Abstract
The functions of miR-126-mediated signal transducers and activators of the transcription 3 (STAT3) signal pathway were investigated in regulating the behavior of cells in non-small cell lung cancer (NSCLC). Cultured NSCLC A549 cells were transfected with empty, miR-126 overexpression or miR-126 knocked-down expression plasmids. After transfection efficiency verification by reverse transcription polymerase chain reaction (RT-PCR) and culture for 24 h, methyl thiazolyl tetrazolium (MTT) was applied to detect cell proliferation rate, migration distance was measured in scratch assays, cell cycle was determined through flow cytometry, the mRNA expression level of caspase-3 in cells was detected using RT-PCR and protein expression levels of STAT3 were detected using western blotting. Our results showed the cell proliferation rate was significantly higher in cells of the overexpression group than that in those of the control group (p<0.05) and the rate in the cells of the low-expression group was the lowest among the three groups (p<0.05). The migration distance of the overexpression group cells was significantly longer than that in the control group cells and the shortest migration distance was found in the low-expression group cells (p<0.05). The amount of cells in mitotic phase in the overexpression group was significantly higher than that in the control group and the same amount in the low-expression group was the lowest (p<0.05). The mRNA expression level of caspase-3 of cells in the overexpression group was significantly lower than that of cells in the control group and the highest expression level was found in the low-expression group (p<0.05). Finally, the protein expression levels of STAT3 in cells in the overexpression group were significantly lower than those in the control group and the highest expression levels were identified in the low-expression group (p<0.05). Based on our findings, the cancer-promoting miR-126 can mediate the activation of the STAT3 signal pathway to regulate the malignant biological behavior of NSCLC cells affecting their proliferation, migration, cycle and apoptosis susceptibility.
Keywords: miR-126, STAT3 signal pathway, non-small cell lung cancer, malignant biological behavior
Introduction
Lung cancer is the number one malignant tumor threatening the life of human beings. The survival chance of patients is strongly diminished even after surgical resection, neoadjuvant chemotherapy, postoperative routine enhanced chemotherapy and radiotherapy, or targeted gene therapy (1) and this is mainly due to the still mostly unknown pathogenesis of lung cancer and the lack of effective early-stage detection methods. Molecular research suggests the cancer is caused by the coeffect of external environment and the genetic changes (2). Epigenetic modifications, including the methylation of DNA and histone modification, inactivation of chromosome X and the regulation of non-coding RNA (miRNAs) are major factors increasing the occurrence of tumors. miRNAs regulate the transcription and translation of effector proteins in approximately 30% of the total mRNA pool and thus interfere in the physiological and pathological behavior of cells (3). Bioinformatic surveys have shown that miR-126 is abnormally expressed in many malignant tumors, such as lung, breast and colorectal cancers and it is also involved in a variety of biological behavior, including cell proliferation, differentiation, invasion, metastasis and apoptosis (4,5). Clinical studies have also confirmed miR-126 is highly expressed in non-small cell lung cancer (NSCLC) patients, who invariably suffer from poor clinical prognoses (6). Signal transducers and activators of the transcription 3 (STAT3) transcription factor protein can promote cell proliferation and inhibit cell apoptosis and the STAT3 signal pathway is also activated by many extracellular signals, such as non-receptor tyrosine kinase, cytokines and growth factors. Once STAT3 is delivered to the nucleus, it binds with the promoters on the target genes of vascular epithelial growth factor (VEGF) and the relative receptor (VEGFR), as well as the epithelial growth factor (EGF) and the relative receptor (EGFR); thereby regulating cell proliferation, angiogenesis and apoptosis and directly influencing the occurrence and progression of tumors (7,8). In this study, hypothesized miR-126 is involved in induction of the STAT3 signal pathway regulating the malignant behavior of NSCLC cells. Our results identified new potential targets for NSCLC intervention.
Materials and methods
Experimental materials
A549 cells purchased from Sangon Biotechnology (Shanghai, China) were cultivated in RPMI-1640 media containing 10% fetal bovine serum (FBS; Invitrogen-Life Technologies, Carlsbad, CA, USA) in a CO2 incubator (HyClone, Logan, UT, USA), and the medium was refreshed every other day. Each culture was terminated upon 85% confluence of cells by digestion using trypsin. Cells were harvested and washed using phosphate-buffered saline (PBS) and cell suspensions were prepared (2×106 cells/ml) and split for subsequent experiments.
Amplification of plasmids
Empty miR-126 overexpression and low-expression plasmids were purchased from Applied Biosystems (Foster City, CA, USA) and E. coli DH5α was purchased from Bio-Rad Laboratories (Hercules, CA, USA). Amplification was carried out for bacteria containing different plasmids followed by centrifugation at 10,000 × g for 5 min to precipitate the bacteria. Then, 250 µl ZL-I/RNAse A mixture was added followed by vibration to fully suspend the cells into solution. ZL-II (250 µl) was subsequently added to the cell suspension to obtain a clear cell lysate. Next, 350 µl ZL-III buffer was added to produce a white flocculent sediment centrifuged at 10,000 × g for 10 min. A Mu-Pu plasmid micro-separation column (Corning Inc., Corning, NY, USA) was used to extract the supernatant for centrifugation at 10,000 × g for 5 min and the flow-through was discarded. Then, 500 µl ZL buffer was added followed by centrifugation at 10,000 × g for 5 min and DNA was obtained after washing with 720 µl ethanol buffer. The substrate in the column was dried through centrifugation at 10,000 × g for 5 min. Finally, 100 µl sterile deionized water was added and the plasmid was eluted with a last centrifugation at 10,000 × g for 5 min.
G418 experiment
A549 cells were inoculated onto 12-well plates (1 ml/well) and cultivated for 24 h to adhere to the wall. Then the culture medium was removed and in each well, 1-ml RPMI-1640 culture medium amounts containing G418 and 10% FBS were added, in which serial gradient concentrations of G418 were within 100 and 1,000 µg/ml (at 100 µg/ml increments). Cells in a well without G418 were set as the control group. The plates was transferred into the incubator (37°C, 5% CO2) for culture. After the 2nd week of treatment, the screening dosage was that doubling the minimal and effective lethal dosage and the sustaining dosage was set at 200 µg/ml less than the screening dosage.
Cell transfection
Cell transfections were performed in accordance with the manufacturer's instructions in the Lipofectamine 2000 (Invitrogen Life Technologies; Thermo Fisher Scientific, Inc., Waltham, MA, USA) insert and the transfection efficiency was assayed using reverse transcription polymerase chain reaction (RT-PCR). The primers to amplify a 256 bp fragment of miR-126 were synthesized by Beijing Zhongshan Golden Bridge Biotechnology (Beijing, China): Forward 5′-GCCAGTCAGATGTGGATGAA-3′ and reverse, 5′-CCCAACACTGGCACCAGTAA-3′. The primers for a 225 bp fragment of reduced glyceraldehyde-phosphate dehydrogenase (GAPDH) to use as the internal gene reference were: Forward, 5′-CGCGAGAAGATGACCCAGAT-3′ and reverse, 5′-GCACTGTGTTGGCGTACAGG-3′. An RT-PCR kit (Takara, Otsu, Japan) was used to synthesize cDNA and the PCR amplification system was set as follows: 5 µl cDNA + 2 µl 10X Ex Buffer + 1.6 µl dNTP mixture (10 mM) + 0.2 µl polymerase Ex Taq HS + 1 µl upstream primer and 1 µl downstream primer and the system was diluted to 20 µl by adding water. The amplification conditions were set up with an initial 94°C for 5 min denaturation step; then 35 cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec; and a final extension at 72°C for 5 min. Each PCR product was identified by 2% agarose gel electrophoresis and the images obtained by ultraviolet spectrometry were analyzed by a gel imaging system. Digital photographs were used for analysis of gray values.
Determination of cell cycle status, proliferation, migration capabilities and apoptosis susceptibility
After 24 h cultures, methyl thiazolyl tetrazolium (MTT) from R&D Systems (Minneapolis, MN, USA) was applied to detect cell proliferation rate using a standard assay protocol. Cell migration distance was measured performing conventional scratch assays viewed under a microscope from Olympus (Tokyo, Japan). The cell cycle status of cells was determined through flow cytometry, using a FACSCaliber flow cytometer from BD Biosciences (Franklin Lakes, NJ, USA). The caspase-3 mRNA expression levels in cells were detected using standard RT-PCR methods (see below) and the STAT3 protein expression levels were detected using western blotting assays as detailed below.
Detecting the mRNA expression level of caspase-3 through RT-PCR
Primer sequences of caspase-3 were synthesized by Beijing Zhongshan Golden Bridge. The primer sequences are: Forward, 5′-GGTTTCATCCAGGATCGAGCAGG-3′ and reverse, 5′-ACAAAGATGGTCACGGTCTGCC-3′, yielding a 325 bp fragment. GAPDH: Forward, 5′-CGCGAGAAGATGACCCAGAT-3′ and reverse, 5′-GCACTGTGTTGGCGTACAGG-3′, amplifying a 225 bp fragment. The reaction system consisted of 2 µl cDNA + 1 µl upstream primer and 1 µl downstream primer + 0.5 µl polymerase Ex Taq HS + 1 µl 10X Ex Buffer and then was diluted to 25 µl. Reaction conditions were set as follows: 95°C for 5 min; 30 cycles of 95°C for 30 sec, 72°C for 30 sec and 72°C for 60 sec; with a final extension at 72°C for 5 min.
Detecting the protein expression level of STAT3 through western blot analysis
Western blotting experiments were carried out according to the kit manufacturer's instructions. Mouse anti-human STAT3 and internal reference β-actin monoclonal antibodies were used as the primary antibodies (dilution, 1:2,000; cat. nos. sc-71792 and sc-130300) and a goat anti-mouse immunoglobulin G polyclonal antibody as the secondary antibody (dilution, 1:500; cat. no. sc-362267). All antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). The total protein extraction kit was purchased from Kangchen Biotech (Shanghai, China), a bicinchoninic acid (BCA) kit for assay of protein concentration and an enhanced chemiluminescent (ECL) kit were purchased from Guangdong Bochuan Biotechnology (Guangdong, China). The Lab Works 4.5 gel imaging software was purchased from Invitrogen Life Technologies.
Statistical analysis
SPSS 20.0 software (IBM Corp., Armonk, NY, USA) was used for statistical analyses. The measurement data are presented as mean ± standard deviation (SD). One-way analysis of variance was adopted for intergroup comparisons and least significant difference test (LSD-t) was performed for paired comparisons. P<0.05 was considered to indicate a statistically significant difference.
Results
Cell proliferation rates in the three groups
In the overexpression group, the cell proliferation rate was significantly higher than that in the control group (p<0.05) and the rate in the low-expression group was the lowest among the three groups (p<0.05); the difference had statistical significance (Fig. 1).
Figure 1.
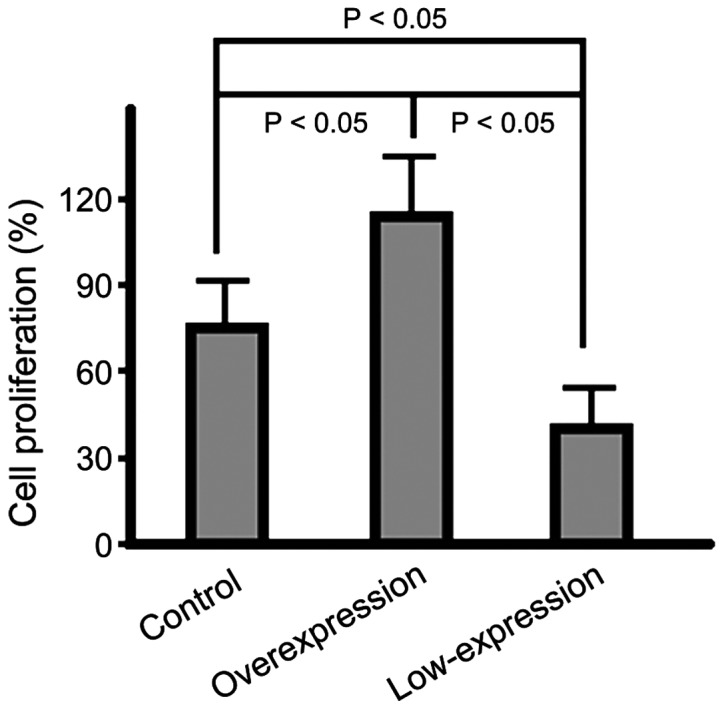
Graph depicting the differences in MTT cell proliferation rates in each group. MTT, methyl thiazolyl tetrazolium.
Distance of cell migration in scratch assays in the three groups
The migration capabilities of the cells in the overexpression group were significantly increased compared to those of cells in the control group. The cells that migrated the shortest distance were those in the low-expression group (p<0.05) (Fig. 2).
Figure 2.
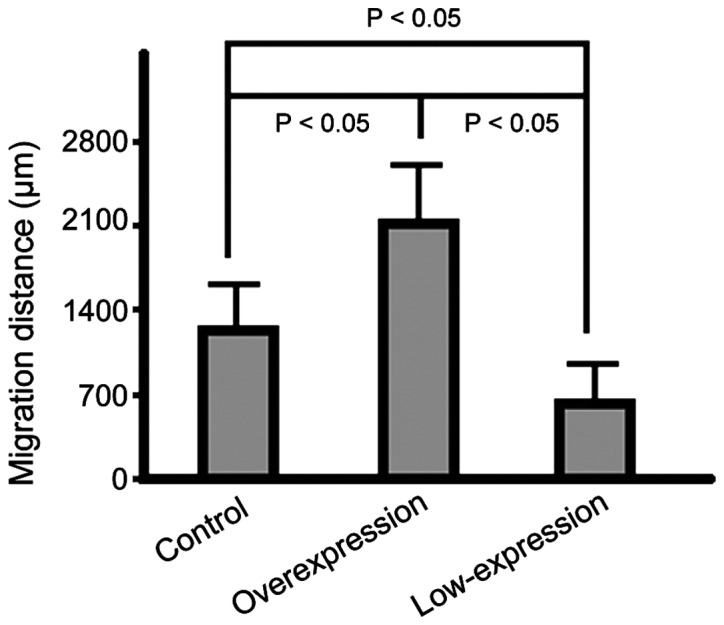
Graphic representation of differences in the distance of cell migration obtained by scratch assays in each group.
Cell cycle status of cells in the three groups
The percentage of cells in mitotic phase in the overexpression group was significantly higher than that in the control group and the percentage in the low-expression group was the lowest (p<0.05) (Fig. 3).
Figure 3.
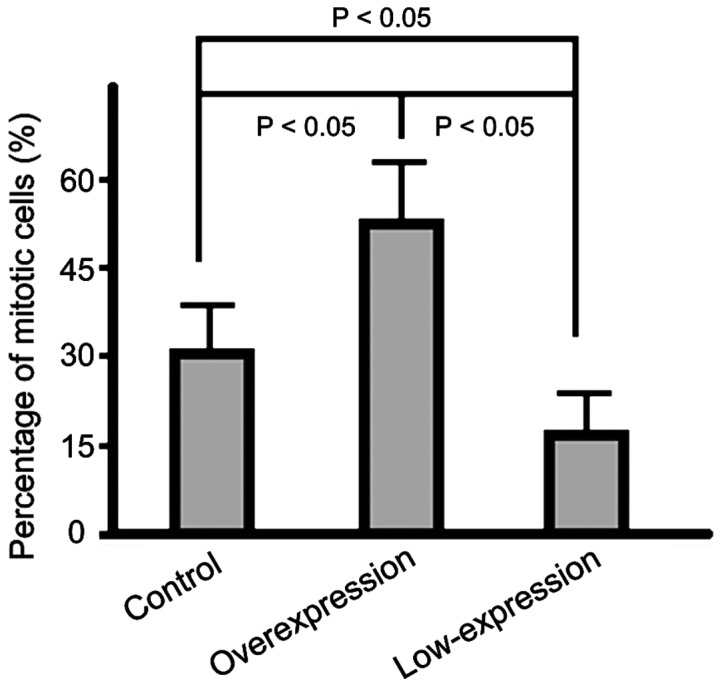
Graphic showing the percentages of cells in mitotic phase in each of the studied groups determined by flow cytometry.
mRNA expression level of caspase-3 in cells of the three groups
The mRNA expression levels of caspase-3 of cells in the overexpression group were significantly lower than those in the control group cells and the highest expression level was found in the cells of the low-expression group (p<0.05) (Fig. 4).
Figure 4.
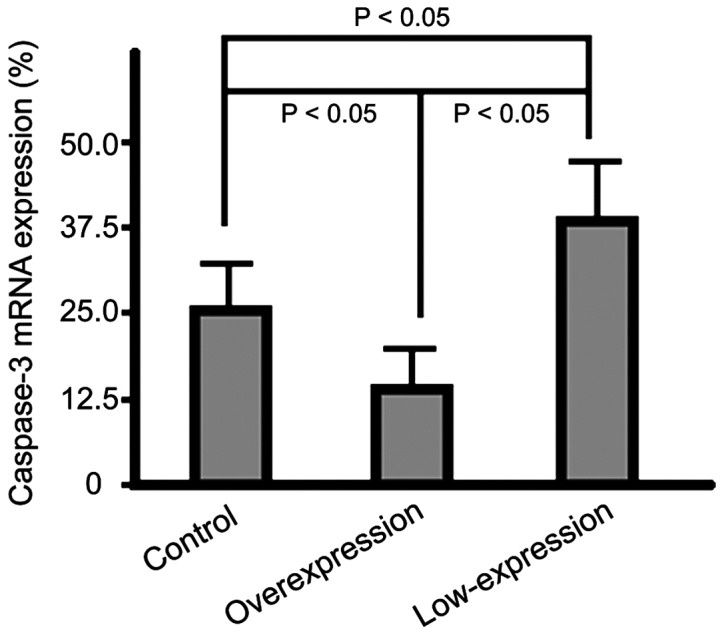
Graph showing average caspase-3 mRNA expression levels in each of the studied groups after RT-PCR. RT-PCR, reverse transcription polymerase chain reaction.
Protein expression level of STAT3 in cells
The STAT3 protein expression in cells of the overexpression group were significantly lower than those of cells in the control group and the highest expression level was identified in the low-expression group cells (p<0.05) (Fig. 5).
Figure 5.
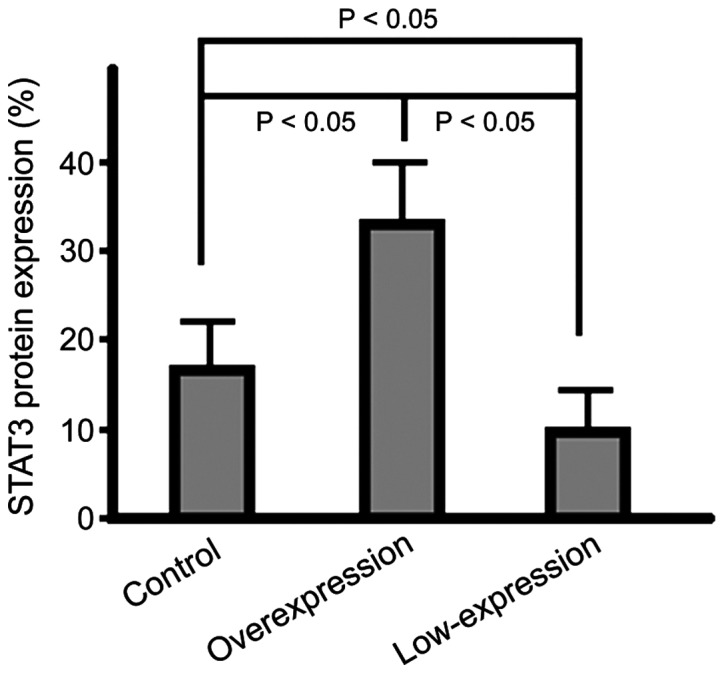
Graphic representation of the average STAT3 protein expression levels in each group through quantitative western blot analysis. STAT3, signal transducers and activators of the transcription 3.
Discussion
The miRNAs are high evolutionarily conserved non-coding single-chain nucleotide chains with lengths of 18 to 24 nucleotides, that can bind to proteins to form RNA-induced silencing complexes (RISCs) that degrade mRNAs or silence their expression through direct mRNA binding interactions (9). Each miRNA can regulate multiple target genes and potentially all of the genetic pathways are regulated by miRNAs. The miR17/20 regulate the interleukin-8, the miR17-92 regulate the p21 and miR181 regulates the B-cell lymphoma-2 (Bcl-2) pathways, thereby exerting a major regulatory role in the occurrence and progression of malignant tumors (10,11).
It has been reported that the expression of miR-126 varies during different stages of development in mouse and human pulmonary tissues and that it is mainly expressed in the endothelial cells of lung tissues and the epithelial cells of bronchi (12).
Through this study, we found that compared with the control group, the cell proliferation rate in cells in miR-126 overexpression group was significantly elevated, the distance of cell migration was extended, the ratio of cells in mitotic phase was increased, the mRNA expression level of caspase-3 was remarkably reduced and the protein expression level of STAT3 was elevated. Furthermore, the results in the low-expression group were contrary to those in the overexpression group and the differences had statistical significance. These results suggest that miR-126, is a cancer-promoting gene, which can mediate the activation of the STAT3 signaling pathway and thus regulate the malignant behavior of NSCLC. Nevertheless, a different study suggested that miR-126 may have cancer suppressing effects (13) and the fact that miR-126 can suppress the expression of target genes SPRED1 and PIK3R2 (negatively regulating the Erk and PIK3R2 signal pathways), that it can inhibit expression of chemotactic factor CXCL12 and block the apoptosis of vascular endothelial cells indicates that miR-126 could regulate embryonic development and tissue growth and sustain the vascular homeostasis. The apparent contradiction with our results may be associated with the variance between the in vivo and in vitro environment of tumor cells and the characteristics of expression in different types of tumor cells. For example, another study confirmed that in peripheral blood of patients with acute myelogenous leukemia, the level of miR-126 expression is elevated due to the decrease in methylation in its promoter region, leading to inhibition of cell apoptosis (14), a consequence more in agreement with our own findings.
It has been shown that STAT3 expression in lung cancer tissues is significantly higher than that in normal para-carcinoma tissues and that the expression levels significantly correlate with the differentiation, lymphatic metastasis and TNM staging of NSCLC tumors (15). Even more, the STAT3 signaling pathway is closely associated with the persistent activation of EGFR (16). Importantly, Tang et al (17) showed that the expression of STAT3 in EGFR-mutant lung cancer cells is significantly higher than that in the wild-type cells and the sensitivity to the target therapy of gefitinib is also improved. EGFR, is a member of the erbB family (a receptor of tyrosine protein kinase, TPK), involved in the occurrence of tumors through activation of multiple downstream target genes by enabling the acidification site of tyrosine at the C terminal domain of the STAT3 protein (18,19).
In conclusion, abnormal expression of miR-126 is closely associated with the malignant behavior of NSCLC cells and the STAT3 signaling pathway is probably the main target leading to that effect. Thus, futher studies should confirm our results that suggest intervening in the activity or expression of miR-126 and STAT3 may be an effective treatment against NSCLC.
Acknowledgements
Not applicable.
Funding
This study was supported by the Shandong Provincial Natural Science Foundation, China (ZR2016HM80).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
ZZ contributed significantly to writing the manuscript and cell culture. JW analyzed and interpreted RT-PCR. JC performed and analyzed western blotting. XY performed G418 experiment and helped the conception of the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Fang W, Ruan W. Advances in surgical treatment of early stage non-small cell lung cancer. Asian Pac J Surg Oncol. 2017;2:1–10. [Google Scholar]
- 2.Lee PN, Forey BA, Coombs KJ. Systematic review with meta-analysis of the epidemiological evidence in the 1900s relating smoking to lung cancer. BMC Cancer. 2012;12:385. doi: 10.1186/1471-2407-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leidinger P, Galata V, Backes C, Stähler C, Rheinheimer S, Huwer H, Meese E, Keller A. Longitudinal study on circulating miRNAs in patients after lung cancer resection. Oncotarget. 2015;6:16674–16685. doi: 10.18632/oncotarget.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Del Vescovo V, Grasso M, Barbareschi M, Denti MA. MicroRNAs as lung cancer biomarkers. World J Clin Oncol. 2014;5:604–620. doi: 10.5306/wjco.v5.i4.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tai HC, Chang AC, Yu HJ, Huang CY, Tsai YC, Lai YW, Sun HL, Tang CH, Wang SW. Osteoblast-derived WNT-induced secreted protein 1 increases VCAM-1 expression and enhances prostate cancer metastasis by down-regulating miR-126. Oncotarget. 2014;5:7589–7598. doi: 10.18632/oncotarget.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu W, Zhou K, Zha Y, Chen D, He J, Ma H, Liu X, Le H, Zhang Y. Diagnostic value of serum miR-182, miR-183, miR-210, and miR-126 levels in patients with early-stage non-small cell lung cancer. PLoS One. 2016;11:e0153046. doi: 10.1371/journal.pone.0153046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun CC, Li SJ, Zhang F, Zhang YD, Zuo ZY, Xi YY, Wang L, Li DJ. The novel miR-9600 suppresses tumor progression and promotes paclitaxel sensitivity in non-small-cell lung cancer through altering STAT3 expression. Mol Ther Nucleic Acids. 2016;5:e387. doi: 10.1038/mtna.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu F, Dai C, Fu Y, Loo JF, Xia D, Gao SP, Ma Z, Chen Z. Physalin A exerts anti-tumor activity in non-small cell lung cancer cell lines by suppressing JAK/STAT3 signaling. Oncotarget. 2016;7:9462–9476. doi: 10.18632/oncotarget.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou J, Meng F, Chan LW, Cho WC, Wong SC. Circulating plasma MicroRNAs as diagnostic markers for NSCLC. Front Genet. 2016;7:193. doi: 10.3389/fgene.2016.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu Z, Willmarth NE, Zhou J, Katiyar S, Wang M, Liu Y, McCue PA, Quong AA, Lisanti MP, Pestell RG. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci USA. 2010;107:8231–8236. doi: 10.1073/pnas.1002080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong J, Zheng S, Zhang L, Wang Y, Meng J. Induction of K562 cell apoptosis by As4S4 via down-regulating miR181. Med Sci Monit. 2017;23:144–150. doi: 10.12659/MSM.899214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, Lan H, Huang X, Liu B, Tong Y. MicroRNA-126 inhibits tumor cell growth and its expression level correlates with poor survival in non-small cell lung cancer patients. PLoS One. 2012;7:e42978. doi: 10.1371/journal.pone.0042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin Z, Zhang Y, Li Y, Lv T, Liu J, Wang X. Prognostic significance of STAT3 expression and its correlation with chemoresistance of non-small cell lung cancer cells. Acta Histochem. 2012;114:151–158. doi: 10.1016/j.acthis.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Shi Y, Yuan Q, Liu X, Yan B, Chen L, Tao Y, Cao Y. Epstein-Barr virus encoded LMP1 regulates cyclin D1 promoter activity by nuclear EGFR and STAT3 in CNE1 cells. J Exp Clin Cancer Res. 2013;32:90. doi: 10.1186/1756-9966-32-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang J, Guo F, Du Y, Liu X, Qin Q, Liu X, Yin T, Jiang L, Wang Y. Continuous exposure of non-small cell lung cancer cells with wild-type EGFR to an inhibitor of EGFR tyrosine kinase induces chemoresistance by activating STAT3. Int J Oncol. 2015;46:2083–2095. doi: 10.3892/ijo.2015.2898. [DOI] [PubMed] [Google Scholar]
- 18.Jaganathan S, Yue P, Paladino DC, Bogdanovic J, Huo Q, Turkson J. A functional nuclear epidermal growth factor receptor, SRC and Stat3 heteromeric complex in pancreatic cancer cells. PLoS One. 2011;6:e19605. doi: 10.1371/journal.pone.0019605. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Lo HW, Cao X, Zhu H, Ali-Osman F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 2010;8:232–245. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.


