Abstract
Background: Widespread clinical implementation of next-generation sequencing (NGS)-based cancer in vitro diagnostic tests (IVDs) highlighted the urgency to establish reference materials which could provide full control of the process from nucleic acid extraction to test report generation. The formalin-fixed, paraffin-embedded (FFPE) tissue and blood plasma containing circulating tumor deoxyribonucleic acid (ctDNA) were mostly used for clinically detecting onco-relevant mutations.
Methods: We respectively developed multiplex FFPE and plasma reference materials covering three clinically onco-relevant mutations within the epidermal growth factor receptor (EGFR) gene at serial allelic frequencies. All reference materials were quantified and validated via droplet digital polymerase chain reaction (ddPCR), and then were distributed to eight domestic manufacturers for the collaborative evaluation of the performance of several domestic NGS-based cancer IVDs covering four major NGS platforms (NextSeq, HiSeq, Ion Proton and BGISEQ).
Results: All expected mutations except one at extremely low allelic frequencies were detected, despite some differences in coefficient of variation (CV) which increased with the decrease of allelic frequency (CVs ranging from 18% to 106%). It was worth noting that the CV value seemed to correlate with a particular mutation as well. The repeatability of determination of different mutations was L858R>T790M>19del.
Conclusions: The results indicated our reference materials would be pivotal for quality control of NGS-based cancer IVDs and would guide the further development of reference materials covering more onco-relevant mutations.
Keywords: reference materials, next-generation sequencing, circulating tumor DNA, formalin-fixed paraffin-embedded.
Introduction
NGS technology has great potential to provide detailed profiling of cancer-related mutations which could improve the diagnosis, prognosis, and treatment of patients 1-3. Although the improvements in NGS have decreased the turnaround time and associated cost, it also presented challenges for test development and validation, proficiency testing, and quality management of these cancer IVDs 4. Therefore, appropriate reference materials and validation approaches were required to ensure the clinical accuracy and efficiency of the novel NGS assays 5,6. Many entities offered well-characterized cell lines or genomic DNA (gDNA) containing cancer-related mutations. However, the clinical-grade reference materials which could provide the whole process quality control from nucleic acid extraction to sequencing report were still publicly unavailable.
There are several types of commercially available reference materials 7-10. In silico sequences were computationally created which were used for optimization of test performance and to guide the selection of proper bioinformatics tools 11. However, computationally generated and mutagenized sequence could not mimicked essential characteristics of real data or cover pre-sequencing steps. Synthetic DNA had recently become a convenient option to supplement the standard use of real gDNA since relevant cancer-related mutations could be easily spiked into human gDNA at desired allelic frequency 12. This approach allowed the establishment of highly multiplexed mutants that could be individually prepared and pooled easily. However, synthetic DNA posed the major challenge that the step of extracting gDNA or ctDNA could not be monitored. This limitation could be overcome by using well-characterized cell lines or artificially modified cell lines as reference material 13, although cell line samples would be limited by multiplexing range and dynamic allelic frequency during passages. The ideal reference materials for NGS-based cancer IVDs should be the real or similar clinical samples. However, it's impractical to get enough quantity or samples at desired allelic frequency from clinical samples.
Currently, most DNA samples used for the detection of cancer-related mutations were obtained from FFPE tumor tissue and the peripheral blood. Therefore, we herein established two types of reference materials, FFPE and plasma reference materials, for the performance evaluation of NGS-based cancer IVDs detecting three mutations (19del, L858R, and T790M) of the EGFR gene. In this study, the reference materials were distributed to eight major domestic manufacturers in China and evaluated using their in-house protocol. Since the aim of the reference materials is to assure the quality of NGS testing process, small-scale collaborative study with current/potential service providers is necessary which can ensure the applicability of the reference materials.
Materials and Methods
Preparation of FFPE reference materials
FFPE reference materials were prepared by spiking EGFR mutant cells into cells with wildtype EGFR (Figure S1). The wildtype A549 cell line, mutant H1650 cell line containing EGFR exon 19 deletion mutation (19del) and H1975 cell line containing both EGFR exon 21 L858R substitution mutation (L858R) and EGFR exon 20 T790M substitution mutation (T790M) were purchased from the Chinese Academy of Sciences (Table 1). Cells were cultured using the Ham's F-12K (Kaighn's) medium (ThermoFisher Scientific Inc., MA, USA) for A549 and the RPMI 1640 medium (ThermoFisher) for H1650 and H1975 supplemented with 10% fetal bovine serum (ThermoFisher). The cell cultures were incubated at 37°C in a humidified atmosphere of 5% CO2 and passaged every two to three days. For the passage process, cells were digested with 0.25% trypsin-EDTA following the spin at 1000rpm for 5min for medium renewal and the sub-cultivation ratio was 1:4.
Table 1.
Characteristics of the wildtype and mutant cell lines.
| Cell lines | EGFR status | DNA change | AA change | Cosmic ID | Variant type | Exon |
|---|---|---|---|---|---|---|
| A549 | WT | N/A | N/A | N/A | N/A | N/A |
| H1650 | mutant | 2235_2249del15 | E746_A750del | 6223 | Deletion | 19 |
| (19del) | ||||||
| H1975 | mutant | 2573T>G | L858R | 6224 | Missense | 21 |
| 2369C>T | T790M | 6240 | Missense | 20 |
Abbreviations: EGFR, epidermal growth factor receptor; DNA, deoxyribonucleic acid; AA, amino acid; WT, wildtype; del, deletion; T, threonine; M, methionine; L, leucine; R, arginine; N/A, not applicable.
For the preparation of FFPE reference materials FFPE-Q1 and FFPE-Q2 at desired allelic frequencies of 1.0% and 5.0% respectively, 2×105 wildtype A549 cells were spiked with 2×103 cells of each mutant H1650 and H1975 cell lines for preparing 1.0% allelic frequency and with 1×104 cells of each H1650 and H1975 for preparing 5.0% allelic frequency. The prepared mixtures were centrifuged at 600g for 10min following resuspension by 1mL 4% paraformaldehyde and fixation at 4°C for 20min to simulate the conditions of FFPE samples. Cells were then washed and resuspended in adequate amount of phosphate buffer serum with 0.2% bovine serum albumin at the final concentration of 2×105cells/mL. After preparation, the FFPE reference materials were aliquoted into 2mL tubes (500μL for each) stored at -70°C or below. The final allelic frequencies of three EGFR mutations of each reference material were validated and quantified by the ddPCR assay described below.
Preparation of plasma reference materials
The plasma reference materials containing three EGFR mutations 19del, L858R, and T790M were prepared by spiking cell line-derived PCR fragments mimicking ctDNA into the negative blood plasma pooled from 300 patients provided by Shanghai Chest Hospital (Figure S1). To obtain the plasma, the whole blood samples were centrifuged at 1000g for 10min at 4°C and then the supernatants were centrifuged at 1600g for 10min at 4°C. The supernatant plasma were pooled for detection of relevant EGFR mutations. The cell-free DNA (cfDNA) was extracted from pooled plasma using the GenoPrep DNA extraction kit (Genosaber Co. Ltd., Shanghai, China) following the manufacture's instruction. In brief, 2mL plasma was mixed with 10μL Carrier RNA, 50μL protease K, and 2mL lysis buffer. Then the mixture was vortexed and incubated at 56°C for 20min. After a short spin down, 1mL isopropanol was added and vortexed. The mixture was transferred to a new spin column and four times sequential washing process was performed using the washing buffer. At last, purified DNA was eluted using elution buffer then was tested by the ddPCR assay for detection of relevant EGFR mutations.
To prepare mimic ctDNA fragments, gDNA of H1650 and H1975 cell lines were isolated by QIAamp DNA FFPE Tissue Kit (Qiagen, CA, USA) according to the manufacturer's instruction. Nucleic acid fragments containing 19del, L858R or T790M mutations were amplified from the gDNA of H1650 and H1975 cell lines using corresponding primers (Table S1). The PCR assay was conducted in quadruplicate for each mutant fragment with ABI 7500 Real-Time PCR Systems (ThermoFisher). Reaction was performed in 50μL volume containing 5μL 10×PCR Buffer for KOD-Plus-Neo, 5μL 2mM dNTPs, 3μL 25mM MgSO4, 1.5μL target primers (10μM for each), 1μL DNA template, 1μL KOD-Plus-Neo (1U/μL) and 33μL water. The thermocycling condition was hot start at 94°C for 2min following by 50 cycles of 98°C for 10s, corresponding annealing temperature (58°C for 19del, 61°C for L858R-1, 60°C for L858R-2, and 63°C for T790M, respectively) for 30s, and 68°C for 30s and holding at 4°C. After amplification PCR process, 200μL PCR products of each mutant fragments were purified through gel electrophoresis. The ddPCR assay was performed for the quantification of the allelic frequency of each mutation in the corresponding DNA fragments.
According to the results of quantification of ctDNA in pooled plasma, a corresponding amount of mutant fragments were spiked to prepare the plasma reference materials Plasma-Q1 and Plasma-Q2 at desired allelic frequencies of 0.5% and 1.0% respectively. The pooled plasma without spiking mutant fragments was served as Plasma-Q0 at desired allelic frequencies of 0.1%. After preparation, the plasma reference materials were aliquoted into 2mL tubes (2mL for each) stored at -70°C or below. The final allelic frequencies of three EGFR mutations of each reference material were validated and quantified by the ddPCR assay described below.
Validation and quantification of the reference materials by ddPCR
For extraction of DNA from FFPE reference materials (FFPE-Q1 and FFPE-Q2), the QIAamp DNA FFPE Tissue Kit (Qiagen) was used following the manufacturer's instruction. In brief, cells were pelleted and resuspended with 180μL buffer ATL and 20μL protease K, vortexed, and incubated at 56°C for 1 hour. Samples were incubated at 90°C for 1 hour for DNA renaturation and then 200μL buffer AL and 200μL anhydrous ethanol were added. After vortex, samples were transferred to the spin column and sequential washing was performed for three times with washing buffer. At last, purified DNA was collected by adding elution buffer. For extraction of DNA from plasma reference materials (Plasma-Q0, Plasma-Q1, and Plasma-Q2), the GenoPrep DNA extraction kit (Genosaber) was used as previously described.
The DNA samples extracted from both the FFPE and plasma reference materials were detected by ddPCR to validate and quantify the allelic frequencies of three EGFR mutations (19del, L858R, and T790M) using the QX200 Droplet Digital PCR System (Bio-Rad Laboratories Inc., CA, USA).
The reaction mixture was 20μL volumes containing 10μL 2×ddPCR Supermix for Probes (no dUTP), 1μL 20×target primers/probe (FAM) and 1μL 20×reference primers/probe (HEX), 6μL DNA template and 2μL nuclease-free water. Three ddPCR probe assays consisting of unlabeled PCR primers and a dual labeled fluorescent probe were purchased, the PrimePCR ddPCR Mutation Assay EGFR p.E746_A750del Human (Bio-Rad), the PrimePCR ddPCR Mutation Assay EGFR p.L858R Human (Bio-Rad), and the PrimePCR ddPCR Mutation Assay EGFR p.T790M Human (Bio-Rad), for validation and quantification of 19del, L858R, and T790M, respectively. After loading the reaction mixture into the well of a droplet generator cartridge, 70μL droplet generation oil was added to each well and then the sample-containing cartridge was placed into the droplet generator for the droplets generation. Once this process was complete, droplets of individual sample were transferred into the wells of a 96-well PCR plate, sealed, and loaded into the thermal cycler. The thermocycling condition was described as below: hot start at 95°C for 10min; 40 cycles of 94°C for 30s following corresponding annealing temperature (58°C for 19del, 61°C for L858R-1, 60°C for L858R-2, 63°C for T790M, respectively) for 1min; 98°C for 10 min and holding at 4°C. After completion of the PCR, the sealed plate was loaded into the droplet reader for determination of the complete ddPCR reactions in droplets. Data analysis was processed using the QuantaSoft v1.7.4.
Determination of allelic frequencies by ARMS-PCR
The amplification refractory mutation system PCR (ARMS-PCR) array was conducted for determination of allelic frequencies of the reference materials using the Human EGFR Gene Mutation Quantitative Detection Kit (Fluorescence qPCR) (Genosaber) following the manufacturer's instruction. For extraction of DNA from FFPE and plasma reference materials, all samples were processed as described above and the extracts were then used for the ARMS-PCR assay. Briefly, the reaction mixture in 0.2mL thin well 6-tube strip was melted at room temperature and then centrifuged at 3000g for 1 min. Then 5μL DNA extract for each reference material and the control samples provided within the kit were added to the 45μL reaction mixture respectively. Before loading, strips containing samples, control materials and calibrators (pre-mixed) were vortexed for 10-20s and centrifuged at 3000g for 4min. All three mutations of the same sample and an external control within the conserved region of EGFR exon to monitor the run quality were analyzed in the same array in order to standardize the detection conditions. The PCR was conducted using the ABI 7500 Real-Time PCR Systems (ThermoFisher). The thermocycling condition was hot start at 95°C for 4min and 50 cycles of 95°C for 10s, 61°C for 30s with fluorescence reading (FAM). Data analysis was processed using the ABI 7500 software v2.3.
Collaborative evaluation of performance of NGS-based cancer IVDs
The prepared reference materials were distributed to eight manufacturers to evaluate the general performance of NGS-based cancer IVDs. Seven participants developed separate NGS-based cancer IVDs for detection of cancer-related mutations from FFPE and plasma samples respectively and one could only test the plasma samples. All reference materials were tested and analyzed using NGS panels and bioinformatics tools developed in house by the participants respectively following the manufacturers' instructions or the standard operation procedures (Table S2-4). Genomic coordinates were based on the hg19 reference sequence. The reference materials were required to be treated as real clinical samples that should be tested only once and the allelic frequencies for each mutation were needed.
Statistical analysis
The ddPCR assay was performed in quadruplicate and the results were assessed by one-way ANOVA test for calculation of uniformity of each mutation. The differences between the results of ddPCR and ARMS-PCR were analyzed by Wilcoxon signed rank test. The general performance of NGS-based cancer IVDs detecting the reference material at a particular allelic frequency was evaluated by calculating the CV of the results of all participants testing the same sample. The CV was defined as the ratio of the standard deviation to the mean expressing the dispersion of the distribution of allelic frequencies detected by the participants. One sample t-test was performed to examine the differences of allelic frequencies detected by the NGS assay compared with those by the ddPCR assay. Significance was defined when P value was less than 0.05. Statistical analyses and data visualization were performed using SPSS v19.0, GraphPad Prism v5.0, and SigmaPlot v13.0.
Results
Preparation, validation, and quantification of reference materials
In this study, two FFPE and three plasma reference materials were prepared mimicking the real clinical FFPE tissue and blood plasma samples. For the preparation of FFPE candidates, different ratios of A549, H1650, and H1975 cells were mixed and fixed with paraformaldehyde. When 1% of H1650 and H1975 cells (2x103 cells) were mixed with of A549 cells (2x105 cells), the final allelic frequency as determined by ddPCR for 19del, L858R and T790M were 1.18%, 1.61%, and 1.70%, respectively. While when 5% of mutant cells (1x104 cells) were mixed with of A549 cells (2x105 cells), the final allelic frequency for 19del, L858R and T790M were 5.09%, 7.19%, and 7.30%, respectively (Table 2). The ratio of cell mixing and the final allelic frequency were similar. For the preparation of plasma candidates, different amount of mutant PCR fragments with similar length as natural ctDNA, around 150bp to 200bp, were spiked into pooled plasma. The ddPCR assay was performed for validation and quantification of each candidate in quadruplicate. The results showed that we established five reference materials of different allelic frequencies for three EGFR mutations 19del, L858R, and T790M (Figure 1). The allelic frequencies of FFPE-Q1, FFPE-Q2, Plasma-Q0, Plasma-Q1, and Plasma-Q2 were shown in Table 2. The Plasma-Q0 was natural blood plasma without spiking of the EGFR mutated PCR fragments. It served as a reference material with low allelic frequency to precisely evaluate the performance of the NGS-based cancer IVD. No significant differences were observed among repeated measurements of allelic frequencies of each mutations, ANOVA P=0.26 for 19del, ANOVA P=0.16 for L858R, and ANOVA P=0.19 for T790M, respectively, indicated that allelic frequencies of reference materials were stably determined by the ddPCR assay.
Table 2.
Results of determination of allelic frequencies of each reference materials by different methods.
| Method | Mutation | Variant allelic frequency (%) | ||||
|---|---|---|---|---|---|---|
| Plasma-Q0 | Plasma-Q1 | Plasma-Q2 | FFPE-Q1 | FFPE-Q2 | ||
| ddPCR | 19del | 0.35 ± 0.13 | 0.85 ± 0.26 | 1.31 ± 0.40 | 1.18 ± 0.14 | 5.09 ±0.37 |
| L858R | 0.53 ± 0.16 | 1.20 ± 0.43 | 1.89 ± 0.20 | 1.61 ± 0.36 | 7.19 ± 0.71 | |
| T790M | 0.03 ± 0.06 | 0.61 ± 0.05 | 1.43 ± 0.36 | 1.70 ± 0.29 | 7.30 ± 0.37 | |
| ARMS-PCR | 19del | 0.32 ± 0.09 | 0.49 ± 0.09 | 0.65 ±0.09 | 0.61 ± 0.06 | 3.10 ± 0.42 |
| L858R | 0.27 ± 0.09 | 0.93 ± 0.44 | 1.94 ± 0.65 | 1.25 ± 0.18 | 5.26 ± 1.23 | |
| T790M | 0.02 ± 0.02 | 0.23 ± 0.08 | 0.50 ± 0.14 | 0.66 ± 0.08 | 3.31 ± 0.50 | |
| NGS | ||||||
| Participant #1 | 19del | 0.58 | 1.55 | 2.17 | 1.34 | 5.59 |
| L858R | 1.02 | 1.85 | 3.43 | 2.38 | 7.33 | |
| T790M | 0.82 | 1.90 | 4.25 | 1.91 | 7.55 | |
| Participant #2 | 19del | †ND | 0.23 | 0.96 | 1.46 | 4.42 |
| L858R | 0.65 | 0.94 | 2.01 | 1.44 | 6.84 | |
| T790M | 0.24 | 0.60 | 1.51 | 2.04 | 7.09 | |
| *Participant #3 | 19del | 0.78 | 0.60 | 1.45 | / | / |
| L858R | 0.57 | 1.53 | 3.25 | / | / | |
| T790M | 0.71 | 0.92 | 2.75 | / | / | |
| Participant #4 | 19del | 0.20 | 0.55 | 1.70 | 1.24 | 3.64 |
| L858R | 1.79 | 3.10 | 2.48 | 1.85 | 7.66 | |
| T790M | 0.23 | 1.59 | 4.81 | 2.19 | 7.05 | |
| Participant #5 | 19del | 0.20 | 0.46 | 0.77 | 0.89 | 2.62 |
| L858R | 0.40 | 1.02 | 2.09 | 1.60 | 6.38 | |
| T790M | 0.23 | 0.49 | 1.31 | 1.48 | 5.18 | |
| Participant #6 | 19del | 1.09 | 1.04 | 0.49 | 1.55 | 3.04 |
| L858R | 0.86 | 1.64 | 6.09 | 1.51 | 5.35 | |
| T790M | 0.14 | 2.42 | 6.70 | 1.94 | 7.30 | |
| Participant #7 | 19del | †ND | 0.04 | †ND | 0.57 | 1.99 |
| L858R | 1.30 | 2.19 | 2.64 | 1.57 | 8.20 | |
| T790M | 0.51 | 1.07 | 1.81 | 2.10 | 7.85 | |
| Participant #8 | 19del | †ND | †ND | †ND | †ND | 3.64 |
| L858R | 0.26 | 0.72 | 0.96 | 1.18 | 5.02 | |
| T790M | 0.22 | 0.34 | 1.00 | 1.43 | 5.79 | |
* Participant #3 didn't develop appropriate NGS assay to deal with FFPE tissue samples.
† All ND results were confirmed by repeated tests.
The results of the ddPCR and ARMS-PCR are shown as mean ± standard deviation from four times repeated measurements. Every reference material was treated as real clinical sample that was detected by the participants using their own developed NGS-based cancer IVDs. All data are expressed as percentage (%) of allelic frequencies of the EGFR mutations.
Abbreviations: ddPCR, droplet digital polymerase chain reaction; ARMS-PCR, amplification refractory mutation system polymerase chain reaction; NGS, next-generation sequencing; del, deletion; T, threonine; M, methionine; L, leucine; R, arginine; FFPE, formalin-fixed, paraffin-embedded; ND, not detected.
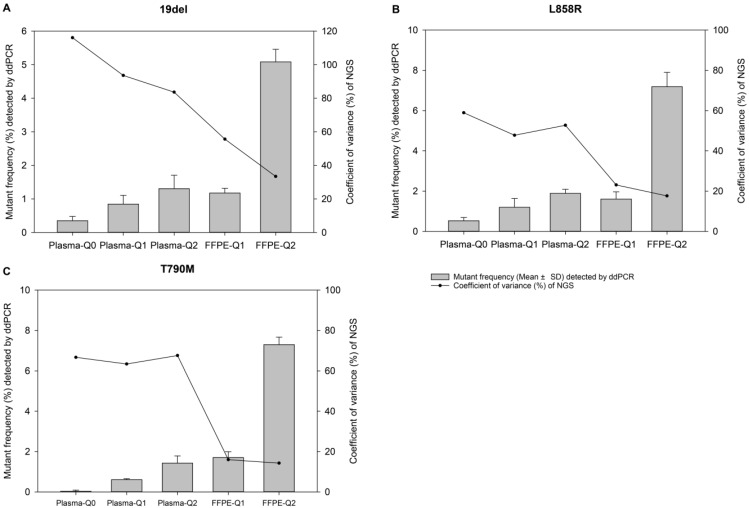
Figure 1.
The relevance of allelic frequency and coefficient of variation. Bars indicate values of mean ± standard deviation of allelic frequencies of reference materials determined by the ddPCR assay in quadruplicate for EGFR (A) 19del, (B) L858R, and (C) T790M mutations. The dots represent the coefficient of variation (%) of the allelic frequencies of reference materials determined by the NGS-based cancer IVDs from the eight participants.
Determination of reference materials by the ARMS-PCR assay
Before distribution for the collaborative evaluation of NGS-based cancer IVDs, the ARMS-PCR assay was performed for determination of variants frequencies of reference materials because it was the most widely used technique in clinical molecular diagnostic. Each reference material was tested in quadruplicate. The allelic frequencies of FFPE-Q1, FFPE-Q2, Plasma-Q0, Plasma-Q1, and Plasma-Q2 were shown in Table 2. No significant differences were observed among repeated measurements of allelic frequencies for each mutation, Wilcoxon P=0.06 for 19del, Wilcoxon P=0.13 for L858R, and Wilcoxon P=0.0625 for T790M, respectively, indicated that allelic frequencies of reference materials were stably determined by the ARMS-PCR assay. These results demonstrated that allelic frequencies determined by the ARMS-PCR assay and the ddPCR assay were comparable, despite the ARMS-PCR results were generally lower than the ddPCR results.
Performance of NGS-based cancer IVDs
After validation and quantification by both ddPCR and ARMS-PCR assay, the FFPE and plasma reference materials were distributed to eight manufacturers to evaluate the performance of their in-house developed NGS-based cancer IVDs (except for participant #3 testing only plasma samples). Four major NGS platforms, NextSeq and HiSeq of Illumia, Ion Proton of ThermoFisher, and BGISEQ-500 of BGI, were used by the participants. The DNA amount of the samples were sufficient for qualified tests (more than 500ng gDNA for each FFPE sample and approximately 100ng cfDNA for each plasma sample). These samples were required to be tested only once to simulate the real clinical application after the sequencing libraries were successfully established. The results showed that 19del mutation failed to be detected by participants #2, #7, and #8 for Plasma-Q0, by participant #8 for Plasma-Q1, and by participant #7 and #8 for Plasma-Q2, respectively (Table 2). The negative results were repeated for confirmation using the new sample to exclude potential operational error. The failure of detection of 19del in Plasma-Q0 and FFPE-Q1 could be due to the low allelic frequencies at 0.32% and 0.61%, respectively. The failure of detection of 19del in Plasma-Q1 and Plasma-Q2 could be due to the mismatch between the designed mimic ctDNA fragments and the primers or probes used by the participants.
For evaluation of repeatability of NGS-based cancer IVDs in detecting different types of samples of different allelic frequencies, CVs of each mutation of each reference material were calculated. It was showed that the CVs of 19del, T790M, and L858R placed from high to low indicating the repeatability of determination of different mutations was L858R>T790M>19del (Table 3). For samples at different allelic frequencies, it was shown that CVs of reference materials decreased as allelic frequency increased (Figure 1). The results demonstrated that the repeatability of determination increased as the allelic frequency of reference material increased. Moreover, the repeatability of determination of FFPE reference materials was better than plasma reference materials (Figure 2).
Table 3.
Coefficient of variations of allelic frequencies of each reference material determined by the NGS-based IVDs
| Reference materials | Coefficient of variations (%) | ||
|---|---|---|---|
| 19del | T790M | L858R | |
| Plasma-Q0 | 106 | 81 | 61 |
| Plasma-Q1 | 89 | 66 | 48 |
| Plasma-Q2 | 81 | 72 | 52 |
| FFPE-Q1 | 58 | 26 | 27 |
| FFPE-Q2 | 30 | 21 | 18 |
Abbreviations: EGFR, epidermal growth factor receptor; del, deletion; T, threonine; M, methionine; L, leucine; R, arginine; FFPE, formalin-fixed, paraffin-embedded.
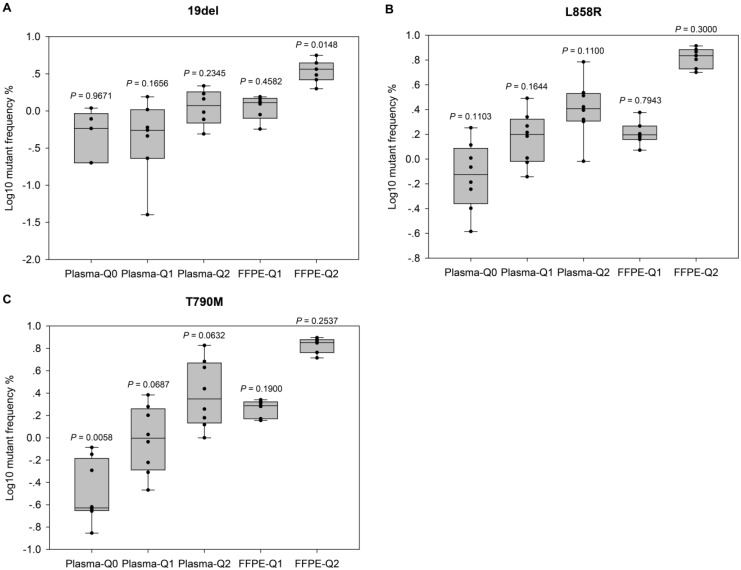
Figure 2.
Evaluation of the performance of NGS-based cancer IVDs. Evaluation of the performance of NGS in detecting EGFR (A) 19del, (B) L858R, and (C) T790M mutations. Allelic frequencies are expressed as Log10 values. Each dot represents the result of a single participant. The inter-quartile ranges are shown as box-and-whisker plot. Values shown above each column indicate the P value of one-sample t-test (with the mean value of ddPCR results as the theoretical value).
For further evaluation of the performance of different NGS-based cancer IVDs in detecting FFPE and plasma samples respectively, one sample t-tests were implemented to examine the differences of allelic frequencies determined by the NGS assays compared with those by the ddPCR assay (Figure 2). The results indicated that most of allelic frequencies detected by the NGS assays were comparable to those by the ddPCR assay (P>0.05), except for that of T790M in Plasma-Q0 (P=0.01) and that of 19del in FFPE-Q2 (P=0.01). The discordance of determination of T790M in Plasma-Q0 could be explained by the limitation of the NGS assay in detecting mutations at extremely low allelic frequency of less than 0.1%. It was noted that allelic frequencies of 19del in FFPE-Q1 and FFPE-Q2 (0.57% and 1.99% respectively) determined by participant #7 were relatively lower than those by other participants. This could probably explain the discordance of 19del in FFPE-Q2. In addition, the results of determination of FFPE reference materials generally showed a smaller inter-quartile range than those of plasma reference materials.
Discussion
Recently, more and more manufacturers developed and provided in-house NGS-based cancer IVDs for determination of cancer-related mutations in FFPE tissues or blood plasma samples. However, their clinical performance was not fully evaluated under a universally accepted standard. Previous studies demonstrated that the consistency of NGS results between companies was discouraging 14,15. The National Institutes for Food and Drug Control (NIFDC) took the responsibility to develop national reference materials for IVD control 16. Therefore we designed this pilot study for the investigation of preparation of reference materials for the quality control of NGS-based cancer IVDs.
Determination of EGFR mutations had important clinical significance. 19del and L858R mutations would predict clinical response to tyrosine kinase inhibitors (TKIs) in non-small cell lung cancers (NSCLC), while T790M mutation could lead to TKIs resistance 17-19. Therefore, these three common EGFR mutations were introduced in our pilot study. The ideal reference material should monitor the whole process of the NGS assay from nucleic acid extraction to result reporting 20. However, most current publicly available reference materials could only monitor a part of testing process. To overcome this limitation, we prepared the FFPE and plasma samples mimicking real clinical samples to fully evaluate the whole process of the NGS-based cancer IVDs.
The allelic frequencies were designed at different levels for different types of reference materials. According to real clinical samples, the lowest allelic frequencies were 0.1% and 1.0% for FFPE and plasma samples respectively. The actual values of allelic frequencies were quantified and validated by the ddPCR assay in quadruplicate, which was a widely accepted method for analyzing samples with low allelic frequencies. Before distribution for the collaborative evaluation, all reference materials were determined by the ARMS-PCR assay since it was the mostly used technique for clinical molecular diagnostic and could serve as the preliminary evaluation of our samples. The results showed that the allelic frequencies determined by the ARMS-PCR were slightly less than that by the ddPCR assay, despite there was no significant difference.
After quantification and validation, the reference materials were used for the collaborative evaluation of performance of the NGS-based cancer IVDs. The performance of the NGS assay was generally acceptable in this study. For the FFPE samples, 9 out of 14 tests had an average depth of more than 1000x. While for the plasma samples, 22 out of 24 tests had an average depth of more than 1000x. In general, all three mutations were successfully determined except the 19del in plasma reference materials by some participants. After communication with manufacturers, the negative results probably were caused by the mismatch of the primers or probes against the targets. As spiked mutant fragments were generated by PCR, the sequences were identical. When the primers/probes used by the test did not covered the specific region, 19del could not be identified by the test, which led to the false negative results. Moreover, the results of detecting 19del, L858R, and T790M mutations in FFPE reference materials showed a smaller inter-quartile range than those of plasma samples, suggesting that NGS-based cancer IVDs had a better reliability in detecting FFPE samples. In the other hand, CVs of detecting the plasma samples were generally higher than FFPE samples, and CVs increased as allelic frequency decreased, indicating that NGS-based cancer IVDs detecting samples at high allelic frequency had a better repeatability than those at lower allelic frequency.
To the best of our knowledge, this is the first study examining the performance of real plasma spike-in as quality control materials for NGS-based cancer IVDs in multi-centers. In contrast to other types of reference materials, real plasma spike-in could better mimic the actual clinical samples. In fact, quality control material that developed base on this study has been announced by the NIFDC, named as “National Reference Materials of Gene Mutations of Lung Cancer Circulating Deoxyribonucleic Acid for Next Generation Sequencing (lot No. 370024-201701)” and is now publicly available to purchase 21. However, as a pilot study, there were still a few limitations in this work. For the preparation of plasma reference materials, only the PCR products containing known mutations were used as spike-in, which could lead to the false negative determination. A better choice for spike-in DNA could be the fragmented gDNA of cell lines containing relevant mutations. Also, considering the cost-effectiveness of preparing FFPE reference materials, cells were only treated with paraformaldehyde, lacking several major steps for FFPE preparation such as dehydration and infiltration with paraffin. In addition, each reference material was required to be tested only once in our collaborative evaluation study because of the attempt to mimic the real clinical testing condition and the high cost. Therefore, this might hinder us from acquiring enough data for comprehensive statistical analysis. It was better to conduct more independent tests in the further study.
In conclusion, we established five reference materials mimicking real clinical FFPE tumor tissue and peripheral blood samples, respectively, which could monitor the whole process of the NGS assay from nucleic acid extraction to result reporting. Then we conduct the collaborative evaluation study for evaluation of performance of NGS-based cancer IVDs detecting both FFPE and plasma samples and our data indicated that their general performance would vary according to different types of samples and different levels of allelic frequencies.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
The authors thank all the manufacturers that participated in this study, which include Annoroad Gene Technology Co., Ltd., Berry Genomics Co., Ltd., BGI-Shenzhen, Genecast Precision Medicine Technology Institute, Geneplus-Beijing, GenoSaber Biotech Co. Ltd., HaploX Biotechnology Co., Ltd., and Novogene Bioinformatics Technology Co., Ltd.
Abbreviations
- 19del
EGFR exon 19 deletion mutation
- ARMS-PCR
amplification refractory mutation system polymerase chain reaction
- cfDNA
cell-free deoxyribonucleic acid
- ctDNA
circulating tumor deoxyribonucleic acid
- CV
coefficient of variation
- ddPCR
droplet digital polymerase chain reaction
- EGFR
epidermal growth factor receptor
- FFPE
formalin-fixed, paraffin-embedded
- gDNA
genomic deoxyribonucleic acid
- IVDs
in vitro diagnostic tests
- L858R
EGFR exon 21 L858R substitution mutation
- NGS
next-generation sequencing
- NIFDC
The National Institutes for Food and Drug Control
- NSCLC
non-small cell lung cancers
- T790M
EGFR exon 20 T790M substitution mutation
- TKIs
tyrosine kinase inhibitors.
References
- 1.Vogelstein B, Papadopoulos N, Velculescu VE. et al. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieg-Bourne CC, Millis SZ, Piccioni DE. et al. Next-generation sequencing in the clinical setting clarifies patient characteristics and potential actionability. Cancer Res. 2017;77:6313–20. doi: 10.1158/0008-5472.CAN-17-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–51. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Endrullat C, Glökler J, Franke P. et al. Standardization and quality management in next-generation sequencing. Appl Transl Genom. 2016;10:2–9. doi: 10.1016/j.atg.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aziz N, Zhao Q, Bry L. et al. College of American Pathologists' laboratory standards for next-generation sequencing clinical tests. Arch Pathol Lab Med. 2014;139:481. doi: 10.5858/arpa.2014-0250-CP. [DOI] [PubMed] [Google Scholar]
- 6.Rehm HL, Bale SJ, Bayrak-Toydemir P. et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–47. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gargis AS, Kalman L, Berry MW. et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012;30:1033–6. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sims DJ, Harrington RD, Polley EC. et al. Plasmid-based materials as multiplex quality controls and calibrators for clinical next-genreation sequencing assays. J Mol Diagn. 2016;18:336–49. doi: 10.1016/j.jmoldx.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lih CJ, Sims DJ, Harrington RD. et al. Analytical Validation and Application of a Targeted Next-Generation Sequencing Mutation-Detection Assay for Use in Treatment Assignment in the NCI-MPACT Trial. J Mol Diagn. 2016;18:51–67. doi: 10.1016/j.jmoldx.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin G, Zhang K, Han Y. et al. Quality control materials for pharmacogenomic testing in the clinic. Clin Chem Lab Med. 2017;55:926–33. doi: 10.1515/cclm-2016-0755. [DOI] [PubMed] [Google Scholar]
- 11.Duncavage EJ, Abel HJ, Pfeifer JD. In silico proficiency testing for clinical next-generation sequencing. J Mol Diagn. 2017;19:35–42. doi: 10.1016/j.jmoldx.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Zhang R, Peng R, Li Z. et al. Synthetic circulating cell-free DNA as quality control materials for somatic mutation detection in liquid biopsy for cancer. Clin Chem. 2017;63:1465–75. doi: 10.1373/clinchem.2017.272559. [DOI] [PubMed] [Google Scholar]
- 13.Yang L, Yang JL, Byrne S. et al. CRISPR/Cas9-Directed Genome Editing of Cultured Cells. Curr Protoc Mol Biol. 2014;107:31.1.1–17. doi: 10.1002/0471142727.mb3101s107. [DOI] [PubMed] [Google Scholar]
- 14.Kuderer NM, Burton KA, Blau S. et al. Comparison of 2 commercially available next-generation sequencing platforms in oncology. JAMA Oncol. 2017;3:996–8. doi: 10.1001/jamaoncol.2016.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torga G, Pienta KJ. Patient-paired sample congruence between 2 commercial liquid biopsy tests. JAMA Oncol. 2017 doi: 10.1001/jamaoncol.2017.4027. DOI: 10.1001/jamaoncol.2017.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CFDA: Beijing, China. Provisions for In-vitro Diagnostic Reagent Registration, in Decree No.5 of China Food and Drug Administration. Revised 30 July 2014. http://eng.sfda.gov.cn/WS03/CL0768/144300.html.
- 17.Paez JG, Jänne PA, Lee JC. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 18.Pao W, Miller V, Zakowski M. et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl AcadSci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi S, Boggon TJ, Dayaram T. et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 20.FDA: Silver Spring, MD, USA. Use of Standards in FDA Regulatory Oversight of Next Generation Sequencing (NGS)-Based In vitro Diagnostics (IVDs) Used for Diagnosing Germline Diseases-Draft Guidance for Stakeholders and Food and Drug Administration Staff. Revised 8 July 2016. https://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidanguidancedoc/ucm509838.pdf.
- 21.NIFDC: Beijing, China. Lists of National Standard Materials and Reference Materials for Registered In vitro Diagnostic Test (4th edition). Revised 1 November 2017. http://www.nifdc.org.cn/bzwz/CL0276/10115.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.