Abstract
Peptidic natural products (PNPs) include many antibiotics and other bioactive compounds. While the recent launch of the Global Natural Product Social (GNPS) molecular networking infrastructure is transforming PNP discovery into a high-throughput technology, PNP identification algorithms are needed to realize the potential of the GNPS project. GNPS relies on the assumption that each connected component of a molecular network (representing related metabolites) illuminates the “dark matter of metabolomics” as long as it contains a known metabolite present in a database. We reveal a surprising diversity of PNPs produced by related bacteria and show that, contrary to the “comparative metabolomics” assumption, two related bacteria are unlikely to produce identical PNPs (even though they are likely to produce similar PNPs). Since this observation undermines the utility of GNPS, we developed a PNP identification tool VarQuest that illuminates the connected components in a molecular network even if they do not contain known PNPs and only contain their variants. VarQuest revealed an order of magnitude more PNP variants than all previous PNP discovery efforts and demonstrated that GNPS already contains spectra from 41% of currently known PNP families. The enormous diversity of PNPs suggests that biosynthetic gene clusters in various microorganisms constantly evolve to generate a unique spectrum of PNP variants that differ from PNPs in other species.
After a decline in the pace of antibiotics discovery in the 1990s, antibiotics and other natural products are again in the center of attention as exemplified by the recent discovery of teixobactin1. Previous studies of natural products mainly relied on low-throughput NMR-based technologies requiring large amounts of highly purified material that are often difficult to obtain. The key condition for enabling the renaissance of the antibiotics research is development of high-throughput computational discovery pipelines such as the recently launched Global Natural Products Social (GNPS) molecular network2 that already contains over a billion of tandem mass spectra, a gold mine for bioactive compounds discovery. However, natural products identification algorithms are needed to transform antibiotics discovery into a high-throughput technology and to realize the promise of the GNPS project.
This study focuses on Peptidic Natural Products (PNPs), an important class of natural products with an unparalleled track record in pharmacology: many antibiotics, antiviral and antitumor agents, immunosuppressors are PNPs. PNPs are produced by Non-Ribosomal Peptide Synthetases (NRPS)3 and Ribosomally synthesized and Posttranslationally modified Peptides Synthases (RiPPS)4. NRPS and RiPPS synthesize Non-Ribosomal Peptides (NRPs) and Ribosomally synthesized and Posttranslationally modified Peptides (RiPPs), respectively. NRPs are not directly inscribed in genomes but instead are encoded by NRPSs using non-ribosomal code, with each A-domain in NRPS responsible for a single amino acid in NRP5, 6. While RiPPs are encoded in the genome, the RiPP-encoding genes are often short, making it difficult to annotate them7.
PNP identification
Given a spectrum and a peptide database, peptide identification refers to finding a peptide in the database that generated the given spectrum. Identification of spectra derived from PNPs8–11 is more difficult than traditional peptide identification in proteomics because many PNPs are non-linear peptides (e.g., cyclic or branch-cyclic) that contain non-standard amino acids and complex modifications.
Identification of non-linear peptides is only one of the two major challenges in PNP identification. In many cases, a PNP is absent in the database of known PNPs, but its modified or mutated variant is present in this database. Identification of an unknown PNP from its known variants is called variable identification (as opposed to standard identification when a PNP is present in the database). Similarly to the problem of variable identification of modified peptides in traditional proteomics12–15, variable PNP identification is difficult because the computational space of this problem is several orders of magnitude larger than for standard PNP identification.
Since most PNPs form families of related peptides, variable identification is crucial for PNP discovery. Finding variants of known PNPs is important since these variants are sometimes more effective than the most abundant representatives of PNP families that currently dominate the PNP databases. The antifungal drug Cancidas16 or modified variants of vancomycin17 are some examples of variant PNPs that proved to be effective in clinical applications.
Spectral networks
Given a set of PNPs P1, . . . , Pm, their peptide network is a graph with nodes P1, . . . , Pm, and edges connecting two PNPs if they differ by a single modification or mutation (substitution, insertion or deletion)18. Each component in the peptide network defines a PNP family. In reality, we are not given PNPs P1, . . . , Pm but only their spectra S1, . . . , Sm. Nevertheless, one can approximate the peptide network by constructing the spectral network on nodes S1, . . . , Sm, where spectra Si and Sj are connected by an edge if they are similar, e.g., can be aligned against each other using the spectral alignment algorithm13.
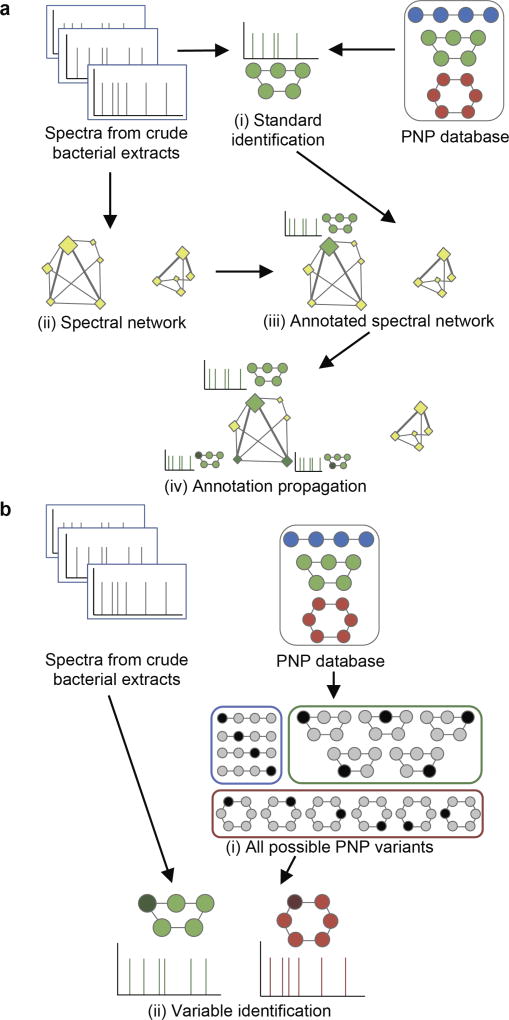
Although spectral networks19 reveal spectra of related peptides without knowing what these peptides are, they have an important limitation—they work only when one of the spectra (nodes) in the connected component of the network corresponds to an unmodified peptide from a database (referred to as an unmodified parent). As the result, orphan components in the spectral network (components without annotated nodes) represent the “dark matter of PNPs” since the spectral network propagation approach18, 19 lacks ability to interpret them in the absence of an unmodified parent (Figure 1a).
Figure 1. Network-based and network-independent strategies for variable PNP identification.
Variant PNPs are colored by the same color as their known compounds in the database; modified/mutated amino acids are highlighted by darker color. (a) Network-based PNP identification starts from the standard identification of spectra (i) and construction of a spectral network (ii). Next, the network is annotated (iii) using the identified PNPs via the spectral network propagation approach. In this example, the network component on the left has a single unmodified parent colored green as the related PNP, while the component on the right is an orphan. Annotation propagation (iv) through the network results in two variable PNP identifications represented by additional green nodes. (b) Network-independent PNP identification relies on an efficient enumeration of all PNP variants (i) and further matching of spectra against these variants using the standard identification strategy (ii).
PNP identification strategies
The DEREPLICATOR algorithm11 identified many PNPs in the GNPS dataset through standard identification (without modifications) and variable identification via spectral networks (Figure 1a). However, the limitation of the spectral network approach prevents DEREPLICATOR from finding many PNP variants. Indeed, variable identification via spectral networks works only when there exists an unmodified parent in a given connected component. Since PNPs vary across diverse related bacteria, this condition does not hold for many GNPS datasets because a PNP identified in one bacterium (and present in a database) is often represented by its modified variant in another bacterium. This limitation raises the challenge of developing methods for variable PNP identification that, in difference from DEREPLICATOR, do not rely on spectral networks.
Modification-tolerant search reveals diverse PNP variants
Since PNP databases are dominated by the most abundant representatives of PNP families, existing algorithms, focusing on identification of known PNPs, annotate only a small fraction of GNPS spectra. To address this limitation, we developed a network-independent VarQuest algorithm for modification-tolerant PNP identification (Figure 1b).
VarQuest revealed that a vast majority of PNP families (78%) identified in GNPS were not represented by any non-modified known PNP and thus are not detectable using the spectral network approach. This observation suggests that not only PNPs are extremely diverse across evolutionary distant microbial species but that also PNP families rapidly evolve so that PNP variants present in one species are often mutated/modified even in closely related species. This evolution of PNP families may reflect adaptation to unique ecological niches under various pressures, not unlike evolution of skyllamycins in Pseudomonas aeruginosa20.
The great diversity of PNP variants underscores the importance of variable PNP identification via VarQuest and reveals a limitation of the spectral network approach implemented in DEREPLICATOR (most components in the GNPS spectral network turned out to be orphans). VarQuest has now fixed this unanticipated limitation. We benchmarked VarQuest and identified an order of magnitude more PNP variants as compared to existing PNP identification strategies.
Results
Brute-force approach
A novel PNP is called a variant of a known PNP if it has the same topology and sequence of amino acid, except for a single modified/mutated amino acid. We focus on identification of PNP variants with mass offset up to MaxMod (the default value MaxMod=300 Da).
The brute-force approach to variable identification (BruteForce) is based on enumeration of all possible modifications/ mutations for each peptide from the database21. Given a spectrum S and each PNP P from the database (with mass difference δ < MaxMod), it considers a modification of mass δ on all possible amino acids in P, forms a list CandidatePeptides(S) containing all such modified PNPs, and finds a PNP in CandidatePeptides(S) with the best match to S. Since the resulting list CandidatePeptides(S) contains a large portion of the entire PNP database, this approach is prohibitively time-consuming. Various database filtering strategies and spectral alignment algorithms were developed to speed-up the brute-force approach in traditional proteomics12–14, 22, 23. However, extending variable identification algorithms from linear peptides to non-linear PNPs remains a challenge.
Spectral network approach
The spectral network approach (SpecNets) constructs the spectral network of all spectra and identifies the connected component of the spectral network that contains a given spectrum S (denoted Component(S)). It further forms the set CandidatePeptides(S) as the set of identifications of all spectra in Component(S) that were discovered using the standard identification method. Afterwards, it applies the spectral network propagation approach to CandidatePeptides(S) to perform variable identification of S. Although this approach is fast (since the CandidatePeptides(S) is typically small), it fails when Component(S) is an orphan, i.e., does not contain any spectra originating from known PNPs.
VarQuest algorithm
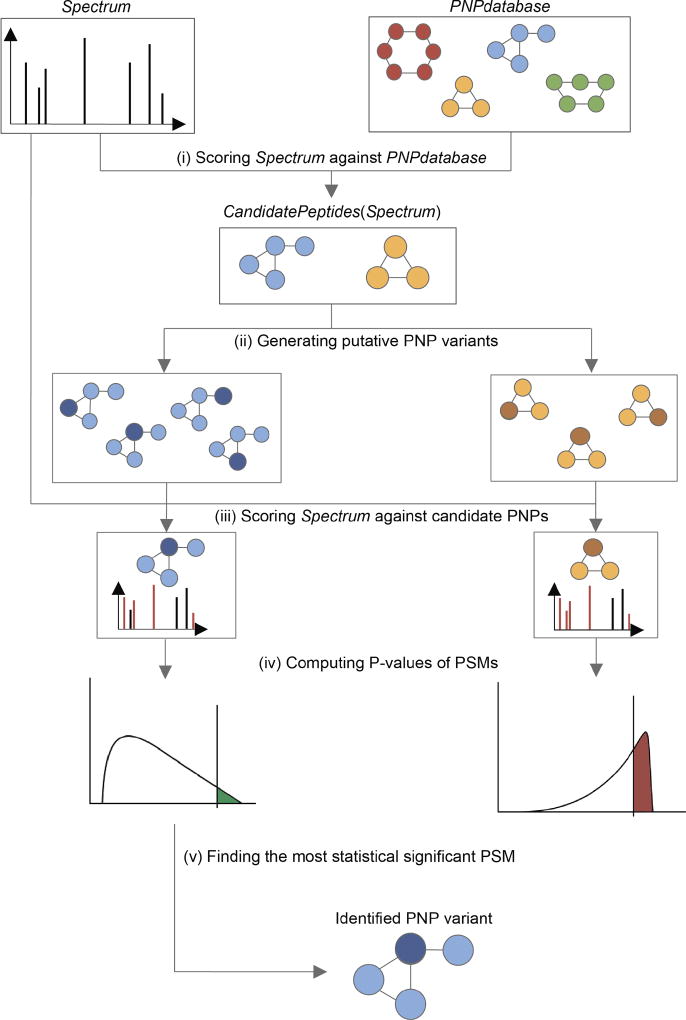
VarQuest pipeline for a single spectrum S (Figure 2) starts with selection of a short list CandidatePeptides(S) from the PNP database. Afterwards, VarQuest scores S against each PNP (with a single modification) in CandidatePeptides(S) and computes P-values of resulting PNP-spectrum matches (PSMs)24. A peptide with the lowest P-value among all PNPs in CandidatePeptides(S) is reported as a candidate PNP that gave rise to the spectrum S.
Figure 2. VarQuest pipeline.
For a spectrum and a PNP database, VarQuest starts from scoring the spectrum against the entire database (i) to form the list of candidate PNPs. All possible modifications are considered for each candidate (ii) and the spectrum is scored against all variants (iii) to select the highest scoring variant per candidate PNP. Statistical significance of the scores is computed (iv) and the most statistically significant PSM is reported (v).
Efficient selection of the small list CandidatePeptides(S) is the key step of VarQuest. The standard identification approaches include a peptide P into CandidatePeptides(S) as long as Mass(S) ≈ Mass(P) with error up to Δ. Since Δ is small for high-resolution spectra, the list CandidatePeptides(S) is much smaller than the number of PNPs in the database, enabling fast standard identification but preventing detection of novel PNP variants. Var-Quest detects novel PNP variants by constructiong a short list CandidatePeptides(S) (in difference from a long list constructed by BruteForce) as described in the Methods section.
Although VarQuest searches for unknown PNPs with a single modification (as compared to a known PNP), it has ability to find PNPs with multiple modifications. However, in such cases, instead of reporting multiple modifications, it reports a single modification with combined mass equal to the total mass of multiple modifications. Below we illustrate how further analysis allows one to infer the positions and masses of multiple modifications.
Benchmarking VarQuest
We benchmarked VarQuest on five high-resolution GNPS datasets: SpectraPSEUD (≈400,000 spectra from 260 Pseudomonas isolates25), SpectraSTREP1 (≈200,000 spectra from Streptomyces7), SpectraSTREP2 (≈500,000 spectra from Streptomyces11, 26), SpectraCYANO (≈11 million spectra from Cyanobacteria27), and SpectraGNPS (≈130 million spectra from GNPS11). Details on these datasets are provided in Supplementary Table 1.
We matched all spectral datasets against the PNPdatabase constructed by combining all PNPs from Anti-Marin28, DNP29, MIBiG30, and StreptomeDB31 (5021 PNPs forming 1582 PNP families). The results were compared with BruteForce, SpecNets, and the standard identification (Standard) algorithms (Table 1). Time and memory requirements of these methods are described in Supplementary Table 2. Although BruteForce can find all VarQuest identifications (for a given P-value threshold) on the same set of spectra, it becomes prohibitively time-consuming even on moderately-sized spectral datasets such as SpectraCYANO. Note that all methods are based on DEREPLICATOR11 and that BruteForce and SpecNets failed to process the SpectraCYANO and SpectraGNPS datasets due to large memory requirements.
Table 1.
Comparison of four PNP identification approaches on various spectral datasets against the PNPdatabase with 5021 PNPs (1582 PNP families). Spectral networks were constructed using the GNPS interface (the Molecular Networking workflow2) with precursor and fragment ion tolerance set to 0.02 Da and all other parameters set to the default values. # PSMs stands for the number of identified PSMs, # variants stands for the number of identified PNP variants, and # PNPs (families) stands for the number of unique PNP identifications (PNP families). P−10, FDR0 and FDR5 stand for the number of identified PSMs, variants, unique PNPs or PNP families with P-value below 10−10, at 0%, and 5% False Discovery Rates, respectively. When processing SpectraCYANO, the SpecNets tool crashed after a week of execution, and the BruteForce approach crashed due to exceeding memory limit (25 GB RAM).
| Method | # PSMs | # variants | # PNPs (families) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| P−10 | FDR0 | FDR5 | P−10 | FDR0 | FDR5 | P−10 | FDR0 | FDR5 | |
| SpectraPSEUD | |||||||||
| Standard | 1029 | 997 | 1029 | 42 | 36 | 42 | 42 (13) | 36 (12) | 42 (13) |
| SpecNets | 1837 | 1837 | 1837 | 164 | 164 | 164 | 41 (15) | 41 (15) | 41 (15) |
| BruteForce | 7545 | 261 | 3822 | 1042 | 60 | 479 | 436 (197) | 33 (13) | 242 (117) |
| VarQuest | 6055 | 224 | 6055 | 871 | 54 | 871 | 341 (148) | 30 (13) | 341 (148) |
|
| |||||||||
| SpectraSTREP1 | |||||||||
| Standard | 228 | 171 | 228 | 20 | 11 | 20 | 20 (10) | 11 (5) | 20 (10) |
| SpecNets | 337 | 265 | 337 | 57 | 37 | 57 | 20 (10) | 9 (3) | 20 (10) |
| BruteForce | 2648 | 57 | 841 | 1698 | 26 | 360 | 621 (293) | 6 (1) | 133 (69) |
| VarQuest | 2266 | 128 | 716 | 1397 | 51 | 287 | 496 (225) | 8 (3) | 95 (49) |
|
| |||||||||
| SpectraSTREP2 | |||||||||
| Standard | 85 | 85 | 85 | 13 | 13 | 13 | 13 (5) | 13 (5) | 13 (5) |
| SpecNets | 101 | 22 | 101 | 23 | 5 | 23 | 13 (5) | 4 (2) | 13 (5) |
| BruteForce | 258 | 25 | 31 | 134 | 8 | 11 | 95 (60) | 5 (2) | 7 (3) |
| VarQuest | 208 | 12 | 147 | 100 | 3 | 56 | 67 (45) | 1 (1) | 37 (25) |
|
| |||||||||
| SpectraCYANO | |||||||||
| Standard | 353 | 146 | 353 | 42 | 23 | 42 | 42 (20) | 23 (14) | 42 (20) |
| VarQuest | 3573 | 226 | 3573 | 2083 | 68 | 2083 | 702 (315) | 25 (14) | 702 (315) |
|
| |||||||||
| SpectraGNPS | |||||||||
| Standard | 14757 | 7464 | 14746 | 420 | 279 | 420 | 420 (143) | 279 (110) | 420 (143) |
| VarQuest | 379089 | 5661 | 179448 | 65204 | 1695 | 19619 | 2673 (835) | 675 (256) | 2025 (648) |
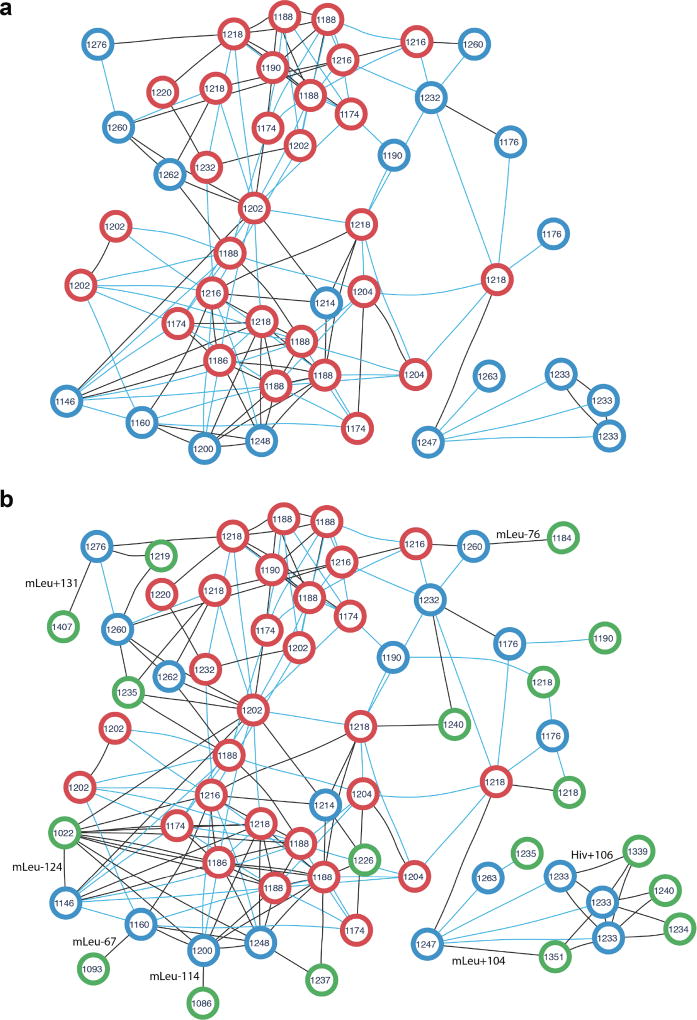
We compared the number of identified PSMs and unique peptides for all methods at 0%and 5%False Discovery Rate (FDR) levels. To compute the FDR, VarQuest uses the concept of a decoy database extended to nonlinear peptides (see the Methods section). All PSMs with P-values above 10−10 were removed beforehand and the FDR was conservatively computed for the remaining PSMs. Table 1 shows more than ten times increase in the number of PSMs and five times increase in the number of PNPs and PNP families identified in GNPS via variable identification with VarQuest compared to the standard DEREPLICATOR at 5% FDR. Figure 3 shows the peptide network of the largest PNP family (cyclosporins) in the PNPdatabase identified by VarQuest (Supplementary Table 3).
Figure 3. Peptide network of the cyclosporin family in the PNPdatabase (a) extended by the newly identified cyclosporin variants (b).
The peptide network was constructed for 47 known cyclosporins using the GNPS interface (the Molecular Networking workflow2). Each node represents theoretical spectrum of a cyclosporin variant, the number inside each node stands for monoisotopic mass in Da rounded to integers. Two nodes are connected by an edge if the corresponding theoretical spectra are similar (cosine score at least 0.8). Blue edges corresponds to characteristic mass shifts of 14, 16, 28, 32, and 42 Da, the remaining edges are black. (a) Peptide network of cyclosporin variants present in the PNPdatabase. Red nodes are 29 cyclosporins identified in SpectraGNPS as known PNPs (both by DEREPLICATOR and VarQuest). Blue nodes are theoretical spectra of the rest 18 PNPs which are not present in GNPS in their known form and added to the network for the sake of completeness. (b) Peptide network of known and novel cyclosporin variants. Green nodes are theoretical spectra of 18 novel variants identified by VarQuest in GNPS. Each novel variant is the most statistically significant identification of the corresponding blue node (an absent known cyclosporin PNP). Likely insertions/deletions are shown on corresponding edges, Hiv stands for Hydroxyisovaleric acid, mLeu stands for methylated leucine.
While DEREPLICATOR revealed spectra corresponding to only 8% of peptides in the PNPdatabase, VarQuest increased this number to 40%. 1605 out of 2025 PNPs identified by VarQuest in SpectraGNPS (at 5% FDR) have their unknown variants present in SpectraGNPS, while their known variants are absent and thus can not be detected by the standard identification strategies.
Our analysis of the entire GNPS dataset at 5% FDR identified 648 PNP families (41% of all known PNP families). At the same time, only a small fraction of identified PNP families (143 out of 648) were identified as an unmodified parent, i.e., a vast majority of identified PNP families (78%) were not represented by any non-modified peptides in the PNPdatabase and thus are not detectable using the spectral network approach.
Modifications/mutations identified by VarQuest
Supplementary Table 4 shows the most common mass offsets identified by VarQuest in the SpectraGNPS dataset at 5% FDR. For each mass offset we identified its most likely position in the PNP. As expected, the most common offsets are −14 Da (demethylation), +14 Da (methylation), +28 Da (dimethylation), +18 Da (hydration), and +16 Da (hydroxylation) corresponding to either modifications/adducts or mutations. In addition, Supplementary Table 4 reveals many surprising offsets such as −95 Da offset (primarily at Leucine/Isoleucine), −81 Da offset (primarily at Valine), and others. These offsets may correspond to the combination of the amino acid loss (−113 Da for Leucine/Isoleucine and −99 Da for Valine) and hydration (+18 Da). The abundance of such offsets suggests that the recently described phenomenon of amino acid deletions/insertions in NRPs25 (due to A-domain stuttering and skipping in NRPSs32) may be more prevalent than previously thought. Genome mining efforts typically rule out such events due to the consecutive arrangements of A-domains in NRP synthetases.
To conservatively estimate the number of indels revealed by spectra in SpectraGNPS, we considered identified mass offsets that matched monoisotopic masses of proteinogenic amino acids within a 0.02 Da error. This analysis revealed 217 (169) putative insertions (deletions) out of 19619 PNP variants identified by VarQuest in SpectraGNPS at 5% FDR. Our confidence in the deletions is higher than in the insertions since for each of them we also checked that the deleted amino acid is present in the known PNP structure (which reduced the initial number of potential deletions by 30% from 242 to 169).
Analysis of PNP diversity
VarQuest identified 19619 PNP variants related to 2025 distinct PNPs in SpectraGNPS at 5% FDR. More than 70% of the identified PNPs (1489) were found in at least two different forms. Each identified PNP was found in 9.7 various PNP forms on average with the maximum value equal to 239 for tolybyssidin A. Our analysis adds a “chemical” dimension to the recently revealed PNP diversity at the biosynthetic gene cluster (BGC) level33, 34. We further revealed that related bacteria are likely to produce similar PNP variants rather than identical PNPs (see the Methods section).
Validation of VarQuest identifications
Our analysis revealed that about 60% of the PNP variants identified by VarQuest in Pseudomonas and Streptomyces datasets are missed by DEREPLICATOR and SpecNets. We validated the most statistically significant VarQuest hits using literature search for identified PNP origin which should correlate with the sample origin (see the Methods section) and searched for BGCs by genome mining35 whenever the genome of the analyzed species is available. We further analyzed three identified PNP variants (referred to as Massetolide-1252, Venepeptide-2154, and Surugamide-769) in more details (Supplementary Figure 1).
Massetolide-1252
Massetolide A is a known NRP from Pseudomonas36 that consists of a cycle TISLSLI and a branch EL (along with 3-hydroxydecanoic acid lipid tail of mass 171 Da) attached to the cycle via a bond connecting T in the cycle with E in the branch. We represent branch-cyclic peptides as a concatenate of its cyclic sequence and its branch sequence, both starting from their attachment points, e.g., massetolide A is represented as TISLSLI*EL. VarQuest identified massetolide A and its novel variant Massetolide-1252 (sequence TISL+113SLI*EL and mass 1252.8 Da) with P-value 4.2 · 10−19 using a spectrum from P. synxantha. The +113 Da offset corresponds to insertion of Leucine or Isoleucine residue and matches the recently identified poaeamide B with sequence TISLLSLI*EL and mass 1252.8 Da. Note that a single run of VarQuest instantly achieved the same goal as the time-consuming semi-manual discovery of poaeamide B25.
VarQuest also rediscovered bananamides, a family of PNPs discovered in the same study25. Bananamide (referred to as Bananamide-1093) was identified with P-value 4.3 · 10−10 as a variant of massetolide A (sequence TIS−46LSLI*EL and mass 1093.7 Da) using a spectrum from P. fluorescens. While the recent study25 did not derive the amino acid sequence from this spectrum, it purified and sequenced a related PNP (named bananamide 2) with sequence TLLQLI*DL (along with C12 3-hydroxy unsaturated acid lipid tail of mass 197 Da) and mass 1105.7 Da (amino acids differing from massetolide A are highlighted except for a change between amino acids I and L with identical masses). While amino acid sequences TIS−46LSLI*EL and TLLQLI*DL appear to be rather different, note that S−46LS has the same mass as LQ suggesting that TIS−46LSLI*EL may actually correspond to TILQLI*EL with a single deleted amino acid as compared to massetolide A. Note that there is only one difference with respect to masses of amino acids between this sequence (TILQLI*EL) and the sequence of bananamide 2 (TLLQLI*DL).
Our analysis of Bananamide-1093 suggests that bananamides emerged from the massetolides family after deletion of a single amino acid (or alternatively, massetolides emerged from bananamides after insertion of a single amino acid). Interestingly, while the PSM for Bananamide-1093 is statistically significant, PSMs for bananamides 1, 2, and 3 identified in25 have rather high P-values that did not pass the VarQuest P-value threshold. Remarkably, the manual analysis in25 missed the most statistically significant PSM for bananamides identified by VarQuest, illustrating the power of automated approaches to PNP identification. Moreover, after identifying Bananamide-1093, VarQuest identifies spectra of bananamides 1, 2, and 3 against Bananamide-1093 as statistically significant PSMs with P-values 1.1 · 10−13, 9.8 · 10−12, and 1.1 · 10−16, respectively.
Surugamide-769
Surugamides are cyclic NRPs from marine streptomyces11, 37. VarQuest identified both known PNP surugamide B with sequence IAIVKIFL and its novel variant IAIVK−128IFL using a spectrum from S. albus (P-value 1.7 · 10−19). The SpecNets approach missed this compound because its connected component does not contain known surugamides.
The amino acid sequence IAIVK−128IFL of Surugamide-769 corresponds to a loss of Lysine. This annotation is consistent with the arrangement of the genes in the surugamide BGC since the deleted Lysine corresponds to the last A-domain in one of two genes in this BGC (Supplementary Figure 2). Thus, Surugamide-769 represents the second evidence of the same NRP synthetase producing two cyclic peptides with different numbers of amino acids, similar to poaeamide B and massetolide A25. However, further experimental validation of this hypothesis and many other likely insertions and deletions listed in Supplementary Table 4 is needed.
Venepeptide-2154
Venepeptide is a linear ribosomal peptide M+28NVITNLLAGVVHFLGWLV that was identified from S. venezuelae38. VarQuest identified its variantM+28NVITN+31LLAGVVHFLGWLV (mass 2154.1 Da) with P-value 3.2 · 10−15 using a spectrum from S. lividans. DEREPLICATOR missed this compound because GNPS does not contain a spectrum corresponding to the known venepeptide. Sequence similarity search39 of this peptide against the genome of S. lividans revealed the sequence MNLLTDILAGLVHFVGWLV (the differences with venepeptide are highlighted). A match of the spectrum against this sequence resulted in a PSM with P-value 8.5 · 10−24 and suggested modification +44 Da on the M residue. Note that while VarQuest is limited to finding variants with a single modified amino acid, it was able to identify that a spectrum from S. lividans has arisen from a variant of venepeptide. The further manual analysis revealed that Venepeptide-2154 structure differs from venepeptide in four amino acids.
Discussion
Although the launch of high-throughput natural products discovery pipelines, such as the GNPS molecular network, is an important step towards future discoveries, the lack of computational approaches is still a bottleneck for spectral identifications in the GNPS infrastructure. Currently, the GNPS spectral library, a collection of identified spectra from GNPS, represents a minuscule fraction of all GNPS spectra. While molecular networks2, 19 have already resulted in discoveries of various PNPs and their variants25, 40, these discoveries still requires time-consuming manual follow-up analysis. Here we demonstrated how the same goal can be achieved in a single push-of-a-button VarQuest run, replicating recent PNP discoveries and finding previously unknown PNP variants. Moreover, variable dereplication of the entire GNPS revealed both surprising diversity of PNPs and limitations of the spectral networks approach. In particular, we demonstrated that the recently discovered phenomenon of insertions and deletions of amino acids is widespread among NRPs.
There is a yet another reason why variable identification is important. Recent Genomic Encyclopedia of Bacteria and Archaea study41 revealed many BGCs with PNPs representing the largest group of secondary metabolites encoded by these BGCs. However, the vast majority of the predicted BGC products remained unknown, reflecting the limited information available for characterized natural products and the lack of genome mining and peptidogenomics tools for matching BGCs and spectra.
While databases in traditional proteomics consist of known peptides, the ongoing genome mining efforts for PNP discovery35 generate vast databases of still uncharacterized putative PNPs7, 42, 43. Since predicting an NRP encoded by an NRPS is a difficult problem, various tools for predicting specificities of A-domains44 output multiple rather than a single candidate amino acid for each A-domain. Supplementary Figure 2 presents three top candidate amino acids for each of eight A-domains in suragamide-encoding NRPS resulting in 38 candidate NRPs. As the result, genome mining efforts typically generate large databases of error-prone putative PNPs, and matching spectra against such databases is prohibitively time-consuming. Thus, development of fast algorithms for variable PNP identification is important for the success of genome mining efforts.
We have presented VarQuest algorithm for variable PNP identification via database search of mass spectra, the only modification-tolerant approach capable of searching the entire GNPS spectral network. Our method revealed an order of magnitude more PNPs than the standard search by DEREPLICATOR illuminating the “dark matter of PNPs”45. It also greatly increased the spectral library of PNPs in GNPS by identifying 41% of all known PNP families in the PNPdatabase. Iterative run of VarQuest has a potential to identify even more PNP variants with multiple modifications.
VarQuest revealed a surprising diversity of PNPs that may reflect evolutionary adaptation of various bacterial species to changing environment and competition, e.g., a continuous change of the repertoire of variants of peptidic antibiotics in response to developing antibiotic resistance. It also revealed a limitation of existing NRP mining tools that were developed based on “NRPS – a single NRP” pairs as the training datasets44 aimed at predicting a single NRP. A more biologically adequate approach would be to use the training datasets “NRPS – NRP network” that have recently become available. With growing availability of paired genomics and mass spectrometry datasets, it is now possible to generate such training datasets using VarQuest.
Methods
Scoring PNP-spectrum matches
A PNP graph of a PNP P is defined as a graph with nodes corresponding to amino acids in P and edges corresponding to generalized peptide bonds11. The mass of a PNP graph (referred to as mass(P)) is defined as the total mass of its amino acids and TheoreticalSpectrum(P) is defined as the set of masses (theoretical peaks) of all connected components of the PNP graph resulting from removal of two edges (a 2-cut in cyclic and branch-cyclic PNPs) or a single edge (a bridge in a branch-cyclic PNP)11. Note that each such removal results in two peaks with total mass equal to Mass(P).
Given a peptide P and a spectrum S, SPCScore(P, S) is defined as the Shared Peak Count, the number of peaks shared between TheoreticalSpectrum(P) and S. Two peaks are shared if their masses are within a threshold ε (0.02 Da for high-resolution spectra). We compute this score only if the precursor mass of the spectrum, denoted as Mass(S), matches Mass(P) with error up to Δ (0.02 Da for high-resolution spectra).
If (A1, . . . , An) is the list of amino acid masses in a PNP P, we define Variant (P, i, δ) as (A1, . . . , Ai + δ, . . . , An), where P and Variant (P, i, δ) have the same topology and Ai + δ ≥ 0. VariableScore(P, S) is defined as
| (1) |
where ω is Mass(P) − Mass(S) and i varies from 1 to |P| (|P| stands for the number of amino acids in the peptide P). We define a variant of peptide P derived from a spectrum S (referred to as Variant(P, S)) as Variant(P, i, ω) of peptide P that maximizes SPCScore(Variant(P, i, ω), S) across all positions i in P.
Selecting candidate peptides
Consider a peptide P and its variant P* = Variant(P, i, δ). TheoreticalSpectrum(P) and TheoreticalSpectrum(P*) share approximately half of their peaks while the remaining peaks in TheoreticalSpectrum(P* are shifted by δ with respect to the corresponding peaks in TheoreticalSpectrum(P) (Supplementary Figure 3). Thus, if an experimental spectrum S is produced by a peptide P* and shares N peaks with TheoreticalSpectrum(P*), we expect that . However, this condition often does not hold in practice due to many noisy and missing peaks in experimental spectra. In practice, we reduce the size of CandidatePeptides(S) by retaining all PNPs that satisfy the condition:
| (2) |
for a small value η. To select the threshold η, we analyzed the values of SPCScore for peptides reported by the brute-force method at various significance levels (Supplementary Table 5). Since the vast majority of statistically significant PSMs (P-value ≤ 10−10) share at least 5 peaks with the corresponding known peptides (74%, 80% and 72% for SpectraPSEUD, SpectraSTREP1 and SpectraSTREP2, respectively), we set the default value η = 5.
For a given spectrum S, VarQuest forms the list CandidatePeptides(S) by selecting all PNPs satisfying the equation 2 (among all PNPs with mass differing from Mass(S) by at most MaxMod). Checking this condition requires computing SPCScore(P, S) values for each peptide P from the PNP database Peptides. Since a naive approach (computing SPCScore for each PNP) is time consuming, VarQuest preprocesses the PNP database, and scores a spectrum S against the entire database at once.
Preprocessing a PNP database
For a given PNP database Peptides, the preprocessing starts from generation of theoretical spectra for each PNP in the database (Stage 1 in Supplementary Figure 4). All peaks from all theoretical specta are combined altogether and sorted to form the array SortedPeaks(Peptides) (Stage 2). The peaks in SortedPeaks(Peptides) are partitioned into M bins of size θ (the default values M = 20000 and 8 = 0.2 Da). Afterwards, VarQuest constructs an indexing table Index(Peptides, M, θ) (Stage 3). The table is designed in such way that the i-th cell Index[i] contains a pointer to the smallest peak p, such that p ≥ i · θ for all i ∈ [0..M − 1].
Scoring a spectrum against a PNP database
To score a given spectrum S against all PNPs in a PNP database Peptides, VarQuest iterates through all the peaks in S and stores PNPs matching the peak into a counting set FeasiblePeptides(S) (Supplementary Figure 5). To match a peak s against all the PNPs, VarQuest counts all the matching theoretical peaks in the interval (s − ε, s + ε) by finding plower (the smallest matched peak) and pupper (the largest matched peak). VarQuest sets and uses binary search to search for the smallest matching peak in the interval between Index[ilower] and Index[ilower + 1] (pupper is found in a similar way). Since the interval is small, the binary search is much faster than the search on the entire array SortedPeaks(Peptides).
After processing all peaks in the spectrum S, the number of occurrences of a PNP P in FeasiblePeptides(S) corresponds to the number of shared peaks between P and S. Thus, the list FeasiblePeptides(S) contains information about SPCScore(P, S) for all PNPs P sharing at least one theoretical peak with S.
Computing false discovery rate
The target-decoy approach46 for estimating FDR is based on generating a decoy database DecoyPeptides from a target database Peptides and searching all spectra against combined DecoyPeptides and Peptides databases. The target-decoy approach further uses the numbers of PSMs found in both databases to evaluate FDR. We refer to the set of all PSMs found in Peptides (DecoyPeptides) and having P-values below τ as PSMτ(Peptides, Spectra) (PSMτ(DecoyPeptides, Spectra)). As the decoy database consists of randomly generated peptides, we expect to find very few PSMs in PSMτ(DecoyPeptides, Spectra) for an appropriately chosen τ. Note that the size of DecoyPeptides is not necessary equal to the size of Peptides. We consider the situation when the frequencies of target and decoy peptides in the combined database are t and d, respectively (t + d = 1). We define the decoy ratio D as and compute FDR as follows:
| (3) |
Since VarQuest algorithm is linear with respect to the size of the PNP database, larger DecoyPeptides lead to increased running time. On the other hand, small database DecoyPeptides may result in an inaccurate estimate of FDR. We thus benchmarked VarQuest with various values of D to show that D = 1 is a good trade-off (Supplementary Table 6).
Generating decoy database
A popular method for generating decoy databases in traditional proteomics is random shuffling of amino acids for each target protein. However this strategy (Supplementary Figure 6b, further referred as Classical) is not suitable for PNPs because (i) PNPs are much smaller than proteins, (ii) many PNPs are cyclic or branch-cyclic, and (iii) many PNPs contain multiple copies of the same amino acid (Supplementary Figure 7). This results in decoy peptides that are similar to the target peptides after the shuffling procedure, resulting in an inflated FDR.
To address this challenge, DEREPLICATOR11 randomly redistributes the total mass of a peptide over the nodes of its PNP graph (Supplementary Figure 6c, DEREPLICATOR strategy). This strategy is motivated by the fact that PNPs often contain non-standard amino acids with a wide range of masses.
VarQuest uses a novel decoy generation approach based on amino acid shuffling and random bond displacement (Supplementary Figure 6d, VarQuest strategy). For each target PNP, VarQuest first generates a decoy PNP by rearranging amino acids. Afterwards, it randomly selects an edge in the PNP graph and substitutes it by a new edge, connected to a randomly selected position, such that the resulting decoy structure represents a connected graph. This strategy takes into account the complex structures present in many PNPs, resulting in a more diverse decoy database.
To compare accuracy of the FDR estimation using these methods we conducted the following experiment. We took 200 top-scoring unique PNP identifications from DEREPLICATOR run on the entire GNPS11. These annotations were manually curated and validated as reliable. In Experiment 1, we ran VarQuest on the spectra with the same PNP database as in11. In Experiment 2, we excluded 200 target peptides and all their known variants from the PNP database and ran the VarQuest again. We expect FDR around 0% in Experiment 1 (all mass spectra are highly trustable) and around 50% in Experiment 2 (the correct peptides are missing from the database, and matches to the target and decoy PNPs are equally likely). Supplementary Table 7 shows FDR estimations for both experiments computed based on various decoy generation approaches. Classical strategy overestimates FDR in Experiment 1 while DEREPLICATOR method underestimates FDR in Experiment 2. VarQuest decoy generation strategy has an acceptable performance in both cases (0.5% and 55.0% respectively).
Constructing the PNPdatabase
We combined all compounds with at least 4 generalized peptide bonds from AntiMarin28, DNP29, MIBiG30, and StreptomeDB31 into a single non-redundant database with 10067 distinct compounds (Supplementary Table 8). These chemical entities were classified into chemical classes using ClassyFire47 software tool. Compounds related to peptidic classes were included into our target database (referred to as PN-Pdatabase) that consists of 5021 distinct PNPs forming 1582 PNP families (Supplementary Table 9). Supplementary Tables 10–14 show distributions of PNP origins, PNP family sizes, PNP structures, number of peptide bonds and the most frequent amino acids in the PNPdatabase.
Revealing PNP diversity in related bacteria
Spectral libraries in metabolomics2, 48 rely on the comparative metabolomics assumption that assumes that two related bacteria are likely to produce identical metabolites. Our analysis revealed that, in the case of PNPs, such cases are relatively rare and that related bacteria are likely to produce similar rather than identical PNPs. To illustrate this point, we visualized strain relations based on DEREPLICATOR (identical known PNPs) and VarQuest (PNP variants of the same origin) identifications in SpectraCYANO (the largest) and SpectraSTREP1 (the less contaminated) datasets at 5% FDR. To illustrate the diversity of PNPs across related bacteria, we introduced the concept of the strain graph with nodes representing strains and edges connecting two strains if they produce variants of the same known PNP (see Supplementary Figure 8)
SpectraCYANO
DEREPLICATOR identified 42 known PNPs in 68 out of 352 Cyanobacteria strains. The strain graph constructed on these PNPs has 284 edges (two strains share identical PNP) and consist of 13 connected components (Supplementary Figure 8a). VarQuest detected PNP variants of 334 known PNPs in the same set of 68 strains. The strain graph has 618 edges (two strains produce PNP variants of the same known PNP) and a single connected component (Supplementary Figure 8b). VarQuest strain graph on the entire SpectraCYANO contains 272 nodes, 3791 edges and 19 connected components.
SpectraSTREP1
DEREPLICATOR identified 20 PNPs in 10 out of 17 Streptomyces strains. Its strain graph has only 6 edges and consists of 6 connected components (Supplementary Figure 8c). In contrast, VarQuest identified 78 PNP variants in these 10 strains and enlarged the graph by 29 additional edges (35 total) turning it into a single connected component (Supplementary Figure 8d). Moreover, VarQuest was able to identify PNP variants in all 17 Streptomyces strains in this dataset. The full strain graph has 73 edges and 3 connected components.
Validating VarQuest identifications using literature search
Supplementary Table 15 shows the list of 244 peptide variants identified by VarQuest in Pseudomonas and Streptomyces datasets. We considered all PNP variants at 5% FDR (871, 287, and 56 for SpectraPSEUD, SpectraSTREP1, and SpectraSTREP2, respectively), and excluded identifications of known PNPs with zero mass offsets (resulting in 662, 239, and 43 remaining peptide variants, respectively). Afterwards, we analyzed 100 peptide variants with the lowest P-values per dataset (for SpectraSTREP2 we considered all 43 variants). Origin of each PNP family was determined based on literature search. The most contaminated dataset is SpectraPSEUD where only 52% of variants have Pseudomonas origin. Four large non-Pseudomonas families (Surfactins, Xentrivalpeptides, Bacillomycins and SNA-60-367) cover 26 variants in this dataset. SpecNets identified 19 out of these 26 variants which indirectly suggests that these spectra are true contaminants rather than VarQuest false positives. Half of the singleton (PNP families with a single identified member of the family) contaminants (10 out of 22) are also reported by SpecNets. Both Streptomyces datasets have higher rate of PNPs originally found in Streptomyces (94% for SpectraSTREP1 and 65% for SpectraSTREP2).
There are a few reasons why spectra from SpectraPSEUD dataset form PSMs with PNPs from other bacterial sources apart from being false PSMs, e.g., laboratory contamination and morphology misidentification as many laboratory collections contain organisms that are misidentified11. Also, Luria Broth growth media prior to autoclaving is not sterile, e.g., surfactins are commonly found in the growth media (even freshly opened bottles).
Running VarQuest iteratively
While VarQuest is limited to searching for PNP variants with a single modification, this limitation can be potentially addressed by an iterative run of VarQuest. In this case, PNP variants identified in the initial VarQuest run are iteratively used as an input PNP database for the subsequent run on the same spectral dataset. Supplementary Table 16 presents results of iterative VarQuest run on SpectraCYANO. On the initial run on this dataset, VarQuest identified 2083 and 95 PNP variants in the target and decoy versions of the PNPdatabase, respectively. For the second iteration, we selected PNP variants with the most reliable mass offsets equal to ±14 Da (methylation), ±28 Da (dimethylation), ±18 Da (hydration), and ±16 Da (hydroxylation) and ended up with a new PNP database with 81 PNP variants (78 targets and 3 decoys) representing 53 unique PNPs. We further refer to this database as FirstIterationDB.
Out of 385 PNP variants identified on the second iteration, 69 were already identified among 2083 PNP variants reported in the first VarQuest run and the remaining 284 are novel (14% increase). We investigated why 353 PNP variants identified at the 2nd iteration were not reported on the first run of VarQuest (Supplementary Figure 9). It turned out that a large fraction of newly identified PNP variants (114 out of 353) were actually identified (but not reported) by VarQuest since they have P-values slightly above the default P-value threshold of 10−10 (varying from 10−10 to 10−7). Thus, FirstIterationDB indeed presents a better PNP database for identifying PNPs with multiple modifications as compared to the original PNP database.
To provide additional evidence that the newly found PNP variants represent correct rather than erroneous modifications, we further checked if the most frequent offsets identified in the 2nd iteration are consistent with the most frequent offsets identified in the initial run of VarQuest (Supplementary Table 17). It turned out that most of these offsets correlate with the most common offsets identified by VarQuest in SpectraGNPS (Supplementary Table 4).
Code availability
VarQuest is available both as a command line tool (http://cab.spbu.ru/software/varquest) and as a web application at the GNPS website (http://gnps.ucsd.edu).
Data availability
LC–MS/MS data are publicly accessible under MassIVE accession nos. MSV000079450 (SpectraPSEUD), MSV000078604 (SpectraSTREP1), MSV000078839 (SpectraSTREP2), and MSV000078568 (SpectraCYANO) at http://gnps.ucsd.edu/ProteoSAFe/datasets.jsp. The list of 120 MassIVE accessions numbrers for SpectraGNPS is available at http://cab.spbu.ru/software/varquest. The PNPdatabase is available at http://cab.spbu.ru/software/varquest.
Supplementary Material
Acknowledgments
We thank Kira Vyatkina for fruitful discussions and Andrey Prjibelski for help on the manuscript preparation. The work of A.G., A.M., A.S., A.K. and P.A.P. was supported by Russian Science Foundation (grant 14-50-00069). The work of H.M. and P.A.P. was supported by the US National Institutes of Health (grant 2-P41-GM103484).
Footnotes
Additional information
Supplementary information is available in the online version of the paper. Correspondence and requests for materials should be addressed to P.A.P.
Author contributions
A.G. implemented VarQuest algorithm. A.S. and A.K. improved and sped up DEREPLICATOR software. A.G., A.M. and H.M. designed the webserver. A.G. and A.M. did VarQuest benchmarking. H.M. and P.A.P. designed and directed the work. A.G., H.M. and P.A.P. wrote the manuscript.
Competing financial interests
P.A.P. has an equity interest in Digital Proteomics, LLC, a company that may potentially benefit from the research results. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
References
- 1.Ling LL, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marahiel MA, Stachelhaus T, Mootz HD. Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis. Chem. Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 4.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stachelhaus T, Mootz HD, Bergendahl V, Marahiel MA. Peptide bond formation in nonribosomal peptide biosynthesis. Catalytic role of the condensation domain. J. Biol. Chem. 1998;273:22773–22781. doi: 10.1074/jbc.273.35.22773. [DOI] [PubMed] [Google Scholar]
- 6.von Dohren H, Dieckmann R, Pavela-Vrancic M. The nonribosomal code. Chem. Biol. 1999;6:R273–279. doi: 10.1016/s1074-5521(00)80014-9. [DOI] [PubMed] [Google Scholar]
- 7.Mohimani H, et al. Automated genome mining of ribosomal peptide natural products. ACS Chem. Biol. 2014;9:1545–1551. doi: 10.1021/cb500199h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng J, et al. Dereplication and de novo sequencing of nonribosomal peptides. Nat. Methods. 2009;6:596–599. doi: 10.1038/nmeth.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim A, et al. Dereplicating nonribosomal peptides using an informatic search algorithm for natural products (iSNAP) discovery. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19196–19201. doi: 10.1073/pnas.1206376109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohimani H, Pevzner PA. Dereplication, sequencing and identification of peptidic natural products: from genome mining to peptidogenomics to spectral networks. Nat Prod Rep. 2016;33:73–86. doi: 10.1039/c5np00050e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohimani H, et al. Dereplication of peptidic natural products through database search of mass spectra. Nat. Chem. Biol. 2017;13:30–37. doi: 10.1038/nchembio.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pevzner PA, Mulyukov Z, Dancik V, Tang CL. Efficiency of database search for identification of mutated and modified proteins via mass spectrometry. Genome Res. 2001;11:290–299. doi: 10.1101/gr.154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsur D, Tanner S, Zandi E, Bafna V, Pevzner PA. Identification of post-translational modifications by blind search of mass spectra. Nat. Biotechnol. 2005;23:1562–1567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- 14.Tanner S, et al. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 15.Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, Nesvizhskii AI. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods. 2017;14:513–520. doi: 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balkovec JM, et al. Discovery and development of first in class antifungal caspofungin (CANCIDAS® )–a case study. Nat. Prod. Reports. 2014;31:15–34. doi: 10.1039/c3np70070d. [DOI] [PubMed] [Google Scholar]
- 17.Okano A, Isley NA, Boger DL. Peripheral modifications of vancomycin with added synergistic mechanisms of action provide durable and potent antibiotics. Proc. Natl. Acad. Sci. U.S.A. 2017 doi: 10.1073/pnas.1704125114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohimani H, et al. Multiplex de novo sequencing of peptide antibiotics. J. Comput. Biol. 2011;18:1371–1381. doi: 10.1089/cmb.2011.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bandeira N. Spectral networks: a new approach to de novo discovery of protein sequences and posttranslational modifications. BioTechniques. 2007;42:687–695. doi: 10.2144/000112487. [DOI] [PubMed] [Google Scholar]
- 20.Navarro G, et al. Image-based 384-well high-throughput screening method for the discovery of skyllamycins A to C as biofilm inhibitors and inducers of biofilm detachment in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014;58:1092–1099. doi: 10.1128/AAC.01781-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yates JR, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 22.Pevzner PA, Dancik V, Tang CL. Mutation-tolerant protein identification by mass spectrometry. J. Comput. Biol. 2000;7:777–787. doi: 10.1089/10665270050514927. [DOI] [PubMed] [Google Scholar]
- 23.Na S, Bandeira N, Paek E. Fast multi-blind modification search through tandem mass spectrometry. Mol. Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.010199. M111.010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohimani H, Kim S, Pevzner PA. A new approach to evaluating statistical significance of spectral identifications. J. Proteome Res. 2013;12:1560–1568. doi: 10.1021/pr300453t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen DD, et al. Indexing the Pseudomonas specialized metabolome enabled the discovery of poaeamide B and the bananamides. Nat Microbiol. 2016;2:16197. doi: 10.1038/nmicrobiol.2016.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duncan KR, et al. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem. Biol. 2015;22:460–471. doi: 10.1016/j.chembiol.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luzzatto-Knaan T, et al. Digitizing mass spectrometry data to explore the chemical diversity and distribution of marine cyanobacteria and algae. Elife. 2017;6 doi: 10.7554/eLife.24214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blunt J, Munro M, Laatsch H. AntiMarin database. Univ. Canterbury; Christchurch, New Zealand: Univ. Gottingen; Gottingen, Ger. 2007 [Google Scholar]
- 29.Gozalbes R, Pineda-Lucena A. Small molecule databases and chemical descriptors useful in chemoinformatics: an overview. Comb. Chem. High Throughput Screen. 2011;14:548–458. doi: 10.2174/138620711795767857. [DOI] [PubMed] [Google Scholar]
- 30.Medema MH, et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucas X, et al. StreptomeDB: a resource for natural compounds isolated from Streptomyces species. Nucleic Acids Res. 2013;41:D1130–1136. doi: 10.1093/nar/gks1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Challis GL, Naismith JH. Structural aspects of non-ribosomal peptide biosynthesis. Curr. Opin. Struct. Biol. 2004;14:748–756. doi: 10.1016/j.sbi.2004.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt EW. The hidden diversity of ribosomal peptide natural products. BMC Biol. 2010;8:83. doi: 10.1186/1741-7007-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hadjithomas M, et al. IMG-ABC: new features for bacterial secondary metabolism analysis and targeted biosynthetic gene cluster discovery in thousands of microbial genomes. Nucleic Acids Res. 2017;45:D560–D565. doi: 10.1093/nar/gkw1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medema MH, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerard J, et al. Massetolides A-H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. J. Nat. Prod. 1997;60:223–229. doi: 10.1021/np9606456. [DOI] [PubMed] [Google Scholar]
- 37.Takada K, et al. Surugamides A-E, cyclic octapeptides with four D-amino acid residues, from a marine Streptomyces sp.: LC-MS-aided inspection of partial hydrolysates for the distinction of D- and L-amino acid residues in the sequence. J. Org. Chem. 2013;78:6746–6750. doi: 10.1021/jo400708u. [DOI] [PubMed] [Google Scholar]
- 38.Kodani S, Sato K, Hemmi H, Ohnish-Kameyama M. Isolation and structural determination of a new hydrophobic peptide venepeptide from Streptomyces venezuelae. J. Antibiot. 2014;67:839–842. doi: 10.1038/ja.2014.81. [DOI] [PubMed] [Google Scholar]
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Watrous J, et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1743–1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukherjee S, et al. 1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life. Nat. Biotechnol. 2017 doi: 10.1038/nbt.3886. [DOI] [PubMed] [Google Scholar]
- 42.Mohimani H, et al. Cycloquest: identification of cyclopeptides via database search of their mass spectra against genome databases. J. Proteome Res. 2011;10:4505–4512. doi: 10.1021/pr200323a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohimani H, et al. NRPquest: Coupling Mass Spectrometry and Genome Mining for Nonribosomal Peptide Discovery. J. Nat. Prod. 2014;77:1902–1909. doi: 10.1021/np500370c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rottig M, et al. NRPSpredictor2–a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011;39:W362–367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.da Silva RR, Dorrestein PC, Quinn RA. Illuminating the dark matter in metabolomics. Proc. Natl. Acad. Sci. U.S.A. 2015;112:12549–12550. doi: 10.1073/pnas.1516878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 47.Djoumbou Feunang Y, et al. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J Cheminform. 2016;8:61. doi: 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith CA, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LC–MS/MS data are publicly accessible under MassIVE accession nos. MSV000079450 (SpectraPSEUD), MSV000078604 (SpectraSTREP1), MSV000078839 (SpectraSTREP2), and MSV000078568 (SpectraCYANO) at http://gnps.ucsd.edu/ProteoSAFe/datasets.jsp. The list of 120 MassIVE accessions numbrers for SpectraGNPS is available at http://cab.spbu.ru/software/varquest. The PNPdatabase is available at http://cab.spbu.ru/software/varquest.