Abstract
Poly(γ-4-((2-(piperidin-1-yl)ethyl)aminomethyl)-benzyl-l-glutamate) (PPABLG), a cationic helical polypeptide, has been recently developed by us as an effective non-viral gene delivery vector. In attempts to elucidate the effect of molecular architecture on the gene delivery efficiencies and thereby identify a potential addition to PPABLG with improved transfection efficiency and reduced cytotoxicity, we synthesized PEG-PPABLG copolymers with diblock, triblock, graft, and star-shaped architectures via a controlled ring opening polymerization. The PPABLG segment in all copolymers adopted helical structure; all copolymers displayed structure-related cell penetration properties and gene transfection efficiencies. In HeLa and HepG-2 cells, diblock and triblock copolymers exhibited reduced membrane activities and cytotoxicities but uncompromised gene transfection efficiencies compared to the non-PEGylated, homo-PPABLG. The graft copolymer revealed lower DNA binding affinity and membrane activity presumably due to the intramolecular entanglement between the grafted PEG segments and charged side chains that led to reduced transfection efficiency. The star copolymer, adopting a spherical architecture with high density of PPABLG, afforded the highest membrane activity and relatively low cytotoxicity, which contributed to its potent gene transfection efficiency that outperformed the non-PEGylated PPABLG and Lipofectamine™ 2000 by 3–5 and 3–134 folds, respectively. These findings provide insights into the molecular design of cationic polymers, especially helical polypeptides towards gene delivery.
Keywords: Non-viral gene delivery, Cationic helical polypeptide, PEGylation, Macromolecular architecture, Cell penetration peptide (CPP)
1. Introduction
Gene therapy has been widely regarded as a promising modality that can potentially treat various inquired or congenital diseases in a highly specific manner [1–3]. The success of gene therapy largely depends on the delivery efficiency of the therapeutic gene. Although viral vectors enable highly efficient gene delivery, they cause associated immune responses and oncogenic transformations with several reported fatal cases in clinical trials [4]. Non-viral gene delivery mediated by cationic liposomal and polymeric vectors has emerged as an attractive alternative because of their low toxicity and low immunogenicity [5–7].
Cell penetrating peptides (CPPs), such as oligoarginine, HIV-TAT, and penetratin, are sequence-specific oligopeptides that have excellent membrane activities and can effectively mediate membrane transduction to facilitate cellular internalization of exogenous materials, such as DNA, RNA, and proteins [8,9]. Hence, they have been extensively applied in non-viral gene delivery [6,7,10]. However, CPPs are often too short (less than 25 amino acid residues) and lack adequate cationic charges to efficiently condense and deliver genes. As such, they often function as membrane-active ligands incorporated or conjugated to delivery vehicles to improve the delivery efficiency [6,7].
We have recently developed a high molecular weight (MW), cationic, α-helical polypeptide, termed poly(γ-(4-((2-(piperidin-1-yl)ethyl)aminomethyl)benzyl-l-glutamate) (PPABLG) [11,12], as an effective non-viral gene delivery vector. We demonstrated that the stabilized helical structure of PPABLG markedly contributed to its membrane penetrating capacity via the pore formation mechanism; its relatively high MW and cationic charge density facilitated the efficient condensation of DNA, making it a more efficient vector than traditional CPPs for gene delivery to mammalian cells [12,13]. However, excessive membrane activity of PPABLG may cause irreversible damage to cell membranes. We thus seek to find a strategy to further improve the gene transfection efficiency of PPABLG and reduce its cytotoxicity.
Polymer physico-chemical properties, such as molecular weight, charge density, three-dimensional structure, chain flexibility, and hydrophilicity/hydrophobicity balance, have significant impact on the efficiency and safety of non-viral gene vectors [14–22]. Therefore, design features to control over these properties may render promising strategy for the identification of an ideal non-viral gene delivery vector with maximized transfection efficiency and minimized toxicity. Incorporation of poly(ethylene glycol) (PEG) moieties in the macromolecular design has been demonstrated an effective strategy to not only reduce the cytotoxicity of the gene delivery vectors but also improve the stability of the polymer/DNA complexes [14,16]. Additionally, PEGylation provides an opportunity to modulate the architecture of the PEG-polymer conjugates, which in turn alters their complexation capacities with DNA, interaction with cellular/endosomal membranes, and ultimately the transfection efficiencies [14,16].
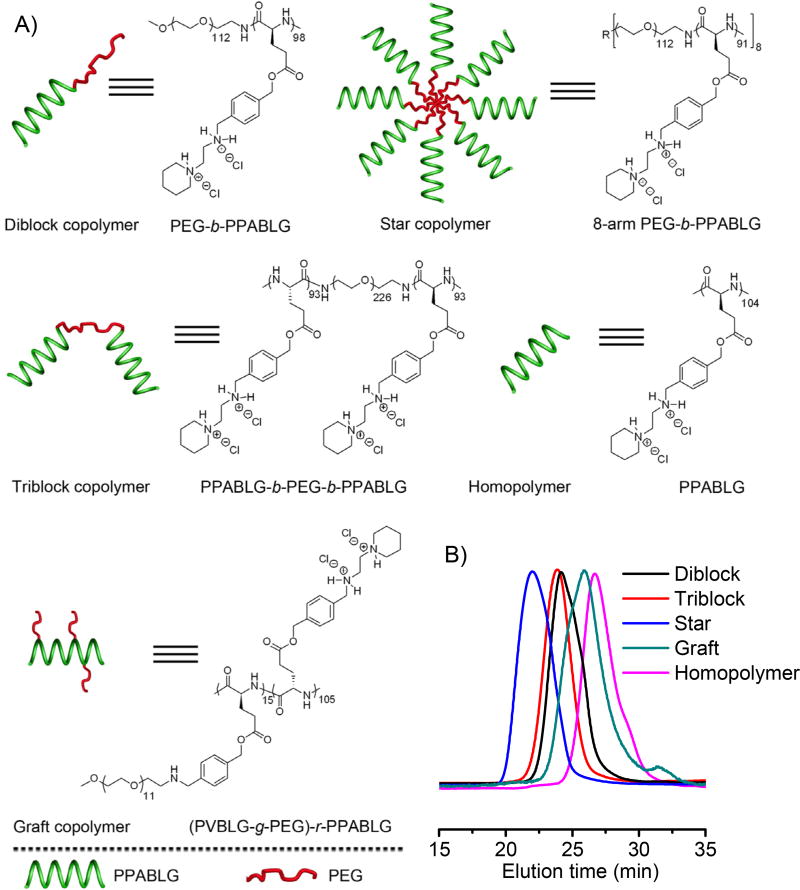
Motivated by these understandings, we designed and synthesized various PEG-PPABLG copolymers with different molecular architectures, attempting to elucidate the effect of polymer topology on the gene delivery efficiencies and thus identify desired PEGylated PPABLG derivative with high gene transfection efficiency and low cytotoxicity. Amine-terminated PEG served as the macroinitiator for the ring-opening polymerization (ROP) of γ-(4-vinylbenzyl)-l-glutamate N-carboxylanhydride (VB-l-glu-NCA, Supplementary Scheme S1) [23], thus achieving the PEG-PPABLG block polymers. By using mPEG-NH2 (5 kDa), H2N-PEG-NH2 (10 kDa), and 8-arm PEG-NH2 (40 kDa) as initiators, diblock, triblock, and star copolymers were obtained (Fig. 1A). Furthermore, mPEG-NH2 (550 Da) was grafted onto PPABLG, yielding the graft copolymer (Fig. 1A). The degree of polymerization (DP) of PPABLG was maintained constant at ~100, along with a comparable weight fraction of PEG segments of ~11%, thus allowing the synthesis of compositionally equivalent while topologically different PEG-PPABLG copolymers. The effect of polymer architecture on the physicochemical properties of the polymer/DNA complexes, gene transfection efficiencies, intracellular kinetics, and cytotoxicity was comprehensively explored, which provided insights into the molecular design of PEGylated cationic helical polypeptides for gene delivery.
Fig. 1.
(A) Schematic representation of PEG-PPABLG conjugates with different architectures. (B) GPC traces of the precursors (PVBLG or PEG-PVBLG conjugates) of various polypeptides.
2. Materials and methods
2.1. Materials and cell culture
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as received unless otherwise specified. Anhydrous tetrahydrofuran (THF), hexane, and dimethylformamide (DMF) were dried by a column packed with 4Å molecular sieves and stored in a glovebox. Dry nitrobenzene (NB) was prepared by treating regular NB with CaH2 followed by distillation under reduced pressure. Hexamethyldisilazane (HMDS) and 1,5,7-triazabicyclo[4.4.0]dec-5-ene (TBD) were dissolved in DMF in a glovebox. VB-l-glu-NCA was synthesized according to published literature [11]. mPEG-NH2 (550 Da and 5 kDa) and H2N-PEG-NH2 (10 kDa) were purchased from Laysan Bio Inc. (Arab, AL, USA). 8-Arm PEG-NH2 (40 kDa) was purchased from Nanocs Inc. (New York, NY, USA). Plasmid DNA encoding luciferase (pCMV-Luc) was purchased from Elim Biopharmaceutics (Hayward, CA, USA). Lipofectamine™ 2000 (LPF2000), YOYO-1, and 3-(4,5-dimethylthiahiazol-2-yl)-2,5-di-phenyl-2H-tetrazolium bromide (MTT) were purchased from Invitrogen (Carlsbad, CA, USA).
HepG-2 (human hepatocellular carcinoma) and HeLa (human cervix adenocarcinoma) cells were purchased from the American Type Culture Collection (Rockville, MD, USA), and cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS).
2.2. Polypeptide synthesis
2.2.1. Synthesis of PPABLG homopolymer
PPABLG was synthesized as described previously [11]. Briefly, in a glovebox, VB-l-glu-NCA (120 mg, 0.415 mmol) was dissolved in a mixture of DMF (1.0 mL) and nitrobenzene (30 µL), followed by addition of HMDS (41.5 µL, 0.1 mol/L, M/I = 100) and TBD (41.5 µL, 0.01 mol/L) solution in DMF. The reaction mixture was stirred at room temperature for 48 h (monomer conversion > 99%) to obtain the poly(γ-(4-vinylbenzyl)-l-glutamate) (PVBLG) homopolymer. Benzyl chloroformate (100 µL) and N,N-diisopropylethylamine (DIEA, 100 µL) were then added to cap the amino end groups. DMF was removed under vacuum. PVBLG was precipitated using cold ether and collected by centrifugation. 1H NMR (500 mHz, CDCl3/TFA-d (85:15, v/v)): δ 7.33 (d, 2H, ArH), 7.19 (d, 2H, ArH), 6.64 (dd, 2H, -CH=CH2), 5.71 (d, 2H, -CH=CH2), 5.23 (d, 2H, -CH=CH2), 5.03 (m, 2H, ArCH2O-), 4.58 (m, 1H, α-H), 2.44 (t, 2H, -COCH2CH2-), 2.09 (m, 1H, -COCH2CH2-), 1.90 (m, 1H, -COCH2CH2-) (Supplementary Fig. S2).
PVBLG (100 mg) was dissolved in chloroform (60 mL) and oxidized by O3 at −78 °C for 3 min. Dimethyl sulfide (1.0 mL) was then added, and the solution was stirred at room temperature overnight followed by removal of the solvent under vacuum. The product was precipitated by methanol and collected by centrifugation. It then reacted with 1-(2-aminoethyl)piperidine (440 µL) in DMF (4 mL) at 50 °C for 24 h; borane pyridine (390 µL) was added and the solution was stirred at 50 °C for another 24 h; 5 mol/L HCl (2 mL) was added and stirred for 10 min. The final product PPABLG was then dialyzed against DI water (MWCO = 1 kDa) and lyophilized. 1H NMR (500 mHz, TFA-d): δ 7.55 (m, 4H, ArH), 5.36 (br s, 2H, ArCH2O-), 4.87 (br s, 2H, ArCH2NH-), 4.55 (s, 1H, α-H), 4.01-3.80 (br m, 6H, -HNCH2CH2N- and -NCH2CH2CH2CH2CH2-), 3.16 (m, 2H, -HNCH2CH2N-), 2.80 (s, 2H, -COCH2CH2-), 2.50-2.20 (br d, 2H, -COCH2CH2-), 2.20-1.60 (br m, 6H, -NCH2CH2CH2CH2CH2-) (Supplementary Fig. S4).
2.2.2. Synthesis of PEG-b-PPABLG diblock copolymer
In a glovebox, VB-l-glu-NCA (120 mg, 0.415 mmol) was dissolved in a mixture of DMF (1.0 mL) and nitrobenzene (30 µL), followed by addition of mPEG-NH2 (5 kDa, 207 µL, 0.02 mol/L, M/I = 100) and TBD (41.5 µL, 0.01 mol/L) solution in DMF. The reaction mixture was stirred at room temperature for 72 h (monomer conversion > 99%) to obtain the PEG-b-PVBLG diblock copolymer. 1H NMR (500 mHz, CDCl3/TFA-d (85:15, v/v)): δ 7.32 (d, 2H, ArH), 7.19 (d, 2H, ArH), 6.63 (dd, 2H, -CH=CH2), 5.70 (d, 2H, -CH=CH2), 5.22 (d, 2H, -CH=CH2), 5.03 (m, 2H, ArCH2O-), 4.58 (m, 1H, α-H), 3.80 (s, 4.61H, -OCH2CH2O-), 2.43 (t, 2H, -COCH2CH2-), 2.09 (m, 1H, -COCH2CH2-), 1.90 (m, 1H, -COCH2CH2-) (Supplementary Fig. S3).
PEG-b-PPABLG was synthesized via ozonolysis of PEG-b-PVBLG and subsequent reductive amination as described above for the synthesis of PPABLG. 1H NMR (500 mHz, TFA-d): δ 7.54 (m, 4H, ArH), 5.32 (br s, 2H, ArCH2O-), 4.86 (br s, 2H, ArCH2NH-), 4.54 (s, 1H, α-H), 4.00 (s, 4H, -OCH2CH2-), 4.00-3.80 (br m, 6H, -HNCH2CH2N- and - NCH2CH2CH2CH2CH2-), 3.14 (m, 2H, -HNCH2CH2N-), 2.78 (s, 2H, -COCH2CH2-), 2.50-2.16 (br d, 2H, -COCH2CH2-), 2.20-1.60 (br m, 6H, -NCH2CH2CH2CH2CH2-) (Supplementary Fig. S5).
2.2.3. Synthesis of PPABLG-b-PEG-b-PPABLG triblock copolymer
In a glovebox, VB-l-glu-NCA (120 mg, 0.415 mmol) was dissolved in a mixture of DMF (1.0 mL) and nitrobenzene (30 µL), followed by addition of NH2-PEG-NH2 (10 kDa, 415 µL, 0.01 mol/L, M/I = 100) and TBD (41.5 µL, 0.01 mol/L) solution in DMF. The reaction mixture was stirred at room temperature for 72 h (monomer conversion > 99%) to obtain the PVBLG-b-PEG-b-PVBLG triblock copolymer. 1H NMR (CDCl3/TFA-d (85:15, v/v), δ, ppm): 7.32 (d, 2H, ArH), 7.18 (d, 2H, ArH), 6.63 (dd, 2H, -CH=CH2), 5.70 (d, 2H, -CH=CH2), 5.22 (d, 2H, -CH=CH2), 5.02 (m, 2H, ArCH2O-), 4.57 (m, 1H, α-H), 3.77 (s, 4.88H, -OCH2CH2O-), 2.43 (t, 2H, -COCH2CH2-), 2.08 (m, 1H, -COCH2CH2-), 1.90 (m, 1H, -COCH2CH2-).
PPABLG-b-PEG-b-PPABLG was synthesized via ozonolysis of PVBLG-b-PEG-b-PVBLG and subsequent reductive amination as described above for the synthesis of PPABLG. 1H NMR (TFA-d, δ, ppm): 7.54 (m, 4H, ArH), 5.33 (br s, 2H, ArCH2O-), 4.86 (br s, 2H, ArCH2NH-), 4.54 (s, 1H, α-H), 4.01 (s, 4H, -OCH2CH2-), 4.00-3.80 (br m, 6H, -HNCH2CH2N- and - NCH2CH2CH2CH2CH2-), 3.15 (m, 2H, -HNCH2CH2N-), 2.78 (s, 2H, -COCH2CH2-), 2.50-2.16 (br d, 2H, -COCH2CH2-), 2.20-1.60 (br m, 6H, -NCH2CH2CH2CH2CH2-) (Supplementary Fig. S6).
2.2.4. Synthesis of 8-arm PEG-b-PPABLG star copolymer
Inside a glovebox, VB-l-glu-NCA (120 mg, 0.415 mmol) was dissolved in a mixture of DMF (1.0 mL) and nitrobenzene (30 µL), followed by addition of 8-arm PEG-NH2 (40 kDa, 415 µL, 0.01 mol/L, M/I = 100) and TBD (41.5 µL, 0.01 mol/L) solution in DMF. The reaction mixture was stirred at room temperature for 120 h to obtain 8-arm PEG-b-PVBLG star copolymer (monomer conversion > 90%). 1H NMR (500 mHz, CDCl3/TFA-d (85:15, v/v)): δ 7.31 (d, 2H, ArH), 7.18 (d, 2H, ArH), 6.62 (dd, 2H, -CH=CH2), 5.69 (d, 2H, -CH=CH2), 5.22 (d, 2H, -CH=CH2), 5.02 (m, 2H, ArCH2O-), 4.55 (s, 1H, α-H), 3.79 (s, 4.97H, -OCH2CH2O-), 2.44 (s, 2H, -COCH2CH2-), 2.09 (s, 1H, -COCH2CH2-), 1.92 (s, 1H, -COCH2CH2-).
8-arm PEG-b-PPABLG was synthesized via ozonolysis of 8-arm PEG-b-PVBLG and subsequent reductive amination as described above for the synthesis of PPABLG. 1H NMR (500 mHz, TFA-d): δ 7.54 (m, 4H, ArH), 5.33 (br s, 2H, ArCH2O-), 4.87 (br s, 2H, ArCH2NH-), 4.53 (s, 1H, α-H), 4.01 (s, 4H, -OCH2CH2-), 4.00-3.80 (br m, 6H, -HNCH2CH2N- and - NCH2CH2CH2CH2CH2-), 3.16 (m, 2H, -HNCH2CH2N-), 2.78 (s, 2H, -COCH2CH2-), 2.50-2.16 (br d, 2H, -COCH2CH2-), 2.20-1.60 (br m, 6H, -NCH2CH2CH2CH2CH2-) (Supplementary Fig. S7).
2.2.5. Synthesis of (PVBLG-g-PEG)-r-PPABLG graft copolymer
PVBLG homopolymer was synthesized and the ozonolysis of PVBLG was conducted as mentioned above (M/I = 110). The resulting polymer was reacted with mPEG-NH2 (550 Da, 37.7 mg) in DMF (4 mL) at 50 °C for 24 h; 1-(2-aminoethyl)piperidine (440 µL) was added and stirred for another 24 h; borane pyridine (390 µL) was then added, and the solution was stirred at 50 °C for 24 h before addition of 5 mol/L HCl (2 mL). The final product (PVBLG-g-PEG)-r-PPABLG was dialyzed against DI water (MWCO = 1 kDa) and lyophilized. 1H NMR (500 mHz, TFA-d): δ 7.55 (m, 4H, ArH), 5.33 (br s, 2H, ArCH2O-), 4.87 (br s, 2H, ArCH2NH-), 4.54 (s, 1H, α-H), 4.01 (s, 6H, -OCH2CH2-), 4.00-3.80 (br m, 5.25H, -HNCH2CH2N- and - NCH2CH2CH2CH2CH2-), 3.14 (m, 1.75H, -HNCH2CH2N-), 2.78 (s, 2H, -COCH2CH2-), 2.50-2.16 (br d, 2H, -COCH2CH2-), 2.20-1.60 (br m, 5.25H, -NCH2CH2CH2CH2CH2-) (Supplementary Fig. S8).
2.3. Characterization of polypeptides
1H NMR spectra were recorded on a Varian U500 MHz or a VXR-500 MHz spectrometer. Chemical shifts were reported in ppm and referenced to the solvent proton impurities. Gel permeation chromatography (GPC) experiments were performed on a system equipped with an isocratic pump (Model 1100, Agilent Technology, Santa Clara, CA, USA), a DAWN HELEOS multi-angle laser light scattering detector (MALLS) detector (Wyatt Technology, Santa Barbara, CA, USA), and an Optilab rEX refractive index detector (Wyatt Technology, Santa Barbara, CA, USA). The detection wavelength was set at 658 nm. Separations were performed using serially connected size exclusion columns (100 Å, 500 Å, 103Å, and 104 Å Phenogel columns, 5 µm, 300 × 7.8 mm, Phenomenex, Torrance, CA, USA) at 60 °C using DMF containing 0.1 M LiBr as the mobile phase. The MALLS detector was calibrated using pure toluene with no need for calibration using polymer standards and can be used for the determination of the absolute molecular weights (MWs). The MWs of polypeptides were determined based on the dn/dc value of each polymer sample calculated offline by using the internal calibration system processed by the ASTRA V software (version 5.1.7.3, Wyatt Technology, Santa Barbara, CA, USA). Circular dichroism (CD) measurements were carried out on a JASCO J-700 CD spectrometer. The polypeptide samples were prepared in DI water at concentrations of 0.02–0.2 mg/mL unless otherwise specified. The solution was placed in a quartz cell with a path length of 0.2 cm. The mean residue molar ellipticity of each polymer was calculated based on the measured apparent ellipticity by following the reported formulas: Ellipticity ([θ] in deg·cm2·dmol−1) = (millidegrees × mean residue weight)/(pathlength in millimeters × concentration of polypeptide in mg·ml−1). The helicity of the polypeptides were calculated by the following equation: helicity = (−[θ222] + 3,000)/39,000 [24]. To evaluate the helical stability of polypeptides at different pH, pH of the polypeptide solution was adjusted using 1 M NaOH or HCl solution (polypeptide concentration fixed at 0.1 mg/mL). To evaluate the stability of helicity at different ionic strength, polypeptides were prepared in NaCl solutions at different NaCl concentrations (polypeptide concentration fixed at 0.1 mg/mL).
2.4. Polyplex formation and characterization
Polypeptide and pCMV-Luc were separately dissolved in water at 0.2 mg/mL and mixed at various N/P ratios. The mixture was vortexed for 5 s and incubated at 37 °C for 30 min to allow polyplex formation. The polyplexes were subjected to electrophoresis in 1% agarose gel at 100 mV for 45 min to evaluate DNA condensation by the polypeptides in terms of DNA migration. To quantitatively monitor the DNA condensation level, the ethidium bromide (EB) exclusion assay was adopted [25]. Briefly, EB solution was mixed with DNA at the DNA/EB ratio of 10:1 (w/w) and incubated at room temperature for 1 h. Polypeptide was then added to the mixture at various N/P ratios, and the mixture was further incubated at room temperature for 30 min before quantification of the fluorescence intensity on a microplate reader (λex=510 nm, λem=590 nm). A pure EB solution and the DNA/EB solution without any polypeptide were used as negative and positive controls, respectively. EB exclusion efficiency (% DNA condensed) was defined as:
Where FEB, F, and F0 denote the fluorescence intensity of pure EB solution, DNA/EB solution with polypeptide, and DNA/EB solution without any polypeptide, respectively.
Particle size and zeta potential of freshly prepared polyplexes were also evaluated by dynamic laser scanning (DLS) on a Malvern Zetasizer (Herrenberg, Germany).
2.5. In vitro gene transfection
Cells were seeded in 96-well plates at 1 × 104 cells/well and incubated for 24 h prior to transfection studies. The medium was replaced by serum-free DMEM, into which polyplexes were added at 0.1 µg DNA/well. After incubation for 4 h, the medium was replaced by serum-containing DMEM and cells were further cultured for 20 h. Luciferase expression was assayed in terms of luminescence intensity using a Bright-Glo Luciferase assay kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol; cellular protein level was determined using a BCA kit (Pierce, Rockford, IL, USA). Results were expressed as relative luminescence unit (RLU) associated with 1 mg of cellular protein.
2.6. Intracellular uptake studies
To allow quantification of the cellular uptake level, DNA (1 mg/mL) was labeled with YOYO-1 (20 µM) at one dye molecule per 50 bp DNA [26]. The resultant YOYO-1-DNA was then allowed to form complexes with the polypeptide at various N/P ratios as described above. LPF2000/DNA complexes were formed according to the manufacture’s protocol.
HeLa and HepG-2 cells were seeded on 96-well plates at 1×104 cells/well, and cultured in serum-containing DMEM for 24 h to reach confluence. The medium was replaced by fresh serum-free DMEM and complexes were added at 0.1 µg YOYO-1-DNA/well. After incubation at 37 °C for 4 h, cells were washed with cold PBS containing 20 U/mL heparin for three times to remove membrane-bound complexes [27] and were thereafter lysed with 100 µL of RIPA lysis buffer. YOYO-1-DNA content in the lysate was quantified by spectrofluorimetry (λex=485 nm, λem=530 nm) and protein content was measured using the BCA kit. Uptake level was expressed as ng YOYO-1-DNA associated with 1 mg of cellular protein.
To elucidate the mechanisms underlying the cellular internalization of polymer/DNA complexes, we performed the uptake study at 4 °C or in the presence of various endocytic inhibitors for 2 h. Briefly, cells were preincubated with endocytic inhibitors including chlorpromazine (10 µg/mL), genistein (200 µg/mL), methyl-β-cyclodextrin (mβCD, 50 µM), dynasore (80 µM), and wortmannin (50 nM) for 30 min prior to the addition of complexes and throughout the 2-h uptake experiment at 37 °C. Results were expressed as percentage uptake level of control cells that were incubated with complexes at 37 °C for 2 h.
To observe the intracellular distribution of the polymer/DNA complexes, HeLa cells were incubated with polymer/YOYO-1-DNA complexes in serum-free DMEM at 0.5 µg DNA/well (6-well plate) for 1 h and 4 h, respectively. Cells were then stained with Lysotracker®-Red (Invitrogen, Carlsbad, CA, USA) and DAPI according to the manufacturer’s protocol, and were visualized by confocal laser scanning microscopy (CLSM, LSM 700, Zeiss, Germany).
2.7. Membrane activity measurement
The membrane disruption capacity of the polypeptides was evaluated in terms of the cell uptake of a membrane-impermeable dye (FITC) [8]. HeLa cells were seeded on 24-well plate at 5×104 cells/well and incubated for 24 h. The medium was changed to serum-free DMEM, into which polypeptides were added at 2 µg/well and FITC was added at 1 µg/well. After incubation for 2 h, cells were washed with cold PBS for three times and lysed with the RIPA lysis buffer. FITC content in the cell lysate was quantified by spectrofluorimetry (λex=488 nm, λem=530 nm), and protein content was determined using the BCA kit. Cells incubated with free FITC in the absence of polypeptide served as the control. The uptake level was expressed as ng FITC associated with 1 mg of cellular protein. Commercial CPPs, Arg9 and HIV-TAT, were used as internal controls.
2.8. Cell viability
HeLa and HepG-2 cells were seeded in 96-well plate at 1×104 cells/well and incubated for 24 h. The medium was changed to serum-free DMEM, and polymer/DNA complexes (N/P ratio of 20) were added at 2, 1, 0.5, 0.2, 0.1, and 0.05 µg DNA/well, respectively. After incubation for 4 h, the medium was replaced by serum-containing DMEM and cells were further incubated for 20 h. Cell viability was then evaluated by the MTT assay. Cells without complex treatment served as the control and results were expressed as percentage viability of control cells.
3. Results
3.1. Synthesis and characterization of polypeptides
PPABLG homopolymer was synthesized via ROP of VB-l-glu-NCA as initiated by HMDS and subsequent side-chain modifications [23]. HMDS allowed a controlled ROP, thus obtaining well-defined polymer with a relatively low molecular weight distribution (MWD) (Table 1). With mPEG-NH2, NH2-PEG-NH2, and 8-arm PEG-NH2 to initiate ROP of VB-l-glu-NCA by their terminal amino groups, we also synthesized diblock, triblock, and star (8-arm) copolymers, respectively (Fig 1A and Supplementary Scheme S1). By calculating the integral ratio of PEG methylene protons to the benzylic ester protons in the 1H NMR spectra (Supplementary Fig. S3), the DP of diblock, triblock, and star copolymers were determined to be 98, 93 and 91 respectively, which accorded well with the theoretical values (Table 1). GPC analysis demonstrated similar MWs of the copolymers, along with narrow MWDs (Table 1 and Fig. 1B). These results collectively indicated that amino-terminated PEG also ensured controlled ROP of VB-l-glu-NCA. By calculating the ratio of PEG methylene protons to the benzylic ester protons in 1H NMR spectra, the PEG content was determined to be 10.6%, 11.1%, and 11.3% in diblock, triblock, and star copolymers, respectively. To make the graft copolymer containing PEG side chains, we co-grafted mPEG-NH2 and 1-(2-aminoethyl)piperidine (AEP) onto the PVBLG backbone via the reductive amination of the aldehyde group. 1H NMR analysis on the resultant (PVBLG-g-PEG)-r-PPABLG revealed that 14.4% PEG was incorporated onto the backbone (Table 1). In this case, we were able to develop compositionally equivalent while topologically different PEG-PPABLG conjugates, thus allowing direct comparison on the architecture-associated gene delivery performance.
Table 1.
Characterization of PEG-PPABLG with different architecturesa.
| Polymer |
Mn* × 10−4 (g/mol)b |
Mn × 10−4 (g/mol)c |
Mw/Mnc | Final Compositiond | PEG ratio (%)d |
|---|---|---|---|---|---|
| Homopolymer | 2.45 | 2.55 | 1.22 | PPABLG104 | 0 |
| Diblock Copolymer | 2.95 | 3.32 | 1.19 | PEG113-b-PPABLG98 | 10.6 |
| Triblock Copolymer | 5.91 | 6.12 | 1.18 | PPABLG93-b-PEG227-b-PPABLG93 | 11.1 |
| Star Copolymer | 23.62 | 23.04 | 1.31 | 8-arm PEG113-b-PPABLG91 | 11.3 |
| Graft Copolymer | 2.70 | 2.94 | 1.21 | (PVBLG-g-PEG12)15-r-PPABLG105 | 14.4 |
All polymerization were conducted in DMF (VB-l-glu-NCA/Initiator = 100/1 for homopolymer, diblock, triblock and star copolymers; VB-l-glu-NCA/Initiator = 110/1 for graft polymer. CM = 0.28–0.37 mol/L) at room temperature. Monomer conversions were above 99% for all copolymers except the star copolymer (above 90%).
Theoretical molecular weight of PEG-PVBLG calculated by M/I ratio.
Molecular weight and molecular weight distribution (MWD) obtained from GPC.
Final PEG-PPABLG composition and PEG weight ratio were calculated from 1H NMR and GPC results.
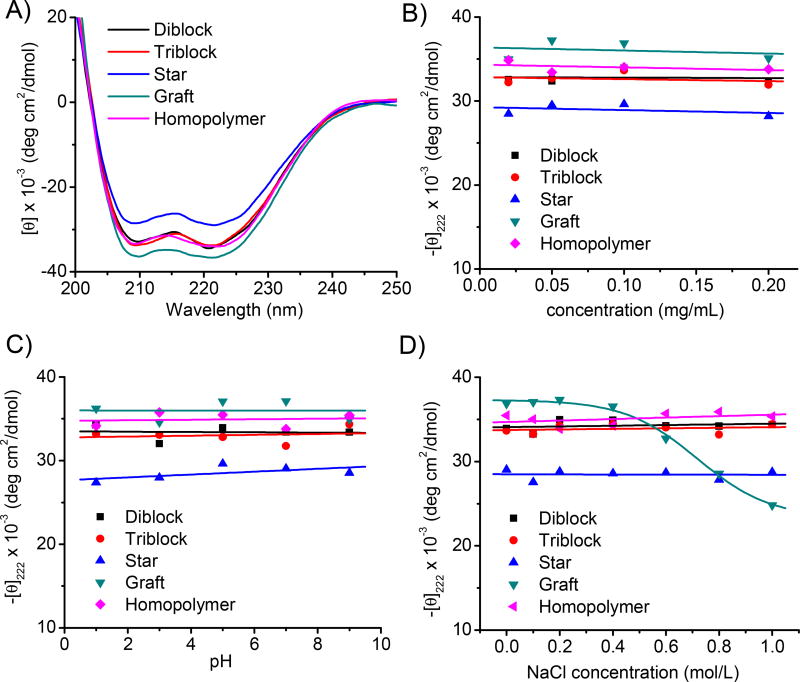
The helical structures of the polymers were evaluated by CD. Homopolymer, diblock, triblock, and graft copolymers all adopted typical helical structures as verified by the characteristic negative ellipticity of minima at 208 and 222 nm (Fig. 2A). Their molar ellipticities at 222 nm were 34.0 ×103, 33.9 ×103, 33.7 ×103, and 35.9 ×103 deg·cm2·dmol−1 for homopolymer, diblock, triblock, and graft copolymers, respectively, showing minimum 95% helicities for all polymers. It therefore indicated that introduction of PEG molecules did not influence the helical structure of PPABLG. Comparatively, star polymer exhibited slightly decreased molar ellipticity at 222 nm (28.9 ×103 deg·cm2·dmol−1 and helicity of 88%), which might be attributed to the steric effect as well as the charge repulsion among each arm that led to the interruption of the helical structure. The helicities of all test polymers remained stable at different concentrations (Fig. 2B), suggesting that they stayed as non-aggregated in the aqueous solution [11]. Their helical conformations were also very stable against pH change from 1 to 9 (Fig. 2C). Such unique property indicated that, as a gene delivery vector, the polymers were able to maintain their helicity-dependent membrane activities at both the neutral extracellular pH and the acidic endosomal/lysosomal pH, which allowed them to trigger effective intracellular internalization as well as endosomal escape [12]. The helicities were stable towards ionic strength increment except an appreciable reduction in the helicity of graft copolymer when the ionic strength reached 0.6 M (Fig. 2D). Hence, all the test polymers were expected to remain stable helical structures in the physiological fluids with ionic strength of approximately 0.15 M. The reduced helicity of graft copolymer at high NaCl concentration could be attributed to the partial shielding of the side charges by adjacent PEG segments, because the electrostatic repulsion among side charges served as driving forces for the stabilization of the α-helix [11].
Fig. 2.
CD spectra of polypeptides in aqueous solution at pH = 7 (0.1 mg/mL) (A). Helicity of polypeptides at different concentrations (B), pH (C), and NaCl concentrations (D) as demonstrated by the molar ellipticity at 222 nm.
3.2. Preparation and characterization of polyplexes
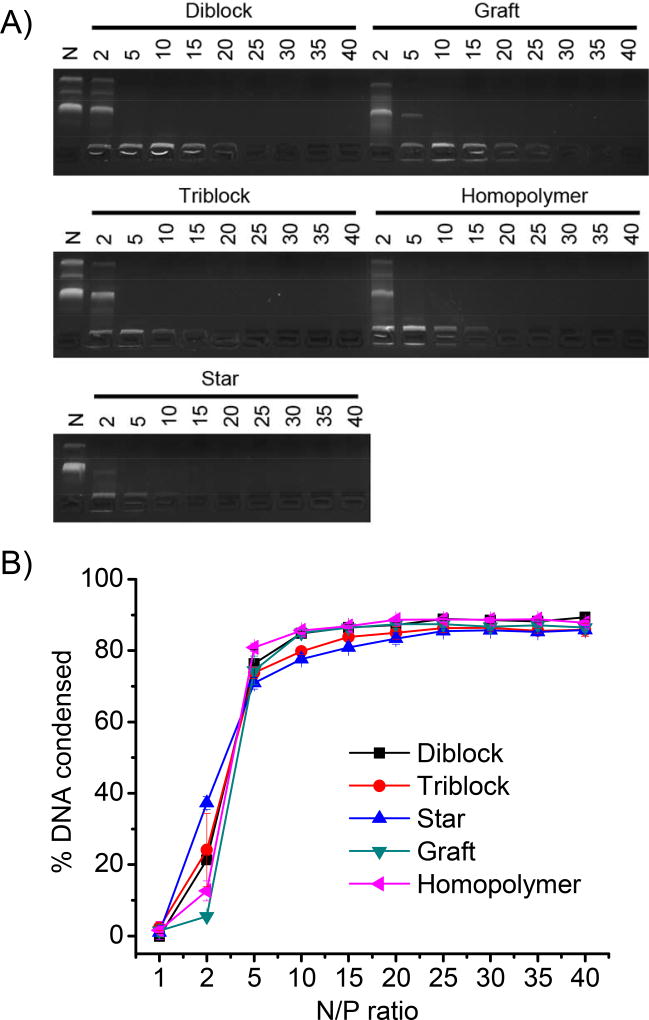
DNA condensation by cationic polymers was evaluated by the gel retardation assay. As evidenced by the restricted DNA migration in gel electrophoresis (Fig. 3A), all test polymers well condensed DNA at the N/P ratio higher than 2 except that the graft copolymer condensed DNA at the N/P ratio higher than 5. Such results were further confirmed by a quantitative EB exclusion assay (Fig. 3B), showing relatively higher DNA condensation capacity of the star copolymer while lower capacity of the graft copolymer, evidenced by the difference of their EB exclusion efficiencies at the N/P ratio of 2.
Fig. 3.
DNA condensation by polypeptides at different N/P ratios as evaluated by the gel retardation assay (A) and EB exclusion assay (B). N represents naked DNA.
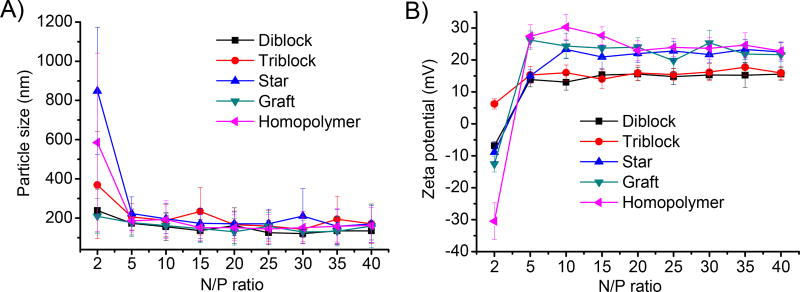
The nature of the polymer-DNA complexes was further probed by DLS. As shown in Fig. 4A, all test polymers were able to form 100~200-nm complexes with DNA at the N/P ratio higher than 5. Diblock and triblock copolymers had lower zeta potentials (~15 mV) than the graft and star copolymers as well as the homopolymer (20–30 mV, Fig. 4B), suggesting that PEG moieties in the diblock and triblock copolymers might have served as a hydrophilic corona on the complex surface to partly shield the cationic charges of the PPABLG segment.
Fig. 4.
Particle size (A) and zeta potential (B) of polyplexes at different N/P ratios as determined by DLS measurement.
3.3. In vitro transfection
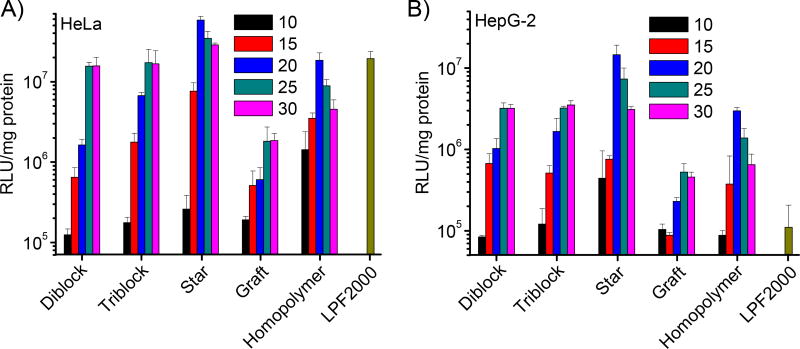
The transfection efficiencies of polymer/DNA complexes at various N/P ratios were evaluated in HeLa and HepG-2 cells by monitoring the luminescence intensity. As shown in Fig. 5, star copolymer and homopolymer reached the highest transfection efficiency at the N/P ratio of 20, while maximal transfection efficiencies were noted at N/P ratios of 25 for diblock, triblock, and graft copolymers. Star copolymer, among all test materials, demonstrated the highest transfection efficiency, outperforming LPF2000 by 3 and 134 folds in HeLa and HepG-2 cells, respectively. Such result thus suggested the superiority of the star-shaped PEG-PPABLG towards LPF2000 as the commercial transfection reagent, especially in HepG-2 cells that have been reported to be difficult-to-transfect [28]. Diblock and triblock copolymers showed comparable efficiency to the homopolymer, while graft copolymer demonstrated notably reduced transfection efficiency.
Fig. 5.
Transfection efficiencies of polyplexes in HeLa (A) and HepG-2 (B) cells at various N/P ratios. Results were indicated as mean ± SD (n=3).
3.4. Intracellular kinetics
The gene transfection efficiency of non-viral vectors is dominantly related to their intracellular kinetics, such as the internalization pathway, endosomal escape mechanism, and nuclear transport [29]. We therefore mechanistically probed the intracellular kinetics of the polymer/DNA complexes in HeLa and HepG-2 cells.
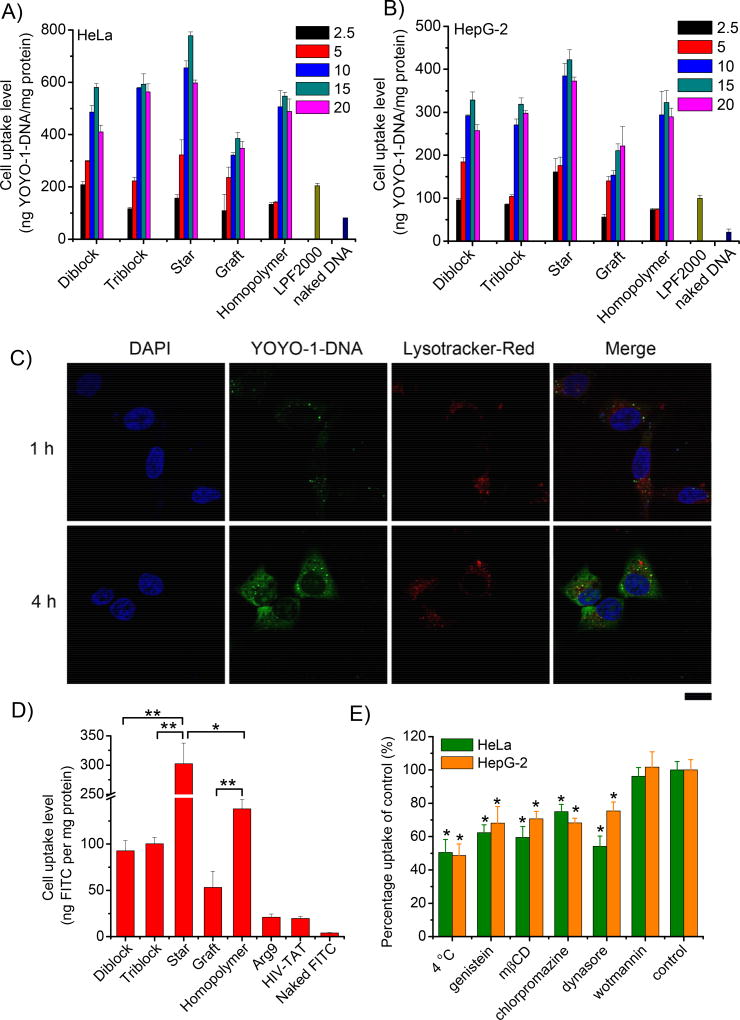
As shown in Fig. 6A and 6B, all polymers were able to promote the internalization of DNA, and the maximal uptake level was noted at the N/P ratio of 15, outperforming LPF2000 by 2–4 folds. Similar to the transfection studies, the uptake levels decreased in the following order: star copolymer > diblock copolymer ≈ triblock copolymer ≈ homopolymer > graft copolymer. CLSM was used to observe the intracellular distribution of the complexes, with star copolymer as the example. Punctated spots of DNA (green fluorescence) were noted in the cytoplasm post 1-h incubation, which correlated to the endocytosed complexes (Fig. 6C) [8]. When the incubation time was prolonged to 4 h, green fluorescence spread to the entire cytoplasm in a permeated manner, which was an indicator for the passive diffusion/permeation of the complexes [8]. We also noted nuclear distribution of DNA post 4-h incubation, suggesting that internalized DNA could be transported to the nuclei to initiate effective gene transcription. Internalized complexes partly separated from the Lysotracker® Red-stained endosomes/lysosomes, suggesting that they were able to mediate effective escape from endosomal entrapment, one of the most critical barriers to effective gene transfection.
Fig. 6.
Uptake levels of YOYO-1 labeled pDNA in HeLa (A) and HepG-2 (B) cells when complexed with polypeptides at different N/P ratios. (C) CLSM images of HeLa cells following treatment with 8-arm PEG-PPABLG/YOYO-1-DNA complexes (N/P ratio of 15) at 37 °C for 1 h and 4 h. Bar represented 20 µm. (D) Cell uptake level of FITC in HeLa cells following co-incubation with polymers for 2 h at 37 °C. (E) Mechanistic probes of the intracellular kinetics of 8-arm PEG-PPABLG/YOYO-1-DNA complexes (N/P ratio of 15) in HeLa and HepG-2 cells by monitoring the cell uptake level in the presence of various endocytic inhibitors. Results were indicated as mean ± SD (n=3).
Motivated by the above findings that a large amount of the polymer/DNA complexes were internalized via passive permeation, we then investigated the pore formation on cell membranes, an important membrane penetration mechanism induced by the helical PPABLG [12]. The cellular uptake of FITC, a hydrophilic and membrane-impermeable fluorescent molecule, was monitored following treatment with different copolymers. As shown in Fig. 6D, FITC was negligibly taken up by HeLa cells while treatment with polymers led to an increase in the FITC uptake level by 1–2 orders of magnitude, substantiating that the polypeptides were able to markedly trigger pore formation on cell membranes to allow extensive diffusion of FITC into the cytoplasm. For all the test polymers, the FITC uptake level was represented by the order of star copolymer > homopolymer > diblock copolymer ≈ triblock copolymer > graft copolymer, which correlated to their capabilities in triggering membrane pore formation. To allow direct comparison with widely used CPPs, Arg9 and HIV-TAT were selected as controls [8]. PEG-PPABLG demonstrated notably stronger membrane activities than Arg9 and HIV-TAT, making these materials better candidates for membrane permeation.
Upon identification of star copolymer as the top-performing membrane penetrating and gene delivery vector, we further probed the mechanisms underlying the cellular internalization of star copolymer/DNA complexes by performing the cell uptake study at lower temperature (4 °C) or in the presence of various endocytic inhibitors. Energy-dependent endocytosis was completely blocked at 4 °C; chlorpromazine inhibited clathrin-mediated endocytosis (CME) by triggering the dissociation of the clathrin lattice; genisteine and mβCD inhibited caveolae by inhibiting tyrosine kinase and depleting cholesterol, respectively; dynasore inhibited both CME and caveolae by inhibiting dynamin; wortmannin inhibited macropinocytosis by inhibiting phosphatidyl inositol-3-phosphate [29,30]. As shown in Fig. 6E, cell uptake was decreased by ~50% at 4 °C, suggesting that half of the complexes entered cells via energy-dependent endocytosis, while the remaining was internalized via energy-independent diffusion or translocation. Genistein, mβCD, chlorpromazine, and dynasore all significantly inhibited the cell uptake level while wortmannin exerted unappreciable inhibitory effect, indicating that the complexes were endocytosed via both the caveolae- and clathrin-mediated pathways rather than macropinocytosis.
3.5. Cytotoxicity
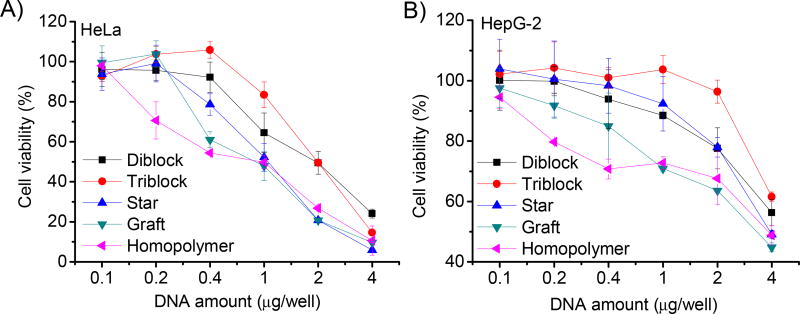
Cytotoxicity of the polymer/DNA complexes was evaluated in both HeLa and HepG-2 cells using the MTT assay. A dose- and cell line-dependent cytotoxicity was noted. In HeLa cells, the cytotoxicity decreased in the order of homopolymer > graft copolymer > star copolymer > diblock copolymer ≈ triblock copolymer (Fig. 7A). While in HepG-2 cells, the cytotoxicity decreased in the order of homopolymer > graft copolymer > star copolymer ≈ triblock copolymer > diblock copolymer (Fig. 7B). Such results suggested that the molecular structure of the PEG-PPABLG conjugates altered the polymer-cell interactions, hence leading to the differences in the cytotoxicity profiles for these compositionally equivalent while topologically different polymers. For each individual polymer, their toxicity to HeLa cells was higher than to HepG-2 cells, which was attributed to the different tolerability of each cell type towards polymer-induced cytotoxicity.
Fig. 7.
Cytotoxicity of polymer/DNA complexes (N/P ratio of 20) towards HeLa (A) and HepG-2 (B) cells as determined by the MTT assay. Results were indicated as mean ± SD (n=3).
4. Discussion
Here we elucidated the effect of molecular architecture of PEG-polypeptide conjugates on their gene delivery efficiencies, mechanistically probing into the various aspects that dominate effective gene transfection, such as DNA condensation capacities, physico-chemical properties of the polymer/DNA complexes, membrane activities, intracellular kinetics, and cytotoxicity. The controlled ROP of VB-l-glu-NCA allowed facial preparation of cationic, helical, PEG-PPABLG conjugates with various architectures (diblock, triblock, star, and graft) when amine-terminated PEG molecules were used as macroinitiators or covalently grafted onto the polymer side chain. By fixing DP of PPABLG at 100 and a comparable weight fraction of PEG chains, we were thus allowed to evaluate the architecture-dependent gene delivery efficiency and compare to the PPABLG homopolymer.
Diblock and triblock polymers exhibited comparable DNA condensation capacities to the non-PEGylated homopolymer, while the polymer/DNA complexes demonstrated lower zeta potentials. It therefore suggested that incorporation of PEG in the diblock and triblock configuration did not compromise the ability of the cationic PPABLG segment to condense and complex oppositely charged DNA. However, PEG molecules on the surface of the polymer/DNA complexes partly shielded the cationic charge by forming a hydrophilic corona, hence leading to reduced zeta potential. As such, the hydrophilic PEG layer prevented the stiff rod-like PPABLG segment from interacting with cell membranes and penetrating into the phospholipid bilayers, which resulted in reduced membrane activity (Fig. 6D) and cytotoxicity (Fig. 7). Due to a proper balance between the membrane activity and cytotoxicity, diblock and triblock polymers could still achieve comparable gene transfection efficiencies to the non-PEGylated homopolymer at a relatively higher N/P ratio (Fig. 5). These findings suggested that diblock and triblock PEG-PPABLG conjugates might be better candidates for in vivo gene delivery, in that the PEG layers could prevent opsonization during circulation and prolong the in vivo circulation time [31].
Graft copolymer, obtained by grafting PEG molecules onto the PPABLG side chains, demonstrated lower DNA binding affinity than the homopolymer, which could be attributed to the intramolecular entanglement between the PEG segment and the cationic side chains that prevented the polymer to access anionic DNA. Similarly, such entanglement prevented effective penetration of the helical PPABLG segment into cell membranes, thus diminishing the membrane activity and DNA delivery efficiency. Zeta potential analysis revealed that these PEG segments on the side chains were unable to effectively shield the cationic charge of the polymer/DNA complexes. Therefore, the graft polymers showed only slightly improved cell tolerability compared to the non-PEGylated homopolymer.
Star copolymer, obtained via ROP of VB-l-glu-NCA initiated by the 8-arm PEG-NH2, showed slightly higher DNA condensation capacities than the homopolymer, which may be attributed to its relatively higher charge density as multiple PPABLG molecules were assembled together in a small region by the PEG core. Such densely charged material appeared like a “multivalent cationic sphere”, which thus notably benefited the interactions with cell membranes to allow membrane permeation and trigger promoted intracellular delivery of DNA (Fig. 6A and 6D). Although showing higher membrane activity, the star polymer demonstrated lower cytotoxicity than the non-PEGylated homopolymer, which might be attributed to the less amount of individual PPABLG moiety in direct contact with the cell membranes. By achieving the optimal balance among these parameters, star copolymer turned out to display the highest gene transfection efficiencies and relatively low cytotoxicity in HeLa and HepG-2 cells, making it an ideal alternative as non-viral delivery vectors.
5. Conclusion
By taking advantage of a controlled ROP strategy, we synthesized PEG-PPABLG conjugates with different topologies, and probed the effect of polypeptide architecture on the gene delivery efficiencies. Diblock and triblock copolymers exhibited lower membrane activity and cytotoxicity while uncompromised gene transfection efficiencies compared to the non-PEGylated homopolymer; graft copolymer revealed lower DNA binding affinity and membrane activity, thus showing reduced gene transfection efficiency; star copolymer displayed the highest membrane activity yet relatively low cytotoxicity, hence showing potent gene transfection efficiency that outperformed the non-PEGylated homopolymer and LPF2000 by 3–5 and 3–134 folds, respectively. These understandings on the structure-property relationship of PEGylated cationic helical polypeptides provide insight into the design of synthetic gene vectors. The star copolymer that adopts a spherical architecture with high density of PPABLG has the highest membrane activity, presumably due to the multivalent polypeptide-membrane interactions. With its highest transfection efficiency and low cytotoxicity, the star copolymer may provide a promising addition to existing non-viral vectors
Supplementary Material
Acknowledgments
J.C. acknowledges support from the NSF (CHE-1153122) and the NIH (Director's New Innovator Award 1DP2OD007246 and 1R21EB013379).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Candolfi M, Xiong WD, Yagiz K, Liu CY, Muhammad AKMG, Puntel M, et al. Gene therapy-mediated delivery of targeted cytotoxins for glioma therapeutics. P Natl Acad Sci USA. 2010;107:20021–6. doi: 10.1073/pnas.1008261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leuschner F, Dutta P, Gorbatov R, Novobrantseva TI, Donahoe JS, Courties G, et al. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat Biotechnol. 2011;29:1005–10. doi: 10.1038/nbt.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paul A, Binsalamah ZM, Khan AA, Abbasia S, Elias CB, Shum-Tim D, et al. A nanobiohybrid complex of recombinant baculovirus and Tat/DNA nanoparticles for delivery of Ang-1 transgene in myocardial infarction therapy. Biomaterials. 2011;32:8304–18. doi: 10.1016/j.biomaterials.2011.07.042. [DOI] [PubMed] [Google Scholar]
- 4.Giacca M, Zacchigna S. Virus-mediated gene delivery for human gene therapy. J Control Release. 2012;161:377–88. doi: 10.1016/j.jconrel.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Cho SK, Kwon YJ. Simultaneous gene transduction and silencing using stimuli-responsive viral/nonviral chimeric nanoparticles. Biomaterials. 2012;33:3316–23. doi: 10.1016/j.biomaterials.2012.01.027. [DOI] [PubMed] [Google Scholar]
- 6.Johnson RN, Chu DSH, Shi J, Schellinger JG, Carlson PM, Pun SH. HPMA-oligolysine copolymers for gene delivery: optimization of peptide length and polymer molecular weight. J Control Release. 2011;155:303–11. doi: 10.1016/j.jconrel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saw PE, Ko YT, Jon S. Efficient liposomal nanocarrier-mediated oligodeoxynucleotide delivery involving dual use of a cell-penetrating peptide as a packaging and intracellular delivery agent. Macromol Rapid Comm. 2010;31:1155–62. doi: 10.1002/marc.200900861. [DOI] [PubMed] [Google Scholar]
- 8.Ter-Avetisyan G, Tuennemann G, Nowak D, Nitschke M, Herrmann A, Drab M, et al. Cell entry of arginine-rich peptides is independent of endocytosis. J Biol Chem. 2009;284:3370–8. doi: 10.1074/jbc.M805550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon EJ, Liong S, Pun SH. A Truncated HGP peptide sequence that retains endosomolytic activity and improves gene delivery efficiencies. Mol Pharmaceut. 2010;7:1260–5. doi: 10.1021/mp1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakase I, Akita H, Kogure K, Graslund A, Langel U, Harashima H, et al. Efficient intracellular delivery of nucleic acid pharmaceuticals using cell-penetrating peptides. Accounts Chem Res. 2012;45:1132–9. doi: 10.1021/ar200256e. [DOI] [PubMed] [Google Scholar]
- 11.Lu H, Wang J, Bai YG, Lang JW, Liu SY, Lin Y, et al. Ionic polypeptides with unusual helical stability. Nat Commun. 2011;2:206. doi: 10.1038/ncomms1209. [DOI] [PubMed] [Google Scholar]
- 12.Gabrielson NP, Lu H, Yin LC, Li D, Wang F, Cheng JJ. Reactive and bioactive cationic α-helical polypeptide template for nonviral gene delivery. Angew Chem Int Ed. 2012;51:1143–7. doi: 10.1002/anie.201104262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabrielson NP, Lu H, Yin LC, Kim KH, Cheng JJ. A cell-penetrating helical polymer for siRNA delivery to mammalian cells. Mol Ther. 2012;20:1599–609. doi: 10.1038/mt.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshpande MC, Davies MC, Garnett MC, Williams PM, Armitage D, Bailey L, et al. The effect of poly(ethylene glycol) molecular architecture on cellular interaction and uptake of DNA complexes. J Control Release. 2004;97:143–56. doi: 10.1016/j.jconrel.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Parelkar SS, Chan-Seng D, Emrick T. Reconfiguring polylysine architectures for controlling polyplex binding and non-viral transfection. Biomaterials. 2011;32:2432–44. doi: 10.1016/j.biomaterials.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Venkataraman S, Ong WL, Ong ZY, Loo SCJ, Ee PLR, Yang YY. The role of PEG architecture and molecular weight in the gene transfection performance of PEGylated poly(dimethylaminoethyl methacrylate) based cationic polymers. Biomaterials. 2011;32:2369–78. doi: 10.1016/j.biomaterials.2010.11.070. [DOI] [PubMed] [Google Scholar]
- 17.Synatschke CV, Schallon A, Jerome V, Freitag R, Muller AHE. Influence of polymer architecture and molecular weight of poly(2-(dimethylamino)ethyl methacrylate) polycations on transfection efficiency and cell viability in gene delivery. Biomacromolecules. 2011;12:4247–55. doi: 10.1021/bm201111d. [DOI] [PubMed] [Google Scholar]
- 18.Hinton TM, Guerrero-Sanchez C, Graham JE, Le T, Muir BW, Shi SN, et al. The effect of RAFT-derived cationic block copolymer structure on gene silencing efficiency. Biomaterials. 2012;33:7631–42. doi: 10.1016/j.biomaterials.2012.06.090. [DOI] [PubMed] [Google Scholar]
- 19.Ahmed M, Narain R. The effect of polymer architecture, composition, and molecular weight on the properties of glycopolymer-based non-viral gene delivery systems. Biomaterials. 2011;32:5279–90. doi: 10.1016/j.biomaterials.2011.03.082. [DOI] [PubMed] [Google Scholar]
- 20.Eltoukhy AA, Siegwart DJ, Alabi CA, Rajan JS, Langer R, Anderson DG. Effect of molecular weight of amine end-modified poly(β-amino ester)s on gene delivery efficiency and toxicity. Biomaterials. 2012;33:3594–603. doi: 10.1016/j.biomaterials.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu YM, Reineke TM. Poly(glycoamidoamine)s for gene delivery. Structural effects on cellular internalization, buffering capacity, and gene expression. Bioconjugate Chem. 2007;18:19–30. doi: 10.1021/bc060029d. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed M, Jawanda M, Ishihara K, Narain R. Impact of the nature, size and chain topologies of carbohydrate-phosphorylcholine polymeric gene delivery systems. Biomaterials. 2012;33:7858–70. doi: 10.1016/j.biomaterials.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Lu H, Bai YG, Wang J, Gabrielson NP, Wang F, Lin Y, et al. Ring-opening polymerization of γ-(4-vinylbenzyl)-l-glutamate N-carboxyanhydride for the synthesis of functional polypeptides. Macromolecules. 2011;44:6237–40. doi: 10.1021/ma201164n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, Rupp B, et al. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry. 2000;39:11657–66. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X, Yin L, Ding J, Tang C, Gu S, Yin C, et al. Thiolated trimethyl chitosan nanocomplexes as gene carriers with high in vitro and in vivo transfection efficiency. J Control Release. 2010;144:46–54. doi: 10.1016/j.jconrel.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 26.Gabrielson NP, Cheng JJ. Multiplexed supramolecular self-assembly for non-viral gene delivery. Biomaterials. 2010;31:9117–27. doi: 10.1016/j.biomaterials.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNaughton BR, Cronican JJ, Thompson DB, Liu DR. Mammalian cell penetration, siRNA transfection, and DNA transfection by supercharged proteins. P Natl Acad Sci USA. 2009;106:6111–6. doi: 10.1073/pnas.0807883106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamano S, Dai JS, Moursi AM. Comparison of transfection efficiency of nonviral gene transfer reagents. Mol Biotechnol. 2010;46:287–300. doi: 10.1007/s12033-010-9302-5. [DOI] [PubMed] [Google Scholar]
- 29.Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 30.Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, et al. The effect of particle design on cellular internalization pathways. P Natl Acad Sci USA. 2008;105:11613–8. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owens DE, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharmaceut. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.