Abstract
Large-intergenic noncoding RNAs (lincRNAs) cooperate with core transcription factors to coordinate the pluripotency network of embryonic stem cells. The mechanisms by which lincRNAs affect chromatin structure and gene transcription remain mostly unknown. Here, we identified that a lincRNA (linc1614), occupied by pluripotency factors at its promoter, was indispensable for both maintenance and acquisition of pluripotency. Linc1614 served as a specific partner of core factor Sox2 in maintaining pluripotency, primarily by mediating the function of Sox2 in the repression of developmental genes. Moreover, Ezh2, an essential subunit of polycomb repressive complex 2 (PRC2), physically interacted with linc1614 and contributed to lincRNA-mediated transcriptional silencing. Thus, we propose that the interplay of linc1614 with Sox2 implicates this lincRNA as a recruitment platform that mediates transcriptional silencing by guiding the PRC2 complex to the loci of developmental genes.
Keywords: lincRNA, Sox2, pluripotency maintenance, PRC2, reprogramming
Introduction
Embryonic stem cells (ESCs) are able to maintain self-renewal and pluripotency in vitro, providing a powerful model system for elucidation of the mechanisms that control cell fate determination (Okita et al., 2007). It is relatively well understood that the gene regulatory circuitry of core transcription factors (Oct4, Sox2, and Nanog) maintains pluripotency by simultaneously maintaining the expression of ESC-specific genes and repressing the expression of developmental genes (Boyer et al., 2005; Niwa, 2007; Jaenisch and Young, 2008; Chen and Daley, 2008; Kim et al., 2008). Somatic cells can be reprogrammed to generate induced pluripotent stem cells (iPSCs), which is useful for clarifying the transcriptional networks involved in pluripotency (Takahashi and Yamanaka, 2006). In particular, the core factor Sox2 primarily interacts with Oct4 to control the transcription network in ESCs (Stefanovic et al., 2009). Recent studies demonstrated that Sox2 participates in the pluripotency regulation independently and also responsibly drives the reprogramming of somatic cells to pluripotent cells. However, it is still challenging to determine the exact partners of Sox2 in the pluripotency maintenance and acquisition.
Long noncoding RNAs (lncRNAs) have recently been found in ESCs and are considered to be novel regulators of the transcriptional networks that maintain pluripotency and control lineage differentiation (Mercer et al., 2009; Guttman et al., 2011). Thus, characterization of these lncRNAs may be of great importance for better understanding the complicated regulation of pluripotency in ESCs. Recent studies have suggested that Sox2 may execute its distinct functions to promote pluripotency (Ng et al., 2012) or to co-regulate neurogenesis (Ng et al., 2013) via associating with lncRNAs. These results suggest that lncRNAs might serve as co-regulators of Sox2 that would determine the regulation of the pluripotency circuitry or lineage commitment. It remains incompletely understood whether lncRNAs functionally participate in the Sox2-mediated transcriptional networks and even mediate the locus specificity of Sox2.
PRC2 is responsible for H3K27me3 modification, which is primarily correlated with gene repression in ESCs (Sparmann and van Lohuizen, 2006). Investigation of genome-wide co-binding targets for PRC2 components and core factors (Oct4, Sox2, and Nanog) has demonstrated that the PRC2 complex is served as the key co-repressor of the core factors in the suppression of developmental genes, which is critical for pluripotency maintenance (Surface et al., 2010). Notably, RNA immunoprecipitation (RIP) sequencing using PRC2 components indicated that nearly one-fifth of the lncRNAs were able to associate with the PRC2 complex in ESCs (Guttman et al., 2009). Recent studies have highlighted the critical roles of lncRNAs in the recruitment of PRC2 complex to repress gene expression (Morris, 2009; Koziol and Rinn, 2010; Ponting et al., 2009). However, the mechanisms by which lncRNAs influence the position of the PRC2 complex at specific gene loci and cooperatively mediate the silencing of these genes in ESCs remain to be fully elucidated.
In the present study, we investigated a lincRNA (linc1614) that was potentially regulated by multiple pluripotency factors in ESCs and identified as a key regulator in both pluripotency maintenance and acquisition. Linc1614 was shown to specifically interact with Sox2 and acted as a key partner mediating the Sox2 function in repressing a large proportion of developmental genes. Investigation of the mechanism underlying this effect showed that Sox2/linc1614 recruited the PRC2 complex to the promoters of co-targeted developmental genes and repressed their expression.
Results
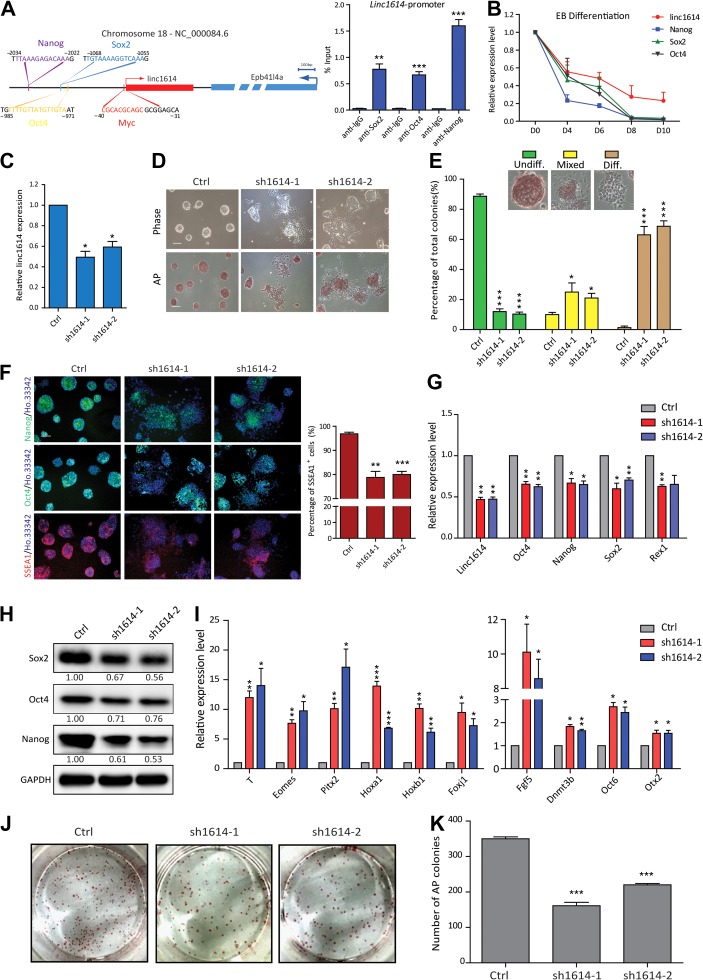
Linc1614 is critical for the pluripotency of ESCs
The core transcription factors (Oct4, Sox2, and Nanog) control the gene regulatory circuitry of pluripotency by simultaneously maintaining the expression of ESC-specific genes and repressing the expression of developmental genes (Boyer et al., 2005; Niwa, 2007; Jaenisch and Young, 2008). Thousands of lincRNAs have been addressed and integrated into the molecular circuitry of ESCs via regulating by key transcription factors (Guttman et al., 2011). To determine the key downstream mediators of core factors in the pluripotency maintenance, we first collected the published ChIP-seq data of transcriptional factors (Oct4, Sox2, Nanog, Klf4, n-Myc, c-Myc, and Tcf3) and comprehensively analyzed the downregulated lincRNAs upon knocking-down of pluripotency factors (Guttman et al., 2011). We preferred the lincRNAs were directly regulated by multiple pluripotency factors and identified a lincRNA (linc1614) transcribed from chromosome 18 (adjacent to the coding gene Epb41l4a), which was co-bound by Myc (−31/−40), Oct4 (−971/−985), Sox2 (−1055/−1068), and Nanog (−2022/−2034) according to bioinformatics prediction (Figure 1A). We further performed ChIP-qPCR assay and verified that the pluripotency factors Oct4, Sox2, Nanog co-bound at the promoter of linc1614 (Figure 1A). Then we performed the RNA hybridization with linc1614 probes to investigate the distribution of linc1614 in undifferentiated and differentiated cells. Our results showed that linc1614 was high-expressed in undifferentiated cells and mainly distributed in the nucleus, which was confirmed by cytoplasmic and nuclear fractionation experiment (Supplementary Figure S1A and B). To explore the exact roles of linc1614 in regulating ESC pluripotency, we first examined the expression level of linc1614 under the differentiation condition, which showed that the expression of linc1614 was gradually decreased along with the core factors (Sox2, Oct4, and Nanog) during the ESC differentiation in embryoid bodies (EBs) (Figure 1B). We further established two independent linc1614-knockdown ESC lines (sh1614-1 and sh1614-2) by introducing shRNAs (Figure 1C), and knockdown of linc1614 had no significant effect on the expression of adjacent gene Epb41l4a (Supplementary Figure S1C). Importantly, the colony morphology of the linc1614-knockdown ESCs was highly dispersed compared with that of the control ESCs (Figure 1D). Then, we evaluated the percentages of three colony states: undifferentiated (undiff.), mixed, and differentiated (diff.). The statistical results showed that knockdown of linc1614 significantly increased the percentages of both the mixed and differentiated colonies while notably decreasing the proportion of undifferentiated colonies (Figure 1E). These results indicated that knockdown of linc1614 led to dysfunction in pluripotency maintenance. Consistent with these findings, the proportion of AP-positive ESC colonies was significantly decreased in parallel with linc1614 knockdown (Figure 1D). Moreover, our results showed that linc1614 knockdown resulted in lower expression of SSEA1, Oct4, and Nanog (Figure 1F), reduced percentage of SSEA1 positive cells (Figure 1F), significantly decreased the expression levels of pluripotency-related genes (Sox2, Oct4, Nanog, etc.) (Figure 1G and H), and upregulated the expression of developmental genes (T, Eomes, Pitx2, etc.) and epiblast stem cell (EpiSC) markers (Fgf5, Dnmt3b, etc.) (Figure 1I). Importantly, these developmental genes including T, Eomes, Pitx2, Hoxa1, Foxj1, and Dnmt3b expressed significantly sooner or higher upon linc1614 knockdown during ESC differentiation (Supplementary Figure S1D). We further performed an in vitro colony formation assay to evaluate the self-renewal ability of the sh1614 ESCs. Compared with the control ESCs, knockdown of linc1614 led to a lower colony formation efficiency (Figure 1J and K). Together, these findings indicated that linc1614 plays critical roles in the pluripotency maintenance of ESCs.
Figure 1.
Linc1614 is critical for the pluripotency of ESCs. (A) Schematic of the mouse linc1614 locus at chr.18 (left). Arrows mark transcription start sites (TSSs), and linc1614 and its adjacent gene Epb41l4a are depicted in red and blue, respectively. Potential binding sites for the transcription factors Myc (−31/−40), Oct4 (−971/−985), Sox2 (−1055/−1068), and Nanog (−2022/−2034) are indicated. ChIP-qPCR for Sox2, Oct4, and Nanog binding at the promoter regions of linc1614 (right). Data were presented as mean ± SD (n = 3). Student’s t-test (**P < 0.01 and ***P < 0.001) was performed relative to IgG. (B) The expression level of linc1614 and core factors (Sox2, Oct4, and Nanog) was decreased on the indicated days (0, 4, 6, 8, and 10) of ESC differentiation in embryoid bodies. (C) qPCR analysis of two distinct shRNAs targeting linc1614 (sh1614-1/sh1614-2) in ESCs. Ctrl represented scramble shRNA with no specific target on the genome. (D) ESC morphology (top) and alkaline phosphatase (AP) activity (bottom) of control and sh1614 ESCs. Scale bar, 100 μm. (E) Statistical analysis of the colony morphology of control and sh1614 ESCs. The colonies were scored into three categories (undifferentiated, mixed, and differentiated) as indicated. Data were presented as mean ± SD from three independent experiments, with >250 individual colonies counted in each independent experiment. (F) Immunofluorescence detection of Oct4, Nanog, and SSEA1 in control and sh1614 ESCs (left). Scale bar, 100 μm. FACS analysis for the percentage of SSEA1-positive cells in control and sh1614 ESCs (right). (G and H) The expression of pluripotency-related genes such as Oct4, Sox2, Nanog, and Rex1 was significantly decreased in sh1614 ESCs, as measured by qPCR (G) and western blotting (H). (I) qPCR analysis of developmental genes such as T, Eomes, Pitx2, Hoxa1, Hoxb1, and Foxj1 and EpiSC markers such as Fgf5, Dnmt3b, Oct6, and Otx2 following the knockdown of linc1614. (J and K) Knockdown of linc1614 attenuated the capability of ESCs to form colonies. The colonies were stained for AP activity on Day 5 of culture under ESC culture conditions. Data were presented as mean ± SD (n = 3). Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001) was performed relative to control ESCs.
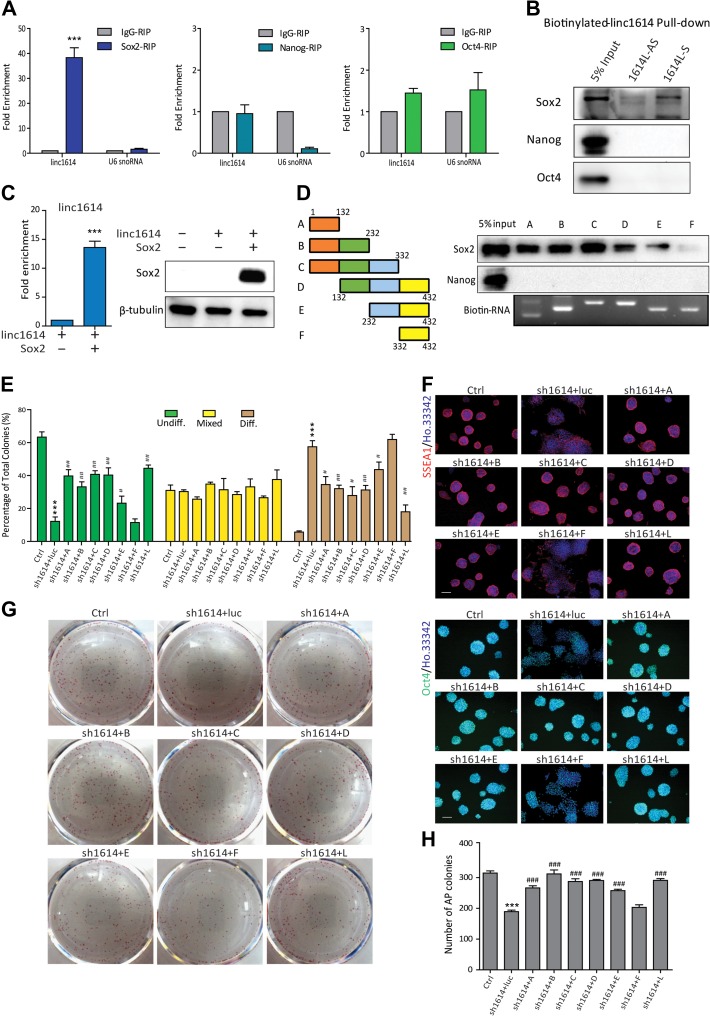
Linc1614 specifically cooperates with Sox2 to maintain pluripotency
The maintenance of pluripotency in ESCs is primarily based on the core regulatory circuitry of transcription factors Oct4, Sox2, and Nanog especially (Boyer et al., 2005; Niwa, 2007). It is still necessary to determine the exact partners of pluripotency factors. We assumed linc1614 potentially served as a novel partner of pluripotency factors and performed an RIP assay followed by qPCR to identify the linc1614-associated core transcription factors in ESCs. Our results showed that linc1614 specifically interacted with the core factor Sox2 but not with Nanog or Oct4 (Figure 2A). To verify these results, we performed biotinylated RNA pull-down experiments to further indicate that linc1614 uniquely enriched the Sox2 protein, but this enrichment was not observed for the Nanog and Oct4 protein (Figure 2B). Moreover, we performed Sox2 and Nanog RIP following exogenous expression of linc1614 with Sox2 or Nanog, respectively, in 293T cells. The results confirmed that linc1614 exclusively bound to Sox2 rather than to Nanog (Figure 2C and Supplementary Figure S1E). We then constructed six linc1614 deletion mutants, including linc1614-A (1–132 nt), linc1614-B (1–232 nt), linc1614-C (1–332 nt), linc1614-D (132–432 nt), linc1614-E (232–432 nt), and linc1614-F (332–432 nt), to map the regions of linc1614 that interacted with Sox2 (Figure 2D). Importantly, we performed a biotin-labeled RNA pull-down assay and identified a 332-nt region in the 5′ region of linc1614 that was essential for interaction with Sox2. Nanog, which was used as a negative control, did not bind to any of the linc1614 deletion mutants (Figure 2D). These results suggested that linc1614 might serve as a specific partner of Sox2 in ESCs.
Figure 2.
Linc1614 specifically cooperates with Sox2 to maintain pluripotency. (A) RIP was conducted to confirm the specific interaction of linc1614 with the core transcription factor Sox2, but not with Nanog or Oct4. U6 snoRNA was indicated as negative control. Data were presented as mean ± SD (n = 3). ***P < 0.001 was performed relative to IgG. (B) Biotinylated linc1614 RNA pull-down showed that linc1614 was associated with Sox2 but not with Nanog or Oct4, as determined by western blotting. Antisense linc1614 (1614L-AS) RNA served as the negative control. (C) RIP analysis for the exogenous expression of linc1614 with Sox2 in 293T cells. Western blotting analysis demonstrated the ectopic expression of Sox2 in 293T cells. Data were presented as mean ± SD (n = 3). ***P < 0.001 was performed relative to control. (D) Schematic representation of the deletion mutants of linc1614 (left). Sox2, but not Nanog, bound to the 5′ region of linc1614 as shown by RNA pull-down experiments (right). (E) Analysis of colony morphology following overexpression of a series of linc1614 deletion mutants in linc1614-knockdown ESCs. The three deletion mutants (linc1614-D, linc1614-E, and linc1614-F) and linc1614 full transcript (linc1614-L) were designed to resist linc1614 shRNAs. More than 250 individual colonies were counted in each of the three independent experiments. (F) Overexpression of linc1614 deletion mutants bound to Sox2 restored the expression levels of these pluripotency genes in linc1614-knockdown ESCs, as determined by immunofluorescence of SSEA1 and Oct4. Scale bar, 100 μm. (G and H) The colony formation assay showed that the self-renewal defect of sh1614 ESCs was significantly rescued by the ectopic expression of the linc1614 fragments that bound to Sox2. Data were presented as mean ± SD (n = 3). Student’s t-test was performed for significance (*,#P < 0.05, **,##P < 0.01, and ***,###P < 0.001). * relative to Ctrl and # relative to sh1614 + luc.
To further define the roles of the Sox2/linc1614 complex in the pluripotency maintenance, we attempted to rescue the phenotypes of ESC colonies upon the knockdown of linc1614 by introducing shRNA-resistant linc1614 mutants into these ESCs (Supplementary Figure S1F). Interestingly, the highly disperse colony morphology observed in the linc1614-knockdown ESCs was significantly rescued by adding the linc1614 mutants that interacted with Sox2. In contrast, the deletion mutant (linc1614-F) that did not bind to Sox2 was not able to reverse the phenotype of the linc1614-knockdown ESCs (Figure 2E and Supplementary Figure S1G). Consistent with these results, the AP activity of the ESCs was also restored by the introduction of linc1614 mutants that associated with Sox2, but not by linc1614-F (Supplementary Figure S1H). These results indicated that the direct interaction of linc1614 and Sox2 was necessary for full reversal of the colony morphology. Further, introduction of the linc1614 mutants that interacted with Sox2 resulted in significant and consistent increases in the protein levels of pluripotency genes, including Oct4 and SSEA1 (Figure 2F). qPCR analysis of pluripotency genes (Nanog and Rex1) and developmental genes (Csf1 and Eomes) showed consistent results (Supplementary Figure S1I and J). More importantly, we performed in vitro colony formation assays which showed that introduction of the linc1614 mutants that could associate with Sox2 led to a higher colony formation efficiency compared with sh1614 ESCs (Figure 2G and H). Taken together, these results demonstrated that linc1614 directly binds with Sox2 to participate in the Sox2-driven pluripotency maintenance in ESCs.
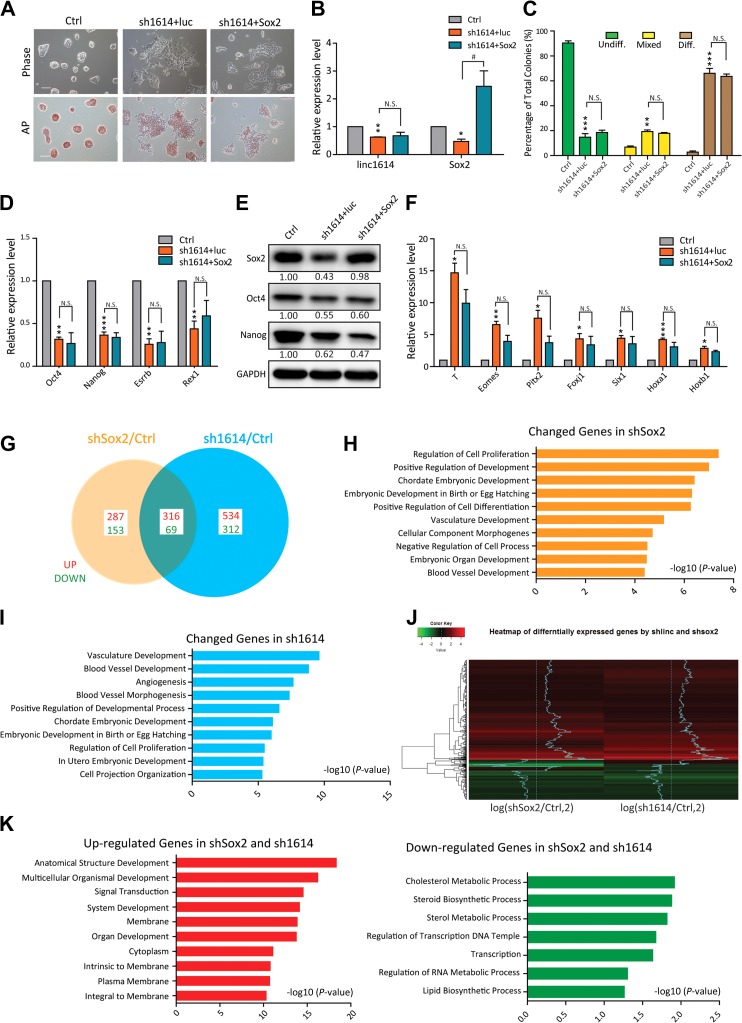
Linc1614 mainly mediates Sox2 function in the repression of developmental genes
The function of the key transcription factor Sox2 is to activate the pluripotency network and repress the expression of developmental genes (Lee et al., 2006; Adachi et al., 2010; Wang et al., 2012). Our data confirmed that knockdown of Sox2 resulted in massive differentiation of ESCs with highly dispersed colony morphology (Supplementary Figure S2A and B), consistent with the loss of pluripotency induced by linc1614 knockdown, and decreased expression of Nanog, Oct4 and linc1614 (Supplementary Figure S2C and D). We firstly introduced exogenous Sox2 into shSox2 cells and our results showed that Sox2 overexpression significantly did reverse the highly dispersed colony morphology and downregulation of Oct4 and Nanog induced by knockdown of Sox2 (Supplementary Figure S2E and F). Consistently, the statistics regarding colony morphology (undiff., mixed, and diff.) were significantly rescued by the introduction of Sox2 (Supplementary Figure S2G). Due to the reduced expression of Sox2 observed in linc1614-knockdown ESCs, it was necessary to determine whether the disperse colony morphology upon linc1614 knockdown was caused by downregulation of Sox2. We then introduced exogenous Sox2 into sh1614 cells and our results showed that Sox2 overexpression did not reverse the highly disperse colony morphology and poor AP activity induced by linc1614 knockdown (Figure 3A and B). Consistently, the statistics regarding colony morphology (undiff., mixed, and diff.) (Figure 3C), the expression levels of both pluripotency genes (Figure 3D and E) and differentiation-related genes (Figure 3F) were not significantly changed by the introduction of Sox2. These results demonstrated that knockdown of linc1614 leads to loss of pluripotency by disrupting the specific interaction with Sox2, rather than by downregulating Sox2 expression.
Figure 3.
Linc1614 mainly mediates Sox2 function in the repression of developmental genes. (A) Representative images following the overexpression of Sox2 in sh1614 ESCs. The cell morphology under bright-field microscopy (top) and AP-stained colonies (bottom) are shown. Scale bar, 100 μm. (B) qPCR analysis showed the restoration of Sox2 expression in linc1614-knockdown ESCs. Data were presented as mean ± SD (n = 3). Student’s t-test was performed for significance (*,#P < 0.05 and **P < 0.01). * relative to Ctrl and # relative to sh1614 + luc. (C) Statistical analysis of the colony morphology for the overexpression of Sox2 in sh1614 ESCs. More than 250 individual colonies were counted in each of the three independent experiments. (D and E) The expression of pluripotency genes upon overexpression of Sox2 in sh1614 ESCs was analyzed by qPCR (D) and western blotting (E). (F) qPCR analysis of the expression of developmental genes upon overexpression of Sox2 in sh1614 ESCs. Data were presented as mean ± SD (n = 3). Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001) was performed relative to Ctrl. n.s. represented no significance. (G) Pairwise comparisons of the number of genes showing changes in regulation (>1.5-fold change compared with that of the control cells) in sh1614 and shSox2 ESCs. The numbers of upregulated (red) and downregulated (green) genes are shown. (H) GO term analysis for genes that were differentially expressed in shSox2 ESCs compared with control ESCs. GO enrichment was assessed using the DAVID gene ontology functional annotation tool. The 10 most significant GO terms are shown. (I) GO term analysis of genes that were differentially expressed in sh1614 ESCs compared with control ESCs. (J) Heat maps of the overlap of the differentially expressed genes following knockdown of Sox2 or linc1614 are shown. The scale indicated the fold change compared with control ESCs. (K) GO term analysis of upregulated and downregulated genes in both shSox2 and sh1614 ESCs are shown.
Sox2 plays pivotal roles throughout the core pluripotency network of ESCs (Wang et al., 2012; Tapia et al., 2015). We next focused on the genome-wide variations that resulted from Sox2 or linc1614 knockdown. We performed microarray analyses of ESCs that were infected with sh1614 or shSox2 viruses for 48 h. The genome-wide gene expression variation data were normalized to the control cells (Figure 3G). Analysis of gene ontology (GO) terms for the genes that were differentially expressed in the shSox2 or sh1614 cells versus the control cells showed that most of the differentially expressed genes were related to various developmental processes and cell differentiation (Figure 3H and I). Notably, approximately half of the differentially expressed genes (385/825) in the Sox2-knockdown ESCs were also dysregulated in sh1614 ESCs (Figure 3G). To further evaluate the reliability of the microarray data, we examined a series of the differentially expressed genes through qPCR (Supplementary Figure S2H and I) and indicated that linc1614 might partner with Sox2 and regulated a common set of genes in ESCs. More importantly, a large proportion of the differentially expressed genes (316/385) were remarkably co-upregulated in linc1614- and Sox2-knockdown ESCs, as shown by the heat map (Figure 3G and J). GO term analyses for these common upregulated genes revealed that these genes were enriched in developmental processes such as anatomical structure development and multicellular organismal development, while the population of genes that were downregulated in both types of cells (69 genes) showed poor enrichment for all terms (Figure 3K). This analysis suggested that the main effect of the interaction of linc1614 with Sox2 might be to repress the expression of large numbers of developmental genes in ESCs.
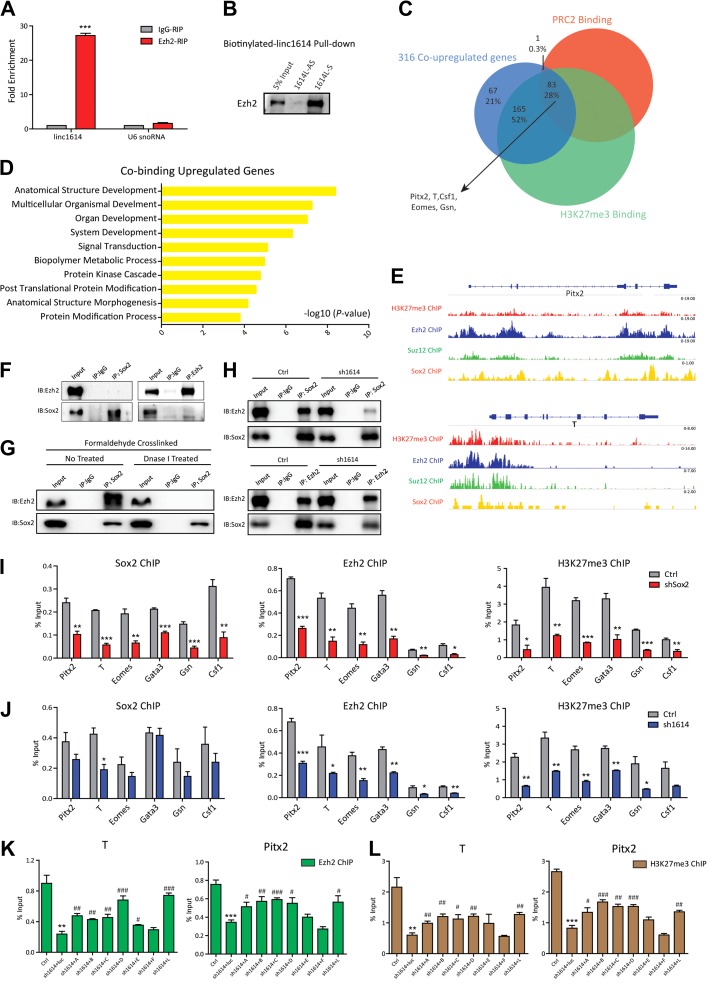
Linc1614/Sox2 recruits the PRC2 complex to suppress the expression of developmental genes
Polycomb group (PcG) proteins contribute to the repression of developmental genes by co-occupying with the core transcription factors Oct4, Sox2, and Nanog, thereby blocking the exit of ESCs from the pluripotent state (Lee et al., 2006; Walker et al., 2010). This raised the possibility that PcG proteins were involved in the linc1614/Sox2 complex in the repression of developmental genes. Accordingly, we performed RIP experiments to explore the relationship between linc1614 and the PRC2 subunits (Ezh2 and Suz12). Our results showed that linc1614 interacted with PRC2 subunits in ESCs (Figure 4A and Supplementary Figure S3A). Purified biotinylated linc1614 RNA also significantly retrieved the Ezh2 protein (Figure 4B). These results suggested that the PRC2 complex directly interacted with linc1614 and might cooperate with the linc1614/Sox2 complex in the regulation of developmental genes. By comparing the genome-wide binding profiles (ChIP-seq) of the PRC2 components (Ezh2 and Suz12) and the corresponding H3K27me3 modification, we found that 83 of the 316 genes, which were upregulated by both sh1614 and shSox2, were occupied by both PRC2 components and H3K27me3. These genes included Pitx2, T, Eomes, Gata3, Gsn, and Csf1 (Figure 4C). GO term analysis for the 83 co-targeted genes showed prominent roles in development processes (Figure 4D). The binding loci for Sox2, Ezh2, Suz12, and H3K27me3 in these genes including Pitx2, T, Csf1, Eomes, and Gata3 (Mikkelsen et al., 2007; Ku et al., 2008; Goren et al., 2010; Lodato et al., 2013) are shown in Figure 4E and Supplementary Figure S3B. Sox2 was previously reported to co-localize extensively with the PRC2 components at the promoter regions of developmental genes (Lee et al., 2006). We performed Co-IP experiments and showed that endogenous Sox2 did not directly interact with Ezh2 (Figure 4F). By using our heterologous system to exogenously expressing Sox2, Ezh2, and linc1614, we further found that exogenously expressed Sox2, Ezh2, and linc1614 did not bind in 293T cells (Supplementary Figure S3C). In addition, formaldehyde-crosslinked Sox2-IP showed that Sox2 associated with the PRC2 component Ezh2 in a DNA-dependent manner (Figure 4G). Furthermore, we performed Sox2 and Ezh2 immunoprecipitation experiments upon fixation in control or linc1614 knockdown ESCs, and the results showed that interaction of Sox2 and Ezh2 was mainly disrupted after linc1614 knockdown (Figure 4H). To further explore whether linc1614 located at these promoters of developmental genes, we introduced the biotin-labeled linc1614 RNA into the ESCs and performed the UV-assisted capture assay. Our results showed that linc1614 was presented at the promoters of co-targeted developmental genes (Supplementary Figure S3D). Subsequently, we performed ChIP assays for Sox2, Ezh2, and H3K27me3 in Sox2-knockdown ESCs followed by qPCR. As expected, Sox2, Ezh2, and H3K27me3 occupancy was significantly decreased at the promoter regions of most developmental genes upon Sox2 knockdown (Figure 4I), whereas there was no significant change in Sox2 enrichment at the promoter regions of developmental genes in shEzh2 ESCs (Supplementary Figure S3E), indicated that Sox2 was required for Ezh2 binding at the promoters of developmental genes. The enrichment of Ezh2 and H3K27me3 was similarly decreased in linc1614-knockdown ESCs, whereas Sox2 enrichment was also not significantly changed upon linc1614 knockdown (Figure 4J). These results showed that Sox2 drove the assembly of the Sox2/linc1614/PRC2 complex at developmental gene promoters and that linc1614 was mainly responsible for the recruitment of PRC2 to developmental gene loci. Furthermore, the introduction of the linc1614 mutants that associated with Sox2 into the linc1614-knockdown ESCs indicated that the linc1614/Sox2 interaction was responsible for rescuing PRC2 recruitment at gene promoter regions (Figure 4K and L, Supplementary Figure S3F and G). Taken together, these findings suggested that PRC2 complex could be recruited to these regions of developmental genes by linc1614/Sox2 to repress expression by catalyzing the repressive H3K27me3 modification.
Figure 4.
Linc1614/Sox2 recruits the PRC2 complex to suppress the expression of developmental genes. (A) The RIP assay confirmed the interaction of linc1614 with the core subunit Ezh2 of the PRC2 complex. U6 snoRNA was indicated as negative control. Data were presented as mean ± SD (n = 3). ***P < 0.001 was performed relative to IgG. (B) The biotinylated linc1614 RNA pull-down assay showed a band corresponding to Ezh2 following sense linc1614 (1614L-S) pull-down. (C) Overlap of the 316 co-upregulated genes in sh1614 and shSox2 ESCs with the genes co-occupied by both PRC2 and H3K27me3 (Mikkelsen et al., 2007; Ku et al., 2008; Goren et al., 2010). (D) GO term analysis of the 83 overlapping genes. The 10 most significant GO terms are shown. (E) ChIP-seq tracks of H3K27me3, Ezh2, Suz12, and Sox2 in the vicinity of the TSSs of Pitx2 and T genes in ESCs are shown (Mikkelsen et al., 2007; Ku et al., 2008; Goren et al., 2010; Lodato et al., 2013). (F) Co-IP experiments showed no direct binding between Sox2 and Ezh2 in ESCs. (G) Immunoprecipitation of formaldehyde-crosslinked ESCs using an anti-Sox2 antibody indicated that the co-occupation of Sox2 with Ezh2 relied on the DNA context by treatment with or without Dnase I. (H) Sox2 and Ezh2 immunoprecipitation experiments upon fixation in formaldehyde-crosslinked control or linc1614 knockdown ESCs. (I) The extent of Sox2, Ezh2, and H3K27me3 binding at the promoter regions of the indicated genes (Pitx2, T, Eomes, Gata3, Gsn, and Csf1) in control or shSox2 ESCs, as measured by ChIP-qPCR. (J) The extent of Sox2, Ezh2, and H3K27me3 binding at the promoter regions of the indicated genes in control or sh1614 ESCs. Data were presented as mean ± SD (n = 3). Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001) was performed relative to Ctrl. (K and L) The extent of Ezh2 (K) and H3K27me3 (L) binding at the promoter regions of the indicated genes (T and Pitx2) in control ESCs, sh1614 ESCs, and sh1614 ESCs plus linc1614 deletion mutants. Data were presented as mean ± SD (n = 3). Student’s t-test was performed for significance (*,#P < 0.05, **,##P < 0.01, and ***,###P < 0.001). * relative to Ctrl and # relative to sh1614 + luc.
Linc1614 is required for somatic cell reprogramming
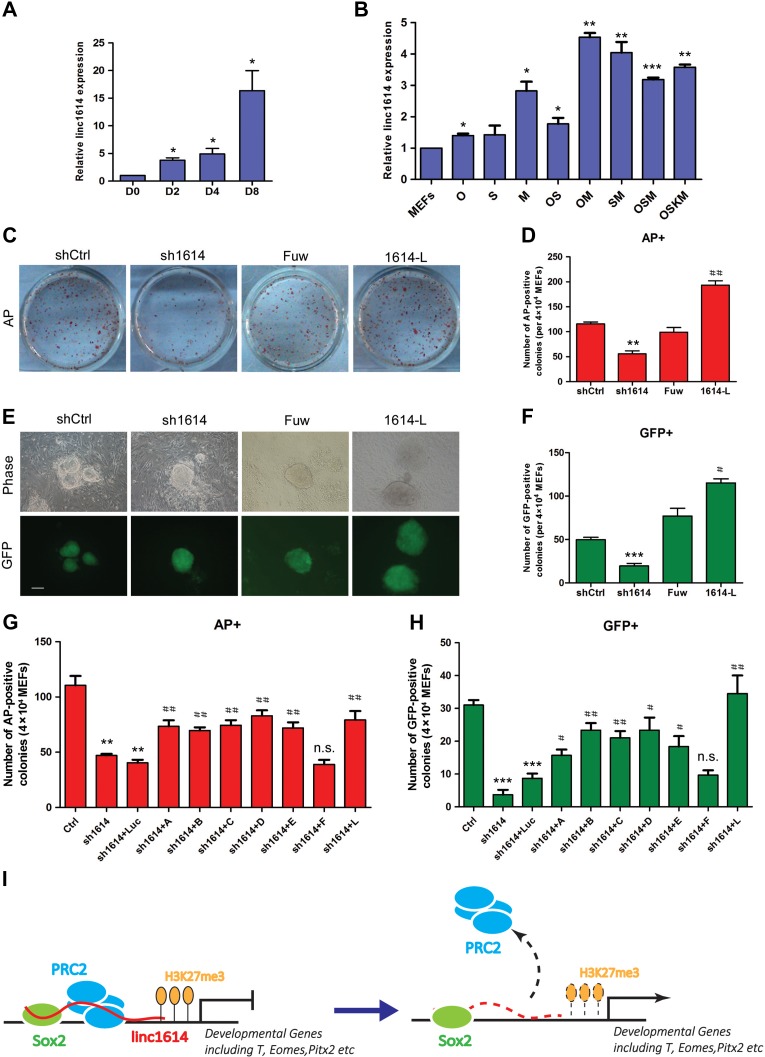
It is known that somatic cells can be reprogrammed to pluripotency by introducing the core factors critical for pluripotency maintenance, which suggests that there are specific common mechanisms underlying both pluripotency maintenance and acquisition (Fang et al., 2014). First, our results showed that linc1614 was remarkably upregulated in OSKM (for the reprogramming factors Oct4, Sox2, Klf4, and c-Myc)-driven iPSC generation (Figure 5A). We also analyzed the changes of linc1614 expression level upon overexpression of transcription factors Oct4, Sox2, and c-Myc (potentially bound at the promoter of linc1614) alone and combination of factors (Oct4 + Sox2, OS; Oct4 + c-Myc, OM; Sox2 + c-Myc, SM; Oct4 + Sox2 + c-Myc, OSM; Oct4 + Sox2 + Klf4 + c-Myc, OSKM) for 48 h. Our results showed that the expression of linc1614 was increased upon the overexpression of Oct4 and c-Myc alone in OG-MEFs (derived from Oct4::GFP transgenic mice), and significantly activated by combination of transcription factors (Figure 5B). These results suggested a potential role of linc1614 in mediating the somatic cell reprogramming.
Figure 5.
Linc1614 is required for somatic cell reprogramming. (A) The change of linc1614 expression level was gradually increased during the days (0, 2, 4, and 8) of iPSC induction. *P < 0.05 relative to D0. (B) Analysis for changes of linc1614 expression level upon overexpression of defined factors (O, Oct4; S, Sox2; M, c-Myc) alone or in combination (OS, OM, SM, OSM, and OSKM). *P < 0.05, **P < 0.01, and ***P < 0.001 relative to MEFs. (C and D) AP staining (on reprogramming Day 8) and statistics for AP-positive colonies from OG-MEFs (per 4 × 104 cells) transduced with OSKM in combination with linc1614 knockdown (sh1614) or linc1614 overexpression (1614-L). shCtrl represented scramble shRNA with no specific target. Fuw represented empty vector. (E and F) Representative images and statistics for GFP-positive iPSC colonies on Day 10 of OSKM-induced reprogramming in combination with sh1614 or 1614-L. Scale bar, 100 μm. Data were presented as mean ± SD (n = 3). Student’s t-test was performed for significance (*,#P < 0.05, **,##P < 0.01, and ***,###P < 0.001). * relative to shCtrl and # relative to Fuw. (G and H) The statistics for the AP-positive colonies (G) and GFP-positive colonies (H) showed that introduction of the linc1614 fragments that bound to Sox2 significantly rescued the efficiency of iPSC induction blocked by the knockdown of linc1614. Ctrl represented scramble shRNA. Data were presented as mean ± SD (n = 3). Student’s t-test was performed for significance (*,#P < 0.05, **,##P < 0.01, and ***,###P < 0.001). * relative to Ctrl and # relative to sh1614 + luc. (I) Schematic diagram of the mechanism through which Sox2 cooperates with linc1614 to recruit the PRC2 complex and repress the expression of developmental genes such as T, Eomes, and Pitx2 in pluripotent cells.
Then, we infected OG-MEFs with the OSKM factors in combination with linc1614 shRNA or overexpression vectors to generate iPSCs, identified by a series of assays (Supplementary Figure S4A–D). The numbers of alkaline phosphatase-positive (AP+) and Oct4-GFP-positive (GFP+) colonies upon OSKM-induced reprogramming were significantly increased when linc1614 was included (Figure 5C–F). In contrast, the introduction of linc1614 shRNA obviously decreased the efficiency of AP+ and GFP+ colony formation during the OSKM-induced reprogramming (Figure 5C–F), which indicated that linc1614 likely played positive roles in iPSC generation. Furthermore, our results showed that the linc1614 mutants that bound Sox2 could rescue the induction efficiency of iPSCs via counting the number of AP+ and GFP+ colonies (Figure 5G and H, Supplementary Figure S4E), suggesting that linc1614 could cooperate with the specific factor Sox2 to drive the reprogramming process.
In conclusion, our data showed that linc1614 could participate in the transcriptional repression of specific developmental genes by interacting with the key regulator Sox2 and recruiting the PRC2 complex, resulting in repression of those genes such as T, Eomes, and Pitx2 in ESCs (Figure 5I).
Discussion
A number of recent papers have revealed that lincRNAs are crucial for the maintenance of pluripotency in ESCs (Sheik Mohamed et al., 2010; Bertani et al., 2011), or emerge as key regulators in early differentiation (Ponting et al., 2009; Pauli et al., 2011). We performed a series of assays to analyze the cellular functions of linc1614, a previously undescribed lincRNA that is highly expressed in ESCs, potentially regulated by core factors (Figure 1A) and decreases along with pluripotency factors during the ESC differentiation (Figure 1B), in modulating the pluripotency of ESCs. Our data provided strong evidences that knockdown of linc1614 led to alterations in ESC morphology, suppression of pluripotency genes, and upregulation of developmental genes, which suggested that linc1614 was critical for maintaining the pluripotency and repressing the differentiation of ESCs. Additionally, linc1614 was gradually upregulated during reprogramming (Figure 5A) and activated by combination of defined factors (Figure 5B), but its knockdown significantly blocked the generation of OSKM-induced iPSCs, whereas ectopic expression of linc1614 increased the efficiency of iPSC generation. Together, these data confirmed that linc1614 is a novel intrinsic regulator of pluripotent cells in terms of both pluripotency maintenance and acquisition.
The maintenance of ESC pluripotency relies on a complicated network of transcription factors, including Oct4, Sox2, Nanog, and Tcf3 (Ivanova et al., 2006; Chen and Daley, 2008; Kim et al., 2008; Tam et al., 2008). It is widely believed that Sox2 mainly interacts with the master transcription factor Oct4 to maintain pluripotency (Aksoy and Stanton, 2013). Recently, Sox2 has also been reported to associate with lncRNAs to regulate the pluripotency of hESCs (Ng et al., 2012). These previous literatures indicated that the action of Sox2 was mainly dependent on its partners. Our data showed that Sox2, not Oct4 or Nanog, specifically interacted with linc1614, which suggested that linc1614 served as an unrevealed partner of Sox2 and might be involved in the Sox2-mediated transcriptional regulation. More importantly, the introduction of linc1614 segments that were able to interact with Sox2 significantly rescued the ESC phenotype and gene expression following linc1614 knockdown, whereas ectopic expression of Sox2 in linc1614-knockdown ESCs had no significant effect on the ESC phenotype or gene expression. Notably, we further introduced the linc1614 segments that interacted with Sox2 into somatic cells subjected to the reprogramming process, and these segments significantly rescued the blocking of iPSC formation that was induced by linc1614 knockdown. These results suggested that Sox2/linc1614 association simultaneously plays roles in pluripotency maintenance and reprogramming.
It has been well established that Sox2 plays distinct roles in transcriptional networks by promoting the expression of ESC-specific genes and suppressing differentiation (Boyer et al., 2005; Chen and Daley, 2008; Kim et al., 2008). Several studies show that lincRNAs can bind with epigenetic modulators (Ng et al., 2012; Klattenhoff et al., 2013) or guide the specific binding of transcription factors (Ng et al., 2013), suggesting that lincRNAs also execute their complicated functions with distinct partners as well as transcription factors. In the current study, perturbation of the factor Sox2 collapsed the circuitry of self-renewal and triggered differentiation of multiple lineages (Ivanova et al., 2006) (Supplementary Figure S2A), which was consistent with the loss of the ESC phenotype upon linc1614 knockdown. Microarray analysis showed that more than 1200 genes were differentially expressed upon linc1614 knockdown, which was comparable to the number affected by Sox2 knockdown (almost 820 genes). GO term analysis of differentially expressed genes under conditions of linc1614 and Sox2 knockdown showed that knockdown of either linc1614 or Sox2 led to abnormal expression of developmental genes. A cross-over analysis showed that 46.7% of the differentially expressed genes detected upon Sox2 knockdown were also differentially expressed under linc1614 knockdown, which suggested that linc1614 specifically partnered with Sox2 to co-regulate a large number of downstream genes. GO term analysis for the genes that were upregulated under both linc1614 and Sox2 knockdown conditions (almost 316 genes) showed that the linc1614/Sox2 co-targeted genes were associated with cell differentiation and organ development, whereas the co-downregulated genes exhibited no significant relationship with the development processes of ESCs. Although a certain number of genes differentially expressed upon knockdown of linc1614 as compared to that of Sox2, suggesting that linc1614 might be involved in the additional pathways independent of Sox2, our present results indicated that linc1614 is a transcriptional regulatory lincRNA that specifically mediates Sox2 function in repressing the expression of downstream developmental genes.
Transcriptional regulation depending on PcG proteins for histone modification is one of the key regulatory mechanisms involved in X-inactivation, genomic imprinting, carcinogenesis, and cell fate determination (Bracken and Helin, 2009; Gieni and Hendzel, 2009). In the most recently established models, the functions of lincRNAs are to interact with chromatin-modifying complexes and determine the target specificity of regulatory complexes as epigenetic modifiers (Bertani et al., 2011; Klattenhoff et al., 2013; Prensner et al., 2013). Our data showed that 26.3% of the co-upregulated genes upon Sox2/linc1614 knockdown were also occupied by Ezh2 and the H3K27me3 modification. The enrichment of Ezh2 and H3K27me3 at the promoters of downstream developmental genes was significantly decreased upon linc1614 knockdown, which suggested that linc1614 might be acting to target the PRC2 complex to the promoters of the developmental genes and control the repressive epigenetic modification. The core factor Sox2 likely collaborates with the PRC2 and co-occupies a significant proportion of developmental genes in ESCs (Lee et al., 2006). The histone variant H2A.Z may serve as a bridge to link Sox2 and PRC2 through binding to Sox2 (Zhou et al., 2016) and Suz12 (a PRC2 component), respectively (Creyghton et al., 2008). However, the relevance of the Sox2 and PRC2 complex at developmental gene loci remains unclear. Our results indicated that Sox2 and Ezh2 co-occupied at these genomic loci, rather than by direct binding (Zhou et al., 2016) (Figure 4F and G). We found that linc1614 was presented at the promoters of co-targeted developmental genes and its knockdown significantly disrupted the interaction of Sox2 and Ezh2 at the genomic loci, suggesting that the formation of Sox2, Ezh2 and linc1614 tripartite complex was dependent on the DNA context of the targeted developmental gene regions. In the present study, the enrichment of Ezh2 and H3K27me3 at the promoters of co-regulated genes was significantly decreased upon knockdown of Sox2 or linc1614, whereas knockdown of either Ezh2 or linc1614 had no obvious effect on Sox2 binding at the promoters of co-regulated genes. Moreover, introducing the linc1614 segments that interacted with Sox2 significantly rescued the enrichment level of Ezh2 and H3K27me3 at the promoters of co-targeted developmental genes. These findings indicated that Sox2 drives the assembly of the transcription factor-lincRNA-epigenetic modulator complex at developmental gene promoters and that linc1614 is mainly responsible for the recruitment of the PRC2 complex.
On the basis of the data shown here, we propose an integrated transcriptional regulatory model in which lincRNAs serve as indispensable partners of the key transcription factors that are required for the both maintenance and acquisition of pluripotency. The discovery of a novel regulatory model that governs the recruitment of transcription factor−lincRNA−epigenetic modulator regulatory complexes to downstream target sites might facilitate efforts to elucidate the complicated mechanisms by which lincRNAs combine with transcriptional regulators to control gene expression in pluripotent cells and to better understand the gene regulatory circuits and mechanisms of lincRNAs in cell fate determination.
Materials and methods
Cell culture
Mouse ESCs (E14) were maintained in DMEM supplemented with 15% FBS (Gibco), 1× nonessential amino acids (NEAA) (Gibco), 1× Glutamax (Gibco), 1× sodium pyruvate (Gibco), and 55 μM β-mercaptoethanol (Gibco), with leukemia inhibitory factor (LIF; Millipore), on gelatin-coated plates. Oct4::GFP MEFs (OG-MEFs) were derived from transgenic mice (C57BL/6) at E13.5 and cultured in DMEM containing 10% FBS. iPSCs were maintained in KOSR medium consisting of knockout DMEM (Gibco) containing 20% knockout serum replacement (Gibco), 1× NEAA, 1× Glutamax, and 55 μM β-mercaptoethanol with LIF on feeder cells. The 293T, Plat-E, and feeder cells were maintained in DMEM containing 10% FBS.
RNA immunoprecipitation
RIP was performed as previously described (Ng et al., 2012). In each immunoprecipitation for the RIP assay, 3 μg of the appropriate antibody was used. For the exogenous RIP assay, an anti-Sox2 (Abcam, ab59776) or anti-Nanog (Abcam, ab80892) antibody was used to bind to the RNA following ectopic expression of linc1614 with Sox2 or Nanog in 293T cells. For the native RIP assay, an anti-Sox2, anti-Nanog, anti-Oct4 (Abcam, ab19857), anti-Ezh2 (Abcam, ab3748), or anti-Suz12 (Abcam, ab12073) antibody was used to pull down the RNA associated with the corresponding proteins. The immunoprecipitated RNA was extracted using RNAiso and analyzed via qPCR. The fold enrichment was calculated relative to the corresponding control IgG sample.
Biotinylated RNA pull-down
Briefly, biotin-labeled RNAs were obtained using RNA Labeling Mix (Roche, 11685597910) and T7/T3 RNA polymerase (Roche, 10881767001/11031171001). A total of 3 μg of the RNA was heated to 90°C for 2 min, held on ice for 2 min in RNA structure buffer (10 mM Tris-HCl pH 7.0, 0.1 M KCl, and 10 mM MgCl2) to allow proper secondary structure formation. ESCs (5 × 106 cells) were lysed with RIP lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM Hepes pH 7.0, and 0.5% NP-40) for 30 min on ice to facilitate lysis. Immunoprecipitation was subsequently performed using streptavidin beads coated with 3 μg of the biotin-labeled RNAs, according to the manufacturer’s instructions. The RNA-binding proteins were treated with SDS lysis buffer for subsequent western blotting analysis.
Fluorescent in situ hybridization assay
ESCs were rinsed briefly in PBS and then fixed in 4% paraformaldehyde for 10 min at RT. Cells were permeabilized in PBS containing 0.5% Triton X-100 for 5 min at 4°C, then washed in PBS for 5 min. Hybridization was carried out with a fluorescent in situ hybridization (FISH) probe in a moist chamber at 37°C in the dark overnight according to the protocol, provided by the manufacturer. Linc1614-cy3 FISH probes were designed and synthesized by RiboBio Co., Ltd. Mouse U6 FISH probes (LNC110103, RiboBio) and mouse 18S FISH probes (LNC110104, RiboBio) were used as the nuclear and cytoplasmic controls respectively. All images were obtained with a confocal microscope.
UV-assisted triplex capture assay
For UV-assisted triplex capture assays, ESCs were transfected with 3′-biotin-modified RNA oligonucleotides with FuGENE HD Transfection Reagent (Roche). After 6 h, isolated nuclei were irradiated for 15 min with UV (365 nm), and chromatin lysate from 5 × 106 ESCs were incubated with streptavidin-coupled agarose (Invitrogen). Upon binding to streptavidin beads, associated DNA was extracted and analyzed by qPCR using promoter-specific primers.
Microarray analysis
Total RNA was extracted from three independently derived ESCs (Control, sh1614, and shSox2) using the RNAiso reagent. Following the manufacturer’s instructions, RNA integrity was examined using an Agilent Bioanalyzer 2100 (Agilent Technologies) to determine the RIN number. Gene expression was analyzed with Affymetrix GeneChip 430 2.0 arrays. The slides were scanned using a GeneChip® Scanner 3000 (Cat# 00-00212, Affymetrix) and analyzed using Command Console Software 4.0 (Affymetrix) with the default settings. The raw data were normalized with the MAS 5.0 algorithm and Gene Spring Software 11.0 (Agilent Technologies). The statistical properties of all arrays after the pre-processing step were examined and confirmed to be similar. Heatmaps were employed to cluster the expression data for differentially expressed genes. GO term analyses were performed using DAVID V6.7.
Statistical analysis
The data presented in this study are shown as mean ± SD from three independent experiments and analyzed using Student's t-test. P < 0.05 was considered statistically significant.
Supplementary Material
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work was supported by grants from the Ministry of Science and Technology (2016YFA0101300), the National Natural Science Foundation of China (81530042, 31210103905, 31371510, 31571529, 31571519, 31471250, and 31571390), the Science and Technology Commission of Shanghai Municipality (15JC1403201), and the Fundamental Research Funds for the Central Universities (2000219136 and 1500219106).
Conflict of interest: none declared.
References
- Adachi K., Suemori H., Yasuda S.Y., et al. (2010). Role of SOX2 in maintaining pluripotency of human embryonic stem cells. Genes Cells 15, 455–470. [DOI] [PubMed] [Google Scholar]
- Aksoy I., and Stanton L.W. (2013). Pluripotency-regulating networks provide basis for reprogramming. Curr. Mol. Med. 13, 695–706. [DOI] [PubMed] [Google Scholar]
- Bertani S., Sauer S., Bolotin E., et al. (2011). The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol. Cell 43, 1040–1046. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Boyer L.A., Lee T.I., Cole M.F., et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken A.P., and Helin K. (2009). Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer 9, 773–784. [DOI] [PubMed] [Google Scholar]
- Chen L., and Daley G.Q. (2008). Molecular basis of pluripotency. Hum. Mol. Genet. 17, R23–R27. [DOI] [PubMed] [Google Scholar]
- Creyghton M.P., Markoulaki S., Levine S.S., et al. (2008). H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135, 649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Zhang L., Wei W., et al. (2014). A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol. Cell 55, 537–551. [DOI] [PubMed] [Google Scholar]
- Gieni R.S., and Hendzel M.J. (2009). Polycomb group protein gene silencing, non-coding RNA, stem cells, and cancer. Biochem. Cell Biol. 87, 711–746. [DOI] [PubMed] [Google Scholar]
- Goren A., Ozsolak F., Shoresh N., et al. (2010). Chromatin profiling by directly sequencing small quantities of immunoprecipitated DNA. Nat. Methods 7, 47–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Amit I., Garber M., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Donaghey J., Carey B.W., et al. (2011). lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N., Dobrin R., Lu R., et al. (2006). Dissecting self-renewal in stem cells with RNA interference. Nature 442, 533–538. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., and Young R. (2008). Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132, 567–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., et al. (2008). An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C.A., Scheuermann J.C., Surface L.E., et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol M.J., and Rinn J.L. (2010). RNA traffic control of chromatin complexes. Curr. Opin. Genet. Dev. 20, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M., Koche R.P., Rheinbay E., et al. (2008). Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 10, e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., et al. (2006). Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato M.A., Ng C.W., Wamstad J.A., et al. (2013). SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLoS Genet. 9, e1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T.R., Dinger M.E., and Mattick J.S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K.V. (2009). RNA-directed transcriptional gene silencing and activation in human cells. Oligonucleotides 19, 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.Y., Bogu G.K., Soh B.S., et al. (2013). The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359. [DOI] [PubMed] [Google Scholar]
- Ng S.Y., Johnson R., and Stanton L.W. (2012). Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. (2007). How is pluripotency determined and maintained? Development 134, 635–646. [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., and Yamanaka S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317. [DOI] [PubMed] [Google Scholar]
- Pauli A., Rinn J.L., and Schier A.F. (2011). Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 12, 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P., Oliver P.L., and Reik W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. [DOI] [PubMed] [Google Scholar]
- Prensner J.R., Iyer M.K., Sahu A., et al. (2013). The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 45, 1392–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik Mohamed J., Gaughwin P.M., Lim B., et al. (2010). Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 16, 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A., and van Lohuizen M. (2006). Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6, 846–856. [DOI] [PubMed] [Google Scholar]
- Stefanovic S., Abboud N., Desilets S., et al. (2009). Interplay of Oct4 with Sox2 and Sox17: a molecular switch from stem cell pluripotency to specifying a cardiac fate. J. Cell Biol. 186, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surface L.E., Thornton S.R., and Boyer L.A. (2010). Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell 7, 288–298. [DOI] [PubMed] [Google Scholar]
- Takahashi K., and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Tam W.L., Lim C.Y., Han J., et al. (2008). T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells 26, 2019–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia N., MacCarthy C., Esch D., et al. (2015). Dissecting the role of distinct OCT4-SOX2 heterodimer configurations in pluripotency. Sci. Rep. 5, 13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E., Chang W.Y., Hunkapiller J., et al. (2010). Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell 6, 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Oron E., Nelson B., et al. (2012). Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10, 440–454. [DOI] [PubMed] [Google Scholar]
- Zhou C., Yang X., Sun Y., et al. (2016). Comprehensive profiling reveals mechanisms of SOX2-mediated cell fate specification in human ESCs and NPCs. Cell Res. 26, 171–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.