Abstract
Genetic testing has been a routine part of paediatic medicine for decades. Over time, the number of genetic tests available for children presenting with features thought to be explained by an underlying genetic aetiology has expanded considerably. Genome-wide sequencing approaches (e.g., whole-exome sequencing, whole-genome sequencing) are now emerging as the most comprehensive approaches to genetic diagnosis that we have seen to date; multiple serial tests that were once required for a child under diagnostic investigation can now be accomplished in a single assay. Moreover, the performance of this single assay appears to be superior to the sum of its parts. Despite this promise, technical, ethical and access-related complexities require considerable attention prior to the implementation of these tools in mainstream paediatrics. To ready paediatricians for the eventual transition to genome-based diagnostics, herein we review both the elements and delivery considerations of this emerging technology.
Keywords: Diagnostic testing, Genome-wide sequencing, Paediatrics, Rare disease
Paediatric Progress: How should it change your practice? is new Paediatrics & Child Health column that contains summaries of recent developments that have practical implications for clinicians. For more information, visit www.cps.ca/en/pch/manuscripts
WHAT’S NEW?
Genome-wide sequencing approaches are emerging as powerful diagnostic tools in paediatrics. Compared to 10% to 15% detection rates associated with conventional genetic testing, genome-wide sequencing yields a diagnosis in 30% to 40% of the patients.
Despite its diagnostic potential, analytic challenges and ethical controversies remain.
Paediatricians will be called upon to initiate appropriate referrals to genetics for children with complex clinical presentations of suspected genetic aetiology and understand the types of results that can emerge genome-wide sequencing.
WHY THE PRIMER?
Genetic diseases are individually rare, but common in aggregate, particularly in paediatrics (1). Up to 34% of paediatric hospitalizations are for children with a genetic disorder and genetic disease is a common cause of disability and death in children (2). The diagnosis of genetic disease is an essential part of clinical care—while often not leading directly to a specific treatment for the child, a diagnosis is important to exclude treatable disorders, provide accurate recurrence risks for family members, inform prognosis and end the diagnostic odyssey (3,4). Genome-wide sequencing approaches are emerging as a powerful diagnostic tool. For example, compared to chromosome microarray, the currently recommended first-tier test for children with developmental delay/intellectual disability (5), genome-wide sequencing achieves a threefold increase in diagnostic yield (6). Unlike the traditional approach of ordering multiple serial genetic tests, this improved diagnostic yield is achieved in a single assay. As such, genome-wide sequencing will change the way the diagnostic process for a suspected rare genetic disease is delivered in Canada. While these changes are beginning to take place in tertiary care genetics centres, it will be some time before test ordering capabilities will be available to community-based paediatricians. Overcoming complexities related to timing of and indication for testing, interpretation, ethics and access is warranted. However, an understanding of the current landscape of genome-wide diagnostics is necessary to prepare for the receipt of increasingly sophisticated genomic sequencing reports on patients referred to tertiary genetics centres. Here, we provide a brief overview of the basic and more complex features of genome-wide sequencing.
CASE EXAMPLE
Patient A is a 7-year-old nondysmorphic boy with medically intractable epilepsy with onset at age 4 months. He presented with nonfebrile status epilepticus repeatedly between ages of 4 and 8 months and failed five antiepilepsy medications. He stopped seizing altogether after starting the ketogenic diet, but remained significantly developmentally delayed. He has autistic features with a nonverbal, nonambulatory phenotype. Over a 6-year period, extensive genetic and metabolic testing using traditional methods was performed (e.g., chromosome microarray, Prader-Willi/Angelman testing, epilepsy gene panel, Rett syndrome sequencing, SCN1A, CDKL5, ATP7A sequencing, plasma amino acids, urine organic acids). Given the high probability of a genetic cause, genome-wide sequencing was performed and a single mutation in the phosphatidylinositol glycosylation protein A (PIGA) gene was identified in the patient and his mother. These findings are consistent with the diagnosis of X-linked recessive phosphatidylinositol glycosylation protein A deficiency (PIGA deficiency) (7).
GENOME-WIDE SEQUENCING: GENERATING THE SEQUENCE
Next-generation sequencing is a contemporary technology that performs sequencing of millions of small fragments of DNA in parallel. Of the entire genome, only 1% codes for proteins; this 1% is called the exome. The exome is organized into ~22,000 genes and is thought to contain 85% of known or potential disease-causing variants. In contrast, an individual’s entire DNA content (protein coding and noncoding regions) is called the genome. Genome sequencing refers to sequencing the entire genetic code of a person and exome sequencing refers to sequencing only the parts of the genome that contain protein-coding genes; both are forms of genome-wide sequencing. Neither type of sequencing is readily available, clinically, in Canada yet. However, efforts are currently underway to deliver publically funded exome sequencing for specific indications in various provinces (8). Genome sequencing is only available on a research basis in Canada and most other countries.
For both approaches, the laboratory process begins with extracting DNA from cells. After extraction, the DNA is broken into short fragments (i.e., 100 to 150 base pairs) and the fragments are put through a process called library preparation. For exome sequencing, an additional enrichment procedure is needed to ‘capture’ only the protein-coding information contained within the exons. The sequencing instrument ‘reads’ the genetic code of these short sequences multiple times in parallel. Using bio-informatics tools, these short sequence reads are aligned and matched to specific positions in the human genome reference sequence. A computerized annotation of the patient’s genotype (consisting of nucleic acids labelled A, T, C, G) at each position in the exome or genome is then created and compared to the reference genome. Similarities and differences between the patient’s sequence and the reference sequence can be identified (4,9,10).
GENOME-WIDE SEQUENCING: INTERPRETING THE VARIANTS
Once analytic accuracy is verified, an interpretation team needs to determine which variants in the patient’s genome may be clinically significant. While generating the raw sequence data is ‘hypothesis free’, understanding its meaning requires clinically-derived hypotheses that reflect possible associations between the patient’s phenotype and potentially relevant variants in genes that are identified in the individual’s sequence data. For this reason and at this time, variant interpretation is best conducted by a multidisciplinary team that includes a bio-informatician (i.e., develops variant pipeline), a genomicist (i.e., analyzes detected variants using relevant software) and a medical geneticist or other subspecialist (i.e., expertise in rare diseases and direct knowledge of the patient being analyzed) (11). First, the raw sequence data are filtered for rare variants (i.e., variants seen in <1% of the population). An exome sequence typically generates ~500 variants whereas a genome sequence can generate up to 20,000 (6). Based upon the patient’s clinical features and the suspected mode of inheritance (i.e., or determined mode of inheritance via parental testing), the variant interpretation team then searches the published literature and various databases of genomic variation (e.g., Human Gene Mutation Database, ClinVar, Leiden Open-source Variation Database) for evidence of an association between suspected genes and the clinical presentation of the patient (4,9–11).
Based upon the current American College of Medical Genetics (ACMG) variant classification system (12), genome-wide sequencing can generate the following categories of results: (i) primary diagnosis, (ii) possible diagnosis, (iii) uninformative test, (iv) dual diagnosis, (v) predictive secondary variant and (vi) pharmacogenomic variant (Table 1).
Table 1.
Types of results that can be generated by genome-wide sequencing
| Variant type | Clinical example |
|---|---|
| Primary diagnosis | Detection of sequence-level variant in the NSD1 gene that is diagnostic of Sotos syndrome, in the presence of Marfan-like features (6) |
| Possible diagnosis | Detection of one sequence level variant in the ZFYVE26 gene in a child with spastic paraplegia. Homozygous or compound heterozygous mutations in this gene are associated with autosomal recessive spastic paraplegia type 15. However, with only one variant detected, the molecular diagnosis cannot be confirmed. Further investigations could identify a second variant (e.g., a deletion) which would confirm a diagnosis (12). |
| Uninformative test result | Detection of biallelic variants in TRIT1 gene in a child with microcephaly, profound developmental delay, hypotonia, epilepsy, and brain anomalies. This gene has never been reported to be a disease-causing gene in humans. The identification of genotypically and phenotypically similar children combined with more extensive analyses identified that mutations in this gene explain the phenotype among these children (14). |
| Dual diagnosis | Detection of a variant in the ITPR1 gene responsible for spinocerebellar ataxia type 29 in a 2-year-old female presenting with ataxia, motor, and language delay. Pathogenic mutation was inherited from her father. At age 5 she presented with seizures which had never been reported to be associated with SCA 29. Her mother had a childhood diagnosis of Landau-Kleffner syndrome with seizures. Analysis revealed a de novo pathogenic mutation in the GRIN2A gene in the proband and her mother, known to be associated with Landau- Kleffner syndrome. The family was counseled regarding the 2 separate autosomal dominant diseases that were identified in the proband, each inherited from a different affected parent (21). |
| Predictive risk result | Detection of sequence-level variant in the KCNH2 gene which is associated with risk for Long QT syndrome. Potential for medical actionability would typically prompt a laboratory to report this variant (22,23). |
| Pharmacogenomic result | Detection of polymorphisms in cytocrome P450 (CYP) enzymes (CYP2C9, CYP2C19) that can lead to differences in serum concentrations and anti-epileptic drug clearance with a greater risk of concentration-dependent adverse effects (30). |
A primary diagnosis refers to a variant that is identified in a disease-causing gene that is the likely cause of the child’s health problem(s). The detection rate for primary diagnoses using genome-wide sequencing approaches 30% to 40% for children with developmental delay and congenital anomalies (6,7).
A possible diagnosis occurs when variants of uncertain significance (VUS) are identified; variants that neither confirm nor disconfirm a genetic aetiology for a set of presenting clinical features (12). VUS have posed longstanding challenges in molecular genetics but with the emergence of genome-wide sequencing, these variants are identified at greater frequencies (9–11). The ACMG advises that certain variant characteristics be used to guide decision making about the pathogenicity of a VUS. These include the mutation type, the frequency of the variant in control and patient databases, its predicted pathogenicity based upon in silico computer programs and its inheritance pattern (13). For example, if a variant is not inherited from a parent (so is de novo) and is associated with a dominant condition, pathogenicity is favoured. Large-scale data repositories of variants as well as novel research tools are emerging as additional strategies for characterizing these variants, the latter including RNA expression, enzyme analysis, protein localization and animal modeling (9,10).
An uninformative test result occurs when no variants are detected, when a variant is detected that is not relevant to the child’s presenting features, or when a variant is detected that has not been reported to be associated with human disease (14). Children who receive uninformative results might benefit from re-analysis of their sequence data at a later point in time or may benefit from access to research initiatives that aim to ‘solve the unsolved’ (e.g., Care4Rare Canada, Matchmaker Exchange, Undiagnosed Disease Network) (15–17). By sharing genotypic and phenotypic data through international data platforms, the likelihood of finding a patient ‘match’, leading to a shared and understandable diagnosis increases substantially. As a result of such international collaboration, gene discovery for rare disease has occurred at a rapid pace over the past 5 years (18,19). As such, re-interrogating an individual’s ‘null’ sequence may generate new information, even after only 1 year. For example, upon re-evaluating the exomes of 40 ‘unsolved’ cases, a genetic diagnosis was identified in 10%, due to new gene discoveries over the course of one preceding year (20). Children for whom a possible diagnosis is received may also be good candidates for sequence re-analysis.
A dual diagnosis occurs when mutations in more than one disease-causing genes are identified and each variant is thought to provide a partial explanation of the individual’s composite presentation. Dual diagnoses have been reported in 4% to 15% of the patients (6,21).
A predictive secondary variant, also known as an incidental finding, refers to variants that are medically actionable but unrelated to the primary indication for testing. While the rate at which these are detected depends upon the bioinformatics filter used, it is estimated that secondary variants will be identified in up to 3.5% of children when the ACMG’s recommended 59-gene list is interrogated (22,23). Mixed views on whether these variants should be actively sought and reported has resulted in conflicting professional guidance on this issue (4,24,25). In part, the controversy stems from varied views on the ethics of proactively searching for unsolicited information (26,27) and in part it stems from the absence of robust evidence on the actual positive predictive value of the secondary variants themselves (e.g., mutation of equivocal association with longQT syndrome detected in the absence of relevant family history) (11,28). This controversy is particularly charged with respect to children, given that traditional guidance in genetics recommends against predispositional testing in children (29). Some experts in the field retain the traditional view of preserving the child’s autonomy related to knowing adult-relevant health information while others advocate that it is in the best interest of the child to generate this information so that parents and other implicated relatives can make preventive health care decisions (26). Against this backdrop, the ACMG recommends that laboratories actively search for and report variants in 59 genes (i.e., associated with hereditary cancer syndromes and cardiac dieases) but that parents and children should be given a choice about whether to receive results on variants deteced in these dynamics (24). In contrast, the European Society of Human Genetics (ESHG) recommends that laboratories analyze only sequence data relevant to the primary indication for testing to reduce the likelihood of generating secondary variants. Where inadvertently identified, however, ESHG advises that medically actionable variants should be reported to the family (25). Similar to the ESHG, the Canadian College of Medical Genetics (CCMG) does not endorse the intentional interrogation of a list of secondary genes, but recommends that if identified childhood-onset medically actionable mutations be reported and that competent adults be offered a choice about receiving secondary variants related to themselves (4).
A pharmacogenomic result occurs when a variant associated with a known drug response is identified (30,31). Genome-wide pharmacogenomics testing has been proposed as a tool for pre-emptive screening to provide anticipatory guidance to families. In one cohort, 95 of 98 children had at least one clinically actionable pharmacogenomic variant, suggesting that pre-emptive screening may in fact act as a patient safety measure embedded within diagnostic genome-wide sequencing for children (31).
GENOME-WIDE SEQUENCING: ANALYTIC LIMITATIONS
Certain analytic limitations of genome-wide sequencing warrant elaboration. First, coverage of the exome/genome using next-generation sequencing technology is estimated to be 85% to 92% (9,10,32). As well, variability in depth of coverage across the exome and genome leads to the possibility of gaps in sequencing and missing or uninterpretable data (32). While the diagnostic yield estimates for exome sequencing are robust (~30%), expansions, structural rearrangement and mutations in regulatory or intergenic regions of the genome cannot be detected (10). For genome sequencing, diagnostic yield estimates suggest it is slightly higher (6,33), but certain regions remain uninterpretable and expansions greater than sequence read lengths remain undetectable (e.g., expansions associated with Fragile X syndrome) (9,10,32). In addition, disease-causing variants reported in the literature and in large-scale databases can be incorrect; error rates have been reported to range from 4% to 23% (34–36). Current software tools for predicting variant pathogenicity should not be used in clinical decision making (37), and functional studies that characterize the effect of the variant on gene expression or protein–protein interaction, for example, are generally not available in a clinical setting (38).
GENOME-WIDE SEQUENCING: IMPLICATIONS FOR PRACTICE
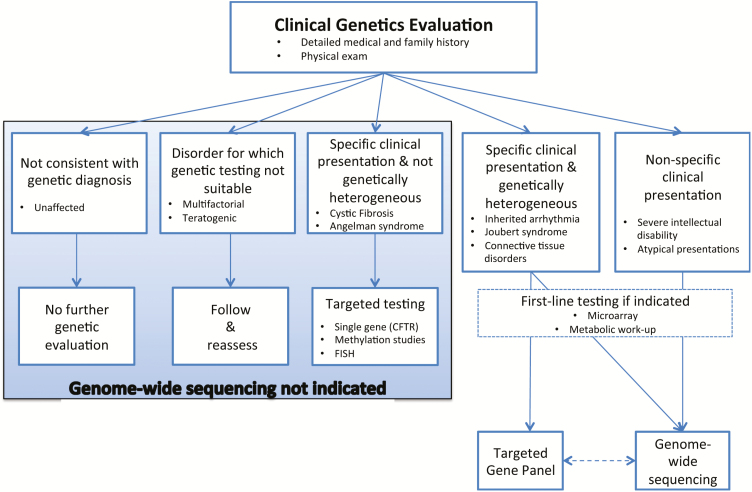
Guidance is emerging to assist clinicians with determining appropriate clinical indications and timing for genome-wide sequencing. With respect to clinical indications, Table 2 presents broad factors that increase the likelihood of identifying a molecular cause for a given condition using genome-wide sequencing (4). Currently, the diagnostic yield using genome-wide sequencing is highest for specific conditions that are known to be genetically heterogeneous (e.g., ataxia) or for cases of nonspecific and unexplained clinical presentation (e.g., moderate-to-severe intellectual disability) (4,32). At the time of CCMG’s position statement on the clinical use of these tools, there was insufficient evidence to recommend their use for children with nonsyndromic autism, learning disabilities and neuropsychiatric disease without additional features, or in the prenatal setting (Figure 1) (4). With respect to timing, a recent prospective study investigated the diagnostic yield and cost of exome sequencing in 44 Australian children at various timepoints in their diagnostic trajectory. The total health care expenditure on all diagnostic testing conducted on this cohort was A$568141(US$430873). The cost per patient of the standard diagnostic pathway (i.e., without exome sequencing) was A$9901 (US$7509). The cost per patient of the standard diagnostic pathway plus exome sequencing was A$12 912 (US$9792). However, exome sequencing performed at initial tertiary presentation had the lowest cost per patient (A$5186 [US$3933]), followed by exome sequencing performed at the first genetics appointment (A$7047[US$5347]) (39).
Table 2.
Factors that increase the likelihood of monogenic disease and/or facilitate the interpretation of genome-wide data
| Family history | Similarly affected individuals |
| Recognizable pattern of inheritance | |
| Consanguinity | |
| Phenotype | Severity of phenotype |
| Specificity of clinical presentation (e.g., neuropathy, metabolic disease) | |
| Clinical interpretation | Careful patient phenotyping (e.g., detailed physical exam, imaging, chemistry) |
| Normal chromosomal microarray analysis and other relevant laboratory testing | |
| Exclusion of acquired causes (e.g., infection) |
Data taken from ref. (4).
Figure 1.
Decision aid to facilitate the diagnostic evaluation of patients with rare disease of suspected monogenic aetiology. This decision aid highlights where genome-wide sequencing may prove useful in the evaluation process. The conditions listed in each box are representative examples only. For specific clinical presentations associated with genetic heterogeneity, the decision regarding the use of a targeted panel versus genome-wide sequencing is dependent on a number of factors, including the availability of the testing options and the yield of such panels. Patients with negative targeted gene panels may benefit from subsequent clinical genome-wide sequencing. Conversely, consideration of a targeted panel subsequent to uninformative clinical genome-wide sequencing would be dependent on the depth of coverage achieved in the latter instance. NB: Fluorescence in situ Hybridization has now been replaced by multiplex ligation-dependent probe amplification in many centres. Reproduced with permission from ref. (4).
Currently, professional guidelines stipulate that genome-wide sequencing can only be ordered by clinicians with specific training in genetics (4,32). Table 2 and Figure 1 can guide paediatricians’ thinking about appropriate indications for referral. Prior to ordering genome-wide sequencing tests themselves, academic and nonacademic paediatricians will require proficiencies in conducting clinical genetics evaluations, understanding the indications for and strengths and limitations of genome-wide sequencing approaches, result interpretation and strategies for cascade testing in family members (4,32). In the not too distant future, efforts to enhance paediatricians’ capacity for ordering genome-wide sequencing themselves will be warranted. When this time comes, we anticipate that providing diagnostic evaluation for phenotypically and genotypically complex cases, disease and disease-modifier gene discovery and the application of these data to tailor patient care will remain the core business of medical genetics experts.
While genome-wide sequencing promises to be a powerful tool for identifying the causes of genetic diseases in children and Canadian nondiscrimination legislation is favourable (40), interpretation, availability and ethics remain core challenges. However, a robust understanding of the aforementioned components of genome-wide sequencing approaches will equip paediatricians to play an active role in genome diagnostics when current analytic and ethical challenges resolve and accessibility improves, guiding patients and their families into the era of genomic medicine.
Research Ethics Board: Not applicable
Work Originated: Hospital for Sick Children Research Institute
References
- 1. Yoon PW, Olney RS, Khoury MJ, Sappenfield WM, Chavez GF, Taylor D. Contribution of birth defects and genetic diseases to pediatric hospitalizations. A population-based study. Arch Pediatr Adolesc Med 1997;151(11):1096–103. [DOI] [PubMed] [Google Scholar]
- 2. McCandless SE, Brunger JW, Cassidy SB. The burden of genetic disease on inpatient care in a children’s hospital. Am J Hum Genet 2004;74(1):121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nathaniel RH. It does matter: The importance of making the diagnosis of a genetic syndrome. Curr Opin Pediatr 2006;18(6):595–7. [DOI] [PubMed] [Google Scholar]
- 4. Boycott K, Hartley T, Adam S et al. ; Canadian College of Medical Geneticists The clinical application of genome-wide sequencing for monogenic diseases in canada: Position statement of the canadian college of medical geneticists. J Med Genet 2015;52(7):431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller DT, Adam MP, Aradhya S et al. . Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010;86(5):749–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stavropoulos DJ, Merico D, Jobling R et al. . Whole-genome sequencing expands diagnostic utility and improves clinical management in paediatric medicine. NPJ Genom Med 2016;1:15012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Joshi C, Kolbe DL, Mansilla MA, Mason SO, Smith RJ, Campbell CA. Reducing the cost of the diagnostic odyssey in early onset epileptic encephalopathies. Biomed Res Int 2016;2016:6421039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Working Group commissioned by the Ontario Genetic Testing Advisory Committee. Use of Genome Wide Sequencing for Undiagnosed Rare Genetic Disease in Ontario http://www.health.gov.on.ca/en/pro/programs/gtac/docs/gtac_report_use_of_gws_for_undiagnosed_rare_genetic_diseases.pdf (Accessed June 15, 2017).
- 9. Warman Chardon J, Beaulieu C, Hartley T, Boycott KM, Dyment DA. Axons to exons: The molecular diagnosis of rare neurological diseases by next-generation sequencing. Curr Neurol Neurosci Rep 2015;15(9):64. [DOI] [PubMed] [Google Scholar]
- 10. Thiffault I, Lantos J. The challenge of analyzing the results of next-generation sequencing in children. Pediatrics 2016;137(Suppl 1):S3–7. [DOI] [PubMed] [Google Scholar]
- 11. Bowdin S, Gilbert A, Bedoukian E et al. . Recommendations for the integration of genomics into clinical practice. Genet Med 2016;18(11):1075–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goizet C, Boukhris A, Maltete D et al. . SPG15 is the second most common cause of hereditary spastic paraplegia with thin corpus callosum. Neurology 2009;73(14):1111–9. [DOI] [PubMed] [Google Scholar]
- 13. Richards S, Aziz N, Bale S et al. ; ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genet Med 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kernohan KD, Dyment DA, Pupavac M et al. ; Care4Rare Consortium Matchmaking facilitates the diagnosis of an autosomal-recessive mitochondrial disease caused by biallelic mutation of the tRNA isopentenyltransferase (TRIT1) gene. Hum Mutat 2017;38(5):511–6. [DOI] [PubMed] [Google Scholar]
- 15. Care4Rare Canada. Children’s Hospital Eastern Ontario Research Institute http://care4rare.ca/ (Accessed June 15, 2017).
- 16. Matchmaker Exchange http://www.matchmakerexchange.org/ (Accessed June 15, 2017).
- 17. Undiagnosed Disease Network https://undiagnosed.hms.harvard.edu/ (Accessed June 15, 2017).
- 18. Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: Discovery to translation. Nat Rev Genet 2013;14(10):681–91. [DOI] [PubMed] [Google Scholar]
- 19. Sawyer SL, Schwartzentruber J, Beaulieu CL et al. ; FORGE Canada Consortium Exome sequencing as a diagnostic tool for pediatric-onset ataxia. Hum Mutat 2014;35(1):45–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wenger AM, Guturu H, Bernstein JA, Bejerano G. Systematic reanalysis of clinical exome data yields additional diagnoses: Implications for providers. Genet Med 2017;19(2):209–14. [DOI] [PubMed] [Google Scholar]
- 21. Balci TB, Hartley T, Xi Y et al. . Debunking Occam’s razor: Diagnosing multiple genetic diseases in families by whole-exome sequencing. Clin Genet 2017. 92(3):281–9. [DOI] [PubMed] [Google Scholar]
- 22. Kalia SS, Adelman K, Bale SJ et al. . Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the american college of medical genetics and genomics. Genet Med 2017;19(2):249–55. [DOI] [PubMed] [Google Scholar]
- 23. Berg JS, Amendola LM, Eng C et al. ; Members of the CSER Actionability and Return of Results Working Group Processes and preliminary outputs for identification of actionable genes as incidental findings in genomic sequence data in the clinical sequencing exploratory research consortium. Genet Med 2013;15(11):860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Green RC, Berg JS,Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O’Daniel JM, Ormond KE, Rehm HL, Watson MS, Williams MS, Biesecker LG;American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van El CG, Cornel MC, Borry P et al. ; ESHG Public and Professional Policy Committee Whole-genome sequencing in health care. Recommendations of the european society of human genetics. Eur J Hum Genet 2013;21(Suppl 1):S1–5. [PMC free article] [PubMed] [Google Scholar]
- 26. Anderson JA, Hayeems RZ, Shuman C et al. . Predictive genetic testing for adult-onset disorders in minors: A critical analysis of the arguments for and against the 2013 ACMG guidelines. Clin Genet 2015;87(4):301–10. [DOI] [PubMed] [Google Scholar]
- 27. Anderson JA, Meyn MS, Shuman C et al. . Parents perspectives on whole genome sequencing for their children: Qualified enthusiasm? J Med Ethics. 2017;43(8):535–9. [DOI] [PubMed] [Google Scholar]
- 28. Johnston JJ, Rubinstein WS, Facio FM et al. . Secondary variants in individuals undergoing exome sequencing: Screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet 2012;91(1):97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clayton EW, McCullough LB, Biesecker LG, Joffe S, Ross LF, Wolf SM; Clinical Sequencing Exploratory Research (CSER) Consortium Pediatrics Working Group Addressing the ethical challenges in genetic testing and sequencing of children. Am J Bioeth 2014;14(3):3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Striano P, Vari MS, Mazzocchetti C, Verrotti A, Zara F. Management of genetic epilepsies: From empirical treatment to precision medicine. Pharmacol Res 2016;107:426–9. [DOI] [PubMed] [Google Scholar]
- 31. Cohn I, Paton TA, Marshall CR et al. . Genome sequencing as a platform for pharmacogenetic information: A cohort study in children. NPJ Genomic Medicine (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bowdin SC, Hayeems RZ, Monfared N, Cohn RD, Meyn MS. The sickkids genome clinic: Developing and evaluating a pediatric model for individualized genomic medicine. Clin Genet 2016;89(1):10–9. [DOI] [PubMed] [Google Scholar]
- 33. Lionel ACCG, Monfared N, Walker S et al. . Improved diagnostic yield and comparable coverage to targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Gen in Med 2017. doi: 10.1038/gim.2017.119. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tong MY, Cassa CA, Kohane IS. Automated validation of genetic variants from large databases: Ensuring that variant references refer to the same genomic locations. Bioinformatics 2011;27(6):891–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bell CJ, Dinwiddie DL, Miller NA et al. . Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med 2011;3(65):65ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cassa CA, Tong MY, Jordan DM. Large numbers of genetic variants considered to be pathogenic are common in asymptomatic individuals. Hum Mutat 2013;34(9):1216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 2012;40(Web Server issue):W452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gray VE, Kukurba KR, Kumar S. Performance of computational tools in evaluating the functional impact of laboratory-induced amino acid mutations. Bioinformatics 2012;28(16):2093–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tan TY, Dillon OJ, Stark Z et al. . Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr. 2017. doi:10.1001/jamapediatrics.2017.1755. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bill S-201Genetic Non-Discrimination Act. An Act to Prohibit and Prevent Genetic Discrimination. 2017. https://openparliament.ca/bills/42-1/S-201/ (Accessed Aug 31, 2017). [Google Scholar]