SUMMARY
Several microsatellite-expansion diseases are characterized by the accumulation of RNA foci and RAN proteins raising the possibility of a mechanistic connection. We explored this question using myotonic dystrophy type 2, a multisystemic disease thought to be primarily caused by RNA gain-of-function effects. We demonstrate the DM2 CCTG•CAGG expansion expresses sense and antisense tetrapeptide poly-(LPAC) and poly-(QAGR) RAN proteins, respectively. In DM2 autopsy brains, LPAC is found in neurons, astrocytes and glia in grey matter and antisense QAGR proteins accumulate within white matter. LPAC and QAGR proteins are toxic to cells independent of RNA gain of function. RNA foci and nuclear sequestration of CCUG transcripts by MBNL1 is inversely correlated with LPAC expression. These data suggest a model, that involves nuclear retention of expansion RNAs by RNA-binding proteins (RBPs) and an acute phase in which expansion RNAs exceed RBP sequestration capacity, are exported to the cytoplasm and undergo RAN translation.
INTRODUCTION
Myotonic dystrophy (DM), one of the most common forms of muscular dystrophy, can be caused by a CTG expansion in the 3’ UTR of DMPK (myotonic dystrophy type 1, DM1) (Brook et al., 1992; Fu et al., 1992; Mahadevan et al., 1992) or an intronic CCTG expansion in CNBP (myotonic dystrophy type 2, DM2) (Liquori et al., 2001). Although both diseases have marked effects on multiple organ systems, including skeletal muscle, the heart, the eye and the endocrine system, the clinical significance of CNS involvement cannot be overstated (Charizanis et al., 2012; Harper, 1989; Meola, 2010; Minnerop et al., 2011). The DM1 and DM2 mutations differ in their effects on the brain. While a subset of DM1 patients with developmental features have mental retardation not found in DM2, a late-onset CNS phenotype involving executive function deficits and white matter abnormalities is common to both disorders. Since CNS abnormalities in DM significantly impact quality of life, there is great clinical need to understand the pathophysiological basis for these changes and to target pathways to slow or reverse the CNS effects. The striking clinical parallels of DM1 and DM2, combined with the apparent non-coding locations of the expansion mutations, helped to establish that CUGEXP and CCUGEXP RNAs cause RNA gain of function (GOF) effects (Liquori et al., 2001). Additionally, the accumulation of CUG- or CCUG- expansion RNAs dysregulate RNA-binding proteins including MBNL and CELF proteins, which leads to RNA processing abnormalities (Kanadia et al., 2003; Liquori et al., 2001; Mankodi et al., 2001; Miller et al., 2000; Ranum and Cooper, 2006; Savkur et al., 2004).
Although substantial data demonstrate that RNA processing abnormalities are found in DM, recent discoveries that change our understanding of how microsatellite expansion mutations are expressed, must also now be considered. First, much of the genome (Katayama et al., 2005) and a growing number of expansion mutations have been shown to be bidirectionally transcribed, including the DM1 CTG•CAG expansion (Cho et al., 2005; Ladd et al., 2007; Libby et al., 2008; Moseley et al., 2006). Additionally, the discovery of repeat associated non-ATG (RAN) translation (Zu et al., 2011) and its growing involvement in neurodegenerative diseases caused by microsatellite expansions (Ash et al., 2013; Banez-Coronel et al., 2015; Ishiguro et al., 2017; Krans et al., 2016; Mori et al., 2013; Todd et al., 2013; Zu et al., 2013) raises the possibility that RAN proteins contribute to the CNS features of myotonic dystrophy.
Here we show the tetranucleotide DM2 CCTG•CAGG expansion mutation is bidirectionally transcribed and the resulting RNAs are RAN translated producing tetrapeptide expansion proteins with Leu-Pro-Ala-Cys (LPAC) from the sense strand or Gln-Ala-Gly-Arg (QAGR) repeats from the antisense strand, and that these proteins accumulate in DM2 patient brains. Additionally, we show nuclear sequestration of CCUG transcripts by MBNL proteins prevents the expression of the LPAC RAN protein suggesting a two-phase model to explain the roles of toxic RNAs and proteins in DM2 and potentially other microsatellite repeat expansion diseases.
RESULTS
Bidirectional transcription and RAN translation across the DM2 expansion mutation
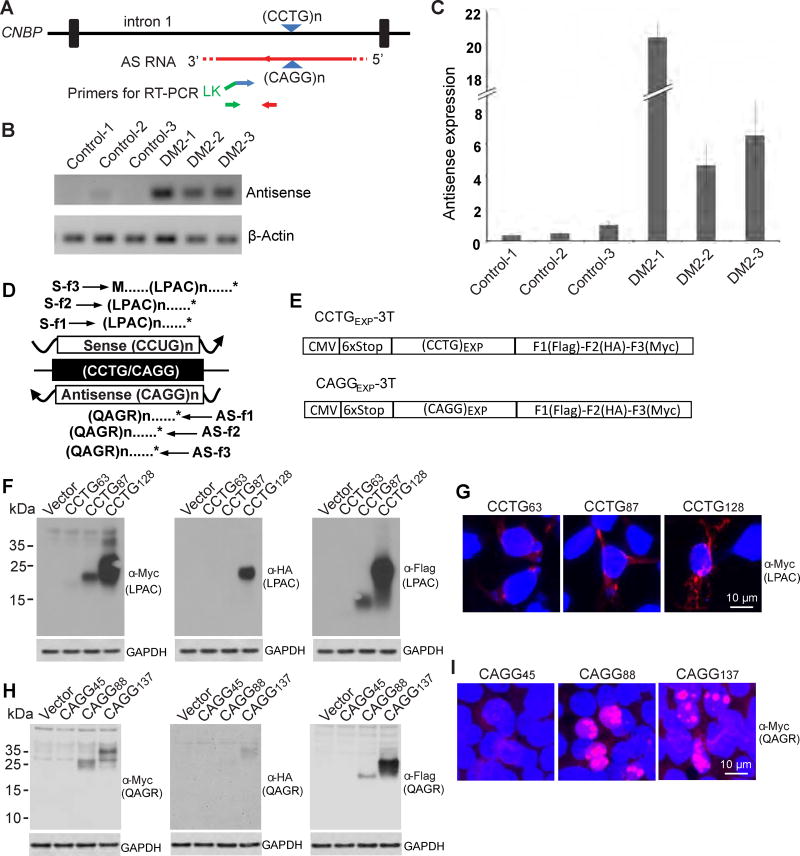
Because a growing number of expansion mutations are bidirectionally transcribed and undergo RAN translation we performed experiments to test if antisense RNAs and RAN proteins play a role in DM2. First, we performed RT-PCR on frontal cortex from DM2 and control human autopsy brains to test if CAGG antisense expansion transcripts are expressed. Strand-specific RT-PCR performed using primers downstream of the antisense CAGG expansion (Figure 1A) by semi-quantitative- and qRT-PCR show antisense transcripts are dramatically increased (~5->20-fold) relative to β-actin in DM2 cases compared to controls (Figures 1B, 1C, and S1A).
Figure 1.
Antisense transcripts in DM2 and tetrapeptide RAN proteins expressed across CCUG and CAGG expansion RNAs. (A) Schematic diagram of CAGG antisense transcripts and relative location of primers for strand-specific RT-PCR. (B, C) qRT-PCR showing elevated antisense mRNA relative to β-actin in DM2 compared with controls. n=3 samples/group, experiments performed at least three times, error bars show standard deviation (SD) with at least three technical replicates (D) Diagram of putative proteins translated from sense and antisense DM2 transcripts. (E) Non-ATG CCTG and CAGG constructs with 6X stop-codon cassette, two stops in each frame, upstream of the CCTG expansion with C-terminal tags in all three reading frames. Immunoblots of transfected HEK293T cells show expanded LPAC proteins (F) and QAGR proteins (H) are expressed in all three frames. IF detection of (LPAC)EXP proteins (G) and (QAGR)EXP proteins (I) in transfected HEK293T cells. At least three independent transfections were performed for panels F-I. See also Figure S1.
The discovery that CAGG antisense expansion transcripts are expressed in DM2 brain, raises the possibility that RAN proteins may be generated from both sense and antisense transcripts. RAN proteins expressed across the CCUG and CAGG expansion transcripts would result in the production of expansion proteins containing Leu-Pro-Ala-Cys (LPAC) or Gln-Ala-Gly-Arg (QAGR) repeat motifs, respectively (Figure 1D). While sense and antisense RAN proteins would be predicted to express LPAC or QAGR repeat motifs in all three reading frames, the C-terminal regions of the proteins produced in each reading frame would be unique.
To test if the DM2 tetranucleotide expansion RNAs can undergo RAN translation, we generated CCTG and CAGG expansion constructs with C-terminal epitope-tags in all three reading frames. These constructs have upstream stop codons in all three reading frames and no ATG-initiation codons upstream of the repeat (Figure 1E). Protein blots from transfected cells show LPAC and QAGR RAN proteins are expressed from these minigenes in all three reading frames (Figures 1F and 1H) without an ATG-initiation codon. Constructs with 128 CCTGs robustly express (LPAC)EXP proteins in all three frames with lower and only trace amounts detected with 87 and 63 CCTG repeats, respectively (Figure 1F). The molecular weight differences of proteins expressed in different frames are consistent with differences in repeat lengths and the lengths of the predicted frame specific C-terminal regions of these individual proteins. Repeat length dependent accumulation of QAGR is also found by protein blot from constructs containing CAGG expansions with detectable levels seen with 137, lower levels and 88 and no detectable expression with 45 repeats (Figure 1H).
Immunofluorescence of transfected cells using antibodies against the C-terminal epitopes show LPAC RAN proteins are mainly cytoplasmic showing both diffuse and punctate staining (Figures 1G and S1B) with the strongest LPAC staining in cells transfected with 128 CCTG repeats, moderate staining with 87 CCTGs and occasional staining with 63 CCTGs. In contrast, QAGR proteins show a distinct pattern of nuclear staining detected in cells transfected with 137 and 88 but not 45 repeats (Figures 1I and S1C).
In summary, we show evidence that antisense CAGG expansion transcripts are upregulated in DM2 patient brains. Additionally, we demonstrate in transfected cells that RAN translation can occur in all three reading frames across both CCUG and CAGG expansion transcripts producing LPAC and QAGR tetrapeptide proteins respectively, and that protein accumulation increases with longer repeat tracts.
LPAC and QAGR RAN proteins accumulate in human DM2 brains
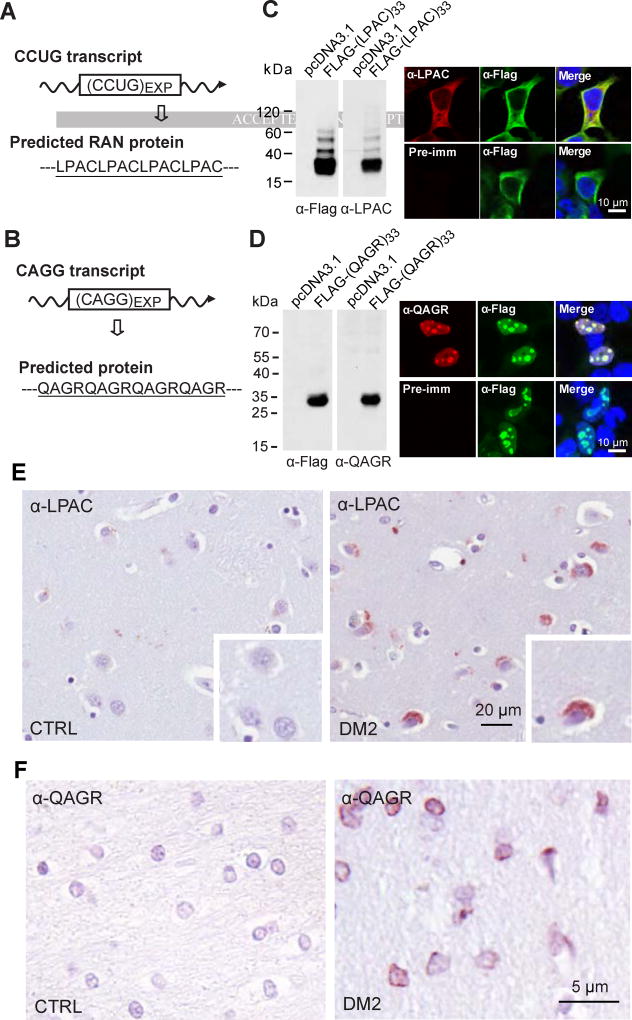
To determine if novel DM2 RAN proteins are expressed in vivo, we generated polyclonal antibodies to the putative DM2 LPAC and QAGR repeat motifs using (LPAC)4 or (QAGR)4 peptides (Figures 2A and 2B). The specificity of these antibodies was demonstrated by showing that α-LPAC and α-QAGR antibodies recognize recombinant flag-tagged proteins expressed in transfected cells by both immunoblot and immunofluorescence (Figures 2C and 2D). Similar to the staining seen with the epitope antibodies, α-LPAC showed diffuse cytoplasmic staining and α-QAGR showed nuclear staining in transfected cells. These validated antibodies were used to test if LPAC and QAGR RAN proteins are expressed in DM2 patients. Immunohistochemistry (IHC) using the α-LPAC and α-QAGR antibodies showed positive staining in multiple brain regions of human DM2 but not DM1 or control autopsy tissue (Figures 2E and 2F; Tables 1 and 2) or with preimmune serum (Figures S2A and 2B). Similar to the staining patterns found in transfected cells, in DM2 autopsy tissue α-LPAC shows cytoplasmic staining with small punctate aggregates and α-QAGR shows primarily nuclear staining with small punctate aggregates located at or close to the nuclear membrane.
Figure 2.
LPAC and QAGR RAN proteins accumulate in human DM2 brain. DM2 pre-mRNA and predicted RAN protein with underlined peptide used for antibody generation for sense (A) and antisense (B) RAN proteins. Antibody validation by detection of recombinant FLAG-tagged LPAC (C) or QAGR protein (D) by α-Flag and the respective custom antibody (α-LPAC or α-QAGR) by protein blot (left) or IF (right). Negative pre-immune controls are shown for IF. IHC staining of human DM2 and control autopsy tissue using α-LPAC (E) or α-QAGR (F) antibodies. At least three independent experiments were performed for panels C-D and panels E-F show examples based on the analysis of 4 DM2 and 3 control cases. See also Figure S2.
Table 1.
Expression of RAN LPAC proteins in DM2 brain. (−) no RAN-positive cells, (+) occasional, (++) moderate, (+++) numerous RAN-positive cells.
| Autopsy Cases | Cortex | Hippocampus | Striatum |
|---|---|---|---|
| (DM2) Case 1 | + | + | +++ |
| (DM2) Case 2 | +/− | +/− | |
| (DM2) Case 3 | ++ | + | +++ |
| (DM2) Case 4 | + | +++ | +/− |
| Control 1 | − | − | − |
| Control 2 | − | − | − |
| Control 3 | − | − | − |
Table 2.
Expression of RAN QAGR proteins in DM2 brain. (−) no RAN-positive cells, (+) occasional, (++) moderate, (+++) numerous RAN-positive cells.
| Autopsy Cases | Cortex | Hippocampus | Striatum |
|---|---|---|---|
| (DM2) Case 1 | +++ | ++ | |
| (DM2) Case 2 | +/− | + | |
| (DM2) Case 3 | + | ++ | |
| (DM2) Case 4 | +++ | − | − |
| Control 1 | − | − | − |
| Control 2 | − | − | − |
| Control 3 | − | − | − |
Cytoplasmic LPAC proteins accumulate primarily in grey matter regions of the brain
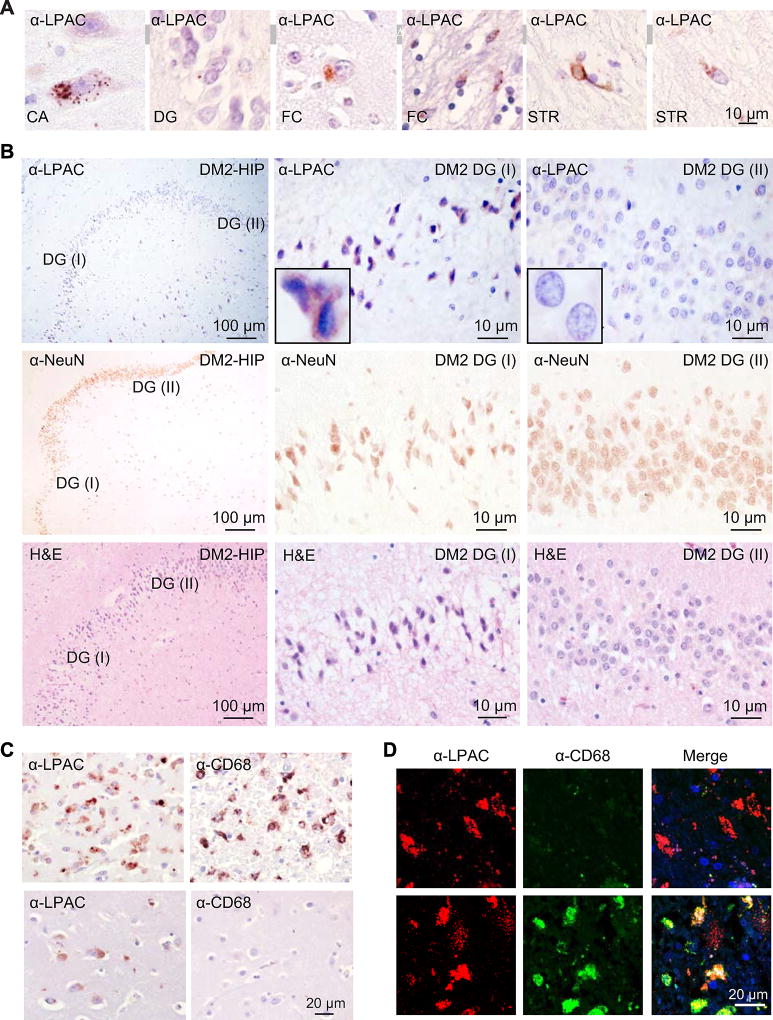
LPAC staining was consistently detected in grey matter regions of the brain. In the hippocampus, α-LPAC staining was found in the pyramidal cells throughout the cornu ammonis (CA) and in granular neurons of the dentate gyrus (DG) regions (Figure 3A). In cortex and striatum, LPAC staining was found in the neurons and occasionally glia (Figure 3A) with staining typically showing punctate cytoplasmic aggregates that can be perinuclear and can also be seen in neuronal processes.
Figure 3.
LPAC RAN protein accumulation, neurodegeneration and microglial activation in DM2 brain. (A) Representative examples of LPAC RAN proteins detected by IHC in DM2 frontal cortex (FC), hippocampus (DG and CA), and striatum (STR). (B) IHC staining of human DM2 hippocampus shows neurodegeneration, confirmed by NeuN and H&E staning (middle and bottom panel), in regions of the dentate gyrus (DG) expressing high levels of LPAC proteins (DG-I top panel) but not in the regions with low levels of LPAC proteins (DG-II top panel). (C) IHC staining of human DM2 brain using α-CD68 or α-LPAC. In DM2 sections, variable levels of staining were observed with high levels of CD68 and LPAC staining in region of organizing necrosis (top) and low levels of CD68 in areas with moderate neuronal LPAC staining (bottom). (D) LPAC RAN protein accumulation in both microglial cells and neurons. Double immunofluorescence shows colocalization of α-CD68 and α-LPAC suggesting that LPAC proteins are expressed in macrophages around blood vessels. LPAC proteins are also detected in CD68-negative neurons. See also Figure S3.
LPAC staining varies substantially between brain regions. Clusters of cells with frequent punctate LPAC aggregates were seen in some regions with little to no staining seen in other regions, even on the same section. For example, abundant clusters of cells with staining were found in the DG and CA4 regions of the hippocampus in the DM2 section shown in Figure 3B. Pyknotic nuclei were found in regions with intense α-LPAC staining in the granule cell layer. In contrast, tissue within neighboring regions of the same section with minimal LPAC staining appeared healthy. Similarly, NeuN staining to label neuronal cells was reduced in regions with intense (DG I) but not with rare (DG II) LPAC staining in the granule cell layer of dentate gyrus (Figure 3B, middle panel). The fact that this hippocampal lesion is focal and all cells in this LPAC positive region appear to be in the same stage of early degeneration by H&E (Figure 3B, lower panel), suggests the possibility that the neuronal changes in this region resulted from a recent focal event. Variability of LPAC staining was found in all DM2 autopsy cases that were examined (Table 1). The variability in RAN protein staining in DM2 is comparable to the variable staining previously reported in C9ORF72 ALS/FTD autopsies (Zu et al., 2013). Taken together, these findings demonstrate that LPAC RAN proteins accumulate in neurons and glia within the grey matter regions, and that LPAC RAN proteins are found in brain regions with focal neurodegeneration.
RAN proteins accumulate in macrophages
One of the DM2 autopsy cases we studied had a history of hypertension with widespread small vessel cerebrovascular disease and localized areas of infarction. In this patient, we found intense staining of LPAC protein aggregates in a region of infarction and organizing necrosis within the basal ganglia (Figure 3C, upper left). This region also showed widespread infiltration of CD68-positive macrophages/microglia (Figure 3C, upper right). LPAC staining was also observed at lower intensity in neighboring neurons in both the caudate nucleus and putamen, but neither of these regions showed macrophage infiltration (e.g. Figure 3C, lower panels). As a control for LPAC antibody specificity we examined three DM2 negative control autopsy cases with similar infarct regions and showed these controls had no similar staining (e.g. Figures S3A and S3B).
Because neuroinflammation involving monocyte and microglial recruitment to sites of pathology has been implicated in several neurological disorders including Huntington’s disease (Bjorkqvist et al., 2008), it is possible that a subset of CD68(+) macrophages/microglia were recruited to sites of LP AC protein accumulation and phagocytosed dead or dying LPAC positive cells. Consistent with phagocytosis by macrophages/microglia, double immunofluorescence staining using α-CD68 and α-LPAC showed cells with LPAC alone (Figure 3D, upper panels) and a subset of CD68 (+) cells also positive for the LPAC protein (Figure 3D, bottom panels). In addition, we found infiltrating CD68 (+) macrophages that were not LPAC positive (Figure S3C, right panels). In summary, we show that macrophages/microglia can be found at sites of LPAC RAN protein accumulation and that a subset of these cells are positive for LPAC protein. Additional work will be required to determine if CD68 positive macrophages/microglia can express LPAC protein or if a subset of these cells become positive because they have phagocytosed the protein.
FISH-IF detection of CCUG RNA foci and LPAC RAN proteins in human autopsy brains
RNA gain of function effects and CCUG RNA foci have been previously reported in DM2 (Liquori et al., 2001). RNA FISH was performed and detected the presence of CCUG RNA foci in all three DM2 brains examined. FISH combined with IF detected CCUG RNA foci as a consistent feature of the disease whereas LPAC protein staining was more variable (Figure S3D). An example of an RNA FISH-IF experiment using frontal cortex from one patient shows marked variation in the number and size of CCUG RNA foci and of LPAC RAN protein staining in various cell types (Figure S3D).
Antisense QAGR RAN proteins found in white matter brain regions
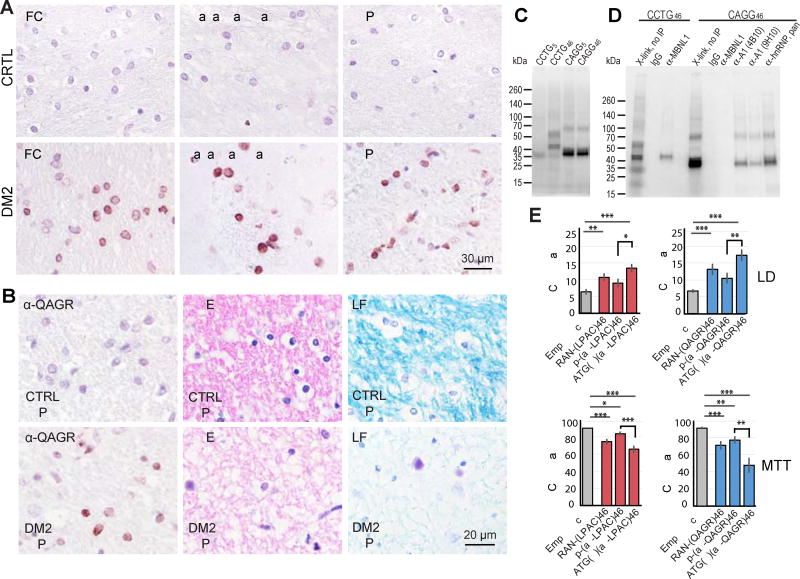
In contrast to LPAC, IHC shows that QAGR RAN proteins have a distinct distribution pattern and primarily accumulate in the white matter regions of the brain. QAGR staining was detectable in the frontal cortex, basal ganglia, and hippocampus from DM2 but not control autopsy brains (Figure 4A; Table 2). While staining in the white matter regions is a consistent feature found in the DM2 cases we examined, staining is patchy with focal areas showing QAGR protein accumulation that differ between patients. Positive staining in all regions appeared as diffuse or punctate nuclear aggregates often located at or near the nuclear membrane and staining was primarily seen in oligodendrocytes. In the frontal cortex, QAGR staining is found in the subcortical white matter, but in contrast to LPAC, was not detected in cortical layers I to V. At the grey-white-matter boundary, in layer VI, immunoreactive QAGR positive glial cells were detected at low frequency (Figure S4A). Consistent with neuroinflammatory changes, H&E staining of the serial sections showed activated microglia and astrocytes are found in QAGR positive white matter regions in the frontal lobe S4B).
Figure 4.
QAGR RAN protein accumulation in human DM2 autopsy brains, identification of hnRNP A1 as CAGG RNA-binding protein and cellular toxicity assays. (A) IHC showing QAGR immunostaining in frontal cortex, basal ganglia and hippocampus in DM2 but not normal control autopsy tissue. (B) IHC showing QAGR immunostaining (left), hematoxylin and eosin (H&E) staining (middle panels), luxol fast blue (LFB) staining of white matter (right panels). (C) UV crosslinking assay of HEK293T cell extracts with CCUG and CAGG RNA probe. CCUG46, but not CCUG5, forms two predominant RNA-protein complexes. Both CAGG5 and CAGG46 also form two predominant RNA-protein complexes. (D) Coupled Crosslinking and Immunoprecipitation show that CCUG46-protein complexes contain MBNL1 and CAGG46-protein complexes contain hnRNP A1. (E) Cell death measured by lactate dehydrogenase (LDH) assay and cell viability (MTT) assays of T98 cells transfected with vectors expressing CCTG repeats or alternative codons encoding LPAC (red) and vectors carrying CAGG repeats or alternative codon repeats encoding QAGR (blue). Bars show average ± SEM, n = 10). (asterisks: *p<0.0125, **p ≤ 0.0025, ***p ≤ 0.00025). See also Figure S4 and S5.
In the basal ganglia, QAGR RAN proteins were detected in three of four DM2 cases tested, with abundant nuclear staining found in the white matter bundles of the striatum, but not in the internal capsule. In areas of the caudate nucleus and the putamen, QAGR RAN proteins were found in glial cells. In the hippocampus, QAGR staining was found in two of the four cases examined. In one of these cases, intense staining and atrophy was found in the white matter tracts of the alveus and fimbria of the fornix (Figure 4B). This region represents the major efferent pathways to other brain areas, especially the limbic system. In another case, QAGR RAN staining was detected in the inferior longitudinal fasciculus, a region that connects the temporal lobe and occipital lobes (data not shown).
Because QAGR protein staining is prominently found in white matter regions and because white matter abnormalities are a consistent feature of DM2 seen by MRI and pathological examination (Minnerop et al., 2011; Ogata et al., 1998) we examined white matter integrity by H&E and luxol fast blue (LFB) staining. Consistent with the possibility that QAGR proteins contribute to DM2 white matter abnormalities, H&E and LFB staining of QAGR-positive regions using serial sections frequently showed rarefaction of fibers suggesting either axonal loss or increased tissue-water intercalating among the fibers (Figure 4B).
In summary, we show QAGR proteins accumulate in a pattern completely distinct from LPAC and are found in various white matter regions throughout the brain. Protein accumulation is primarily found in the nuclei of oligodendrocytes. Additionally, we show specific sites of QAGR RAN protein staining are found in regions with neuropathological changes including the accumulation of activated microglia, increased astrocytes and white-matter rarefaction.
CAGG repeats bind hnRNP A1 but not MBNL1 by CLIP
To further explore the role of CAGG expansion transcripts in DM2 autopsy tissue we performed RNA FISH but found no convincing RNA foci. To identify potential CAGG RNA-binding proteins we performed UV crosslinking and CLIP experiments. UV crosslinking and PAGE shows CAGG and CCUG transcripts bind distinct proteins (Figure 4C). CLIP using α-MBNL1 confirms that CCUG transcripts bind MBNL1 (Figure 4D). In contrast, CAGG expansions do not crosslink to MBNL1, the prominent 38 kDa crosslinked protein and the binding motif suggested hnRNP A1 as a candidate CAGG RBP. CLIP using two anti-hnRNP A1 monoclonal antibodies, as well as an anti hnRNP pan antibody, identifies hnRNP A1 as a novel CAGG RNA-binding protein (Figure 4D).
LPAC and QAGR proteins are toxic independent of RNA effects
To determine if LPAC and QAGR proteins are toxic independent of RNA gain of function effects in neuronal cells we generated (+/−) ATG-initiated codon-replacement constructs, ATG(+)(alt-LPAC)46 and ATG(+)(alt-QAGR)46 (Figure S5A) and compared them with CCUG RAN-(LPAC)46 and CAGG RAN-(QAGR)46. These alternative codon constructs express LPAC or QAGR expansion proteins using an ATG initiation codon from non-hairpin-forming transcripts. Using these constructs, we tested the toxicity of the LPAC and QAGR proteins independent of RNA gain of function effects. The lactate dehydrogenase (LDH) assay measures LDH release in cells with damaged membranes (cell death) and the methylthiazol tetrazolium (MTT) assay measures metabolic activity (cell viability). For the LDH assay, T98 cells expressing LPAC and QAGR with alternative codons showed significant increases in cell death compared to empty vector (p<0.00025) and (−)ATG alternative-codon controls (p<0.0125 for LPAC and p<0.0025 for QAGR) (Figure 4E). Additionally, the MTT assay shows alternative codon constructs expressing LPAC and QAGR decrease cell viability compared to empty vector (p<0.00025) and (−)ATG alternative codon controls (p<0.00025 for LPAC and 0.0025 QAGR) (Figure 4E). Additional controls include: 1) protein blots showing levels of LPAC and QAGR protein made from the various constructs; 2) protein blot confirmation that alternative-codon constructs for LPAC and QAGR that lack an ATG initiation codon do not make protein (Figures S5B and S5C); 3) qRT PCR showing levels of transcripts are comparable for all constructs (Figure S5D).
Taken together, these data show LPAC and QAGR proteins are toxic when overexpressed in neural cells independent of CCUG or CAGG RNA gain of function effects. The greater loss of cell viability for QAGR (52%) compared to LPAC RAN (34%) protein is consistent with previous studies showing that arginine-containing dipeptides are more toxic to cells (Mizielinska et al., 2014; Yang et al., 2015).
RAN translation of CCUG expansions is modulated by MBNL1
Sequestration of MBNL by CCUG expansion transcripts results in the formation of nuclear RNA foci and RNA processing abnormalities (Goodwin et al., 2015). Although this is considered a detrimental process, we wondered if RNA foci could also be protective because nuclear sequestration of expansion RNAs would prevent RAN translation. To explore this question, we performed a series of experiments to understand the relationship between MBNL expression, RNA foci formation and RAN translation of CCUG expansion RNAs.
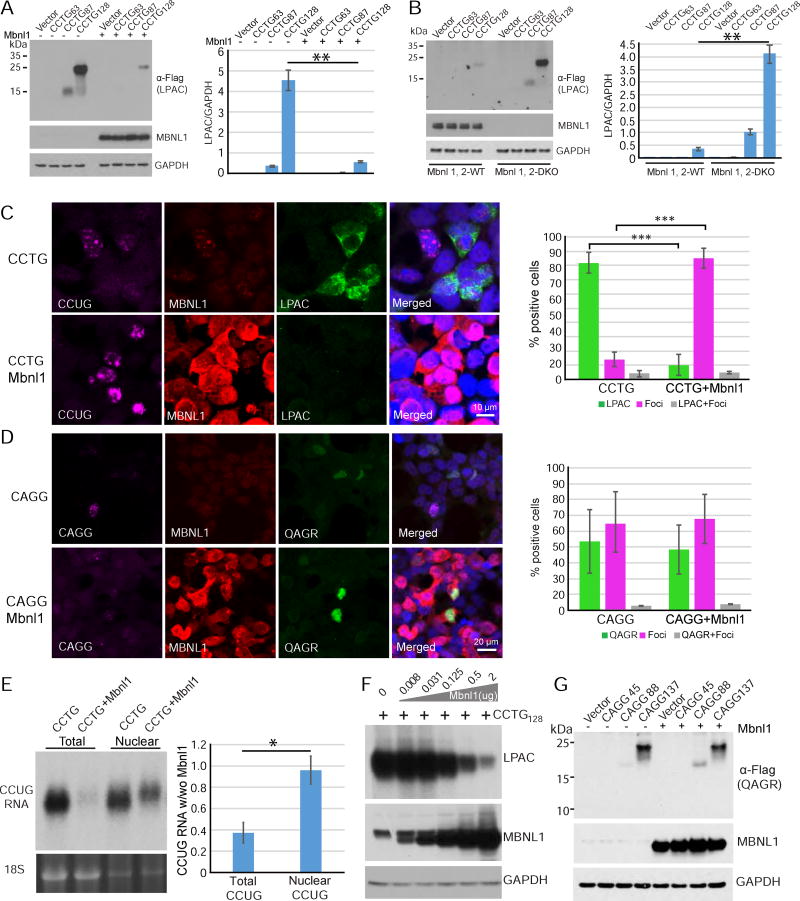
First, we tested if RAN translation across CCUG expansion transcripts is modulated by MBNL proteins and show that LPAC RAN protein expression was substantially decreased in cells co-transfected with the MBLN1 construct (Figure 5A). Next, we show RAN translation is upregulated in mouse embryonic fibroblasts (MEFs) generated from knockout mice lacking Mbnl1 and Mbnl2 compared to control MEFs (Figure 5B). Because loss of Mbnl1 gene expression leads to upregulation of Mbnl2 (Lee et al 2013; Wang et al 2012), Mbnl1−/−; Mbnl2−/− double knock-out (DKO) MEFs were used for these experiments to ensure muscleblind depletion. In general, MEFs express lower levels of RAN proteins than HEK293T cells and comparisons of LPAC RAN protein expression in WT and Mbnl DKO MEFs shows that a reduction of Mbnl proteins leads to increases in RAN protein levels by protein blot and IF (Figures 5B and S6A).
Figure 5.
RAN translation across CCTG•CAGG expansion is regulated by Mbnl1 proteins. (A) LPAC RAN proteins are decreased by protein blot analysis in HEK293T cells cotransfected with CCTG expansion and Mbnl1 constructs. (B) LPAC RAN proteins are increased in Mbnl1, 2 DKO MEFs transfected with constructs containing varying number of CCTG repeats. (C) IF labeling with CAGG RNA probe (purple) and antibodies against MBNL1 (red) and tagged-LPAC proteins (green) in transfected HEK293T. (D) IF labeling with CCTG RNA probe (purple) and antibodies against MBNL1 (red) and tagged-QAGR proteins (green) in transfected HEK293T. (C,D) Cells positive for RAN LPAC or QAGR protein staining are inversely related to cells positive for nuclear CCUG or CAGG RNA foci with Pearson’s correlation coefficients of r = −0.999 and −0.998, respectively. Percentage of cells expressing RAN only, foci only or RAN and foci are shown on the right. Levels of RAN LPAC but not QAGR are affected by MBNL1 overexpression. (E) RNA blot showing enrichment of nuclear CCUG transcripts compared to total RNA in cells overexpressing MBNL1 with 18S RNA as loading control (lower panel). (F) Protein blot showing dose-dependent decreases in steady state levels of LPAC protein with increasing MBNL1 protein expression. (G) Protein blot showing QAGR RAN protein levels are not affected by MBNL overexpression. Error bars show S.E.M. n≤3/group with * p<0.05, ** p<0.01 and *** p<0.001. See also Figures S6–8.
Surprisingly, qRT-PCR analyses showed CCUG transcripts in HEK293T cells overexpressing MBNL1 were consistently reduced (Figure S6B). In contrast MBNL1 OE did not reduce CAGG expansion transcripts (Figure S6C) and CCUG transcripts were roughly comparable in WT and MBNL1,2 DKO MEFs (Figure S6D). These results suggest two possibilities: 1) steady state levels of CCUG transcripts are reduced by MBNL1 overexpression through decreased transcription or increased RNA turnover or 2) MBNL1 proteins sequester CCUG expansion transcripts into extraction resistant complexes as has been previously reported for some RBPs (Gagnon et al., 2014). Because MBNL1 is known to bind to and sequester CCUG expansion RNAs (Liquori et al., 2001), we performed additional RNA extraction experiments to better solubilize the CCUG expansion transcripts. The use of a Trizol protocol with heat treatment (Gagnon et al., 2014) increased the amount of extractable CCUG expansion RNA in cells overexpressing MBNL1 consistent with MBNL1-CCUG RNA forming extraction resistant complexes (Figure S6E).
Because Mbnl proteins sequester CCUG RNAs in nuclei as RNA foci and nuclear sequestration of the RNA would be predicted to decrease RAN, we tested the relationship between RNA foci formation, Mbnl expression and RAN translation. We performed triple-label FISH-IF using a CAGG RNA probe and antibodies against MBNL1 and tagged-LPAC proteins. As expected, MBNL1 co-localized with CCUG-containing nuclear foci in HEK293T cells transfected with a CCTG-3T construct containing 128 repeats. In general, cells showing nuclear CCUG RNA foci were RAN negative and cells that were positive for LPAC did not show distinct nuclear CCUG RNA foci. Similarly, overexpression of MBNL1 protein decreases RAN protein accumulation and increases nuclear sequestration of CCUG expansion transcripts with a Pearson’s correlation coefficient of −0.999 indicating a strong negative correlation between RNA foci and LPAC RAN protein staining (Figure 5C). Additionally, RNA FISH experiments are consistent with the CCUG RNAs being sequestered in the nucleus by MBNL1 rather than being reduced by degradation or decreased synthesis (Figure 5C). In contrast, no correlation between MBNL1 and CAGG expansion transcript localization was found (Figure 5D). Northern analyses of HEK293T extracts shows CCUG transcripts shift to a predominantly nuclear localization with MBNL1 overexpression (Figure 5E). In a separate control experiment, we show knockdown of CCUG transcripts, using siRNAs to target cytoplasmic CCUG RNAs, leads to decreased steady state levels of LPAC RAN proteins (Figure S6F). Figure 5F shows that steady state levels of LPAC RAN protein show dose-dependent decreases with increasing MBNL1 levels. Similarly, steady state levels of RAN proteins expressed across CUG expansion RNAs decrease in all three reading frames with MBNL1 overexpression (Figures S7A and S7C).
Additionally, MBNL1 also decreases RAN protein levels expressed across CUG, and similar to a previous report (Sun et al., 2015) CAG expansion RNAs (Figures S7). These results are consistent with our CCUG findings because muscleblind proteins have been shown to bind to CAG and CUG expansion RNAs (Ho et al., 2005; Kino et al., 2004; Mykowska et al., 2011; Yuan et al., 2007). In contrast, RAN translation across DM2 CAGG antisense transcripts and C9ORF72 GGGGCC expansion transcripts was not affected by MBNL1 overexpression (Figures 5G and S8A). Similarly, MBNL1 overexpression did not affect canonical ATG-initiated translation of a GFP control (Figure S8B).
DISCUSSION
Myotonic dystrophy is widely considered to be an RNA gain of function disease in which CUG or CCUG expansion RNAs dysregulate key RNA-binding proteins that result in RNA processing abnormalities. In this study, we demonstrate that the DM2 expansion is bidirectionally transcribed and that CCUG and CAGG transcripts produce tetrapeptide LPAC and QAGR RAN proteins in all three reading frames, respectively. Both LPAC and QAGR RAN proteins accumulate in DM2 human autopsy brains in distinct patterns. For LPAC cytoplasmic aggregates are found in neurons, astrocytes and glia in the grey matter regions of the brain. In contrast, QAGR RAN protein accumulation, which is nuclear, is found primarily in oligodendrocytes located in white matter regions of the brain. Both LPAC and QAGR RAN proteins are toxic to cells in culture and are found in regions of the brain that show pathologic changes. Additionally, we identify hnRNPA1 as a novel CAGG RBP. Further, we provide evidence that RAN protein accumulation can be modulated by MBNL1 levels and that nuclear sequestration of CCUG, CUG or CAG RNAs decrease steady-state levels of RAN proteins. Taken together, our data suggest a two-phase model of disease, which initially involves nuclear retention of expansion RNAs and RBP depletion followed by a later disease phase in which cytoplasmic expansion RNAs undergo RAN translation and exacerbate disease.
Similar to other RAN expansion diseases (C9ORF72 ALS/FTD, SCA8, HD and FXTAS) RAN protein accumulation across both CCUG and CAGG expansion transcripts is length dependent (Ash et al., 2013; Banez-Coronel et al., 2015; Mori et al., 2013; Todd et al., 2013; Zu et al., 2011; Zu et al., 2013). In contrast to polyAla and polySer RAN proteins in SCA8 and HD (Banez-Coronel et al., 2015; Zu et al., 2011), the LPAC and QAGR proteins expressed in transfected cells migrate as one or a few prominent bands suggesting that these proteins are relatively soluble and that translation is likely to initiate at one or a few sites near the beginning of the repeat tract. Similarly, the detection of three different C-terminal epitope tags in cells transfected with constructs of a single repeat length indicates that RAN translation across the CCUG and CAGG expansion transcripts can occur in all three reading frames. In patient samples these tetrapeptide proteins are predicted to contain C-terminal sequences from all three reading frames. Finally, unlike tri- or hexanucleotide repeats, repeat length changes of the DM2 tetranucleotide expansion will shift the reading frame. For example, a (CCTG)90 that ends at the first stop codon in reading frame 1 will end in frame 2 or frame 3 if repeat instability increases the length of the repeat tract from 90 to 91 or 92.
A similarity between DM1 and DM2 that has previously been highlighted is that both diseases involve the accumulation of similar CUG and CCUG expansion transcripts that lead to the depletion of muscleblind proteins and RNA processing abnormalities (Ranum and Cooper, 2006; Scotti and Swanson, 2016). We now show that both sense and antisense transcripts are expressed and that the CAGG antisense expansion transcripts are upregulated in DM2 frontal cortex versus control autopsy brains. Although antisense transcripts were not detectable by RNAseq in muscle from one DM2 patient (Wagner et al., 2016) or by polyA-seq of cerebellum (Goodwin et al., 2015), additional work will be needed to understand if the upregulation of DM2 antisense transcripts is limited to the CNS or even specific subregions of the brain. The upregulation of antisense transcripts in affected tissue is also seen in C9ORF2 ALS/FTD patient brains (Zu et al., 2013) suggesting that the longer repeats in both diseases may increase antisense transcription or increase the stability of antisense transcripts.
Many diseases are characterized by the death of specific subsets of neurons. For example, ataxia is best known as a disease involving the loss of cerebellar Purkinje cells (Nelson et al., 2013). In contrast, Parkinson’s is characterized by the loss of dopaminergic neurons in the substantia nigra (Shulman et al., 2011) and Huntington disease for loss of medium spiny neurons in the striatum (Vonsattel et al., 2011). Although CNS features in DM, including executive function deficits, are highly problematic for patients and white matter abnormalities are detected by MRI and pathological examination (Minnerop et al., 2011; Ogata et al., 1998), routine pathological findings are diffuse and previous reports suggest findings are non-specific (Conforti et al., 2016; Wozniak et al., 2013; Wozniak et al., 2014; Wozniak et al., 2011; Zanigni et al., 2016). The discovery that LPAC and QAGR tetrapeptide proteins accumulate in DM2 provides a new opportunity to understand the impact of the DM2 expansion mutation on the brain and to look for pathological findings at sites of RAN protein accumulation.
Our findings show cytoplasmic accumulation of LPAC in neurons and glia in grey matter regions of the cortex, hippocampus and brainstem. LPAC staining is primarily cytoplasmic and is found to accumulate at focal sites in the grey matter regions. The focal uneven distribution of LPAC staining we observe is similar to the focal sites of RAN protein accumulation found in C9ORF2 ALS/FTD and HD (Banez-Coronel et al., 2015; Zu et al., 2013). LPAC RAN protein accumulation is found throughout the grey matter regions but is dramatically increased in regions that appear to have undergone acute ischemic injury. Regions with intense LPAC staining were characterized by microglial/monocyte infiltration, decreased NeuN staining, pyknotic nuclei and spongiform changes. These results suggest LPAC is toxic or that damage or stress results in increased LPAC expression. In contrast, nearby regions without LPAC staining appeared healthy. These data, and similar observations from C9ORF72 ALS/FTD and HD, suggest a model in which RAN proteins drive disease at multiple and varied sites within the brain such that pathology found in one patient differs from the pathology found in other patients. It is possible that focal regions of RAN protein accumulation are caused by templating and cell to cell spreading of aggregates through a prion-like mechanism. Alternatively, it is possible that there are local increases in RAN translation caused by stress or localized perturbations in protein homeostasis that lead to the patchy clustering of LPAC RAN protein accumulation. Similar to findings recently reported in HD (Banez-Coronel et al., 2015), microglia/monocytes are recruited to sites of LPAC RAN protein accumulation. The observation that a subset of these cells are positive for LPAC suggests these microglia/monocytes are either recruited to sites of LPAC RAN protein accumulation and have phagocytosed the protein or that a subset of these cells express the mutant LPAC protein.
QAGR RAN proteins show a distinct pattern of accumulation within the white matter regions of the brain. Punctate nuclear staining is found in oligodendrocytes with tiny aggregates primarily located at the internal side of the nuclear membrane. Similar to LPAC and other RAN protein staining (Banez-Coronel et al., 2015; Zu et al., 2013), QAGR staining is not uniform, but positive QAGR regions found in the white matter are more frequent and generally cover larger areas. White matter lesions have been previously detected throughout the brain by DTI/MRI in both DM1 and DM2. Until now, the leading hypothesis is that clinical and pathological disease features shared by DM1 and DM2 are caused by the RNA gain of function effects of the similar CUG and CCUG expansion transcripts. Our demonstration that antisense QAGR proteins accumulate in white matter regions of DM2 autopsy brains and that QAGR proteins are toxic to cells independent of RNA GOF effects, suggests that that QAGR RAN proteins cause the white matter abnormalities found in DM2 patient brains. While polyGln RAN proteins were previously reported in DM1 blood, heart and skeletal muscle (Zu et al., 2011), more work is needed determine if RAN proteins also accumulate in DM1 brains. Because RAN proteins in both DM2 and HD accumulate in white matter regions, a top priority will be to determine if RAN proteins are also found in the white matter regions of DM1 autopsy brains.
RNA foci and RAN proteins are found in a growing number of diseases including SCA8, C9ORF72 ALS/FTD (Cleary and Ranum, 2014) and now DM2. This raises the question of whether RNA GOF and RAN mechanisms are linked. Our data show that expression of LPAC RAN proteins can be blocked by MBNL1 overexpression and that RNA foci are inversely correlated with RAN protein expression in transfected cells. These data suggest an RNA nuclear sequestration failure model of disease (Figure 6). Based on our in vitro data, we predict that cells expressing sufficient levels of MBNL sequester CCUG expansion RNAs within the nucleus and these cells will not express LPAC RAN proteins. Additionally, we predict that RAN translation will increase when nuclear sequestration fails and the expansion RNAs are exported to the cytoplasm and produce LPAC proteins. This sequestration failure is predicted to occur under conditions in which the concentration of RNA-binding sites on expansion RNAs exceeds the binding capacity of the RBPs. This model predicts conditions that favor RAN translation could include: 1) cells expressing high levels of expansion RNAs; 2) cells expressing low levels of specific RBPs; 3) cells where the binding capacity of the repeat exceeds that of the relevant RBP. In the brain, which has many different cell types, this model predicts that the patterns of RNA foci and LPAC RAN protein accumulation will be highly variable because the capacity to sequester CCUG expansion RNAs or undergo LPAC RAN protein expression will vary from cell to cell. Consistent with this predicted variability, IF-FISH of DM2 autopsy brains shows examples of cells (Figure S3D) with: 1) a single or several RNA foci and no RAN proteins; 2) numerous RNA foci and no RAN proteins; 3) high levels of RNA and LPAC RAN proteins suggesting that the CCUG expansion RNAs in these cells is not completely sequestered in foci (end stage). Further analysis of animal models and additional autopsy tissue will be required to better understand the relationship between MBNL, CCUG foci, LPAC RAN proteins and toxicity in vivo and over time.
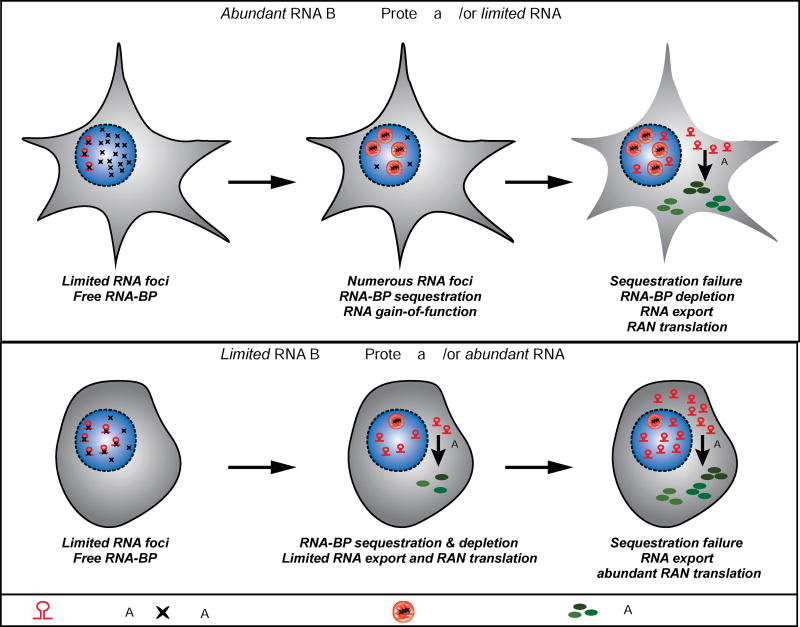
Figure 6.
Top Panel: RNA sequestration failure model of RAN translation. In cells with abundant RNA-binding proteins or limited RNA, expression of repeat RNA begins sequestering RBP (e.g. MBNL proteins) (Stage I). With time, increased repeat length and/or increased expression, the majority of RBP is sequestered triggering functional failure and RNA gain-of-function toxicity (Stage II). Further increases in RNA or repeat length cause sequestration failure and export of expanded RNA into the cytoplasm where RAN translation occurs resulting in the accumulation of toxic homopolymeric proteins (Stage III). Lower Panel: In cells with limited RBP or abundant RNA (bottom panel), expression of expanded repeat RNAs leads to rapid RBP sequestration with limited foci formation (Stage I). Increased repeat length or RNA expression results in rapid failure of RNA sequestration by the RBP and the presence of cytoplasmic expanded repeat RNAs and RAN translation (Stage II). In this situation, large amounts of RAN proteins can be produced from a cell with very little or no RNA foci (Stage III).
Additionally, we show MBNL1 overexpression decreases steady-state levels of RAN proteins made from CUG and CAG repeats. Because these repeats are found in multiple diseases including HD, DM1, SCA1, 2 ,3, 6, 7, 8, 17 and because RAN translation has been reported in a growing number of disorders (Ash et al., 2013; Banez-Coronel et al., 2015; Mori et al., 2013; Todd et al., 2013; Zu et al., 2011; Zu et al., 2013), our MBNL-RNA sequestration model may be relevant to these diseases and other diseases with discrete RNA foci such as C9ORF72 ALS/FTD (Cleary and Ranum, 2017) and SCA36 (Kobayashi et al., 2011). Although the repeat motif and the specific RNA-binding proteins will differ for each disease, this is a general model that could explain the variability in disease features and the severe late-onset features of diseases such as ALS/FTD, which may be precipitated by an increase in RAN protein expression over time.
The distribution difference between the LPAC and QAGR proteins with LPAC primarily expressed in the grey matter and QAGR in the white matter is surprising. This finding suggests several possibilities. One is that the distribution of CAGG and CCUG expansion RNAs differ with CCUG and CAGG expansion transcripts being more abundant in grey or white matter regions, respectively. A second possibility is that differences in RNA-binding protein (MBNL vs hnRNP A1) levels and sequestration capacity differs among brain regions, such that MBNL depletion and LPAC expression is favored in grey matter regions, and hnRNP A1 nuclear sequestration is more effective in grey vs. white matter regions. Consistent with this possibility, previously reported RNAseq data in mice showed relative levels of hnRNP A1 transcripts are low in oligodendrocytes, which are frequently positive for QAGR RAN protein staining (Zhang et al., 2014). A third possibility is that factors required for RAN translation across CCUG expansion RNAs differ from factors required for CAGG translation and the distributions of cells expressing these factors differs among specific cell types expressed in various brain regions.
In summary, we provide compelling evidence that tetrapeptide RAN proteins accumulate in myotonic dystrophy type 2 autopsy brains. These DM2 RAN proteins may prove to be useful biomarkers to predict disease onset and progression and responses to future therapeutic treatments. The discovery that RAN translation occurs in the brains of DM2 patients provides new insight into the CNS aspects of the disease, which significantly impact quality of life. Additionally, we show RNA GOF and RAN mechanisms are temporally and mechanistically linked and that nuclear sequestration of expansion RNAs into nuclear foci protects against RAN protein production. This RNA sequestration failure model of RAN translation may be a common theme with RNA foci and RAN translation in a growing number of diseases including SCA8, DM1, FXTAS, C9ORF2 ALS/FTD and HD. Finally, the expression of sense and antisense expansion RNAs and RAN proteins in DM2 human autopsy brains suggests strategies targeting both transcripts may be required to effectively treat disease in humans.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and fulfilled by the Lead Contact, Laura P.W. Ranum (ranum@ufl.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines included HEK293T cells for studies of RAN translation and RNA foci and T98 cells for toxicity assays
HEK293T and T98 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum and incubated at 37°C in a humid atmosphere containing 5% CO2. Sex of the HEK293T cells is female and the sex of the T98 cells is male. The cells lines used were obtained from ATCC have not been authenticated. The plasmids were grown in HB101 E.coli.
Human subjects
Human subjects included adult males and females with myotonic dystrophy type 2 and control samples. Control and DM autopsy tissue was collected at the University of Minnesota and the University of Florida with informed consent of patients or their relatives and approval of local institutional review boards.
METHOD DETAILS
cDNA constructs
Triple 3x-epitope (frame 1–3xFlag, frame 2–3xHA, and frame 3–3xMyc) fragments were synthesized by Integrated DNA Technologies and subcloned into pcDNA3.1 vector. CCTG repeat expansion were amplified by PCR from constructs with varying number of repeats using the 5’primer (CTAAGTAACTAGAATTCGCCCGGGCCT) containing a XmaI restriction site and the 3’ primer (GGATCCTGAATTCGCCCTCGAGGCA) containing a XhoI restriction site and cloned into pCR4Blunt-TOPO vector. The XmaI/XhoI CCTG fragments were subcloned into pcDNA-6Stops-3xTags. In the resulting constructs, CCTGEXP-3xT, six stop codons (two in each frame) were placed prior to the 5’ end of the CCTG expansion and a 3xFlag epitope is placed 3’ of the expanded CCUG repeat in frame 1, followed by 3xHA in frame2 and 3xMyc in frame3. The CAGGEXP-3xT constructs were made by inserting CAGG repeat fragments into the pcDNA-6Stops-3xTags. This construct contains 6Stop codon cassette upstream of the CAGG expansion and three different C-terminal 3xTags. To generate the FLAG-LPAC and FLAG-QAGR constructs, CCTG or CAGG repeat fragments were placed downstream of N-terminal 3xFlag epitope tag in p3XFLAG vector. Alternative sequences (CTTCCCGCCTGT)46 for poly(LPAC) or (CAAGCGGGAAGG)46 for poly(QAGR) were synthesized by GenScript and subcloned into p3XFLAG vector. The resulting constructs, 3xFlag-LPAC-Ate and 3xFlag-QAGR-Ate, express the transcripts that do not form hairpin structure and are translated into poly(LPAC) and poly(QAGR), respectively. The integrity of all constructs was confirmed by sequencing. We developed a modified triple-3x-epitopes (3xFlag, 3xHA, and 3xMyc) tagged construct to monitor protein expression in all three reading frames. Three copies of each tag significantly enhance the detection of fusion proteins compared to single copy epitope. In this construct a 3xFlag epitope is placed 3’ of the expanded CCUG repeat in frame 1, followed by 3xHA in frame2 and 3xMyc in frame 3.
Production of polyclonal antibodies
The polyclonal antibodies were generated by New England Peptide (Gardner, MA). The α-LPAC antisera were raised against a synthetic peptide corresponding to the (LPAC)EXP motif (H2N-LPACLPACLPACLPAC-OH). The α-QAGR antisera were raised against a synthetic peptide corresponding to the (QAGR)EXP motif (Ac-CQAGRQAGRQAGRQAGR-amide).
Cell culture and transfection
HEK293T and T98 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum and incubated at 37°C in a humid atmosphere containing 5% CO2. DNA transfections were performed using Lipofectamine 2000 Reagent (Invitrogen) according to the manufacturer’s instructions.
Immunofluorescence
The subcellular distribution of polymeric proteins was assessed in transfected HEK293T cells by immunofluorescence. Cells were plated on 8 well tissue-culture chambers and transfected with plasmids the next day. Forty-eight hours post-transfection, cells were fixed in 4% paraformaldehyde (PFA) in PBS for 30 min and permeabilized in 0.5% triton X-100 in PBS for 15 min on ice. The cells were blocked in 1% normal goat serum (NGS) in PBS for 30 min. After blocking, the cells were incubated for 1 hour at RT in blocking solution containing the rabbit anti-Myc (Abcam), mouse anti-HA (Covance), mouse anti-Flag (Sigma), rabbit α-LPAC primary antibodies at a dilution of 1:400. The slides were washed three times in PBS and incubated for 1 hour at RT in blocking solution containing Goat anti-rabbit conjugated to Cy3 (Jackson ImmunoResearch, PA) and goat anti-mouse conjugated to Alexa Fluor 488 (Invitrogen) secondary antibodies at a dilution of 1:200. The slides were washed three times in PBS and mounted with mounting medium containing DAPI (Invitrogen).
FISH and immunofluorescence
Cells were fixed in 4% paraformaldehyde for 10 min at room temperature and immersed in pre-chilled 70% ethanol for 30 min on ice and then rehydration in 40% formamide in 2× SSC for 10 min. After being blocked in hybridization buffer (40% formamide, 2× SSC, 20 (µg/ml BSA, 100 mg/ml dextran sulfate, and 200 (µg/ml yeast tRNA, 2 mM vanadyl sulfate ribonucleosides) for 15 min at 37 °C, cells were incubated with denatured RNA probe (400 ng/ml) in hybridization buffer at 37 °C for 2 hours. After being washed three times in a pre-warmed solution of 40% formamide and 2× SSC three times at 37 °C for 20 min each wash and then washed once in PBS at room temperature for 10 min, IF was performed. The cells were blocked in 1% normal goat serum (NGS) in PBS for 30 min. After blocking, the cells were incubated with rabbit anti-MBNL1 (Abcam) and mouse anti-HA (Covance) at a dilution of 1:500 for overnight at 4 °C. The slides were washed three times in PBS and incubated with goat anti-rabbit conjugated to Alexa Fluor 545 (Invitrogen) and goat anti-mouse conjugated to Alexa Fluor 488 (Invitrogen) secondary antibodies at a dilution of 1:500 for 1 hour at RT. The slides were washed three times in PBS and mounted with mounting medium containing DAPI (Invitrogen).
FISH-IF in autopsy tissue
Autopsy tissues were embedded with OCT. 10µm frozen sections were cut on the cryostat. Frozen sections were fixed in 4% PFA in PBS for 20 min and incubated in prechilled 70% ethanol for 30 min or longer at 4°C Following rehydration in 40% formamide in 2× SSC for 10 min, the slides were blocked with hybridization solution (40% formamide, 2× SSC, 20 (µg/mL BSA, 100 mg/mL dextran sulfate, and 250 (µg/mL yeast tRNA, 2 mM Vanadyl Sulfate Ribonucleosides) for 30 min at 37 °C and then incubated with 200 ng/mL of denatured DNA probe (CAGGx4-Cy-3 for sense foci in the hybridization solution at 37 °C for 3 h. After hybridization, the slides were washed three times with 40% formamide in 2× SSC and briefly washed one time in PBS. The slides were then permeabilized in 0.5% triton X-100 in PBS for 15 min on ice and blocked in 2% normal goat serum in PBS for 1 hour. After blocking, the slides were incubated overnight at 4°C with α-LPAC antisera (1:500) diluted in the blocking solution. The slides were then washed three times in PBS and incubated for 1 h at room temperature with goat anti-rabbit conjugated to Alexa Fluor 488 (Invitrogen) secondary antibodies at a dilution of 1:500. Slides were washed and autofluorescence of lipofuscin was quenched by 1% of Sudan Black B in 70% ethanol for 5min. Slides were mounted with mounting medium containing DAPI (Invitrogen) and imaged in 10 z-stacks (0.6 µm between stacks) on a Leica confocal using a 63× water objective. Three independent DM2 and three control samples were analyzed and evaluated by at least three different scorers including scorers who were blinded.
Immunoblotting
Transfected cells in each well of a six-well tissue-culture plate were rinsed with PBS and lysed in 300 µL RIPA buffer with protease inhibitor cocktail for 45 min on ice. DNA was sheared by passage through a 21-gauge needle. The cell lysates were centrifuged at 16,000 × g for 15 min at 4 °C, and the supernatant was collected. The protein concentration of the cell lysate was determined using the protein assay dye reagent (Bio-Rad). Twenty micrograms of protein were separated in a 4–12% Bis-Tris gel (Bio-Rad) and transferred to a nitrocellulose membrane (Amersham). The membrane was blocked in 5% dry milk in PBS containing 0.05% Tween-20 (PBS-T) and probed with the anti-Flag (1:2000), anti-Myc (1:1000), anti-HA (1:1000), or rabbit polyclonal antibodies (1:1000) in blocking solution. After the membrane was incubated with anti-rabbit or anti-mouse HRP-conjugated secondary antibody (GE Heathcare), bands were visualized by the ECL plus Western Blotting Detection System (PerkinElmer).
Immunohistochemistry
Ten-micrometer sections were deparaffinized in xylene and rehydrated through graded alcohol, incubated with 95–100 % formic acid for 5 min, and washed with distilled water for 10 min. Heat-Induced Epitope Retrieval (HIER) was performed by steaming sections in citrate buffer, pH 6.0, at 90 °C for 30 min. To block nonspecific immunoglobulin binding, a serum-free block (Biocare Medical) was applied for 30 min. Rabbit polyclonal antibodies were applied at a dilution of from 1:5000 to 1:15,000 in serum-free block (Biocare Medical) and incubated overnight at 4 °C. Linking reagent (streptavidin and/or alkaline phosphatase, Covance) was applied for 30 min at room temperature. These sections were incubated in 3% H2O2 for 15 min to bleach endogenous peroxidase activity. Then labeling reagent (HRP, Covance) was applied for 30 min at room temperature. Peroxidase activity was developed with NovaRed substrate (Vector) and sections were counterstained with hematoxylin. IHC results summarized in Table 1 and 2 were based on the staining and evaluation of more than six slides per patient, which were analyzed by at least three independent investigators, including investigators blinded to the genotypes.
Histology
The human brain sections (5 µm) were examined by haematoxylin and eosin (H&E) and luxol fast blue (LFB) staining. For luxol fast blue (LFB) staining, the sections were deparaffinized in xylene and hydrated to 95% ethyl alcohol. The sections were left in 0.1% luxol fast blue solution at 56 °C overnight. The slides were rinsed in 95% ethyl alcohol and distilled water. Subsequently, the slides were differentiated in 0.05% lithium carbonate solution for 30 seconds and in 70% ethyl alcohol for 30 seconds and washed in distilled water. The slides were then counterstained in 0.1% cresyl violet solution for 40 seconds and rinsed in distilled water. The slides were finally dehydrated and mounted with Cytoseal 60 (Electron microscopy sciences).
Histological results summarized in Figures 3 and 4 were based on the staining and evaluation of three patients and three controls with staining of at least three slides per sample. Results were analyzed by at least three independent investigators, including investigators blinded to the genotypes.
Cell-toxicity assays
Cells were transfected at ~ 60% cell confluence. Next, 500 ng of each vector was transfected in 35-mm wells (35 ng per well in 96 well plates). Cell death was determined by measuring LDH cell release, using CytoTox 96 nonradioactive cytotoxicity assay (Promega) according to the manufacturer’s instructions. Absorbancewas recorded at 490 nm and total LDH release was measured by lysing the cells with 1% Triton X-100.
Cell viability was assayed using the 3-(4,5- dimethythiazol-. 2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. MTT is reduced by metabolically active cells, resulting in the intracellular accumulation of a purple formazan product. MTT was added to cell culture media at 0.5 mg/mL final concentration and incubated for 45 minutes at 37°C. Cell media was removed and MTT-derived formazan was solubilized in 75 µL of DMSO, and quantified by spectrophotometry at 550 nm.
In each experiment “n” refers to completely independent replicates. For each independent experimental replicate, determinations were performed in quintuplicates for each experimental condition and average data calculated. Statistical significance was determined using the two-tailed unpaired Student t test (p<0.05. p-values withstand Bonferroni correction for multiple pair-wise comparisons.
RT-PCR and quantitative RT-PCR
To ensure that RT-PCR product was amplified from the specific strand, strand specific primer containing a linker sequence (LK) was used to generate linker-tailed first strand products that can only be subsequently amplified after a primer complimentary to the primed sequence makes a complimentary copy of the specific LK. For antisense strand, upstream of CAGG expansion, LK RT-PCR products were amplified from the CAGG but not the CCUG strand, using LK-AS-Up-R primer for RT reaction and AS-Up-F and the LK for PCR to specifically amplify antisense cDNAs. Similar strategies were used to amplify downstream of CAGG repeats. Total RNA from cells was extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. cDNA was generated from total RNA using random-hexamer primers and the SuperScript III system (Invitrogen). qRT-PCR was performed on a StepOnePlus Real-Time PCR Systems (Applied Biosystems) using Power SYBR Green PCR Master Mix and 3xTag primer sets. Human β-actin expression was used as a reference gene amplified with primer set ACTB3 and ACTB4. Primer sequences are listed below.
Northern analysis
Total RNA from cells was extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. For isolation of total RNA using guanidinium method, cells were lysed in thiocyanate buffer (4 M guanidinium isothiocyanate, 20 mM sodium acetate, pH 5.2, 0.1 mM DTT, 0.5% N-lauroylsarcosine), and RNA was pelleted through a 5.7 M CsCl cushion. An equal amount of RNA was separated on a glyoxal gel, blotted to a nylon membrane, and probed with a [32P]ATP-labeled oligonucleotide complementary to the 3’ end of the expanded repeat-containing transcripts. Blots subsequently were probed with a [32P]dCTP-labeled GAPDH cDNA probe.
In vitro transcription and probe purification
Plasmid DNAs were linearized with BamHI and used as templates for in vitro transcription using T7 RNA polymerase (Ribomax T7; Promega). Probes were synthesized using 1 mM GTP, 1 mM ATP, 1 mM UTP, 0.04 mM CTP, and 25 µCi of 32P-labeled UTP (800Ci/mmol) for 1.5 h at 37°C. The transcription reactions were treated with DNase I for 20 min and then phenol:chloroform-extracted. The aqueous phase was passed through a Micro Bio-Spin P30 column according to the manufacturer’s instructions (Bio-Rad). Equal volumes of Gel Loading Buffer II (Ambion) were added to the samples and they were furthered purified using 15% polyacrylamide/7.5 M Urea denaturing gel electrophoresis. The probes were extracted from the gel by adding 400 ul of G50 buffer (20 mM Tris-HCl, pH 7.5, 300 mM sodium acetate, 2 mM EDTA, 0.25% SDS), freezing on dry ice, thawing and then crushing with a pestle in a microcentrifuge tube (BioMasher II, Kimble). The samples were then left overnight to elute from the gel, while rotating at room temperature. The samples were filtered (UltraFree MC, Millipore), phenol:cholorform:isoamyl alcohol extracted and ethanol precipitated. The probes were resuspended in nuclease-free water, and stored at −80°C.
Nuclear extract preparation
For preparation of nuclear and cytosolic extract, HEK293T cells were incubated with 2 packed cell volumes (PCV) of Buffer A (10 mM HEPES pH 7.4, 10 mM KCl, 0.75 mM MgCl2, 0.5 mM DTT, Complete EDTA-free protease inhibitor cocktail tablet (Roche Diagnostics) for 15 minutes on ice. Cells were lysed 5 times using a 25-gauge needle and the extract was centrifuged at 14000 rpm. The supernatant was removed and saved for further processing into cytosolic extracts. The pellet was resuspended in 0.5 PCV of Buffer B (20 mM HEPES pH 7.8, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT) with 20 mM KCl, and followed by drop-wise addition of one half PCV of Buffer B with 1.2 M KCl. After incubation on ice for 30 min, the extract was centrifuged at 14000 rpm at 4°C. This supernatant constitutes the nuclear extract and was aliquoted and stored at −80°C. Protein concentrations were determined with the Pierce 660 nM Protein Assay using BSA as a standard.
UV Crosslinking
Crosslinking with HEK293T cell extracts was done as follows. Protein extracts (55 ug) were incubated with 1.5×105 cpm of each in vitro transcribed 32P-labelled RNA probe, as indicated. The final reaction conditions were 20 mM HEPES(4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), pH 7.6, 44 mM KCl, 1.4 mM MgCl2, 0.9 mM EDTA, 10% glycerol, 8 mM creatine phosphate, 0.375 mM ATP, 200 ng tRNA and 12 U RNAsin Plus RNase Inhibitor (Promega) in a 25 ul reaction. The protein-RNA interactions were allowed to equilibrate for 30 minutes at 30°C. The reactions were exposed to UV light (254 nm, 125 mJoules) for 10 minutes, followed by treatment with 20 µg RNase A (Fermentas) for 60 minutes at 37°C. Samples were analyzed by electrophoresis by SDS-PAGE on 4–15% gradient gels. The gels were dried and proteins were visualized with a PhosphorImager screen.
Coupled Crosslinking and Immunoprecipitation
The crosslinking reactions were carried out as above, then 18 ul of Buffer RSB (10mM Tris-HCl, 10 mM NaCL, 1.5 mM MgCl2) and SDS was added to a final concentration of 1%. The samples were heated to 95°C for three minutes to promote dissociation of any complexes, then 500 uL of RIPA buffer was added along with the antibodies as indicated. The MBNL1 polyclonal A2764 was a generous gift of Dr. Charles Thornton. The hnRNPA1 antibodies 4B10 and 9H10 were generous gifts from Dr. Gideon Dreyfuss. The pan-hnRNP antibody 7A9 was a generous gift from Dr. Sefarin Piñol-Roma. The immunoprecipitation was incubated at room temperature with rotation for one hour. For each reaction, 20 uL of Protein G magnetic beads (Life Technologies) were washed, and then added to the reactions, with a further incubation of 2 hours. Samples were then washed three times with 400 ul of RIPA buffer, and once with buffer RSB. The beads were then resuspended in 4x Laemmli dye and the proteins were eluted by heating to 95°C. The samples were analyzed by electrophoresis by SDS-PAGE on 4–15% gradient gels and visualized with a PhosphorImager screen.
QUANTIFICATION AND STATISTICAL ANALYSIS
Toxicity studies
In each experiment “n” refers to completely independent replicates. For each independent experimental replicate, determinations were performed in quintuplicates for each experimental condition and average data calculated. Statistical significance was determined using the two-tailed Student t test (p<0.05). When applicable p-values withstand Bonferroni correction for multiple pair-wise comparisons.
Densitometry
Each experiment was done at least three independent times and results were quantified using ImageJ Software. Differences between groups were determined by Student’s t-test (p<0.05) and when applicable Bonferroni correction was applied for multiple pair-wise comparisons.
RNA Foci and LPAC RAN protein staining
Excel software was used to calculate Pearson’s Correlation coefficient. Samples were run in at least three independent experiments.
RT-PCR
Samples were run using the Applied Biosystems StepOnePlus Real-time PCR System. Results were analyzed using Excel and the 2-delta delta Ct method. Samples were run in triplicate with at least three independent replicates.
Supplementary Material
Highlights.
DM2 expanded CAGG antisense transcripts are elevated in DM2 autopsy brains
Poly-(LPAC) and poly-(QAGR) tetrapeptide RAN proteins expressed in DM2
RAN translation in DM2 is modulated by MBNL levels though nuclear sequestration
Nuclear sequestration failure of CCUG repeats increases RAN protein levels
Acknowledgments
We would like to thank myotonic dystrophy family members who contributed to this study, John Day and the Stanford Tissue Repository and the Center for NeuroGenetics at the University of Florida for autopsy tissue. This work was funded by the NIH NINDS PO1-NS058901 (L.P.W.R., M.S.S., H.B.C), the Muscular Dystrophy Association (L.P.W.R.) and a Postdoctoral Fellowship from the Myotonic Dystrophy Foundation (J.D.C). T.Z. and L.P.W.R. have patents pending on RAN translation. No other authors have conflicts. A.T.Y. receives partial support from the 1-Florida ADRC (P50 AG047266-01A1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Author contributions: T.Z., J.D.C., M.S.S. and L.P.W.R. designed research; T.Z., J.D.C., Y.L., M.B.-C., J.B., F.A., performed research; T.A., G.X., H.B.C., A.T.Y. contributed new reagents/analytic tools/samples; T.Z., J.D.C. Y.L., M.B.-C., J.L.B., F.A., H.B.C., A.T.Y., and L.P.W.R. analyzed data; and T.Z. and L.P.W.R. wrote the paper with input from all authors.
References
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, et al. Unconventional translation of C90RF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, Pletnikova O, Borchelt DR, Ross CA, Margolis RL, et al. RAN Translation in Huntington Disease. Neuron. 2015;88:667–677. doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkqvist M, Wild EJ, Thiele J, Silvestroni A, Andre R, Lahiri N, Raibon E, Lee RV, Benn CL, Soulet D, et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J Exp Med. 2008;205:1869–1877. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook JD, McCurrah ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion J-P, Hudson J, et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- Charizanis K, Lee KY, Batra R, Goodwin M, Zhang C, Yuan Y, Shiue L, Cline M, Scotti MM, Xia G, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Cleary JD, Ranum LP. Repeat associated non-ATG (RAN) translation: new starts in microsatellite expansion disorders. Curr Opin Genet Dev. 2014;26:6–15. doi: 10.1016/j.gde.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Ranum LP. New developments in RAN translation: insights from multiple diseases. Curr Opin Genet Dev. 2017;44:125–134. doi: 10.1016/j.gde.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti R, de Cristofaro M, Cristofano A, Brogna B, Sardaro A, Tedeschi G, Cirillo S, Di Costanzo A. Brain MRI abnormalities in the adult form of myotonic dystrophy type 1: A longitudinal case series study. Neuroradiol J. 2016;29:36–45. doi: 10.1177/1971400915621325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y-H, Pizzuti A, Fenwick RGJ, King j, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, De Jong P, et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- Gagnon KT, Li L, Janowski BA, Corey DR. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat Protoc. 2014;9:2045–2060. doi: 10.1038/nprot.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin M, Mohan A, Batra R, Lee KY, Charizanis K, Fernandez Gomez FJ, Eddarkaoui S, Sergeant N, Buee L, Kimura T, et al. MBNL Sequestration by Toxic RNAs and RNA Misprocessing in the Myotonic Dystrophy Brain. Cell Rep. 2015;12:1159–1168. doi: 10.1016/j.celrep.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper PS. Myotonic Dystrophy. 2. London: W.B. Saunders; 1989. [Google Scholar]
- Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Clocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- Ishiguro T, Sato N, Ueyama M, Fujikake N, Sellier C, Kanegami A, Tokuda E, Zamiri B, Gall-Duncan T, Mirceta M, et al. Regulatory Role of RNA Chaperone TDP-43 for RNA Misfolding and Repeat-Associated Translation in SCA31. Neuron. 2017;94:108–124. el07. doi: 10.1016/j.neuron.2017.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Urbinati CR, Crusselle VJ, Luo D, Lee YJ, Harrison JK, Oh SP, Swanson MS. Developmental expression of mouse muscleblind genes Mbnll, Mbnl2 and Mbnl3. Gene Expr Patterns. 2003;3:459–462. doi: 10.1016/s1567-133x(03)00064-4. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai L, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum Mol Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Abe K, Matsuura T, Ikeda Y, Hitomi T, Akechi Y, Habu T, Liu W, Okuda H, Koizumi A. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am J Hum Genet. 2011;89:121–130. doi: 10.1016/j.ajhg.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krans A, Kearse MG, Todd PK. Repeat-associated non-AUG translation from antisense CCG repeats in fragile X tremor/ataxia syndrome. Ann Neurol. 2016;80:871–881. doi: 10.1002/ana.24800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, Hagerman RJ, Tassone F, Tapscott SJ, Filippova GN. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- Libby RT, Hagerman KA, Pineda VV, Lau R, Cho DH, Baccam SL, Axford MM, Cleary JD, Moore JM, Sopher BL, et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 2008;4:el000257. doi: 10.1371/journal.pgen.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C, Narang M, Barcelo j, O’Hoy K, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3’ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- Meola G. Myotonic dystrophies as a brain disorder. Neurol Sci. 2010;31:863–864. doi: 10.1007/s10072-010-0414-2. [DOI] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO. 2000;J19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnerop M, Weber B, Schoene-Bake JC, Roeske S, Mirbach S, Anspach C, Schneider-Gold C, Betz RC, Helmstaedter C, Tittgemeyer M, et al. The brain in myotonic dystrophy 1 and 2: evidence for a predominant white matter disease. Brain. 2011;134:3530–3546. doi: 10.1093/brain/awr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- Mykowska A, Sobczak K, Wojciechowska M, Kozlowski P, Krzyzosiak WJ. CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res. 2011;39:8938–8951. doi: 10.1093/nar/gkr608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL, Orr HT, Warren ST. The unstable repeats-three evolving faces of neurological disease. Neuron. 2013;77:825–843. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata A, Terae S, Fujita M, Tashiro K. Anterior temporal white matter lesions in myotonic dystrophy with intellectual impairment: an MRI and neuropathological study. Neuroradiology. 1998;40:411–415. doi: 10.1007/s002340050613. [DOI] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Savkur RS, Philips AV, Cooper TA, Dalton JC, Moseley ML, Ranum LP, Day JW. Insulin receptor splicing alteration in myotonic dystrophy type 2. Am J Hum Genet. 2004;74:1309–1313. doi: 10.1086/421528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman JM, De Jager PL, Feany MB. Parkinson’s disease: genetics and pathogenesis. Annu Rev Pathol. 2011;6:193–222. doi: 10.1146/annurev-pathol-011110-130242. [DOI] [PubMed] [Google Scholar]
- Sun X, Li PP, Zhu S, Co hen R, Marque LO, Ross CA, Pulst SM, Chan HY, Margolis RL, Rudnicki DD. Nuclear retention of full-length HTT RNA is mediated by splicing factors MBNL1 and U2AF65. Sci Rep. 2015;5:12521. doi: 10.1038/srep12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, Renoux AJ, Chen KC, Scaglione KM, Basrur V, et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440–455. doi: 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP, Keller C, Cortes Ramirez EP. Huntington’s disease - neuropathology. Handb Clin Neurol. 2011;100:83–100. doi: 10.1016/B978-0-444-52014-2.00004-5. [DOI] [PubMed] [Google Scholar]
- Wagner SD, Struck AJ, Gupta R, Farnsworth DR, Mahady AE, Eichinger K, Thornton CA, Wang ET, Berglund JA. Dose-Dependent Regulation of Alternative Splicing by MBNL Proteins Reveals Biomarkers for Myotonic Dystrophy. PLoS Genet. 2016;12:el006316. doi: 10.1371/journal.pgen.1006316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak JR, Mueller BA, Bell CJ, Muetzel RL, Lim KO, Day JW. Diffusion tensor imaging reveals widespread white matter abnormalities in children and adolescents with myotonic dystrophy type 1. J Neurol. 2013;260:1122–1131. doi: 10.1007/s00415-012-6771-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak JR, Mueller BA, Lim KO, Hemmy LS, Day JW. Tractography reveals diffuse white matter abnormalities in Myotonic Dystrophy Type 1. J Neurol Sci. 2014;341:73–78. doi: 10.1016/j.jns.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak JR, Mueller BA, Ward EE, Lim KO, Day JW. White matter abnormalities and neurocognitive correlates in children and adolescents with myotonic dystrophy type 1: a diffusion tensor imaging study. Neuromuscul Disord. 2011;21:89–96. doi: 10.1016/j.nmd.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanigni S, Evangelisti S, Giannoccaro MP, Oppi F, Poda R, Giorgio A, Testa C, Manners DN, Avoni P, Gramegna LL, et al. Relationship of white and gray matter abnormalities to clinical and genetic features in myotonic dystrophy type 1. Neuroimage Clin. 2016;11:678–685. doi: 10.1016/j.nicl.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. RAN proteins and RNA foci from antisense transcripts in C90RF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.