCMU (20 μm final concentration) was added to the cultures 10 min before the onset of light (A and B) and at 6 h of light (6L) (C). Light (L) and dark (D) time points are indicated by white and black bars above the autoradiograms (time point 23D corresponds to 11 h of darkness). MU7 and MU31 are chloroplast transformants harboring rbcL:GUS reporter genes that differ in the sequences upstream of the GUS coding region (see ”Materials and Methods“). C, Control without DCMU.
When MU7 cells were grown in alternating 12-h light/12-h dark cycles, GUS transcripts fell to relatively low levels within an hour after the onset of light (Fig. 1A). The addition of DCMU (final concentration 20 μm) to a culture of MU7 cells 10 min before the onset of light prevented the light-induced decrease in GUS transcript levels (Fig. 1A). GUS transcripts remained abundant in the light for at least 9 h in the presence of DCMU (Fig. 1B). Because the light-induced decrease in levels of GUS transcripts in transformant MU7 (Fig. 1A) is known to be caused by accelerated degradation (Salvador et al., 1993a), the prevention of the decrease by DCMU strongly suggests that non-cyclic photosynthetic electron transport is required for light-dependent degradation of GUS transcripts in MU7 cells.
To find out whether the effect of DCMU on GUS transcript stability is related to the specific GUS gene or the genetic background of transformant MU7, we analyzed the effect of DCMU in another C. reinhardtii chloroplast transformant designated MU31, which carries a variant rbcL:GUS gene (see “Materials and Methods”) whose transcripts are much more stable in the light than transcripts of the chimeric rbcL:GUS gene in transformant MU7 (Fig. 1B). In the presence of DCMU, GUS transcript levels remained unchanged in MU31 for 9 h into the light period, whereas in control cells without DCMU, GUS transcript levels fell to about 60% of the dark level during this time period (Fig. 1B), showing that stabilization of the GUS mRNA pool by DCMU is independent of the transformant and the chimeric GUS gene analyzed.
DCMU added to MU7 cells in the middle of the 12-h light period caused the pool of GUS transcripts to increase compared with the pool of GUS transcripts in cells maintained without the electron transport inhibitor (Fig. 1C). Thus, in the presence of DCMU, GUS transcript levels behave in the light as they would in the dark, suggesting that blocking non-cyclic photosynthetic electron transport with DCMU simulates dark conditions with regard to regulation of transcript levels in the C. reinhardtii chloroplast.
Effect of DCMU on Levels of Transcripts of Endogenous C. reinhardtii Chloroplast Genes
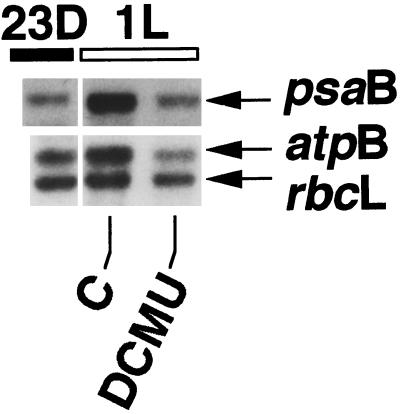
Transformant MU7 was chosen for the analyses described above, because in this transformant the stability of chimeric rbcL:GUS transcripts is known to be about 15 times lower in the light than in the dark, making it easier to detect putative factors that influence transcript stability. Previous work (Salvador et al., 1993b) has shown that not only the stability of GUS transcripts, but also that of all chloroplast transcripts studied, was lower in the light period than in the dark period in light/dark-grown C. reinhardtii. However, the light/dark difference in transcript stability was found to be much smaller for transcripts of endogenous genes than for transcripts of the GUS reporter gene (Salvador et al., 1993a). To determine whether DCMU also has a measurable influence on transcript levels of endogenous chloroplast genes, levels of transcripts of the endogenous psaB, atpB, and rbcL genes were determined in the presence and absence of DCMU (Fig. 2).
Figure 2.
Effect of DCMU on levels of transcripts of endogenous C. reinhardtii chloroplast genes. Total RNA was isolated from the transformant MU7 at the time points indicated. RNA gel blots were hybridized to specific radiolabeled psaB, atpB, and rbcL gene probes. DCMU (20 μm) was added to the culture at 11 h of darkness (23D).
As in the analyses of GUS gene expression, DCMU added at the end of the dark period kept pools of transcripts of the psaB, atpB, and rbcL genes at dark levels (Fig. 2), while control cells showed the typical increase in RNA levels at 1 h of light (see Salvador et al., 1993b). This shows that photosynthetic electron transport is not only involved in regulating fluctuations of levels of transcripts of the GUS reporter gene, but also that of transcripts of endogenous chloroplast genes. In addition, because the normal increase in mRNA levels at 1 h of light is known to be due to an increase in rates of transcription (Salvador et al., 1993b), prevention of the increase by DCMU points to an effect of DCMU not only on degradation of transcripts but also on transcript synthesis (gene transcription). This was confirmed for the genes analyzed by determining their relative transcription rates in the presence of DCMU (not shown). Again, the results indicate that blocking non-cyclic electron transport with DCMU signals dark conditions to the chloroplast regulatory machinery, causing transcription, transcript degradation, and, as a result, transcript levels to change from a light to a dark state.
Changes in Redox Conditions Affect Rates of Chloroplast RNA Degradation
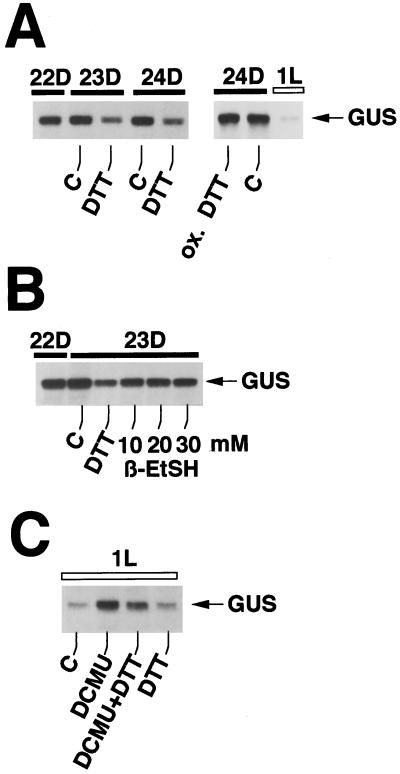
To test the notion that the redox state within the chloroplast is important for rates of degradation of chloroplast transcripts we tried to manipulate the internal redox conditions by adding DTT, a dithiol reductant, mercaptoethanol, a monothiol reductant, or diamide, an oxidizing agent, to MU7 transformants of C. reinhardtii in the dark and in the light. The half-life of rbcL:GUS transcripts in MU7 cells has been determined previously (Salvador et al., 1993b) to be around 5 h in the dark. In the presence of DTT (20 mm), levels of rbcL:GUS transcripts fell about 70% within an hour of incubation in the dark (Fig. 3A). Oxidized DTT had no effect on GUS transcript levels (Fig. 3A), showing that the ability of DTT to lower GUS transcript levels depends on its reducing capability. Mercaptoethanol, a reductant containing one sulfhydryl group, lowered levels of GUS transcripts in MU7 cells in the dark about 20% (Fig. 3B). Compared with the two-sulfhydryl-reductant DTT, which was applied at a concentration of 5 mm and which at this concentration lowered GUS transcript levels about 40%, mercaptoethanol was far less effective, even when used at concentrations up to 30 mm (Fig. 3B). The three concentrations of β-mercaptoethanol (10, 20, and 30 mm) all lowered levels of GUS transcripts about 25% compared with the control sample.
Figure 3.
Reducing conditions accelerate the degradation of GUS transcripts. Cultures of transformant MU7 were grown in 12-h dark/12-h light cycles and total RNA was isolated at the time points indicated. In A and B, the reductant DTT was added at 10 h of dark (22D) at concentrations of 20 mm (A) and 5 mm (B), respectively. β-Mercaptoethanol was added at 22 d at the concentrations indicated. In C, DTT and/or DCMU were added at 0 h of light (0L = 24D). ox. DTT, Oxidized DTT; β-EtSH, β-mercaptoethanol.
At the highest concentration tested (20 mm), DTT did not affect the viability of the cells, as determined by measuring rates of oxygen uptake in the dark (respiration) and oxygen evolution in the light (photosynthesis), both measured polarographically with an oxygen electrode. Viability was also determined by comparing the number of colonies growing on plates on which DTT-treated cells had been spread with the number of colonies on control plates.
DTT was also able to induce a decrease in GUS transcript levels in the light in cells in which transcript levels were first stabilized by the addition of DCMU (Fig. 3C). In Figure 3C, GUS transcript levels at 1 h of light (1L) were about 500%, 250%, and 75% of the control for cultures treated with DCMU, DCMU plus DTT, and DTT, respectively. Control experiments (not shown) indicated that DTT does not block transcription of chloroplast genes in the dark, indicating that the DTT-induced decrease in levels of GUS transcripts is caused by accelerated degradation of GUS RNA. Even if transcription were completely blocked by DTT in the dark, the longevity of GUS transcripts, which have a half-life of 5 h in the dark, must be significantly shortened to account for the 40% decrease (Fig. 3B) in abundance of GUS transcripts within an hour of incubation with DTT.
Effect of DTT on Levels of Transcripts of Endogenous Chloroplast Genes
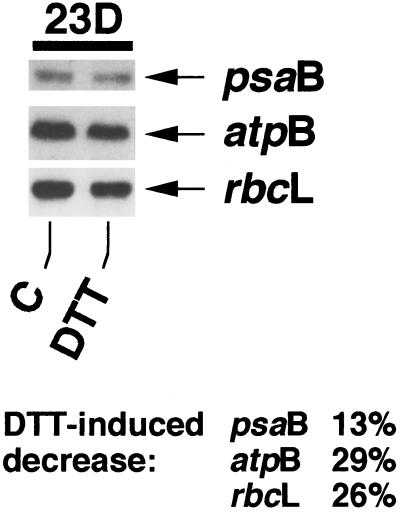
Levels of transcripts of the endogenous chloroplast genes psaB, atpB, and rbcL decreased about 13%, 29%, and 26%, respectively, in the dark within an hour in the presence of DTT (Fig. 4). The decrease occurred in a time period (22–23 h darkness) in which chloroplast transcript levels normally increase (Salvador et al., 1993b). Although the decrease was not as conspicuous as seen for GUS transcript levels, it shows that the influence of DTT is general and not specific to GUS transcripts.
Figure 4.
The effect of DTT on levels of transcripts of endogenous C. reinhardtii chloroplast genes. DTT (10 mm) was added to a light/dark-grown culture of transformant MU7 at 10 h of darkness (22D). Samples were processed as described previously (see legend to Fig. 1 and “Materials and Methods”).
The Oxidant Diamide Delays Degradation of GUS Transcripts in the Light
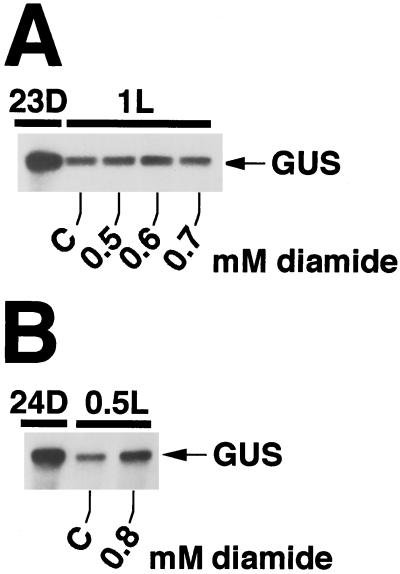
Diazenedicarboxylic acid bis(N,N-dimethylamide) (diamide) has been shown to alter the redox state of cells by irreversibly oxidizing the glutathione pool (Kosower and Kosower, 1995). Added to C. reinhardtii cultures at the end of the dark period, it caused a measurable delay in the light-induced breakdown of the GUS transcript pool (Fig. 5, A and B) but did not prevent it. Of the diamide concentrations and incubation periods tested, an incubation for 30 min at a concentration of 0.8 mm diamide resulted in a significant inhibition of GUS transcript degradation (Fig. 5B). Longer incubation periods and/or higher concentrations of diamide damaged the cells noticeably, as seen by the decrease in total RNA levels, halted growth, and physical disintegration. The relatively small effect of diamide on the GUS transcript pool in the light in transformant MU7 was probably due to its specificity to oxidize glutathione. The redox state of the glutathione pool might not link directly to the regulation of molecular processes in chloroplasts, and any effect of diamide on RNA levels is likely to be indirect and less pronounced. Nevertheless, the short-term effect of diamide on GUS transcript levels, together with the effects of DTT and mercaptoethanol, supports the notion that redox poise is important for the regulation of RNA degradation in the C. reinhardtii chloroplast.
Figure 5.
The oxidant diamide delays degradation of GUS transcripts in transformant MU7. Diamide at the final concentrations indicated was added to a light/dark-grown culture of transformant MU7 at 0 h of light (0L = 24D). In A, levels of GUS transcripts were about 108%, 128%, and 100% of transcript levels in the control sample for diamide concentrations of 0.5, 0.6, and 0.7 mm, respectively. In B, in the presence of 0.8 mm diamide, the level of GUS transcripts at 30 min of light (0.5L) was about 50% higher than in the control sample.
DISCUSSION
The redox state has long been known to be an important factor for control of the activities of chloroplast enzymes. For example, the flow of intermediates through the reductive pentose phosphate (Calvin) cycle is in part controlled by the reduction state of key enzymes of the cycle (Buchanan, 1991), which is linked to light-driven non-cyclic electron flow via photosystem II and photosystem I in the thylakoid membrane. Likewise, the activities of other chloroplast enzymes, e.g. ATP synthase (Nalin and McCarty, 1984) and Glc-6-P dehydrogenase (Wenderoth et al., 1997), depend on photosynthetic electron flow. Chloroplast thioredoxins play a central role as redox carriers in this regulatory network (Holmgren, 1985; Buchanan, 1991).
Recent analyses indicate that not only photosynthetic carbon and energy metabolism but also basic molecular processes in plant cells are affected by changes in redox poise. Rates of gene transcription (Escoubas et al., 1995; Pfannschmidt et al., 1999), RNA processing (Deshpande et al., 1997; Liere and Link, 1997), translation (Danon and Mayfield, 1994a), and protein degradation (Garcia-Ferris and Moreno, 1994) have all been reported to be influenced by the redox state. Our results indicate that the stability of chloroplast transcripts is also affected.
It is tempting to speculate that redox regulation of transcript stability is mediated by redox carriers such as thioredoxins that link photosynthetic electron transport to the molecular mechanism of transcript degradation. The fact that the reducing agent DTT, with its two vicinal sulfhydryl groups, seems to have a much stronger effect on RNA stability than β-mercaptoethanol (Fig. 3), which has only one sulfhydryl group, points to a role of a compound with two sulfhydryl groups (such as thioredoxin) as the redox carrier in this process. We did not attempt to identify putative components of the signal transduction chain, but it is likely that at the end of the chain binding of regulatory proteins to chloroplast transcripts is altered by a change in redox poise. Redox-influenced binding of regulatory proteins to mRNA has been found before in chloroplasts (Danon and Mayfield, 1994a; Hayes et al., 1996; Liere and Link, 1997) and in mammalian systems (Müllner et al., 1992; Rondon et al., 1995). Most sequence elements that have been identified as binding sites for regulatory proteins are located in 5′ and/or 3′ sequences of transcripts (Dickey et al., 1992; Hayes et al., 1996; Liere and Link, 1997).
Phosphorylation/dephosphorylation of RNA binding proteins seems to be an additional regulatory mechanism in chloroplasts coupling the energy state (ATP level) to gene expression (Allen, 1992; Danon and Mayfield, 1994b; Lisitsky and Schuster, 1995; Liere and Link, 1997). We believe that ATP-dependent protein phosphorylation is not the major regulatory process in the control of chloroplast mRNA stability, because DCMU had the same effect on GUS transcript levels in cells growing heterotrophically in a medium with acetate as the carbon and energy source (not shown) as on GUS transcript levels in autotrophically growing cells (Fig. 1). Considering that acetate metabolism supplies chloroplasts with enough ATP to allow normal chloroplast functioning, even in the absence of photophosphorylation (Boschetti and Schmid, 1998), the results indicate that ATP levels are not as crucial as the redox state in the control of RNA degradation.
DCMU has been reported to affect the light-dependent accumulation (Petracek et al., 1997) and stability (Petracek et al., 1998) of transcripts of a nuclear Fed-1 transgene in tobacco, suggesting a role of photosynthetic electron flow in control of Fed-1 mRNA stability. Sequences in the 5′ untranslated region and at the beginning of the coding region of Fed-1 transcripts have been found to be essential for light/dark regulation of Fed-1 mRNA levels (Dickey et al., 1992, 1998). Similarly, in C. reinhardtii, 5′ sequences were identified as important stability determinants of chloroplast transcripts (Kuchka et al., 1989; Rochaix et al., 1989; Salvador et al., 1993a; Nickelsen et al., 1994). Thus, it does not seem unusual that stability-regulating sequence elements are located in the 5′ portions of transcripts, which suggests that 5′ sites are involved in the initiation of mRNA degradation in plants.
Because a number of different processes involved in transcriptional and posttranscriptional control of chloroplast gene expression have been found to be sensitive to changes in redox poise (Levings and Siedow, 1995), it is not surprising to find that the redox state influences the regulation of RNA degradation too. Thus, redox control of RNA degradation seems to be a component of a larger regulatory network that includes all levels of chloroplast gene expression, suggesting that this type of control plays an important role in plant metabolism. The physiological importance of redox control of gene expression in chloroplasts probably lies in its ability to directly couple non-cyclic electron flow and the availability of reducing power to regulation of the chloroplast molecular machinery. This allows plants and algae to respond at the level of gene expression to changes in non-cyclic electron flow as they occur diurnally during day and night, or in cases in which the components of the electron transport chain are degraded during development, for example, in the course of leaf or fruit development.
ACKNOWLEDGMENT
We thank Dr. Joaquin Moreno for critically reading the manuscript.
Footnotes
The work was supported by grants from the Dirección General de Investigacion Cientifica y Technica (grant no. PB 95–1075 to M.L.S.) and the Norwegian Research Council (grant no. 100946/410 to U.K.).
LITERATURE CITED
- Allen JF. Protein phosphorylation in regulation of photosynthesis. Biochim Biophys Acta. 1992;1098:275–335. doi: 10.1016/s0005-2728(09)91014-3. [DOI] [PubMed] [Google Scholar]
- Blowers AD, Bogorad L, Shark KB, Sanford JC. Studies on Chlamydomonas chloroplast transformation: foreign DNA can be stably maintained in the chromosome. Plant Cell. 1989;1:123–132. doi: 10.1105/tpc.1.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blowers AD, Ellmore GS, Klein U, Bogorad L. Transcriptional analysis of endogenous and foreign genes in chloroplast transformants of Chlamydomonas. Plant Cell. 1990;2:1059–1070. doi: 10.1105/tpc.2.11.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blowers AD, Klein U, Ellmore GS, Bogorad L. Functional in vivo analyses of the 3′ flanking sequences of the Chlamydomonas chloroplast rbcL and psaB genes. Mol Gen Genet. 1993;238:339–349. doi: 10.1007/BF00291992. [DOI] [PubMed] [Google Scholar]
- Boschetti A, Schmid K. Energy supply for ATP-synthase deficient chloroplasts of Chlamydomonas reinhardii. Plant Cell Physiol. 1998;39:160–168. [Google Scholar]
- Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, Klein TM, Shark KB, Sanford JC. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Buchanan BB. Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin-thioredoxin system. Arch Biochem Biophys. 1991;288:1–8. doi: 10.1016/0003-9861(91)90157-e. [DOI] [PubMed] [Google Scholar]
- Danon A. Translational regulation in the chloroplast. Plant Physiol. 1997;115:1293–1298. doi: 10.1104/pp.115.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. Light regulated translational activators: identification of chloroplast gene specific mRNA binding proteins. EMBO J. 1991;10:3993–4001. doi: 10.1002/j.1460-2075.1991.tb04974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science. 1994a;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. ADP-dependent phosphorylation regulates RNA-binding in vitro: implications in light-modulated translation. EMBO J. 1994b;13:2227–2235. doi: 10.1002/j.1460-2075.1994.tb06500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande NN, Bao Y, Herrin DL. Evidence for light/redox-regulated splicing of psbA pre-RNAs in Chlamydomonas chloroplasts. RNA. 1997;3:37–48. [PMC free article] [PubMed] [Google Scholar]
- Dickey LF, Gallo-Meagher M, Thompson WF. Light regulatory sequences are located within the 5′ portion of the Fed-1 message sequence. EMBO J. 1992;11:2311–2317. doi: 10.1002/j.1460-2075.1992.tb05290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey LF, Petracek ME, Nguyen TT, Hansen ER, Thompson WF. Light regulation of Fed-1 mRNA requires an element in the 5′ untranslated region and correlates with differential polyribosome association. Plant Cell. 1998;10:475–484. doi: 10.1105/tpc.10.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elich TD, Edelman F, Mattoo AK. Dephosphorylation of photosystem II core proteins is light-regulated in vivo. EMBO J. 1993;12:4857–4862. doi: 10.1002/j.1460-2075.1993.tb06175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubas J-M, Loumas M, LaRoche J, Falkowski PG. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ferris C, Moreno J. Oxidative modification and breakdown of ribulose-1,5-bisphosphate carboxylase induced in Euglena gracilis by nitrogen starvation. Planta. 1994;193:208–215. [Google Scholar]
- Gillham NW, Boynton JE, Hauser CR. Translational regulation of gene expression in chloroplasts and mitochondria. Annu Rev Genet. 1994;28:71–93. doi: 10.1146/annurev.ge.28.120194.000443. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Coordination of nuclear and chloroplast gene expression in plant cells. Int Rev Cytol. 1998;177:115–180. doi: 10.1016/s0074-7696(08)62232-9. [DOI] [PubMed] [Google Scholar]
- Gruissem W, Tonkyn JC. Control mechanisms of plastid gene expression. Crit Rev Plant Sci. 1993;12:19–55. [Google Scholar]
- Hayes R, Kudla J, Schuster G, Gabay L, Maliga P, Gruissem W. Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J. 1996;15:1132–1141. [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- Kim J, Mayfield SP. Protein disulfide isomerase as a regulator of chloroplast translational activation. Science. 1997;278:1954–1957. doi: 10.1126/science.278.5345.1954. [DOI] [PubMed] [Google Scholar]
- Klein RR, Mullet JE. Light-induced transcription of chloroplast genes: psbA transcription is differentially enhanced in illuminated barley. J Biol Chem. 1990;265:1895–1902. [PubMed] [Google Scholar]
- Kosower NS, Kosower EM. Diamide: an oxidant probe for thiols. Methods Enzymol. 1995;251:123–133. doi: 10.1016/0076-6879(95)51116-4. [DOI] [PubMed] [Google Scholar]
- Kuchka MR, Goldschmidt-Clermont M, van Dillewijn J, Rochaix J-D. Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PSII. Cell. 1989;58:869–876. doi: 10.1016/0092-8674(89)90939-2. [DOI] [PubMed] [Google Scholar]
- Levings CS, III, Siedow JN. Regulation by redox poise in chloroplasts. Science. 1995;268:695–696. doi: 10.1126/science.268.5211.695. [DOI] [PubMed] [Google Scholar]
- Liere K, Link G. Chloroplast endoribonuclease p54 involved in RNA 3′-end processing is regulated by phosphorylation and redox state. Nucleic Acids Res. 1997;25:2403–2408. doi: 10.1093/nar/25.12.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky I, Schuster G. Phosphorylation of a chloroplast RNA-binding protein changes its affinity to RNA. Nucleic Acids Res. 1995;23:2506–2511. doi: 10.1093/nar/23.13.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A, Jansson C. Photosynthetic electron transport controls degradation but not production of psbA transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol. 1991;16:891–897. doi: 10.1007/BF00015080. [DOI] [PubMed] [Google Scholar]
- Mullet JE. Dynamic regulation of chloroplast transcription. Plant Physiol. 1993;103:309–313. doi: 10.1104/pp.103.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müllner EW, Rothenberger S, Müller AM, Kühn LC. In vivo and in vitro modulation of the mRNA-binding activity of iron-regulatory factor: tissue distribution and effects of cell proliferation, iron levels and redox state. Eur J Biochem. 1992;208:597–605. doi: 10.1111/j.1432-1033.1992.tb17224.x. [DOI] [PubMed] [Google Scholar]
- Nalin CM, McCarty RE. Role of a disulfide bond in the gamma subunit in activation of the ATPase of chloroplast coupling factor 1. J Biol Chem. 1984;259:7275–7280. [PubMed] [Google Scholar]
- Nickelsen J, van Dillewijn J, Rahire M, Rochaix J-D. Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J. 1994;13:3182–3191. doi: 10.1002/j.1460-2075.1994.tb06617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad I, Adir N, Koike H, Kyle DJ, Inoue Y. Mechanism of photoinhibition in vivo: a reversible light-induced conformational change of reaction center II is related to an irreversible modification of the D1 protein. J Biol Chem. 1990;265:1972–1979. [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Huber SC, Thompson WF. Light-regulated changes in abundance and polyribosome association of ferredoxin mRNA are dependent on photosynthesis. Plant Cell. 1997;9:2291–2300. doi: 10.1105/tpc.9.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Nguyen TT, Gatz C, Sowinski DA, Allen GC, Thompson WF. Ferredoxin-1 mRNA is destabilized by changes in photosynthetic electron transport. Proc Natl Acad Sci USA. 1998;95:9009–9013. doi: 10.1073/pnas.95.15.9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- Rochaix J-D. Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol Biol. 1996;32:327–341. doi: 10.1007/BF00039389. [DOI] [PubMed] [Google Scholar]
- Rochaix J-D, Kuchka M, Mayfield S, Schirmer-Rahire M, Girard-Bascou J, Bennoun P. Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. EMBO J. 1989;8:1013–1021. doi: 10.1002/j.1460-2075.1989.tb03468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon IJ, Scandurro AB, Wilson RB, Beckman BS. Changes in redox affect the activity of erythropoietin RNA binding protein. FEBS Lett. 1995;359:267–270. doi: 10.1016/0014-5793(95)00066-i. [DOI] [PubMed] [Google Scholar]
- Salvador ML, Klein U, Bogorad L. 5′ Sequences are important positive and negative determinants of the longevity of Chlamydomonas chloroplast gene transcripts. Proc Natl Acad Sci USA. 1993a;90:1556–1560. doi: 10.1073/pnas.90.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador ML, Klein U, Bogorad L. Light-regulated and endogenous fluctuations of chloroplast transcript levels in Chlamydomonas: regulation by transcription and RNA degradation. Plant J. 1993b;3:213–219. doi: 10.1046/j.1365-313x.1993.t01-13-00999.x. [DOI] [PubMed] [Google Scholar]
- Sexton TB, Christopher DA, Mullet JE. Light-induced switch in barley psbD-psbC promoter utilization: a novel mechanism regulating chloroplast gene expression. EMBO J. 1990;9:4485–4494. doi: 10.1002/j.1460-2075.1990.tb07899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1960;46:83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M, Sugiura M. Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol. 1996;32:315–326. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- Wenderoth I, Scheibe R, von Schaewen A. Identification of the cysteine residues involved in redox modulation of plant plastidic glucose-6-phosphate dehydrogenase. J Biol Chem. 1997;272:26985–26990. doi: 10.1074/jbc.272.43.26985. [DOI] [PubMed] [Google Scholar]
- Yohn CB, Cohen A, Danon A, Mayfield SP. A poly(A)-binding protein functions in the chloroplast as a message-specific translation factor. Proc Natl Acad Sci USA. 1998a;95:2238–2243. doi: 10.1073/pnas.95.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn CB, Cohen A, Rosch C, Kuchka MR, Mayfield SP. Translation of the chloroplast psbA mRNA requires the nuclear-encoded poly(A)-binding protein, RB47. J Cell Biol. 1998b;142:435–442. doi: 10.1083/jcb.142.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]