Abstract
The rise in genomic knowledge over the past decade has revealed the molecular etiology of many diseases, and has identified intricate signaling network activity in human cancers. Genomics provides the opportunity to determine genome structure and capture the activity of thousands of molecular events concurrently, which is important for deciphering highly complex genetic diseases such as cancer. In this review, we focus on genomic efforts directed towards one of cancer’s most frequently mutated networks, the RAS pathway. Genomic tools such as gene expression signatures and assessment of mutations across the RAS network enable the capture of RAS signaling complexity. Due to this high level of interaction and cross-talk within the network, efforts to target RAS signaling in the clinic have generally failed, and we currently lack the ability to directly inhibit the RAS protein with high efficacy. We propose that the use of expression data can identify effective treatments that broadly inhibit the RAS network as this approach measures pathway activity independent of mutation status or any single mechanism of activation. Here we review the genomic studies that map the complexity of the RAS network in cancer, and that show how genomic measurements of RAS pathway activation can identify effective RAS inhibition strategies. We also address the challenges and future directions for treating RAS-driven tumors. In summary, genomic assessment of RAS signaling provides a level of complexity necessary to accurately map the network that matches the intricacy of RAS pathway interactions in cancer.
Keywords: RAS, genomics, gene expression signature, targeted therapy, cancer
1. Introduction
High-throughput genomic analysis has benefitted the study of signal transduction over the past decade [1]. Genomic sequencing techniques are now routinely used in many research laboratories, and are steadily becoming adopted in clinical settings [2]. The scientific community has used these technologies to better understand the genetic basis of many human diseases, to help diagnose disease and predict disease progression, and to pioneer personalized healthcare initiatives [3,4]. Cancer is one of the diseases that has been impacted greatly by the implementation of genomics [4]. Large-scale cancer sequencing projects have allowed us to view the cancer genome using multiple genomic profiling strategies including whole-genome and transcriptome sequencing, proteomics, genome-wide DNA methylome analysis, and DNA copy number analysis, all collectively defined as “omics” [5–7]. These strategies have reshaped how we view the cancer genome and have shown that individual tumors harbor their own unique genetic makeup containing mutations, copy number changes, epigenetic modifications, and aberrant expression of hundreds to thousands of genes; therefore highlighting that multidimensional genomic data contributes to understanding cancer [5,8]. Genomics has been successfully applied to oncology in many different contexts [1,9] including the identification of cancer subgroups such as the intrinsic subtypes in breast cancer [10–12], the development of breast cancer prognostic tools such as Oncotype DX and MammaPrint to predict the risk of cancer recurrence [13], and the identification of KRAS mutations as predictors of poor drug response in lung cancer [14]. Although approximately 140 driver mutations have been discovered in human cancer, most of these mutations converge on roughly 12 pathways that regulate three vital cellular processes: cell growth, cell survival, and genome maintenance [8]. Thus, tumors tend to rely on a subset of signaling phenotypes to maintain growth and survival.
The RAS pathway is one of the most frequently dysregulated pathways in cancer, with approximately 30% of all patient tumors expressing activating RAS gene mutations [15]. Of the three main isoforms of oncogenic RAS, KRAS is the most frequently mutated, affecting ~90% of pancreatic cancers, ~35% of colon cancers, and ~18% of lung cancers, while NRAS is mutated in ~15% of melanomas, and HRAS is rarely mutated in cancer [16]. Aberrations in RAS genes themselves contribute to RAS pathway activation, but aberrations of genes up- and downstream of RAS can also activate the pathway (Figure 1), highlighting the need for genomics to broadly measure RAS pathway activation [17]. Cancers with RAS gene mutations are associated with drug resistance, poor prognosis, shorter survival, and enhanced metastasis [18–23]. Extensive efforts have been made towards the development of RAS protein inhibitors but, to date, no effective direct RAS inhibitors are available in the clinic. Thus, targeting this pathway effectively has a high potential for patient benefit.
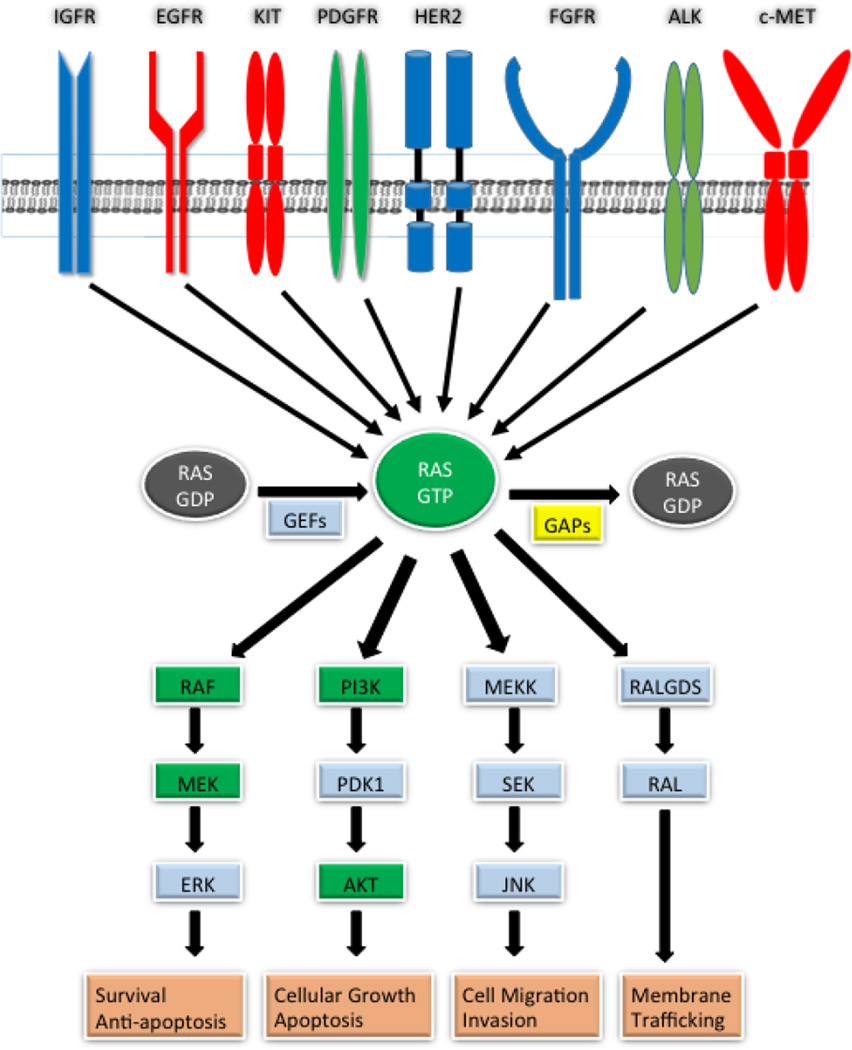
Figure 1.
RAS pathway aberrations in human cancers. The RAS pathway can be activated by mutation (green) or by overexpression (blue) of pathway proteins. In some cancers, proteins are both mutated and overexpressed (red). Dysregulation can occur in downstream effector molecules including RAF, MEK, PI3K, and AKT. RAS is also activated by the loss of function of RAS regulators such as GAPs (yellow).
In this review we discuss the role that genomics plays in deciphering the RAS signaling network and its mediators and how the use of genomics has led to a better understanding of RAS network complexity. Also, as omic-level measurement captures RAS activity in both RAS-mutant and RAS-wild type tumors, these approaches may enable identification of novel RAS pathway inhibitors not specific to mutant RAS. Overall, we expect genomics will continue to lead to discoveries that will aid in the treatment of RAS-driven cancers in the near future.
2. Genomics provides insight into the RAS pathway
2.1 Why study RAS at the genomic level?
The RAS pathway is an intricate signaling cascade consisting of numerous up- and downstream proteins and interconnecting pathways [24]. Due to the complexity of this pathway, a genomics framework is necessary in order to study its activities concurrently as a network. While extracellular growth signals normally activate the RAS pathway, in cancer, activating mutations in RAS pathway genes lead to sustained pathway signaling, resulting in the aberrant activation of downstream oncogenic processes such as cellular proliferation, cell survival, metabolic changes, and metastasis [22,25–29]. The RAS pathway is not linear and can activate multiple downstream pathways such as the RAF/MEK/ERK pathway, the phosphoinositide 3-kinase (PI3K)/AKT/mTOR pathway, and the RAL-GDS pathway, all leading to various oncogenic events. Adding further complexity, RAS can activate additional proteins including AF-6, CANOE, TIAM1, MEKK1, p120GAP, NF1, RIN1, PKC-ζ, and NORE1, illustrating the far-reaching roles of RAS [30]. In cancer, the RAS pathway can become activated by aberrations in either upstream growth factor receptors such as EGFR and IGF1R, or in downstream pathway proteins such as GAPs, GEFs, RAF, MEK, and ERK, by loss of function of RAS negative regulators (SPRY, SPRED, DUSPs, RASA1, NF1), and through activation of alternative pathways (PI3K, PTEN, RALGDS, MEKK1) [25,27,31–35] (Figure 1). Therefore, the RAS pathway is a complex network requiring a genomic approach that matches that complexity.
2.2 Genomics shows dysregulation of RAS pathway components across cancers
The availability of genomic sequencing has enabled the mass profiling of various cancer types using multi-omic data [7]. One such effort has been pioneered by The Cancer Genome Atlas research network (TCGA), a large international research effort that has produced omic data for over twenty different cancers, including both DNA- and RNA-sequencing for over 11,000 tumors [36]. Here, we highlight the spectrum of RAS pathway aberrations from the TCGA’s findings in several cancer types including lung adenocarcinoma, colorectal carcinoma, and head and neck squamous cell carcinoma (HNSCC).
Upon profiling colorectal carcinoma, the TCGA found that 55% of non-hypermutated colorectal carcinomas, a molecular subtype accounting for 84% of the studied samples, demonstrated KRAS, NRAS, or BRAF alterations; mutations in these genes were found to be significantly mutually exclusive [37]. Interestingly, the TCGA also found a co-occurrence of RAS pathway and PI3K pathway mutations in one-third of colorectal tumors, indicating the need to target both pathways to effectively inhibit tumor growth in cancers of this type. Furthermore, genomic analysis of lung adenocarcinoma revealed that 62% of these cancers bear a canonical oncogenic driver mutation in the RAS/RAF/MEK pathway [38]. Upon expanding this analysis to include focal amplifications of upstream receptor tyrosine kinases (RTKs), as well as loss of function mutations in tumor suppressor genes in the RAS pathway, such as NF1, the number of lung adenocarcinomas with driver mutations in the RAS pathway increased to 76%. Importantly, this study also used reverse phase protein array (RPPA) data to demonstrate that both KRAS-mutant lung adenocarcinoma samples and a subset of KRAS-wild type samples exhibited high MAPK pathway activity. These results highlight the importance of understanding pathway-level activation beyond single gene mutational status when assessing a tumor’s dependency on a pathway for survival. Subsequent investigation of HNSCC demonstrated that in this cancer type, 5% of HPV-negative cancers contain an HRAS mutation [39]. It is important to note, however, that the study also found mutation or amplification in EGFR (15% of HPV-negative samples), FGFR1 (10%), ERBB2 (5%), IGF1R (4%), and several other RTKs (3% or less), thus contributing to a wider spectrum of RAS pathway aberrations than HRAS mutation alone. Therefore, by implementing whole-genome sequencing, the TCGA research network confirmed the high prevalence of somatic mutations and amplifications contributing to RAS pathway activation in RAS-driven cancers.
Publically available TCGA datasets have also enabled further discoveries that have provided additional insight into RAS pathway aberrations. For example, Raphael and Fabio developed a pathway linear progression model to determine the temporal order of somatic driver mutation in key pathways during oncogenesis [40]. Using the TCGA colorectal cancer dataset, they showed that mutations in the RAS pathway occur late in tumorigenesis--mutations in APC or FBXW7 and either TP53 or PIK3CA generally occur before members of the RAS pathway are mutated in colorectal cancers. Additionally, Want and colleagues integrated the TCGA breast cancer data consisting of somatic mutations, copy number variations, transcriptomics, and DNA methylomics, into “risk pathways” by mapping alterations in genes at each tested omic level to pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) to determine pathways altered in breast cancer [31]. Additionally, these risk pathways were constructed into pathway cross-talk networks based on protein-protein interaction data from the Human Protein Reference Database (HPRD). Want and colleagues identified KRAS as a major connector between multiple risk pathways, thus supporting the importance of targeting RAS dependence as a significant therapeutic opportunity [31]. Thus, not only has TCGA genomic data provided unprecedented insight into the omic landscape of cancer, it has also enabled a broader understanding of both mutational progression during oncogenesis and of pathways dysregulated in cancers.
2.3 Gene expression signatures can quantify RAS network activity independent of the mechanism by which the pathway is activated
A gene expression signature is defined as a group of genes whose combined expression patterns are uniquely characteristic of a biological phenotype, or in the context of this review, a biological pathway [9]. In the early 2000s researchers began developing gene expression signatures to predict the activity of various oncogenic signaling pathways using microarray data [3,41,42]. Gene expression signatures have the capability to measure cellular signaling events because whether or not the signaling event directly modulates transcription factors, as cellular signaling eventually results in gene-expression changes [43]. Understanding that the RAS pathway could be activated by RAS gene mutations, and by multiple other mechanisms, led researchers to generate RAS gene expression signatures as a method to better determine RAS pathway activation [42].
One of the first RAS-specific gene expression signatures was generated by overexpressing the Hras gene in mouse embryonic fibroblast cells using recombinant adenoviruses [44]. This signature accurately reflected the activation state of Hras, acting as a proof of principle that overexpressing oncogenes in cells could result in specific gene expression changes, which could then represent specific pathway activity [44]. An additional RAS gene expression signature was derived by Sweet-Cordero et al. [45] from a sporadically-activated Kras2 mouse model. This signature was generated by comparing gene expression differences between activated Kras2 tumors and normal lung tissue, was validated, and able to predict KRAS activity in lung adenocarcinoma, a RAS-activated cancer. This approach suggested that signatures generated in mouse tumors could accurately reflect human biology, and provided a strategy for using genomic analysis of animal models to probe human disease [45].
Bild et al. [46] built upon the work of Huang et al. [44] in generating a pathway-based gene expression signature by overexpressing mutant HRAS in human primary epithelial cells. The group used supervised clustering to generate gene expression changes indicative of RAS pathway activation. This signature accurately predicted RAS pathway activation in mice and human tumors with RAS mutations, such as in human non-small cell lung carcinoma. Interestingly, the study found that higher RAS pathway activity correlated with decreased survival in lung cancer.
Chang and colleagues also developed a novel approach for utilizing gene expression signatures by deconstructing a RAS gene expression signature into “modules,” which represent smaller components of the pathway [47]. This study found that particular modules from the RAS gene expression signature were able to distinguish high- and low-risk survival groups in lung adenocarcinoma better than using the entire gene expression signature. These results further demonstrate the benefits of using gene expression signatures to deconstruct and better understand the RAS network. Other important uses of RAS gene expression signatures include, but are not limited to, the prediction of RAS activity in gastric cancer by Ooi et al. [48], and the generation of a “KRAS dependency” signature in lung cancer by Singh et al. [49]. Overall, methods for using gene expression signatures to measure RAS pathway activity transcend the traditional use of single genes to measure RAS activation, which, as shown here, does not always represent pathway activity [3].
2.4 KRAS, EGFR, and RAF gene expression signatures show RAS pathway complexities across multiple cancers
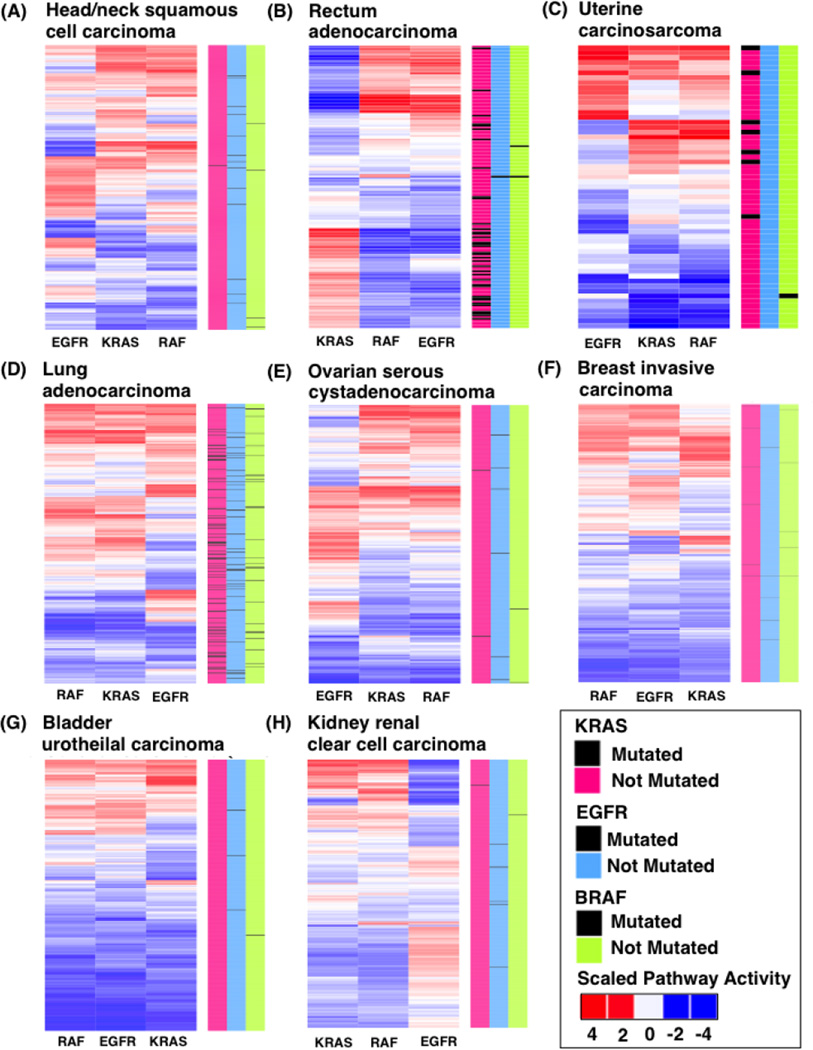
Genomics has facilitated the understanding that many different RAS pathway components contribute to RAS pathway activation, and that RAS mutations do not always correlate with activation of the pathway [46,50,51]. This illustrates the need for higher level genomic measurements of the RAS pathway. To further explore pathway activation in relation to mutational status, we measured pathway activity and mutational status for key RAS pathway components EGFR, KRAS, and RAF across 8 different cancers in TCGA [6] which express varying levels of KRAS, EGFR, or BRAF mutations. Specifically, we used our previously generated gene expression signatures that measure the activity of the EGFR, KRAS, and RAF1 pathway components using our published pathway modeling toolkit, Adaptive Signature Selection and InteGratioN (ASSIGN) [46,52,53] (see methods section). Unsupervised hierarchical clustering of the pathway activity estimates for all cancer types and pathway signatures revealed distinct patterns of pathway activation across cancer types (Figure 2).The pathway activity for EGFR, KRAS, and RAF1 and mutational status for KRAS (pink), BRAF (blue), and EGFR (green) for each TCGA cancer and patient are represented in Figure 2 for (A) head and neck squamous cell carcinoma, (B) rectum adenocarcinoma, (C) uterine carcinoma, (D) lung adenocarcinoma, (E) ovarian serous cystadenocarcinoma, (F) breast invasive carcinoma, (G) bladder urothelial carcinoma, and (H) kidney renal clear cell carcinoma.
Figure 2.
Scaled pathway activation scores for the EGFR, RAF, and RAS pathway from patient TCGA data. (A) head and neck squamous cell carcinoma, (B) rectum adenocarcinoma, (C) uterine carcinoma, (D) lung adenocarcinoma, (E) ovarian serous cystadenocarcinoma, (F) breast invasive carcinoma, (G) bladder urothelial carcinoma, and (H) kidney renal clear cell carcinoma. Red values indicate high pathway activity and blue represent low pathway activity. Color bars on the right side represent different gene mutations in KRAS (pink), EGFR (light blue), and BRAF (green). Black bars in gene columns indicate presence of mutations.
To illustrate the ability of gene expression signatures to accurately predict pathway activation in patient tumors, we highlight situations in which gene mutations complement pathway activation. For example, 81% of all rectum adenocarcinoma patients harboring KRAS mutations also have high KRAS activation scores (Figure 2B). We also found high EGFR activation scores (51% of patients) in head and neck squamous cell carcinoma (Figure 2A), a cancer in which EGFR is known to be overexpressed [54], and lung adenocarcinoma (44% of patients), a cancer with high EGFR mutation rate (Figure 2D). While gene mutations are generally reflective of pathway activation, there were cases in which gene mutational status did not alone correlated to activation of the pathway. For example, in lung adenocarcinoma, which is known to be a RAS-driven cancer subtype, a high proportion of patients have RAS pathway activation independent of mutation status (Figure 2D). We observed additional instances in which gene mutations were not found, but the pathways were activated. For example, in bladder urothelial carcinoma (Figure 2G), only 3 patients had EGFR mutations, and no mutations were found in KRAS or RAF, but pathway activation was found in 42%, 22%, and 38% of cases for EGFR, KRAS, and RAF pathways, respectively, thus highlighting that the absence of mutations does not always mean the pathway is inactivated. Few mutations and low pathway activation were also observed in breast invasive carcinoma (Figure 2F), kidney renal clear cell carcinoma (Figure 2H), and ovarian serous cystadenocarcinoma (Figure 2E). These results support the idea that pathway activation can occur due to other mechanisms such as mutations or amplifications in other genes or crosstalk/compensation within the RAS pathway [55]. Using expression signatures to measure pathway activity, we also found that each cancer had its own unique and heterogeneous pattern of EGFR, KRAS, and RAF1 activation (Figures 2B–H). Overall, these results demonstrate how the use of multiple mechanisms to measure pathway activity uncovers patterns that are not simply a reflection of mutation status. These results also show how the complexity of signaling network interactions in tumors cannot be generalized to all cancer types.
3. The impact of genomics on RAS pathway driven cancer therapeutics
3.1 Genomics helps guide the use of targeted therapies towards RAS pathway components
Since the initial characterization of RAS as an oncogene in 1982, [56,57] various initiatives have been taken to target RAS genes, proteins, and, more recently, downstream members of the RAS pathway. For example, in the early 1990s, researchers attempted to target RAS proteins directly with small molecule inhibitors and with farnesyltransferase inhibitors (FTIs) [58]. While FTIs efficiently inhibited farnesylation in HRAS mutant cancers [59,60], they failed to show efficacy in KRAS and NRAS mutant cancers as these isoforms can undergo alternative methods of membrane association [61]. Similarly, attempting to directly target the guanine nucleotide binding site of RAS using small molecule inhibitors failed due to the protein’s lack of allosteric regulatory sites and its picomolar affinity for GTP [62]. Therefore, few effective treatment options are currently available for patients with RAS-driven cancers, which has led to the characterization that RAS is “undruggable” [63,64]. However, recent studies have identified compounds capable of either binding to mutant RAS proteins directly or interfering with RAS’s ability to bind to the guanine nucleotide exchange factor Son of Sevenless (SOS) [65–68]. Nevertheless, these novel RAS-targeting compounds require further development before they can be implemented into clinical trials.
The discovery of RAS effector proteins and recurrent oncogenic mutations in downstream RAS pathway components (BRAF, MEK, ERK, and PI3K pathway members) [69–72], led to the development of several inhibitors including sorafenib, vemurafenib, and dabrafenib for RAF-mutated cancers, and trametinib and cobimetinib for MEK-mutated cancers [26,73]. More recently, ERK inhibitors and PI3K pathway inhibitors, such as the FDA-approved PI3K inhibitor idelalisib, have also been developed [74–76]. Combination treatments targeting the RAF/MEK/ERK pathway and PI3K pathway are now under different phases of clinical evaluation in various advanced solid tumors [77–79].
Measuring the mutational status of RAS pathway genes has provided clinical benefits such as guiding the use of targeted therapies, and selecting appropriate patient populations for clinical trials in particular cancers. For example, KRAS mutations are indicative of resistance to anti-EGFR therapies [80,81], and BRAF V600E mutations are indicative of response to RAF inhibitors [82]. However, determining the mutational status of specific genes is not always beneficial for predicting drug response, as mutations do not always correlate with pathway activation [46,50]. For instance, only 53% of patients with BRAF V600E mutations demonstrate partial or complete response to the RAF inhibitor, vemurafenib [83]. Cancers carrying mutations in the RAS pathway are not always dependent on RAS signaling, and the absence of RAS gene mutations does not always correlate with RAS inactivity as additional components of the network may be activated [41,49]. For example, absence of negative-feedback regulators, such as Sprouty (SPRY) and Sprouty-related (SPRED), and RAS GAPs such as NF1, can also activate the RAS pathway in various cancers [32,84]. These studies support the notion that treatment decisions based solely on RAS mutational analysis may overlook a large population of patients not carrying RAS mutations, but have RAS pathway activation.
3.2 Gene expression signatures aid in predicting response to targeted therapies in RAS-driven cancers
Previously, several groups have demonstrated that RAS gene expression signatures are capable of measuring RAS pathway activation [44–46]. In addition, gene expression signatures can also be used to predict drug response to RAS inhibitors. For example, breast cancer cell lines with high RAS pathway activity responded better to RAS farnesyltransferase inhibitors than cell lines with low RAS pathway activity [46]. The ability to predict drug response in cell lines engendered the idea that gene expression signatures may be capable of predicting response to targeted therapies in patients [85]. Loboda et al. [51] also used a gene expression signature to predict response to PI3K and RAS pathway inhibitors a different approach that leveraged RAS gene expression signatures from multiple datasets [45,46,86] to create a comprehensive RAS gene expression signature. Not only was this signature predictive of KRAS mutation status in lung tumors and cell lines, but also it was superior to KRAS mutation status for predicting RAS signaling dependence and drug response [51].
Dry and colleagues [87] were the first to develop gene expression signatures capable of predicting MEK addiction and drug response to a MEK inhibitor, selumetinib, in a large panel of diverse cell lines. These gene expression-based signatures were able to predict drug response in multiple cancer types and xenograft mouse models and provided a useful tool for studying MEK biology and application of MEK inhibitors [83]. Similarly, Tentler et al. [88] also used gene expression-based signatures to predict response to selumetinib in KRAS-mutant colorectal cancer (CRC) using both in vitro and xenograft models. This study identified 3 gene pairs (PEG10 & CYBRD1, CALB1 & NELL2, and SKAP1 & MIA) which predicted the response to selumetinib with 86% accuracy in an independent set of 14 KRAS mutant CRC cell lines. This study further validated these 3 gene pairs in human CRC explants with 71% accuracy.
With the knowledge that RAS pathway gene expression signatures can predict RAS signaling dependence more effectively than KRAS mutations alone, Sun and colleagues [89] analyzed gene expression patterns from a large number of patients with colorectal cancer and built a model for identifying activated EGFR signaling. This signature consisted of a combination of mutational signatures from patients with KRAS, BRAF, and PIK3CA mutations and characterized response to the EGFR inhibitor cetuximab. This study highlighted the use of combining multiple gene expression signatures together from various nodes in the same pathway to identify which patients will benefit from pathway inhibition [89].
Recently, El-Chaar et al. [50] used the Bild et al. RAS signature [46] to develop a network-based genomic framework for drug discovery. El-Chaar and colleagues projected the RAS signature into non-small cell lung cancer (NSCLC) cell lines to determine RAS pathway activation, then treated cell lines with targeted drug regimens along with a panel of 366 novel compounds. Results showed that combined inhibition of EGFR and MEK was effective at inhibiting RAS pathway-active cancer cells. Also, KRAS pathway activation accounted for the responsiveness to the combined EGFR/MEK inhibition, rather than KRAS mutation status alone, further highlighting the problems with relying on single genes to predict drug response. These results further illustrate the benefits of using genomic signatures to characterize oncogenic pathways in cancer, and to find drugs that target and inhibit a specific network [50]. Of note, the above-mentioned results require further research to explore whether the gene expression-based drug response signatures hold true in patient-derived samples.
4. Conclusion and future prospects
The RAS network is large and complex and consists of many interconnecting pathways that play a major role in cellular growth, evasion of apoptosis, and metastasis [33]. Cancers reliant on RAS signaling for survival are often aggressive and treatment options are limited [90]. RAS-driven tumors are challenging to treat due to the difficulties of measuring RAS-related signaling events in tumors [46], the current inability to directly target RAS proteins [91], and the inevitable development of drug resistance to targeted therapies [92]. Here, we have reviewed genomic studies showing that the RAS pathway can become activated by dysregulation of multiple nodes of the network and that gene expression and mutation signatures can be used to measure activation of the RAS network more broadly. We also highlighted how these genomic tools can predict drug response better than single genes, how genomics can identify drug strategies that target RAS, and how genomic data can be used to determine the probability of patient response to therapy. Thus, these genomics-guided findings have the potential to change how we measure RAS activity and find effective treatments for RAS-driven cancers.
Although genomics methods do hold great promise in cancer, it is also important to note some of the drawbacks and continued challenges inherent to these methods. In relation to gene expression signatures to guide drug response, clinical relevance requires clinical trials and analytical testing to validate their benefit [9]. Therefore, gene expression signatures will need to be made into clinically-relevant biomarkers, similar to OncotypeDX and MammaPrint in breast cancer [93]. Another important point is that pathways function differently depending on the cell type, specific genomic alteration, and organism [94]. For example, BRAF inhibitors work well in melanomas harboring mutations in the BRAF gene, but have no therapeutic effect in colorectal cancer patients harboring the identical BRAF mutations, due to PI3K/AKT activation common in colorectal cancers [95,96]. This highlights the dangers of generalization and the need to measure activation of the various RAS pathway nodes concurrently in individual patients.
Lastly, we would like to note that the use of genomics to capture changes in RAS network activity broadly over time will enable us to combat development of drug resistance. Current methods to assess a patient’s response to therapy, including imaging or blood tests, fail to personalize treatment regimens after drug resistance has been identified. We propose that measuring RAS pathway activation using genomics prior to time points when standard clinical tests such as computerized tomography (CT) scans are actionable will enable “real time” assessment of resistance mechanisms. Importantly, identification of the mechanisms of acquired resistance to drug inhibitors of this network, which will be feasible using genomics, will help us adapt therapy strategies to match the dynamic nature of cancer.
Overall, genomics has contributed greatly to the understanding of cancer, including RAS-driven cancers [5]. We anticipate that genomic discoveries will continue to improve our understanding of the RAS signaling network and inform new strategies for managing treatments, and that in the near future, RAS-driven tumors may no longer be considered “undruggable.”
5. Methods
5.1 EGFR, KRAS (G12V), and RAF1 gene-expression profiling data
EGFR, KRAS (G12V), and RAF1 were overexpressed in primary human mammary epithelial cells (HMECs) using recombinant adenoviruses as detailed by Bild et al. 2006, Rahman et al. 2015, and Rahman et al. [46,53, 97]. Cells were incubated with virus for 18 hours except for KRAS (G12V), which was incubated for 36 hours. KRAS virus was obtained from Vector Biolabs, RAF1 from Cell Biolabs, and EGFR was a gift from Duke University. To validate that infections worked and proteins were overexpressed we extracted protein from EGFR, KRAS (G12V), and RAF1 overexpressing cells and compared to GFP controls using western blotting methods described by Bild et al. 2006 and Rahman et al [46,53]. HMECs were probed with the following primary antibodies: EGFR (#4267), pEGFR ((#2234), KRAS (sc-30), pMEK ((#9154), p-cRAF ((#9427), GAPDH ((#5174), and β-tubulin ((#2146). All antibodies were obtained from Cell Signaling Technology, besides KRAS, which was from Santa Cruz. RNA was extracted using methods by Rahman et al. [97]. cDNA libraries were prepared from extracted RNA using the Illumina Stranded TruSeq protocol (Illumina). cDNA libraries were sequenced at Oregon Health and Sciences University using the Illumina HiSeq 2000 sequencing platform with six samples per lane. Single-end reads of 101 base pairs were generated. Log2TPM gene expression data for the EGFR, KRAS (G12V), and RAF1 pathways were all processed using methods described by Rahman et al. [53,97]. This data is available on Gene Expression Omnibus (GEO) accession numbers: GSE73628 for RAF1, GSE76877 for KRAS (G12V), and GSE59765 for EGFR.
5.2 The Cancer Genome Atlas (TCGA) data
All TCGA gene expression data was obtained from GEO accession number GSE62944 [97]. TCGA gene mutation data for EGFR, BRAF, and KRAS was downloaded from CbioPortal [98]. Any mutations found in KRAS, EGFR, or BRAF were included on heatmaps. We only included TCGA samples which had both gene expression and mutation data. The following TCGA data sets were used: head and neck squamous cell carcinoma, rectum adenocarcinoma, uterine carcinoma, lung adenocarcinoma, ovarian serous cystadenocarcinoma, breast invasive carcinoma, bladder urothelial carcinoma, and kidney renal clear cell carcinoma.
5.3 Generation of gene expression signatures
We used Adaptive Signature Selection and InteGratioN (ASSIGN; Version 1.7.1), to generate gene expression signatures. A formal definition of the ASSIGN model and software implementation was previously described [52]. RNA-Seq data from HMECs overexpressing GFP control were compared to HMECs overexpressing KRAS (G12V), RAF1, and EGFR. ASSIGN uses a Bayesian variable approach [99] to select genes with the highest weights and signal strengths, indicating differential expression. These genes represent oncogenic signatures, and are also found in Rahman et al. [53].
5.4 Batch adjustment of gene expression signatures and TCGA data
We adjusted the batch effects within and between the signatures and TCGA gene expression data using the “ComBat” function from the R package sva (version 3.16.1) [100,101]. ComBat was run using the reference-batch option, which adjusts the data to match an indicated batch. We selected the sequencing batch containing RAF1 as the reference batch. Additionally, we adjusted for background baseline gene expression differences between oncogenic signatures and test samples (TCGA patient data) using ASSIGN’s adaptive background parameter.
5.5 Optimization of single-pathway estimates in TCGA BRCA patient data
To determine the optimum number of genes for each oncogenic signature, we generated signatures with gene lists lengths from 25 to 500 genes, in 25 gene increments in breast cancer, using ASSIGN’s single pathway settings. For all of the signatures that passed internal leave-one-out-cross-validation, pathway estimates were included for further validation in mutation, gene expression, and proteomics data all described by Rahman et al. [53].
5.6 ASSIGN for all other cancers
We applied optimized gene expression signatures to head and neck squamous cell carcinoma (n=504), rectum adenocarcinoma (n=167), uterine carcinoma (n=57), lung adenocarcinoma (n=541), ovarian serous cystadenocarcinoma (n=429), breast invasive carcinoma (n=1119), bladder urothelial carcinoma (n=414), and kidney renal clear cell carcinoma (n=542) to generate pathway predictions using ASSIGN. Pathway predictions generated by ASSIGN are represented as values from zero to one. Values of zero represent no pathway activity, and values of one represent high pathway activity. We adjusted for the variation in magnitude and direction of signature-relevant gene expression between oncogenic signatures training samples and test samples using ASSIGN’s adaptive signature parameter. The code for running this analysis can be found at https://github.com/smacneil1/PANCAN24_Analysis.
Acknowledgments
AHB, SMM, GS, and JAR are funded by the NIH grant 5U01CA164720.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. International Human Genome Sequencing, Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Offit K. Personalized medicine: New genomics, old lessons. Hum. Genet. 2011;130:3–14. doi: 10.1007/s00439-011-1028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garman KS, Nevins JR, Potti A. Genomic strategies for personalized cancer therapy. Hum. Mol. Genet. 2007;16(Spec No) doi: 10.1093/hmg/ddm184. [DOI] [PubMed] [Google Scholar]
- 4.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat. Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 5.Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat. Genet. 2003;33(Suppl):238–244. doi: 10.1038/ng1107. [DOI] [PubMed] [Google Scholar]
- 6.Cancer T, Atlas G. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vucic EA, Thu KL, Robison K, Rybaczyk LA, Chari R, Alvarez CE, Lam WL. Translating cancer “omics” to improved outcomes. Genome Res. 2012;22:188–195. doi: 10.1101/gr.124354.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer Genome Landscapes. Science (80-.) 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itadani H, Mizuarai S, Kotani H. Can Systems Biology Understand Pathway Activation? Gene Expression Signatures as Surrogate Markers for Understanding the Complexity of Pathway Activation. Curr. Genomics. 2008;9:349–360. doi: 10.2174/138920208785133235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 11.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernard PS, Parker JS, Mullins M, Cheung MCU, Leung S, Voduc D, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Győrffy B, Hatzis C, Sanft T, Hofstatter E, Aktas B, Pusztai L. Multigene prognostic tests in breast cancer: past, present, future. Breast Cancer Res. 2015;17:11. doi: 10.1186/s13058-015-0514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raponi M, Winkler H, Dracopoli NC. KRAS mutations predict response to EGFR inhibitors. Curr. Opin. Pharmacol. 2008;8:413–418. doi: 10.1016/j.coph.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–58. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh H, Longo DL, Chabner BA. Improving prospects for targeting RAS. J. Clin. Oncol. 2015;33:3650–3659. doi: 10.1200/JCO.2015.62.1052. [DOI] [PubMed] [Google Scholar]
- 17.Katz M, Amit I, Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim. Biophys. Acta - Mol. Cell Res. 2007;1773:1161–1176. doi: 10.1016/j.bbamcr.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosaka T, Yatabe Y, Onozato R, Kuwano H, Mitsudomi T. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J. Thorac. Oncol. 2009;4:22–29. doi: 10.1097/JTO.0b013e3181914111. [DOI] [PubMed] [Google Scholar]
- 19.Osumi H, Shinozaki E, Suenaga M, Matsusaka S, Konishi T, Akiyoshi T, et al. RAS mutation is a prognostic biomarker in colorectal cancer patients with metastasectomy. Int. J. Cancer. 2016 doi: 10.1002/ijc.30106. [DOI] [PubMed] [Google Scholar]
- 20.Vauthey J-N, Zimmitti G, Kopetz SE, Shindoh J, Chen SS, Andreou A, et al. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann. Surg. 2013;258:619–626. doi: 10.1097/SLA.0b013e3182a5025a. discussion 626–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young A, Lyons J, Miller AL, Phan VT, Alarcón IR, McCormick F. Ras signaling and therapies. Adv. Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]
- 22.Adjei A. Blocking oncogenic Ras signaling for cancer therapy. J. Natl. Cancer Inst. 2001;93:1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 23.Passot G, Chun YS, Kopetz SE, Overman MJ, Conrad C, Aloia TA, Vauthey JN. Prognostic factors after resection of colorectal liver metastases following preoperative second-line chemotherapy: Impact of RAS mutations. Eur. J. Surg. Oncol. 2016 doi: 10.1016/j.ejso.2016.02.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EWT, Chang F, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta - Mol. Cell Res. 2007;1773:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez-Viciana P, McCormick F. RalGDS comes of age. Cancer Cell. 2005;7:205–206. doi: 10.1016/j.ccr.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 26.Samatar AA, Poulikakos PI. Targeting RAS–ERK signalling in cancer: promises and challenges. Nat. Rev. Drug Discov. 2014;13:928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 27.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 28.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J. 2000;351(Pt 2):289–305. [PMC free article] [PubMed] [Google Scholar]
- 30.Ehrhardt A, Ehrhardt GRA, Guo X, Schrader JW. Ras and relatives - Job sharing and networking keep an old family together. Exp. Hematol. 2002;30:1089–1106. doi: 10.1016/s0301-472x(02)00904-9. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Xiao Y, Ping Y, Li J, Zhao H, Li F, Hu J, Zhang H, Deng Y, Tian J, Li X. Integrating multi-omics for uncovering the architecture of cross-talking pathways in breast cancer. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lock R, Cichowski K. Loss of negative regulators amplifies RAS signaling. Nat. Genet. 2015;47:426–427. doi: 10.1038/ng.3299. [DOI] [PubMed] [Google Scholar]
- 33.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Towatari M, Nakao A, Iida H, Kiyoi H, Nakano Y, Tanimoto M, Saito H, Naoe T. Lack of constitutive activation of MAP kinase pathway in human acute myeloid leukemia cells with N-Ras mutation. Leukemia. 1999;13:585–589. doi: 10.1038/sj.leu.2401369. http://ovidsp.ovid.com/ovidweb.cgi?T=JS&PAGE=reference&D=med4&NEWS=N&AN=10214865. [DOI] [PubMed] [Google Scholar]
- 35.Natarajan K, Berk BC. Crosstalk coregulation mechanisms of G protein-coupled receptors and receptor tyrosine kinases. Methods Mol. Biol. 2006;332:51–77. doi: 10.1385/1-59745-048-0:51. [DOI] [PubMed] [Google Scholar]
- 36.Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. advance on. 2014:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence MS, Sougnez C, Lichtenstein L, Cibulskis K, Lander E, Gabriel SB, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raphael BJ, Vandin F. Simultaneous Inference of Cancer Pathways and Tumor Progression from Cross-Sectional Mutation Data. J. Comput. Biol. 2015;22:510–527. doi: 10.1089/cmb.2014.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Downward J. Cancer biology: signatures guide drug choice. Nature. 2006;439:274–275. doi: 10.1038/439274a. [DOI] [PubMed] [Google Scholar]
- 42.Watters JW, Roberts CJ. Developing gene expression signatures of pathway deregulation in tumors. Mol. Cancer Ther. 2006;5:2444–2449. doi: 10.1158/1535-7163.MCT-06-0340. [DOI] [PubMed] [Google Scholar]
- 43.Chibon F. Cancer gene expression signatures-The rise and fall? Eur. J. Cancer. 2013;49:2000–2009. doi: 10.1016/j.ejca.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 44.Huang E, Ishida S, Pittman J, Dressman H, Bild A, Kloos M, et al. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat. Genet. 2003;34:226–230. doi: 10.1038/ng1167. [DOI] [PubMed] [Google Scholar]
- 45.Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat. Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 46.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 47.Chang JT, Carvalho C, Mori S, Bild AH, Gatza ML, Wang Q, et al. A Genomic Strategy to Elucidate Modules of Oncogenic Pathway Signaling Networks. Mol. Cell. 2009;34:104–114. doi: 10.1016/j.molcel.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ooi CH, Ivanova T, Wu J, Lee M, Tan IB, Tao J, et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. A Gene Expression Signature Associated with “K-Ras Addiction” Reveals Regulators of EMT and Tumor Cell Survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El-Chaar NN, Piccolo SR, Boucher KM, Cohen AL, Chang JT, Moos PJ, Bild AH. Genomic classification of the RAS network identifies a personalized treatment strategy for lung cancer. Mol. Oncol. 2014;8:1339–1354. doi: 10.1016/j.molonc.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loboda A, Nebozhyn M, Klinghoffer R, Frazier J, Chastain M, Arthur W, Roberts B, et al. A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC Med. Genomics. 2010;3:26. doi: 10.1186/1755-8794-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen Y, Rahman M, Piccolo SR, Gusenleitner D, El-Chaar NN, Cheng L, et al. ASSIGN: Context-specific genomic profiling of multiple heterogeneous biological pathways. Bioinformatics. 2015:1745–1753. doi: 10.1093/bioinformatics/btv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahman M, MacNeil SM, Jenkins DF, Piccalo SR, Shrestha G, Wyatt SR, et al. Discrete breast cancer growth and survival phenotypes influence apoptosis and drug response. Genome Biol. (Submitted) [Google Scholar]
- 54.Hansen AR, Siu LL. Epidermal growth factor receptor targeting in head and neck cancer: Have we been just skimming the surface? J. Clin Oncol. 2013;31:1381–1383. doi: 10.1200/JCO.2012.47.9220. [DOI] [PubMed] [Google Scholar]
- 55.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Der CJ, Krontiris TG, Cooper GM. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc. Natl. Acad. Sci. U. S. A. 1982;79:3637–3640. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parada LF, Tabin CJ, Shih C, Weinberg RA. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297:474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 58.Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haluska P, Dy GK, Adjei AA. Farnesyl transferase inhibitors as anticancer agents. Eur. J. Cancer. 2002;38:1685–1700. doi: 10.1016/s0959-8049(02)00166-1. [DOI] [PubMed] [Google Scholar]
- 60.Basso AD. Thematic review series: Lipid Posttranslational Modifications. Farnesyl transferase inhibitors. J. Lipid Res. 2005;47:15–31. doi: 10.1194/jlr.R500012-JLR200. [DOI] [PubMed] [Google Scholar]
- 61.Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 62.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission Possible? Nat. Rev. Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chandrashekar R, Adams PD. Prospective Development of Small Molecule Targets to Oncogenic Ras Proteins. Open J. Biophys. 2013;3:207–211. doi: 10.4236/ojbiphy.2013.34025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lim SM, Westover KD, Ficarro SB, Harrison RA, Choi HG, Pacold ME, et al. Therapeutic targeting of oncogenic K-ras by a covalent catalytic site inhibitor. Angew. Chemie - Int. Ed. 2014;53:199–204. doi: 10.1002/anie.201307387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016;6:316–329. doi: 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- 67.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chemie - Int. Ed. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 70.Bromberg-white JL, Andersen NJ, Duesbery NS. Mek genomics in development and disease. Brief. Funct. Genomics. 2012;11:300–310. doi: 10.1093/bfgp/els022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 72.Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br. J. Cancer. 2006;94:455–459. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whittaker SR, Cowley GS, Wagner S, Luo F, Root DE, Garraway LA. Combined Pan-RAF and MEK Inhibition Overcomes Multiple Resistance Mechanisms to Selective RAF Inhibitors. Mol. Cancer Ther. 2015;14:2700–2711. doi: 10.1158/1535-7163.MCT-15-0136-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nissan MH, Rosen N, Solit DB. ERK pathway inhibitors: How low should we go? Cancer Discov. 2013;3:719–721. doi: 10.1158/2159-8290.CD-13-0245. [DOI] [PubMed] [Google Scholar]
- 75.Stark A-K, Sriskantharajah S, Hessel EM, Okkenhaug K. PI3K inhibitors in inflammation, autoimmunity and cancer. Curr. Opin. Pharmacol. 2015;23:82–91. doi: 10.1016/j.coph.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keating GM. Idelalisib: a review of its use in chronic lymphocytic leukaemia and indolent non-Hodgkin’s lymphoma. Target. Oncol. 2015;10:141–151. doi: 10.1007/s11523-015-0359-8. [DOI] [PubMed] [Google Scholar]
- 77.Chappell WH, Steelman LS, Long JM, Kempf RC, Abrams SL, Franklin RA, et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget. 2011;2:135–164. doi: 10.18632/oncotarget.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jokinen E, Koivunen JP. MEK and PI3K inhibition in solid tumors: rationale and evidence to date. Ther. Adv. Med. Oncol. 2015;7:170–180. doi: 10.1177/1758834015571111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jokinen E, Laurila N, Koivunen JP. Alternative dosing of dual PI3K and MEK inhibition in cancer therapy. BMC Cancer. 2012;12:612. doi: 10.1186/1471-2407-12-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 81.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 82.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao Z, Chen CC, Rillahan CD, Shen R, Kitzing T, McNerney ME, et al. Cooperative loss of RAS feedback regulation drives myeloid leukemogenesis. Nat. Genet. 2015;47:539–543. doi: 10.1038/ng.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bild AH, Potti A, Nevins JR. Linking oncogenic pathways with therapeutic opportunities. Nat. Rev. Cancer. 2006;6:735–741. doi: 10.1038/nrc1976. [DOI] [PubMed] [Google Scholar]
- 86.Blum R, Elkon R, Yaari S, Zundelevich A, Jacob-Hirsch J, Rechavi G, Shamir R, Kloog Y. Gene expression signature of human cancer cell lines treated with the Ras inhibitor salirasib (S-farnesylthiosalicylic acid) Cancer Res. 2007;67:3320–3328. doi: 10.1158/0008-5472.CAN-06-4287. [DOI] [PubMed] [Google Scholar]
- 87.Dry JR, Pavey S, Pratilas CA, Harbron C, Runswick S, Hodgson D, et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244) Cancer Res. 2010;70:2264–2273. doi: 10.1158/0008-5472.CAN-09-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tentler JJ, Nallapareddy S, Tan AC, Spreafico A, Pitts TM, Morelli MP, et al. Identification of predictive markers of response to the MEK1/2 inhibitor selumetinib (AZD6244) in K-ras-mutated colorectal cancer. Mol. Cancer Ther. 2010;9:3351–3362. doi: 10.1158/1535-7163.MCT-10-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tian S, Simon I, Moreno V, Roepman P, Tabernero J, Snel M, et al. A combined oncogenic pathway signature of BRAF, KRAS and PI3KCA mutation improves colorectal cancer classification and cetuximab treatment prediction. Gut. 2013;62:540–549. doi: 10.1136/gutjnl-2012-302423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stephen AG, Esposito D, Bagni RG, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 91.Gysin S, Salt M, Young A, McCormick F. Therapeutic strategies for targeting ras proteins. Genes Cancer. 2011;2:359–372. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McCubrey JA, Abrams SL, Fitzgerald TL, Cocco L, Martelli AM, Montalto G, et al. Roles of signaling pathways in drug resistance, cancer initiating cells and cancer progression and metastasis. Adv. Biol. Regul. 2015;57:75–101. doi: 10.1016/j.jbior.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 93.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N. Engl. J. Med. 2009;360 doi: 10.1056/NEJMra0801289. 790–800+752. [DOI] [PubMed] [Google Scholar]
- 94.Grueneberg DA, Degot S, Pearlberg J, Li W, Davies JE, Baldwin A, et al. Kinase requirements in human cells: I. Comparing kinase requirements across various cell types. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16472–16477. doi: 10.1073/pnas.0808019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mao M, Tian F, Mariadason JM, Tsao CC, Lemos R, Dayyani F, et al. Resistance to BRAF inhibition in BRAF-mutant colon cancer can be overcome with PI3K inhibition or demethylating agents. Clin. Cancer Res. 2013;19:657–667. doi: 10.1158/1078-0432.CCR-11-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rahman M, Jackson LK, Johnson WE, Li DY, Bild AH, Piccolo SR. Alternative preprocessing of RNA-Sequencing data in the Cancer Genome Atlas leads to improved analysis results. Bioinformatics. 2015;31:3666–3672. doi: 10.1093/bioinformatics/btv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013;6:pl1–pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.George EI, Mcculloch RE. Approaches for bayesian variable selection. Stat. Sin. 1997;7:339–373. [Google Scholar]
- 100.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]