Abstract
For over 20 years, nuclear resonance vibrational spectroscopy (NRVS) has been used to study vibrational dynamics of iron-containing materials. With the only selection rule being iron motion, 57Fe NRVS has become an excellent tool to study iron-containing enzymes. Over the past decade, considerable progress has been made in the study of complex metalloenzymes using NRVS. Iron cofactors in heme-containing globins; [2Fe2S], [3Fe4S], [4Fe4S] proteins; the [NiFe] and [FeFe] hydrogenases; and nitrogenases have been explored in a fashion not possible through traditional vibrational spectroscopy. In this chapter, we discuss the basics of NRVS, a strategy to perform NRVS, and a discussion of the application of NRVS on the simple FeS site in rubredoxin and [FeFe] hydrogenase.
Keywords: NRVS, FeS, cluster, nuclear, vibrational, spectroscopy, synchrotron, metallocofactors
1. Introduction
Nuclear resonance vibrational spectroscopy (NRVS) has become a staple technique in the study of Mössbauer-active isotopes since its inception over two decades ago (Seto, Yoda, Kikuta, Zhang, & Ando, 1995; Sturhahn et al., 1995). The synchrotron-based technique has particularly enjoyed success with 57Fe, stemming from favorable nuclear parameters that align with contemporary synchrotron technology. Originally, NRVS was used for materials sciences and enzyme model complexes, but in recent years the method has been expanded as a probe of metallocofactors in biological systems.
NRVS is uniquely suited for studying low-frequency Fe-based vibrational modes in biological systems and can solve questions of metal-ligand bond strength and ligand identity. Ultimately, the power of NRVS is derived directly from specificity, selection, and sensitivity. Fe is ubiquitous in biological metallocofactors either as part of electron transfer or catalysis. Studying these cofactors with traditional vibrational spectroscopy, such as infrared (IR) or Raman spectroscopy, is impeded by the background created by the biomolecule proper or even fluorescence of the cofactor. As such, the extreme specificity of 57Fe NRVS for only 57Fe motion, makes the perfect tool to study vibrational dynamics of the critical Fe-containing components of biomolecules. Without contribution from the biological sample background, NRVS enjoys minimal spectroscopic noise; for example, modes of Fe hydrides within enzymes (E. J. Reijerse, 2016; Hideaki Ogata et al., 2015) have been observed which are typically difficult to directly identify with other techniques (Kaesz & Saillant, 1972; Rosenberg, 1989). The lack of conventional selection rules allows for 57Fe NRVS to reveal both IR- and Raman- active modes that contain 57Fe motion – thus providing a complete picture for Fe vibration.
NRVS is related to, and descended from, Mössbauer spectroscopy, which relies on disconnecting the vibrational motions from a Mössbauer nuclear excitation to learn about the electronic structure of the atom – a recoilless excitation.(Mössbauer, 1958a, 1958b, 1959) The logical antithesis of this idea was to realize and differentiate vibrational states that are created or annihilated upon nuclear excitation (Figure 1).(Singwi & Sjölander, 1960; Visscher, 1960) Mössbauer spectroscopy is achieved by mounting a decaying parent isotope to a velocity transducer to sweep an energy range by Doppler effect. When the energy matches a nuclear transition of the daughter isotope in the sample, a nuclear resonance excitation occurs. However, the energy resonances in 57Fe Mössbauer spectroscopy are typically on the order of neV and vibrational quanta observed through NRVS are on the order of meV. This implies a Doppler shifted source would need to move with a velocity 6 orders of magnitude faster than a typical 57Fe Mössbauer experiment in order to perform 57Fe NRVS which is widely considered impractical. To access the 57Fe vibrational modes, a different source with high flux, high monochromation, and high tuneability is necessary. Fortunately, beamlines at third generation synchrotron X-ray light sources meet these requirements and make NRVS possible.(Wang, Alp, Yoda, & Cramer, 2014)
Figure 1:
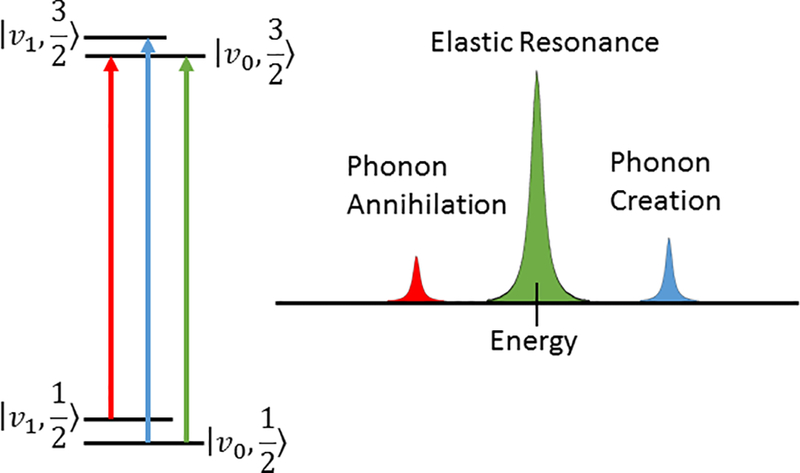
A simplification of the NRVS processes. The green line and peak shape contains the recoilless Mössbauer transition that is beyond the resolving power of NRVS. The red and blue peaks correspond to phonon annihilation and creation respectively – and the ratio of their intensity is temperature dependent.
2. Materials
As described above, NRVS requires the use of a synchrotron radiation light source. Few synchrotron beamlines can accommodate the challenging NRVS requirements for flux, monochromation, and time-sensitive detection. As such, an NRVS experiment can only be performed by applying for beamtime at four synchrotron radiation light sources: The Advanced Photon Source (APS - http://www.aps.anl.gov), The Positron-Electron Tandem Ring Accelerator (PETRA-III - http://petra3.desy.de), The European Synchrotron Radiation Facility (ESRF - http://www.esrf.eu), and the Super Photon Ring – 8 GeV (SPring-8 - http://www.spring8.or.jp/en/).
Despite differences in synchrotron facilities, the principle setup of an NRVS experiment remains the same. Charged particles circulating at relativistic speeds within an ultra-high vacuum (UHV) storage ring pass through a set of magnets called an undulator that generates a tangential beam of photons, called synchrotron radiation, that covers the energy of nuclear resonance (14.4 keV for 57Fe). The “white light” beam (ΔE≈100 eV) travels down a UHV transport channel, with optics to aperture and focus the beam, towards the target beamline and is initially monochromated by a liquid nitrogen, or water, cooled high heat load monochromator (ΔE≈100 meV). The incident light is then monochromated by a tuneable high-resolution monochromator (ΔE≈1 meV) composed of Si or Ge crystals. Finally, the beam interacts with the sample within a cryostat or cryojet stream.
After excitation, 57Fe within the sample will relax either through direct nuclear relaxation (≈14.4 keV photon) or internal conversion followed by, primarily, Kα emission (≈6.4 keV photon) with the internal conversion process favored 8:1. Both relaxation channels are relevant to NRVS and must be detected. The detection system is time resolved, and designed to exclude early-time electronic scattering events while focusing on photons resulting from the 57Fe relaxation (with a τ=141 ns). The sophisticated detection scheme is made possible by the nanosecond response avalanche photodiode (APD) detector analyzed by a constant fraction discriminator (CFD). Multiple “bunches”, or groups, of particles within a storage ring are spaced throughout the period of the synchrotron such that photons arrive at the beamline in a periodic pulsed pattern (the parameters of which define a “bunch mode”). As such, t=0 must be defined to the CFD for each bunch – a process synchronized by the “bunch clock”. The CFD then vetoes photon counts 10–20 ns around t=0 – giving time for the APD to recover from the prompt (incident) pulse of photons and isolating the NRVS emission signal from electronic scattering. Collectively, the properly implemented setup typically allows for dark count rates of less than 5 × 10−3 per second.
Given the low natural abundance of 57Fe (2.14%), 57Fe NRVS requires that biological samples be enriched with 57Fe. Generally for bioinorganic applications, enrichment can be achieved by dissolving elemental 57Fe in aqua regia and adding to an Fe-free growth medium. Likewise, 57Fe enriched salts (e.g. 57FeCl3 or 57FeSO4) can provide an avenue for integration to the protein or enzyme of interest. To study specific Fe atoms or clusters out of many within a protein – care must be taken to integrate 57Fe by targeting steps in the enzyme maturation or reintroducing the enriched cofactor of interest to an unenriched apoprotein.
Although some variants exist, a typical 57Fe NRVS sample cell for proteins has dimensions of 10×1×1 mm. These parameters take into account the dimensions of the synchrotron X-ray beam, the penetration depth of the beam, the solid angle projection from the sample to the APD, and the escape depth of the Fe emission following internal conversion. The cell is sealed liquid-tight with an adhesive tape and a port is available in the cell to inject sample solution, followed by subsequent freezing in liquid nitrogen. The cell and tape should be made from cryo-compatible materials that do not interact with the sample nor the X-ray beam. A commonly used sample cell material is Poly(methyl methacrylate) (often referred to by the eponym “Lucite”) and a common tape material can be polyimide (eponymously “Kapton Tape”). When in doubt, materials can be checked for their X-ray transmission properties at http://henke.lbl.gov/optical_constants/filter2.html.
NRVS experiments for bioinorganic applications are often performed at cold sample temperatures with the sample loaded onto a copper cold finger within a helium cryostat. Although, typically the nominal temperature reading in the cryostat may be near helium temperatures <10 K, the actual sample temperature can vary widely based on the sample properties and the liquid properties used to marry the sample cell to the cold finger. Due to favorable physical properties, particularly a melting point of 147 K, 1-propanol is considered one of the best mounting liquids.(Wang, Yoda, Kamali, Zhou, & Cramer, 2012) The true sample temperature can be obtained by fitting the ratio of phonon annihilation events to creation events using the PHOENIX software and can be approximated by a Maxwell-Boltzmann distribution. To ensure the temperature can be approximated, a standard NRVS spectrum is usually collected slightly below the elastic resonance at −180 cm−1 (annihilation of phonons), through the elastic resonance, and at energies higher than the resonance into the region of interest (creation of phonons).
It is generally favorable to analyze the spectrum in terms of partial vibrational density of states (PVDOS) to isolate single phonon events and remove their energy dependence. Sturhahn created a program called PHOENIX that is the standard to convert raw NRVS data to PVDOS (among many other features not discussed here)(Sturhahn, 2000) using the Fourier-log method.(Johnson & Spence, 1974) The program can be downloaded from: http://www.nrixs.com/downloads.html. Alternatively, data can be converted and analyzed through a web-based graphical user interface for PHOENIX through http://www.spectra.tools/.
The transformation into PVDOS, which is proportional to the normal mode composition factors for 57Fe, allows for a straightforward prescription to estimate the PVDOS intensity through normal mode analysis using density functional theory (DFT) or other in silico methods; the normal mode composition factors and normal mode energies can be extracted from diagonalization of the Hessian force matrix used in typical computational normal mode analysis.(Petrenko, Sturhahn, & Neese, 2008)
Calibration
The monochromator crystals are very sensitive to the experimental environment specifically temperature. As such, the proportion between expected energy and true energy of monochromation can vary between experiments. It is ideal to calculate a linear “calibration factor” for the monochromator using a secondary standard that has been analyzed using a technique with a reliable energy domain. Typically, this can be handled by small 57Fe compounds such as: (Et4N)57FeCl4, Prussian Blue, or 57Fe(CN)6.
For (Et4N)57FeCl4, assuming an idealized Td point group, two T2 modes can be observed with maxima in the FT-IR calibrated NRVS spectrum at 378 cm−1 and 139 cm−1 (asymmetric Fe-Cl stretching modes and Fe-Cl bending modes respectively) – as well as mixed optical and acoustic modes grouped as “lattice modes” below 60 cm−1 (Figure 2).(Smith et al., 2005) Notably absent from the NRVS spectrum, are the A1 Fe-Cl symmetric stretching mode and the, E, Fe-Cl symmetric bend as these modes do not contain Fe motion. A user need only collect an NRVS spectrum for (Et4N)57FeCl4 and align the T2 peaks to the previously calibrated positions with a linear factor and apply this factor to the energy axes of their subsequent spectra.
Figure 2:
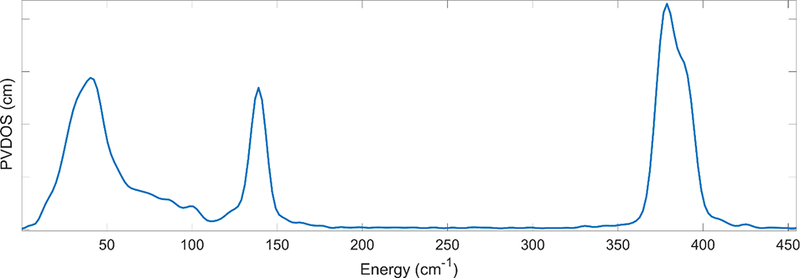
The NRVS spectrum of (et4N)57FeCl4. The low energy intensity is related to lattice phonons and the features at 378 and 139 cm−1 can be approximated as T2 modes in Td symmetry. The energies of these modes are used to calibrate NRVS spectra. Noticeably absent is the A1 symmetric stretch as that mode does not involve 57Fe motion.
Interpretation of NRVS data generally involves fundamental vibrational analysis, particularly the practice of isotopic substitution, group theory, and empirical comparison to identify spectral features. A tool to assist in analysis, called Vibratz,(Dowty, 1987) allows the user to fit a Urey-Bradley forcefield to their spectrum based on a set of internal coordinate force constants. The user may also define the point group for the molecule of interest to connect the irreducible representations of the point group to the features of the spectrum. This type of analysis may lead to new interpretations about the geometry of the molecule of interest when minimal structural data is available.
3. Procedure
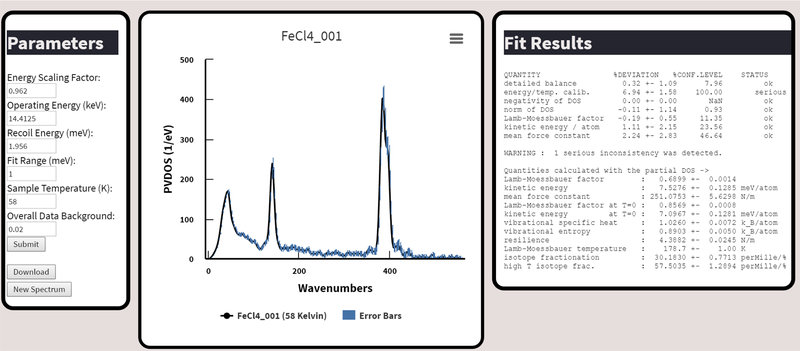
This procedural outline will describe a standard process of collecting and analyzing cryogenic bioinorganic NRVS data using the spectra.tools website (Figure 3).
Figure 3.
The output window of the spectra.tools NRVS data processing. Here parameters (on the left) can be adjusted to achieve desirable statistics (on the right). The graph in the center can be exported in image formats and the data downloaded by the “Download” button in the parameters window.
Obtain beamtime at one of the above listed synchrotron beamlines where NRVS is available.
Follow established protocols to produce the enzyme of interest in an Fe free media.
Add 57Fe as either a pure powder or salt to the media.
- Follow established protocols to purify the enzyme of interest.
- A typical concentration goal is at least 1mM of 57Fe in the final NRVS sample, but this depends on the intensity of the signals of interest.
Obtain a three walled 1×1×10mm cell made of Lucite with an injection port milled through the top.
- Wrap the cell, liquid tight, in Kapton tape that is at most 1 mil in thickness.
- Take care to minimize and flatten the tape on the back of the cell – as this will interfere with thermal contact to a cryostat cold finger.
- Inject the purified enriched enzyme into the cell.
- For air-sensitive Fe-containing proteins, the sample should be injected into the cell anaerobically then immediately frozen in liquid nitrogen.
If the synchrotron is not local, ship a cold, dry, liquid nitrogen shipping dewar containing the sample(s) to the synchrotron. Then travel to the synchrotron.
Assuming the cryogenics are already established at the beamline, at a nominal temperature of 110–120K vent the sample space vacuum with helium and remove the cold finger housing.
Apply 1-propanol to the cold finger surface. Generally, a thin layer is required to establish good thermal contact – too much will cause poor contact.
Attach the NRVS cell to the 1-propanol on the cold finger, with attention paid that the “sample side” is “up” or in position to receive the incident X-ray.
Reattach the cold finger housing, turn off the venting gas, reestablish a vacuum in the sample space, and allow the cryostat to cool.
- At a nominal temperature of 10K, begin data collection.
- This step is beamline dependent, and the local beamline scientist should be able to assist.
- If specific regions of the spectrum are of interest, the beamline scientist should be able to perform specialized region of interest scans.
- As data workup later depends on the elastic region, even region of interest scans need to contain the elastic peak, but the acquisition time for the region of interest will be much higher.
Once the data is satisfactory the sample may be switched following the steps above. If a thin even layer of 1-propanol was applied most of it will come off with the sample cell – otherwise the surface may need to be cleaned slightly with a lab tissue.
Depending on the beamline, there may be inhouse programs/methods to analyze data. Otherwise navigate a web browser to http://www.spectra.tools/.
At the navigation bar towards the top, under the drop down for NRVS, click “NRVS Spectra Processing Tool”.
In the “Data Upload” window select the data source type (supported currently: SPring-8, APS .mda files, and PETRA-III). Click “Choose Files” and navigate to the folder that contains the NRVS scan files – on most systems SHIFT+DRAG (or holding CTRL) can be used to highlight multiple files. Then select the submit button.
- A three-panel window will appear:
- The left panel (“Parameters”) allows the user to alter different NRVS-related parameters. By default, the parameters listed are the most commonly changed, but advanced parameters can be altered by clicking “Advanced Submit”.
- The central window displays the PVDOS spectrum – by popular demand it is displayed as a function of wavenumbers. The name of the final file processed is the title of the graph. The graph is zoomable and exportable to image formats. Error bars calculated by PHOENIX are shown, and can be turned off by clicking “Error Bars”. To obtain the complete data (including everything required by PHOENIX) the “Download” button in the left panel is used.
- The right panel (“Fit Results”). Indicates the results of the statistical analysis performed by PHOENIX.
- Once the spectrum is loaded, adjust the parameters on the left-hand side and resubmit the spectrum for analysis.
- First, ensure that the “Energy Scaling Factor” is appropriate. As mentioned previously this factor is determined by collecting the NRVS spectrum for a secondary standard particularly (Et4N)57FeCl4. The normal value ranges are from 0.92–1.10, are beamline dependent, and should be determined at least once during the experiment.
- Second, minimize the background contribution to the spectrum. This is achieved by increasing “Overall Data Background” until the signal in regions with no expected signal is minimized.
- Third, alter the “Sample Temperature” until the “detailed balance” in the right hand “Fit Results” window is acceptable.
Ideally, once all the “Fit Results” indicate a status of “OK” then the spectral parameters are acceptable. Occasionally this is an unattainable goal, especially at a very high number of scans or if there were significant changes in sample temperature during the experiment. In this case, each scan should be carefully examined.
If everything is satisfactory click the “Download” button in the parameters window. A download of a compressed folder should begin. The file in the folder with the suffix *.dos will contain the final partial vibrational density of states.
4. Application
Rubredoxin
Rubredoxins are small electron transfer proteins. Rubredoxin from P. furiosus is composed of 53 amino acid residues for nearly 6 kDA in mass. Electron transfer is handled by a single Fe redox center coordinated to four cysteinyl thiolates. The two redox states are formally Fe(II) and Fe(III). The simplicity and size made rubredoxin an excellent early candidate for bioinorganic 57Fe NRVS.(Xiao et al., 2005)
The spectrum for the oxidized and reduced proteins is shown in Figure 4. There are three prominent features near 70, 150 and 360 for the Fe(III) site in the oxidized sample, and the high frequency features shifts to 303 cm-1 upon reduction.
Figure 4.
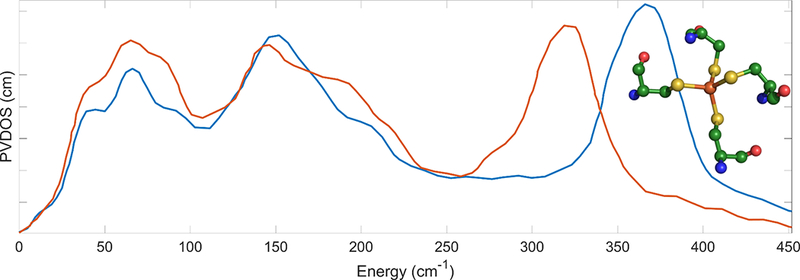
The NRVS spectrum of oxidized (–––) and reduced (–––) rubredoxin in solution. Inset: structure of the Fe site from PDB:1CAD. Figure reproduced from previous work.(Xiao et al., 2005)
Naively speaking, the initial assumption is that the three features are analogous to the Td symmetry FeCl4 modes discussed above (Figure 2). However, the broadening and obvious shouldering of the three distinct features in the spectrum (Figure 4) indicate that interpretation under presumption of a perfect Td symmetry, as with (Et4N)57FeCl4, is not appropriate. To fit the internal coordinate force constants to the collected spectrum, it is effective to use a descent in symmetry (Td→D2d→C1) whilst increasing the complexity of the Vibratz model (Fe(SC)4→Fe(SCC)4→ Fe(SCys)4). For the final model incorporating four cysteines, Xiao and coworkers used the geometries from the pertinent crystal structures (reduced PDB:1CAD,(Day et al., 1992) oxidized PDB:1BQ8(Bau et al., 1998)). This iterative treatment allowed for a reasonable fit of the internal coordinate force constants. The results of modeling the NRVS spectra was a calculated 36% decrease in Fe-S bond force constant (1.24 to 0.92 mdyne/Å) upon reduction of the protein.
The Vibratz program also describes the motion of the components of the fitted spectrum. For the oxidized protein model, the broad region from 345 to 375 cm−1 is composed of three strong components at 375, 358, 350 cm−1 and three weaker contributions at 365, 364, and 340 cm−1 that are principally described as Fe-S asymmetric stretching modes. The intensity in the region from 300 to 340 cm−1 arises from nearly symmetric Fe-S stretch modes – a consequence of the deviation from ideal symmetry. Due to much lower 57Fe motion, the contributions in this so-called “breathing” region are approximately 10% that of the asymmetric stretch components. The area around 150 cm−1 is composed of nearly degenerate S-Fe-S bending modes. The region between 100 and 150 cm−1 are assigned as Fe-S-C bending modes with mixing of S-Fe-S modes. Finally, the low energy region below 100 cm−1 appear analogous to the “lattice modes” described in 57FeCl4, however they are more complex and involved significant dihedral, torsional, acoustic, and delocalized vibrational modes within the greater protein and can provide a measure on the degree of vibrational coupling between the cofactor and protein matrix.(Guo et al., 2012)
When the same analysis is performed for the reduced Fe center, the order of assignments still hold, but the stretching modes downshift by 18%. It is noteworthy that the symmetric mode centroids shift to approximately 270 cm−1 and start to mix with the bending modes leading to enhanced intensity throughout the 150–300 cm−1 region (Figure 4).
These results confirmed the delocalization of the dynamic properties of the redox-active Fe site which at the time was debated.(Hiroshi, Takeo, Kaori, Akio, & Tatsuhiko, 1991) Besides the broadness of the Fe-S stretch region in both redox states, the weaker Fe-S force constant in the reduced rubredoxin leads to even stronger coupling between the Fe site and the motion of the local protein environment, evidenced experimentally by increased intensity below 300 cm−1. This work on rubredoxin laid the framework for a later NRVS and molecular dynamics study that revealed non-Rieske [2Fe2S] clusters can couple even more dramatically to the motion of their environment and possibly induce binding and release of redox partners.(Xiao et al., 2008)
[FeFe] Hydrogenase
The [FeFe] hydrogenase is an enzyme that catalyzes the reversible evolution of H2. It is an FeS cluster containing enzyme with a catalytic [2Fe] cofactor supported by the electron transfer activity of one or more other [FeS] clusters of varying composition depending on the source organism. The conversion of protons to dihydrogen presumably involves at least one Fe-H intermediate state. Unlike the bridging hydride in the related [NiFe] hydrogenase intermediate states,(Hideaki Ogata et al., 2015; Ogata, Nishikawa, & Lubitz, 2015) the consensus was that the intermediate hydride is terminally bound to the cofactor,(Mulder, Guo, Ratzloff, & King, 2017) a direct vibrational observation was recently made for the Fe-H bond by NRVS.(Reijerse et al., 2017)
To trap the hydride-bound intermediate in significant amounts, and to selectively label the relevant diiron site, Reijerse and coworkers inserted a synthetic 57Fe-labeled [2Fe] site into the enzyme. Whereas the natural enzyme contains an azadithiolate bridge between the two irons, proton transport to the synthetic active site was disrupted by an oxadithiolate (ODT) bridge, inhibiting full catalysis and increasing the yield of the hydride-bound intermediate called “Hhyd”.
The experimental NRVS spectrum for the [2Fe]-ODT site in Chlamydomonas reinhardtii [FeFe] hydrogenase in the Hhyd state is shown in Figure 5. The vibrational modes from 0–400 cm−1 are predominantly Fe-S in nature. The modes from 400–600 cm−1 are Fe-CO/CN stretching and bending vibrations. However, the most noteworthy observations are at 727 cm−1 and 670 cm−1 which involve Fe-H bending motions within and perpendicular to the Fe-Fe-H plane respectively. The two hydride features disappear upon deuteration of the sample. Beside a similar Fe-S region, the deuterated enzyme features a general shuffling of the Fe-CO/CN region intensities as the Fe-CO/CN vibrational motions couple differently to the deuteride motion in comparison to the hydride. The deuterated bending modes shift to 625 cm−1 and 564 cm−1 for the in-plane and perpendicular Fe-D bending modes respectively.
Figure 5.
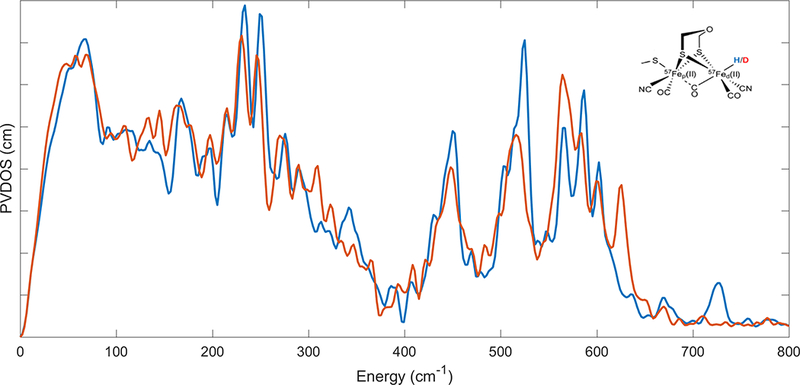
The NRVS spectrum of the hydride (–––) and deuteride (–––) forms of the modified cofactor-inserted [FeFe] hydrogenase Hhyd state. Inset: schematic structure of the modified diiron cofactor. Figure reproduced from previous work.(Reijerse et al., 2017)
These observations, coupled to concomitant work on the natural cofactor in a mutant [FeFe] hydrogenase, (Mulder et al., 2017) provided an unambiguous determination of the Fe-H bonding moiety in the Hhyd state. This work pushes the limits of the technique as hydridic modes normally involve very little Fe motion. Consequently, another frontier for 57Fe NRVS will be the observation of higher energy hydride/deuteride stretching vibrational modes in a biological system.
5. Perspective
Here we have discussed the NRVS spectral acquisition and analysis of bioinorganic FeS systems. Besides rubredoxin (Guo et al., 2012; Xiao et al., 2005); non-Rieske [2Fe2S](Xiao et al., 2008); [3Fe4S](Lauterbach et al., 2016); and [4Fe4S](Mitra et al., 2011) proteins, it is important to note that 57Fe NRVS has been expanded to study electron transfer and catalysis in larger enzyme metallocofactors such as (but not comprehensively) the NiFe site in [NiFe] hydrogenase,(Kamali et al., 2013; Lauterbach et al., 2015; Hideaki Ogata et al., 2015) the diiron site in [FeFe] hydrogenase,(Gilbert-Wilson et al., 2015; Kuchenreuther et al., 2013) the P [8Fe7S]- and M [7Fe9SMo]- clusters of Mo-Nitrogenase,(George et al., 2012; Mitra et al., 2013; Scott et al., 2014; Xiao et al., 2006) heme systems,(Leu et al., 2009; Sage et al., 2001) non-heme systems,(Sutherlin et al., 2016; Wong et al., 2013) and nitric-oxide sensing [4Fe4S] clusters.(Serrano et al., 2016) Further as the systems of interest have become more complicated, with multiple FeS clusters of differing composition, innovative strategies of site-specific 57Fe labeling have been implemented that target maturation or a modified cofactor insertion. (Gilbert-Wilson et al., 2015; Reijerse et al., 2017)
Since its inception over two decades ago, improvements in synchrotron flux, resolution, and sample preparation have allowed for the NRVS investigations of weaker features and more dilute biological samples. The technique is continuing to mature and is solving questions in ways that resonance Raman and infrared spectroscopies cannot. The continued advancement of synchrotron technologies and the development of X-ray free electron lasers ensure that NRVS will become a more prolific tool to solve complex questions in bioinorganic systems.
6. References
- Bau R, Rees DC, Kurtz DM Jr., Scott RA, Huang H, Adams MWW, & Eidsness MK (1998). Crystal structure of rubredoxin from Pyrococcus furiosus at 0.95 Å resolution, and the structures of N-terminal methionine and formylmethionine variants of Pf Rd. Contributions of N-terminal interactions to thermostability. JBIC Journal of Biological Inorganic Chemistry, 3(5), 484–493. doi: 10.1007/s007750050258 [DOI] [Google Scholar]
- Day MW, Hsu BT, Joshua-Tor L, Park JB, Zhou ZH, Adams MW, & Rees DC (1992). X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus. Protein Sci, 1(11), 1494–1507. doi: 10.1002/pro.5560011111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowty E (1987). Fully automated microcomputer calculation of vibrational spectra. Physics and Chemistry of Minerals, 14(1), 67–79. doi: 10.1007/bf00311150 [DOI] [Google Scholar]
- Reijerse EJ, P. CC, Pelmenschikov V, Gilbert-Wilson R, Adamska-Venkatesh A, Siebela JF , Gee LB, Yoda Y, Tamasaku K, Lubitz W, Rauchfuss TB, and Cramer SP. (2016). Direct Observation of an Iron-bound Terminal Hydride in [FeFe]-Hydrogenase by Nuclear Resonance Vibrational Spectroscopy. [DOI] [PMC free article] [PubMed]
- George SJ, Barney BM, Mitra D, Igarashi RY, Guo YS, Dean DR, … Seefeldt LC (2012). EXAFS and NRVS Reveal a Conformational Distortion of the FeMo-cofactor in the MoFe Nitrogenase Propargyl Alcohol Complex. Journal of inorganic biochemistry, 112, 85–92. doi:DOI 10.1016/j.jinorgbio.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert-Wilson R, Siebel JF, Adamska-Venkatesh A, Pham CC, Reijerse E, Wang H, … Rauchfuss TB (2015). Spectroscopic Investigations of [FeFe] Hydrogenase Maturated with [57Fe2(adt)(CN)2(CO)4]2–. J Am Chem Soc, 137(28), 8998–9005. doi: 10.1021/jacs.5b03270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Brecht E, Aznavour K, Nix JC, Xiao Y, Wang H, … Cramer SP (2012). Nuclear resonance vibrational spectroscopy (NRVS) of rubredoxin and MoFe protein crystals. Hyperfine Interactions, 222(S2), 77–90. doi: 10.1007/s10751-012-0643-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroshi S, Takeo I, Kaori W, Akio U, & Tatsuhiko Y (1991). Resonance Raman Active Vibrations of Rubredoxin. Normal Coordinate Analysis of a 423-Atom Model. Bulletin of the Chemical Society of Japan, 64(3), 829–836. doi: 10.1246/bcsj.64.829 [DOI] [Google Scholar]
- Johnson DW, & Spence JCH (1974). Determination of the single-scattering probability distribution from plural-scattering data. Journal of Physics D: Applied …, 771. [Google Scholar]
- Kaesz HD, & Saillant RB (1972). Hydride complexes of the transition metals. Chemical Reviews, 72(3), 231–281. doi: 10.1021/cr60277a003 [DOI] [Google Scholar]
- Kamali S, Wang H, Mitra D, Ogata H, Lubitz W, Manor BC, … Cramer SP (2013). Observation of the Fe-CN and Fe-CO vibrations in the active site of [NiFe] hydrogenase by nuclear resonance vibrational spectroscopy. Angew Chem Int Ed Engl, 52(2), 724–728. doi: 10.1002/anie.201204616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchenreuther JM, Guo YS, Wang HX, Myers WK, George SJ, Boyke CA, … Cramer SP (2013). Nuclear Resonance Vibrational Spectroscopy and Electron Paramagnetic Resonance Spectroscopy of Fe-57-Enriched [FeFe] Hydrogenase Indicate Stepwise Assembly of the H-Cluster. Biochemistry, 52(5), 818–826. doi:Doi 10.1021/Bi301336r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbach L, Gee LB, Pelmenschikov V, Jenney FE, Kamali S, Yoda Y, … Cramer SP (2016). Characterization of the [3Fe-4S]0/1+ cluster from the D14C variant of Pyrococcus furiosus ferredoxin via combined NRVS and DFT analyses. Dalton Transactions, 45(17), 7215–7219. doi: 10.1039/C5DT04760A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbach L, Wang H, Horch M, Gee LB, Yoda Y, Tanaka Y, … Cramer SP (2015). Nuclear resonance vibrational spectroscopy reveals the {FeS} cluster composition and active site vibrational properties of an O 2 -tolerant {NAD} $\mathplus$ -reducing [{NiFe}] hydrogenase. Chem. Sci, 6(2), 1055––1060. doi: 10.1039/c4sc02982h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu BM, Ching TH, Zhao JY, Sturhahn W, Alp EE, & Sage JT (2009). Vibrational Dynamics of Iron in Cytochrome c. Journal of Physical Chemistry B, 113(7), 2193–2200. doi:Doi 10.1021/Jp806574t [DOI] [PubMed] [Google Scholar]
- Mitra D, George SJ, Guo YS, Kamali S, Keable S, Peters JW, … Cramer SP (2013). Characterization of [4Fe-4S] Cluster Vibrations and Structure in Nitrogenase Fe Protein at Three Oxidation Levels via Combined NRVS, EXAFS, and DFT Analyses. J Am Chem Soc, 135(7), 2530–2543. doi:Doi 10.1021/Ja307027n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra D, Pelmenschikov V, Guo Y, Case DA, Wang H, Dong W, … Cramer SP (2011). Dynamics of the [4Fe-4S] cluster in Pyrococcus furiosus D14C ferredoxin via nuclear resonance vibrational and resonance Raman spectroscopies, force field simulations, and density functional theory calculations. Biochemistry, 50(23), 5220–5235. doi: 10.1021/bi200046p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mössbauer R (1958a). Kernresonanzabsorption von Gammastrahlung in Ir191. Naturwissenschaften, 45(22), 538–539. doi: 10.1007/bf00632050 [DOI] [Google Scholar]
- Mössbauer R (1958b). Kernresonanzfluoreszenz von Gammastrahlung in Ir191. Zeitschrift für Physik, 151(2), 124–143. doi: 10.1007/BF01344210 [DOI] [Google Scholar]
- Mössbauer R (1959). Kernresonanzabsorption von γ-Strahlung in Ir191 Zeitschrift für Naturforschung A (Vol. 14, pp. 211). [Google Scholar]
- Mulder DW, Guo Y, Ratzloff MW, & King PW (2017). Identification of a Catalytic Iron-Hydride at the H-Cluster of [FeFe]-Hydrogenase. J Am Chem Soc, 139(1), 83–86. doi: 10.1021/jacs.6b11409 [DOI] [PubMed] [Google Scholar]
- Ogata H, Kramer T, Wang H, Schilter D, Pelmenschikov V, van Gastel M, … Cramer SP (2015). Hydride bridge in [NiFe]-hydrogenase observed by nuclear resonance vibrational spectroscopy. Nature Communications, 6. doi: 10.1038/ncomms8890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Nishikawa K, & Lubitz W (2015). Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase. Nature, 520(7548), 571–574. doi: 10.1038/nature14110 [DOI] [PubMed] [Google Scholar]
- Petrenko T, Sturhahn W, & Neese F (2008). First-principles calculation of nuclear resonance vibrational spectra. Hyperfine Interactions, 175(1–3), 165–174. doi: 10.1007/s10751-008-9600-5 [DOI] [Google Scholar]
- Reijerse EJ, Pham CC, Pelmenschikov V, Gilbert-Wilson R, Adamska-Venkatesh A, Siebel JF, … Cramer SP (2017). Direct Observation of an Iron-Bound Terminal Hydride in [FeFe]-Hydrogenase by Nuclear Resonance Vibrational Spectroscopy. J Am Chem Soc. doi: 10.1021/jacs.7b00686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E (1989). Kinetic deuterium isotope effects in transition metal hydride clusters. Polyhedron, 8(4), 383–405. doi: 10.1016/S0277-5387(00)80733-0 [DOI] [Google Scholar]
- Sage J, Durbin S, Sturhahn W, Wharton D, Champion P, Hession P, … Alp E (2001). Long-Range Reactive Dynamics in Myoglobin. Physical Review Letters, 86(21), 4966–4969. doi: 10.1103/PhysRevLett.86.4966 [DOI] [PubMed] [Google Scholar]
- Scott A, Pelmenschikov V, Guo Y, Wang H, Yan L, George S, … Cramer SP (2014). J. Am. Chem. Soc, 136, 15942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano PN, Wang H, Crack JC, Prior C, Hutchings MI, Thomson AJ, … Cramer SP (2016). Nitrosylation of Nitric-Oxide-Sensing Regulatory Proteins Containing [4Fe-4S] Clusters Gives Rise to Multiple Iron–Nitrosyl Complexes. Angewandte Chemie International Edition, 55(47), 14575–14579. doi: 10.1002/anie.201607033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto M, Yoda Y, Kikuta S, Zhang X, & Ando M (1995). Observation of Nuclear Resonant Scattering Accompanied by Phonon Excitation Using Synchrotron Radiation. Physical Review Letters, 74(19), 3828–3831. doi: 10.1103/PhysRevLett.74.3828 [DOI] [PubMed] [Google Scholar]
- Singwi KS, & Sjölander A (1960). Resonance Absorption of Nuclear Gamma Rays and the Dynamics of Atomic Motions. Physical Review, 120(4), 1093–1102. [Google Scholar]
- Smith MC, Xiao Y, Wang H, George SJ, Coucouvanis D, Koutmos M, … Cramer SP (2005). Normal-Mode Analysis of FeCl4- and Fe2S2Cl42- via Vibrational Mössbauer, Resonance Raman, and FT-IR Spectroscopies. Inorganic Chemistry, 44(16), 5562–5570. doi: 10.1021/ic0482584 [DOI] [PubMed] [Google Scholar]
- Sturhahn W (2000). CONUSS and PHOENIX : Evaluation of nuclear resonant scattering data. 125, 149–172. [Google Scholar]
- Sturhahn W, Toellner T, Alp E, Zhang X, Ando M, Yoda Y, … Dabrowski B (1995). Phonon Density of States Measured by Inelastic Nuclear Resonant Scattering. Physical Review Letters, 74(19), 3832–3835. doi: 10.1103/PhysRevLett.74.3832 [DOI] [PubMed] [Google Scholar]
- Sutherlin KD, Liu LV, Lee Y-M, Kwak Y, Yoda Y, Saito M, … Solomon EI (2016). Nuclear Resonance Vibrational Spectroscopic Definition of Peroxy Intermediates in Nonheme Iron Sites. J Am Chem Soc, 138(43), 14294–14302. doi: 10.1021/jacs.6b07227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher WM (1960). Study of lattice vibrations by resonance absorption of nuclear gamma rays. Annals of Physics, 9(2), 194–210. doi: 10.1016/0003-4916(60)90028-2 [DOI] [Google Scholar]
- Wang H, Alp EE, Yoda Y, & Cramer SP (2014). A Practical Guide for Nuclear Resonance Vibrational Spectroscopy (NRVS) of Biochemical Samples and Model Compounds In Fontecilla-Camps CJ & Nicolet Y (Eds.), Metalloproteins: Methods and Protocols (pp. 125–137). Totowa, NJ: Humana Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yoda Y, Kamali S, Zhou ZH, & Cramer SP (2012). Real sample temperature: a critical issue in the experiments of nuclear resonant vibrational spectroscopy on biological samples. Journal of synchrotron radiation, 19(Pt 2), 257–263. doi: 10.1107/S0909049512001380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SD, Srnec M, Matthews ML, Liu LV, Kwak Y, Park K, … Solomon EI (2013). Elucidation of the Fe(IV)=O intermediate in the catalytic cycle of the halogenase SyrB2. Nature, 499(7458), 320–323. doi: 10.1038/nature12304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Smith MC, Newton W, Case DA, George S, Wang H, … Cramer S (2006). How Nitrogenase Shakes – Initial Information about P-Cluster and FeMo-cofactor Normal Modes from Nuclear Resonance Vibrational Spectroscopy (NRVS). J Am Chem Soc, 128(23), 7608–7612. doi: 10.1021/ja0603655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Tan ML, Ichiye T, Wang H, Guo Y, Smith MC, … Cramer SP (2008). Dynamics of Rhodobacter capsulatus [2FE-2S] ferredoxin VI and Aquifex aeolicus ferredoxin 5 via nuclear resonance vibrational spectroscopy (NRVS) and resonance Raman spectroscopy. Biochemistry, 47(25), 6612–6627. doi: 10.1021/bi701433m [DOI] [PubMed] [Google Scholar]
- Xiao Y, Wang H, George SJ, Smith MC, Adams MW, Jenney FE Jr., … Cramer SP (2005). Normal mode analysis of Pyrococcus furiosus rubredoxin via nuclear resonance vibrational spectroscopy (NRVS) and resonance raman spectroscopy. J Am Chem Soc, 127(42), 14596–14606. doi: 10.1021/ja042960h [DOI] [PubMed] [Google Scholar]