Abstract
Colorectal cancer (CRC) is one of the most frequently occurring primary malignant tumors worldwide. Chemotherapeutic resistance is a major clinical problem in the treatment of CRC. Therefore, it is of great importance to investigate novel biomarkers that may predict chemoresistance and facilitate the development of individualized treatment for patients with CRC. The present study reported that let-7f-5p expression was elevated in chemotherapy-resistant CRC tissues compared with chemotherapy-sensitive tissues. Furthermore, upregulating let-7f-5p increased the expression levels of the anti-apoptotic proteins, B-cell lymphoma 2 (Bcl-2) and B-cell lymphoma-extra large (Bcl-xL), and decreased the activity of caspase-3 and caspase-9 in CRC cells. By contrast, downregulating let-7f-5p yielded the opposite effect. Notably, the results indicated that let-7f-5p promoted chemotherapeutic resistance by directly repressing the expression of several pro-apoptotic proteins, including tumor protein p53, tumor protein p53-inducible nuclear protein 1, tumor protein p53-inducible nuclear protein 2 and caspase-3. Therefore, a novel mechanism by which let-7f-5p enhances the resistance of CRC cells to chemotherapeutics has been revealed, indicating that silencing let-7f-5p may become an effective therapeutic strategy against CRC.
Keywords: colorectal cancer, let-7f-5p, chemotherapeutic resistance, B-cell lymphoma-2, B-cell lymphoma-extra large, tumor protein p53, tumor protein p53-inducible nuclear protein 1, tumor protein p53-inducible nuclear protein 2, caspase-3/9
Introduction
Colorectal cancer (CRC) is one of the most prevalent causes of cancer-associated mortality worldwide (1,2). Clinically, 5-fluorouracil (5-FU)-based chemotherapy serves as the initial first-line chemotherapeutic drug of choice in patients with CRC; however, the response rates to 5-FU are ~15% in patients with advanced CRC (3). Although novel therapeutic approaches, including combination chemotherapy of 5-FU and oxaliplatin (FOLFOX) or irinotecan (FOLFIRI), have presented promising potential, the majority of the patients continue to exhibit inadequate responses (4,5). The failure of chemotherapy in CRC patients is primarily attributed to therapeutic resistance following a long period of treatment (6). These observations emphasize that chemotherapeutic resistance is a major obstacle in the treatment of CRC, and that the molecular mechanisms underlying this issue remain unclear.
Chemotherapeutic resistance poses a major challenge in cancer chemotherapy. Chemoresistance of tumor cells develops primarily due to acquired resistance de novo, following an initially sensitive response, and/or in combination with pre-established cell-intrinsic mechanisms (7). A number of well-established mechanisms are reported to be involved in cancer drug resistance, including failure of the drug to reach or enter the target cell, formation of efflux pumps on the surface of tumor cells, increased expression of anti-apoptotic proteins, induction of drug-detoxifying mechanisms and upregulation of DNA repair enzymes (8,9). However, there is an absence of integral understanding of acquired drug resistance in the context of CRC. Such insight may be crucial for the development of an effective therapeutic approach to overcome chemoresistance and to improve clinical therapeutic response in CRC.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs composed of 19–25 nucleotides, which are involved in numerous biological processes, including survival, apoptosis, the cell cycle and gene regulation (10,11). miRNAs regulate downstream target gene expression at a post-transcriptional level by binding with specific sequences in the 3′-untranslated region (3′-UTR) of downstream target genes, leading to mRNA degradation and/or translational inhibition (12). Several lines of evidence have indicated that aberrant expression of miRNAs is involved in the progression and metastasis of different types of cancer (13–18). Furthermore, research has indicated that upregulation or downregulation of a certain miRNA (let-7a) was directly correlated with the response to chemotherapeutic agents (19). Emerging evidence has demonstrated that miRNAs are key regulators in the chemotherapeutic resistance of CRC. Ectopic expression of miR-125a/b restored paclitaxel sensitivity through downregulation of aldehyde dehydrogenase 1 in HT29 cells (20). Furthermore, Zhang et al (21) revealed that miR-587 promoted the resistance of CRC cells to 5-FU via inhibiting protein phosphatase 2 scaffold subunit Aβ. Notably, silencing miR-587 re-sensitized CRC cells to 5-FU. These observations indicated that miRNAs may function as tumor-suppressors or inducers in CRC.
The present study demonstrated that let-7f-5p expression was elevated in chemotherapy-resistant CRC tissues compared with chemotherapy-sensitive tissues. Furthermore, upregulating let-7f-5p increased the expression of the anti-apoptotic proteins, B-cell lymphoma 2 (Bcl-2) and B-cell lymphoma-extra large (Bcl-xL), and decreased the activity of caspase-3 and caspase-9 in CRC cells. Conversely, downregulating let-7f-5p decreased the expression of Bcl-2 and Bcl-xL, and increased the activity of caspase-3 and caspase-9 in CRC cells. Of note, the results of the present study indicated that let-7f-5p promotes chemotherapy-resistance via directly repressing the expression of several pro-apoptotic proteins, including tumor protein p53 (TP53), TP53-inducible nuclear protein 1, TP53-inducible nuclear protein 2 and caspase-3. Therefore, the present study identified a novel mechanism by which let-7f-5p enhances the resistance of CRC cells to chemotherapeutics, indicating that therapy targeting let-7f-5p in combination with chemotherapeutics may become an effective therapeutic strategy against CRC.
Materials and methods
The Cancer Genome Atlas (TCGA) analysis
The miRNA dataset of colorectal cancer was downloaded from the TCGA and obtained the expression value(s) of the corresponding genes from the Level 3 data of each sample. The unit of miRNA expression levels in TCGA is Reads per kilobase of exon model per million mapped reads (RKPM). Analysis of the log2 value of each sample was performed using Excel 2010 and GraphPad 5 (GraphPad Software, Inc., La Jolla, CA, USA). Furthermore, statistical analysis of the miRNA expression levels within colorectal cancer tissues was performed using paired t-test or unpaired t-test.
Cell lines and cell culture
The human CRC HCT116 and SW480 cell lines were obtained from Type Culture Collection Chinese Academy of Sciences (Shanghai, China) and were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with penicillin G (100 U/ml), streptomycin (100 mg/ml) and 10% fetal bovine serum (Thermo Fisher Scientific, Inc.). HCT116 and SW480 cells were incubated in 10 µmol/l 5-FU for 24 h, as previously described (22,23). The cells were incubated at 37°C in a humidified atmosphere with 5% CO2 and were routinely sub-cultured using 0.25% (w/v) trypsin-ethylenediaminetetraacetic acid solution (Thermo Fisher Scientific, Inc.).
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using an RNA Isolation kit (Qiagen, Inc., Valencia, CA, USA), according to the manufacturer's protocol. mRNA and microRNA (miRNA) were reverse transcribed from total RNA using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. cDNA was amplified and quantified on the CFX96 system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using iQ SYBR-Green (Bio-Rad Laboratories, Inc.). The thermocycling conditions were as follows: 95°C for 3 min, then 35 cycles of 95°C for 5 sec, 60°C for 15 sec, and 72°C for 15 sec followed by 72°C for 10 min. The primer sequences are provided in Table I. Primers for U6 and let-7f-5p were synthesized and purified by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). U6 or GAPDH were used as endogenous controls. Relative fold expression was calculated using the 2−ΔΔCq method (24).
Table I.
Primers used in reverse transcription-quantitative polymerase chain reaction.
| Primer | Sequence (5′-3′) | |
|---|---|---|
| TP53 | Forward | GCTCGACGCTAGGATCTGAC |
| Reverse | GCTTTCCACGACGGTGAC | |
| TP53INP1 | Forward | TATGCTGCCCCATTTCATTT |
| Reverse | CTGTGCATAACTCCTGCCCT | |
| TP53INP2 | Forward | TGAAGAAGAGGCTGGAGAGG |
| Reverse | CTCCCAGCTGTTTGGATCAC | |
| Caspase-3 | Forward | TCGCTTCCATGTATGATCTTTG |
| Reverse | CTGCCTCTTCCCCCATTCT | |
| Bcl-2 | Forward | GTGGATGACTGAGTACCTGAACC |
| Reverse | AGACAGCCAGGAGAAATCAAAC | |
| Bcl-xL | Forward | GGTATTGGTGAGTCGGATCG |
| Reverse | TGCTGCATTGTTCCCATAGA | |
| GAPDH | Forward | ATTCCACCCATGGCAAATTC |
| Reverse | TGGGATTTCCATTGATGACAAG |
TP53, tumor protein p53; TP53INP1, TP53-inducible nuclear protein 1; TP53INP2; TP53-inducible nuclear protein 2; Bcl-2, B-cell lymphoma 2; Bcl-xL, B-cell lymphoma-extra large.
Transfection
let-7f-5p mimics, inhibitors and respective control RNAs, including siRNA scramble and negative controls, (50 nmol/l) were synthesized and purified by Guangzhou RiboBio Co., Ltd. Transfection of miRNA, siRNAs and plasmids was performed using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), 48 h after transfection, cells were harvest and the indicated experiments were performed, according to the manufacturer's protocol.
Western blot analysis
Nuclear/cytoplasmic fractionation was performed using a Cell Fractionation kit (Cell Signaling Technology, Inc., Danvers, MA, USA) according to the manufacturer's protocol, and the whole cell lysates were extracted using radioimmunoprecipitation assay buffer (Cell Signaling Technology, Inc.). The cell lysates were loaded with 10% loading buffer (Beyotime Institute of Biotechnology, Haimen, China) and were heated for 5 min at 100°C. The protein concentrations were determined using the BCA Protein Assay kit (Invitrogen; Thermo Fisher Scientific, Inc.). Equal quantities of denatured protein samples (40 µl) were resolved on 10% SDS-polyacrylamide gels, prior to being transferred onto polyvinylidene difluoride membranes (Roche Diagnostics, Basel, Switzerland). Following 1 h blocking with 5% dry skimmed milk in Tris-buffered saline/0.05% Tween-20 at room temperature, membranes were incubated with primary antibodies against Bcl-2 at a dilution of 1:1,000 (cat. no. ab194583; Abcam, Cambridge, UK) or Bcl-xL at a dilution of 1:1,000 (cat. no. ab32370; Abcam), overnight at 4°C, followed by incubation with a horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. Proteins were visualized using Pierce™ Fast Western Blot kit (Thermo Fisher Scientific, Inc.). The membranes were stripped and reprobed with an anti-α-tubulin antibody at a dilution of 1:1,000 overnight at 4°C (cat. no. T6199; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) as the loading control.
Caspase-9 or −3 activity assays
Activity of caspase-9 or −3 was analyzed by spectrophotometry using a caspase-9 colorimetric assay kit (cat. no. KGA403) or acaspase-3 colorimetric assay kit (cat. no. KGA202; Nanjing KeyGen Biotech, Co. Ltd., Nanjing, China), according to the manufacturer's protocol. In brief, 5×106 cells were washed with cold phosphate-buffered saline, re-suspended in Lysis Buffer (from the two kits) and incubated on ice for 30 min. The 50 µl cell suspension was mixed with 50 µl Reaction Buffer (from the two kits) and 5 µl caspase-3/-9 substrate, prior to being incubated at 37°C for 4 h. The absorbance was measured at 405 nm, and bicinchoninic acid protein quantitative analysis was used as the reference to normalize each of the experimental groups.
Flow cytometric analysis
Flow cytometric analysis of apoptosis was performed using the fluorescein isothiocyanate (FITC)-Annexin V Apoptosis Detection kit I (BD Biosciences, Franklin Lakes, NJ, USA), and was performed, according to the manufacturer's protocol. In brief, cells were dissociated with trypsin and were resuspended at 1×106 cells/ml in binding buffer (Thermo Fisher Scientific, Inc.) with 50 µl/ml FITC-Annexin V and 50 µl/ml propidium iodide (PI). The cells were subsequently incubated for 15 min at room temperature, prior to being analyzed using a Gallios flow cytometer (Beckman Coulter, Inc., Brea, CA, USA). The inner mitochondrial membrane potential (Δψm) of the cells was detected by flow cytometry using a MitoScreen JC-1 staining kit (cat no. 551302; BD Biosciences), according to the manufacturer's protocol. In brief, cells were dissociated with trypsin and resuspended at 1×106 cells/ml in assay buffer (BD Biosciences), prior to being incubated at 37°C for 15 min with 10 µl/ml JC-1. Prior to flow cytometric analysis, cells were washed twice in assay buffer (BD Biosciences), part of the MitoScreen JC-1 staining kit. Flow cytometry data were analyzed using FlowJo 7.6 software (FlowJo LLC, Ashland, OR, USA).
Luciferase assay
A total of 4×104 cells were seeded in triplicate onto 24-well plates and were cultured at 37°C for 24 h. Cells were transfected with 100 ng control pmirGLO, pmirGLO-TP53-3′UTR, -TP53INP1-3′UTR, -TP53INP2-3′UTR, or -caspase3-3′-UTR luciferase plasmid, plus 5 ng pRL-TK Renilla plasmid (Promega Corporation, Madison, WI, USA), using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Luciferase and Renilla signals were measured 36 h after transfection using a Dual Luciferase Reporter assay kit (Promega Corporation) at room temperature, according to the manufacturer's protocol.
Statistical analysis
All values are presented as the mean ± standard deviation. Significant differences were determined using GraphPad 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). The Mann-Whitney U test and χ2 tests were used to determine statistical differences between two groups. One-way analysis of variance, followed by the Student-Newman-Keuls test, was used to determine statistical differences among multiple groups. P<0.05 was considered to indicate a statistically significant difference. All the experiments were repeated three times.
Results
let-7f-5p is upregulated in chemoresistant CRC
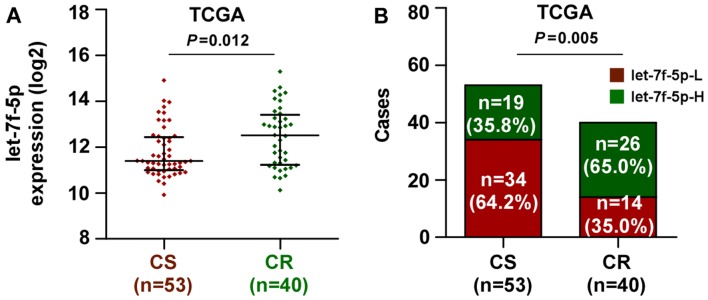
The miRNA sequencing datasets of CRC were analyzed using TCGA, which identified that let-7f-5p expression was elevated in chemoresistant CRC tissues, compared with chemosensitive tissues (Fig. 1A). Notably, the present study demonstrated that the expression level of let-7f-5p was upregulated in patients with CRC exhibiting poor chemotherapeutic response, compared with those with a positive chemotherapeutic response (Fig. 1B). These observations revealed that let-7f-5p may contribute toward chemoresistance in CRC.
Figure 1.
Upregulation of let-7f-5p is associated with poor chemotherapeutic response in CRC. (A) In the TCGA dataset, let-7f-5p expression levels were markedly higher in CRC patients with a poor chemotherapy response (CR) than those with a favorable chemotherapy response (CS). A Mann-Whitney U test was used to determine statistical differences between two groups. The data are presented as the median ± interquartile range. (B) Percentages and numbers of samples exhibiting high or low let-7f-5p expression with different chemotherapeutic responses in the TCGA dataset. The χ2 test was used to determine statistical differences between two groups. CRC, colorectal cancer; TCGA, The Cancer Genome Atlas; CR, chemoresistant; CS, chemosensitive.
Overexpression of let-7f-5p increases the expression of anti-apoptotic proteins Bcl-2 and Bcl-xL in CRC cells
The role of let-7f-5p in the chemoresistance of CRC was additionally examined following treatment with the first-line drug 5-FU. let-7f-5p was upregulated and downregulated following transfection with let-7f-5p mimics and inhibitors, respectively, in CRC HCT116 and SW480 cells (Fig. 2A). The effect of let-7f-5p on the expression of anti-apoptotic proteins, Bcl-2 and Bcl-xL, was further examined. The results demonstrated that upregulating let-7f-5p increased Bcl-2 and Bcl-xL expression, while downregulating let-7f-5p decreased their expression (Fig. 2B). Therefore, these results demonstrated that let-7f-5p promoted the chemoresistance of CRC cells by enhancing the expression of anti-apoptotic proteins, Bcl-2 and Bcl-xL.
Figure 2.
let-7f-5p increases the expression of the anti-apoptotic proteins, Bcl-2 and Bcl-xL, and decreases the activity of caspase-3 and caspase-9 in CRC cells. (A) Reverse transcription-quantitative polymerase chain reaction analysis of let-7f-5p expression in HCT116 and SW480 cells transfected with a let-7f-5p mimic or inhibitor, compared with controls. Transcript levels were normalized to U6 expression. Error bars represent the mean ± standard deviation of three independent experiments. *P<0.05. One-way ANOVA was used to determine statistical differences among multiple groups. (B) Western blot analysis of Bcl-2 and Bcl-xL in the indicated cells. The activities of (C) caspase-3 and (D) caspase-9 were detected by the cleaved forms of these two proteins. Error bars represent the mean ± standard deviation of three independent experiments. *P<0.05. One-way ANOVA was used to determine statistical differences among multiple groups. CRC, colorectal cancer; Bcl-2, B-cell lymphoma 2; Bcl-xL, B-cell lymphoma-extra large; ANOVA, analysis of variance.
Overexpression of let-7f-5p decreases the activity of caspase-3 and caspase-9 in CRC cells
The effect of let-7f-5p on the activity of caspase-3 and-9 was examined. As demonstrated in Fig. 2C and D, upregulating let-7f-5p decreased the activity of caspase-3, while silencing let-7f-5p had the opposite effect. Overexpression of let-7f-5p consistently repressed the activity of caspase-9, while silencing of let-7f-5p increased the activity of caspase-9. Therefore, these observations suggested that let-7f-5p promotes the chemoresistance of CRC cells by inhibiting the activity of caspase-3 and caspase-9 in CRC cells.
Overexpression of let-7f-5p decreases the apoptotic rate and increases the mitochondrial potential of CRC cells
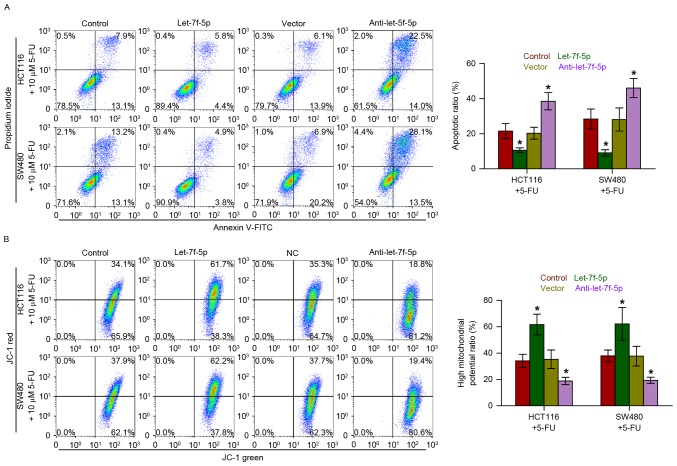
Annexin V-FITC/PI staining demonstrated that overexpression of let-7f-5p decreased the apoptotic rate of HCT116 and SW480 cells treated with 5-FU, while silencing let-7f-5p had the opposite effect (Fig. 3A). Furthermore, the effect of let-7f-5p on the mitochondrial potential of CRC cells was examined, and the results indicated that upregulating let-7f-5p enhanced the mitochondrial potential of HCT116 and SW480 cells following treatment with 5-FU; however, silencing let-7f-5p exhibited the opposite effects on the mitochondrial potential of CRC cells (Fig. 3B). Therefore, these observations demonstrated that let-7f-5p promotes the chemoresistance of CRC cells to 5-FU by decreasing the apoptotic rate and increasing the mitochondrial potential of HCT116 and SW480 cells.
Figure 3.
let-7f-5p decreases the apoptotic rate and increases the mitochondrial potential of CRC cells. (A) Annexin V-FITC/PI staining of the indicated cells treated with 5-FU for 36 h. Error bars represent the mean ± standard deviation of three independent experiments. *P<0.05. One-way ANOVA was used to determine statistical differences among multiple groups. (B) The JC-1 staining revealed that upregulating let-7f-5p enhanced the mitochondrial potential of the CRC cells, while silencing let-7f-5p decreased the mitochondrial potential of these cells. Error bars represent the mean ± standard deviation of three independent experiments. *P<0.05. One-way ANOVA was used to determine statistical differences among multiple groups. CRC, colorectal cancer; FITC, fluorescein isothiocyanate; PI, propidium iodide; 5-FU, fluorouracil; ANOVA, analysis of variance.
let-7f-5p targets multiple pro-apoptotic proteins in CRC cells
Using the publicly available algorithms, TargetScan and miRanda, it was identified that multiple pro-apoptotic proteins, including TP53, TP53INP1, TP53INP2 and caspase-3, may be potential targets of let-7f-5p (Fig. 4). RT-qPCR analysis revealed that let-7f-5p overexpression reduced the mRNA expression levels of TP53, TP53INP1, TP53INP2 and caspase-3. By contrast, silencing of let-7f-5p increased the expression of TP53, TP53INP1, TP53INP2 and caspase-3, suggesting that let-7f-5p negatively regulated the mRNA expression levels of TP53, TP53INP1, TP53INP2 and caspase-3 (Fig. 5). Furthermore, a luciferase assay revealed that let-7f-5p overexpression decreased the reporter activity driven by the 3′-UTRs of these transcripts, while silencing let-7f-5p increased this activity (Fig. 6). Taken together, the results of the present study indicated that let-7f-5p directly targets TP53, TP53INP1, TP53INP2 and caspase-3, which further promotes chemotherapeutic resistance in CRC.
Figure 4.

let-7f-5p targets multiple pro-apoptotic proteins. Predicted let-7f-5p targeting sequence in 3′-UTRs of TP53, TP53INP1, TP53INP2 and caspase-3. TP53, tumor protein p53; TP53INP1, TP53-inducible nuclear protein 1; TP53INP2, TP53-inducible nuclear protein p53 2; UTR, untranslated region.
Figure 5.
let-7f-5p decreases the mRNA expression levels of TP53, TP53INP1, TP53INP2 and caspase-3. Reverse transcription-quantitative polymerase chain reaction analysis of TP53, TP53INP1, TP53INP2 and caspase-3 following transfection with a let-7f-5p mimic or inhibitor in (A) HCT116 and (B) SW480 cells. Transcript levels were normalized to U6 expression. Error bars represent the mean ± standard deviation of three independent experiments. *P<0.05. One-way analysis of variance was used to determine statistical differences among multiple groups. TP53, tumor protein p53; TP53INP1, TP53-inducible nuclear protein 1; TP53INP2, TP53-inducible nuclear protein p53 2; UTR, untranslated region.
Figure 6.
let-7f-5p decreases the luciferase activity of the 3′-UTRs of TP53, TP53INP1, TP53INP2 and caspase-3. Luciferase assay of cells transfected with pmiRGLO-3′UTR reporter of TP53, TP53INP1, TP53INP2 and caspase-3 in let-7f-5p-overexpressing and silenced (A) HCT116 and (B) SW480 cells. One-way analysis of variance was used to determine statistical differences among multiple groups. TP53, tumor protein p53; TP53INP1, TP53-inducible nuclear protein 1; TP53INP2, TP53-inducible nuclear protein p53 2; UTR, untranslated region. Error bars represent the mean ± standard deviation of three independent experiments. *P<0.05 vs. control cells.
Discussion
The results of the present study demonstrated that let-7f-5p expression was elevated in chemotherapy-resistant CRC tissues, compared with chemotherapy-sensitive tissues. Furthermore, upregulating let-7f-5p increased the expression of the anti-apoptotic proteins, Bcl-2 and Bcl-xL, and decreased the activity of caspase-3 and caspase-9 in CRC cells, while downregulating let-7f-5p had the opposite effect. Notably, the observations of the present study indicated that let-7f-5p promotes chemotherapy resistance via directly repressing the expression of a number of pro-apoptotic proteins, including TP53, TP53INP1, TP53INP2 and caspase-3. Therefore, the results of the present study indicate that overexpression of let-7f-5p exhibits an important function in the chemoresistance of CRC.
The transcription factor, p53 is the most characterized tumor suppressor gene and has been reported to be involved in the development and progression of several types of cancer via controlling cell cycle checkpoints, and promoting apoptosis and senescence (25,26). Loss or mutation of the p53 gene has been identified to be positively associated with the recurrent incidence of chemotherapeutic resistance in different types of cancer (27). In CRC, p53 is mutated in ~50% of patients and these mutations of are considered to be inactivating, leaving to TP53 being incapable of regularly exerting its functions, including its pro-apoptotic role (28,29). The present study identified that let-7f-5p decreased the apoptosis of CRC cells induced by 5-FU and conferred the resistance of these cells to 5-FU via inhibiting TP53 expression. Furthermore, the results further demonstrated that let-7f-5p simultaneously targets TP53INP1, TP53INP2 and caspase 3, which cooperatively attenuated the apoptosis induced by 5-FU and promoted chemoresistance in CRC cells. TP53INP1 and TP53INP2, which act as pro-apoptotic and anti-proliferative proteins, have been reported to positively regulate p53, and to stimulate its ability to induce apoptosis and to regulate the cell cycle (30). A previous study reported that miR-182 increased drug resistance in cisplatin-treated hepatocellular carcinoma cells via inhibiting TP53INP1 expression (31). Caspase-3 has been reported to be involved in the activation cascade of caspases responsible for apoptosis, and functions by cleaving and activating caspases-6, −7 and −9, which serve important roles in the chemoresistance of different types of cancer (32–34). Through the use of luciferase assays, the present study identified that let-7f-5p simultaneously repressed the activity of TPp53, TP53INP1, TP53INP2 and caspase-3 by binding to the 3′-UTRs of their mRNAs, resulting in a decrease in the apoptosis induced by 5-FU in CRC cells and the development of chemoresistance.
Members of the Let-7 family have been identified to function as tumor suppressors through regulating multiple oncogenic signals (35). For example, Di Fazio et al (36) reported that let-7b exhibited an important tumor-suppressive role via inhibiting the expression of high-mobility group AT-hook 2, a nuclear non-histone transcriptional co-factor with known oncogenic properties, in liver cancer cell lines. Furthermore, emerging evidence has demonstrated that members of the let-7 family of miRNAs are downregulated in a number of types of human cancer, and that low let-7 expression has been correlated with resistance to chemotherapeutics (37,38). Downregulation of let-7 was also associated with chemoresistance in breast cancer cells (39). In addition, Sugimura et al (38) reported that low expression of let-7b and let-7c was significantly associated with a poor response to chemotherapy in esophageal squamous cell carcinoma, and that transfection of let-7c restored sensitivity to cisplatin and increased the rate of apoptosis following exposure to cisplatin in vitro assays. Additionally, Tsang and Kwok (34) reported that induced expression of let-7a increased the resistance of human squamous carcinoma A431 cells to chemotherapeutic drugs, including interferon-γ, doxorubicin and paclitaxel. These observations revealed that the members of the let-7 family may function as oncomiRs and tumor-suppressive miRNAs, depending on the type of tumor. The present study reported that, as a member of the let-7 family, let-7f-5p was elevated in chemoresistant CRC tissues, compared with chemosensitive tissues. Upregulation of let-7f-5p decreased the apoptosis induced by 5-FU in vitro assays, while silencing let-7f-5p had the opposite effect. These results, in conjunction with those of other studies, indicated that the pro-cancer and anticancer roles of the let-7 family are environmentally and tumor-type dependent.
Previous studies have implicated the altered expression of let-7f-5p in several diseases, including major depression, myasthenia gravis, Alzheimer's disease (40–42), and neoplastic conditions, including laryngeal carcinoma, CRC and thyroid cancer (43,44). Notably, Pathak et al (45) reported that let-7f-5p was highly upregulated in the human colon cancer HCT116 and SN38 cell lines following radiation treatment in a p53-directed manner, indicating that let-7f-5p may contribute to the chemoresistance of CRC. However, the specific role of let-7f-5p in CRC remains unclear. The results of the present study also demonstrated that let-7f-5p expression was elevated in chemoresistant CRC tissues compared with chemosensitive tissues. Furthermore, ectopic expression of let-7f-5p had the effect of reducing the apoptosis induced by 5-FU, while silencing let-7f-5p enhanced the apoptosis induced by 5-FU. Additionally, silencing let-7f-5p enhanced sensitivity to 5-FU. Importantly, the results demonstrated that let-7f-5p overexpression promotes chemotherapeutic resistance in CRC via directly repressing the expression of several pro-apoptotic proteins, including TP53, TP53INP1, TP53INP2 and caspase-3.
In summary, the results of the present study revealed that let-7f-5p serves an important role in the chemotherapy resistance of CRC via directly repressing expression of TP53, TP53INP1, TP53INP2 and caspase-3. Therefore, improved understanding of the specific role of let-7f-5p in the pathogenesis of CRC would facilitate an increase in the available knowledge of CRC development, which will assist in the development of novel therapeutic measures against CRC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science Foundation of Gansu Province (Lanzhou, China; grant no. 096RJZA096).
Availability of data and materials
The datasets generated and analyzed in the present study are included in this published article.
Authors' contributions
YT and CC developed ideas and drafted the manuscript. HY and HW conducted the experiments and contributed to the analysis of data. HT and YP contributed to the analysis of data. BL and XD contributed to the analysis of data and revised the manuscript. YY and ZQ analysed and interpreted the data and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors contributed to revise the manuscript and approved the final version for publication.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing interests.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Ricci-Vitiani L, Fabrizi E, Palio E, De Maria R. Colon cancer stem cells. J Mol Med (Berl) 2009;87:1097–1104. doi: 10.1007/s00109-009-0518-4. [DOI] [PubMed] [Google Scholar]
- 3.Johnston PG, Kaye S. Capecitabine: A novel agent for the treatment of solid tumors. Anticancer Drugs. 2001;12:639–646. doi: 10.1097/00001813-200109000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Giacchetti S, Perpoint B, Zidani R, Le Bail N, Faggiuolo R, Focan C, Chollet P, Llory JF, Letourneau Y, et al. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2000;18:136–147. doi: 10.1200/JCO.2000.18.1.136. [DOI] [PubMed] [Google Scholar]
- 5.Douillard JY, Cunningham D, Roth AD, Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J, Alakl M, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: A multicenter randomised trial. Lancet. 2000;355:1041–1047. doi: 10.1016/S0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- 6.Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275–292. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol. 2007;608:1–22. doi: 10.1007/978-0-387-74039-3_1. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 9.Wu X, Xiao H. MiRNAs modulate the drug response of tumor cells. Sci China C Life Sci. 2009;9:797–801. doi: 10.1007/s11427-009-0114-4. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren D, Wang M, Guo W, Huang S, Wang Z, Zhao X, Du H, Song L, Peng X. Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cancer cells. Cell Tissue Res. 2014;358:763–778. doi: 10.1007/s00441-014-2001-y. [DOI] [PubMed] [Google Scholar]
- 14.Guo W, Ren D, Chen X, Tu X, Huang S, Wang M, Song L, Zou X, Peng X. HEF1 promotes epithelial mesenchymal transition and bone invasion in prostate cancer under the regulation of microRNA-145. J Cell Biochem. 2013;114:1606–1615. doi: 10.1002/jcb.24502. [DOI] [PubMed] [Google Scholar]
- 15.Simerzin A, Zorde-Khvalevsky E, Rivkin M, Adar R, Zucman-Rossi J, Couchy G, Roskams T, Govaere O, Oren M, Giladi H, Galun E. The liver-specific microRNA-122*, the complementary strand of microRNA-122, acts as a tumor suppressor by modulating the p53/mouse double minute 2 homolog circuitry. Hepatology. 2016;64:1632–1636. doi: 10.1002/hep.28679. [DOI] [PubMed] [Google Scholar]
- 16.Ren D, Wang M, Guo W, Zhao X, Tu X, Huang S, Zou X, Peng X. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR-145. Int J Oncol. 2013;42:1473–1481. doi: 10.3892/ijo.2013.1825. [DOI] [PubMed] [Google Scholar]
- 17.Wang M, Ren D, Guo W, Wang Z, Huang S, Du H, Song L, Peng X. Loss of miR-100 enhances migration, invasion, epithelial-mesenchymal transition and stemness properties in prostate cancer cells through targeting Argonaute 2. Int J Oncol. 2014;45:362–372. doi: 10.3892/ijo.2014.2413. [DOI] [PubMed] [Google Scholar]
- 18.Sahu N, Stephan JP, Cruz DD, Merchant M, Haley B, Bourgon R, Classon M, Settleman J. Functional screening implicates miR-371-3p and peroxiredoxin 6 in reversible tolerance to cancer drugs. Nat Commun. 2016;7:12351. doi: 10.1038/ncomms12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Chen Y, Chen Z. MiR-125a/b regulates the activation of cancer stem cells in paclitaxel-resistant colon cancer. Cancer Invest. 2013;31:17–23. doi: 10.3109/07357907.2012.743557. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Talmon G, Wang J. MicroRNA-587 antagonizes 5-FU-induced apoptosis and confers drug resistance by regulating PPP2R1B expression in colorectal cancer. Cell Death Dis. 2015;6:e1845. doi: 10.1038/cddis.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Chen Y, Hao L, Hou A, Chen X, Li Y, Wang R, Luo P, Ruan Z, Ou J, et al. Macrophages induce resistance to 5-fluorouracil chemotherapy in colorectal cancer through the release of putrescine. Cancer Lett. 2016;381:305–313. doi: 10.1016/j.canlet.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Du C, Huang D, Peng Y, Yao Y, Zhao Y, Yang Y, Wang H, Cao L, Zhu WG, Gu J. 5-Fluorouracil targets histone acetyltransferases p300/CBP in the treatment of colorectal cancer. Cancer Lett. 2017;400:183–193. doi: 10.1016/j.canlet.2017.04.033. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 26.Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J Pathol. 2011;223:116–126. doi: 10.1002/path.2784. [DOI] [PubMed] [Google Scholar]
- 27.Alam SK, Yadav VK, Bajaj S, Datta A, Dutta SK, Bhattacharyya M, Bhattacharya S, Debnath S, Roy S, Boardman LA, et al. DNA damage-induced ephrin-B2 reverse signaling promotes chemoresistance and drives EMT in colorectal carcinoma harboring mutant p53. Cell Death Differ. 2016;23:707–22. doi: 10.1038/cdd.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deschoemaeker S, Di Conza G, Lilla S, Martín-Pérez R, Mennerich D, Boon L, Hendrikx S, Maddocks OD, Marx C, Radhakrishnan P, et al. PHD1 regulates p53-mediated colorectal cancer chemoresistance. EMBO Mol Med. 2015;7:1350–1365. doi: 10.15252/emmm.201505492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 30.Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y, Monden M, Nakamura Y. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol Cell. 2001;8:85–94. doi: 10.1016/S1097-2765(01)00284-2. [DOI] [PubMed] [Google Scholar]
- 31.Qin J, Luo M, Qian H, Chen W. Upregulated miR-182 increases drug resistance in cisplatin-treated HCC cell by regulating TP53INP1. Gene. 2014;538:342–347. doi: 10.1016/j.gene.2013.12.043. [DOI] [PubMed] [Google Scholar]
- 32.Harrington HA, Ho KL, Ghosh S, Tung KC. Construction and analysis of a modular model of caspase activation in apoptosis. Theor Biol Med Model. 2008;5:26. doi: 10.1186/1742-4682-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R, Pascual J, Imamura S, Kishi S, Amatruda JF, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsang WP, Kwok TT. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis. 2008;13:1215–1222. doi: 10.1007/s10495-008-0256-z. [DOI] [PubMed] [Google Scholar]
- 35.Chang CJ, Hsu CC, Chang CH, Tsai LL, Chang YC, Lu SW, Yu CH, Huang HS, Wang JJ, Tsai CH, et al. Let-7d functions as novel regulator of epithelial-mesenchymal transition and chemoresistant property in oral cancer. Oncol Rep. 2011;26:1003–1010. doi: 10.3892/or.2011.1360. [DOI] [PubMed] [Google Scholar]
- 36.Di Fazio P, Montalbano R, Neureiter D, Alinger B, Schmidt A, Merkel AL, Quint K, Ocker M. Downregulation of HMGA2 by the pan-deacetylase inhibitor panobinostat is dependent on hsa-let-7b expression in liver cancer cell lines. Exp Cell Res. 2012;318:1832–1843. doi: 10.1016/j.yexcr.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 37.Xu C, Xie S, Song C, Huang L, Jiang Z. Lin28 mediates cancer chemotherapy resistance via regulation of miRNA signaling. Hepatogastroenterology. 2014;61:1138–1141. [PubMed] [Google Scholar]
- 38.Sugimura K, Miyata H, Tanaka K, Hamano R, Takahashi T, Kurokawa Y, Yamasaki M, Nakajima K, Takiguchi S, Mori M, Doki Y. Let-7 expression is a significant determinant of response to chemotherapy through the regulation of IL-6/STAT3 pathway in esophageal squamous cell carcinoma. Clin Cancer Res. 2012;18:5144–5153. doi: 10.1158/1078-0432.CCR-12-0701. [DOI] [PubMed] [Google Scholar]
- 39.Wu J, Li S, Jia W, Deng H, Chen K, Zhu L, Yu F, Su F. Reduced Let-7a is associated with chemoresistance in primary breast cancer. PLoS One. 2015;10:e0133643. doi: 10.1371/journal.pone.0133643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maffioletti E, Cattaneo A, Rosso G, Maina G, Maj C, Gennarelli M, Tardito D, Bocchio-Chiavetto L. Peripheral whole blood microRNA alterations in major depression and bipolar disorder. J Affect Disord. 2016;200:250–258. doi: 10.1016/j.jad.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 41.Punga T, Bartoccioni E, Lewandowska M, Damato V, Evoli A, Punga AR. Disease specific enrichment of circulating let-7 family microRNA in MuSK+ myasthenia gravis. J Neuroimmunol. 2016;292:21–26. doi: 10.1016/j.jneuroim.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Satoh J, Kino Y, Niida S. MicroRNA-Seq data analysis pipeline to identify blood biomarkers for Alzheimer's disease from public data. Biomark Insights. 2015;10:21–31. doi: 10.4137/BMI.S25132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Damanakis AI, Eckhardt S, Wunderlich A, Roth S, Wissniowski TT, Bartsch DK, Di Fazio P. MicroRNAs let7 expression in thyroid cancer: Correlation with their deputed targets HMGA2 and SLC5A5. J Cancer Res Clin Oncol. 2016;142:1213–1220. doi: 10.1007/s00432-016-2138-z. [DOI] [PubMed] [Google Scholar]
- 44.Lu ZM, Lin YF, Jiang L, Chen LS, Luo XN, Song XH, Chen SH, Zhang SY. Micro-ribonucleic acid expression profiling and bioinformatic target gene analyses in laryngeal carcinoma. Onco Targets Ther. 2014;7:525–533. doi: 10.2147/OTT.S59871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pathak S, Meng WJ, Nandy SK, Ping J, Bisgin A, Helmfors L, Waldmann P, Sun XF. Radiation and SN38 treatments modulate the expression of microRNAs, cytokines and chemokines in colon cancer cells in a p53-directed manner. Oncotarget. 2015;6:44758–44780. doi: 10.18632/oncotarget.5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed in the present study are included in this published article.