Abstract
Corticotropin-releasing factor (CRF) and its receptors have been detected in numerous tumors and have an important role in tumorigenesis and proliferation. However, the role of these peptides has not been established in human glioma and malignant glioma cell lines. The present study evaluated for the first time, the expression of CRF receptor 1 (CRFR1) in 35 human glioma samples, 13 normal brain tissues and human U87 glioma cells using immunohistochemistry, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis. Levels of CRFR1 were identified to be significantly increased in human glioma and U87 cells and higher levels of CRFR1 were observed in glioma tissues of higher grade. The biological functions of human CRF (hCRF) on U87 cells glioma cells were investigated by cell counting, a bromodeoxyuridine assay and flow cytometry. The U87 cells under hCRF treatment exhibited reduced proliferation, increased apoptosis and a cell cycle arrest in S and G2/M phase. The tumor protein p53 (p53) gene may participate in the activation of hCRF via CRFR1 in U87 cells, therefore p53 mRNA and protein were evaluated using RT-qPCR and western blot analysis. Finally, the present results suggest that hCRF inhibits proliferation and induces cell-cycle arrest and apoptosis in U87 cells via the CRFR1-mediated p53 signaling pathway. Therefore, the present study also suggests that hCRF may be used therapeutically, and CRFR1 may be a putative therapeutic target for human glioma.
Keywords: corticotrophin releasing factor, proliferation, apoptosis, glioma, U87 cells, tumor protein p53
Introduction
Glioma is the most prevalent and most aggressive primary brain tumor in adults, accounting for >50% of all tumors of the central nervous system (1). Although a combination of microsurgery, radiotherapy and chemotherapy have been widely used to treat glioma, the prognosis of patients with malignant glioma remains poor, with a median survival time in the range of 12 to 15 months (2). Dozens of molecules and signal pathways have been established to be involved in the genetic alterations and chromosome aberrations that occur during tumorigenesis. Therefore, the development of efficient treatment therapies, based on an improved understanding of the pathophysiological and molecular properties of malignant glioma, are required.
Human corticotropin-releasing factor (hCRF), a 41-residue neuropeptide, is produced in the hypothalamus and is an important component in the regulation of the hypothalamic-pituitaryadrenal axis. CRF receptors constitute a family of G-protein-coupled receptors, of which there are two major classes, CRFR1 and CRFR2 (3,4). CRFR1 binds CRF with a higher affinity compared with CRFR2 and is expressed primarily in the brain and pituitary gland. The activation of CRFR1 exerts numerous central and peripheral effects associated with pathological disease (5). In addition to the expression of CRFRs in the central nervous system, receptors are identified in the immune, cardiovascular and reproductive systems, suggesting that the CRF receptors may mediate numerous diverse functions (6,7).
Previous studies have established that CRF and its receptors may contribute to the pathogenesis of neurodegenerative disorders, including brain injury, ischemia, Alzheimer's disease and Parkinson's disease (8–11). CRF-family proteins and their receptors have been detected in a number of tumor types. Numerous epithelial tumors, epithelial tumor cell lines and neuroendocrine tumor cell lines have been demonstrated to express CRF receptors (12,13). CRFR1 has been detected in the MCF7 breast cancer cell line, in human melanomas and in melanoma cell lines (14–16). Even though the expression of CRF and its receptors has been described in different types of tumors and cancer cells, the role of these peptides has not been previously investigated in human glioma and malignant glioma cell lines. However, evidence suggests that hCRF may represent a novel therapeutic target in peritumoral brain edema (17).
In the present study, the expression levels of CRFR1 in human glioma tissues and U87 glioblastoma cell lines were assessed, and its association with pathological grades determined. The U87 glioma cell line was subsequently treated with hCRF to assess the action of the ligand on cell proliferation, cell cycle and apoptosis. hCRF was identified to affect cell proliferation, the cell cycle and apoptosis of U87 glioma cells via targeting p53, demonstrating CRFR1 may be a putative therapeutic target for human glioma treatment.
Materials and methods
Human sample collection
Human glioma samples were obtained from the Department of Neurosurgery, The Second Affiliated Hospital of Hebei Medical University (Hebei, China) in 2014. Of the 35 samples, 18 were from male patients and 17 from females, with a mean age ± standard deviation (SD) 45.15±12.35 years (age range, 3–62). The 35 tumor samples were divided into 4 groups (18–20): Medulloblastoma (n=5); grade II (n=8); grade III (n=12); and grade IV (n=10). All specimens were obtained during initial surgery and patients were not administered preoperative radiotherapy, chemotherapy or immunotherapy. Diagnoses were established using histology staining by two pathologists, according to the WHO classification guidelines (21). Normal cerebral tissues, used as controls, were obtained from 13 patients suffering from severe brain injury who underwent intracranial decompression. All samples were stored at −80°C in liquid nitrogen immediately following resection. Data collection and analysis were approved by the ethics committee of The Second Affiliated Hospital of Hebei Medical University.
Cells and cell culture
Human U87 glioma cells were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). The cells were maintained in Dulbecco's modified Eagle's medium (HyClone, Logan, UT, USA) enriched with 10% fetal bovine serum (HyClone; GE Healthcare Life Sciences, Shanghai, China), 100 U/ml penicillin and 100 µg/ml streptomycin at a 37°C with 5% CO2.
hCRF treatment
hCRF was purchased from Shanghai Shenggong Biology Engineering Technology Service, Ltd. (Shanghai, China). U87 cells were divided into 4 groups, including the control group, U87 cells treated with 3 doses of hCRF (10−7, 10−8 and 10−9 mol/l). All cells in this and the remaining experiments were harvested at 24, 48 and 72 h. The experiment was repeated 3 times.
Immunohistochemistry (IHC) staining
IHC staining of tumor tissue was performed following the standard procedures (22). In brief, paraffin-embedded tumor tissue sections (5-µm) on poly-L-lysine-coated slides were heated for 30 min at 60°C, dewaxed in xylene and rehydrated in an ethanol series of 100 (10 min), 95 (5 min), 80 (5 min) and 75% (5 min), then submerged in water, followed by a 5-min microwave (400 W) antigen retrieval set. Endogenous peroxidase was inactivated with 3% H2O2 for 20 min and sections were incubated in 5% bovine serum albumin (BSA) solution for 20 min to block non-specific binding. Subsequently, sections were incubated overnight at 4°C with goat anti-CRFR1 polyclonal antibodies (cat. no., ab59023; 1:100; Abcam, Cambridge, UK). The membraned were washed 3 times in PBS for 5 min, followed by a biotinylated secondary anti-goat antibody (dilution, 1:200; cat. no., sc-2350; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) incubation for 30 min. 3,3′-Diaminobenzidine was used to reveal the IHC reaction and the slides were counterstained with hematoxylin, and examined using a Leica DM1000 microscope (Leica, Microsystems GmbH, Wetzlar, Germany). Primary antibodies were substituted for phosphate buffered saline (PBS) in the negative control.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the tumor tissues and U87 glioma cells using Trizol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. Total RNA (2 µg) was reverse-transcribed with M-MLV reverse transcriptase (Invitrogen Thermo Fisher Scientific, Inc.,) and oligo (dT) primers (Sangon Biotech Co., Ltd., Shanghai, China). cDNA (1 µl) was used for qPCR, which was performed detect CRFR1, p53 using SYBR Green Master mix (Takara, Bio, Inc., Otsu, Japan) according to the manufacturer's protocol. Sequences of CRFR1, p53 and GAPDH primers were as follows: CRFR1 forward, 5′-TACGACAATGAGAAGTGCTGGT-3′ and reverse, 5′-GGACGATGTAAAGGAAGAT-3′; p53 forward, 5′-CGCCTGAGGTTGGCTCTGACTGTA-3′ and reverse, 5′-GTCTCTCCCAGGACAGGCACAAAC-3′; GAPDH (internal control) forward, 5′-ACGAGGCCCAGAGCAAGAGA-3′ and reverse, 5′-AGGTGTGGTGCCAGATTTTC-3′. qPCR was performed with an initial denaturation step at 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 40 sec. Data were analyzed using GraphPad Prism 4.0 software (GraphPad Software, Inc., La Jolla, CA, USA). The 2−ΔΔCq method was used to quantitate the relative gene expression (23). PCR experiments were repeated 3 times.
Western blot analysis
Western blot analysis was performed to evaluate the expression levels of CRFR1 in glioma tissues and U87 cells and the protein levels of p53 were evaluated in U87 cells. Total proteins were extracted and the protein concentration was determined using a bicinchoninic acid assay kit (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). Equal amounts of protein (50 mg) were separated using 10% SDS-PAGE, transferred onto polyvinylidene fluoride membranes (Roche Diagnostics GmbH, Mannheim, Germany) by a transfer apparatus at 200 mA for 50 min. The membrane was blocked with 5% skimmed milk for 2 h, and incubated with indicated primary antibodies overnight at 4°C, including goat anti-CRFR1 polyclonal antibodies (1:1,000; cat. no., ab59023; Abcam), rabbit anti-p53 polyclonal antibodies (cat. no., ab1431; 1:1,000; Abcam) and rabbit anti-β-actin monoclonal antibody (1:1,000; cat. no., RB-9421-P0; Affymetrix, Inc., Santa Clara, CA, USA). Then membranes were incubated with horseradish peroxidase conjugated anti-rabbit or anti-goat IgG (1:5,000; cat. nos. sc-2370 and sc-2350; Santa Cruz Biotechnology, Inc.) for 2 h at room temperature. Blots were detected using an enhanced chemiluminescence kit and the densitometry signals were quantified using Image Lab (Image Lab v4.1; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The immunoreactive bands of all proteins were normalized to the intensity of corresponding bands for β-actin. Results were analyzed using the National Institutes of Health Image v1.41 software (Bethesda, MD, USA).
Cell proliferation assay using cell counting for quantification
The U87 cells (control group) or treated with hCRF (10−7 mol/l) were cultured for 48 h and were collected in the logarithmic growth phase. The cells were then resuspended, seeded in triplicate in 96-well plates at a density of 2,000 cells/well and cultured at 37°C in an incubator with a 5% CO2 atmosphere. After 24 h, images of the cell plates were captured under a Nikon 90I light microscope (Nikon, Japan) at ×100, magnification and the images were quantified using Cellomics ArrayScan VTI (Thermo Fisher Scientific, Inc.) once a day for 3 days and then a cell growth curve was generated based on data obtained using the Cellomics ArrayScan VTI.
5-bromo-2-deoxyuridine (BrdU) cell proliferation assay
The U87 cells treated with hCRF (10−7 mol/l) were cultured for 48 h. The cells were then resuspended, seeded in 96-well plates at 1×106 cells per 200 µl, and then cultured for 24–72 h. The cell proliferation was assessed using a BrdU Cell Proliferation ELISA kit (Roche Applied Science, Rotkreuz, Switzerland) according to the manufacturer's protocol. During the final 2–24 h, BrdU reagents were diluted at a ratio of 1:100 and added to the cells (10 µl/well). Then, the cells were fixed using 200 µl FixDenat per well for 30 min and blocked with 5% BSA for 30 min at room temperature. Anti-BrdU-POD monoclonal antibody (dilution, 1:100; cat. no. 11444611001; Roche Applied Science, Penzberg, Germany) and added (100 µl/well) to the cells for 90 min at room temperature. Following 3 washes with washing buffer, substrate solution (100 µl/well) (provided in the ELISA kit) was added and the cells were incubated for 5–30 min in the dark. The color reaction was activated with 10% H2SO4 for 30 min and the BrdU quantity was determined at a wavelength of 450 nm using an ELx800 Absorbance Mircoplate Reader (BioTek Instruments, Inc., Winooski, VT, USA).
Flow cytometry
For cell-cycle analysis, the control and hCRF group cells in a log phase of growth were harvested at a density of 5×105 cells and washed with PBS twice, fixed with ice-cold 70% ethanol for >1 h. Subsequent to washing with PBS, the cells were treated with RNase A, 0.2% Triton X-100 and 50 µg/ml propidium iodide stain (PI; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 4°C for 30 min. The cell cycle profiles were then analyzed using flow cytometry. The experiments were performed in triplicate. The apoptosis analysis was performed in control and hCRF group cells by staining with the Annexin V V-APC Apoptosis Detection kit (BioVision, Inc., Milpitas, CA, USA) according to the manufacturer's protocol. In brief, cells were harvested at a density of 1×106 cells/ml, and 5×105 cells were plated into a 6-well plate and treated with 0.1% trypsin and 0.02% EDTA, and were washed twice with PBS at 4°C prior to collection. The cells were mixed with staining buffer and 100 µl of this cell suspension were mixed with 5 µl of annexin V solution at room temperature in the dark for 10–15 min. The level of cell apoptosis was analyzed using flow cytometry (FACSCalibur). All assays were performed in triplicate.
Statistical analysis
Data were analyzed using the statistical package SAS version 8 software (SAS, Inc., NC, USA) and all experiments were repeated >3 times. All the data were expressed as mean ± SD. Significant differences between the groups were analyzed using a χ2 test and the one-way analysis of variance, followed by the Student-Newman-Keuls test. P<0.05 was considered to indicate a statistically significant difference.
Results
High CRFR1 expression is associated with malignant phenotypes of human glioma
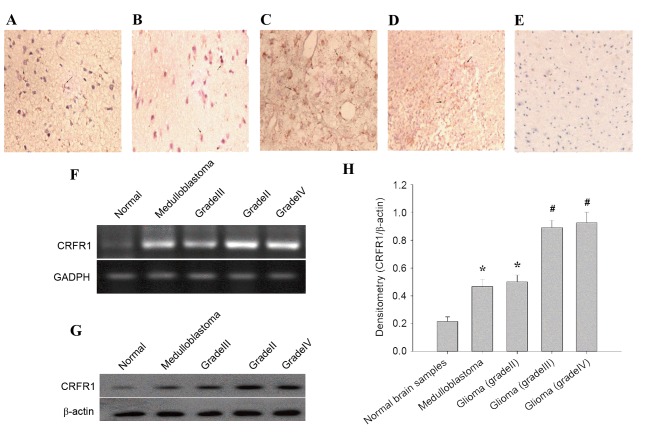
In the present study, the expression pattern of CRFR1 was evaluated in 35 glioma tissues and 13 normal brain tissues using IHC staining. CRFR1 expression was significantly increased in glioma tissues, but minimally detected in the normal brain specimens (Fig. 1A-E). In glioma cells positive for immunostaining, the labeling was primarily cytoplasmic as observed using light microscopy. The positive expression rate of CRFR1 was significantly different (χ2=17.0817, P<0.0001) between the human glioma (74.29%, 26/35) and the normal brain tissues (7.69%, 1/13; Table I). Furthermore, the association between CRFR1 expression and clinicopathological characteristics were analyzed in the current study. There was no significant association observed between CRFR1 expression and age, gender and position (all P>0.05). Notably, a significant difference (χ2=6.4852, P=0.0396) was identified between the low-grade (grade II, 2/8, 25%) and the high-grade (medulloblastoma, 4/5, 80%; grade III and IV, 16/22, 72.72%) glioma (Table II). Semi-quantitative RT-PCR results demonstrated that CRFR1 mRNA levels were significantly higher in human glioma compared with the normal cerebral tissues (Fig. 1F). In addition, the protein levels of CRFR1 was determined using western blot analysis, the results demonstrated that CRFR1 protein levels were significantly increased in human glioma compared with the normal cerebral tissues (Fig. 1G and H). These results suggested that increased CRFR1 expression is associated with high-grade glioma.
Figure 1.
CRFR1 mRNA and protein expression in glioma and normal brain tissues. (A-D) Immunohistochemical staining for CRFR1 in glioma specimens and (E) normal cerebral tissues. The CRFR1-positive cells contain homogeneous brown-yellow areas in the cytoplasm. (A) Medulloblastoma; (B) grade II; (C) grade III; (D) grade IV; and (E) normal cerebral tissue (magnification, ×200). (F) The CRFR1 mRNA levels in human glioma samples and normal cerebral tissues were detected using semi-quantitative reverse transcription-polymerase chain reaction. (G) CRFR1 protein levels were detected using western blot analysis. (H) Densitometry analysis of CRFR1 bands relative to β-actin. *P<0.01, #P<0.001, glioma tissue vs. normal brain tissue. CRFR1, corticotropin releasing factor receptor 1.
Table I.
Expression of CRFR1 in human gliomas and normal brain samples using immunohistochemical staining.
| Expression status of CRFR1 | ||||||
|---|---|---|---|---|---|---|
| Group | Cases, n | Negative | Positive | CRFR1-positive, % | χ2-value | P-value |
| Glioma tissues | 35 | 9 | 26 | 74.29 | 17.0817 | <0.0001 |
| Normal brain tissues | 13 | 12 | 1 | 7.69 | ||
CRFR1, corticotropin releasing factor receptor 1.
Table II.
Association between CRFR1 expression and clinicopathological parameters assessed by immunohistochemical staining.
| CRFR1 expression, n | ||||||
|---|---|---|---|---|---|---|
| Clinicopathological characteristics | Cases, n | Negative | Positive | CRFR1-positive tumors, % | χ2-value | P-value |
| Age, years | 0.0204 | 0.8864 | ||||
| <45 | 24 | 6 | 18 | 75.00 | ||
| ≤45 | 11 | 3 | 8 | 72.73 | ||
| Gender | 0.0826 | 0.7738 | ||||
| Male | 18 | 5 | 13 | 72.22 | ||
| Female | 17 | 4 | 13 | 76.47 | ||
| Position | 0.0374 | 0.8467 | ||||
| Supratentorial | 28 | 7 | 21 | 75.00 | ||
| Subtentorial | 7 | 2 | 5 | 71.43 | ||
| Glioma grade | 6.4852 | 0.396 | ||||
| Medulloblastoma | 5 | 1 | 4 | 80.00 | ||
| II | 8 | 6 | 2 | 25.00 | ||
| III+IV | 22 | 6 | 16 | 72.72 | ||
CRFR1, corticotropin releasing factor receptor 1.
CRFR1 is expressed in human U87 glioma cells
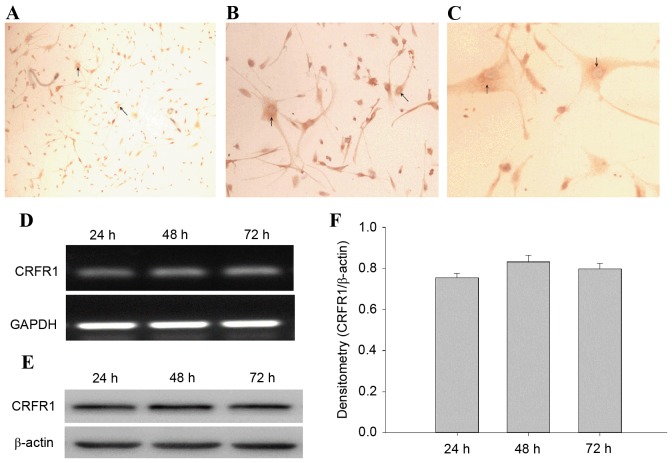
The CRFR1-positive cells were observed and the immunostaining was primarily cytoplasmic (Fig. 2A-C). CRFR1 mRNA and protein levels were determined using semi-quantitative RT-PCR and western blot analysis at 24, 48 and 72 h. The results revealed that the CRFR1 mRNA and protein was expressed in U87 cells at each time point (Fig. 2D and E). However, there was no significant difference between the levels of CRFR1 expression between the 3 time points (Fig. 2F).
Figure 2.
CRFR1 mRNA and protein expression in human U87 glioma cells lines. (A-C) Immunohistochemical analysis of the expression of CRFR1 in U87 cells. The CRFR1-positive cells contain homogeneous brown-yellow areas in the cytoplasm. (A) U87 cells (magnification, ×100); (B) U87 cells (magnification, ×200); (C) U87 cells (magnification, ×400). The arrows indicate expression of CRFR1-positive cells. (D) CRFR1 gene expression was determined in U87 cells at 24, 48 and 72 h using semi-quantitative reverse transcription-polymerase chain reaction. GAPDH was used as an internal control. (E) Protein levels were determined in U87 cells at 24, 48 and 72 h using western blot analysis. (F) Densitometry analysis of CRFR1 band relative to β-actin. CRFR1, corticotropin releasing factor receptor 1.
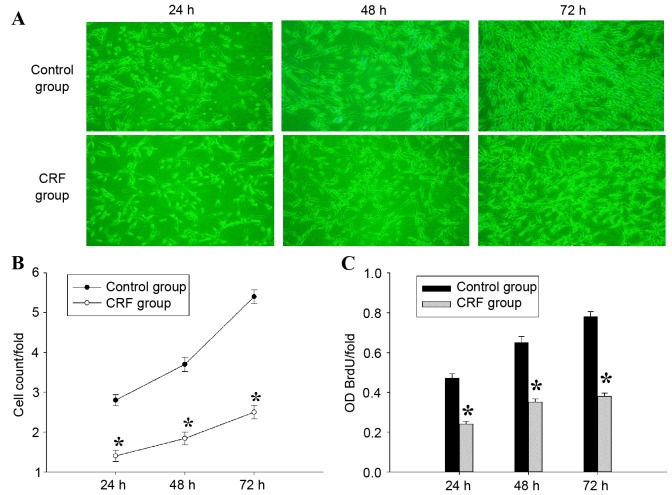
hCRF inhibits the proliferation of U87 glioma cells
In order to determine the effect of hCRF on cell growth, U87 cells were treated with or without hCRF (10−7 mol/l) and cultured for 24, 48 and 72 h. The cell number was measured using Cellomics ArrayScan VTI every day for 3 days, and a significant inhibition of cell proliferation was observed in U87 cells from 24 to 72 h following treatment with hCRF (Fig. 3A and B). To confirm the suppressive effect of hCRF on the proliferation of U87 cells further, a BrdU incorporation assay was performed to assess cell proliferation status. In this assay, cell proliferation was evaluated by measuring a DNA synthesis marker, which was quantified by the BrdU incorporation ratio. Consistent with the results of cell counting, the quantity of newly synthesized DNA decreased significantly at 24, 48 and 72 h following treatment with hCRF, compared with the control group (Fig. 3C). These results indicated that cell proliferation and DNA synthesis were suppressed by hCRF in human U87 glioma cells.
Figure 3.
hCRF treatment inhibits proliferation of U87 glioma cells. (A) Representative images of U87 cell growth in control or hCRF-treated U87 cells at 24, 48 and 72 h. (B) Proliferation of U87 cells was analyzed using Cellomics ArrayScan VTI. Proliferation data are presented as fold-changes of cell numbers relative to the number of control cells at the previous time point. The results are expressed as the mean ± SD of 3 separate experiments. (C) Proliferation of U87 cells treated with hCRF, analyzed using a BrdU incorporation assay. The BrdU incorporation ratio is represented as fold-change in absorbance at 450 nm. The results represent the mean ± SD of 3 separate experiments. *P<0.01 vs. control group. hCRF, human corticotropin releasing factor; SD, standard deviation; BrdU, bromodeoxyuridine; OD, optical density.
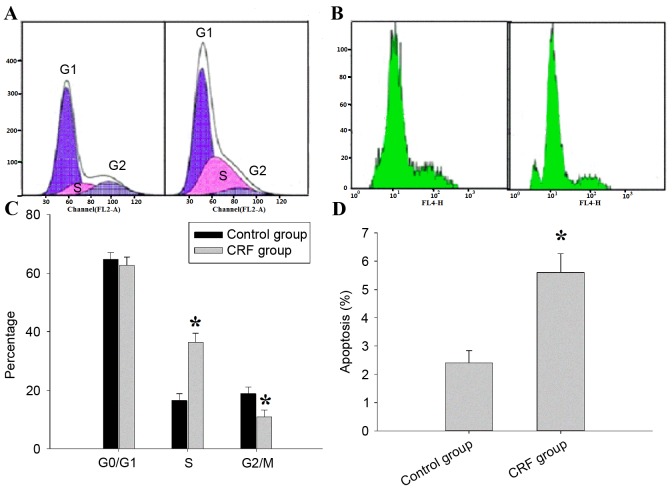
hCRF induces cell cycle arrest and promotes apoptosis in human U87 glioma cells
To investigate the underlying mechanisms of the inhibitory effect of hCRF on U87 cell growth, the role of hCRF in cell cycle progression of the U87 was assessed using PI-FACS (Fig. 4A). In the control group, 65.32±1.15, 16.53±0.78 and 18.81±0.45% cells were found in the G0/G1, S and G2/M phases, respectively; in the hCRF group, 56.18±0.87, 34.62±0.49 and 10.87±0.68% cells were in the G0/G1, S and G2/M phases, respectively. U87 cells that underwent hCRF treatment exhibited an increased percentage of cells in the S phase compared with control (*P<0.01) and concomitantly with a significant decrease in G2/M phase cells (*P<0.01). The level of cell apoptosis was assessed using annexin V staining and flow cytometry (Fig. 4B). Cell apoptosis was significantly increased in hCRF group when compared with the control group (5.41±0.28 vs. 2.64±0.15, *P <0.01). The proportion of cells in S phase increased from 16.53 to 34.62% in the hCRF-treated group, whereas the proportion in the G2/M phase decreased from 18.81 to 10.87% (Fig. 4C). Therefore, the present results indicated that hCRF treatment led to the inhibition of the S-M transition. Apoptosis was observed in 5.68% of U87 cells treated with hCRF, while only being observed in 2.45% of U87 cells in the control group (Fig. 4D). These data suggest that hCRF may promote cell apoptosis in U87 cells.
Figure 4.
hCRF results in cell cycle arrest and induces cell apoptosis in U87 cells line. (A) Effect of hCRF on the U87 cell cycle distribution. Histograms obtained from flow cytometry analysis cells of hCRF and control group (representative of 3 independent experiments). (B) Effect of hCRF on U87 cells apoptosis. Histograms obtained from propidium iodide-annexin V analysis of hCRF and control cells (representative of 3 independent experiments). (C) Percentage of U87 cells at different cell cycle phases following treatment with control or hCRF. (D) Percentage of apoptotic cells following treatment with control or hCRF. *P<0.01 vs. control group. hCRF, human corticotropin releasing factor.
hCRF upregulates the expression of p53 in human U87 glioma cells
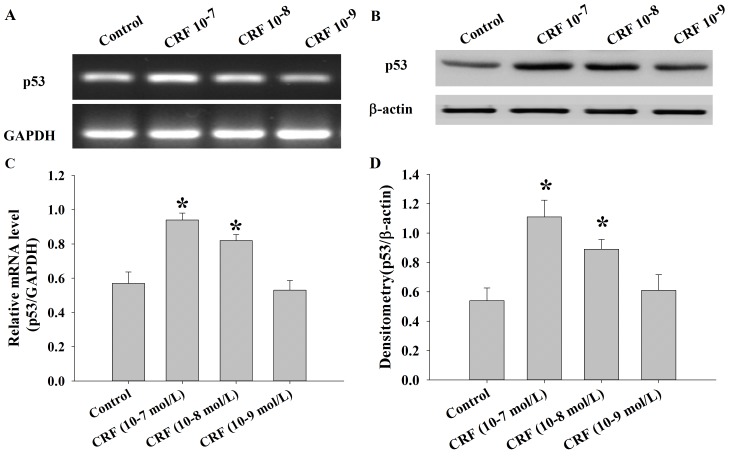
The levels of p53 mRNA and protein were determined in U87 cells following treatment with control or hCRF (at 10−7, 10−8 and 10−9 mol/l) group using semi-quantitative RT-PCR and western blot analysis. The results demonstrated that the expression of p53 mRNA was significantly upregulated in the hCRF treatment group compared with the control group (*P<0.01; Fig. 5A and B). Consistent with these results, the levels of p53 protein were also significantly increased in the groups treated with higher hCRF concentrations (10−7 and 10−8 mol/l) compared with the control or low hCRF concentration (10−9 mol/l) group (*P<0.01; Fig. 5C and D).
Figure 5.
p53 mRNA and protein expression in U87 cells following treatment with hCRF (10−7, 10−8 and 10−9 mol/l) at 24 h. (A) p53 mRNA expression was determined in U87 cells following treatment with hCRF or control using semi-quantitative reverse transcription-polymerase chain reaction. GAPDH was used as an internal control. (B) Protein levels of p53 were determined in U87 cells following treatment with hCRF or control using western blot analysis. (C) The p53 mRNA levels were analyzed by RT-PCR. *P<0.01 vs. control group. (D) Densitometry analysis of CRFR1 bands, relative to β-actin. *P<0.01 vs. control. p53, tumor protein 53; hCRF, human corticotropin release factor; CRFR1, corticotropin release factor receptor 1.
Discussion
Glioma are the most prevalent, aggressive type of tumor observed in the central nervous system and the prognosis remains poor due to their biological characteristics (24). CRF is a major hypothalamic stress-induced neuropeptide that is also expressed in peripheral tissues. CRF and its receptors have been detected in a number of tumor types. In a previous study using an animal tumor model, CRF has been established to inhibit tumor proliferation (25). This action is mediated by the specific CRF receptors, CRFR1 and CRER2 (26). The aim of the present study was to determine the expression of CRFR1 in human glioma tissue and the human U87 glioma cell line, and to evaluate the potential effect of CRF on human U87 glioma cell proliferation, apoptosis and the cell cycle.
Previous studies have demonstrated that CRFR1 is expressed in tumor cells, and CRF and urocortin may reduce tumor cell growth via their interaction with CRFR1 (13,25). A number of malignancies have been established to express high levels of CRFR1 and CRFR2 and to be sensitive to the suppressive effects of CRF and its agonists; however, there is little currently understood about the role of CRF in glioma. To the best of our knowledge, the present study is the first to investigate the specific expression of CRFR1 in human glioma cells. The increased expression of CRFR1 is associated with an increased glioma grade, which provides additional support for the involvement of CRF in the development of glioma. However, CRFR1 expression was minimally detected in normal brain tissue. Following analysis of U87 glioma cells, the expression of the CRFR1 gene and protein were significantly increased in U87 glioma cells at all time points following CRF treatment. These results suggested that the role of CRF in glioma is distinct and essential.
In order to further characterize the effect of CRF in glioma, U87 cells were treated with hCRF [10−7 mol/l; the dosage was determined in accordance with a previous study (25)]. Compared to the control group, U87 cells that underwent hCRF treatment exhibited reduced cell proliferation, were arrested in S-phase and demonstrated an increased level of apoptosis. These results suggest that CRF may be involved in the regulation of cell cycle checkpoints in U87 cells and may demonstrate that CRF promotes a marked reduction in replication initiation in glioma cells. Therefore, CRF has an important role in promoting U87 cell growth and is associated with U87 cell cycle phase distribution. Using primary neuronal cultures from the cerebellum, cerebral cortex and hippocampus, low CRF concentrations (10−8 mol/l) have been reported to exert a brain region-specific neuroprotective effect on amyloid-ß 25–35 toxicity in the cerebellum and hippocampus (27). However, a higher CRF concentration (10−7 mol/l) led to the additional protection of cortical neurons (25). CRF was reported to act on CRFR1 to inhibit the proliferative effects of estrogens on MCF7 cells in a paracrine and autocrine manner (16) and urocortin inhibited the proliferation of melanoma cells in vitro and in vivo, also through CRFR1 (28). Moroz et al (29) reported that hCRF was more effective compared with dexamethasone or temozolomide in the treatment of U87 xenografts, with certain patients reporting an increase in long-term survival and only mild toxicity. Therefore, the results of the present study are in accordance with these previous studies, as hCRF inhibits the proliferation and promotes the apoptosis of U87 cells in vitro via CRFR1.
p53 is one of the most extensively studied molecules in cancer research and molecular biology. p53 has numerous anticancer functions and serves vital roles in DNA repair, apoptosis and inhibition of angiogenesis (30). Previous studies have reported that the wild type p53 gene is present in ~70% of primary glioblastoma (31), however U87 possesses wild type p53 (http://p53.free.fr/Database/Cancer_cell_lines/p53_cell_lines.html). Xu et al (32) reported that histone acetyltransferase inhibitor II inhibits proliferation and induces apoptosis via the caspase-dependent pathway in human U87 and U251 glioma cell lines, potentially by activating the p53 signaling pathway (32). Another study identified that the CRFR1-triggered ERK1/2 pathway is involved in the p53 activation and the suppression of the apoptotic Bax gene in rat liver cells following hypoxia (33). The present study also hypothesizes that the p53 signaling pathway may be influenced by CRFR1 in U87 cells. The results of the current study demonstrated that the expression of p53 mRNA and protein were significantly increased in U87 cells treated with hCRF (10−7 and 10−8 mol/l). It is therefore possible that hCRF may inhibit proliferation and induce cell cycle arrest and apoptosis in U87 glioma cells in a CRFR1-mediated p53-dependent mechanism. Notably, in a phase 1 clinical trial, patients who received hCRF reported improved neurological symptoms and physical results, with little or no toxicity (17).
In conclusion, the results of the present study suggest that hCRF may inhibit cell proliferation and apoptosis in U87 cells via a CRFR1-mediated p53 signaling pathway. Data from previous studies and the present study demonstrate that hCRF is innocuous in humans and that CRFR1 may be a key regulator of human glioma, suggesting it is a novel and attractive therapeutic target for anticancer therapy.
Acknowledgments
Not applicable.
Funding
The present study was supported by a grant from the Outstanding medical talents plan of 2015 Hebei Province (grant no. 361004).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YF performed the experiments. XL and QW assisted the first author in tumor specimen collection, IHC staining, cell culture and related experiments (cell proliferation, flow cytometry, RNA extraction, RT-qPCR, western blotting). FH and HZ analyzed the results and data. LW was responsible for designing the study, summarizing the results and writing the article. XS performed the analysis and offered assistance to the corresponding author in the manuscript writing and its English-language editing.
Ethics approval and consent to participate
Data collection and analysis were approved by the ethics committee of The Second Affiliated Hospital of Hebei Medical University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Stupp R, Tonn JC, Brada M, Pentheroudakis G, ESMO Guidelines Working Group: High-grade malignant glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up Ann Oncol. 2010;21(Suppl):S190–S193. doi: 10.1093/annonc/mdq187. [DOI] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 3.Chen R, Lewis KA, Perrin MH, Vale WW. Expression cloning of a human corticotropin-releasing-factor receptor. Proc Natl Acad Sci USA. 1993;90:8967–8971. doi: 10.1073/pnas.90.19.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perrin M, Donaldson C, Chen R, Blount A, Berggren T, Bilezikjian L, Sawchenko P, Vale W. Identification of a second corticotropin-releasing factor receptor gene and characterization of a cDNA expressed in heart. Proc Natl Acad Sci USA. 1995;92:2969–2973. doi: 10.1073/pnas.92.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dautzenberg FM, Hauger RL. The CRF peptide family and their receptors: Yet more partners discovered. Trends Pharmacol Sci. 2002;23:71–77. doi: 10.1016/S0165-6147(02)01946-6. [DOI] [PubMed] [Google Scholar]
- 6.Audhya T, Jain R, Hollander CS. Receptor-mediated immuno-modulation by corticotropin-releasing factor. Cell Immunol. 1991;134:77–84. doi: 10.1016/0008-8749(91)90332-6. [DOI] [PubMed] [Google Scholar]
- 7.Clifton VL, Owens PC, Ribinson PJ, Smith R. Identification and characterization of a corticotropin-releasing hormone receptor in human placenta. Eur J Endocrinol. 1995;133:591–597. doi: 10.1530/eje.0.1330591. [DOI] [PubMed] [Google Scholar]
- 8.Roe SY, McGowan EM, Rothwell NJ. Evidence for the involvement of corticotropin-releasing hormone in the pathogenesis of traumatic brain injury. Eur J Neurosci. 1998;10:553–559. doi: 10.1046/j.1460-9568.1998.00064.x. [DOI] [PubMed] [Google Scholar]
- 9.Strijbos PJ, Relton JK, Rothwell NJ. Corticotropinreleasing factor antagonist inhibits neuronal damage induced by focal cerebral ischaemia or activation of NMDA receptors in the rat brain. Brain Res. 1994;656:405–408. doi: 10.1016/0006-8993(94)91485-0. [DOI] [PubMed] [Google Scholar]
- 10.Lezoualc'h F, Engert S, Berning B, Behl C. Corticotropinreleasing hormone-mediated neuroprotection against oxidative stress is associated with the increased release of non-amyloidogenic amyloid b precursor protein and with the suppression of nuclear factor-jB. Mol Endocrinol. 2000;14:147–159. doi: 10.1210/mend.14.1.0403. [DOI] [PubMed] [Google Scholar]
- 11.Baram TZ, Hatalski CG. Neuropeptide-mediated excitability: A key triggering mechanism for seizure generation in the developing brain. Trends Neurosci. 1998;21:471–476. doi: 10.1016/S0166-2236(98)01275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutknecht E, Hauger RL, Van der Linden I, Vauquelin G, Dautzenberg FM. Expression, binding, and signaling properties of CRF2 (a) receptors endogenously expressed in human retinoblastoma Y79 cells: Passage-dependent regulation of functional receptors. J Neurochem. 2008;104:926–936. doi: 10.1111/j.1471-4159.2007.05052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoeffter P, Feuerbach D, Bobirnac I, Gazi L, Longato R. Functional, endogenously expressed corticotropin-releasing factor receptor type 1 (CRF1) and CRF1 receptor mRNA expression in human neuroblastoma SH-SY5Y cells. Fundam Clin Pharmacol. 1999;13:484–489. doi: 10.1111/j.1472-8206.1999.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 14.Sato H, Nagashima Y, Chrousos GP, Ichihashi M, Funasak Y. The expression of corticotropin-releasing hormone in melanoma. Pigment Cell Res. 2002;15:98–103. doi: 10.1034/j.1600-0749.2002.1o063.x. [DOI] [PubMed] [Google Scholar]
- 15.Funasaka Y, Sato H, Chakraborty AK, Ohashi A, Chrousos GP, Ichihashi M. Expression of proopiomelanocortin, corticotropinreleasing hormone (CRH), and CRH receptor in melanoma cells, nevus cells, and normal human melanocytes. J Investig Dermatol Symp Proc. 1999;4:105–109. doi: 10.1038/sj.jidsp.5640192. [DOI] [PubMed] [Google Scholar]
- 16.Graziani G, Tentori L, Muzi A, Vergati M, Tringali G, Pozzoli G, Navarra P. Evidence that corticotropin-releasing hormone inhibits cell growth of human breast cancer cells via the activation of CRH-R1 receptor subtype. Mol Cell Endocrinol. 2007;264:44–49. doi: 10.1016/j.mce.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Moliterno JA, Henry E, Pannullo SC. Corticorelin acetate injections for the treatment of peritumoral brain edema. Expert Opin Investig Drugs. 2009;18:1413–1419. doi: 10.1517/13543780903190689. [DOI] [PubMed] [Google Scholar]
- 18.Muleci A, Cervoni L, Delfini R. Medulloblastomas in children and in adults:a comparative study. Acta Neurochir (Wien) 1992;119:62–67. doi: 10.1007/BF01541783. [DOI] [PubMed] [Google Scholar]
- 19.Roberts RO, Lynch CF, Jones MP, Hart MN. Medulloblastoma: A population-based study of 532 cases. J Neuropathol Exp Neurol. 1991;50:134–144. doi: 10.1097/00005072-199103000-00005. [DOI] [PubMed] [Google Scholar]
- 20.Sarkar C, Pramanik P, Karak AK, Mukhopadhyay P, Sharma MC, Singh VP, Mehta VS. Are childhood and adult medulloblastomas different? A comparative study of clinicopathological features, proliferation index and apoptotic index. J Neurooncol. 2002;59:49–61. doi: 10.1023/A:1016357731363. [DOI] [PubMed] [Google Scholar]
- 21.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao J, Wang Z, Wang Y, Liang Z, Zhang X, Zhao Z, Jiao B. Eukaryotic initiation factor 3C silencing inhibits cell proliferation and promotes apoptosis in human glioma. Oncol Rep. 2015;33:2954–2962. doi: 10.3892/or.2015.3881. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 25.Graziani G, Tentori L, Portarena I, Barbarino M, Tringali G, Pozzoli G, Navarra P. CRH inhibits cell growth of human endometrial adenocarcinoma cells via CRH-receptor 1-mediated activation of cAMP-PKA pathway. Endocrinology. 2002;143:807–813. doi: 10.1210/endo.143.3.8694. [DOI] [PubMed] [Google Scholar]
- 26.Grigoriadis DE, Lovenberg TW, Chalmers DT, Liaw C, De Souze EB. Characterization of corticotropin-releasing factor receptor subtypes. Ann N Y Acad Sci. 1996;780:60–80. doi: 10.1111/j.1749-6632.1996.tb15112.x. [DOI] [PubMed] [Google Scholar]
- 27.Bayatti N, Zschocke J, Behl C. Brain region-specific neuroprotective action and signaling of corticotropin-releasing hormone in primary neurons. Endocrinology. 2003;144:4051–4060. doi: 10.1210/en.2003-0168. [DOI] [PubMed] [Google Scholar]
- 28.Carlson KW, Nawy SS, Wei ET, Sadee W, Filov VA, Rezsova VV, Slominski A, Quillan JM. Inhibition of mouse melanoma cell proliferation by corticotropin-releasing hormone and its analogs. Anticancer Res. 2001;21:1173–1179. [PubMed] [Google Scholar]
- 29.Moroz MA, Huang R, Kochetkow T, Shi W, Thaler H, de Stanchina E, Gamez I, Ryan RP, Blasberg RG. Comparison of corticotropin- releasing factor, dexamethasone, and temozolomide: Treatment efficacy and toxicity in U87 and C6 intracranial gliomas. Clin Cancer Res. 2011;17:3282–3292. doi: 10.1158/1078-0432.CCR-10-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee YJ, Chung DY, Lee SJ, Jhon Ja G, Lee YS. Enhanced radiosensitization of p53 mutant cells by oleamide. Int J Radiat Oncol Biol Phys. 2006;64:1466–1474. doi: 10.1016/j.ijrobp.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 31.Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schüler D, Probst-Hensch NM, Maiorka PC, et al. Genetic pathways to glioblastoma: A population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 32.Xu LX, Li ZH, Tao YF, Li RH, Fang F, Zhao H, Li G, Li YH, Wang J, Feng X, Pan J. Histone acetyltransferase inhibitor II induces apoptosis in glioma cell lines via the p53 signaling pathway. J Exp Clin Cancer Research. 2014;33:108. doi: 10.1186/s13046-014-0108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Wang MY, Hao K, Chen XQ, Du JZ. CRHR1 mediates p53 transcription induced by high altitude hypoxia through ERK 1/2 signaling in rat hepatic cells. Peptides. 2013;44:8–14. doi: 10.1016/j.peptides.2013.03.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.