Abstract
The cancer stem cell (CSC) theory implicates a small subpopulation of cells with stem-like properties, which is responsible for tumour initiation, development and metastasis. The unique biological and functional characteristics of CSCs, widely associated with treatment resistance, indicate an association between metastasis and stemness. It was hypothesised that metastatic cell lines may be enriched in CSCs and that this would correlate with a more resistant tumour. In the present study, the SW480 and SW620 paired cell lines derived from a colon adenocarcinoma and its lymph node metastasis, respectively were compared as an in vitro model of cancer progression. Their chemosensitivity and CSC properties were investigated. A range of in vitro assays were performed, including the side population assay, ALDEFLUOR assay, tumoursphere assay and assessment of CSC-associated surface phenotypes. It was determined that the SW480 and SW620 cells exhibited similar growth rates, although the SW480 cells were more migratory in wound healing assays on collagen and fibronectin matrices. SW480 and SW620 cells displayed similar CSC profiles, however, SW480 cells demosntrated significantly greater tumoursphere forming efficiency over SW620 cells. Tumourspheres derived from SW480 and SW620 cells also displayed differential sensitivity to 5-fluorouracil, oxaliplatin, geldanamycin and novobiocin that was not apparent when cells were grown under adherent conditions. Taken together, these results suggest that although the two cell lines have similar levels of putative CSC populations, there are differences in their biology that cannot be explained by these CSC levels. To the best of our knowledge, this is the first study to conduct a detailed analysis of the CSC populations using multiple in vitro assays in a paired cell line model. These results have clinical relevance for the understanding of the differences between primary tumours and their metastatic counterparts.
Keywords: cancer stem cell, colon cancer metastasis, tumoursphere, anti-cancer inhibitors, Hsp90 inhibitors
Introduction
The cancer stem cell (CSC) theory challenges earlier models concerning the development of cancers by implicating a small proportion of cells with stem-like properties in the development, propagation and drug resistance of tumours (1,2). CSCs display unlimited differentiation potential and an ability to self-renew, both of which are features required for tumour initiation and development (2). Recent studies have suggested that the differentiation and self-renewal capabilities unique to CSCs may also regulate the progression and propagation of a tumour, proposing a role for CSCs in the metastatic spread of cancers (2–5). There is cumulative evidence to support the CSC theory in the propagation of leukaemic (blood) and solid (tissue) tumours from CSC of primary cancers and cancer cell lines of the brain, breast, colon, lung, prostate, as well as melanoma and glioblastoma (6–12).
Several functional and phenotypic in vitro assays have been used to identify putative CSC populations. These include the side population (SP) and ALDEFLUOR assays, the detection of specific cell surface markers, and the assessment of the ability of cells to grow as tumourspheres (TS) in suspension (13–15). The SP assay identifies putative CSCs based on the high activity of the ATP-binding cassette transporter protein (ABC)G2, which is also implicated in drug resistance due to its role in the efflux of chemotherapeutics from the cell (16,17). On the other hand, the ALDEFLUOR assay identifies CSCs using another unique CSC marker, aldehyde dehydrogenase (ALDH). The detoxifying effect of ALDH is thought to protect stem cells against oxidative damage and may modulate the proliferative capacity of stem cells (18). As a functional assay, the generation of three-dimensional spheres using serum-free culture methods takes advantage of the stem-like nature of CSC by allowing survival from anoikis and this method has been utilised for the identification and expansion of CSC populations in vitro (19) In colon cancer, putative CSCs have been identified in vitro using a range of the aforementioned techniques, in particular according to the expression of the cell surface protein markers, CD44 and CD133, and to the expression of ALDH and ABCG2 (4,14,19–21).
The most appropriate and accurate method of identifying of CSCs remains a subject of intense debate, Furthermore, many researchers remain sceptical as to the role of this subpopulation in cancer initiation and progression. In particular, whether or not the presence of CSCs determines the metastatic potential of the tumour has yet to be fully elucidated (3).
In this study, we used a paired colon cancer cell line model derived from a single patient, representative of the primary tumour (SW480) and its lymph node metastisis (SW620) (22). As the SW480 and SW620 cell lines developed from the same genetic background, they provide an in vitro model to study the cellular changes that occur during cancer progression and development of a metastatic phenotype. Our analysis focussed on the comparative in vitro analysis of putative CSC populations in these paired lines. We hypothesised that if CSC are responsible for metastasis, then the SW620 cell line may be enriched in CSC compared to the SW480 cell line and therefore should be the more chemoresistant of the two cell lines.
Materials and methods
Adherent cell lines and culture conditions
Paired human colon adenocarcinoma cell lines SW480 (primary/pre-metastatic tumour; cat. no. 87092801) and SW620 (lymph node metastasis; cat. no. 87051203) were purchased from the European Collection of Animal Cell Cultures (Salisbury, UK). SW480 and SW620 cells were maintained in Leibovitz's L-15 medium with GlutaMAX™-I (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 10% (v/v) fetal bovine serum (Biowest, Nuaillé, France) and 100 U/ml PSA (Gibco; Invitrogen) at 37°C. Cell lines used in experiments exhibited passage numbers below 50.
Light and phase contrast microscopy
Cells and TS were analysed by light microscopy using an Olympus CKX41 microscope (Olympus Corporation, Tokyo, Japan), or by phase contrast microscopy using a Zeiss DSZ5000X inverted microscope (Zeiss AG, Oberkochen, Germany). All images were analysed using ImageJ software (National Institute of Health, Bethesda, MD, USA).
Cell adhesion and growth profile
Real-time cell analysis was performed using the xCELLigence™ System (Roche Applied Science, Rotkreuz, Swtizerland). SW480 and SW620 cells were seeded at a density of 3×103 cells per well in 100 µl growth medium in a 96-well E-plate (Roche Applied Science). The E-plate was analysed over 96 h at 37°C using RTCA 1.2.1 software (ACEA Biosciences, San Diego, CA, USA), collecting data from one sweep/h. An increase in cells on the gold electrodes of the well surface was detected by changes in the electrical impedance, producing the cell index (CI) output in relation to the impedance at the beginning of the experiment. An unchanged CI output therefore suggests no change in the number (or size) of cells, while increases in CI output over time are indicative of cell growth/proliferation.
Wound healing assay
SW480 and SW620 cells were seeded into untreated 96-well plates (at a density of 1×106 cells/ml) or plates pre-treated with 1 µg/ml type I collagen or 5 µg/ml fibronectin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and allowed to adhere overnight at 37°C. Wounds to each well were created by the removal of a section of the confluent cell monolayer using a sterile toothpick and changes in the wound size analysed at intervals over 24 h. Images were taken at 0, 4, 8, 12 and 24 h after creating the wound using a Zeiss DSZ5000X inverted microscope. The change in wound area (%) relative to the start of the assay (0 h) was calculated for each time point using ImageJ software (National Institute of Health).
Hoechst efflux assay
The experimental procedure used by Goodell and colleagues was used for the development of the optimised Hoechst 33342 dye exclusion assay (23). The SP is characterised as being negative for both Hoechst blue (Hblue−) and Hoechst red (Hred) staining. This Hblue−/Hred−SP was identified by comparing the signals (% of cells with that phenotype) obtained with or without treatment with verapamil, an inhibitor of the ABCG2 transporter protein that prevents the efflux of Hoechst 33342. Cells were treated in 75 cm3 flasks with 5 µg/ml Hoechst 33342 alone or a combination of 5 µg/ml Hoechst 33342 (Invitrogen) and 50 µM verapamil (Sigma-Aldrich; Merck KGaA) for 1 h at 37°C. Cells were harvested by centrifugation (300 × g for 8 min) and resuspended in PBS (1×106 cells/ml) containing 2 µg/ml PI (Sigma-Aldrich; Merck KGaA) for live/dead cell discrimination. The SP of putative CSC was detected using the FACSAria II (BD Biosciences, Erembodegem, Belgium). The Hoechst 33342 dye was excited at 350 nm and the dual emission of Hoechst blue and Hoechst red analysed at 405/30 and 670/40 nm, respectively. Data were analysed with FlowJo software (version 7.6.5; Tree Star, Inc., Ashland, OR, USA).
ALDEFLUOR® assay
Cell populations with high ALDH enzyme activity were identified by flow cytometry using the ALDEFLUOR® kit (#01700; Stemcell Technologies, Inc., Vancouver, BC, Canada), in which the fluorescent substrate of ALDH, ALDEFLUOR, is cleaved intracellularly by active ALDH to produce a fluorescent product. The assay was carried out as per the manufacturer's instructions. Briefly, cells were resuspended at a density of 1×106 cells/ml in ALDEFLUOR® assay buffer. Activated ALDEFLUOR reagent (150 µM) was added to each sample and 5×105 cells from each sample were treated with 150 µM ALDH inhibitor, diethylaminobenzaldehyde (DEAB), as the inhibited control used to identify the ALDHhigh population. Comparing ALDEFLUOR-stained cells with stained cells treated with DEAB, the percentage of ALDHhigh cells was calculated based on the loss of ALDH activity [% cells ALDHhigh = (% FITChigh cells without DEAB) - (% FITChigh cells with DEAB)]. Staining was carried out at 37°C for 45 min and samples were analysed using the FACSAria II flow cytometer (BD Biosciences), with excitation at 488 nm and emission detected in the 530/30 nm filter channel. Data were analysed using FlowJo software (version 7.6.5; Tree Star, Inc.).
Flow cytometric analysis of CSC markers
Cells were lifted using 1% (w/v) EDTA, centrifuged (800 x g for 2 min) and resuspended in PBS (5×105 cells/ml) containing 0.25 µg of allophycocyanin (APC), phycoerythrin (PE) or FITC directly conjugated antibodies (anti-CD44 APC, anti-CD24 FITC anti-ABCG2 PE and anti-integrin α6 APC (eBioscience, Inc., San Diego, CA, USA), and anti-CD133 PE (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany), or used as unstained controls. Isotype controls (mouse IgG2b PE, Rat IgG2b APC, mouse IgG1 FITC, mouse IgG1 PE; eBioscience, Inc.) were used to account for any non-specific antibody binding to live cells by establishing a gating threshold (limit 0.5%) according to fluorescence intensity and calculated compensation. All antibodies were used at a 1:400 dilution (0.25 µg) and samples were incubated at 4°C for 1 h before harvesting of cells for analysis). The FACSAria II flow cytometer was used to record 50,000 events prior to analysis using FlowJo software. The APC fluorophore was excited at 633 nm and emission recorded in the 620/20 filter channel. Both the FITC and the PE fluorophores were excited at 488 nm and emission recorded in the 530/30 or 585/42 filter channels, respectively. Compensation controls were used to establish values for fluorescent overlap.
Culture of TS
The ability of colon cancer cells to form TS under anchorage-independent conditions was analysed for 14 days using a protocol adapted from that of Dontu et al as well as Kreso and O'Brien (24,25). Cells were harvested, filtered and resuspended as a single-cell suspension in conditioned medium to a density of 2×104 cells/ml. TS were maintained in medium (DMEM) with 1× B-27 supplement (Gibco; Invitrogen), 20 ng/ml basic fibroblast growth factor (bFGF; Thermo Fisher Scientific, Inc.), 20 ng/ml epidermal growth factor (EGF; Sigma-Aldrich; Merck KGaA), 5 µg/ml insulin (Novorapid; Novo Nordisk A/S, Bagsværd, Denmark), 10 µg/ml heparin (Sigma-Aldrich; Merck KGaA) and 100 U/ml PSA (Gibco) in ultra-low attachment culture flasks and/or multiwell plates (Corning Incorporated, Pittsburgh, PA, USA). Sphere-forming efficiency (SFE) was quantified per number of cells seeded and expressed as a percentage.
Cell cycle analysis
Cells were seeded (2×104 cells/ml) in 25 cm3 flasks for TS culture over 5 days, and overnight for adherent conditions. Cells were lifted with Accutase™ (Invitrogen) pelleted at 1,600 x g and fixed with 70% (v/v) ethanol before harvesting and staining for 40 min at 37°C in PI solution [50 µg/ml PI, 100 µg/ml RNAse A, 0.05% (v/v) Triton X-100; Sigma-Aldrich; Merck KGaA]. Doublet discrimination was carried out to eliminate false positive results arising from aggregated cells.
Fluorescence microscopy
Cells seeded on coverslips coated with 5 µg/ml fibronectin were fixed in methanol and incubated with anti-heat shock protein 90 (Hsp90)α/β antibody (sc-13119; Santa Cruz Biotechnology, Inc., Dallas, TX, USA; used at 1:100 dilution) overnight at 4°C, followed by incubation with donkey anti-mouse Alexa Fluor-488 secary antibody (A21202; Thermo Fischer Scientific, Inc., used at 1:500 dilution) for 1 h. Nuclear staining was carried out using 1 µg/ml Hoechst 33342 (Invitrogen). Images were taken using a Zeiss AxioVert.A1 Fluorescence LED inverted microscope with a high resolution AxioCam MRm Rev 3, and analysed with ZEN Lite 2012 1.1.2.0 software (Zeiss AG).
SDS-PAGE and western blot analysis
Cell lysates were resuspended to equal total protein concentrations as determined by a NanoDrop 2000 spectrophotometer (Thermo Fischer Scientific, Inc.) Proteins resolved by SDS-PAGE gels were transferred onto Hybond Supported Nitrocellulose membranes (GE Healthcare Life Sciences, Chalfont, UK) membrane. Hsp90 was detected by chemiluminescence using the ECL Advanced Western Blotting kit (GE Healthcare Life Sciences), after incubation with a horseradish peroxidise-conjugated donkey anti-mouse secondary antibody, using the Chemidoc™ EQ system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Images were analysed using ImageJ software. Anti-Histone H3 (#9715; used at 1:2,500 dilution) and anti-Hsp90α/β (sc-13119; used at 1:1,000 dilution) antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA) and Santa Cruz Biotechnology, Inc., respectively.
Viability studies with anti-cancer drugs
The viability of 3×103 cells/well (adherent) and 1×103 cells/well (TS) in a 96-well plate were assessed following treatment with GA, NOV, 5-fluorouracil (5-FU) or oxaliplatin (Sigma-Aldrich; Merck KGaA) at increasing concentrations in comparison to untreated or DMSO (vehicle control). After the addition of 10 µl/well WST-1 proliferation reagent (Roche Applied Science) and incubation for 3 h, the absorbance at 440 nm was measured using a PowerWave spectrophotometer (BioTek Instruments, Inc., Winooski, VT, USA). The concentration at which a chemotherapeutic agent effectively altered cell viability by 50% (EC50) for each cell line was calculated in GraphPad Prism 4.03 software (GraphPad Software, Inc., La Jolla, CA, USA) using non-linear regression.
Statistical analysis
The statistical analyses of data were performed using GraphPad Prism 4.03 software (GraphPad Software, Inc.). Using two-way ANOVA, Mann Whitney and t-test (as indicated in the figure legends), data were assessed for statistical significance, where P<0.05 was deemed the minimum statistical significance upon comparison of samples.
Results
Comparison of biological characteristics of the paired SW480 and SW620 cancer cell lines
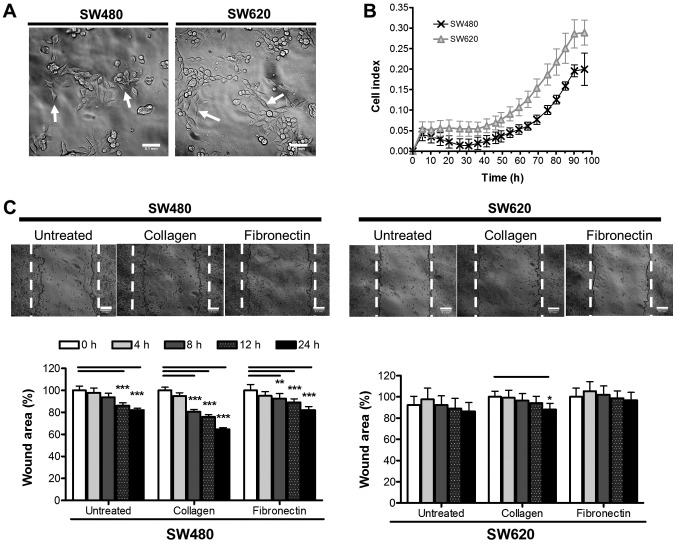
The morphology of SW480 and SW620 cells were analysed by phase contrast microscopy to reveal two cell morphologies (Fig. 1A). The SW480 cell line showed small groupings of irregular-shaped cells (80% of fully adhered cells were irregular in shape, with only 20% representing a more spindle-like morphology), in comparison to the small spindle-like shape of the SW620 cells (53% of fully adhered cells were of a traditional, elongated spindle-shape, while the remaining 47% were either more rounded spindles or irregular in shape). These morphologies are similar to those reported previously in literature (22). SW480 and SW620 cells were next compared in terms of cell growth. Using xCELLigence™, the adhesion and growth of SW480 and SW620 cells over 96 h was followed in real time (Fig. 1B) and assessed based on the reported CI. CI values for the SW620 cells were significantly higher overall than that of the SW480 cells. The higher CI values may reflect the different morphologies of the cells observed, since this value is based on the extent to which the growth surface is covered by the cell monolayer causing electrical impedance on the electrode, and will thus logically be affected by not only the number of cells but also their size and shape (Fig. 1A). Nonetheless, the two cell lines appeared to display similar growth rates as indicated by the similarity in the shapes of the respective curves (Fig. 1B). This data was supported by an assessment of SW480 and SW620 cells for metabolic activity as a measure of proliferation over 96 h by WST-1 assay, further indicating a similar growth trend between cell lines (data not shown).
Figure 1.
Growth and migration characteristics of SW480 and SW620 colon cancer cell lines in vitro. (A) Adherent morphology of cells 24 h after seeding. Images were taken using a DSZ5000X inverted microscope and analysed using ImageJ software (scale bar, 0.1 mm). Arrows illustrate distinct cell morphologies. (B) SW480 and SW620 cell growth assessed over 96 h [biological triplicates using an xCELLigence™ real time cell analyzer (RTCA 1.2.1)]. CI represents a measurement calculated from the difference in impedance at an individual point of time (Zi) from the impedance at the beginning of the experiment (Z0), where CI = . (C) Migration potential of SW480 and SW620 cells over 24 h assessed by wound closure. Wound healing potential was measured according to the area of the wound (%) at 0, 4, 8, 12 and 24 h for SW480 cells and SW620 cells on untreated plastic, 1 µg/ml collagen type I or 5 µg/ml fibronectin. Dotted lines show the wound edges at 0 h. Results are representative of six independent biological replicates. Statistical significance in terms of differences in migration between the two cell lines was assessed in GraphPad Prism by two-way ANOVA with Bonferroni post hoc test. *P<0.05; **P<0.01; ***P<0.001. CI, cell index.
A wound healing assay was used to compare the migratory potential of the SW480 and SW620 cells in vitro and to investigate the effect of different matrices (collagen and fibronectin) on migration of the cells (Fig. 1C). Our data revealed that, overall, SW480 cells have a significantly higher migratory potential than SW620 cells. Furthermore, we report that, in the absence of additional extracellular matrix proteins, limited migration was observed, particularly for SW620 cells, after the 24 h assay period (SW480, 17.94±4.50%; SW620, 10.01±12.96% decrease in wound area; Fig. 1C). Collagen produced the most significant effect on migration in both SW480 and SW620 cells, from as early as 8 h in the SW480 cells (35.23±5.84 and 12.02±3.64% wound closure in SW480 and SW620 cells, respectively; after 24 h, while fibronectin significantly stimulated migration only in the SW480 and not SW620 cells (19.67±4.96 and 6.05±5.24%, respectively) (Fig. 1C).
Comparison of putative cancer stem-like cell populations in the SW480 and SW620 cell lines in vitro
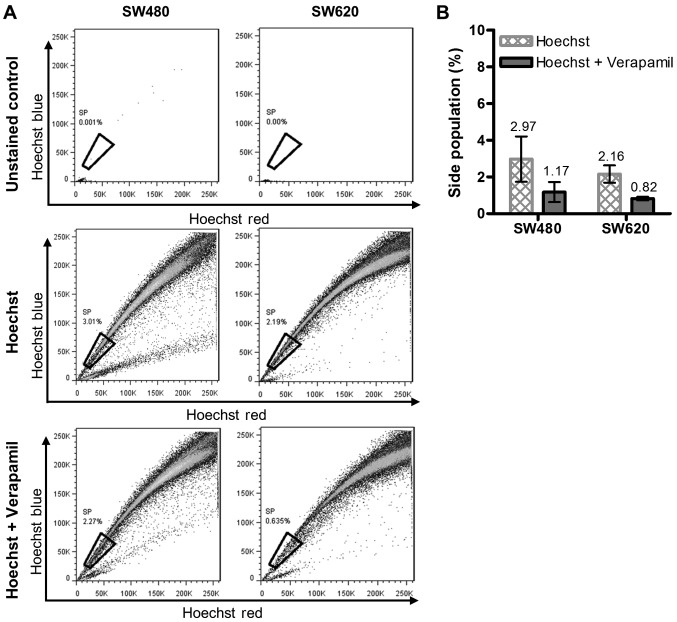
In this study, the presence of putative CSCs was investigated in vitro by analysis of the SP, expression of active ALDH and levels of putative CSC surface protein markers. Given that the SW620 cell line is a metastasis of the primary SW480 cell line, we hypothesised that the SW620 cells may be enriched in CSC-like cells. Hoechst 33342 stained cells treated with or without verapamil were compared with unstained uninhibited (control) cells to determine the proportion of the SP (Fig. 2A). The average results obtained from three independent analyses indicated that there was no significant difference between the sizes of the SP detected within the SW480 and SW620 cell lines (SP of 1.80 and 1.34%, respectively, Fig. 2B).
Figure 2.
Putative cancer stem cell identification by Hoechst 33342 dye exclusion in SW480 and SW620 cells. The proportion of cells representing the SP in the SW480 and SW620 cell lines was determined using Hoechst 33342 dye efflux. (A) Representative dot plots indicating the SP within SW480 and SW620 cells stained with 5 µg/ml Hoechst 33342. The SP was detected by the loss in the fluorescent population (Hblue-/Hred−) upon treatment with the ABCG2 inhibitor, verapamil (50 µM). (B) Putative cancer stem cell populations representative of the average percentage SP observed (mean ± standard deviation) from triplicate independent experiments measured as: SP = (% cells Hblue−/Hred− without verapamil) - (% cells Hblue−/Hred− with verapamil). No significant differences were identified when comparing the SP of the SW480 and SW620 cell lines (t-test with Welch's correction). SP, side population.
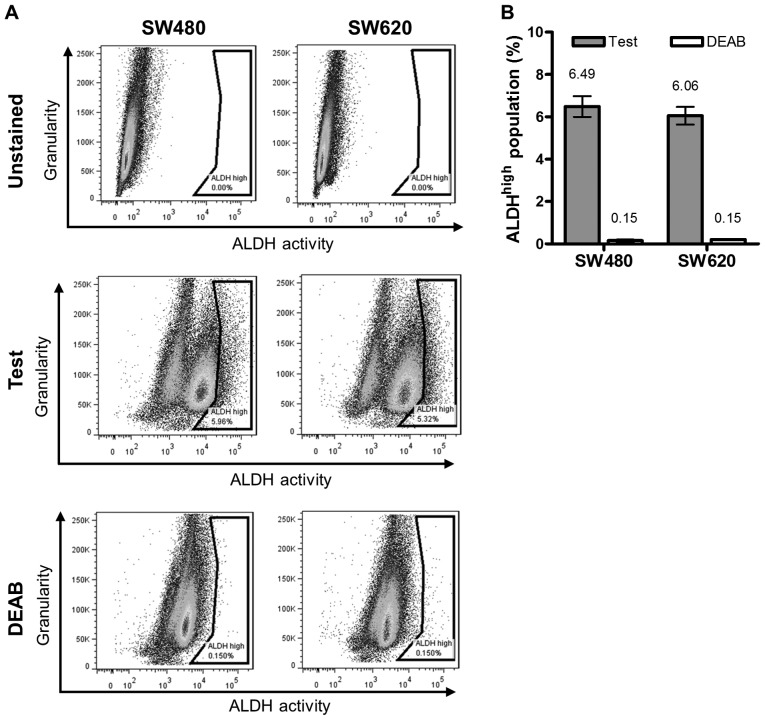
We next compared the proportion of putative CSC between the two lines according to the levels of active ALDH, using the ALDEFLUOR® assay (Fig. 3). The ALDHhigh population. was quantified as the difference in the subpopulation (%) of intensely stained cells (indicated in the gate to the far right of the histograms) between samples with or without DEAB treatment (Fig. 3A). Our data revealed no significant difference in this population between the cell lines (average ALDHhigh population of SW480 and SW620 cells: 6.49 and 6.06%, respectively (Fig. 3B).
Figure 3.
Putative cancer stem cells identified by aldehyde dehydrogenase activity in SW480 and SW620 cells. (A) Representative plots of ALDHhigh populations within SW480 and SW620 cells identified using ALDH inhibitor, DEAB. (B) Average ALDHhigh populations (mean ± standard deviation) were calculated as: % cells ALDHhigh = (% FITChigh cells without DEAB) - (% FITChigh cells treated with DEAB). Results are representative of the average percentage of populations observed from three independent experiments. No significant differences were identified upon comparison between the ALDHhigh populations of SW480 and SW620 cell lines (unpaired t-test with Welch's correction). DEAB, diethylaminobenzaldehyde; ALDH, aldehyde dehydrogenase.
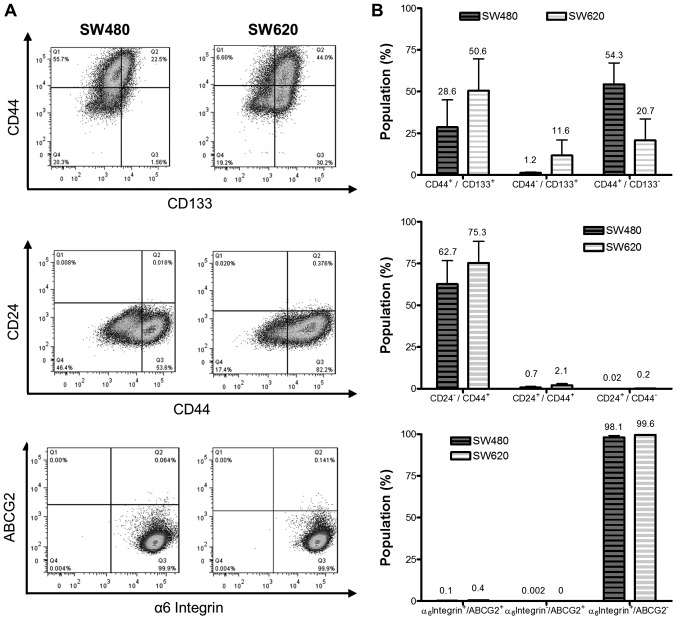
We employed a third strategy to compare putative CSC levels between the cell lines by analyzing the expression of the cell surface markers ABCG2 (CD338), CD44 and CD133, CD24 and the α6 integrin (CD49f) (Fig. 4A). SW620 cells revealed a larger proportion of CD44+/CD133+ cells than SW480 cells (50.6 and 28.6%, respectively; Fig. 4B), while SW480 cells revealed a greater proportion of CD44+/CD133− cells than the SW620 cell line (54.3 and 20.7%, respectively). The CD44+/CD24− population represented 62.7 and 75.3% of SW480 and SW620 cells, respectively α6 integrin was highly expressed by both cell lines, accounting for greater than 98 and 99% integrin α6+/ABCG2− populations of SW480 and SW620 cells, respectively. Co-staining for the α6 integrin+/ABCG2+ marker revealed populations of 0.1 and 0.4% in SW480 and SW620 cells. The differences in expression of these phenotypic markers between the cell lines were not, however, deemed to be significant for any of the cell surface proteins tested, in part due to the high degree of variability in staining between replicates, which was consistently observed in subsequent replicate experiments.
Figure 4.
Comparison of putative cancer stem cell surface marker phenotypes in SW480 and SW620 cell lines. SW480 and SW620 cells analysed for phenotypic subpopulations in terms of CD24, CD44, CD133, α6 integrin and ABCG2 for CSC detection. (A) Representative plots of SW480 and SW620 putative CSC populations analysed by flow cytometry assigned relative to antibody isotype controls. Plots represent the data from one of three independent experiments showing consistent results. (B) Average percentage of CD44+/CD133+, CD24−/CD44+ and ABCG2+/α6 integrin+ cells from triplicate results (mean ± standard deviation). No significant differences were identified when comparing the putative cancer stem populations between SW480 and SW620 cell lines (two-way analysis of variance with Bonferroni post hoc test). CD, cluster of differentiation; CSC, cancer stem cell.
Anchorage independent growth model for putative cancer stem-like cells
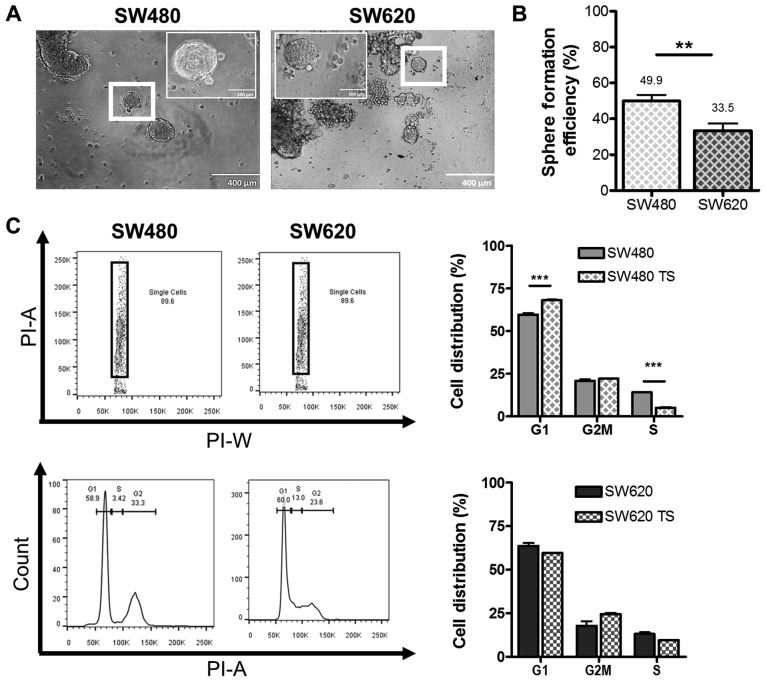
Finally, we compared the levels of putative CSC between the SW480 and SW620 cell lines using a functional assay, where we investigated the ability to form TS under anchorage-independent conditions. Both cell lines were capable of anchorage-independent growth, forming TS, characterized as non-adherent three-dimensional spherical colonies with distinct borders, of approximately 120 µm in diameter within 14 days of culture (Fig. 5A), which correlates with sizes reported in the literature for other primary cancers and cell lines (26–28). We report that SW480 cells displayed significantly higher SFE than SW620 cells after 7 days of culture (49.9 and 33.5%, respectively, Fig. 5B). Since quiescence is a feature of CSC (29), we compared the cell cycle distribution of SW480 and SW620 cells grown adherently and as TS (Fig. 5C). TS derived from SW480 cells displayed a statistically significant (P<0.001) increase in the number of cells in G1/G0 phase and a concomitant decrease in the proportion of cells in S phase compared to SW480 cells cultured under adherent conditions. No significant difference in the distribution of cells between the cell cycle phases was detected on comparison of SW620 cells grown in adherent and TS conditions. The significant increase in the proportion of cells in G1/G0 phase in SW480 TS may account for the greater SFE observed for the SW480 cell line over the SW620 cell line (Fig. 5B).
Figure 5.
Anchorage independent growth of the SW480 and SW620 cell lines. SW480 and SW620 cells were analysed for their ability to survive anchorage-independence and form tumourspheres in vitro. (A) Representative images of SW480 and SW620 tumourspheres after 14 days of anchorage independent growth, taken using an Olympus CKX41 light microscope (magnification ×200 and ×400; scale bar, 0.1 and 0.4 mm) analysed using ImageJ software. (B) SFE was calculated as the number of spheres formed after 7 days of anchorage-independent growth per 1,000 cells seeded and represented as a percentage. Data are representative of the mean ± standard deviation for 10 biological replicates per cell line (unpaired t-test; P<0.01). (C) The elimination of cell doublets (PI-A vs. PI-W) for SW480 and SW620 cells and tumourspheres and the identification of the G1 (inclusive of G0), S and G2M (G2) phases of the cell cycle analysed using propidium iodide staining (50 µg/ml) and flow cytometry. The distribution of cells across the phases of the cell cycle was compared between adherent cells and 7-day old tumourspheres, representative of the mean ± standard deviation for three independent studies. Data were analysed by two-way analysis of variance with Bonferroni post hoc test (**P<0.01; ***P<0.001). SFE, sphere forming efficiency.
Chemosensitivity of SW480 and SW620 cells in vitro
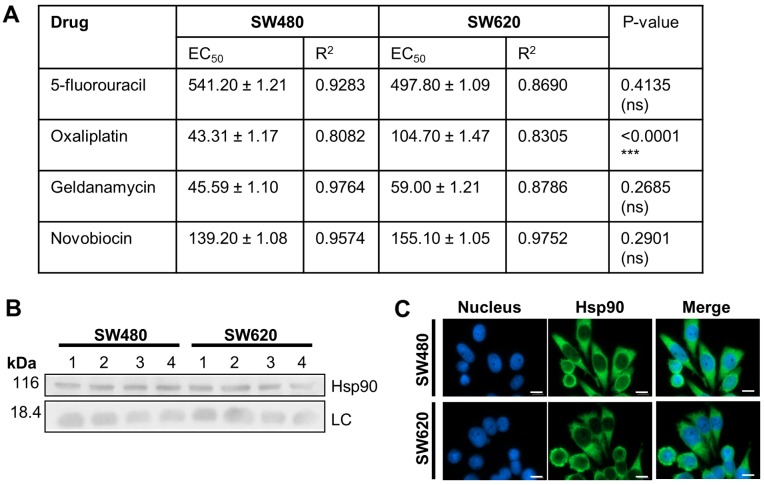
We next compared the chemosensitivity of the SW480 and SW620 cell lines to anti-cancer drugs, including 5-FU and oxaliplatin, used in the treatment of colon cancer (30,31), and the Hsp90 inhibitors, GA and NOV (Fig. 6). SW620 cells displayed greater sensitivity than SW480 cells to 5-FU treatment, although the difference in EC50 values was not significant), while SW480 cells were significantly more sensitive to oxaliplatin. SW480 and SW620 cells displayed similar sensitivities to GA and NOV and demonstrated similar subcellular localisation and expression levels of Hsp90, the target of these drugs (Fig. 6B and C).
Figure 6.
Sensitivity of adherent SW620 and SW480 cells to anti-cancer drugs. (A) Sensitivity of SW480 and SW620 cells exposed to increasing dosage of 5-fluorouracil (0–300 µM), oxaliplatin (0–200 µM), geldanamycin (0–1,000 nM) and novobiocin (0–500 µM) analysed by WST-1 after 72 h (represented as the mean ± standard deviation for triplicate independent analysis). EC50: The concentration at which 50% of cells remain viable. The P-value was determined for the EC50: Values, comparing between the cell lines for each drug, using an unpaired t-test. (B) Western blot analysis of Hsp90 levels in replicate SW480 and SW620 whole cell lysates. (C) Distribution of Hsp90 in SW480 and SW620 cells by fluorescent microscopy (nucleus stained with Hoechst 33342). Images were taken using a Zeiss AxioVert.A1 Fluorescence LED inverted microscope (scale bars, 0.02 mm). ***P<0.0001. LC, histone H3 loading control; Hsp90, heat shock protein 90 kDa.
Response of TS to anti-cancer drugs
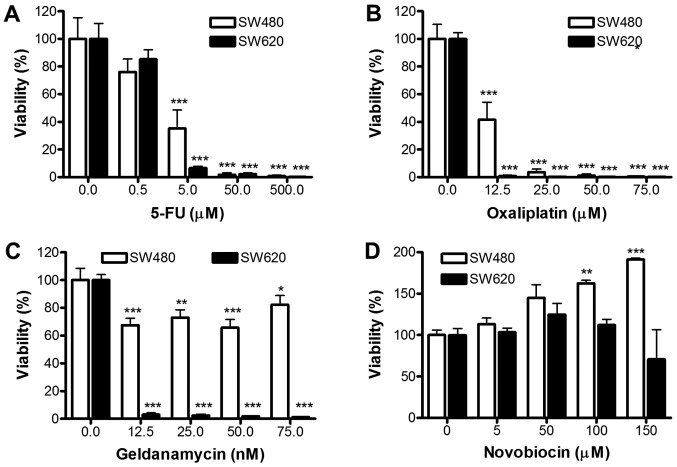
We further assessed whether TS from SW480 and SW620 differ in their response to 5-FU, oxaliplatin, GA or NOV (Fig. 7). Dose-dependent decreases in cellular viability were obtained for both SW480 and SW620 TS following treatment with 5-FU and oxaliplatin at concentrations above 5 and 12.5 µM (P<0.001), respectively. These results suggest that both SW480 and SW620 derived TS exhibit enhanced sensitivity (in comparison to adherent cells) to 5-FU and oxaliplatin treatments (SW620 TS demonstrated greater sensitivity to 5-FU; Fig. 7A), oxaliplatin (Fig. 7B) and GA (Fig. 7C) compared to SW480 TS. Indeed, SW620 derived TS showed significant reductions in viability after treatment with only 12.5 nM GA (P<0.001), which is approximately 4-fold lower that the EC50 for SW480 and SW620 adherent cells. Interestingly, NOV caused a significant dose-dependent increase in viability in SW480 TS (P<0.01 for 100 µM and P<0.001 for 150 µM treatments; Fig. 7D). In contrast, NOV had no significant effect on SW620 TS formation, although the TS were more resistant to NOV compared to the SW620 cells cultured under adherent conditions.
Figure 7.
Comparison of response of SW480 and SW620 tumourspheres to anti-cancer drugs. Viability of SW480 and SW620 tumourspheres analysed by WST-1 assay 7 days after seeding cells in the presence of increasing doses of (A) 5-FU (0–500 µM), (B) oxaliplatin (0–75 µM), (C) geldanamycin (0–75 nM) and (D) novobiocin (0–150 µM). Results represent the mean ± standard deviation of triplicate biological replicates (1,000 cells seeded) that included triplicate technical replicates. Data were analysed by two-way ANOVA with Bonferroni post hoc test. *P<0.05; **P<0.01; ***P<0.001 vs. the untreated control. 5-FU, 5-fluorouracil.
Discussion
The SW480 and SW620 paired cell lines were established from the primary tumour and metastatic counterpart of the same individual and therefore have a shared genetic background, representing an in vitro model for the changes associated with the progression of a colon cancer (32). In this study, the two cell lines were shown to have similar growth rates, while the SW480 cell line was more migratory on collagen and fibronectin in linear wound healing assays than the SW620 cell line. While this may seem counterintuitive, if we consider that the physiology of the pre-metastatic cell line must support metastatic potential, this may account for the enhanced migration potential observed in the SW480 cells.
Our in vitro CSC analysis suggests that, despite having similar levels of cells bearing putative CSC markers, the SW480 and SW620 paired cell lines can be differentiated on the basis of the ability to grow under anchorage independent conditions as TS. Furthermore, the culture of TS revealed differences in chemosensitivity of the SW480 and SW620 cell lines that were not observed when the cells were cultured under adherent conditions. To the best of our knowledge, this is the first study to report an extensive comparison of any paired cell lines using a range of in vitro CSC assays.
In colon cancer, the CD133, CD24 and CD44 surface proteins have been reported to represent putative colorectal CSC populations (20). However, the use of CD44 and CD133 markers for the detection of colon CSC remains controversial (33,34). The proportions of putative CSC identified within the SP of SW480 and SW620 cells in our study were low (~1%) in both cases, similar to reports for other colon cancer lines and gastrointestinal tumours from literature (21,35). Similarly, the proportion of cells expressing ALDH was comparable between the two cell lines, and the CD44+/CD133+ population was also not significantly different between SW480 and SW620 cells. While both paired cell lines formed TS, the SW480 cell line demonstrated significantly greater TS formation efficiency. These data would suggest that the SW480 cell line should be more enriched in cells with a CSC-like phenotype than the SW620 line. If the proportion of CSC is correlated with tumourigenic potential, then these data were unexpected as SW620 cells have demonstrated a greater tumorigenic potential than the SW480 cell line in vivo (32). The CD133+ marker has been used to identify drug-resistant CSC populations in colon cancer, although, there is some indication that both CD133+ and CD133− populations in colorectal cancer can regenerate the growth of a tumour with in vitro serum-free culture (36,37). We originally hypothesised that if there were a link between CSC and metastasis, as suggested in the literature (3–5), then the SW620 cell line might be enriched in cells bearing CSC expression markers and/or CSC properties. Our results do not support this hypothesis, since there was no significant difference in SP, ALDH levels or CSC markers between SW620 and SW480 cells and SW620 cells displayed a lower TS forming ability. However, if we consider the definition of a CSC in terms of their ability to recapitulate a tumour with a similar CSC hierarchy (7,38), this would explain the similarity in CSC profiles, between the SW480 and SW620 cell lines observed in this study in phenotypic marker-based assays.
The SW480/SW620 model was used to explore differential chemosensitivity between cells derived from a primary tumour and its metastasis, as well as to assess the effect of putative CSC-enrichment on drug susceptibility. Under adherent conditions, the SW480 and SW620 cells were similarly sensitive to 5-FU and the Hsp90 inhibitors, GA and NOV, while SW480 cell lines were more sensitive to oxaliplatin than the SW620 cell lines. This demonstrated that differences in the chemosensitivity of primary and secondary tumour-derived cell lines vary between drugs; however, in our study, the overall trend was that of a similar toxicity profile in terms of selected chemotherapeutic agents between the two cell lines. To address whether the presence of a putative CSC subpopulation contributes to drug resistance, the viability of putative CSC-enriched TS grown in the presence of anti-cancer drugs was compared to the aforementioned results obtained under adherent conditions. We report that the anti-cancer drugs 5-FU, oxaliplatin and GA were able to inhibit or reduce TS formation in both lines at concentrations below the EC50 values determined for adherent lines. This is encouraging in the light of the recent focus on developing CSC-targeted therapies (5). In contrast, NOV treatment led to an increase in TS viability, particularly in the SW480 cell line. These data support the hypothesis that the SFE and, by extension, the presence of putative CSCs, influences the sensitivity of cell populations to anti-cancer drugs (16).
The most striking and intriguing trends observed in this study were those observed when comparing the SW480 and the SW620 cell lines in terms of the differences in drug sensitivity of their respective TS. In particular, SW480 TS unlike SW480 cells grown under adherent conditions, revealed a significantly greater tolerance to oxaliplatin, 5-FU, and GA in comparison to SW620 TS. The differences in chemosensitivities of the SW480 and SW620 TS were most prominent for the Hsp90 inhibitors GA and NOV, with SW620 TS showing a dramatic increase in sensitivity to GA and the SW480 TS showing a marked and unexpected increase in viability upon exposure to NOV. These differences may be linked to the increased expression, in SW620 cells, of the oncogene ERBB3 and telomerase (39,40), both of which are Hsp90 clients and have been shown to support anchorage independent growth (41–44). If the SW620 line is dependent on the latter for growth under anchorage independent conditions, then this cell line may be more sensitive to Hsp90 inhibition. The increase in viability of SW480 TS upon NOV treatment was unexpected and is in contrast to other studies showing that NOV and other C-terminal Hsp90 inhibitors were able to block anchorage independent growth of cancer cell lines (45,46), although both of these studies used the soft agar assay, not the TS assay, to measure anchorage independent growth.
In conclusion, our data from the analysis of the SW480 and SW620 paired cell lines do not support a correlation between CSC populations as identified using a range of accepted in vitro assays and the migration potential or drug resistance of the cells since the cell lines have similar levels of putative CSC but show differences in these biological responses.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ABC
ATP-binding cassette transporter protein
- ALDH
aldehyde dehydrogenase
- CSC
cancer stem cells
- Hsp90
heat shock protein 90 kDa
- SP
side population
- SFE
sphere forming efficiency
- 5-FU
5-fluorouracil
- GA
geldanamycin
- NOV
novobiocin
- TS
tumourspheres
Funding
This study was supported by funding from the South African Research Chairs Initiative of the Department of Science and Technology (DST) and National Research Foundation (NRF) of South Africa (grant no. 98566), NRF CPRR and Incentive funding (grant nos. 91523 and 90641), the Cancer Association of South Africa, Medical Research Council South Africa with funds from the National Treasury under its Economic Competitiveness and Support Package and Rhodes University. CS was the recipient of a NRF/DST Innovations postgraduate bursary from the NRF and JAdlM. was the recipient of an Innovations postdoctoral fellowship from the DST/NRF.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
ALE and CS conceived the study. CS and JAdlM conducted the experiments. ALE, JAdlM and CS analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no comepting interests.
References
- 1.Sell S. Stem cell origin of cancer and differentiation therapy. Crit Rev Oncol Hematol. 2004;51:1–28. doi: 10.1016/j.critrevonc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Wicha MS, Liu S, Dontu G. Cancer stem cells: An old idea-A paradigm shift. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 3.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: Migrating cancer stem cells-an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 4.Carpentino JE, Hynes MJ, Appelman HD, Zheng T, Steindler DA, Scott EW, Huang EH. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009;69:8208–8215. doi: 10.1158/0008-5472.CAN-09-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han L, Shi S, Gong T, Zhang Z, Sun X. Cancer stem cells: Therapeutic implications and perspectives in cancer therapy. Acta Pharm Sin B. 2013;3:65–75. doi: 10.1016/j.apsb.2013.02.006. [DOI] [Google Scholar]
- 6.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 7.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 8.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 10.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 11.Dou J, Pan M, Wen P, Li Y, Tang Q, Chu L, Zhao F, Jiang C, Hu W, Hu K, Gu N. Isolation and identification of cancer stem-like cells from murine melanoma cell lines. Cell Mol Immunol. 2007;4:467–472. [PubMed] [Google Scholar]
- 12.Qiang L, Yang Y, Ma YJ, Chen FH, Zhang LB, Liu W, Qi Q, Lu N, Tao L, Wang XT, et al. Isolation and characterization of cancer stem like cells in human glioblastoma cell lines. Cancer Lett. 2009;279:13–21. doi: 10.1016/j.canlet.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Allikmets R, Gerrard B, Hutchinson A, Dean M. Characterization of the human ABC superfamily: Isolation and mapping of 21 new genes using the expressed sequence tags database. Hum Mol Genet. 1996;5:1649–1655. doi: 10.1093/hmg/5.10.1649. [DOI] [PubMed] [Google Scholar]
- 14.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikhail S, Zeidan A. Stem cells in gastrointestinal cancers: The road less travelled. World J Stem Cells. 2014;6:606–613. doi: 10.4252/wjsc.v6.i5.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Wang CY, Liu T, Wu B, Zhou F, Xiong JX, Wu HS, Tao J, Zhao G, Yang M, Gou SM. Persistence of side population cells with high drug efflux capacity in pancreatic cancer. World J Gastroenterol. 2008;14:925–930. doi: 10.3748/wjg.14.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golebiewska A, Brons NH, Bjerkvig R, Niclou SP. Critical appraisal of the side population assay in stem cell and cancer stem cell research. Cell Stem Cell. 2011;8:136–147. doi: 10.1016/j.stem.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida A, Rzhetsky A, Hsu LC, Chang C. Human aldehyde dehydrogenase gene family. Eur J Biochem. 1998;251:549–557. doi: 10.1046/j.1432-1327.1998.2510549.x. [DOI] [PubMed] [Google Scholar]
- 19.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 20.Langan RC, Mullinax JE, Raiji MT, Upham T, Summers T, Stojadinovic A, Avital I. Colorectal cancer biomarkers and the potential role of cancer stem cells. J Cancer. 2013;4:241–250. doi: 10.7150/jca.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sussman RT, Ricci MS, Hart LS, Sun SY, El-Deiry WS. Chemotherapy-resistant side-population of colon cancer cells has a higher sensitivity to TRAIL than the non-SP, a higher expression of c-Myc and TRAIL-receptor DR4. Cancer Biol Ther. 2007;6:1490–1495. doi: 10.4161/cbt.6.9.4905. [DOI] [PubMed] [Google Scholar]
- 22.Leibovitz A, Stinson JC, McCombs WB, III, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–4569. [PubMed] [Google Scholar]
- 23.Goodell BM, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dontu G, Al-Hajj M, Abdallah WM, Clarke MF, Wicha MS. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003;36(Suppl 1):S59–S72. doi: 10.1046/j.1365-2184.36.s.1.6.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreso A, O'Brien CA. Colon cancer stem cells. Curr Protoc Stem Cell Biol Chapter. 2008;3 doi: 10.1002/9780470151808.sc0301s7. Unit 3.1. [DOI] [PubMed] [Google Scholar]
- 26.López J, Poitevin A, Mendoza-Martínez V, Pérez-Plasencia C, García-Carrancá A. Cancer-initiating cells derived from established cervical cell lines exhibit stem-cell markers and increased radioresistance. BMC Cancer. 2012;12:48. doi: 10.1186/1471-2407-12-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao L, Zhou Y, Zhai B, Liao J, Xu W, Zhang R, Li J, Zhang Y, Chen L, Qian H, et al. Sphere-forming cell subpopulations with cancer stem cell properties in human hepatoma cell lines. BMC Gastroenterol. 2011;11:71. doi: 10.1186/1471-230X-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Many AM, Brown AM. Both canonical and non-canonical Wnt signaling independently promote stem cell growth in mammospheres. PLoS One. 2014;9:e101800. doi: 10.1371/journal.pone.0101800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins K, Jacks T, Pavletich NP. The cell cycle and cancer. Proc Natl Acad Sci USA. 1997;94:2776–2778. doi: 10.1073/pnas.94.7.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson EC, Hessman C, Levin TG, Monroe MM, Wong MH. The role of colorectal cancer stem cells in metastatic disease and therapeutic response. Cancers (Basel) 2011;3:319–339. doi: 10.3390/cancers3010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Constant S, Huang S, Wiszniewski L, Mas C. Colon cancer: Current treatments and preclinical models for the discovery and development of new therapies. Drug Discov. 2012:433–458. [Google Scholar]
- 32.Hewitt RE, McMarlin A, Kleiner D, Wersto R, Martin P, Tsokos M, Stamp GW, Stetler-Stevenson WG. Validation of a model of colon cancer progression. J Pathol. 2000;192:446–454. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH775>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 33.Botchkina IL, Rowehl RA, Rivadeneira DE, Karpeh MS, Jr, Crawford H, Dufour A, Ju J, Wang Y, Leyfman Y, Botchkina GI. Phenotypic subpopulations of metastatic colon cancer stem cells: Genomic analysis. Cancer Genomics Proteomics. 2009;6:19–29. [PubMed] [Google Scholar]
- 34.Calvet CY, André FM, Mir L. The culture of cancer cell lines as tumorspheres does not systematically result in cancer stem cell enrichment. PLoS One. 2014;9:e89644. doi: 10.1371/journal.pone.0089644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haraguchi N, Utsunomiya T, Inoue H, Tanaka F, Mimori K, Barnard GF, Mori M. Characterization of a side population of cancer cells from human gastrointestinal system. Stem Cells. 2006;24:506–513. doi: 10.1634/stemcells.2005-0282. [DOI] [PubMed] [Google Scholar]
- 36.Fang DD, Kim YJ, Lee CN, Aggarwal S, McKinnon K, Mesmer D, Norton J, Birse CE, He T, Ruben SM, Moore PA. Expansion of CD133(+) colon cancer cultures retaining stem cell properties to enable cancer stem cell target discovery. Br J Cancer. 2010;102:1265–1275. doi: 10.1038/sj.bjc.6605610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu CS, Tung CY, Yang CY, Lin CH. Response to stress in early tumor colonization modulates switching of CD133-positive and CD133-negative subpopulations in a human metastatic colon cancer cell line, SW620. PLoS One. 2013;8:e61133. doi: 10.1371/journal.pone.0061133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Melcher R, Steinlein C, Feichtinger W, Müller CR, Menzel T, Lührs H, Scheppach W, Schmid M. Spectral karyotyping of the human colon cancer cell lines SW480 and SW620. Cytogenet Cell Genet. 2000;88:145–152. doi: 10.1159/000015508. [DOI] [PubMed] [Google Scholar]
- 40.Kolquist KA, Ellisen LW, Counter CM, Meyerson M, Tan LK, Weinberg RA, Haber DA, Gerald WL. Expression of TERT in early premalignant lesions and a subset of cells in normal tissues. Nat Genet. 1998;19:182–186. doi: 10.1038/554. [DOI] [PubMed] [Google Scholar]
- 41.Gerbin CS, Landgraf R. Geldanamycin selectively targets the nascent form of ERBB3 for degradation. Cell Stress Chaperones. 2010;15:529–544. doi: 10.1007/s12192-009-0166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim RH, Kim R, Chen W, Hu S, Shin KH, Park NH, Kang MK. Association of hsp90 to the hTERT promoter is necessary for hTERT expression in human oral cancer cells. Carcinogenesis. 2008;29:2425–2431. doi: 10.1093/carcin/bgn225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan H, Veldman T, Rundell K, Schlegel R. Simian virus 40 small tumor antigen activates AKT and telomerase and induces anchorage-independent growth of human epithelial cells. J Virol. 2002;76:10685–10691. doi: 10.1128/JVI.76.21.10685-10691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu D, Zhang R, Zhao R, Chen G, Cai Y, Jin J. A novel function of novobiocin: disrupting the interaction of HIF 1α and p300/CBP through direct binding to the HIF1α C-terminal activation domain. PLoS One. 2013;8:e62014. doi: 10.1371/journal.pone.0062014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.