Abstract
The initiation and progression of colorectal cancer (CRC) involves genetic and epigenetic alterations influenced by dietary and environmental factors. Increasing evidence has linked the intestinal microbiota and colorectal cancer. More recently, Fusobacterium nucleatum (Fn), an opportunistic commensal anaerobe in the oral cavity, has been associated with CRC. Several research teams have reported an overabundance of Fn in human CRC and have elucidated the possible mechanisms by which Fn is involved in colorectal carcinogenesis in vitro and in mouse models. However, the mechanisms by which Fn promotes colorectal carcinogenesis remain unclear. To provide new perspectives for early diagnosis, the identification of high risk populations and treatment for colorectal cancer, this review will summarize the relative research progresses regarding the relationship between Fn and colorectal cancer.
Keywords: Fusobacterium nucleatum, colorectal cancer, intestinal microbiota.
Introduction
Colorectal cancer (CRC) is the third most common malignancy in males and the second most common in females, with about 1.4 million new cases in 2012 that lead to 0.69 million deaths according to the World Health Organization1. Many risk factors have been identified for colorectal cancer, mainly including a family history of colorectal cancer, inflammatory bowel disease, excessive consumption of red meat, diabetes and obesity2. Recently, accumulating evidences has suggested that bacteria dysbiosis may be a risk factor for colorectal adenoma and cancer3-7. There is a huge colonization of the human intestine by the microbiome, containing more than 1000 different categories and 1014 microorganisms8, 9. These microorganisms can play an important role in the maintenance of epithelial homeostasis and protection against potential pathogens10, 11 Fusobacterium nucleatum (Fn) is an intestinal and oral microbe that has attracted increasing attention in the field of colorectal cancer12-14. Fn is a gram-negative anaerobic bacterium that mainly resides in the human oral cavity and plays a significant role in the development of periodontal disease and gingivitis15-17. It is also involved in many infectious diseases outside of the oral cavity, such as brain abscesses, liver abscesses, and intrauterine infections associated with pregnancy complications18-20. Fn can adhere to and invade epithelial and endothelial cells via the adhesin FadA, which contributes to its dissemination in hosts17, 21, 22. Several studies have confirmed the enrichment of Fn in CRC5, 12. Moreover, the abundance of Fn in inflammatory bowel disease is significantly higher than that in normal bowels and it becomes a risk factor for inflammation-associated colorectal cancer23. Fn may have great potential to promote colorectal cancer, analogous to the promotion of stomach cancer and MALT lymphoma by Helicobacter pylori24, 25. However, it is unclear whether and how Fn promotes CRC progression.
Clinical Significance of Fn Enrichment in CRC
An enrichment of Fn in CRC tissues compared to the level in controls has been confirmed by many studies, but there are still many inconsistencies in other respects. Several studies demonstrated that the amount of tissue Fn DNA was positively associated with colorectal cancer-specific mortality, which suggested that the enrichment of Fn in CRC tissue might serve as a negative prognostic biomarker12, 26-28. A positive association between Fn abundance and lymph node metastasis has also been observed in CRC5, 27, 29. These findings may suggest Fn-high CRC as a more biologically aggressive cancer subtype. However, other researches did not observe the negative prognostic value of Fn in colorectal cancer5, 30-32. Therefore, it is still unclear whether Fn can be used as an independent prognostic factor for CRC. Fn is enriched in colonic adenoma and stool samples from patients with colorectal adenoma and carcinoma, and the abundance of Fn is consistent with the normal-adenoma-carcinoma sequence model, which suggests that Fn accumulation is an event that occurs at an early stage of colonic tumorigenesis26, 33, 34. Notably, recent studies have demonstrated that Fn may also be involved in the serrated neoplasm pathway30, 35, an alternative pathway of CRC development that contributes to premalignant colorectal lesions36, 37. As we know, identifying colorectal adenomas or early cancer by screening can effectively reduce colorectal cancer mortality38, 39. Traditional colonoscopy as an invasive examination method cannot be widely used in screening for colorectal neoplasia. In contrary, the detection of biomarkers in feces or blood has a broad potential application in screening for colorectal cancer40, 41. Interestingly, several studies have reported the potential value of Fn in improving the diagnosis of colorectal neoplasia. Wong SH et al reported that quantification of fecal Fn combined with fecal immunochemical test can improve the diagnosis of advanced colorectal adenoma and carcinoma42. In this study, the relative abundance of fecal Fn in CRC group was 132 times that of the control group, and was 3.8-fold higher in the advanced adenoma group than in the control group. Liang Q et al found that with fecal Fn alone, the sensitivity and specificity of CRC diagnosis was 80.2% and 80.7%, respectively, while the sensitivity increased to 92.8% when fecal Fn was used in combination with FIT and three other fecal bacteria43. A metagenomic profiling study of the CRC fecal microbiomes indicated that butyryl-CoA dehydrogenase, a gene marker from Fn, can serve as a non-invasive and effective diagnostic biomarker of CRC44. Wang HF et al found that serum anti-Fn-IgA can serve as a potential biomarker for the early diagnosis of CRC45.Collectively, with the detection of Fn, fecal and serum microbiome-based strategies might practical tools for the early diagnosis and treatment of colorectal adenoma and carcinoma.
Location and Translocation Route of Fn in vivo
How Fn is able to colonize the gut mucosa from the oral cavity is still unknown. A recent study reported that the microbial community types observed in the oral cavity and the gut were predictive of each other46, which may suggest an oral-gut translocation route for Fn. Two possible routes might be involved in this process. The first possible route is via swallowing the bacteria, which may further lead to dysbiosis of the gut microbiota, disruption of gut homeostasis, and alteration of the microenvironment that favors the development of CRC47. In the APCmin+ mouse model, daily administration of Fn can increase colon tumor load, and Fn can be isolated from tumors14. Fn is a leading cause of periodontal disease, which may increase the risk of CRC as reported by a retrospective analysis with a large samples size48. Interestingly, in another study, no association was found between Fn abundance in the oral cavity and colorectal cancer history49. Intravenously injected Fn can localize to mouse tumor tissues50, and Fn was detected simultaneously in a pyrogenic liver abscess and colon cancer according to a case report51. Moreover, Fusobacterium can be detected in liver metastasis, and is stably and persistently associated with paired primary-metastatic CRC52, which suggests that Fn might serve as part of the metastatic tissue colonization through a hematogenous route or the lymphatic metastasis pathway. However, different Fusobacterium isolated from colorectal tumor tissues may have different biological features and carry more potential virulence proteins compared to their oral counterparts53. Flanagan et al reported that Fn abundance in stool samples did not correlate with that in paired tissue samples26. Notably, Fn has been detected predominantly in proximal colon cancer, and the proportions of Fn-high CRC gradually increase from rectum to cecum30, 54, this correlation between Fn prevalence and tumor location is consistent with the point of view that microbiota organization is a distinct feature of proximal CRC55. Accordingly, the exact mechanisms of dissemination from oral cavity to colon and the potential links between Fn abundance and tumor location remain to be further elucidated. Once known, those mechanisms will help us reveal the true role of Fn in colorectal carcinogenesis.
Molecular Pathological Features in Fn-positive CRC
Molecular pathological epidemiology, the concept proposed by Ogino and Stampfer, which deeply investigates the phenotypic outcomes of diseases such as cancer using molecular pathological analyses, may represent a new dimension and provide new insights into the carcinogenic mechanism of CRC56-58.It is well-known that all that high microsatellite instability (MSI-H) is an important pathological characteristic of CRC59. Interestingly, many studies have observed a significant correlation between MSI-H and a greater amount of Fn in colorectal cancer tissues30, 31, 60, although the underlying mechanism remains unknown. One possible explanation for this phenomenon is that reactive oxygen species(ROS) and inflammatory cytokines, induced by Fn in colorectal cancer tissues, can attenuate the activity of mismatch repair (MMR) protein leading to MSI-H33, 61. CpG island methylator phenotype (CIMP) is another characteristic feature of chronic inflammation and is closely related to carcinogenesis in many types of cancer. Several studies have reported that the presence of Fn is more often detected in high-CIMP CRC12, 30, 31, 60. In addition, Fusobacterium-high CRC correlates with wild type TP53, mutant CHD7/8 and a high frequency of somatic mutation60. Therefore, Fusobacterium enrichment may be associated with specific molecular subsets of CRCs62.Moreover, the combination of Fusobacterium detection and the molecular pathological features of CRC may provide us with insights into the role of microorganisms in carcinogenesis and better treatment options for patients based on molecular pathological epidemiology56.
Diverse Detection of Fn in CRC
The methods for detecting Fn include conventional quantitative PCR, metagenomic sequencing, Fluorescence in situ hybridization, and 16S ribosomal RNA sequencing63. Among these detection methods, quantitative PCR is the least expensive and most widely used in large case studies. However, different sample types and different detection methods may result in ambiguous results. Therefore, there is an understandable inconsistency in the association between the Fn level and the clinicopathological features and prognosis of CRC among different studies. For example, sample types mainly include Formalin-fixed paraffin-embedded (FFPE) tissue blocks, fresh frozen tissues, stool, mucosal biopsies, and intestinal swabs. The rate of Fn-positive results in FFPE samples has been reported to be 8% in Japan and 13% in the United States12, 32. Mima et al found that the results of Fn detection in paired FFPE and frozen tissue specimens show a high linearity and high reproducibility, which suggests an acceptable level of detection of Fn in FFPE tissues31. Chen et al found that the microbial community of the intestinal lumen is different from that in the cancerous tissue, while the microbial community of cancerous tissue is similar to that in the adjacent noncancerous tissue64. In contrast, the Fn-positive rate is much higher in fresh frozen CRC tissues with the same method, quantitative PCR5, 28, 60. Therefore, the amount of Fn DNA detected in FFPE tissues might not precisely represent the actual abundance of Fn in CRC. However, another study indicated that the qPCR results were consistent when the abundance of Fn in gastroenterological cancer was detected in paired frozen and FFEP tissues65. Loop-mediated isothermal amplification primer sets that, detect two highly conserved genes in Fn, nusG and FadA, could serve as sensitive and rapid molecular tools for Fn detection 66.
Is Fn a Driver or a Passenger?
The role of Fn in CRC carcinogenesis remains uncertain. Whether Fn colonization is a consequence or a cause of CRC is unknown. A bacteria driver-passenger model proposed by Tjalsma et al may help us understand the complex mechanism of polymicrobial interaction in CRC67. In this model, 'driver' bacteria colonization contributes to epithelial DNA damage, creating an altered niche in the colon and leading to the exposure of epithelial cells to microbial compounds from 'passenger' bacteria. Then, 'driver' bacteria are gradually replaced by those 'passenger' bacteria, which can result in inflammation, barrier failure, and a change in the microenvironment that favors the development of CRC. In contrary, 'the alpha-bug' hypothesis proposed by Sears and Pardoll emphasizes the direct pro-oncogenic function of the major pathogenic bacteria, which can remodel the colonic bacteria community and modulate the tumor microenvironment68. It is plausible that the role of Fn may be as a 'driver' rather than a 'passenger' on the basis of the enrichment of Fn in colorectal adenoma. However, some studies have not supported this speculation. Flanagan et al found an over-abundance of Fn in cancerous tissue compared to that in normal tissue, while the Fn level in adenoma was not significantly higher than that in normal tissue26. Similarly, Amitay EL et al found that the abundance of Fn in feces was strongly associated with the presence of CRC, while no association was observed with the presence of colorectal adenoma, leading them to speculate that Fn was just a passenger rather than a driver in CRC development 69. Interestingly, Tomkovich et al found that Fn did not exhibit proinflammatory or pro-tumorigenic activities in preclinical models of colon carcinogenesis, and these authors speculated that only a special group of Fn strains possess carcinogenic abilities when interacting with other members of the microbial community70. Accordingly, further investigation is needed into the molecular mechanism underlying the cross-talk between Fn and host proteins and, the structure and metabolic activities of the intestinal bacterial community, which may provide mechanistic insights into the relationship between Fn and colorectal tumor initiation and progression.
Virulence Proteins of Fn Involved in CRC Carcinogenesis
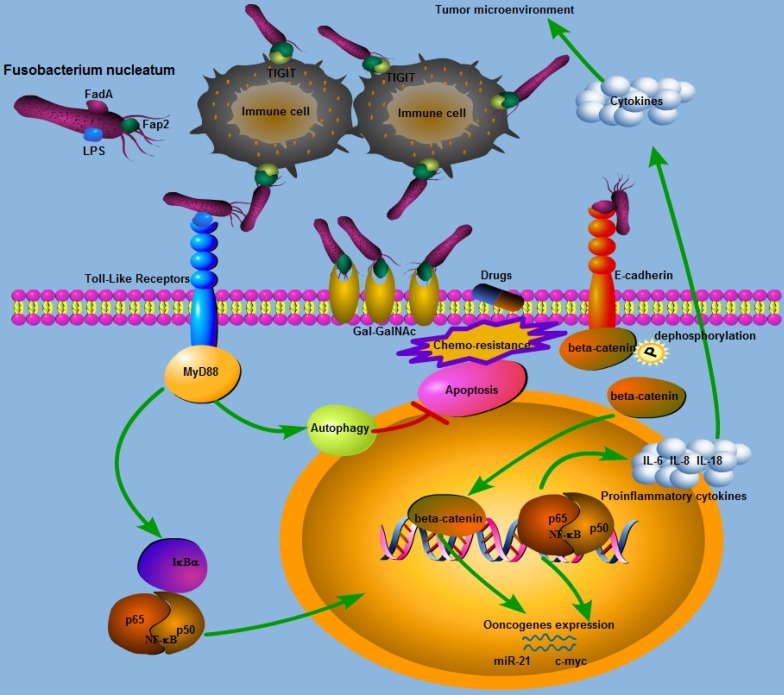
Although Fn has been associated with colorectal cancer (CRC), the underlying mechanisms remain unknown. Recently, two outer membrane proteins from Fn, Fap2 and FadA, have attracted attention. Fap2 is a galactose-sensitive hemagglutinin and adhesion protein, which contributes to the invasive ability of Fn71. Gur et al found that the Fap2 protein of Fn directly interacts with TIGIT expressed on NK cells and tumor-infiltrating lymphocytes, leading to the inhibition of NK cell cytotoxicity and lymphocytes activity72. Meanwhile, tumors infected with Fn were protected from NK-mediated killing and immune cell attack72. Abed et al identified a host polysaccharide, Gal-GalNAc, that is highly expressed in human CRC50. Moreover, Fap2 binding to Gal-GalNAc in human CRC mediates Fn enrichment in adenocarcinoma50. Interestingly, high levels of Gal-GalNAc has also been found in many other types of adenocarcinoma which may suggest that it is a potential target in the treatment of Fn-enriched cancers 73. The FadA gene is unique to Fn, and the FadA protein, a surface adhesin expressed by Fn, plays an essential role in the process of cell attachment21, 74. Rubinstein et al demonstrated that FadA could bind to E-cadherin on CRC cells, mediating Fn attachment to and invasion of the epithelial cells. Binding of FadA to E-cadherin could further activate beta-catenin signaling, leading to elevated expression levels of oncogenes, Wnt genes, and inflammatory genes. In the above-mentioned process, clathrin mediated internalization is involved in activating the Wnt signaling pathway and promoting CRC carcinogenesis75.Wnt pathway activation caused by APC gene mutation can affect the phosphorylation status of beta-catenin and initiate colorectal tumorigenesis 76-78. However, another study has demonstrated that the presence of FadA and Fap2 adhesins in Fn was not sufficient to induce either inflammation or cancer in a preclinical mouse model of colon carcinogenesis70.Toll-like receptor 4 (TLR4), an important receptor for bacteria lipopolysaccharide (LPS),is over-expressed in CRC and may also play a role in tumor promotion79. We have previously demonstrated that infection of CRC cells with Fn increases expression of miR-21 by activating TLR4 in a mouse model80, and miR-21 could serve as a key inhibitor of colitis-associated colon cancer (CAC)81. Chen et al found that both Fn and LPS extracted from Fn could activate beta-catenin signaling in colorectal cancer via the TLR4/P-PAK1 cascade82. Collectively, the Wnt/beta-catenin signaling pathway appears to play an essential role in Fn mediated colorectal carcinogenesis, and more investigations into the three Fn virulence proteins is needed to further determine the mechanisms by which Fn promotes colorectal carcinogenesis.
Fn, Inflammation and Immunity in CRC
It is now well-established that chronic inflammation induced by bacterial infection increases the risk of cancer83. A recent study has provided insights into the relationship between the gut microbiome and inflammatory cytokine production capacity84. Fn infection induces local inflammation and increased expression of inflammatory cytokines, mainly including IL-6, IL-8, TNF-α and cox-2, in the tumor microenvironment, and these cytokines can promote tumorigenesis in CRC33, 53, 75, 85, 86. Ye et al found that Fn can induce the expression of the chemokine CCL20 in colorectal cancer cells when they are co-cultured with Fn87. Moreover, Fn can also stimulate monocyte/macrophage activation and migration which can promote colorectal cancer development87, 88. NF-κB is a transcription factor that is involved in regulating the expression of many genes and that promotes tumor growth and progress89, 90. NF-κB is more frequently activated in Fn-enriched CRC33, 80, 86. Consist with this finding, Rubinstein et al found that only wild-type Fn induced NF-κB expression in HCT116 xenografts75. As NF-κB serves as the central link between inflammation and cancer91, it has been suggested that NF-κB activation may be involved in colorectal carcinogenesis in tumors with Fn enrichment. Kostic et al observed a proinflammatory expression phenomenon in CRC infected with Fn, and determined that Fn may expand myeloid-derived immune cell types, which inhibited the T cell-mediated immune response against colorectal tumors and facilitate tumor progression by modulating the tumor-immune microenvironment33. However, in that study, no association between Fn and inflammation-associated intestinal carcinogenesis was observed in the APCmin+ mouse model of intestinal tumorigenesis33. Fn can also display immunosuppressive activities by causing severe aggregation and apoptosis of immune cells71, 88, 92, 93. In line with these experimental results, Mima et al observed an inverse association between the amount of Fn and CD3+ T cell density31, while another study showed that high levels of Fn were not significantly associated with CD3+ T cell density88. We may infer the immunosuppressive role of Fn in colorectal cancer initiation or development. The gut microbiota has been reported to affect the response to anticancer immunotherapy including PD-L1 and CTLA-494, 95. Based on this, whether Fn affects antitumor immune therapy is very promising route for further investigation, which may provide us with valuable insights into clinical management of CRC patients.
Fn, Lifestyle and Drug Treatment in CRC
It is reported that a 'western' lifestyle characterized by a high consumption of fat, processed meat and red meat, and a low intake of fiber can increase the risk of CRC through the promotion bacterial outgrowth that may lead to a hostile gut environment 96-99. A prospective cohort study demonstrated that prudent diets rich in whole grains and dietary fiber are associated with a lower risk for Fn-positive CRC, which suggests a potential role of Fn in mediating the association between diet and colorectal cancer 100. A recent study has shown that the Fn abundance in fecal samples was not associated with lifestyle or dietary habits69. In addition, different dietary patterns may lead to the variation in Fn abundance observed in different countries32, 98, 101. It has been reported that short-chain fatty acids (SCFA), gut microbiota-derived bacterial fermentation products, can promote colonic homeostasis and health102. Commensal gut microflora and probiotics may exert antimicrobial activity against CRC promoting bacteria by producing colonic fermentation products, such as SCFA and antimicrobial peptides, which can lower intestinal pH, change the immune microenvironment, and eventually create a less hospitable environment for colonization by Fn6, 103-105. These findings may imply that Fn infection in CRC may be closely related to dietary and lifestyle exposure, two important subjects investigated by the evolving field of molecular pathological epidemiology106. Probiotics and prebiotics can increase the production of lactic acid bacteria by adjusting the balance of the intestinal microbiota, which can decrease the risk of CRC107, 108. A recent study showed that treatment of mice bearing a colon cancer xenograft with the antibiotic metronidazole can successfully decrease Fusobacterium load, cancer cell proliferation and tumor growth, suggesting antibiotics as a potential effective intervention for patients with Fn-positive CRC52. Guo et al found that a subunit of the antioxidant protein alkyl hydroperoxide reductase could serve as a potential vaccine candidate to protect against Fn infection, which may add a dimension to our strategy for the prevention of CRC associated with Fn infection109. Interestingly, bacterial metabolism can also differentially affect the host response to chemotherapeutic drugs including 5-fluorouracil (5-Fu) in the interspecies model system C. elegans110, 111. Therefore, we may reason that the efficacy of 5-Fu, the principal anti-colorectal cancer drugs, may be affected by the intestinal microbiota. Consistent with these findings, Yu et al found that the amount of Fn was higher in a CRC recurrence group than that in a non-recurrence group among those who had received 5-Fu-based postoperative chemotherapy112. Moreover, Fn can mediate CRC chemoresistance in response to 5-Fu by selectively targeting miR-18a* and miR-4802 followed by autophagy pathways activation, suggesting that Fn may be a potential risk factor for CRC recurrence112. Taken together, with dietary and lifestyle interventions and appropriate and specific drug treatment, targeting Fn may be a promising new way to address CRC associated with Fn infection, providing us with unique insights for future investigations to further develop our current clinical strategies for CRC.
Future Direction and Concluding Remarks
In this review, we focused on the potential association between Fn infection and colorectal carcinogenesis. The molecular mechanisms involved may include the WNT signaling pathway, the MSI pathway, the epithelial-to-mesenchymal transition (EMT) pathway27, CIMP and the “serrated” pathway, a microRNA pathway and an inflammation pathway, which may help us to build special models linking the intestinal microbiota and CRC and to further understand how CRC initiates and progresses113. Although Fn infection is prevalent in human CRC, there is disagreement about the relationship between Fn and colorectal adenoma. Whether Fn colonization is a consequence or a cause of colorectal carcinogenesis remains unclear. While there is a significant correlation between MSI-H and overabundance of Fn in colorectal cancer tissues, the underlying mechanism remains unknown. Quantification of fecal Fn may serve as a promising biomarker for detecting colorectal neoplasm, although the prognostic value of Fn is controversial. Different sample types and diverse methods may affect the accuracy in detecting the abundance of Fn. FadA, Fap2, and LPS, as virulence proteins of Fn, may play essential roles in mediating Fn invasion and the promotion of CRC. Moreover, miRNA, autophagy and the WNT signaling pathway appear to be involved in Fn-mediated complex networks that promote colorectal cancer. Screening and identification of the virulence proteins of Fn may help us to further investigate the Fn-host cross-talk and to develop a new way to abrogate Fn infections114, 115. The immune suppression role and chemotherapy resistance mechanisms of Fn may alter our therapeutic and preventive strategies for CRC. We can now better understand the intestinal bacteria composition based on the technology of 16S ribosomal DNA sequencing and metagenomic analysis. Nevertheless, we cannot yet establish an in vitro model that efficiently simulates the complex bacterial community and the colonization ecosystem. Thus, further investigation is needed to focus on the host-pathogen interaction partners and the virulence proteins that are involved in colorectal carcinogenesis.
Figure 1.
Schematic representation of the molecular mechanisms of Fn in colorectal cancer carcinogenesis and progression.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet (London, England) 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Castillo V, Sanhueza E, McNerney E, Onate SA, Garcia A. Microbiota dysbiosis: a new piece in the understanding of the carcinogenesis puzzle. J Med Microbiol. 2016;65:1347–1362. doi: 10.1099/jmm.0.000371. [DOI] [PubMed] [Google Scholar]
- 4.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y. et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microbial ecology. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 5.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J. et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome research. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nature reviews Microbiology. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 7.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E. et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):S4586–S4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 11.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS. et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA. et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voigt AY, Zeller G, Bork P. Microbial Biomarkers for Early Cancer Detection. Dtsch Med Wochenschr. 2017;142:267–274. doi: 10.1055/s-0042-110193. [DOI] [PubMed] [Google Scholar]
- 14.Kostic Aleksandar D, Chun E, Robertson L, Glickman Jonathan N, Gallini Carey A, Michaud M. et al. Fusobacterium nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host & Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolstad AI, Jensen HB, Bakken V. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev. 1996;9:55–71. doi: 10.1128/cmr.9.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huttenhower C, Gevers D, Knight R. et al. The Human Microbiome Project Consortium; Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han YW, Shi W, Huang GT, Kinder Haake S, Park NH, Kuramitsu H. et al. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68:3140–3146. doi: 10.1128/iai.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han YW, Fardini Y, Chen C, Iacampo KG, Peraino VA, Shamonki JM. et al. Term stillbirth caused by oral Fusobacterium nucleatum. Obstet Gynecol. 2010;115:442–445. doi: 10.1097/AOG.0b013e3181cb9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kai A, Cooke F, Antoun N, Siddharthan C, Sule O. A rare presentation of ventriculitis and brain abscess caused by Fusobacterium nucleatum. J Med Microbiol. 2008;57:668–671. doi: 10.1099/jmm.0.47710-0. [DOI] [PubMed] [Google Scholar]
- 20.Yoneda M, Kato S, Mawatari H, Kirikoshi H, Imajo K, Fujita K. et al. Liver abscess caused by periodontal bacterial infection with Fusobacterium necrophorum. Hepatol Res. 2011;41:194–196. doi: 10.1111/j.1872-034X.2010.00748.x. [DOI] [PubMed] [Google Scholar]
- 21.Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. The Journal of biological chemistry. 2007;282:25000–25009. doi: 10.1074/jbc.M611567200. [DOI] [PubMed] [Google Scholar]
- 22.Fardini Y, Wang X, Temoin S, Nithianantham S, Lee D, Shoham M. et al. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Molecular microbiology. 2011;82:1468–1480. doi: 10.1111/j.1365-2958.2011.07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R. et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 24.Polk DB, Peek RM Jr. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V. et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology. 2014;33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 27.Yan X, Liu L, Li H, Qin H, Sun Z. Clinical significance of Fusobacterium nucleatum, epithelial-mesenchymal transition, and cancer stem cell markers in stage III/IV colorectal cancer patients. Onco Targets Ther. 2017;10:5031–5046. doi: 10.2147/OTT.S145949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaoka Y, Suehiro Y, Hashimoto S, Hoshida T, Fujimoto M, Watanabe M. et al. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J Gastroenterol. 2017 doi: 10.1007/s00535-017-1382-6. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B. et al. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016;22:3227–3233. doi: 10.3748/wjg.v22.i11.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H. et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137:1258–1268. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 31.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K. et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA oncology. 2015;1:653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H. et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22:557–566. doi: 10.3748/wjg.v22.i2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 35.Park CH, Han DS, Oh YH, Lee AR, Lee YR, Eun CS. Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis. Sci Rep. 2016;6:25271. doi: 10.1038/srep25271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosty C, Hewett DG, Brown IS, Leggett BA, Whitehall VL. Serrated polyps of the large intestine: current understanding of diagnosis, pathogenesis, and clinical management. J Gastroenterol. 2013;48:287–302. doi: 10.1007/s00535-012-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bettington M, Walker N, Clouston A, Brown I, Leggett B, Whitehall V. The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology. 2013;62:367–386. doi: 10.1111/his.12055. [DOI] [PubMed] [Google Scholar]
- 38.Rex DK, Johnson DA, Anderson JC, Schoenfeld PS, Burke CA, Inadomi JM. et al. American College of Gastroenterology guidelines for colorectal cancer screening 2009. Am J Gastroenterol. 2009;104:739–750. doi: 10.1038/ajg.2009.104. [DOI] [PubMed] [Google Scholar]
- 39.Sung JJ, Ng SC, Chan FK, Chiu HM, Kim HS, Matsuda T. et al. An updated Asia Pacific Consensus Recommendations on colorectal cancer screening. Gut. 2015;64:121–132. doi: 10.1136/gutjnl-2013-306503. [DOI] [PubMed] [Google Scholar]
- 40.Xie YH, Gao QY, Cai GX, Sun XM, Zou TH, Chen HM. et al. Fecal Clostridium symbiosum for Noninvasive Detection of Early and Advanced Colorectal Cancer: Test and Validation Studies. EBioMedicine. 2017;25:32–40. doi: 10.1016/j.ebiom.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eklof V, Lofgren-Burstrom A, Zingmark C, Edin S, Larsson P, Karling P. et al. Cancer-associated fecal microbial markers in colorectal cancer detection. Int J Cancer. 2017;141:2528–2536. doi: 10.1002/ijc.31011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G. et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66:1441–1448. doi: 10.1136/gutjnl-2016-312766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J. et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin Cancer Res. 2017;23:2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 44.Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y. et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 45.Wang HF, Li LF, Guo SH, Zeng QY, Ning F, Liu WL. et al. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci Rep. 2016;6:33440. doi: 10.1038/srep33440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flynn KJ, Baxter NT, Schloss PD. Metabolic and Community Synergy of Oral Bacteria in Colorectal Cancer. mSphere; 2016. p. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Momen-Heravi F, Babic A, Tworoger SS, Zhang L, Wu K, Smith-Warner SA. et al. Periodontal disease, tooth loss and colorectal cancer risk: Results from the Nurses' Health Study. Int J Cancer. 2017;140:646–452. doi: 10.1002/ijc.30486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato I, Vasquez AA, Moyerbrailean G, Land S, Sun J, Lin HS. et al. Oral microbiome and history of smoking and colorectal cancer. J Epidemiol Res. 2016;2:92–101. doi: 10.5430/jer.v2n2p92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A. et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shigefuku R, Watanabe T, Kanno Y, Ikeda H, Nakano H, Hattori N. et al. Fusobacterium nucleatum detected simultaneously in a pyogenic liver abscess and advanced sigmoid colon cancer. Anaerobe. 2017;48:144–146. doi: 10.1016/j.anaerobe.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 52.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D. et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K. et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1:16. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y. et al. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol. 2016;7:e200. doi: 10.1038/ctg.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ. et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A. 2014;111:18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogino S, Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: the evolving field of molecular pathological epidemiology. J Natl Cancer Inst. 2010;102:365–367. doi: 10.1093/jnci/djq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogino S, Nishihara R, VanderWeele TJ, Wang M, Nishi A, Lochhead P. et al. Review Article: The Role of Molecular Pathological Epidemiology in the Study of Neoplastic and Non-neoplastic Diseases in the Era of Precision Medicine. Epidemiology. 2016;27:602–611. doi: 10.1097/EDE.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J. et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep. 2017;7:11590. doi: 10.1038/s41598-017-11237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drewes JL, White JR, Dejea CM, Fathi P, Iyadorai T, Vadivelu J. et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes. 2017;3:34. doi: 10.1038/s41522-017-0040-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamamura K, Baba Y, Miyake K, Nakamura K, Shigaki H, Mima K. et al. Fusobacterium nucleatum in gastroenterological cancer: Evaluation of measurement methods using quantitative polymerase chain reaction and a literature review. Oncol Lett. 2017;14:6373–6378. doi: 10.3892/ol.2017.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang S, Yang Z, Zou D, Dong D, Liu A, Liu W. et al. Rapid detection of nusG and fadA in Fusobacterium nucleatum by loop-mediated isothermal amplification. J Med Microbiol. 2016;65:760–769. doi: 10.1099/jmm.0.000300. [DOI] [PubMed] [Google Scholar]
- 67.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nature reviews Microbiology. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 68.Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis. 2011;203:306–311. doi: 10.1093/jinfdis/jiq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amitay EL, Werner S, Vital M, Pieper DH, Hofler D, Gierse IJ. et al. Fusobacterium and colorectal cancer: causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis. 2017;38:781–788. doi: 10.1093/carcin/bgx053. [DOI] [PubMed] [Google Scholar]
- 70.Tomkovich S, Yang Y, Winglee K, Gauthier J, Muhlbauer M, Sun X. et al. Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis. Cancer Res. 2017;77:2620–2632. doi: 10.1158/0008-5472.CAN-16-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, Han YW. et al. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun. 2015;83:1104–1113. doi: 10.1128/IAI.02838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M. et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abed J, Maalouf N, Parhi L, Chaushu S, Mandelboim O, Bachrach G. Tumor Targeting by Fusobacterium nucleatum: A Pilot Study and Future Perspectives. Front Cell Infect Microbiol. 2017;7:295. doi: 10.3389/fcimb.2017.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M. et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. 2005;187:5330–5340. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 78.Pishvaian MJ, Byers SW. Biomarkers of WNT signaling. Cancer Biomark. 2007;3:263–2674. doi: 10.3233/cbm-2007-34-510. [DOI] [PubMed] [Google Scholar]
- 79.Santaolalla R, Sussman DA, Ruiz JR, Davies JM, Pastorini C, Espana CL. et al. TLR4 activates the beta-catenin pathway to cause intestinal neoplasia. PLoS One. 2013;8:63298. doi: 10.1371/journal.pone.0063298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y. et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152:851–866. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shi C, Yang Y, Xia Y, Okugawa Y, Yang J, Liang Y. et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut. 2016;65:1470–1481. doi: 10.1136/gutjnl-2014-308455. [DOI] [PubMed] [Google Scholar]
- 82.Chen Y, Peng Y, Yu J, Chen T, Wu Y, Shi L. et al. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget. 2017;8:31802–31814. doi: 10.18632/oncotarget.15992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Martel C, Franceschi S. Infections and cancer: established associations and new hypotheses. Crit Rev Oncol Hematol. 2009;70:183–194. doi: 10.1016/j.critrevonc.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 84.Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA. et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 2016;167:1125–1136. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun. 2011;79:2597–2607. doi: 10.1128/IAI.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei Z, Cao S, Liu S, Yao Z, Sun T, Li Y. et al. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients' survival? A pilot study on relevant mechanism. Oncotarget. 2016;7:46158–46172. doi: 10.18632/oncotarget.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ye X, Wang R, Bhattacharya R, Boulbes DR, Fan F, Xia L. et al. Fusobacterium Nucleatum Subspecies Animalis Influences Proinflammatory Cytokine Expression and Monocyte Activation in Human Colorectal Tumors. Cancer Prev Res (Phila) 2017;10:398–409. doi: 10.1158/1940-6207.CAPR-16-0178. [DOI] [PubMed] [Google Scholar]
- 88.Park HE, Kim JH, Cho NY, Lee HS, Kang GH. Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma. Virchows Arch. 2017;471:329–336. doi: 10.1007/s00428-017-2171-6. [DOI] [PubMed] [Google Scholar]
- 89.Inoue J, Gohda J, Akiyama T, Semba K. NF-kappaB activation in development and progression of cancer. Cancer Sci. 2007;98:268–274. doi: 10.1111/j.1349-7006.2007.00389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 91.DiDonato JA, Mercurio F, Karin M. NF-kappaB and the link between inflammation and cancer. Immunol Rev. 2012;246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 92.Shenker BJ, DiRienzo JM. Suppression of human peripheral blood lymphocytes by Fusobacterium nucleatum. J Immunol. 1984;132:2357–2362. [PubMed] [Google Scholar]
- 93.Kaplan CW, Ma X, Paranjpe A, Jewett A, Lux R, Kinder-Haake S. et al. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun. 2010;78:4773–4778. doi: 10.1128/IAI.00567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Watson AJ, Collins PD. Colon cancer: a civilization disorder. Dig Dis. 2011;29:222–228. doi: 10.1159/000323926. [DOI] [PubMed] [Google Scholar]
- 97.Song M, Garrett WS, Chan AT. Nutrients, foods, and colorectal cancer prevention. Gastroenterology. 2015;148:1244–1260. doi: 10.1053/j.gastro.2014.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.O'Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K. et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z. et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 100.Mehta RS, Nishihara R, Cao Y, Song M, Mima K, Qian ZR. et al. Association of Dietary Patterns With Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA oncology. 2017;3:921–927. doi: 10.1001/jamaoncol.2016.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM. et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome research. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ostaff MJ, Stange EF, Wehkamp J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol Med. 2013;5:1465–1483. doi: 10.1002/emmm.201201773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roy CC, Kien CL, Bouthillier L, Levy E. Short-chain fatty acids: ready for prime time? Nutr Clin Pract. 2006;21:351–366. doi: 10.1177/0115426506021004351. [DOI] [PubMed] [Google Scholar]
- 105.Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, Calignano A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol. 2011;17:1519–1528. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rescigno T, Micolucci L, Tecce MF, Capasso A. Bioactive Nutrients and Nutrigenomics in Age-Related Diseases. Molecules. 2017;22:105. doi: 10.3390/molecules22010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Coleman OI, Nunes T. Role of the Microbiota in Colorectal Cancer: Updates on Microbial Associations and Therapeutic Implications. Biores Open Access. 2016;5:279–288. doi: 10.1089/biores.2016.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ambalam P, Raman M, Purama RK, Doble M. Probiotics, prebiotics and colorectal cancer prevention. Best Pract Res Clin Gastroenterol. 2016;30:119–131. doi: 10.1016/j.bpg.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 109.Guo SH, Wang HF, Nian ZG, Wang YD, Zeng QY, Zhang G. Immunization with alkyl hydroperoxide reductase subunit C reduces Fusobacterium nucleatum load in the intestinal tract. Sci Rep. 2017;7:10566. doi: 10.1038/s41598-017-11127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Scott TA, Quintaneiro LM, Norvaisas P, Lui PP, Wilson MP, Leung KY. et al. Host-Microbe Co-metabolism Dictates Cancer Drug Efficacy in C. elegans. Cell. 2017;169:442–456. doi: 10.1016/j.cell.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Garcia-Gonzalez AP, Ritter AD, Shrestha S, Andersen EC, Yilmaz LS, Walhout AJM. Bacterial Metabolism Affects the C. elegans Response to Cancer Chemotherapeutics. Cell. 2017;169:431–441. doi: 10.1016/j.cell.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J. et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548–563. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zanzoni A, Spinelli L, Braham S, Brun C. Perturbed human sub-networks by Fusobacterium nucleatum candidate virulence proteins. Microbiome. 2017;5:89. doi: 10.1186/s40168-017-0307-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kumar A, Thotakura PL, Tiwary BK, Krishna R. Target identification in Fusobacterium nucleatum by subtractive genomics approach and enrichment analysis of host-pathogen protein-protein interactions. BMC Microbiol. 2016;16:84. doi: 10.1186/s12866-016-0700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]