Abstract
Developmental tumors in children and young adults carry few genetic alterations, yet they have diverse clinical presentation. Focusing on Ewing sarcoma, we sought to establish the prevalence and characteristics of epigenetic heterogeneity in genetically homogeneous cancers. We performed genome-scale DNA methylation sequencing for a large cohort of Ewing sarcoma tumors and analyzed epigenetic heterogeneity on three levels: between cancers, between tumors, and within tumors. We observed consistent DNA hypomethylation at enhancers regulated by the disease-defining EWS-FLI1 fusion protein, thus establishing epigenomic enhancer reprogramming as a ubiquitous and characteristic feature of Ewing sarcoma. DNA methylation differences between tumors identified a continuous disease spectrum underlying Ewing sarcoma, which reflected the strength of an EWS-FLI1 regulatory signature and a continuum between mesenchymal and stem cell signatures. There was substantial epigenetic heterogeneity within tumors, particularly in patients with metastatic disease. In summary, our study provides a comprehensive assessment of epigenetic heterogeneity in Ewing sarcoma and thereby highlights the importance of considering nongenetic aspects of tumor heterogeneity in the context of cancer biology and personalized medicine.
Ewing sarcoma (EwS) is a developmental cancer defined and diagnosed by the presence of the EWS-FLI1 fusion oncogene1,2. Despite this shared molecular basis, the clinical presentation and disease courses of patients with EwS vary3–5. This heterogeneity is not reflected by the genetics of EwS, which is characterized by few somatic mutations6 and only three genes with recurrent genetic lesions (CDKN2A, STAG2 and TP53)7–9. We hypothesized that the observed clinical heterogeneity might coincide with widespread epigenetic heterogeneity, given that two recent studies established the relevance of epigenetics in EwS by identifying a direct link between the EWS-FLI1 fusion protein and widespread epigenomic reprogramming10,11.
To characterize epigenetic heterogeneity in EwS, we performed DNA methylation sequencing in a large collection of EwS tumors, many of which had previously undergone whole-genome sequencing9. We focused this analysis on DNA methylation as the classic epigenetic mark, which is intricately linked to cancer12 and well-suited for dissecting tumor heterogeneity13,14. On the basis of the resulting data set, we investigated epigenetic heterogeneity on three levels (Fig. 1a): (i) Analysis of inter-cancer heterogeneity identified EwS-specific patterns that accurately distinguished them from other cell types not expressing EWS-FLI1; (ii) analysis of inter-individual heterogeneity identified DNA methylation signatures that were stronger in some patients than in others, reflecting epigenomic differences between tumors; and (iii) analysis of intra-tumor heterogeneity quantified the variability among single cells within the same tumor.
Figure 1. DNA methylation profiling reveals a characteristic epigenomic signature of Ewing sarcoma.
(a) Epigenetic heterogeneity in Ewing sarcoma (EwS) analyzed at three levels: between cancer types (inter-cancer), between EwS tumors (inter-individual), and within EwS tumors (intratumor). (b) Multidimensional scaling plot showing this study’s RRBS profiles, which includes EwS tumors, EwS cell lines, and mesenchymal stem cells (MSCs) derived from bone marrow (BM), umbilical cord (UC), and placenta (PL), in the context of published RRBS profiles for other cancers (Supplementary Table 2). DNA methylation levels were averaged across 5-kb tiling regions. APL, acute promyelocytic leukemia; CHS, chondrosarcoma; CLL, chronic lymphocytic leukemia; CRPC, castration-resistant prostate cancer. (c) Multidimensional scaling plot as shown in b, but focusing specifically on EwS tumors, EwS cell lines, and MSCs. (d) DNA methylation heat map for CpGs with lower DNA methylation levels in EwS tumors as compared to reference profiles for other cancers and for a diverse set of other cell types (Supplementary Table 2). Bar plots indicate significant overlap of EwS-specific hypomethylated regions with public annotation data, based on LOLA analysis19 (Supplementary Table 3). (e) As in d, but focusing on CpGs with higher DNA methylation levels in EwS tumors as compared to the reference profiles. (f) Example of EwS-specific hypomethylation at the CCND1 locus, with substantially lower DNA methylation (and anti-correlated histone H3K27 acetylation) in EwS tumors and EwS cell lines as compared to all reference samples. DNA methylation levels are shown for 50 bins spanning the locus (yellow, high methylation; blue, low methylation; white, no data). H3K27ac profiles include a cross-tissue consensus track from the ENCODE project, as well as ChIP-seq data for an EwS cell line (A673sh) with inducible knock-down of EWS-FLI1 (EWS-FLI1 high/low)11 and this study’s data for three EwS tumors (tumors 119, 120, and 121). (g) As in f, but focusing on EwS-specific hypermethylation of a putative regulatory region at the GATA2 locus.
In contrast to many other cancers, differences between EwS tumors did not uncover discrete subtypes, but instead defined a continuous spectrum along two distinct and biologically interpretable dimensions. Individual tumors differed by the strength of an EWS-FLI1 regulatory signature and along a continuum defined by mesenchymal versus stem cell signatures, which potentially reflects the regulatory state of the cell from which the tumor was originally derived. Together, these two dimensions established an epigenetic disease spectrum underlying EwS, which was associated partially with somatic mutations in STAG2 and TP53. EwS tumors also differed in their intra-tumor heterogeneity, and primary tumors from patients who presented with metastatic disease were more heterogeneous than those of patients with localized disease. In summary, this study provides a comprehensive assessment of DNA methylation heterogeneity in EwS, as well as a resource for studying epigenomic deregulation and tumor heterogeneity in genetically homogeneous cancers.
Results
DNA methylation profiling uncovers a unique and predictive epigenomic signature of Ewing sarcoma
To dissect epigenetic tumor heterogeneity in EwS, we established DNA methylation maps for 140 EwS tumors (Supplementary Table 1). DNA methylation profiling was performed by using reduced representation bisulfite sequencing (RRBS)15,16, which is an accurate and high-throughput assay for DNA methylation profiling17. Data quality was consistently high (Supplementary Fig. 1a–c and Supplementary Table 2), and genomic coverage included not only CpG islands and promoter regions, but also many CpGs located in distal enhancers, CpG island shores and other functional elements (http://sheffield2017.computational-epigenetics.org). For assay validation, we performed whole genome bisulfite sequencing (WGBS) on three representative samples and observed high consistency with the RRBS data (Supplementary Fig. 1d). We also profiled 16 EwS cell lines with RRBS, including six low-passage cell lines derived from tumors that were part of our cohort. Finally, given the proposed role of mesenchymal stem cells (MSCs) as a potential cell-of-origin of EwS18, we generated RRBS data for 32 primary MSC samples obtained from bone marrow (n = 22), placenta (n = 2), and umbilical cord (n = 8) from both patients with EwS and healthy individuals (Supplementary Table 1).
On the basis of this data set, we investigated epigenetic heterogeneity between cancers, between patients, and within individual tumors (Fig. 1a). We compared the EwS tumor profiles to publicly available RRBS data for several other cancers and cell types (Supplementary Table 2). Unsupervised visualization using multidimensional scaling separated the EwS samples from all other cancers (Fig. 1b). To confirm and quantify this observation, we trained a logistic regression classifier on the DNA methylation profiles and found that this classifier could distinguish between EwS tumors and a diverse set of other cell types (Supplementary Table 2) with a cross-validated test-set accuracy of 98.6% (Supplementary Fig. 1e). Focusing on our RRBS data set, we also observed that the MSCs separated by tissue-of-origin independently of donor age (Fig. 1c), whereas the EwS cell lines continued to cluster with the EwS tumors, albeit with a tendency toward the edge. A logistic regression classifier distinguished between EwS tumors and MSCs with 99.4% accuracy (Supplementary Fig. 1f). Low-passage cell lines most closely resembled those tumors from which they were derived (Supplementary Fig. 1g), which highlights that patient-specific DNA methylation characteristics were retained in early-passage EwS cell lines cultured in vitro.
To determine an EwS-specific DNA methylation signature, we compared the EwS tumor profiles to a diverse set of RRBS profiles representing more than 50 different cell types (Supplementary Table 2). We identified 2,917 CpGs that were specifically hypomethylated in EwS (Fig. 1d) and 1,820 CpGs that were specifically hypermethylated (Fig. 1e) The EwS-specific hypomethylated CpGs were exclusive to EwS samples and were heavily methylated in essentially all other cell types, whereas the difference was less pronounced for EwS-specific hypermethylated CpGs. We performed region set enrichment analysis with LOLA19 to test these CpGs for enrichment against the LOLA Core database, which consists of a broad collection of DNaseI hypersensitive elements20, chromatin immunoprecipitation sequencing (ChIP-seq) peaks21,22 and other regulatory region sets23,24. EwS-specific hypomethylated CpGs were enriched for EwS-specific enhancers11 (Fig. 1d and Supplementary Table 3), which validates our use of DNA methylation as a marker of epigenomic reprogramming in EwS tumors. EwS-specific hypomethylated CpGs were also enriched for open chromatin in prostate cancer cell lines, which might be explained by the biological similarity between EWS-FLI1 and the prostate-specific TMPRSS2-ERG fusion protein, both of which include an ETS factor as a fusion partner25. By contrast, EwS-specific hypermethylated CpGs overlapped with developmental regulators of various lineages, including polycomb-repressed regions in pluripotent stem cells, AP-1 binding sites, and binding sites of various developmental transcription factors (Fig. 1e and Supplementary Table 3). The characteristic DNA methylation profiles of EwS samples are also illustrated by epigenome snapshots of individual loci (Fig. 1f,g).
Collectively, these results establish a DNA methylation signature that was shared by all EwS tumors, distinguishing them with remarkable sensitivity and specificity from other cell types in our data set. EwS cells are thus marked by a highly characteristic epigenomic state, which we further investigated by ChIP-seq analysis for seven histone marks in three representative tumors (Supplementary Fig. 2). DNA methylation levels were consistently anti-correlated with histone H3 lysine 27 acetylation (H3K27ac), a defining mark of active enhancers26,27 (Supplementary Fig. 3a,b). Furthermore, patient-specific differences in DNA methylation reflected patient-specific differences in H3K27ac, with 65% of the most variable regions showing a Pearson correlation below –0.8 (Supplementary Fig. 3c). These results confirm that the observed differences in DNA methylation reflect broader epigenomic differences between patients.
Ewing sarcoma is epigenetically heterogeneous in the absence of discrete disease subtypes
To compare inter-individual heterogeneity in different cancer types, we calculated the coefficient of variation (CV, defined as the ratio between s.d. and mean) of DNA methylation levels across the genome, which has been proposed as a measure of global heterogeneity within a DNA methylation data set28. EwS fell in the medium-to-high range of CV values that we observed across several cancers and cell types (Fig. 2a). Specifically, the CV of EwS was on par with that of prostate cancer and chronic lymphocytic leukemia (CLL), two genetically heterogeneous cancers of the elderly (the average age of diagnosis is ~66 years for prostate cancer and ~71 years for CLL, as opposed to ~15 years for EwS). This result is unlikely to be biased by differences in sample purity, given that CV values were similar between primary EwS tumors and EwS cell lines grown in vitro (Fig. 2a), and that there was no correlation between CV contribution and estimated tumor purity among the EwS tumors (Supplementary Fig. 4a). EwS tumors thus seem to be characterized by substantial epigenetic heterogeneity, which contrasts with the genetic homogeneity of EwS.
Figure 2. DNA methylation in EwS shows inter-individual heterogeneity without distinct subtypes.
(a) Bar plot showing the coefficient of variation across samples for median DNA methylation levels per sample as a measure of heterogeneity between samples. The coefficient of variation was calculated separately for EwS cell lines, EwS tumors, and MSCs derived from bone marrow (BM), umbilical cord (UC) and placenta (PL) from this study, and for reference profiles of other cancers. APL, acute promyelocytic leukemia; CHS, chondrosarcoma; CLL, chronic lymphocytic leukemia; CRPC, castration-resistant prostate cancer. (b) DNA methylation profiles for four types of EwS-linked regulatory regions: (i) EwS-specific DNaseI elements based on DNase-seq in the SK-N-MC cell line; (ii) EWS-FLI1 binding sites based on ChIP-seq for EWS-FLI1 in the A673sh cell line; (iii) EWS-FLI1-correlated enhancers based on decreased H3K27ac ChIP-seq signal upon EWS-FLI1 knockdown in A673sh; and (iv) EWS-FLI1-anti-correlated enhancers based on increased H3K27ac ChIP-seq signal upon EWS-FLI1 knockdown in A673sh. 50 randomly selected regions are shown to illustrate DNA methylation variability between tumors (see Supplementary Fig. 4b for all data). (c) Example of epigenetic heterogeneity at one EWS-FLI1-correlated enhancer showing opposing trends in DNA methylation versus H3K27ac among three EwS tumors (119, 120, and 121). Black vertical lines represent CpG sites, with the height indicating their DNA methylation levels. (d) As in c, but focusing on epigenetic heterogeneity at one EWS-FLI1-anti-correlated enhancer. (e) EwS tumor grouping using five alternative methods for sample clustering and unsupervised subtype identification, showing no evidence of epigenetically defined disease subtypes in EwS.
To dissect the biological basis of DNA methylation heterogeneity among EwS tumors, we focused on four types of genomic region with previously reported regulatory relevance in EwS (Fig. 2b and Supplementary Fig. 4b): (i) EwS-specific DNaseI elements based on DNase-seq data for an EwS cell line (SK-N-MC)20; (ii) EWS-FLI1-correlated enhancers, defined as elements that lose H3K27ac signal upon EWS-FLI1 knockdown11; (iii) EWS-FLI1 binding sites based on ChIP-seq for EWS-FLI1 in an EwS cell line (A673)25; and (iv) EWS-FLI1-anti-correlated enhancers, defined as elements that gain H3K27ac signal upon EWS-FLI1 knockdown11. DNA methylation levels were most variable at EwS-specific DNaseI elements and at EWS-FLI1-anti-correlated enhancers (Fig. 2b and Supplementary Fig. 4b,c). Inter-individual differences in DNA methylation in these regions were associated with differences in H3K27ac (Fig. 2c,d), which indicates that the observed patterns of DNA methylation heterogeneity reflect broader epigenomic variability at Ewing-specific regulatory regions.
We expected the inter-individual differences among EwS tumors to group them into a few distinct and epigenomically defined disease subtypes, as observed for other cancers29–33. To test this hypothesis, we applied various unsupervised clustering methods to our DNA methylation data set. However, none of these methods provided convincing evidence of distinct EwS subtypes, but rather identified broadly distributed patterns reminiscent of a continuous disease spectrum (Fig. 2e). The absence of any consistent and reproducible disease subtypes in our data set was confirmed by multiple lines of evidence, including coverage-based data filtering, imputation of missing values, use of pairwise shared CpGs, and averaging across tiling regions (Supplementary Fig. 5), and we also did not observe a consistent association between the data set’s principal components and various clinical variables (Supplementary Fig. 6).
DNA methylation heterogeneity defines an epigenetic disease spectrum in Ewing sarcoma
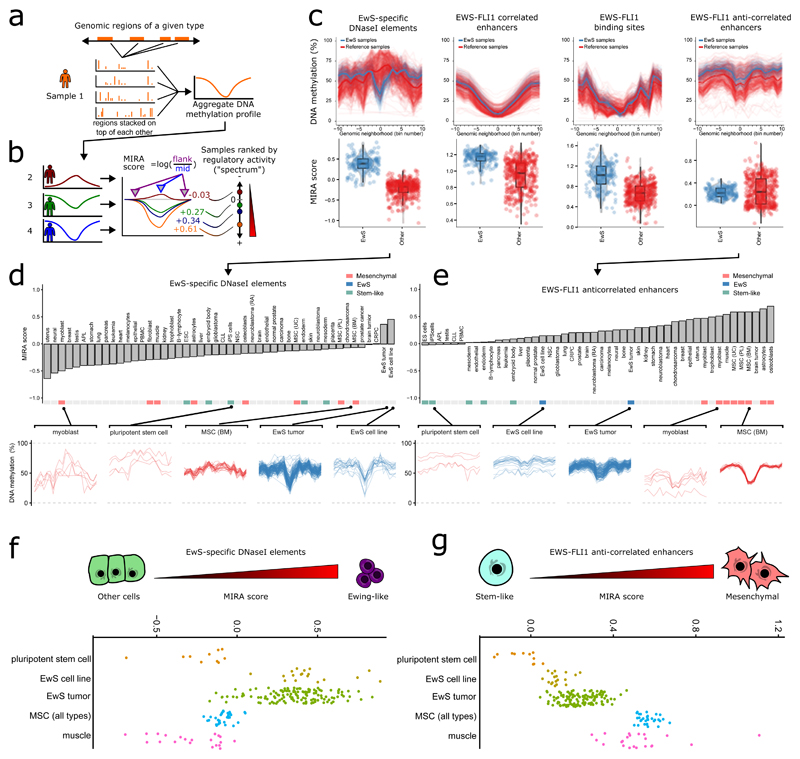
Given widespread inter-individual heterogeneity in the absence of well-defined patient clusters or disease subtypes, we focused once more on the four genomic region sets with regulatory relevance in EwS (introduced in Fig. 2b), and we analyzed how DNA methylation in these regions varied across samples. Overlaying all regions in each of the sets, we calculated four DNA methylation profiles that aggregate the DNA methylation levels of these regions (Fig. 3a). Because DNA methylation levels are anti-correlated with transcription factor occupancy and regulatory activity34–38, we used these aggregate DNA methylation profiles to define the ‘methylation-based inference of regulatory activity’ (MIRA) score as a quantitative measure of the regulatory activity of a given region set in a given sample (Fig. 3b).
Figure 3. DNA methylation at regulatory elements defines an epigenetic disease spectrum underlying EwS.
(a,b) Conceptual outline of the ‘methylation-based inference of regulatory activity’ (MIRA) score. (a) First, all genomic regions of a given annotation type (such as EwS-specific DNaseI elements) are superimposed and their aggregate DNA methylation profiles derived. (b) Next, the MIRA score of a given region type in a given sample is calculated as the logarithm of the ratio between the mean DNA methylation level at the aggregate DNA methylation profile’s flanking regions versus the corresponding value at the region’s center. High MIRA scores correspond to a strong dip in DNA methylation and high inferred regulatory activity, whereas MIRA scores close to zero correspond to flat DNA methylation profiles and little potential for regulatory activity. (c) Aggregate DNA methylation profiles for four types of EwS-linked regulatory regions: EwS-specific DNaseI elements, EWS-FLI1-correlated enhancers, EWS-FLI1 binding sites, and EWS-FLI1-anti-correlated enhancers. Each line corresponds to the aggregate DNA methylation profile of an EwS sample (blue) or non-EwS reference sample (red). Box plots show MIRA scores for the corresponding region sets (boxes represent median and quartiles, and whiskers extend from the box to the most extreme point located within 1.5 times the inter-quartile range). (d) Bar plot for samples grouped by cell type and ordered according to the mean MIRA score for EwS-specific DNaseI elements. Aggregate DNA methylation profiles for myoblasts, pluripotent stem cells, and MSCs are shown to illustrate low regulatory activity in these regions, whereas high MIRA scores for EwS tumors and EwS cell lines indicate high regulatory activity. (e) As in d, but focusing on MIRA scores for EWS-FLI1-anti-correlated enhancers. (f) Distribution of MIRA scores for EWS-specific DNaseI elements, which places the EwS tumors on an epigenetic disease spectrum with different levels of “Ewing-ness”. (g) Distribution of MIRA scores for EWS-FLI1-anti-correlated enhancers, which places the EwS tumors on an epigenetic disease spectrum that is linked to a stem-like regulatory signature on the one end and a mesenchymal regulatory signature on the other end.
By comparing aggregate DNA methylation profiles and MIRA scores in EwS samples with a diverse set of reference profiles (Supplementary Table 2), we observed the most striking differences for the first and the last of the four region sets (Fig. 3c). EwS-specific DNaseI elements showed strong dips and high MIRA scores specifically in EwS tumors and cell lines, indicating the presence of EwS-specific activity at these elements. EWS-FLI1 binding sites and EWS-FLI1-correlated enhancers behaved similarly, although these regions had lower DNA methylation levels in all tissues and a smaller difference in MIRA score between EwS and reference samples. Finally, for EWS-FLI1-anti-correlated enhancers, we observed higher levels of DNA methylation in the EwS samples and MIRA scores that were within the range observed among the reference samples.
Focusing on the EwS-specific DNaseI elements and grouping the reference samples by cell type, only EwS tumors and EwS cell lines had positive MIRA scores (indicative of regulatory activity at these EwS-specific DNaseI elements), whereas all other cell types—including various cancers, primary tissues, and cultured cell lines—had negative MIRA scores (Fig. 3d and Supplementary Fig. 7). By contrast, plotting the MIRA scores for EWS-FLI1-anti-correlated enhancers (Fig. 3e) placed EwS tumors and EwS cell lines in the middle of a continuous spectrum. On the basis of the RRBS profiles and annotations of the reference samples, we found that this spectrum was marked by mesenchymal cells at one end (high MIRA scores, indicating strong regulatory activity) and pluripotent stem cells at the other end (low MIRA scores, indicating little or no regulatory activity).
Our analysis thus uncovered two biologically informative dimensions underlying the observed DNA methylation heterogeneity in EwS. The first dimension is defined by EwS-specific DNaseI hypersensitive elements and seems to measure the degree to which a tumor’s epigenome has been reprogrammed to the characteristic regulatory state of EWS-FLI1 expressing cells (Fig. 3f). The second dimension, which is defined by EWS-FLI1-anti-correlated enhancers, reflects the relative strength of a mesenchymal differentiation signature as opposed to a signature associated with pluripotent stem cells (Fig. 3g). When plotted, the scores of individual EwS tumors along these two dimensions cover a continuous spectrum (Fig. 3f,g), with little correlation between the two (Pearson’s r = –0.23; Supplementary Fig. 8a).
The observed DNA methylation differences along these two dimensions could not be explained as a side effect of technical or biological biases. First, higher tumor purity was positively correlated with higher scores on the Ewing-like dimension mainly because of a few outliers with low tumor purity (r = 0.52 dropping to 0.17 when these samples are removed; Supplementary Fig. 8b). Second, the mesenchymal dimension was largely uncorrelated with tumor purity (r = –0.26, Supplementary Fig. 8c). Third, the distribution of EwS cell lines cultured in vitro (which did not have any adjacent tissue) was similar to the EwS tumors (Fig. 3f,g), which suggests that a sample’s position along these two dimensions is a cell-intrinsic property. Fourth, there was no apparent association between tumor location in the body and either of the two dimensions (Supplementary Fig. 8d). Our results thus establish an epigenetic disease spectrum underlying EwS, defined by ‘Ewing-like’ and ‘mesenchymal versus stem-like’ regulatory signatures as its two dimensions.
Ewing sarcoma tumors are characterized by high and variable levels of intra-tumor heterogeneity
Having investigated DNA methylation heterogeneity between cancers (Fig. 1) and between individuals (Figs. 2 and 3), we next focused on DNA methylation differences between individual cells within the same tumor. RRBS provides a powerful tool for dissecting such intra-tumor heterogeneity, given that DNA methylation is a binary mark, and that each sequencing read captures the DNA methylation status of one allele obtained from one single cell. We used two bioinformatic methods for assessing intra-tumor heterogeneity in EwS: the ‘proportion of discordant reads’ (PDR) and the ‘proportion of sites with intermediate methylation’ (PIM).
The PDR score has been proposed as a measure of locally disordered DNA methylation13. It is calculated as the proportion of discordant sequencing reads among all RRBS reads that cover at least four CpGs, where discordant reads are defined as those that contain both methylated and unmethylated CpGs, and concordant reads contain only methylated or only unmethylated CpGs. High PDR values have been interpreted as an indicator of epigenomic instability in individual cells, which might contribute to clonal evolution13. Calculating PDR scores for our data set, we observed that these values were strongly associated with average DNA methylation levels. They were highest in regions with intermediate DNA methylation levels (Supplementary Fig. 9a) and lowest in regions with DNA methylation levels near 0% or near 100% (Supplementary Fig. 9b). The average PDR score across all EwS tumors was in the same range as those for chondrosarcoma and prostate cancer, but lower than those for acute promyelocytic leukemia, CLL, and glioblastoma (Fig. 4a).
Figure 4. DNA methylation patterns identify widespread intra-tumor heterogeneity in EwS.
(a) Distribution of sample-specific PDR scores of EwS tumors, EwS cell lines, and MSCs derived from bone marrow (BM), umbilical cord (UC), and placenta (PL) from this study, as compared to reference profiles for other cancers. APL, acute promyelocytic leukemia; CHS, chondrosarcoma; CLL, chronic lymphocytic leukemia; CRPC, castration-resistant prostate cancer (boxes represent median and quartiles, and whiskers extend from the box to the most extreme point located within 1.5 times the inter-quartile range). (b) Conceptual outline of the PIM score, which measures the proportion of CpG sites with intermediate DNA methylation levels. Higher PIM scores indicate higher levels of intra-tumor heterogeneity. (c) As in a, but focusing on sample-specific relative PIM scores. (d) Density scatterplot (left) showing the relationship between PDR and PIM scores for 5-kb tiling regions, including only regions with more than 25 CpG dinucleotides. The two scores are correlated (r = 0.76), but there are also many regions with divergent scores, which is illustrated by two conceptual examples (right).
To complement and extend these analyses, we investigated intermediate DNA methylation levels as an alternative measure of epigenetic intra-tumor heterogeneity. The PIM score leverages the binary character of DNA methylation: a single allele in a single cell is either 0% or 100% methylated, and intermediate DNA methylation arises from averaging across a heterogeneous population that comprises both methylated and unmethylated alleles of a given CpG. Intermediate DNA methylation levels thus reflect cell-to-cell heterogeneity. We identified CpGs with intermediate DNA methylation levels in a given sample using a Bayesian binomial credibility interval (Fig. 4b and Supplementary Fig. 9c) and calculated PIM scores for each sample. To compare PIM scores between data sets, we controlled for differential CpG coverage by restricting the analysis to shared CpGs in each pair of samples, calculating relative PIM scores as the ratio of pairwise shared CpG sites with intermediate DNA methylation in one sample versus another (Fig. 4b). All pairwise relative PIM scores for a given sample versus all other samples were averaged, and their sample-specific mean was used as an indicator of the sample’s overall level of intra-tumor heterogeneity.
We observed high relative PIM scores among the EwS tumors (Fig. 4c), which places them in a range of intra-tumor heterogeneity similar to that of acute promyelocytic leukemia and CLL, and above that of prostate cancer. There was substantial variability between EwS tumors, in part owing to differences in tumor purity (r = –0.46, corresponding to 21% variance explained), which we statistically corrected for as described below. The more homogeneous EwS cell lines had lower average relative PIM scores than EwS tumors (Wilcoxon P value < 10–4), but even their PIM scores were higher than those of prostate cancer.
Comparing PDR and PIM scores genome-wide, we observed substantial correlation not only across genomic regions (r = 0.76; Fig. 4d), but also across samples within a given genomic region (median r = 0.51; Supplementary Fig. 9d), which suggests the measures capture related but distinct aspects of intra-tumor heterogeneity. For example, PIM identifies regions with a combination of fully methylated and fully unmethylated reads as heterogeneous, whereas PDR does not; by contrast, PDR identifies regions with consistent and reproducible patterns of methylated and unmethylated CpGs as disordered, whereas PIM considers them homogeneous (Fig. 4d). One practical advantage of PIM over PDR is that it can assess heterogeneity at any covered CpG, not just those in reads spanning at least four CpGs, which resulted in much higher genomic coverage for PIM (Supplementary Fig. 9e). Regions with high average PDR or PIM among the EwS samples (mean score across samples exceeding 80%) were strongly enriched for intronic as well as intergenic regions, highlighting that intra-tumor heterogeneity is most prevalent outside of gene promoters (Supplementary Fig. 9f,g). In summary, our analyses identified high and variable levels of intra-tumor heterogeneity in EwS, which were in the same range as those observed for much more genetically heterogeneous cancers.
DNA methylation heterogeneity in EwS can be linked to genetic and clinical data
On the basis of the pronounced differences in epigenetic heterogeneity that we observed among EwS tumors, we explored associations with genetic as well as clinical data. Focusing on the 79 EwS tumors that had whole-genome sequencing data9 (which allowed us to statistically control for differences in tumor purity), we compared the heterogeneity scores defined above (MIRA, PDR, and PIM) with patient annotations such as age, metastatic status at diagnosis, tumor size, tumor location, relapse status, and mutations for STAG2, TP53, and CDKN2A. After controlling for tumor purity and sex using linear models (Supplementary Fig. 10), we identified significant associations between MIRA scores and somatic mutation status, between PDR scores and tumor location, and between PIM scores and metastatic status at diagnosis (Fig. 5a and Supplementary Figs. 11 and 12).
Figure 5. DNA methylation heterogeneity in EwS is associated with genetic and clinical data.
(a) Heat map illustrating the association between measures of epigenetic heterogeneity and genetic, as well as clinical annotations among the EwS tumors. Brighter colors indicate higher significance according to the Wilcoxon rank-sum test. (b) Violin plot comparing the MIRA score at EWS-FLI1-anti-correlated enhancers (which corresponds to the mesenchymal versus stem-like dimension of the disease spectrum) for EwS tumors with or without mutations in STAG2 (boxes represent median and quartiles, and whiskers extend from the box to the most extreme point located within 1.5 times the inter-quartile range). (c) Violin plot comparing the MIRA score at EwS-specific DNaseI elements (which corresponds to the Ewing-like dimension of the disease spectrum) for EwS tumors with or without mutations in TP53. (d) Violin plot comparing the PIM score between primary EwS tumors of patients whose disease was metastatic at diagnosis versus patients with localized disease. (e) Receiver operating characteristic (ROC) curve and area under curve (AUC) value for predicting whether a patient was metastatic at diagnosis on the basis of the PIM score. Inset, distribution of AUC values and the resulting P value according to permutation testing with randomly shuffled labels.
Comparing 16 tumors with a STAG2 mutation to 63 tumors without such a mutation, we observed significantly lower MIRA scores for EWS-FLI1-anti-correlated enhancers in the STAG2 mutated tumors (Wilcoxon P value < 0.01; Fig. 5b). This result places the STAG2 mutated tumors in the more stem-like area of the EwS spectrum, which is consistent with recent research showing that cohesin mutants enforce stem cell programs39. The deletion of CDKN2A, which is a relatively common genetic lesion in EwS, showed no significant association (Wilcoxon P value > 0.1; Supplementary Fig. 11), but among the seven TP53-mutated tumors in our cohort, we observed increased MIRA scores for EwS-specific DNaseI elements; this places TP53 mutants in the more Ewing-like area of the spectrum (Wilcoxon P value < 0.03; Fig. 5c).
Focusing on intra-tumor heterogeneity, we observed significantly higher PDR scores for tumors whose primary location was in the spine (Wilcoxon P value < 0.02). EwS tumors in the spine also had lower MIRA scores for EWS-FLI1-correlated enhancers and for EwS-specific DNaseI elements (Supplementary Fig. 12). However, given that only six tumors with primary location in the spine were included in our data set, there is limited statistical support for such an association between intra-tumor heterogeneity and tumor location.
Finally, we observed a significant association between PIM scores and metastatic status at diagnosis. On average, primary tumors from patients whose disease was already metastatic at diagnosis had higher PIM scores (indicating higher intra-tumor heterogeneity) than those observed for patients with localized disease (Wilcoxon P value < 0.03; Fig. 5d). A logistic regression model predicting metastatic status at diagnosis solely on the basis of PIM score performed significantly better than expected by chance, with an area under curve (AUC) value of 0.66 and a permutation P value below 0.04 (Fig. 5e).
Discussion
Our study establishes the prevalence and characteristics of epigenetic tumor heterogeneity in EwS on the basis of DNA methylation sequencing and bioinformatic analysis of a large patient cohort. Analyzing DNA methylation patterns across cancer types, we identified patterns of enhancer reprogramming that were shared by all EwS samples. But we also observed substantial epigenetic tumor heterogeneity between patients and within tumors, which stands in stark contrast to the genetic homogeneity of EwS.
We and others have previously reported characteristic changes of the epigenome in EWS-FLI1 expressing cells10,11, yet we were surprised to see how unique and predictive the DNA methylation patterns of EwS were in comparison to a broad range of reference samples. Bioinformatic classification based on our DNA methylation data set resulted in test-set accuracies close to 100% for distinguishing EwS samples from various other cell types (including MSCs, a potential cell-of-origin of EwS). Regions that were demethylated in EwS but methylated in other cell types were strongly enriched for EwS-specific DNaseI elements, most of which were located outside of gene promoters and constitute putative enhancer elements. Our data thus support the conceptualization of EwS as an ‘enhancer disease,’ with widespread epigenomic reprogramming driven by EWS-FLI1.
Epigenetic heterogeneity between patients with EwS followed unexpected patterns. Rather than identifying a small number of distinct subtypes, as observed for many other cancers29–33, we found that DNA methylation differences in EwS gave rise to a continuous disease spectrum along two dimensions. First, the EWS-FLI1 regulatory signature was stronger in some EwS tumors than in others, and also slightly stronger in cell lines than in tumors. Second, the EwS tumors were broadly scattered across a continuum, with a mesenchymal regulatory signature on one end and a pluripotent stem cell signature on the other end. We speculate that the latter dimension might reflect the differentiation state of the cell-of-origin from which a specific EwS tumor has been derived, whereas the former dimension might reflect the depth and degree with which the epigenome of the cancer cells has been reprogrammed to the characteristic EwS-specific enhancer state.
We also observed substantial epigenetic heterogeneity within individual tumors, which we bioinformatically quantified on the basis of the RRBS data. Primary tumors from patients that were metastatic at diagnosis had higher PIM scores (indicating higher intra-tumor heterogeneity) than tumors of patients with localized disease. This observation is consistent with the emerging view that tumor heterogeneity tends to be higher in patients with more aggressive disease40. However, at this stage, we can only speculate whether the observed patterns of epigenetic heterogeneity might have any causal role in EwS (for example, by fueling clonal evolution41) and to what degree the patterns are caused by other regulatory mechanisms, such as EWS-FLI1 binding to the DNA.
Finally, our study describes broadly applicable methods for dissecting epigenetic heterogeneity, which contribute to ongoing research into the biological and medical relevance of tumor heterogeneity42. Focusing on DNA methylation as a measure of epigenetic heterogeneity has important advantages, including its correlation with other epigenomic marks and with transcription factor binding (we observed a clear footprint of EWS-FLI1 binding in our DNA methylation maps), high accuracy and robustness of clinical DNA methylation assays43, and existing proof of concept that DNA methylation biomarkers can help to inform personalized cancer therapy44,45.
Online Methods
Ewing sarcoma tumors
EwS tumor samples from 140 patients were included in the analysis (Supplementary Table 1). In all cases, the EwS diagnosis was ascertained by testing for the presence of an EWS-ETS fusion. Of the 140 EwS tumors, 96 were provided by the tumor bank at the Institut Curie (Paris, France), 25 tumors by CCRI (Vienna, Austria), 11 by Biobank Graz (Graz, Austria), and eight by the biobank of the European Intergroup Cooperative Ewing’s Sarcoma Study (Münster, Germany). Most of the French samples (79 out of 96) have recently undergone whole-genome sequencing9, thus providing comprehensive maps of genetic lesions that were included in the analysis. Genome-sequencing data also established estimates of tumor purity for these samples, which were obtained from the supplementary material of the previous study9. Specifically, tumor purity was estimated on the basis of the loss of heterozygosity, copy number change, and mutated allele fraction of single-nucleotide variants using established methodology46. Most patients with EwS were treated according to the EuroEwing protocol47 or slight variations thereof. Informed consent was obtained according to the Declaration of Helsinki, and the study was approved and overseen by the ethics committees of the contributing institutions.
Ewing sarcoma cell lines
EwS cell lines were obtained from several sources. The STA-ET series was established at the CCRI (Vienna, Austria)48. STA-ET-2.1 and STA-ET-2.2 were generated from a biopsy of the primary tumor and a bone marrow infiltrate from one patient; STA-ET-7.1 and STA-ET-7.3 were generated from the primary tumor and a distant metastasis; STA-ET-8.1 (primary tumor) and STA-ET-8.2 (pleural effusion) were also established from the same patient. STA-ET-5, STA-ET-7.1, STA-ET-9, STA-ET-10, STA-ET-21, and STA-ET-22 were established from tumors that were included in the cohort of 140 tumor samples selected for this study. SK-N-MC cells were obtained from J. Beidler (Memorial Sloan Kettering Cancer Center, New York, USA). WE68 and WE68M2 were established from the same patient and provided by F. Van Valen (University Hospital Münster, Germany). CHLA-9 and CHLA-10 were established from the same patient and provided by P. Sorensen (British Columbia, Canada). Additional annotations are listed in Supplementary Table 1.
Mesenchymal stem cells
Low-passage human MSCs were provided by the Centro de Investigación del Cáncer (Salamanca, Spain) and Paracelsus Medical University (Salzburg, Austria). They were obtained from the bone marrow of patients with EwS, as well as from bone marrow, umbilical cord, and placenta of healthy individuals. MSCs were cultured as previously described49,50. Additional annotations are listed in Supplementary Table 1.
DNA extraction
DNA was isolated from 10 mg to 25 mg of snap-frozen tumors, cell lines, and MSCs by standard proteinase K digestion and phenol/chloroform extraction. DNA was quantified using a Qubit 2.0 Fluorometer (ThermoFisher Scientific, Q32866) and the Qubit dsDNA BR Assay Kit (ThermoFisher Scientific, Q32850).
RRBS
RRBS was performed as described previously51,52, starting with 100 ng of genomic DNA per sample. Custom-designed methylated and unmethylated oligonucleotides were added at a concentration of 0.1% to serve as spike-in controls for monitoring bisulfite conversion efficiency. After adaptor ligation, RRBS libraries were quantified by qPCR and pooled in combinations of six. For library enrichment, the number of PCR cycles was determined by qPCR and never exceeded 18 cycles. The library was purified twice using Agencourt AMPure XP beads (Beckman Coulter, A63880). Quality control for the final library was performed by measuring the DNA concentration with the Qubit dsDNA HS assay (ThermoFisher Scientific, Q32851) on Qubit 2.0 Fluorometer (ThermoFisher Scientific, Q32866) and by determining library fragment sizes with the Experion DNA 1K Analysis kit (Bio-Rad, 700-7107) on the Experion Automated Electrophoresis Station (Bio-Rad, 701-7000). Libraries were sequenced on Illumina HiSeq 2000/2500 machines.
WGBS
WGBS for three primary human tumors was performed using the µWGBS workflow described previously53, starting with 50 ng of genomic DNA per sample. Bisulfite conversion followed the EZ DNA Methylation-Direct Kit (Zymo Research, D5020), with the modification of eluting the DNA in only 9 μl of elution buffer. Custom-designed methylated and unmethylated oligonucleotides were added at a concentration of 0.1% to serve as spike-in controls for monitoring bisulfite conversion efficiency. Libraries for next-generation sequencing were prepared using the EpiGnome Methyl-Seq kit (Epicentre, EGMK81312). The library was purified twice using Agencourt AMPure XP beads (Beckman Coulter, A63880). Quality control for the final library was performed by measuring the DNA concentration with the Qubit dsDNA HS assay (Life Technologies, Q32851) on Qubit 2.0 Fluorometer (Life Technologies, Q32866) and by determining library fragment sizes with the Experion DNA 1K Analysis kit (Bio-Rad, 700-7107) on the Experion Automated Electrophoresis Station (Bio-Rad, 701-7000). Libraries were sequenced on Illumina HiSeq 2000/2500 machines.
ChIP-seq
ChIP-seq for three primary human tumors was done as described previously11. Chromatin was prepared from 20 to 50 sections (25 µm each) of snap-frozen tumors obtained by microtome sectioning. The following antibodies were used: H3K4me3 (1 µg/ChIP; Diagenode, C15410003-50), H3K27me3 (1 µg/ChIP; Diagenode, C15410195), H3K4me1 (1 µg/ChIP; Diagenode, C15410194), H3K27ac (1 µg/ChIP; Diagenode, C15410196), H3K56ac (4 µl/ChIP; Active Motif, 39281), H3K9me3 (1 µg/ChIP; Diagenode, C15410193), and H3K36me3 (1 µg/ChIP; Diagenode, C15410192). Library preparation for ChIP DNA and input control DNA was performed using the NEBNext Ultra kit (New England Biolabs, E7370S/L) following the manufacturer’s instructions. Quality control for the final libraries was done by measuring the DNA concentration with the Qubit dsDNA HS assay (Life Technologies, Q32851) on Qubit 2.0 Fluorometer (Life Technologies, Q32866) and by determining library fragment sizes with the Experion DNA 1K Analysis kit (Bio-Rad, 700-7107) on the Experion Automated Electrophoresis Station (Bio-Rad, 701-7000). Libraries were sequenced on Illumina HiSeq 2000/2500 machines.
RRBS/WGBS data processing
Bisulfite sequencing data were processed with a custom pipeline (http://sheffield2017.computational-epigenetics.org), which was based on Pypiper (http://databio.org/pypiper) and Looper (http://databio.org/looper). Read sequences were trimmed using Trimmomatic with ILLUMINACLIP settings “:2:40:7 SLIDINGWINDOW:4:15 MAXINFO:20:0.50 MINLEN:18”. Reads were aligned to the GRCh38 assembly of the human genome, using BSMAP in its RRBS mapping mode for RRBS54,55 and Bismark v.0.12.2 for WGBS56. DNA methylation levels for individual CpGs were calculated using custom Python scripts. Bisulfite conversion efficiency was estimated by aligning unmapped reads to the spike-in genome for methylated or unmethylated control sequences. CpGs located in repetitive regions according to the UCSC RepeatMasker track were excluded from further analysis. RRBS reference data from public databases (Supplementary Table 2) were downloaded from GEO as raw sequence data and processed with the same pipeline.
ChIP-seq data processing
ChIP-seq reads were aligned to the GRCh38 assembly of the human genome using the Bowtie2 short-read aligner (version 2.2.4)57 in end-to-end mode and processed with SAMtools (version 1.2)58, followed by peak finding with MACS2 (version 2.1.0.20150420)59 with factor-specific parameter settings as provided by the developers60. The bdgcmp algorithm of MACS2 was used in subtraction mode to generate signal plots, which were incorporated into a UCSC Genome Browser track hub for interactive visualization61. For samples that had two or more replicates, the Bioconductor package DiffBind (version 1.16.3)62 was used to characterize significantly differentially occupied peak regions on the basis of the MACS2 peak calls.
Bioinformatic analysis of heterogeneity between cancer types
To assess how well DNA methylation discriminates between EwS and non-EwS samples, we calculated the mean DNA methylation values of 5-kb tiling regions across the genome in each sample, and we used these data as attributes for predicting whether or not a sample was an EwS sample (tumor or cell line). Logistic regression classifiers were trained using liblineaR (version 1.94-2)63, and their performance was evaluated by fivefold cross-validation. For comparison, we measured the prediction performance on randomized data in 100 repetitions with randomly shuffled class labels. Receiver operating characteristic (ROC) curves were plotted using ROCR64. To assess EwS-specific differential DNA methylation, we focused on individual CpGs and used Wilcoxon rank-sum tests to compare DNA methylation values of all EwS tumors to those of three independent reference sets: The MSCs included in this study, a cancer diversity panel assembled from public data, and a cell-type diversity panel that was composed primarily of the ENCODE RRBS data set65 (Supplementary Table 2). Individual CpGs were classified as differentially methylated if the Wilcoxon P value was less than 0.01 and the absolute DNA methylation difference was at least 0.15 in each of these comparisons, and we restricted the analysis to CpGs covered by at least 30 EwS tumors and at least 30 samples from the cell-type diversity panel. We performed enrichment analysis of differentially methylated genomic regions using the LOLA Bioconductor package v1.1.3 and the LOLA Core database19. To visualize DNA methylation differences at individual regions, we produced locus-specific DNA methylation plots (Fig. 1f,g), averaging DNA methylation levels across 50 bins spanning the region of interest.
Bioinformatic analysis of heterogeneity among tumor samples
For the remaining analyses, the 11 formalin-fixed paraffin-embedded (FFPE) from Graz were excluded because of their lower data quality when compared to the 129 fresh-frozen samples. Global DNA methylation heterogeneity between tumor samples was quantified by the coefficient of variation (CV), as described previously28. For each set of samples, the CV was calculated from the distribution of genome-wide average DNA methylation levels in 5-kb tiling regions across samples, dividing the s.d. by the mean of the distribution. Region-specific DNA methylation heterogeneity was analyzed for sets of genomic regions with characteristic regulatory dynamics in EWS-FLI1 expressing cell lines11. First, EwS-specific DNaseI elements were defined by using ENCODE DNaseI data across 112 cell types21. We downloaded the processed DNaseI data matrix from an earlier study20 and selected elements with chromatin-accessibility scores consistently above 0.4 in SK-N-MC cells (an EwS cell line) and below 0.1 in all other samples. Second, EWS-FLI1 binding sites were derived from raw ChIP-seq data for EWS-FLI1 in the A673 cells (an EwS cell line)25. Third and fourth, we included EWS-FLI1-correlated enhancers and EWS-FLI1-anti-correlated enhancers, as defined in our previous study looking at the effect of EWS-FLI1 knockdown on H3K27ac peaks11. Heterogeneity between tumor samples was quantified as follows: for each region set, we aggregated DNA methylation levels in each region and retained only regions with at least ten sequencing reads. We then calculated the CV across samples for each region in the set. We used the median of these region-specific CV values as the score for each region set.
Bioinformatic analysis of sample clustering
To assess whether the DNA methylation profiles could be grouped into any consistent and reproducible clusters, we used several combinations of filtering, imputation, aggregation and clustering methods. All analyses were done in R (https://www.r-project.org/). These analyses were based on single CpG methylation calls without any binning, as well as mean DNA methylation values for 5-kb tiling regions in each sample. We calculated distance and similarity matrices on the basis of all CpGs with a coverage of at least 10 reads in a pairwise manner to minimize the loss of coverage due to missing values. To calculate these matrices, we used the dist and the cor functions of the stats package in R with the parameter use = “pairwise.complete.obs”. Correlation matrices as obtained with the cor function were converted into distance-like matrices by subtracting the correlation values from 1. We then used these distance matrices as input for clustering. We tried five methods for sample clustering and unsupervised subtype identification: multidimensional scaling, principal component analysis, independent component analysis, non-negative matrix factorization, and hierarchical clustering. As a measure of cluster quality, we calculated the mean silhouette width using the silhouette function of the cluster package. For non-negative matrix factorization (NMF), the NMF package was used66. Independent component analysis (ICA) was done using the fastICA package, which implements the algorithm of Hyvarinen and Oja67.
Bioinformatic definition of the epigenetic disease spectrum
To define the dimensions of the epigenetic disease spectrum, we focused on DNA methylation profiles for the four regulatory region types described above. We first split each region into 21 bins and averaged DNA methylation values in each of these bins. We then aggregated each of the 21 bins across all regions of that type, yielding a single vector with 21 values for each of the four region sets in each sample. Composite plots were prepared for each sample, providing aggregate DNA methylation patterns across all regions of a given type in each sample. The MIRA score reduces the dimensionality of the vector by summarizing the profile into a single number, calculated as the log ratio between DNA methylation values for the center bin (bin 0) and the average of two flanking bins (bins –5 and +5). For each sample and region set, the MIRA score represents the inferred sample-specific activity of the corresponding set of regulatory regions. The R functions used to calculate the MIRA scores are available from http://sheffield2017.computational-epigenetics.org.
Bioinformatic analysis of heterogeneity within individual tumors
The proportion of discordant reads (PDR) score was calculated as previously described13. Aligned RRBS read containing at least four CpGs were classified as concordant if all CpGs had the same DNA methylation state or discordant if they had different states. We determined the proportion of discordant reads for each CpG covered by at least ten reads. To assess the relationship between DNA methylation levels and PDR scores, we averaged both on 5-kb tiling regions and then binned all regions by their DNA methylation levels in increments of 5 percentage points. Consistent with the definition of PDR, low PDR levels were observed for regions with high (>75%) and low (<25%) levels of DNA methylation (Supplementary Fig. 9b).
To complement the PDR score, which focuses on co-methylation of neighboring CpGs in the same sequencing read, we developed the proportion of sites with intermediate methylation (PIM) score as a CpG-centric (rather than read-centric) measure of intra-tumor heterogeneity. For all CpGs covered by at least ten reads, we first classified each CpG as uniformly or intermediately methylated as follows: if a 95% binomial Bayesian credibility interval estimate of the CpG’s DNA methylation level was completely above 75% or below 25%, the CpG was considered uniformly methylated, whereas all other CpGs were considered intermediately methylated (Supplementary Fig. 9c). We then calculated the proportion of intermediately methylated CpGs to obtain the PIM score for a given sample. In pairwise comparisons between samples, we controlled for differential RRBS coverage by restricting the analysis to the subset of CpGs that pass coverage filtering in both samples being compared. The final PIM score of each sample is the mean log ratio of relative PIM scores between that sample and any other sample in the data set. The R functions used to calculate PIM scores are available from http://sheffield2017.computational-epigenetics.org.
Association with clinical data
To explore links between DNA methylation and clinical or genetic annotations, we divided samples according to annotation status and compared their corresponding measures of DNA methylation heterogeneity using the Wilcoxon rank-sum test. We also investigated links with the primary site of tumor location by grouping tumors according to their locations and testing for pairwise differences between tumors of each location against all others. To remove the confounding effect of tumor purity and gender, which were significantly associated with several measures of DNA methylation heterogeneity (Supplementary Fig. 10), we statistically corrected for these associations. This adjustment restricts the comparison to a subset of tumors (n = 76) from the French cohort for which genome sequencing data and quantitative estimates of tumor purity were available1. To quantify the association between PIM scores and metastatic status at diagnosis, we built logistic regression models predicting the clinical annotation from the PIM score on all data and assessed their model performance on the data set using the ROC area under curve (AUC) metric in comparison to 1,000 permutations with shuffled class labels.
Supplementary Material
Acknowledgments
We would like to thank all patients who have donated samples for this study. We also thank the team of the Biomedical Sequencing Facility at CeMM for support with next generation sequencing; the members of the Delattre, Kovar, and Bock labs for discussions; A. Rendeiro and C. Dietz for contributing to the analysis pipelines; K. Clement for sharing his implementation of the PDR score; A. Lankester for providing MSCs; and the following physicians for providing tumor samples: J.M. Guinebretière, L. Brugières, A. de Muret, R. Tichit, N. Sirvent, F. Millot, F. Guilhot, J.P. Vannier, C. Michot, E. Plouvier, A. Gomez-Brouchet, J. Rivel, B. Petit, F. Dijoud, F. Larousserie, A. Kurt, A. Foulet, A.S. Desfachelles, H. Sartelet, I. Quintin Roue, J. Otten, J. Chasles, C. Bouvier, C. Soler, M. Peuchmaur, and X. Rialland. This study was funded by a grant from the Austrian National Bank’s Jubiläumsfonds to E.M.T. (OeNB project number: 15714) and by a peer-reviewed institutional grant to E.M.T., which was based on a charitable donation of the Kapsch group (http://www.kapsch.net/kapschgroup) to St. Anna Kinderkrebsforschung. The French samples were collected in the context of the Plateforme Hospitalière de Génétique Moléculaire des Cancers of the Institut Curie and Centre Hospitalier de Versailles, with support by grants from INSERM within the framework of the International Cancer Genome Consortium program and from the Ligue Nationale Contre Le Cancer (Equipe labellisée), and the Société Française des Cancers de l’Enfant. The following associations supported this work: Courir pour Mathieu, Dans les pas du Géant, Olivier Chape, Les Bagouzamanon, Enfants et Santé, and les Amis de Claire. The study was performed in the context of the following European Union consortia: Euro Ewing (grant agreement no. 602856), BLUEPRINT (grant agreement no. 282510), PROVABES (grant agreement no. 01KT1310), ASSET (grant agreement no. 259348), and TECHNOBEAT (grant agreement no. 668724). N.C.S. was supported by a long-term fellowship of the Human Frontier Science Program (LT000211/2014). J.K. was supported by a DOC Fellowship of the Austrian Academy of Sciences. D.S. was supported by the Institut Curie-SIRIC (Site de Recherche Intégrée en Cancérologie) program. E.d.A. was supported by Ministry of Economy and Competitiveness of Spain-FEDER grants (RD12/0036/0017, PI14/01466), María García-Estrada Foundation, and Pablo Ugarte Association. C.B. was supported by a New Frontiers Group award of the Austrian Academy of Sciences and by a European Research Council (ERC) Starting Grant (European Union’s Horizon 2020 research and innovation program; grant 679146). E.M.T. was supported by fellowships of the Austrian Science Fund (FWF, Lise Meitner Fellowship M1448-B13; and Elise Richter Fellowship V506-B28).
Footnotes
Data availability
All raw and processed data produced for this study are available for download at NCBI GEO (accession: GSE88826 and GSE89026). DNA methylation and ChIP-seq profiles are also available for interactive browsing and download from http://sheffield2017.computational-epigenetics.org. Genome sequencing for 79 samples are available from EBI EGA (accession code: EGAS00001000855).
Author Contributions
N.C.S., O.D., H.K., C.B., and E.M.T. designed the study. N.C.S. performed the data analysis with contributions from J.K., A.S., and M.S. G.P., D.Su., D.G., E.L., P.F., J.C., R.B., I.M.A., C.H., E.S., A.T.A., E.d.A., K.S., D.St., B.R., B.L.-A., B.H., A.L., G.d.P., P.T., V.L., J.M., R.L., W.H., R.W., U.D., P.F.A., and O.D. provided materials such as tumor samples, clinical data, cell lines, and MSC samples. P.D., J.H., D.W., and E.M.T. performed the experiments. N.C.S., C.B., and E.M.T. wrote the manuscript with contributions from all authors.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.de Álava E, Lessnick SL, Sorensen PHB. Ewing sarcoma. In: Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F, editors. WHO Classification of tumours of soft tissue and bone. 2013. pp. 306–309. [Google Scholar]
- 2.Delattre O, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 3.Parham DM, et al. Neuroectodermal differentiation in Ewing’s sarcoma family of tumors does not predict tumor behavior. Hum Pathol. 1999;30:911–918. doi: 10.1016/S0046-8177(99)90244-7. [DOI] [PubMed] [Google Scholar]
- 4.Pinto A, Dickman P, Parham D. Pathobiologic markers of the ewing sarcoma family of tumors: state of the art and prediction of behaviour. Sarcoma. 2011;2011 doi: 10.1155/2011/856190. 856190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt D, Herrmann C, Jürgens H, Harms D. Malignant peripheral neuroectodermal tumor and its necessary distinction from Ewing’s sarcoma. A report from the Kiel Pediatric Tumor Registry. Cancer. 1991;68:2251–2259. doi: 10.1002/1097-0142(19911115)68:10%3c2251::AID-CNCR2820681025%3e3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 6.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brohl AS, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;10:e1004475. doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crompton BD, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4:1326–1341. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 9.Tirode F, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4:1342–1353. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riggi N, et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;26:668–681. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomazou EM, et al. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Reports. 2015;10:1082–1095. doi: 10.1016/j.celrep.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landau DA, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26:813–825. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med. 2016;22:792–799. doi: 10.1038/nm.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu H, et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7:133–136. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meissner A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bock C, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin PP, Wang Y, Lozano G. Mesenchymal stem cells and the origin of Ewing’s sarcoma. Sarcoma. 2011;2011:276463. doi: 10.1155/2011/276463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheffield NC, Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2016;32:587–589. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheffield NC, et al. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res. 2013;23:777–788. doi: 10.1101/gr.152140.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu T, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sánchez-Castillo M, et al. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res. 2015;43:D1117–D1123. doi: 10.1093/nar/gku895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bilke S, et al. Oncogenic ETS fusions deregulate E2F3 target genes in Ewing sarcoma and prostate cancer. Genome Res. 2013;23:1797–1809. doi: 10.1101/gr.151340.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agirre X, et al. Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers. Genome Res. 2015;25:478–487. doi: 10.1101/gr.180240.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe M, et al. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Res. 2005;65:828–834. [PubMed] [Google Scholar]
- 30.Hovestadt V, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510:537–541. doi: 10.1038/nature13268. [DOI] [PubMed] [Google Scholar]
- 31.Johann PD, et al. Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell. 2016;29:379–393. doi: 10.1016/j.ccell.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Kulis M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 33.Mazor T, et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell. 2015;28:307–317. doi: 10.1016/j.ccell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aran D, Hellman A. Unmasking risk loci: DNA methylation illuminates the biology of cancer predisposition: analyzing DNA methylation of transcriptional enhancers reveals missed regulatory links between cancer risk loci and genes. BioEssays. 2014;36:184–190. doi: 10.1002/bies.201300119. [DOI] [PubMed] [Google Scholar]
- 35.Bock C, et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burger L, Gaidatzis D, Schübeler D, Stadler MB. Identification of active regulatory regions from DNA methylation data. Nucleic Acids Res. 2013;41:e155. doi: 10.1093/nar/gkt599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hon GC, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 39.Mazumdar C, et al. Leukemia-associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell. 2015;17:675–688. doi: 10.1016/j.stem.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabassum DP, Polyak K. Tumorigenesis: it takes a village. Nat Rev Cancer. 2015;15:473–483. doi: 10.1038/nrc3971. [DOI] [PubMed] [Google Scholar]
- 41.Mazor T, Pankov A, Song JS, Costello JF. Intratumoral heterogeneity of the epigenome. Cancer Cell. 2016;29:440–451. doi: 10.1016/j.ccell.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alizadeh AA, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21:846–853. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bock C, et al. BLUEPRINT consortium Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol. 2016;34:726–737. doi: 10.1038/nbt.3605. [DOI] [PubMed] [Google Scholar]
- 44.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 45.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell. 2013;24:710–724. doi: 10.1016/j.ccr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ladenstein R, et al. Primary disseminated multifocal Ewing sarcoma: results of the Euro-EWING 99 trial. J Clin Oncol. 2010;28:3284–3291. doi: 10.1200/JCO.2009.22.9864. [DOI] [PubMed] [Google Scholar]
- 48.Ambros IM, et al. MIC2 is a specific marker for Ewing’s sarcoma and peripheral primitive neuroectodermal tumors. Evidence for a common histogenesis of Ewing’s sarcoma and peripheral primitive neuroectodermal tumors from MIC2 expression and specific chromosome aberration. Cancer. 1991;67:1886–1893. doi: 10.1002/1097-0142(19910401)67:7%3c1886::AID-CNCR2820670712%3e3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 49.Amaral AT, et al. Characterization of human mesenchymal stem cells from ewing sarcoma patients. Pathogenetic implications. PLoS One. 2014;9:e85814. doi: 10.1371/journal.pone.0085814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reinisch A, et al. Epigenetic and in vivo comparison of diverse MSC sources reveals an endochondral signature for human hematopoietic niche formation. Blood. 2015;125:249–260. doi: 10.1182/blood-2014-04-572255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klughammer J, et al. Differential DNA methylation analysis without a reference genome. Cell Reports. 2015;13:2621–2633. doi: 10.1016/j.celrep.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veillard A-C, Datlinger P, Laczik M, Squazzo S, Bock C. Diagenode premium RRBS technology: cost-effective DNA methylation mapping with superior CpG resolution and coverage. Nat Methods. 2016;13 (Application Note), •••. [Google Scholar]
- 53.Farlik M, et al. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Reports. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xi Y, et al. RRBSMAP: a fast, accurate and user-friendly alignment tool for reduced representation bisulfite sequencing. Bioinformatics. 2012;28:430–432. doi: 10.1093/bioinformatics/btr668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Speir ML, et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44(D1):D717–D725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ross-Innes CS, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan R-E, Chang K-W, Hsieh C-J, Wang X-R, Lin C-J. LIBLINEAR: A library for large linear classification. J Mach Learn Res. 2008;9:1871–1874. [Google Scholar]
- 64.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 65.Varley KE, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hyvärinen A, Oja E. Independent component analysis: algorithms and applications. Neural Netw. 2000;13:411–430. doi: 10.1016/S0893-6080(00)00026-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.