Abstract
Detailed characterization of the cell types in the human brain requires scalable experimental approaches to examine multiple aspects of the molecular state of individual cells, and computational integration of the data to produce unified cell-state annotations. Here we report improved high-throughput methods for single-nucleus Droplet-based sequencing (snDrop-seq) and single-cell transposome hypersensitive-site sequencing (scTHS-seq). We used each method to acquire nuclear transcriptomic and DNA accessibility maps for >60,000 single cells from the human adult visual cortex, frontal cortex, and cerebellum. Integration of these data revealed regulatory elements and transcription factors that underlie cell-type distinctions, providing a basis for studying complex processes in the brain, such as genetic programs coordinating adult remyelination. We also mapped disease-associated risk variants to specific cellular populations, providing insights into normal and pathogenic cellular processes in the human brain. This integrative multi-omics approach permits more detailed single-cell interrogation of complex organs and tissues.
The human brain is an enormously complex network comprising ~100 billion spatially organized and functionally interconnected neurons embedded in an even larger population of glia and non-neural cells. Producing a complete cell atlas of the human brain requires highly scalable and unbiased single-cell approaches that are neither constrained by availability of fresh biopsies, nor the dissociation methods required to isolate living whole cells. Cell nuclei isolates provide a viable alternative, as they can be derived from fresh or archived tissues, provide sufficient RNA for accurate prediction of cellular expression levels1–4, and are free of artifacts associated with tissue dissociation5. We have recently demonstrated that single-nucleus transcriptome sequencing (SNS) can resolve neuronal subtype diversity across multiple human cortical brain regions4, at a relative high sequencing depth (~8 million reads per cell). However, scaling-up was limited by throughput (maximally 96 cells per microfluidic chip), high cost and sampling bias arising from poor capture of smaller non-neuronal nuclei on microfluidic chips. Higher-throughput single-nucleus RNA-seq approaches specifically applicable to archived human tissues were needed.
Although transcriptomic profiling permits identification of functionally distinct cell types that make up complex tissues, overlaying epigenetic information can provide a more complete picture on how these expression profiles are regulated or maintained. Genome wide studies have mapped regulatory sites to open or hyper-accessible chromatin located within gene promotor and enhancer regions, revealing shared cis-regulatory sites that can distinguish cell types and lineages6, 7. Identification of such cell-type specific regulomes will improve our understanding of the genetic programs defining cellular differentiation, commitment and functionality. Furthermore, because common genetic variants associated with diverse traits and diseases fall mostly within intronic or intergenic regions8, with enrichment within tissue-specific regulatory sites6, 7, generation of cell-type specific regulome maps could provide additional valuable insights into the underlying mechanisms of disease. As with transcriptomic studies, a major limitation of available epigenetic assays has been the requirement for large cell numbers. Recent methods have improved sensitivity down to hundreds of cells9 and even to the single-cell level10–13, however, application of such single-cell methods have yet to be demonstrated at a large scale on highly heterogeneous archived human tissues, such as the brain.
Ultimately, the comprehensive mapping of human brain cell types and their overall phenotypic potential necessitates more efficient methods for nuclear RNA sequencing and co-profiling epigenomic attributes using archived tissues. Given that nuclear isolates are quite amenable to single-cell genomic studies14, 15, we have developed two parallel high-throughput methods for quantifying nuclear transcripts and measuring DNA accessibility at the single-cell level that are applicable to the same pool of nuclei. This provides a means for integrative analysis of gene expression and regulation within the same archived human tissue. Here, we have resolved extensive cellular diversity in defined regions of the human cortex and cerebellum, identified region-specific neuronal and non-neuronal cell types and identified their defining transcription factor activities and target gene expression profiles on a large scale. Finally, through mapping disease risk variants to cell-type-specific regulatory regions, we provide proof-of-concept identification of possible pathogenic cell types underlying multiple brain-related diseases.
Results
Single-Cell Interrogation of Human Cortex and Cerebellum
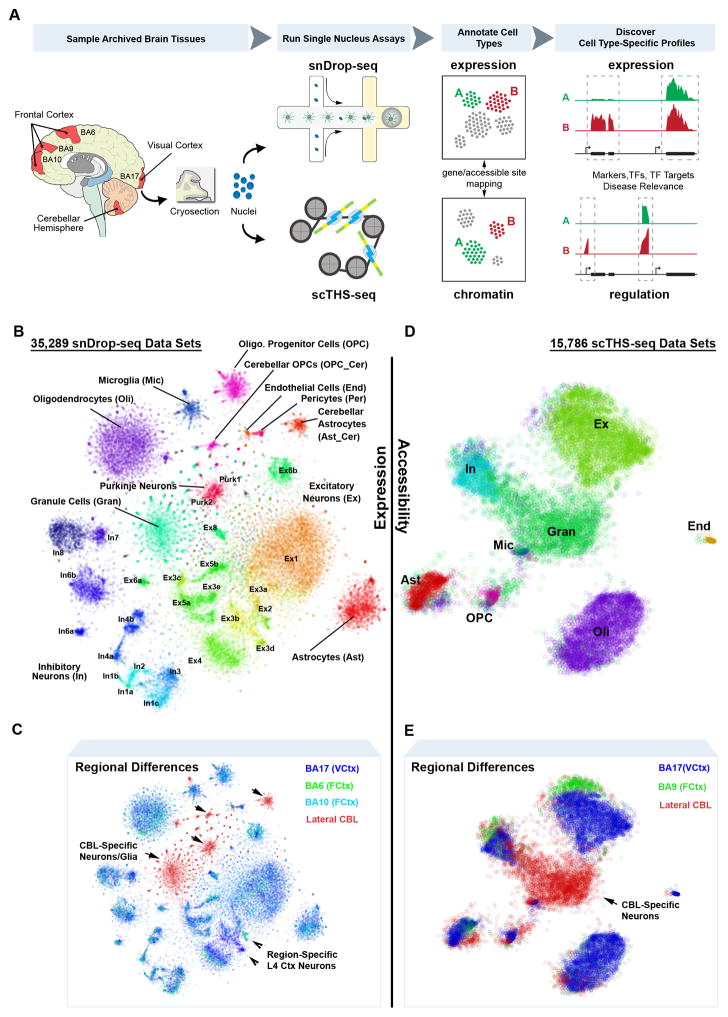
Recent advances in droplet-based technologies have greatly enhanced the throughput of single-cell RNA-sequencing (RNA-seq)16–18, enabling simultaneous transcriptomic profiling on the order of tens of thousands of single cells. Although these methodologies reduce depth of coverage, they enable extensive cell type and state classification, providing unique expression signatures to resolve functional heterogeneity existing within tissues. We have adapted a droplet-based methodology17 to analyze single nuclei, termed snDrop-seq (Fig. 1A, Fig. S1) to permit larger-scale assessment of gene expression dynamics in live and archived human tissue. Our method specifically addressed the challenge of disrupting nuclear membranes in micro-droplets without introducing excessive RNA degradation. We applied snDrop-seq to adult human post-mortem brain samples encompassing the visual cortex (Brodmann Area 17 (BA17) or V1), the frontal cortex (BA10 and BA6) and the lateral cerebellar hemisphere from six different individuals (Table S1).
Fig. 1.
Integrative single-cell analyses resolve intra- and inter-regional cellular diversity in the adult human brain. A. Overview of single-nucleus isolation from the visual cortex (BA17), frontal cortex (BA6, BA9, BA10) and cerebellum (CBL) for snDrop-seq, scTHS-seq and downstream expression/regulation analyses. B. Combined expression (snDrop-seq) data showing distinct cell type and subtype clustering visualized using t-distributed Stochastic Neighbor Embedding (t-SNE). C. Regional origination of data sets shown in (B). D. Combined chromatin accessibility (scTHS-seq) data showing major cell type clusters visualized (Table S2) using t-SNE. E. Regional origination of data sets shown in (D).
To co-investigate epigenetic configurations, we developed a single-cell DNA accessibility assay that combines our previously developed THS-Seq assay9, with combinatorial cellular indexing11 using customized barcoded transposomes (Fig. 1A, Fig. S2). scTHS-seq takes advantage of linear amplification by in vitro transcription and an engineered super-mutant of Tn5 transposase19 to achieve higher sensitivity than ATAC-seq20, including better coverage of distal enhancers found to be highly cell-type specific9, 21. Applying both methodologies, we have profiled expression and regulation signatures from the same brain regions, permitting independent and unbiased discovery of cellular diversity, and, through integrative analyses, gene expression and regulation profiles distinctive to these cellular specializations (Fig. 1A).
We generated 36,166 single-nucleus expression measurements after quality filtering, of which 35,289 from the visual (19,368 nuclei) and frontal (10,319 nuclei) cortices and the cerebellar hemisphere (5,602 nuclei) were resolved into neuronal or non-neuronal cell types (Fig. 1B, Fig. S3, Table S2). Analysis of cross-species mixing confirmed a low percentage of doublets, comparable with that found for whole-cell measurements (2–11%, Fig. S1)17. These libraries were sequenced to an average of 6,200 usable reads per nucleus (Table S1) with the majority of mapped reads falling within intronic regions and predominantly to the 3′ ends of transcripts (Fig. S1), consistent with poly-A capture of both mRNA transcripts as well as pre-mRNAs which are abundant in nuclei22. In comparison with whole-cell RNA-seq methodologies (Fig. S4), nuclear and whole-cell Drop-seq17 showed shallow coverage, but remained highly comparable in terms of median transcript or unique molecular identifier (UMI) counts and gene detection rates. Whereas nuclear and whole-cell expression levels have proven highly consistent23, nuclei data did show a systematic bias for longer genes (Fig. S4), likely reflecting differential transcript processing and export rates associated with genic length and intron fraction22. Overall, we detected a median of 928 unique transcripts and 719 genes per nucleus (Fig. S4), a depth expected to resolve effectively both cell-type diversity and gene expression dynamics given an increased sampling size24. Indeed, analysis of transcriptional heterogeneity within our data (see Methods) resolved 35 distinct cellular clusters including excitatory (Ex) and inhibitory (In) neuronal subtypes detected in the cortex4, distinct cerebellar granule (Gran) cells and Purkinje (Purk) neurons, as well as non-neuronal cells, including endothelial cells (End), smooth muscle cells or pericytes (Per), astrocytes (Ast), oligodendrocytes (Oli) and their precursor cells (OPCs), and microglia (Mic) (Fig. 1B, Fig. S3). We also resolved regional differences in these populations, including cerebellar-specific astrocytes (Ast_Cer) and OPCs (OPC_Cer) as well as different excitatory neuronal populations detected between the visual and frontal cortices (Fig. 1C).
To identify corresponding regulatory signatures, we generated scTHS-seq data from 32,869 single nuclei, of which 27,906 from the visual (13,232) and frontal (4,753) cortices and the cerebellum (9,921) were resolved into neuronal or non-neuronal cell types after clustering each region independently (Table S2). Of these, 15,786 data sets could further be resolved into combined cell type profiles (Fig. 1D). Overall, we identified 287,381 peaks associated with DNA accessibility regions covering 144 million base pairs showing unique genomic alignments. This gave a median of 10,168 unique reads per cell that were confirmed to represent accessible regions (Fig. S5, Table S1). Analysis of human-mouse species mixing confirmed a low proportion of doublets at rates that were expected from combinatorial indexing protocols11 (Fig. S5). To identify epigenetically-distinct subpopulations within the scTHS-seq data, we first used an unbiased clustering strategy modeling the probability of observing reads from a genomic site in each cell as a censored Poisson process (see Methods). This approach accounts for the fact that the scTHS-seq signal from even the most accessible site will saturate after only a few reads.
Characterizing the identity of epigenetically-defined subpopulations is more challenging than in the case of transcriptionally-defined subpopulations, because functional roles of most regulatory sequences remain poorly annotated. However, based on the functional annotation of the genes neighboring differentially-accessible sites, we could distinguish broad cell types across the cortical and cerebellar regions representing Ex, In and Gran neurons, as well as Ast, Oli, OPC, Mic, and End cell populations (Fig. 1D, Fig. S6). Therefore, our accessibility data resolved epigenetic signatures associated with the major cell types common between frontal and visual cortices as well as a previously not described neuronal signature specific to cerebellar Gran neurons (Fig. 1E).
Cell type and Regional Heterogeneity from snDrop-seq Data
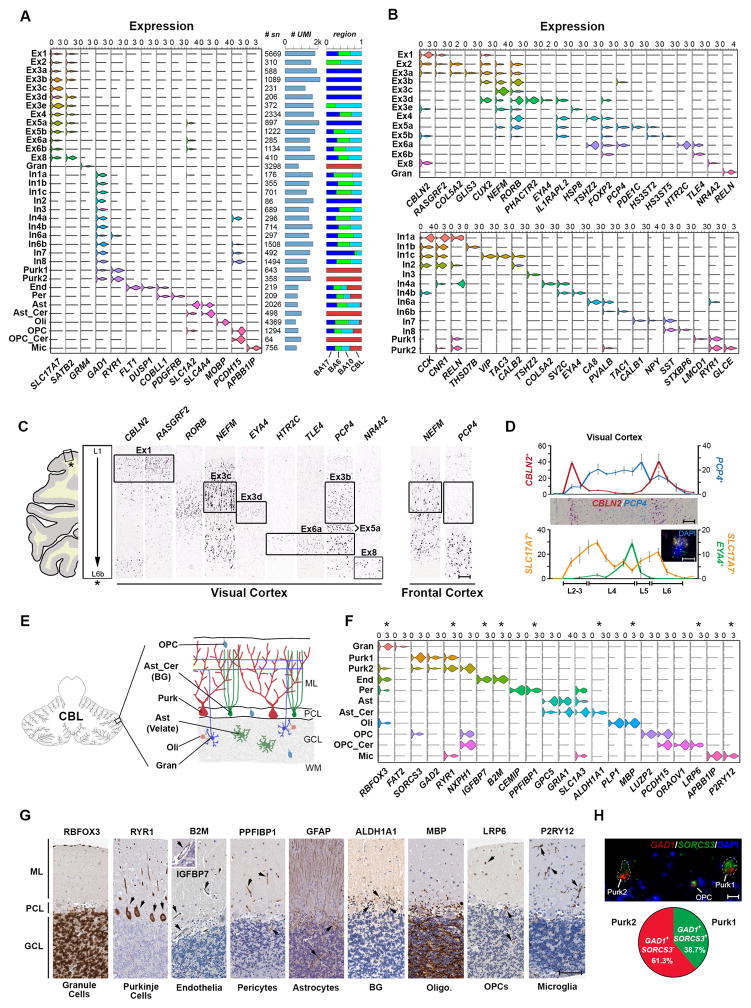
Using transcriptome data initially to define and characterize cellular identities within the different brain regions, we found expected expression of cell type or subtype marker genes (Fig. 2A–B, Table S3) and profiles that were highly consistent with pooled cell populations from the mouse25 and human (temporal lobe)26 cerebral cortex (Fig. S7). Comparison with single-cell data generated from the mouse visual cortex27 and human temporal lobe28 further confirmed broad cell type classification and consistency between nuclear and whole cell data (Fig. S7). However, we observed an over-representation of neurons at the expense of non-neuronal types such as astrocytes and endothelial cells (Fig. S7). Therefore, there likely remains some technical biases in cell type proportion estimates from snDrop-seq studies. This may stem from a bias in sample processing or uneven detection rates for the cell types with lower total transcription levels (Fig. 2A). Ex and In subpopulations were annotated based upon correlation values with subtypes previously identified from SNS in six cortical regions4 (Fig. S7). In addition to the high correspondence, snDrop-seq permitted finer resolution of these into sub-populations (e.g. Ex3 to Ex3a–d of the visual cortex). This demonstrates the high sensitivity of snDrop-seq in resolving neuronal subtype diversity though shallow profiling of a larger cell cohort compared with our previous SNS efforts4.
Fig. 2.
Expression data permits identification and classification of molecularly and spatially distinct cell types and subtypes. A. Violin plots of expression values for type-specific marker genes. Number of data sets, average transcript (UMI) counts and relative proportion across regions sampled (Fig. 1C) are indicated for each cluster. B. Top panel: violin plots showing gene expression values of layer specific4, 29 and subtype-enriched markers for excitatory neuronal subtypes. Bottom panel: violin plots showing expression values for classical interneuron marker genes4 and subtype-enriched transcripts. C. RNA in situ hybridization (ISH) stains (Allen Human Brain Atlas59, Table S9) of the visual cortex for select marker genes shown in (B). Frontal cortex stains demonstrate absence of associated layer 4 subpopulations. Scale = 200 μm. D. Top panel: RNA ISH counts showing number of positive cells for CBLN2 and PCP4 (chromogenic image shown) in image fields spanning the pial layer to the white matter. Scale = 200 μm. Lower panel: RNA ISH counts for SLC17A7 single positive cells and SLC17A7 and EYA4 double positive cells (as shown in inset). Error bars represent standard deviation for four separate layer cross-sections (replicate regions). Scale = 10 μm. E. Schematic of the cerebellar cytoarchitecture. ML = molecular layer, PCL = purkinje cell layer, GCL = granule cell layer, WM = white matter. F. Violin plots of expression values for type-specific marker genes specifically for cerebellar data. Asterisks indicate markers shown in (G). G. Protein staining (Human Protein Atlas60, Table S10) for select cell-type specific markers shown in (F). Scale = 100 μm. H. Fluorescent RNA ISH image (adjusted for visualization, see Methods) showing representative GAD1 positive Purk1 (SORCS3+) and Purk2 (SORCS3− or low) neurons. OPCs showing low expression of GAD1 were also SORCS3+. Scale = 20 μm. Pie chart shows proportions of GAD1+/SORCS3+ and GAD1+/SORCS3− populations quantified from imaged Purk neurons (Fig. S9D).
Excitatory neurons (Ex, Gran) marked by expression of SLC17A7 and GRM4 (Fig. 2A) were resolved into 14 distinct subtypes, showing enriched marker gene profiles (Fig. S8, Table S3) and that could be distinguished by their spatial orientation29 (Fig. 2B). In addition to resolving further subpopulations located within cortical layers, including the distinct HS3ST5−PCP4+ (Ex5a), HS3ST5+PCP4− (Ex5b) and HTR2C+PCP4+TLE4+ (Ex6a) subpopulations in layer 5, the latter bordering on a HTR2C−TLE4+ (Ex6b) layer 6 population (Fig. 2C). We were also able to resolve substantial regional heterogeneity in layer 4 Ex neurons, with a clear expansion in the number of visual cortex-specific subtypes (Fig. 2A), including: the RORB+PCP4+ Ex3b, RORB+NEFMhi Ex3c, and RORB+PHACTR2+EYA4+ Ex3d sub-populations (Fig. 2C). We further confirm EYA4+ Ex3d neurons as specific to layer 4 of the visual cortex (Fig. 2D), but not to the frontal cortex (Fig. S8). Inhibitory (In) and Purk neurons, marked by shared expression of GAD1 (Fig. 2A), were resolved into 13 subtypes showing enriched marker gene expression (Fig. S8, Table S3), that showed distinct profiles of canonical interneuron markers (e.g. VIP, RELN, PVALB, SST) as well as sub-type restricted expression (e.g. THSD7B, CA8, GLCE) (Fig. 2B). We were further able to resolve spatially distinct inhibitory neuron subpopulations, including: layer 1 RELN+CCK+CNR1+ In1a; upper layer VIP+CALB2+TAC3+ In1d; PVALB+CA8+ In6a concentrated around layer 4, as well as the more peripheral PVALB+TAC1+ In6b; and two distinct SST positive subtypes, including the upper layer SST+CALB1+ (In7) and lower layer SST+CALB1− (In8) subpopulations (Fig. S8).
In the cerebellum, which shows a distinct cytoarchitecture compared to the cerebral cortex (Fig. 2E), we resolved multiple major cell populations, including the Gran and Purk neurons, and their supportive cell types (Fig. 2F–G). Notably, we find two distinct Purk neuron populations expressing inhibitory markers GAD1/GAD2 (Fig. 2F, Fig. S9) distinguishable by expression of SORCS3 (Fig. 2H). Our expression data also identified two populations of astrocytes known to exist within this region: the velate astrocytes (or Ast) that show transcriptomic signatures resembling cortical astrocytes and which play a supportive role for Gran neurons; and Bergmann Glia (Ast_Cer), representing specialized astrocytes that play important roles in the laminar development of the cerebellum and which support and modulate synaptic activities of Purk neurons30 (Fig. 2F). Consistently, Ast_Cer, marked by expression of ALDH1A1 (Fig. 2G), showed enriched expression of the AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptor-encoding gene GRIA1 (GluA1 in mouse) and SLC1A3 (or GLAST) characteristic of Bergmann Glia31. In addition to this, we resolved two distinct populations of OPCs, a LUZP2+CASK+ population showing a general transcriptomic signature resembling the cortical OPCs, and an ORAOV1+LRP6+RCN2+ population (OPC_Cer) specifically found in the cerebellum (Fig. 2F–G, Fig. S9). This is consistent with the majority of the cerebellar Oli originating from outside the cerebellum and only a minority being derived from local progenitors30. Additional morphologically distinct cell types have been found in the cerebellar hemisphere32 that were not resolved in this study, likely due to their more limited quantities and the oversampling of granule neurons (Table S2) which represent the most abundant cell type within this tissue. However, we did demonstrate extensive cellular expression profiling and subtype resolution by snDrop-seq for both cortical and cerebellar regions using human postmortem tissues.
An Integrated Transcriptome and Accessibility Model
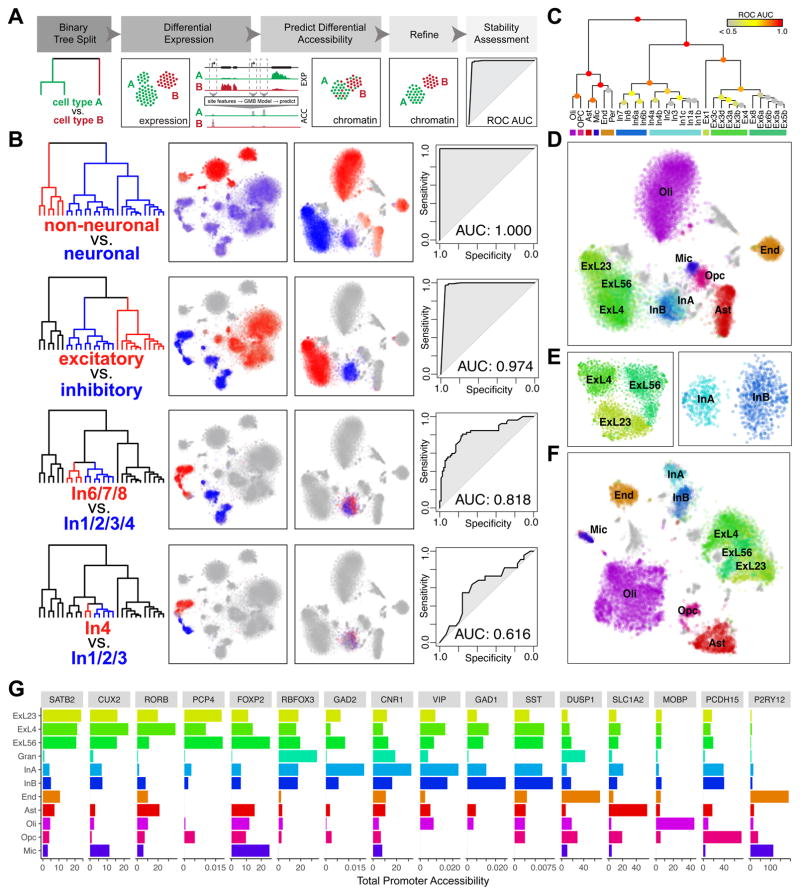
To establish a more precise correspondence between transcriptional and epigenetic states of different subpopulations, we sought to identify cells corresponding to transcriptional subpopulations in the chromatin accessibility data and cells corresponding to epigenetic subpopulations in the transcriptional data. To do so, we trained a gradient boosting model (GBM) to predict differentially accessible genomic sites based on the differential expression patterns, and a separate GBM to predict differential expression based on differential accessibility (Fig. 3), using features that include the distance of a site to a gene and the degree of differential expression or accessibility of the site or gene (see Methods). Although the ability to predict differential expression or differential accessibility of any individual gene or site is limited, joint consideration of multiple genes or sites allows for confident cell type classifications (Fig. S10).
Fig. 3.
Integrative mapping of transcriptional and epigenetic subtypes. A. Overview. First, a taxonomy of cell types is constructed based on the expression data. For each binary split in the transcriptional taxonomy, a set of genes differentially expressed between the two branches is identified. A GBM model is used to predict a set of differentially accessible chromatin sites corresponding to the identified differential expression signature, to classify scTHS-seq cells as belonging to either branch. Predicted branch annotations are refined by identifying differentially accessible sites using scTHS-seq data. Stability of the branch annotations is assessed using cross-validation (see Methods). B. Identification of In neuron subpopulations using the integrative approach. In the top binary split of transcriptional taxonomy, neuronal cells are separated from non-neuronal cells. Differentially expressed genes (Z > 1.96) are identified. Average expression of genes significantly upregulated in each branch is shown, with red corresponding to high expression in the red branch and blue corresponding to high expression in the blue branch. Predicted differentially accessible sites are visualized in the same way. Prediction performance, as assessed by ROC curves and AUC, demonstrates high stability of split for non-neuronal vs. neuronal, Ex vs. In, and In1,2,3,4 vs. In6,7,8 but not In4 vs. In1,2,3. C. Summary of stability for each binary split of transcriptional taxonomy. D. Final cell type predictions from the integrated analysis projected onto the original visual cortex scTHS-seq data t-SNE embedding. E. Refinement of the visual cortex scTHS-seq data t-SNE embedding for Ex (left) and In (right) subpopulations only, integrating predicted differentially accessible sites. F. Refinement of the complete visual cortex scTHS-seq data t-SNE integrating predicted differentially accessible sites. G. Accessibility of select marker genes. Read mapping to promoters of each gene for all cells within each epigenetic subpopulation from (F) are averaged for number of sites and cells for comparison across subpopulations.
Given the higher resolution of the transcriptional data, we sought to apply this model to further partition chromatin accessibility clusters by identifying chromatin accessibility signatures associated with the observed transcriptional subpopulations (Fig. 3A). Briefly, using transcriptional data, we first performed hierarchical clustering of the identified cell types based on their cumulative expression signature to establish a cell type relationship dendrogram. We then iteratively performed binary splits on this dendrogram and identified differentially expressed genes between the two branches. We then applied our GBM classifier to predict differentially accessible genomic sites. Joint consideration of all predicted differentially accessible sites was then used to classify the cells measured by scTHS-seq as corresponding to either branch based on the pattern of their chromatin accessibility. Based on this initial classification, we built a refined differential chromatin accessibility signature, which was used to determine the final branch assignment and to assess stability of the branch annotations through cross-validation (see Methods).
In this way, we first identified differentially expressed genes between non-neuronal and neuronal cell types from transcriptional data. The predicted differentially accessible sites allowed us to confidently resolve non-neuronal and neuronal cell types in the chromatin accessibility data. Having resolved neuronal cell types, we then repeated the procedure to distinguish excitatory from inhibitory cells. The procedure was then applied to resolve different inhibitory neuron subtypes (Fig 3B–C), and so on. In this manner, we were able to identify epigenetic differences relevant to inhibitory neuron subtypes (InA, InB) distinguished by their developmental origin from subcortical regions of the medial or lateral/caudal ganglionic eminences4, 33, 34 (Fig. 3B). However, attempts to further resolve additional inhibitory subtypes within InA and InB resulted in low stability of cell identities, suggesting a lack of differentially accessible sites sufficient to consistently distinguish between the two predicted groups (Fig 3B–C). Similarly, although layer 4 excitatory neurons (ExL4 = Ex2–4) were not distinguishable from layer 5 and 6 excitatory neurons (ExL5/6 = Ex5–8) using an unbiased analysis of chromatin accessibility data alone, integrating differential expression information from the higher resolution transcriptional data allowed us to identify relevant differentially accessible sites to further partition chromatin accessibility clusters (Fig. 3D–F). We could confidently resolve all major cell types such as Oli, OPC, Ast, End, In, and Ex cells as expected from the visual cortex (Fig. 3C), Ast, Oli, In, and Ex cells in the frontal cortex and Gran, Oli, and End cells in the cerebellum (Fig S6). We further confirmed that the resolved cell types and subtypes exhibit enriched accessibility at promoters of marker genes (Fig. 3G, Table S4). Thus, despite lower intrinsic cell type resolution of accessibility data compared to transcriptional data, computational integration of both scTHS-seq and snDrop-seq results allowed us to reconstruct detailed epigenetic profiles of fine-grained cell types within the brain, enabling investigations of the regulatory processes active within each cell type.
Transcription Factor Activities in Remyelination
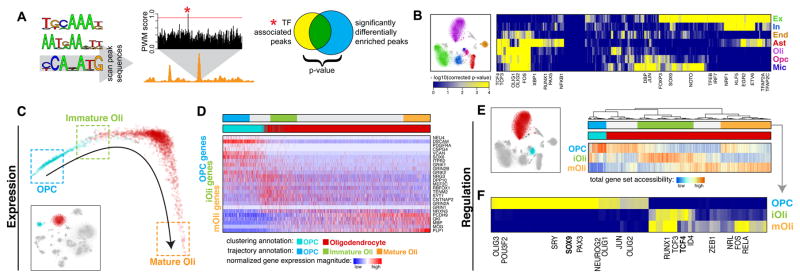
Having established the cell-type identity of each epigenetically-distinct subpopulation, we sought to identify transcription factors (TFs) relevant to the regulatory states of each cell type. To do so, we looked for TFs whose predicted binding sites are over-represented within regions of differential chromatin accessibility distinguishing a given cell type (Fig. 4A). Screening a set of 379 TFs with position weight matrices from the JASPAR database35, we identified TFs showing statistically significant association with at least one of the major cell types in the visual cortex (Fig. 4B, Table S5). We further find TF activities potentially specific to spatially distinct excitatory neuron subpopulations (L2/3 vs L4 vs L5/6) as well as inhibitory neurons derived from different subcortical regions (Table S5). As an independent validation, we cross-validated with snDrop-seq data to confirm that the TFs showing significant association with a particular cell type or subtype also tend to show higher expression levels within that cell type (Fig. S11).
Fig. 4.
Mapping transcription factor (TF) activities to specific cell types to resolve remyelination programs. A. Schematic of TF analysis. Briefly, putative TF binding sites (TFBS) were identified within all hypersensitive sites based on matching position weight matrices (PWMs). To identify relevant factors for a given cell type, sites showing differential accessibility within that cell type were tested for statistical enrichment of different TFBS. B. Heatmap of TF association to epigenetic subpopulations (right). Each column is a TF. Each row is an epigenetic subpopulation from the visual cortex (left). Select TFs are annotated. C. Diffusion map pseudotime trajectory for OPCs and Oli snDrop-Seq datasets from the visual cortex (shown as inset). Datasets are colored by the original dataset annotations from clustering analysis. Refined annotations based on the inferred pseudotime trajectory are shown as boxes. D. Heatmap of select genes involved in remyelination program. Columns are datasets ordered by the pseudotime trajectory in (C). Rows are genes ordered by association with OPCs, iOli, and mOli based on significance of differential upregulation in each group. E. Accessibility of genes involved in remyelination programs for OPCs and Oli scTHS-Seq datasets from the visual cortex (left). Heatmap of total promoter accessibility (right). Each column is a cell. Each row represents accessibility for genes differentially upregulated in OPCs, iOli, and mOli respectively. F. Heatmap of TF association to stages of Oli maturation. Each column is a TF. Each row is an epigenetic subpopulation inferred from (E). Select TFs are annotated.
To further demonstrate the utility of an integrative approach in uncovering relevant biology, we focused on the transition of OPCs to Oli in the adult brain. Myelin regeneration occurs through neuronal activation and differentiation of OPCs into myelinating oligodendrocytes that re-sheath neuronal axons to restore saltatory conduction and normal functionality. Dysregulation of this process can lead to severe neurologic disorders including multiple sclerosis (MS)36, 37. Notably, we find specific transcription factor signatures distinguishing OPC and Oli populations (Fig. 4B, Table S5). To determine whether these can reveal key regulatory processes underlying adult remyelination, we assessed differentiation states and associated gene expression signatures within these lineages in the visual cortex. Using diffusion mapping with Destiny38, we could orient OPC and Oli cells along a developmental trajectory (Fig. 4C, Fig. S12) and assess differential expression among cells in the beginning, middle, and end. In doing so, we found intermediary cells (immature or iOli), independent of experimental batch, that showed a unique expression signature (Fig. 4C–D, Fig. S12, Table S6) that could provide insight into the early mechanisms of human adult Oli maturation. Consistent with findings in the mouse39, our human OPC population expressed markers associated with mouse OPCs (PDGFRA, CSPG4, SOX6, VCAN), yet they also expressed markers for more committed mouse progenitors (ITPR2, NEU4), indicating the inability to distinguish these subtle states in our human data (Fig 4D, Table S6). Furthermore, the mature Oli (mOli) population, expressed markers associated with myelin formation (PLP1, MBP, MOG) (Fig. 4D, Table S6) and did not resolve into further sub-populations as seen in the mouse, possibly accounting for the absence of juvenile states in the adult human brain.
However, the progressive expression signatures found in OPC, iOli and mOli, which were conserved across brain regions and independent of the ordering method, could be further refined into stages of an OPC glutamate-activation response40 (Fig. S13). Recent studies propose that AMPA and kainate receptors mediate an initial axon-OPC synaptic response to glutamate that directs OPCs to exposed axonal sites where NMDA receptor activation directs remyelination40–44. Consistent with this finding, our data showed AMPA and kainate receptor-encoding genes (GRIN/GRIK) enriched in OPCs and iOli, and NMDA receptor-encoding genes (GRIN2A, GRIN2B) enriched only in the iOli subpopulation (Fig. 4D, Table S6). Functional ontogenies for gene sets identified in OPC maturation progressed along six stages: (I) neurogenesis (progenitor marker expression), (II) glutamate receptor activities, (III) synaptic transmission, (IV) ion channel activities, (V) membrane assembly, to lastly (VI) axon ensheathment (Fig. S13). These results provide independent support for mechanisms of neuronal-activity in remyelination.
To understand regulatory mechanisms defining these gene expression dynamics, we jointly assessed accessibility of the differentially upregulated genes within OPC and Oli subpopulations in the visual cortex from our scTHS-seq data. Consistent with our expression data, regulatory sites for OPC, iOli and mOli gene sets revealed differential accessibility (Fig. 4E). Further, OPC and iOli gene accessibilities showed nearly complete mutual exclusivity, indicating active regulatory mechanisms that might maintain these two states. Most significant TF activities within OPC accessible sites were associated with SOX9 (Fig. 4F, Table S7), known to be required for mouse OPC specification45, survival and migration46. Moreover, we find that the iOli-specific accessible sites showed significant enrichment for TCF4 TF binding (Fig. 4G, Table S7), which plays an important role in modulating Wnt/β-catenin to promote remyelination in the mouse47, 48. Thus, our TF analyses implicate conserved regulatory mechanisms that maintain adult oligodendrocyte progenitors and coordinate their maturation for remyelination.
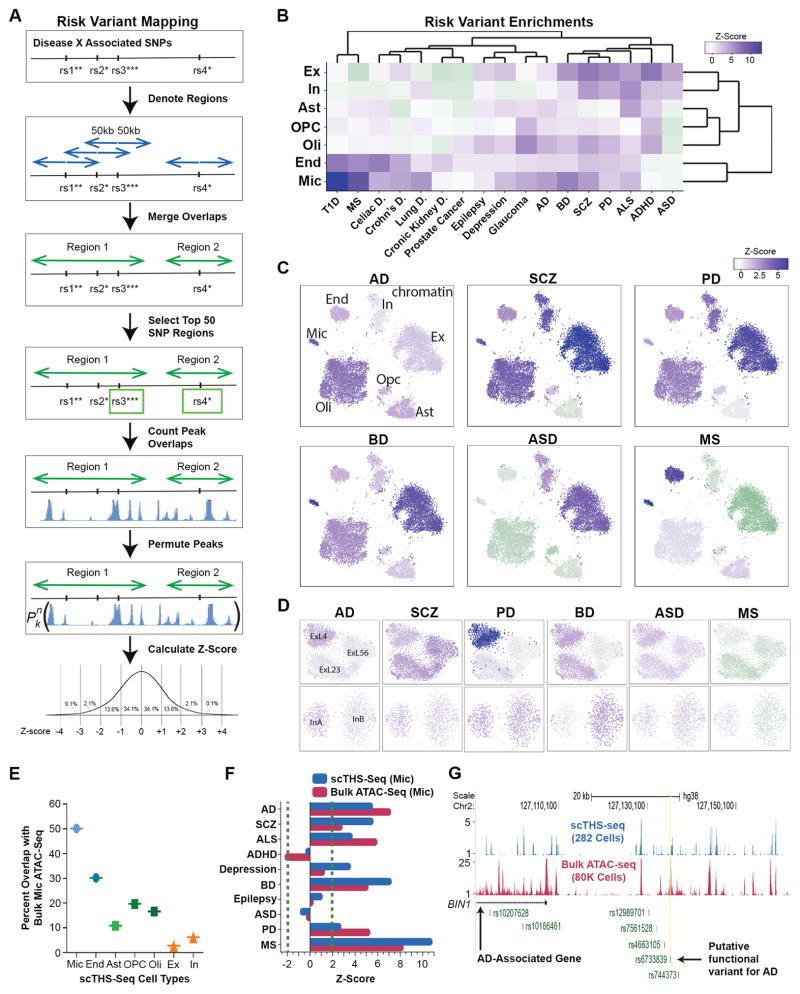
Mapping of Pathogenic Risk to Specific Brian Cell Types
Cell-type specific epigenomic information has been highly valuable for identifying pathogenic cell types and specific regulatory mechanisms underlying many common genetic diseases49–51, yet brain diseases remain inadequately understood. Towards filling this gap, we used NIH GRASP significant SNPs (p-values < 10−6) identified from genome-wide association studies (GWAS) from ten brain related disorders, and seven non-brain related diseases as controls. Given that causal variants are often located at different positions in linkage disequilibrium with the GWAS SNPs, we searched for enrichment of DNA accessibility regions in 100kb windows centered on all GWAS SNPs of a given disease, and assessed the significance by random permutations (Fig 5A, see Methods). This analysis identified strong disease-specific enrichments in multiple cell types and sub-types, contrasting with an alternative possibility of uniformity (Fig. 5B–D, Fig. S14, Table S8). Notably, we find a highly significant enrichment for Alzheimer’s Disease (AD) risk variants in Mic (Z score = 5.41, Table S8), which is in line with the significant microglia signatures found activated in the late-onset AD cortex52 and for AD-risk variants showing higher expression in microglia53. Comparisons with bulk ATAC-seq data53 demonstrated the sensitivity of our single-cell data to both predict microglia regulatory sites and their associated disease-specific risk variant enrichments (Fig. 5E–F, Table S8). No significant enrichments for non-brain related disease variants were found in neurons, further supporting our disease pathogenic cell type predictions. In fact, a majority of non-brain related enrichments were for cell types closely related to those implicated in these diseases, such as Mic and End in autoimmune diseases (Crohn’s, Celiac and Type I diabetes or T1D). Therefore, our single-cell regulatory maps were highly consistent with bulk studies and may permit linkage to cell-type specific disease risks. Although further validation involving much larger samples sizes, other disease datasets, and mechanistic studies should be pursued in the future, our chromatin maps provide a cell type or subtype-specific dataset through which new aspects of brain diseases can be understood.
Fig. 5.
Mapping of common disease risk variants to specific brain cell types. A. Method overview. Briefly, GWAS SNPs were obtained for each disease, extended to 100KB, merged, the top 50 most significant SNPs selected, number of peaks in overlaps counted, peaks permuted and the number of peaks counted in each region for each permutation, and then lastly Z-scores were calculated. B. Heat map representing the enrichment Z-scores across 7 cell clusters (rows) for 10 brain diseases (columns) and 7 unrelated diseases (Table S8). T1D = Type 1 Diabetes, MS = Multiple Sclerosis, AD = Alzheimer’s Disease, BD = Bipolar Disorder, SCZ = Schizophrenia, PD = Parkinson’s Disease, ALS = Amyotrophic Lateral Sclerosis, ADHD = Attention Deficit Hyperactivity Disorder, ASD = Autism Spectrum Disorder. Dark purple and purple represent a significant Z-score over 1.96, whereas light purple, gray and light green represent an insignificant Z-score, and green represents a significant negative association with a Z-score less than −1.96. C. Z-scores for the enrichment of GWAS SNPs in the open chromatin of Ex, In, Oli, OPC, Ast, End, Mic, populations were overlaid onto the cell clusters. Six brain disorders are shown. D. Z-scores for the enrichment of GWAS SNPs in open chromatin of three excitatory sub-clusters and two inhibitory sub-clusters. Z-score color representation as in (B). E. Percent overlap of published bulk microglia ATAC-seq53 data with differential peaks for each cell population identified from scTHS-seq data. F. Comparison of GWAS SNPs enrichment in open chromatin from published bulk microglia ATAC-seq data and differential open chromatin regions from scTHS-seq microglia data. G. Visualization of combined scTHS-seq data and published bulk ATAC-seq data on microglia over the gene and promoter region of Alzheimer’s disease associated gene of BIN1. The putative AD causal SNP located in a PU.1 binding footprint53 is also denoted.
Discussion
Reconstruction of cellular composition is an important goal towards understanding the normal function of the human brain as well as mechanisms of dysfunction and disease. This study provides a demonstration of a large scale, integrative transcriptomic and epigenomic single-cell analysis on the human adult brain, utilizing two highly scalable methods applicable to post-mortem tissues: snDrop-Seq and scTHS-seq. Using nuclei isolation to overcome challenges associated with live tissues or the processing of archived tissues, we recovered 35 subpopulations of non-neuronal and neuronal cell types in human adult cortex and cerebellar hemisphere. Our results underscore the power of sparse sampling of single cells in complex tissues at a massive scale: as long as the data from individual cells are informative enough for clustering and “virtual sorting” into different groups54, 55, they can be combined into aggregate profiles that are as rich as bulk sequencing of different cell populations. This further applies to accessibility data, accounting for the greater cell type discovery observed for the larger visual cortex dataset (Fig. S6). However, despite increased coverage allowed by scTHS-seq, chromatin accessibility data, on its own, showed lower power to resolve finer cellular subtypes, reflecting the need for improvements in sensitivity. Further, although snDrop-seq permits more wide-ranging tissue profiling compared to our previously published method4, we were unable to distinguish subpopulations of cortical astrocytes and oligodendrocytes found in mouse studies56, 57. It remains unclear whether this might be attributed to: technical artefacts associated with nuclear isolation; the more limited detection of transcripts in glia; differences in the tissues or regions sampled; differences associated with tissue archiving; or biologically limited heterogeneity in the mature adult human brain. On the other hand, our expression data extensively resolved neuronal and non-neuronal subpopulations, as well as distinct subtypes found between the cerebral and cerebellar cortices. Furthermore, our combined transcriptomic and epigenomic profiles were able to detect evolutionarily conserved expression and regulation dynamics underling adult remyelination, demonstrating the sensitivity of our methods to resolve the cellular heterogeneity and genetic programs that exist in the adult human brain.
We have additionally outlined a computational strategy for mapping between corresponding transcriptional and epigenetic states that can be used to reconstruct aggregate epigenetic profiles for fine-grained cell types. Such profiles provide valuable insights into the regulatory processes and elements shaping the identity of different cell types, as well as their relevance to human disease. Whereas previous studies have identified pathogenic cell types for several human common diseases, our analysis provides proof-of-concept data to assess common genetic risk alleles in multiple cell types of an organ, particularly the brain. It provides a coherent framework to consolidate previous GWAS findings, such as the relative contributions of glia, microglia and neurons to sporadic AD58, and could potentially extend to single-neuron genomic mosaicism that also becomes altered in this disease14. Generating multiple types of –omics maps from single cells en mass leverages the strength of each method to synergistically increase the confidence of cell type assignment to enrich cell annotations. This combined approach thus represents a strategy for systematic construction of atlases composed of single-cell data for human organs like the brain and eventually, for the full human body.
Supplemental Methods
Sample Origin and Nuclei Preparation
All human tissue protocols were approved by the Office for Human Research Protection at Sanford Burnham Prebys Medical Discovery Institute and conform to National Institutes of Health guidelines. Nuclei were prepared using nuclear extraction buffer (NEB) as described previously4. Briefly, fresh frozen post-mortem brain tissue was sectioned at 50 μm using a cryostat and placed in 1 ml of ice-cold NEB for 10 minutes. Nuclei were extracted using a glass dounce homogenizer with Teflon pestle using 10–12 up-and-down strokes in 1 ml of NEB. Samples were passed through a 50 μm filter (Sysmex Partec), incubated on ice for 10 minutes. Samples were spun for 5 minutes at 250–300 × g, washed in PBS + 2 mM EGTA (PBSE), and resuspended in PBSE supplemented with 1% fatty-acid free bovine serum albumin (FAF-BSA, Gemini) containing 4′,6-diamidino-2-phenylindole (DAPI). DAPI+ single nuclei were purified by flow cytometry using MoFlo Astrios (Beckman Coulter) or FACSAria Fusion (Becton Dickinson), concentrated at 900 × g for 10 minutes and used directly for droplet encapsulation.
Nuclei encapsulation, mRNA-Seq Library Preparation and Sequencing
Drop-seq was performed as described previously17, but with modifications optimized for processing nuclei, now termed snDrop-seq. Before droplet generation, connecting tubing and syringes were coated with 1% bovine serum albumin (BSA) to prevent non-specific binding of nuclei to the surface, and then rinsed with PBS. To reduce nuclei settling, Ficoll PM-400 was added to nuclei suspension buffer, rather than the lysis buffer. Nuclei were loaded at the concentration of 100 nuclei/μl, and co-encapsulated in droplets with barcoded beads purchased from ChemGenes Corporation, Wilmington MA (Cat. # Macosko201110). When encapsulation was complete, the droplet-collecting falcon tubes were added with a layer of mineral oil, and then transferred to 72°C water bath. After 5 minutes of incubation, the tubes were removed from the water bath to ice and droplets were broken by perfluorooctanol, following which beads were harvested, and hybridized RNA was reverse transcribed. cDNA was then PCR amplified for 16 cycles with primer, buffer and cycle conditions identical to those described previously17. A total of 46 libraries were prepared from 20 experiments (Table S1), and cDNA from each replicate was prepared and tagmented by Nextera XT and indexed with different Nextera index 1 primers. cDNA libraries were pooled and sequenced on the Illumina Hiseq 2500 using Read1CustSeqB17 for priming of read 1 (read 1 was 30 bp; read 2 (paired end) was 120 bp).
snDrop-seq data processing
Paired-end sequencing reads were processed largely as described (http://mccarrolllab.com/wp-content/uploads/2016/03/Drop-seqAlignmentCookbookv1.2Jan2016.pdf) with additional correction steps. First, paired-end reads were filtered out if read 1 had more than 4 non-T bases in the last 10 bases (to remove all non-poly T-captured contaminated reads), or had one or more bases with poor quality score (less than 10). And cell barcode and UMI information were then inferred from the first 12 bases and the next 8 bases of read 1 respectively. The right mate of each read pair was trimmed to remove any portion of the SMART adaptor sequence or large stretches of polyA tails (6 consecutive bp or greater). The trimmed reads were then aligned to the human genome (GENCODE GRCH38) using STAR v2.5 with the default parameter settings. Reads mapping to intronic or exonic regions of genes as per the GENCODE gene annotation were both recorded. One further correction step to fix barcode synthesis errors was performed by inserting N at last base of the cell barcode for reads which had identical first 11 bases of the cell barcode and same last T base of UMI. Read mapping statistics are listed in Table S1. Useful reads were calculated by adding together all un-collapsed mapped genic reads (generated by Dropseq pipeline) from each cell barcode that passed filter [approximately equal to: (total raw reads) * (proportion of reads containing poly T signal) * (proportion of reads mapping to the genome as generated by STAR aligner) * (the proportion of reads mapping to genic (exon + intron) regions as generated from RSeQC) * (the proportion of reads associated with cell barcodes passing filter)]. The digital expression matrix was then generated with genes as rows and cells as columns. UMI counts were assigned for each gene of each cell by collapsing UMI reads which had only 1 edit distance.
snDrop-seq Data Clustering and Analyses
UMI matrix cell barcodes were tagged by their associated sequencing library batch ID (Table S1) and combined across independent experiments. Mitochondrial genes not expressed in nuclei were excluded and only UMI counts associated with protein-coding genes were used for clustering analyses. Nuclei with fewer than 300 molecules or more than 5,000 molecules (outliers) were omitted. Molecular counts were normalized using the total number of reads, as the estimate of library size for each cell. Variance normalization and clustering was performed using the PAGODA2 package (https://github.com/hms-dbmi/pagoda2). Clustering and analysis were first performed separately for the visual cortex, frontal cortex, and cerebellum datasets. Briefly, the expression values were rescaled so that mean expression of a gene within each measurement batch was equal to the dataset-wide average. Winsorization procedure was used to cap the magnitude of 10 most extreme values for each gene. To estimate the residual variance for each gene, variance dependency on the expression magnitude (log scale) was modeled as a smoothed generalized additive model with smoothing term k=10 (mgcv package in R). The observed to expected variance ratio for each gene was modeled using F distribution using the degrees of freedom corresponding to the number of successful gene observations. To normalize the contribution of each gene in the subsequent PCA analysis, the variance of each gene was rescaled to match the tail probability obtained from the F distribution under a standard normal sampling process. Cell clusters were determined using approximate k-nearest neighbor graph based on a cosine distance of the top 150 principal components (PCs) derived from the top 2000 variable genes from the variance-adjusted expression matrix, using the infomap community detection algorithm (as implemented in the igraph R package). Cell clusters with fewer than 30 cells were omitted from further analysis. A preliminary round of clustering grouped low-depth cells that could not be confidently assigned to other clusters, and was omitted. Resulting cells were re-clustered and visualized using t-Distributed Stochastic Neighbor Embedding (t-SNE) on the 150 PCs. Cell clusters were annotated manually based on known markers for the frontal cortex, visual cortex, and cerebellum separately. For combined visualization, all datasets from the frontal cortex, visual cortex, and cerebellum were pooled and reclustered using the same general approach as described previously. The R script is provided for additional information on parameters used for each individual and combined dataset (Occ.R, Fcx.R, Cer.R, Combined.R) at https://github.com/JEFworks/Supplementary-Code.
Violin plots and differential gene expression analyses were performed using Seurat software (V1.4.0.5) in R (https://github.com/satijalab/seurat). For normalization, UMI counts for all annotated nuclei were scaled by total UMI counts (excluding mitochondrial genes), multiplied by 10,000 and transformed to log space. Technical effects associated with UMI coverage and batch identity were regressed from scaled data using the RegressOut function in Seurat. Genes differentially expressed between cell types and subtypes were identified (Seurat software) using a likelihood-ratio test on all genes to identify 0.25 fold (log scale) enriched genes detected in at least 25% of cells in the cluster. Differential expression analyses were performed for all clusters, for excitatory or inhibitory neuron subtypes separately, for cerebellar data sets separately or for all oligodendrocyte lineage cells separately (Table S3).
Comparison of snDrop-seq Data with Published Data
Control bulk RNA-seq data (FPKM values) from the mouse cerebral cortex and human temporal lobe were obtained from: http://web.stanford.edu/group/barres_lab/brain_rnaseq.html and http://web.stanford.edu/group/barres_lab/brainseqMariko/brainseq2.html, respectively. Top 50 cell type enriched genes were derived from comparison of averaged expression values of each cell type against an average of the remaining cell types (with the exception of oligodendrocyte sub-populations which were compared only against non-oligodendrocyte lineages). Type enriched genes from bulk data sets were used for correlation of log averaged FPKM values of the associated bulk RNA-seq data with log transformed average expression values from snDrop-seq data.
For comparison with single-cell RNA-seq data from the human temporal lobe28, gene count data was obtained from GEO (GSE67835), normalized using Seurat as mentioned above using a minimum cutoff of 1000 genes detected. Highly variable genes were identified from a mean variability plot (average expression versus dispersion (Variance/mean) assigned to 20 bins based on average expression) using a log(Variance/mean) cutoff of 1 to identify 2235 genes. Principle component analysis (PCA) was performed on these highly variable genes then projected to the entire dataset. Statistically significant PCs (p value < 0.05) were identified using a jack straw approach. Cell identities from the original publication were maintained and the top 50 genes from the statistically significant principal components differentiating these cell types, as well as the top 10 differentially expressed genes associated with each cell type, were identified using Seurat and used for correlation of log transformed averaged expression values from scRNA-seq and snDrop-seq data. For comparison with single cell RNA-seq data from the mouse visual cortex27, gene RPKM data was obtained (GSE71585), log transformed and loaded into Seurat using published cluster annotations. Neuronal subtypes were combined into a single group and average cluster expression values were obtained across cell types using previously described marker genes present in each cluster27 and a correlation heatmap of log transformed averaged expression values was generated. SNS data generated on the Fluidigm C1 platform4 (dbGaP accession phs000833.v3.p1) were used for correlation of log transformed subtype-averaged expression values for differentially expressed genes (greater than 2-fold) underlying previous subtype clustering and classifications4. For pairwise sample correlations, all differentially expressed genes (greater than 2-fold) identified during clustering of all data sets were used.
For comparison of UMI counts and genes detected with scDrop-seq data from the mouse retina17, the full UMI count table for 44808 annotated samples was obtained from GEO (GSE63472). For comparison with 9k brain cell data sets from an E18 mouse generated on the 10X platform (Cell Ranger 1.3., v2 Chemistry), filtered gene matrices were downloaded from the company website. For comparison with human embryonic midbrain single cell data sets generated using the Fluidigm C1 platform61, annotated UMI count matrices were obtained from GEO (GSE76381). Each data set was analyzed using Seurat for t-SNE visualization of clusters.
RNA In Situ Hybridization (ISH) and Protein Expression Data
Combinatorial RNA ISH experiments (Fig. 2E, Fig. 2H, Fig. S8D, Fig. S9D) were performed using RNAscope Multiplex Fluorescence Kit (SLC17A7, EYA4, GAD1, SORCS3) or the RNAscope Brown Chromogenic Kit (CBLN, PCP4) according to manufacturer’s instructions (Advanced Cell Diagnostics) and as previously described4 and outlined in Table S11. RNAscope counts were obtained for four separate layer cross-sections (replicate regions) (Table S11) and averaged values and standard deviation (error bars) were plotted. For improved visualization of GAD1/SORCS3 stains in Fig. 2H, images were further adjusted for contrast in ImageJ, however, counts were performed on representative images shown in Fig. S9D. For single gene RNA ISH from the visual or frontal cortex (Fig. 2C, Fig. S8E), representative images were obtained from the Allen Human Brain Atlas (http://human.brain-map.org) and corresponding links are provided in Table S9. For individual protein stains (Fig. 2G, Fig. S9C), representative images were obtained from The Human Protein Atlas (http://www.proteinatlas.org) and referenced in Table S10. All image panels were assembled in Adobe Illustrator and/or Adobe Photoshop.
scTHS-seq Sample Origin and Nuclei Preparation
The human tissue samples used for each scTHS-seq experiment are listed in Table S1. After flow cytometry, nuclei were kept on ice, spun down at 500xg for 5 minutes at 4°C, then supernatant was removed, and the pellet was resuspended in 1X lysis buffer (1X concentration: 10 mM Tris-Hcl pH 7.4, 10 mM NaCl, 3 mM MgCl2 0.1% NP-40, 2% bovine serum albumin, one roche protease inhibitor tablet per 10 mL, in PBS) and chilled at 4°C for 5 minutes without shaking. Then nuclei were spun down at 500xg for 5 minutes, supernatant was removed, and the pellet resuspended in 1.5X tagmentation buffer (1.0X concentration: 33 mM Tris-OAc, pH 7.8, 66 mM K-OAc, 10 mM Mg-OAc, 16% Dimethylformamide). Now the nuclei sample was ready for nuclei counting and species/species sample mixing. For running scTHS-seq, a mouse nuclei sample and a human nuclei sample was always mixed so we could perform assay quality control and calculate collision rates, ensuring low collision rates are achieved. This is further discussed in “scTHS-seq collision rate determination”. The mouse nuclei sample was prepared the same way after flow cytometry. For species/species mixing, both the human nuclei and mouse nuclei samples were counted on a Bio-Rad TC20 cell counter, and diluted with 1.5X tagmentation buffer, or further concentrated by spinning down nuclei at 500×g for 5 minutes at 4°C and resuspending in a lower volume with 1.5X tagmentation buffer, so the samples cell counts were within 10% of each other. A cell concentration of ~2.4×105 nuclei/mL was obtained for each sample with the optimal range being 2.0×105 – 5.0×105 nuclei/mL with ~1 million total nuclei for each sample. Next, equal volumes mouse and human samples were combined, and mixed gently. The sample was now ready for transposition and combinatorial indexing.
scTHS-seq transposon generation
Each transposon consisted of two oligos synthesized by IDT and kept at 100 μM stock solutions in TE buffer, the 74 bp barcoded transposon and 19 bp universal 5′ phosphorylated mosaic end. In total, there were 384 barcoded r5 transposons each with a unique 6 bp barcode and all barcodes had a minimum edit distance of 2 (Table S12). To generate annealed transposons: 10 μL of each 100 μM oligo was added to each well of a 384-well plate (final concentration of 50μM), incubated at 95°C for two minutes, cooled to 14°C at 0.1°C/second, diluted to 8.4 μM in TE buffer with a final concentration of 50% glycerol, then stored at −20°C.
scTHS-seq barcoded transposome complex generation
Tn5059 was generated and normalized for activity at Illumina. To independently generate Tn5059, the mutations and methods of protein expression and purification for Tn5059 have been published19. Because complexed Tn5059 and transposons slowly lose activity over time, with noticeable loss in data quality after a few weeks, r5 transposome complexes were generated fresh for each scTHS-seq run, and used within a few days. First, Tn5059 was diluted to 4.2 μM in standard storage buffer (Illumina) and 1 μL added to each well of 384 well plate. Next, 1 μL of 8.4 μM annealed barcoded r5 transposon was added to each well and the 384 well plate was incubated at room temperature for 30 minutes. For custom nXTv2_i7 Tn5059 transposome generation, the annealed nXTv2_i7 transposon (50 μM) was generated as described in “scTHS-seq transposon generation” (Table S12). To generate a complexed transposome solution, 7 μM Tn5059 was incubated with 10 μM annealed transposon for 30 minutes at room temperature, and diluted to 0.7 μM Tn5059 transposome complex with standard storage buffer (Illumina). These custom i7 transposome complexes were stored at −20°C and used within a few days.
scTHS-seq nuclei tagmentation and barcoding
To the 384 well plate of freshly generated uniquely barcoded Tn5059 r5 transposome complexes, 4 μL of human/mouse mixed cell sample was added for a total of ~960 nuclei/well (optimally ~2000 nuclei/well) and final concentration of 0.7 μM Tn5059 r5 transposome complex. Each sample was mixed gently 5X by the electronic pipettor and incubated at 37°C for 30 minutes. To stop the reaction, 4.0 μL of 50 mM EDTA was added to each well and mixed gently 5X by the electronic pipettor, and incubated at 37°C for 15 minutes before storing at −20°C overnight. The next day, samples were thawed, one volume of cold 2X FACS buffer (1X FACS buffer: 2 mM EDTA, 1% BSA in PBS) was added to each well and samples were mixed gently 3X by the electronic pipettor and pooled into one tube on ice, which was spun down at 500xg for 5 minutes at 4°C. Supernatant was removed, and tagmented nuclei resuspended in 1.5 mL cold 1X FACS buffer. Next, 75 μL Propidium Iodide (PI, eBioscience) was added and nuclei were sorted by flow cytometry into 96 well plates containing 10 μL PBS/well at 100 nuclei/well, and kept on ice. Doublets were removed based on forward and side scatter plots, and PI stained events selected.
scTHS-seq library preparation
Each 96 well plate of nuclei was processed individually. First, 11 μL guanidine hydrochloride was added to each well and mixed by lightly vortexing. Reactions were purified by addition of 40 μL (1.8X) AMPure SPRI beads and lightly vortexed, then bead pelleting and 80% ethanol washes were performed with the “flick and blot” method and magnetic plate from V&P Scientific. After 80% ethanol washes were complete the plate was quickly spun down at 500xg and leftover 80% ethanol removed by pipetting. 10 μL 1X NEB Taq polymerase was added to each reaction and the plate was lightly vortexed to resuspend the beads (SPRI beads left in reaction), followed by running the reactions at 72°C for 3 minutes for end fill in and placed on ice. For in vitro transcription (IVT) amplification the NEB HiScribe T7 high yield synthesis kit was used. To each reaction a mastermix of 2 μL of 10X transcription buffer, 2 μL ATP, 2 μL CTP, 2 μL GTP, 2 μL UTP was directly added to the end fill in reactions, lightly vortexed, and incubated at 37°C for 19 hours. After incubation, a couple samples were run on a TBU gel to check that IVT amplification had occurred. Reactions were purified by addition of 44 μL (2.0X) SPRI binding buffer (20% PEG 8000, 2.5 M NaCl, 10 mM Tris–HCl, 1 mM EDTA) to each reaction and the plate vortexed thoroughly. 80% ethanol washes and leftover 80% ethanol removal were performed as described above, and SPRI beads were resuspended in 9 μL nuclease free water. For reverse transcription, first 2.5 μL of 20 μM random hexamers was added to each reaction, the plate vortexed lightly and then heated to 70°C for 3 minutes, and immediately cooled on ice. Then Clontech SMART® MMLV Reverse Transcriptase kit was used with addition of 4 μL 5X first strand synthesis, 2 μL dNTP mix, 2 μL 100 mM DTT, 0.5 μL SMART MMLV RT in a mastermix to each reaction and the plate vortexed lightly. Reactions were incubated at 22°C for 10 minutes, then 42°C for 60 minutes, and terminated at 70°C for 10 minutes. To degrade RNA in cDNA-RNA hybrids 1 μL of 0.5 units Enzymatics RNase H was added to each reaction, the plate was vortexed lightly and incubated at 37°C for 20 minutes. For second strand synthesis, first 2.5 μL of 20 μM sss_scnXTv2 (Table S12) was added to each reaction and lightly vortexed, then incubated for 2 minutes at 65°C and immediately cooled on ice. Then 5.9 μL of NEB taq5X was added to each reaction and incubated at 72°C for 8 minutes. After cooling on ice, 60 μL (2.0X) SPRI binding buffer was added to each sample. The plate was vortexed thoroughly and 80% ethanol washes and leftover 80% ethanol removal performed as described previously, and SPRI beads resuspended by light vortexing in 7 μL of nuclease free water. Double stranded cDNA fragments then underwent simultaneous fragmentation and 3′ adaptor addition with a custom nXTv2_i7 Tn5059 transposome (Table S12). To 7 μL of sample, 2 μL of 5X tagmentation buffer was added to each sample, followed by addition of 2 μL of prepared 0.7 μM custom nXTv2_i7 Tn5059 transposomes (final transposome concentration of 0.14 μM), vortexed lightly, then incubated at 55°C for 6 minutes, and cooled briefly on ice. Immediately after cooling, 19 μL of 6.32M guanidine hydrochloride, for a final guanidine hydrochloride concentration of 4M was added to each reaction and briefly vortexed. Then 60 μL (2.0X) SPRI binding buffer was added to each sample. The plate was vortexed thoroughly and 80% ethanol washes and leftover 80% ethanol removal was performed as described previously. However, for this purification SPRI beads were resuspended in 16 μL nuclease free water and the plate was placed back onto the magnetic plate. Sample was eluted off SPRI beads held by the magnetic plate, and transferred to a qPCR plate. Standard Illumina Nextera XT v2 barcoding in an 8×12 (i5xi7) format was performed with qPCR, using custom scTHS-seq i5 indexes and standard Illumina i7 indexes (Table S12). In total, 20 μL of KAPA SYBR Fast, 2 μL of 10 μM scT7_S5XX index primer and 2 μL of 10 μM nXTv2_i7XX index primer were added to each reaction for a total volume of 40 μL, and mixed well. qPCR was run at 72°C for 3 minutes, 95°C for 30 seconds, followed by cycling for (95°C for 10 seconds, 63°C for 30 seconds, 72°C for 1 minute) until curves reach saturation, typically 9–12 cycles. Plates were stored at −20°C.
scTHS-seq library validation, pooling and sequencing
To validate libraries, 1 μL of each qPCR reaction was run on 6% Tris-borate-EDTA (TBE) gels stained with SYBR Gold. For pooling, 2 μL (4 μL or 6 μL if yields were low) of each uniquely barcode qPCR reaction was combined and size selection was performed as described9. Resultant size selected libraries were quantified with Qubit, sequenced on Illumina MiSeq (50+32+32 single-end reads) for validation, then Illumina HiSeq 2500 high throughput (50+8+32 single-end reads) for data generation.
scTHS-seq data processing
Raw BCL files were demultiplexed to fastq files Read1, Index1, and Index2 files using bcl2fastq v2.17.1.14, then used as input to deindexder (https://github.com/ws6/deindexer) with 0 mismatch barcode demultiplexing. Barcode combinations associated with each read were appended to each reads header with in house Perl scripts and all fastq files were combined and mapped to a hg38 no alternative loci plus decoy reference genome (GCA_000001405.15_GRCh38_no_alt_plus_hs38d1_analysis_set) and mm10 no alternative loci reference genome (GCA_000001635.5_GRCm38.p3_no_alt_analysis_set) using BWA 0.7.12-r1039. Mapped sam outputs were re-demultiplexed by barcodes, converted to bam files and clonal reads removed with samtools 1.3.1, while gathering read statistics for each barcode combination. To determine which uniquely barcoded nuclei was suitable for downstream analyses, nuclei were filtered requiring log10(total reads + 1) > 3.
Joint peak calling was performed on pooled bam files using SPP (v1.13; https://github.com/hms-dbmi/spp). In total, all 32,869 cells (Table S2) were pooled. Reads mapping within 100bps of known repeat regions according to annotations from Repeat Masker (http://www.repeatmasker.org) were removed. A smoothed density of pooled reads was generated using window tag counts with a window size of 500bps and a window step of 100bps. DNA accessible regions (peaks) were called based on the smoothed density with a minimum threshold of 5 reads and minimum span of 5 steps between each peak. Peaks were filtered using a permutation based FDR of 10e-8 and filtered for presence in at least 30 cells from the visual cortex, resulting in 52694 final peaks called. Called peaks were then assessed for reads in each individual cell to generate a matrix of peaks versus cells for downstream clustering and analysis. The used R script (spp_comb.R) is provided for additional information at https://github.com/JEFworks/Supplementary-Code.
scTHS-seq Data Clustering and Analysis
The molecular count matrix was binarized for further analysis. 52,694 sites that were observed in 30 or more cells of the visual cortex were selected. Variance normalization and clustering was performed using a modified model on the PAGODA2 package to better represent the limited dynamic range of scTHS-seq data. Clustering and analysis were first performed separately for the visual cortex, frontal cortex, and cerebellum datasets. Briefly, data was modeled as a right-censored Poisson process (observing at most 1 molecule per site). To determine cell depth and batch-specific site observation probabilities, an EM algorithm was used, with each iteration fitting MLE values for library size and batch-specific site probabilities sequentially. To evaluate over-dispersion of each site, total deviance calculated across all observations was calculated for a given site under the censored Poisson process. The relationship between the total deviance and mean site occurrence frequencies was modeled using generalized additive model (mgcv R package, smoothing term k=10). The observed / expected deviance difference was scored using variance gamma distribution. To cluster and visualize the cells in the visual cortex, the top 30 PCs were determined on the censored Poisson deviance residual matrix. The negative deviance residuals associated with non-observed sites were ignored. Cell clusters were determined on a k-nearest neighbor graph (k=50) using multilevel community detection method (igraph R package). Cell clusters with fewer than 30 cells were omitted from further analysis. Two-dimensional visualization was achieved by applying t-Distributed Stochastic Neighbor Embedding (t-SNE) on the 30 PCs using a perplexity of 50. The 30 PCs derived from the visual cortex were also used to project cells from the frontal cortex and cerebellum. Due to there being fewer cells in the frontal cortex and cerebellum datasets than the visual cortex dataset, a smaller k=30 and perplexity of 30 was used for the k-nearest neighbor graph and t-SNE embedding respectively. Cell clusters were annotated based on accessibility of marker genes as well as by the scTHS-seq and snDrop-Seq joint analysis described below. Upon inspection, three smallest clusters appeared to represent poorly-resolved cells or doublets mixing signals from two or more subpopulations (based on artificial mixing of cells from another populations) and were annotated or examined in further analysis. The used R script for analysis of the visual cortex (scTHSSeq_Occ.R) and projection of other datasets (scTHSSeq_other.R) is provided for additional information at https://github.com/JEFworks/Supplementary-Code.
scTHS-seq collision rate determination
For each unique barcode combination, the proportion of unique reads that map to either mouse or human genome was calculated (Table S1). A unique barcode combination was determined to belong to one species if 89% of the reads mapped to one genome, otherwise the barcode combination was determined to be a detectable collision. This calculation excludes the possibility of three nuclei collisions, which would represent extremely rare events. For visualization, results were then graphed in R using a X,Y scatter plot and density plot.
scTHS-Seq and snDrop-Seq joint analysis
To map between transcriptional and epigenetic space, we trained a generalized boosted regression model (GBM) to predict the probability of differential expression from patterns of nearby accessibility differences, and a separate GBM in reverse, predict probability of differential accessibility given the differential expression observations. GBM was implemented using the caret (V6.0-72) package in R. The prediction GBM utilized the following features: mean expression of the associated gene, distance of the site to the gene’s transcription start site, differential expression Z-score of the gene, fold enrichment of the gene, boolean representations of whether the site in a promoter, exon, distal intergenic region, five prime UTR, genic region, intergenic region, immediately downstream of the gene end, in an intron, in a three prime UTR, and whether the gene is most highly expressed in one cluster compared to all others. Models were trained on Astrocyte and Oligodendrocyte data from the visual cortex only to learn relevance of features as weights (Table S13). Models were fit using 10x cross-validation. Joint scores (across multiple genes or sites) were calculated as probability means of individual elements (sites or genes).
We applied our classifier to identify epigenetic subpopulations from our scTHS-seq data integrating information from the finer resolution snDrop-seq data. To do this, we first perform hierarchical clustering on cell type similarities based on expression of all genes to establish a cell type relationship dendrogram. We then iteratively perform binary splits on this dendrogram and identify significantly differentially upregulated genes (Z-score > 1.28) in each branch by Fisher’s exact test. We apply our GBM model to predict differentially accessible genomic sites.
To classify scTHS-seq cells as corresponding to either branch, we assessed for accessibility in the predicted accessible sites for each branch, normalized by the number of accessible sites observed in total for each cell. Thus, cells with high accessibility of sites predicted to be accessible in branch A will be assigned as such. Ties were randomly broken. Having identified putative corresponding subpopulations in scTHS-seq data, we then refine the predicted branch annotations by identifying significantly differentially accessible sites (Z-score > 1.28) using a Fisher’s exact test, and reassessing each cell’s joint accessibility. Refinement is repeated until convergence ie. until cell branch annotations no longer change by more than 10%. This typically requires 2 to 5 repeats. Finally, we assess stability of the branch annotations by using, randomly, 90% of cells from each group to identify differentially accessible sites that are used to derive joint accessibility scores for the remaining 10% of cells. Stability is quantified as the area under the ROC curve from joint accessibility scores with the original annotations.
To enhance separation of refined subpopulations in our data visualization, we identified differentially accessible sites for each refined subpopulation and computed the joint accessibility scores for each cell and each refined subpopulation. We applied t-SNE on the joint accessibility scores in addition to the original 30 PCs to achieve a refined 2D embedding that better segregates our refined subpopulations (Fig. 3E,F).
scTHS-seq transcription factor analysis
To infer relevant transcription factors (TFs) and transcription factor binding sites (TFBSs), we obtained DNA sequences corresponding to scTHS-seq peaks and position weight matrices (PWMs) 379 TFs from the JASPAR database. A sliding window was used to identify the maximum PWM score for each peak, taking into consideration both the plus and minus strands. PWM scores within each peak were normalized by subtracting the theoretical minimum and dividing by the maximum score possible for each PWM using the PWMscoreStartingAt() function from the matchPWM package in R assuming a uniform prior distribution on all nucleotides. Scores for each peak as well as TF were then standardized to Z-scores by subtracting the mean and dividing by the standard deviation of scores for each TF to control for background rates of binding and non-specificity. TFBSs were inferred as corresponding to peaks with Z-score > 1.96 for each TF. We assess the overlap of inferred TFBSs with previously identified cell type specific peaks using a Fisher’s exact test. TFs with TFBSs significantly overlapping with cell type specific peaks (Bonferroni corrected P-value < 0.2) were inferred to be relevant to the cell type. We integrate snDrop-Seq data to assess the expression fold change of these TFs in each cell type, assessing significance by using rank-based gene set enrichment analysis (GSEA). Specifically, TF expression was averaged across cells for each cell type. A log2 fold change comparing the average expression in oligodendrocyte versus neuronal cell types was used to assess for enrichment of expression for predicted oligodendrocyte-related TFs. GSEA was performed using the LIGER package in R (https://github.com/JEFworks/liger).
scTHS-seq GWAS data analysis
GWAS SNPs were downloaded from the GRASP database, using categories with any trait for selection with p-values <1×10−6. Categories that were selected Alzheimer’s disease, Schizophrenia, Parkinson’s disease, Bipolar disorder, Autism, Multiple sclerosis, ADHD, ALS, Epilepsy, Depression, Glaucoma, Crohn’s disease, Celiac disease, Type I diabetes, Lung disease, Chronic kidney disease, Prostate cancer. For the rest of the analysis in house python and shells scripts were used. For each category, all SNPs were extended at each end to encompass a 100 KB region. Any SNP regions overlapping each other were merged with bedtools to generate a larger SNP region containing both SNPs. Next, for each SNP region, the most significant p-value SNP was selected. This removed any multiple instances of linked variants for the same trait, and ensured there were no variants in linkage disequilibrium. Next, the top 50 most significant p-value SNPs and their gene regions were selected for further analysis. To determine overlap of accessible regions in each cluster defined during cell clustering and identification, first peak calling with SPP v1.2 was performed on merged data of each cluster to generate list of peak regions, and then lists of differential peaks (peaks present in one or more cell types and not others) were generated for all the cell types. Peaks with Z-scores <400 were removed to generate a final peak list for each cluster. Next, those peaks were overlapped with the top 50 SNP regions for each disease category and the number of overlaps counted. To determine if enrichment was significant, Z-scores were calculated. First, 20,000 permutations of the peak regions over the hg38 reference genome using only autosomes was performed, and for each permutation overlaps of peaks for each cluster with SNP regions was counted. From the permutations, averages and standard deviation were calculated, and in conjunction with previously calculated total overlaps, the Z-score for each cluster was calculated. For visualization, R was used to overlap Z-scores onto the clusters, and generate a heat map of similarity between cell types and diseases. For the excitatory and inhibitory sub-clusters, the same analysis was performed with slight modifications. All peaks were kept for analysis (instead of removing peaks with Z-scores <400), because there were overall less differential peaks between the sub-clusters. This is due to the exclusion of differential peaks that would define the main excitatory and inhibitory clusters, and are not differential between the sub clusters.
Bulk ATAC-seq microglia dataset comparison
Raw bulk microglia ATAC-seq fastq files were obtained from Gosselin et al.53, and mapped with BWA 0.7.12–r1039 to Hg38. Peak calling was performed with Dfilter 1.0, and peaks were overlapped with the differential peaks files for each cluster from visual cortex data. For GWAS risk variant enrichment analysis, the peaks file was run though the same pipeline with the same parameters as the visual cortex differential peaks clusters files.
Developmental Ordering of Oligodendrocyte Lineage Data Sets
To order cells according to their developmental trajectory along the oligodendrocyte lineage, 3064 snDrop-seq datasets for cells from the visual cortex identified as OPC (644 datasets) or Oli (2420 datasets) by the previous PAGODA2 clustering-based approaches were selected. A diffusion map approach applied using the Destiny package38 in R was applied to normalized counts with parameter k = 100 and otherwise default parameters. Cells were ordered according to their value along the first eigenvector. To identify OPC, immature Oli, and mature Oli genes along the developmental trajectory, the first 400 cells were selected as representative OPC, the 700th to 1100th cells were selected as representative immature Oli, and the 2664th to 3064th cells were selected as representative mature Oli. Differentially upregulated genes from each group were identified using PAGODA2. GO annotations for each gene set (Table S6) were obtained from toppgene.cchmc.org. To establish a corresponding trajectory according to accessibility, 5077 scTHS-seq datasets for cells from the visual cortex as identified as OPC (505 datasets) or Oli (4572 datasets) by the previous PAGODA2 clustering-based approaches were selected. All peaks were annotated using the ChIPseeker package62 with annotations from the TxDb.Hsapiens.UCSC.hg38.knownGene package. For differentially upregulated genes from each group, joint accessibility was quantified as the average accessibility of all sites corresponding to said genes multiplied by 1e6. In this manner, a joint accessibility score was derived from each cell for OPC, immature Oli, and mature Oli genes. Joint accessibility scores were scaled and clustered with hierarchical clustering and a ward.D2 linkage for visualization. The used R script (Destiny.R) is provided for additional information at https://github.com/JEFworks/Supplementary-Code.
Statistics
Combined snDrop-seq analyses were performed on 35,442 single-nucleus data sets generated over 20 experiments, each split into 1–6 libraries for 46 libraries in total (Table S1). For brain regions analyzed, biological replicates included: visual cortex – 5 individuals; frontal cortex – 4 individuals; cerebellar hemisphere – 4 individuals). For analyses on individual regions, 19,368 (visual cortex), 10,319 (frontal cortex) and 5,602 (cerebellar hemisphere) single-nucleus data sets were used (Table S2). Differentially expressed genes between cell type clusters (number of data sets per cluster are listed in Table S2) was performed using “bimod” likelihood-ratio test using Seurat, p values and false discovery rates (FDR < 0.05) are listed in Table S3.
Likewise, to identify gene expression signatures associated with remyelination in the visual cortex, we performed differential expression analysis on a limited set of 400 OPCs, 400 immature Oli, and 400 mature Oli cells identified based on pseudotime ordering from the Destiny analysis (Table S6).
For scTHS-seq analyses, 32,869 single cell data sets were generated from 3 experiments, each split into two (visual and frontal cortex) or three (cerebellum) libraries for sequencing. For each region, data sets (13,232 – visual cortex; 4,753 – frontal cortex; 9,921 – cerebellar hemisphere) were generated from a single individual, with a different individual for each region, for a total of three individuals (Table S2). For analyses across regions, 15,786 combined data sets were used (Table S2).
To identify potentially important cell-type specific TFs using scTHS-seq data, we screened a set of 379 TFs with known position weight matrices from the JASPAR database for significant over-representation within differentially accessible peaks associated with each cell type (Ex vs. In vs. End vs. Ast vs. Oli vs. Opc vs. Mic) or cell type subpopulation (ExL23 vs. ExL4 vs. ExL56, InA vs. InB, OPC vs. Immature Oli vs. Mature Oli) in the visual cortex (number of data sets per group are listed in Table S2). Significance of over-representation was assessed using a Fisher’s exact test (n = 13,232 data sets total) with Bonferonni multiple-testing correction (Table S5, Table S7).
To identify cell type specific risk variant enrichments for common genetic diseases, we defined the top 50 most significant SNP regions for each disease using SNPs from the GRASP database. For each cell type within the visual cortex (number of data sets per group are listed in Table S2), a list of differential peaks was defined (peaks present in one or more cell types and not others, only peaks with Z-score >400). To determine Z-scores, differential peaks from each cell type were overlapped with the top 50 SNP regions for a disease, and the number of overlaps counted. Next, 20,000 permutations of the peak regions on the hg38 reference genome using only autosomes was performed, and overlaps within the top 50 SNP regions counted for each permutation. From this, averages and standard deviations were calculated, and in conjunction with previously calculated total overlaps, the Z-score for each cell type was calculated. For the excitatory and inhibitory sub-clusters, the same analysis was performed with the exception that all peaks were kept for analysis (instead of removing peaks with Z-scores <400). For the published bulk microglia ATAC-seq data, the same analysis was performed, with the exception that all peaks were kept for analysis. Z-scores are listed in Table S8.
Data Availability
Raw sequencing data, annotated snDrop-seq and scTHS-seq count matrices, and DNA accessibility peak files are all available from the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/), SuperSeries accession code GSE97942.
A Life Sciences Reporting Summary is available for this publication.
Supplementary Material
Acknowledgments
Raw data and annotated count matrices are available from the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/), accession code GSE97942. Flow cytometry was performed both at the UCSD Human Embryonic Stem Cell Core and TSRI Flow Cytometry Core. We thank T.F. Osothprarop and M.M. He for providing Tn5059 transposase, N. Salathia for assistance in sequencing, Y. Wu, T. Pakozdi and Z. Chiang for assistance in sequencing analysis, and G. Kennedy for help on RNAscope. Funding support was from the NIH Common Fund Single Cell Analysis Program 1U01MH098977 (K.Z, J.C.). T.E.D. is a Fellow in the Pediatric Scientist Development Program and is supported by Award Number K12-HD000850 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. G.E.K. was additionally supported by Neuroplasticity of Aging Training Grant (5T32AG000216-24). P.V.K. was supported by NIH Centers for Excellence in Genomic Science (P50MH106933), NIH 1R01HL131768, and NSF CAREER (NSF-14-532) awards. J.F. was supported by NIH grant F31 CA206236. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The GTEx samples used for the analyses described in this study were obtained from the University of Miami Brain Endowment Bank.
Footnotes
Author’s Contributions
K.Z, P.V.K. and J.C. oversaw the study. B.B.L., S.C., B.C.S., G.E.K., Y.C.Y., T.E.D. and D.G. performed experiments. J.F., B.B.L., S.C. and B.C.S performed bioinformatics analyses, B.B.L., S.C., B.C.S, J.F., J.C., P.V.K and K.Z. wrote the manuscript with inputs from all co-authors.
Competing Financial interests
The authors declare no competing financial interest.
References
- 1.Grindberg RV, et al. RNA-sequencing from single nuclei. Proc Natl Acad Sci U S A. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Habib N, et al. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krishnaswami SR, et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc. 2016;11:499–524. doi: 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacar B, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7:11022. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roadmap Epigenomics C et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sos BC, et al. Characterization of chromatin accessibility with a transposome hypersensitive sites sequencing (THS-seq) assay. Genome Biol. 2016;17:20. doi: 10.1186/s13059-016-0882-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cusanovich DA, et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin W, et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature. 2015;528:142–146. doi: 10.1038/nature15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rotem A, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bushman DM, et al. Genomic mosaicism with increased amyloid precursor protein (APP) gene copy number in single neurons from sporadic Alzheimer’s disease brains. Elife. 2015;4 doi: 10.7554/eLife.05116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gole J, et al. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat Biotechnol. 2013;31:1126–1132. doi: 10.1038/nbt.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kia A, et al. Improved genome sequencing using an engineered transposase. BMC Biotechnol. 2017;17:6. doi: 10.1186/s12896-016-0326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ameur A, et al. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 23.Lake BB, et al. A comparative strategy for single-nucleus and single-cell transcriptomes confirms accuracy in predicted cell-type expression from nuclear RNA. Sci Rep. 2017;7:6031. doi: 10.1038/s41598-017-04426-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heimberg G, Bhatnagar R, El-Samad H, Thomson M. Low Dimensionality in Gene Expression Data Enables the Accurate Extraction of Transcriptional Programs from Shallow Sequencing. Cell Syst. 2016;2:239–250. doi: 10.1016/j.cels.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tasic B, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darmanis S, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng H, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–496. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buffo A, Rossi F. Origin, lineage and function of cerebellar glia. Prog Neurobiol. 2013;109:42–63. doi: 10.1016/j.pneurobio.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Saab AS, et al. Bergmann glial AMPA receptors are required for fine motor coordination. Science. 2012;337:749–753. doi: 10.1126/science.1221140. [DOI] [PubMed] [Google Scholar]
- 32.Butts T, Green MJ, Wingate RJ. Development of the cerebellum: simple steps to make a ‘little brain’. Development. 2014;141:4031–4041. doi: 10.1242/dev.106559. [DOI] [PubMed] [Google Scholar]
- 33.Hansen DV, et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16:1576–1587. doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma T, et al. Subcortical origins of human and monkey neocortical interneurons. Nat Neurosci. 2013;16:1588–1597. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- 35.Mathelier A, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi JW, et al. FTY720 (fingolimod) efficacy in an animal model of multiple sclerosis requires astrocyte sphingosine 1-phosphate receptor 1 (S1P1) modulation. Proc Natl Acad Sci U S A. 2011;108:751–756. doi: 10.1073/pnas.1014154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groves A, Kihara Y, Chun J. Fingolimod: direct CNS effects of sphingosine 1-phosphate (S1P) receptor modulation and implications in multiple sclerosis therapy. J Neurol Sci. 2013;328:9–18. doi: 10.1016/j.jns.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angerer P, et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 39.Marques S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gautier HO, et al. Neuronal activity regulates remyelination via glutamate signalling to oligodendrocyte progenitors. Nat Commun. 2015;6:8518. doi: 10.1038/ncomms9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hines JH, Ravanelli AM, Schwindt R, Scott EK, Appel B. Neuronal activity biases axon selection for myelination in vivo. Nat Neurosci. 2015;18:683–689. doi: 10.1038/nn.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lundgaard I, et al. Neuregulin and BDNF induce a switch to NMDA receptor-dependent myelination by oligodendrocytes. PLoS Biol. 2013;11:e1001743. doi: 10.1371/journal.pbio.1001743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mensch S, et al. Synaptic vesicle release regulates myelin sheath number of individual oligodendrocytes in vivo. Nat Neurosci. 2015;18:628–630. doi: 10.1038/nn.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wake H, Lee PR, Fields RD. Control of local protein synthesis and initial events in myelination by action potentials. Science. 2011;333:1647–1651. doi: 10.1126/science.1206998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pozniak CD, et al. Sox10 directs neural stem cells toward the oligodendrocyte lineage by decreasing Suppressor of Fused expression. Proc Natl Acad Sci U S A. 2010;107:21795–21800. doi: 10.1073/pnas.1016485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finzsch M, Stolt CC, Lommes P, Wegner M. Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor alpha expression. Development. 2008;135:637–646. doi: 10.1242/dev.010454. [DOI] [PubMed] [Google Scholar]
- 47.Zhao C, et al. Dual regulatory switch through interactions of Tcf7l2/Tcf4 with stage-specific partners propels oligodendroglial maturation. Nat Commun. 2016;7:10883. doi: 10.1038/ncomms10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rocha H, Sampaio M, Rocha R, Fernandes S, Leao M. MEF2C haploinsufficiency syndrome: Report of a new MEF2C mutation and review. Eur J Med Genet. 2016;59:478–482. doi: 10.1016/j.ejmg.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 49.Trynka G, et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang B, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gosselin D, et al. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356 doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan HC, Fu GK, Fodor SP. Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347:1258367. doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- 55.Ramani V, et al. Massively multiplex single-cell Hi-C. Nat Methods. 2017;14:263–266. doi: 10.1038/nmeth.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marques S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 58.Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 61.La Manno G, et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell. 2016;167:566–580. e519. doi: 10.1016/j.cell.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data, annotated snDrop-seq and scTHS-seq count matrices, and DNA accessibility peak files are all available from the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/), SuperSeries accession code GSE97942.
A Life Sciences Reporting Summary is available for this publication.