Abstract
The hepatitis B virus (HBV) is an important global human pathogen and represents a major cause of hepatitis, liver cirrhosis and liver cancer. The HBV capsid is composed of multiple copies of a single viral protein, the capsid or core protein (HBc), plays multiple roles in the viral life cycle, and has emerged recently as a major target for developing antiviral therapies against HBV infection. Although several systems have been developed to study HBV capsid assembly, including heterologous overexpression systems like bacteria and insect cells, in vitro assembly using purified protein, and mammalian cell culture systems, the requirement for non-physiological concentrations of HBc and salts and the difficulty in manipulating host regulators of assembly presents major limitations for detailed studies on capsid assembly under physiologically relevant conditions. We have recently developed a mammalian cell-free system based on the rabbit reticulocyte lysate (RRL), in which HBc is expressed at physiological concentrations and assembles into capsids under near-physiological conditions. This system has already revealed HBc assembly requirements that are not anticipated based on previous assembly systems. Furthermore, capsid assembly in this system is regulated by endogenous host factors that can be readily manipulated. Here we present a detailed protocol for this cell-free capsid assembly system, including an illustration on how to manipulate host factors that regulate assembly.
Keywords: Hepatitis B virus, Cell-free capsid assembly system, Rabbit reticulocyte lysate, Phosphorylation, RNA binding
Background
The hepatitis B virus (HBV) is an important global human pathogen that chronically infects hundreds of millions of people worldwide and represents a major cause of viral hepatitis, liver cirrhosis, and liver cancer ( Seeger et al., 2013 ; Trepo et al., 2014 ). HBV replicates its genomic DNA, a relaxed circular, partially duplex DNA (RC DNA), via reverse transcription of an RNA intermediate, the so-called pregenomic RNA (pgRNA), within a nucleocapsid (NC) (Summers and Mason, 1982; Hu and Seeger, 2015; Hu, 2016), which packages a copy of pgRNA together with the virally encoded reverse transcriptase (RT) protein (Bartenschlager and Schaller, 1992; Hu and Lin, 2009). It is within NCs that the RT converts pgRNA into RC DNA.
The icosahedral HBV capsid shell consists of multiple copies of a single viral protein, the HBV core (capsid) protein (HBc). HBc is composed of an N-terminal domain (NTD, aa 1-140) and a C-terminal domain (CTD, 150-183 or 185 depending on strains), which are connected by a linker region (141-149). In heterologous overexpression systems including bacteria and insect cells and in vitro assembly reactions using high concentrations of purified HBc and/or salt, NTD alone, without CTD, is sufficient for capsid assembly; thus it is also called the assembly domain ( Gallina et al., 1989 ; Birnbaum and Nassal, 1990; Lanford and Notvall, 1990; Wingfield et al., 1995 ). Though not required for capsid assembly in these systems, the highly basic and arginine-rich CTD shows non-specific nucleic acid binding activities ( Hatton et al., 1992 ) and plays important roles in viral RNA packaging (Nassal, 1992), DNA synthesis (Nassal, 1992; Yu and Summers, 1994), and nuclear import of capsids (Liao and Ou, 1995; Liu et al., 2015 ), all of which is further regulated by the dynamic phosphorylation state of CTD controlled by host factors (Kann and Gerlich, 1994; Kann et al., 1999 ; Daub et al., 2002 ; Ludgate et al., 2012 ; Liu et al., 2015 ). Whether CTD, and its state of phosphorylation, play a role in capsid assembly under physiological conditions remained unclear.
To study HBV capsid assembly under more physiological conditions, we have recently developed a mammalian cell-free assembly system based on the commonly used mammalian cell extract, rabbit reticulocyte lysate (RRL), in which HBc is expressed at physiological (low) concentrations (25-50 nM, monomer) and assembles into capsids under near-physiological conditions ( Ludgate et al., 2016 ). This system allowed us to reveal an unexpected role of CTD in capsid assembly, which is further subjected to regulation by the state of CTD phosphorylation as controlled by endogenous host factors ( Ludgate et al., 2016 ). This protocol is adapted from Ludgate et al. (2016) and more detailed information on this cell-free capsid assembly system is included, and different treatments are applied to address the roles of viral and host factors, such as RNA-binding activities of CTD and host phosphatases, in HBV capsid assembly under near-physiological conditions. This protocol will facilitate detailed studies on capsid assembly and host regulation under physiological conditions and identification of novel antiviral agents targeting HBc.
Materials and Reagents
Pipette tips (Denville Scientific, catalog numbers: P1096-FR, P1121, P1122, P1126)
1.5 ml microcentrifuge tube (Denville Scientific, catalog number: C2170)
Gloves and lab coat (Denville Scientific, catalog number: G4162; Medline Industries, catalog number: 83044QHW)
Proteinase K, RNA grade (Thermo Fisher Scientific, InvitrogenTM, catalog number: 25530049)
Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L4509-1KG)
Phenol solution (Sigma-Aldrich, catalog number: P4557)
Chloroform (Fisher Scientific, catalog number: BP1145-1)
Sodium acetate (Sigma-Aldrich, catalog number: S2889-1KG)
Ethyl alcohol (EtOH) (AmericanBio, catalog number: AB00515-00100)
UltraPureTM DNase/RNase-Free Distilled Water (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10977015)
Agarose (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15510027)
TNT® Coupled Rabbit Reticulocyte Lysate (RRL) (Promega, catalog number: L4610)
EasyTagTM L-[35S]-Methionine (PerkinElmer, catalog number: NEG709A001MC)
RNasin® Plus Ribonuclease Inhibitor (Promega, catalog number: N2611)
RNaseZapTM RNase Decontamination Solution (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9780)
RNase A, DNase and protease-free (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EN0531)
NEBuffer 3 (New England Biolabs, catalog number: B7003S)
Alkaline Phosphatase, Calf Intestinal (CIAP) (New England Biolabs, catalog number: M0290S)
Tris Base (Fisher Scientific, catalog number: BP152-10)
Ethylenediaminetetraacetic acid, EDTA (Sigma-Aldrich, catalog number: E5134)
Sodium fluoride (Sigma-Aldrich, catalog number: S7920)
Sodium pyrophosphate tetrabasic decahydrate (Sigma-Aldrich, catalog number: S6422)
β-Glycerophosphate (Sigma-Aldrich, catalog number: G6251)
Sodium orthovanadate (Sigma-Aldrich, catalog number: S6508)
cOmpleteTM, EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics, catalog number: 04693132001)
TE buffer (see Recipes)
10x phosphatase inhibitors (PPI) (see Recipes)
25x protease inhibitor (see Recipes)
Equipment
Pipettes (Gilson, P1000, P200, P20, P2)
Fume hood (e.g., Protector Xstream Laboratory Hood, Labconco)
30 °C/37 °C Oven (SciGene, Robbins Scientific, model: Model 400)
Microcentrifuge (Fisher Scientific, FisherbrandTM, model: accuSpinTM Micro 17)
Spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM1000)
Procedure
-
Make RNase-free plasmids for in vitro translation in rabbit reticulocyte lysate
-
Treat plasmid DNA with Proteinase K and SDS to remove any contaminating RNases in plasmid DNA. As shown in the table below, incubate HBc-expressing plasmids with SDS (final concentration 0.6% w/v) and Proteinase K (final concentration 0.4 µg/µl) at 50 °C for 30 min.
Component volume for 50 µg of the construct TE buffer (µl) Add to adjust final volume to 200 µl 20% SDS (µl) 6 µl 20 mg/ml Proteinase K (µl) 4 µl Total Vol. (µl) 200 µl Notes:
Always dilute 20% of SDS in the TE buffer first before adding Proteinase K.
Any plasmid vector containing a bacteriophage promoter (T7 in the example below) that can be used for in vitro transcription using the cognate phage RNA polymerase can be used for coupled transcription and translation in this system.
Add an equal volume (200 µl) of phenol to the digestion reaction, mix well and spin at the maximal speed (17,000 × g) for 5 min at room temperature. Transfer the aqueous (top) layer to a new tube and repeat this step.
Add an equal volume of chloroform and extract once as described in Step A2.
Measure the aqueous solution and add an appropriate volume of 3 M of sodium acetate (final concentration at 0.3 M) and 100% of ethanol (final concentration at 70%), mix well and incubate at -20 °C for 30 min. Centrifuge at the maximal speed (17,000 × g) for 20 min at 4 °C and wash the pellet 3 times each with 1 ml of 70% ethanol, with spinning at the maximal speed for 10 min at 4 °C.
Resuspend DNA with 30 µl of RNase-free water. Measure DNA concentration with a spectrophotometer (NanoDropTM 1000) and adjust the DNA concentration to 1 µg/µl and then verify DNA integrity by agarose gel electrophoresis.
-
-
In vitro translation in rabbit reticulocyte lysate
A TNT® Coupled Rabbit Reticulocyte Lysate (RRL) in vitro translation system (Promega) is used to express the HBc protein as recommended by the manufacturer.
-
Preparation of the translation kit
Put TNT® RNA Polymerase (T7) on ice.
Quickly thaw the TNT® Rabbit Reticulocyte Lysate by hand-warming and immediately put on ice.
Thaw the other components at RT (25 °C) and put on ice.
Notes:
The TNT® Reaction Buffer may contain a precipitate after thawing and sitting on ice. Redissolve the precipitate by vortexing at room temperature for 30 sec.
Do not freeze-thaw the lysate more than twice.
Any unused lysate should be refrozen in a dry ice/ethanol bath as soon as possible after thawing to minimize loss of translational activity.
-
Assemble the reaction components in a 1.5 ml microcentrifuge tube following the table below.
Component Reaction with [35S]methionine Reaction without [35S]methionine TNT® Rabbit Reticulocyte Lysate 50 µl 50 µl TNT® Reaction Buffer 4 µl 4 µl TNT® RNA Polymerase (T7) 2 µl 2 µl Amino Acid Mixture, Minus Leucine, 1 mM - 1 µl Amino Acid Mixture, Minus Methionine, 1 mM 2 µl 1 µl [35S]methionine (> 1,000 Ci/mmol at 10 mCi/ml) 4 µl - RNasin® Ribonuclease Inhibitor (40 U/μl) 2 µl 2 µl DNA template(s) (1 μg/μl) 2 µl 2 µl Nuclease-free dH2O 34 µl 38 µl Total reaction volume 100 µl 100 µl Note: Small-scale reactions may be performed by reducing all components proportionally.
Incubate reaction at 30 °C for 90 min.
-
Put the reactions on ice and proceed to the assembly steps or store the reactions at -80 °C for future use.
Notes:
Except for the actual translation incubation, all handling of the lysate components should be done at 4 °C or on ice.
-
A ribonuclease-free environment is required during all of the operations:
Add RNasin® Ribonuclease Inhibitor to all TNT® Lysate reactions to prevent RNA degradation.
Microcentrifuge tubes and pipette tips should be RNase-free.
Use a cleaning agent (such as RNaseZAPTM) to clean the bench and gloves to remove RNase.
-
-
In vitro capsid assembly in RRL
Our recent studies have shown that both HBc NTD and CTD are required for HBV capsid assembly at physiologically low HBc concentrations translated in RRL ( Ludgate et al., 2016 ). CTD likely facilitates assembly under these conditions via RNA binding and protein-protein interactions. Moreover, CTD is subject to phosphorylation and dephosphorylation by endogenous host kinase(s) and phosphatase(s) in RRL, which further regulates capsid assembly.
To confirm the roles of CTD in capsid assembly in RRL, especially its RNA binding activity and regulation of assembly by phosphorylation state, the following treatments, as shown in Figure 1, are applied to the HBc in vitro assembly assay in RRL.
-
Mock incubation
-
Assemble the reaction components in a 1.5 ml microcentrifuge tube following the table below.
Component Volume NEBuffer 3 (10x) 1 µl 25x RNase-free PI (25x) 0.4 µl RNasin RNase Inhibitor (40 U/µl) 0.2 µl RRL translation reactions (µl) 3 µl Nuclease-free dH2O (µl) 5.4 µl Total (µl) 10 µl Note: PI, a mixture of protease inhibitors (see Recipes below).
Incubate the reaction at 37 °C for 16 h.
Put the reactions on ice and proceed to the analysis steps or store the reactions at -80 °C for future use.
-
-
CIAP incubation
-
Assemble the reaction components in a 1.5 ml microcentrifuge tube following the table below.
Component Volume NEBuffer 3 (10x) 1 µl 25x RNase-free PI (25x) 0.4 µl RNasin RNase Inhibitor (40 U/µl) 0.2 µl RRL translation reactions (µl) 3 µl Nuclease-free dH2O (µl) 5.4 µl CIAP (10 U/µl) 1 µl Total (µl) 10 µl Incubate the reaction at 37 °C for 16 h.
Put the reactions on ice and proceed to the analysis steps or store the reactions at -80 °C for future use.
-
-
CIAP followed by RNase incubation
-
Assemble the reaction components in a 1.5 ml microcentrifuge tube following the table below.
Component Volume NEBuffer 3 (10x) 1 µl 25x RNase-free PI (25x) 0.4 µl RNasin RNase Inhibitor (40 U/µl) 0.2 µl RRL translation reactions (µl) 3 µl Nuclease-free dH2O (µl) 4.4 µl CIAP (10 U/µl) 1 µl Total (µl) 10 µl Incubate the reaction at 37 °C for 16 h.
Add 0.1 µl of RNase A (10 mg/ml) to the assembly reaction and incubate at 37 °C for another 1 h.
Put the reactions on ice and proceed to the analysis steps or store the reactions at -80 °C for future use.
-
-
RNase followed by CIAP incubation
-
Assemble the reaction components in a 1.5 ml microcentrifuge tube following the table below.
Component Volume NEBuffer 3 (10x) 1 µl 25x RNase-free PI (25x) 0.4 µl RRL translation reactions (µl) 3 µl Nuclease-free dH2O (µl) 5.5 µl RNase A (10 mg/ml) 0.1 µl Total (µl) 10 µl Incubate the reaction at 37 °C for 1 h.
Add 1 µl of CIAP (10 U/µl) to the assembly reaction and incubate at 37 °C for another 16 h.
Put the reactions on ice and proceed to the analysis steps or store the reactions at -80 °C for future use.
-
-
PPI incubation
-
Assemble the reaction components in a 1.5 ml microcentrifuge tube following the table below.
Component Volume NEBuffer 3 (10x) 1 µl 25x RNase-free PI (25x) 0.4 µl RNasin RNase Inhibitor (40 U/µl) 0.2 µl RRL translation reactions (µl) 3 µl Nuclease-free dH2O (µl) 4.4 µl PPI (10x) 1 µl Total (µl) 10 µl Incubate the reaction at 37 °C for 1 h.
Put the reactions on ice and proceed to the analysis steps or store the reactions at -80 °C for future use.
-
-
Figure 1. Conditions for in vitro capsid assembly in RRL.
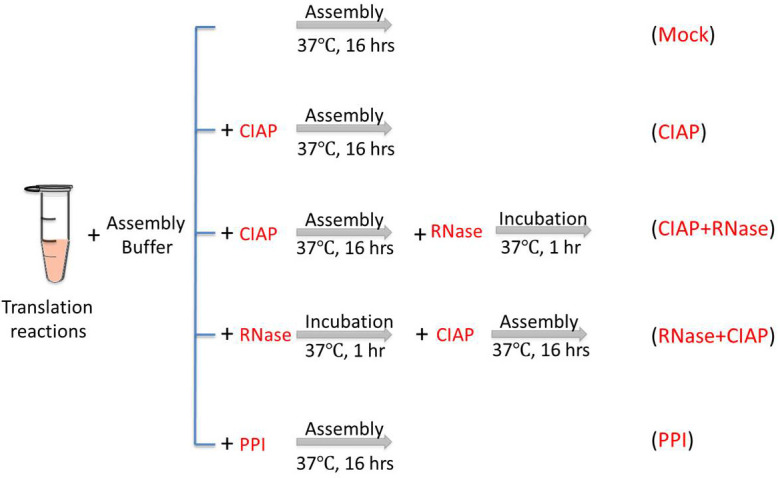
Mock, the reaction is incubated at 37 °C for 16 h without any treatment. CIAP, CIAP (10 U/µl) is added in the reaction during the assembly incubation. CIAP + RNase, RNase A (0.1 mg/ml) is added and incubated for another 1 h after the CIAP incubation. RNase + CIAP, the reaction is treated with RNase A before the CIAP incubation. PPI, PPI (1x) is added during the assembly incubation. PPI, a mixture of phosphatase inhibitors (see Recipes below), CIAP, calf intestinal alkaline phosphatase.
Notes
All work with [35S]methionine should be done with appropriate precautions and personal protective equipment like gloves and lab coat during all the operations. It is recommended to open the stock in a fume hood as [35S]methionine is volatile. Tools and equipment, such as PIPETMAN, centrifuge and incubators should be checked for contamination after using [35S]methionine.
Data analysis
The expression of HBc and capsid assembly in this cell-free system can be examined using SDS-PAGE, agarose gel electrophoresis, and Western blot, as shown in Figure 2 and Ludgate et al., 2016 .
Figure 2. HBV capsid assembly in RRL under different conditions.
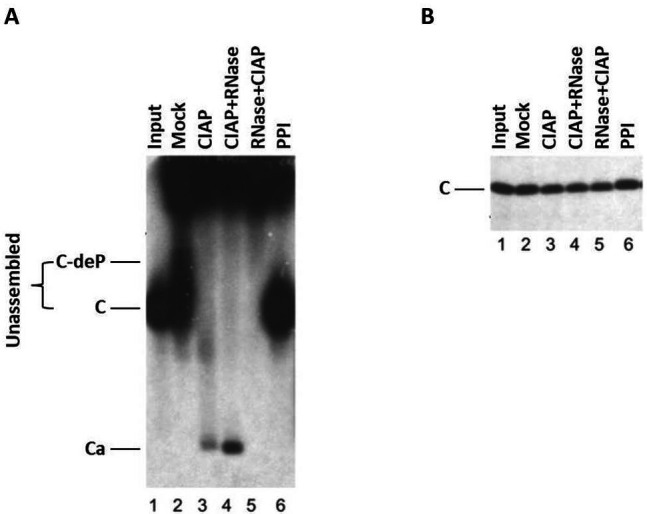
The WT HBc proteins were translated in RRL, and the translation reaction mixtures were resolved by agarose gel (1%) electrophoresis (A) or SDS-PAGE (12.5%) (B) without any further treatment (Input) (lane 1) or were treated with NEBuffer 3 alone overnight at 37 °C (Mock) (lane 2), with NEBuffer 3 plus CIAP overnight at 37 °C (CIAP) (lane 3), with NEBuffer 3 plus CIAP overnight at 37 °C followed by RNase treatment for one additional hour (CIAP + RNase) (lane 4), with RNase for 1 h followed by NEBuffer 3 plus CIAP overnight at 37 °C (RNase + CIAP) (lane 5), or with the mixture of phosphatase inhibitors overnight at 37 °C (PPI) (lane 6). All lanes contained 2 µl translation products. 35S-labeled HBc proteins were detected by autoradiography. C, HBc subunits; C-deP, dephosphorylated HBc subunits; Ca, capsids. (Adapted from Ludgate et al., 2016 )
Recipes
-
TE buffer
10 mM Tris-HCl pH 8.0
1 mM EDTA
-
10x phosphatase inhibitors (PPI)
100 mM sodium fluoride
500 mM β-glycerophosphate
100 mM sodium pyrophosphate
20 mM sodium orthovanadate
-
25x protease inhibitor
1 tablet of cOmpleteTM EDTA-free Protease Inhibitor Cocktail dissolved in 2 ml of DNase/RNase free dH2O
Acknowledgments
This protocol is adapted from Ludgate et al., 2016 . This work was supported by a Public Health Service grant (R01 AI043453 to J.H.) from the National Institutes of Health. K. L. is supported by a Scientific Research Staring Foundation (15042169-Y) from Zhejiang Sci-Tech University. The authors declare no conflicts of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Bartenschlager R. and Schaller H.(1992). Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J 11(9): 3413-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Birnbaum F. and Nassal M.(1990). Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol 64(7): 3319-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daub H., Blencke S., Habenberger P., Kurtenbach A., Dennenmoser J., Wissing J., Ullrich A. and Cotton M.(2002). Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J Virol 76: 8124-8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gallina A., Bonelli F., Zentilin L., Rindi G., Muttini M. and Milanesi G.(1989). A recombinant hepatitis B core antigen polypeptide with the protamine-like domain deleted self-assembles into capsid particles but fails to bind nucleic acids. J Virol 63: 4645-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hatton T., Zhou S. and Standring D.(1992). RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their role in viral replication. J Virol 66: 5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu J.(2016). In: Hepatitis B virus in human diseases. In: Liaw, Y. F. and Zoulim, F.(Eds.). Humana Press, Springer; Chap. 1, pp: 1-34. [Google Scholar]
- 7. Hu J. and Lin L.(2009). RNA-protein interactions in hepadnavirus reverse transcription. Front Biosci(Landmark Ed) 14: 1606-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu J. and Seeger C.(2015). Hepadnavirus genome replication and persistence. Cold Spring Harb Perspect Med 5(7): a021386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kann M. and Gerlich W. H.(1994). Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J Virol 68: 7993-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kann M., Sodeik B., Vlachou A., Gerlich W. H. and Helenius A.(1999). Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J Cell Biol 145: 45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lanford R. E. and Notvall L.(1990). Expression of hepatitis B virus core and precore antigens in insect cells and characterization of a core-associated kinase activity. Virology 176(1): 222-233. [DOI] [PubMed] [Google Scholar]
- 12. Liao W. and Ou J. H.(1995). Phosphorylation and nuclear localization of the hepatitis B virus core protein: significance of serine in the three repeated SPRRR motifs. J Virol 69: 1025-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu K., Ludgate L., Yuan Z. and Hu J.(2015). Regulation of multiple stages of hepadnavirus replication by the carboxyl-terminal domain of viral core protein in trans. J Virol 89(5): 2918-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ludgate L., Liu K., Luckenbaugh L., Streck N., Eng S., Voitenleitner C., Delaney W. E. 4th and Hu, J. (2016). Cell-free hepatitis B virus capsid assembly dependent on the core protein C-terminal domain and regulated by phosphorylation. J Virol 90: 5830-5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ludgate L., Ning X., Nguyen D. H., Adams C., Mentzer L. and Hu J.(2012). Cyclin-dependent kinase 2 phosphorylates s/t-p sites in the hepadnavirus core protein C-terminal domain and is incorporated into viral capsids. J Virol 86: 12237-12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nassal M.(1992). The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol 66: 4107-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seeger C., Zoulim F., Mason W. S.(2013). In: Fields virology. In: Knipe, D. M. and Howley, P. M.,(Eds.). Lippincott, Williams& Wilkins, Philadelphia 2185-2221. [Google Scholar]
- 18. Summers J. and Mason W. S.(1982). Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell 29(2): 403-415. [DOI] [PubMed] [Google Scholar]
- 19. Trepo C., Chan H. L. and Lok A.(2014). Hepatitis B virus infection. Lancet 384(9959): 2053-2063. [DOI] [PubMed] [Google Scholar]
- 20. Wingfield P. T., Stahl S. J., Williams R. W. and Steven A. C.(1995). Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly . Biochemistry 34: 4919-4932. [DOI] [PubMed] [Google Scholar]
- 21. Yu M. and Summers J.(1994). Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J Virol 68: 4341-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]


