Abstract
Hematopoiesis is an essential and highly regulated biological process that begins with hematopoietic stem cells (HSCs). In healthy organisms, HSCs are responsible for generating a multitude of mature blood cells every day, yet the molecular pathways that instruct HSCs to self-renew and differentiate into post-mitotic blood cells are not fully known. To understand these molecular pathways, we investigated novel genes expressed in hematopoietic-supportive cell lines from the zebrafish (Danio rerio), a model system increasingly utilized to uncover molecular pathways important in the development of other vertebrate species. We performed RNA sequencing of the transcriptome of three stromal cell lines derived from different stages of embryonic and adult zebrafish and identified hundreds of highly expressed transcripts. For our studies, we focused on isthmin 1 (ism1) due to its shared synteny with its human gene ortholog and because it is a secreted protein. To characterize ism1, we performed loss-of-function experiments to identify if mature blood cell production was disrupted. Myeloid and erythroid lineages were visualized and scored with transgenic zebrafish expressing lineage-specific markers. ism1 knockdown led to reduced numbers of neutrophils, macrophages, and erythrocytes. Analysis of clonal methylcellulose assays from ism1 morphants also showed a reduction in total hematopoietic stem and progenitor cells (HSPCs). Overall, we demonstrate that ism1 is required for normal generation of HSPCs and their downstream progeny during zebrafish hematopoiesis. Further investigation into ism1 and its importance in hematopoiesis may elucidate evolutionarily conserved processes in blood formation that can be further investigated for potential clinical utility.
Introduction
Hematopoiesis is an essential cellular process in which hematopoietic stem cells (HSCs) differentiate into the multitude of different cell lineages that comprise mature blood[1–3]. HSCs must self-renew and persist for an organism’s lifespan to replenish the mature, post-mitotic blood cells that are constantly being recycled, ensuring that the system is never depleted[4, 5]. The control of this recycling and replacement of blood cells is regulated by an intricate set of signaling molecules and molecular pathways, many of which are still enigmatic. Improper regulation of hematopoiesis can result in serious diseases such as anemia, thrombocytopenia, neutropenia, and leukemia, so understanding these signaling pathways is of clinical relevance.
In vertebrates, HCSs first arise from hemogenic endothelium located in the floor of the dorsal aorta[6–13]. This occurs at embryonic day (E) 10.5 in mice[6], between E27-40 in humans[14], and between 36–52 hours post fertilization (hpf) in the developing zebrafish embryo[7, 8]. Lineage tracing studies in zebrafish[7] and mice[15, 16] indicate that the HSCs that arise during this time give rise to all hematopoietic cells for the organism’s lifespan. Importantly, studies in mice and humans indicate that HSCs don’t directly differentiate into mature blood cells. Instead, they differentiate into populations of restricted hematopoietic stem and progenitor cells (HSPCs); common lymphoid progenitors (CLPs)[17, 18], which eventually produce T, B, and natural killer (NK) cells, and common myeloid progenitors (CMPs)[19, 20] that eventually generate granulocytes, erythrocytes, macrophages, and platelets. Downstream of CMPs are megakaryocyte erythroid progenitors (MEPs) that generate erythrocytes and platelets, and granulocyte macrophage progenitors (GMPs) that generate basophils, eosinophils, neutrophils, and macrophages[19, 20]. Together these HSPCs help maintain the multitude of blood cells in healthy adult organisms.
HSPC differentiation is a developmentally restrictive process, controlled by a multitude of cytokines. These small, extracellular proteins influence HSPCs to self-renew and/or undergo stepwise differentiation into mature blood cell lineages and are secreted in hematopoietic niches, mainly by stromal cells that are found in hematopoietic-supportive tissues and organs (reviewed in [21–23]). These factors then bind to receptors on the surface of HSPCs to mediate a multitude of different downstream cellular responses. Identification and elucidation of the downstream molecular events activated by cytokines is of key interest due to their essential role in hematopoietic regulation; improper differentiation of HSPCs can lead to an accumulation of immature cells, causing the development of lymphoma and leukemia.
To study hematopoiesis and HSPC biology, many laboratories utilize Danio rerio (zebrafish), which have become a promising model system for many reasons (reviewed in [24, 25]). First, they are the phylogenetically lowest vertebrate model system that has a similar circulatory and hematopoietic system to humans, including adaptive immunity (reviewed in [26]). Secondly, zebrafish are transparent and develop ex utero; within 48 hpf functional HSPCs are present[7, 8, 27–30]. Due to the fact that a multitude of hematopoietic-specific fluorescent transgenic zebrafish lines currently exist (reviewed in [24, 31]), HSPCs can be visualized, isolated, and studied in early embryos, a feat not possible in mammals. The fact that zebrafish are fecund, generating hundreds of embryos per clutch, allows large sample sizes and experimental replication. Finally, because zebrafish develop quickly and externally, they are excellent for mutagenesis studies[32–36] and screening compounds that have utility for treating human diseases[37–41]. For all of these reasons, the zebrafish has become a powerful model for studying normal hematopoiesis and its dysregulation during disease.
HSPCs, the cells mutated during the onset of hematopoietic disease, are not easily grown with traditional cell culture techniques because they require specific microenvironments and cytokine signals to keep them proliferating and to encourage their differentiation into mature blood cells. Studies performed in mice[42–45] and humans[46, 47], isolating and growing HSPCs on stromal cell lines, were important for elucidating cytokines and other signaling molecules required for HSPC proliferation and differentiation. The recent generation of hematopoietic-supportive zebrafish stromal cell lines from sites of embryonic[48, 49] and adult[50] hematopoiesis now allows functional testing of HSPCs in zebrafish. Putative hematopoietic-supportive factors expressed in zebrafish cell lines have been identified with RNA sequencing (RNA-seq), comparing the transcriptome of these cells to that of non-hematopoietic supportive zebrafish stroma[48]. Additionally, comparison of their transcriptome to mammalian hematopoietic-supportive stroma allows investigation of transcripts conserved throughout vertebrate evolution[51].
One interesting transcript uniquely upregulated in all hematopoietic-supportive zebrafish cell lines was isthmin 1 (ism1), a gene first identified in the midbrain–hindbrain boundary (MHB) of Xenopus[52]. ism1 is expressed in lymphocytes,[53] bone marrow[54], and in embryonic blood islands[52], and encodes a secreted 60 kDa protein containing a copy of the thrombospondin repeat (TSR) region, a domain involved in cell migration and tissue remodeling[55]. Importantly, it is also expressed in mouse and chicken lateral plate mesoderm, tissue which gives rise to blood in the developing embryo[54]. ism1 is also implicated in angiogenesis; addition of ISM1 protein into matrigel plugs with murine tumors resulted in decreased endothelial capillary networks and decreased overall tumor growth[55]. Additionally, ism1 morphant zebrafish exhibit decreased inter-segmental vessels (ISVs)[55]. Importantly, ism1 levels are increased in response to the upregulation of Wnt signaling[56], implicating its role in early embryonic processes such as cell fate specification, migration, and the beginning of definitive hematopoiesis. Finally, ism1 is co-expressed with fibroblast growth factor ligands that are essential for HSC specification during this developmental period[52]. ism1’s high expression within hematopoietic-supportive stromal cell lines coupled with its expression during development in blood-forming tissues and co-expression with essential hematopoietic factors indicated that ism1 was potentially involved in the formation and modulation of HSPCs.
To understand ism1’s role within developmental hematopoiesis, we performed loss-of-function experiments, which indicate that ism1 morphants have reduced mature erythroid and myeloid cells. Additionally, ism1 morphants show reduced numbers of HSPCs present in developing fish. Overall, these data indicate that ism1 is an important gene for the formation of the embryonic zebrafish hematopoietic system.
Materials and methods
Zebrafish
Wildtype (AB) and transgenic zebrafish lines (gata1a:DsRed[57], mpx:EGFP[58], and kdrl:EGFP[59]) used in these studies were raised and maintained in accordance with California State University, Chico IACUC guidelines. All experiments were approved by the IACUC committee before being performed.
ism1 sequence read counts
All sequenced libraries were processed and analyzed as previously described[48].
Generation of ism1 mRNA
ism1 transcript was amplified from zebrafish kidney cDNA using the following ism1 primers: FWD 5’-ATGGTGCGTCTGGCGGCGGAG-3’ and REV 5’-TCAAAACTCCCGGGCCTCTTCA-3’. ism1 transcript was cloned into a TOPO-TA vector (Invitrogen, Carlsbad CA) and validated by Sanger sequencing. ism1 was than subcloned into pCS2+ and linearized with Not1. ism1 mRNA was generated using a mMessage SP6 kit (Ambion, Austin, TX).
Morpholino and ism1 mRNA injections
ism1 antisense morpholino (MO) was designed against the 5’ untranslated region (UTR) and start codon to prevent translation of ism1 mRNA (Gene Tools, Philomath, OR). The MO sequence is as follows: 5’-CCAGACGCACCATCCTCTTCACC-3’. For microinjection into embryos, a mix of 8 μL of 7.6 mg/mL of ism1 MO was mixed with 0.6 μL of phenol red for a final concentration of 7.0 ng/nL of ism1 MO. 1 μL of this mix was loaded into a needle made with a PM102 micropipette puller (MicroData Instrument, Plainfield, NJ). Single-cell stage embryos were collected, placed onto a 1% agarose microinjection chamber plate with troughs, and injected with 1 nL (7.0 ng) of ism1 MO with a PM 1000 Cell Microinjector (MicroData Instrument, Plainfield, NJ). For rescue injections, phenol red was reduced to 0.2 μL, and 0.4 μL of 44.7 ng/μL ism1 mRNA was added. In this way, rescued embryos received 7.0 ng of ism1 MO and 17.88 ng of ism1 mRNA.
Microscopic visualization of the hematopoietic system
To discern hematopoietic phenotypes and to quantitate cell lineages, florescent microscopy was utilized. Transgenic zebrafish were visualized under a Leica M165C (Leica, Wetzlar, Germany) fluorescent dissecting microscope at time points correlated with the emergence of specific hematopoietic cell lineages. Erythrocytes were visualized at 48 hpf with gata1a:DsRed transgenic animals and further examined by flow cytometry. Neutrophils and macrophages were visualized and individual cells were counted under the microscope at 48 hpf with mpx:GFP transgenic animals. Zebrafish and their fluorescently labeled cells were imaged using a Leica FireCam camera (Leica, Wetzlar, Germany), scored, and enumerated. For myeloid quantitation, images were labeled by a reference number and the numbers of mpx:GFP+ cells were counted in each animal by several undergraduate students to insure no bias in results.
Flow cytometry
To enumerate the percentage of fluorescent cells in an embryo, we used transgenic zebrafish in combination with flow cytometry. 72 hpf transgenic embryos were grouped in samples of three and washed 3x with E3[60]. After the last wash, the E3 was removed, leaving 100 μL. 1 mL of 10 mM dithiothreitol (DTT) in E3 was added, and samples were incubated for 25 mins. Samples were than washed 3x with Dulbecco's phosphate-buffered saline (DPBS) containing Ca2+ and Mg2+. After the last wash, 500 μL of DPBS and 5 μL of 5 mg/mL (26U/mL) Liberase TM (Roche, Upper Bavaria, Germany) were added. Samples were incubated at 37°C on a horizontal orbital shaker at 180 rpm for 60 mins. Samples were than triturated with a P-1000 to ensure proper dissociation and transferred to a 5 mL polystyrene round bottom tube with cell strainer cap. 1 μL of SytoxRed (ThermoFisher Scientific, Waltham, MA) was added to each sample to mark dead cells. Samples were run through a BD Accuri C6 flow cytometer (BD Biosciences, San Jose, CA) and enumerated. Data were analyzed using FloJo software (FloJo LLC, Ashland, Oregon) to quantitate total percentage of positive fluorescent cells.
Quantitation of HSPCs in developing zebrafish embryos
HSPC isolation and culture was performed as previously described[61]. Samples were given carp serum, Gcsf, and Epo to stimulate myeloid and erythroid differentiation[62, 63]. They were incubated at 32°C and 5% CO2 for 7–10 days and imaged with an Olympus IX53 inverted microscope (Olympus, Center Valley, PA) at 40x to enumerate colony forming units (CFUs).
RT-PCR
mRNA was extracted from ZKS, ZEST, CHEST, and whole kidney using a Qiagen RNAeasy kit (Qiagen, Hilden, Germany). To obtain myeloid, lymphoid, and precursor cells, fluorescence-activated cell sorting (FACS) was performed on whole kidney marrow (WKM)[31]; these populations are easily separated based on their size and granularity[57]. cDNA was then generated with the iScript cDNA synthesis kit (Biorad, Hercules, CA), and PCR was performed with Jumpstart ReadyMix Taq (Sigma-Aldrich, St. Louis, MO).
Quantitative RT-PCR (qRT-PCR)
mRNA was extracted from embryos at 48 and 72 hpf using a Qiagen RNAeasy kit (Qiagen, Hilden, Germany). cDNA was then generated with the iScript cDNA synthesis kit (Biorad, Hercules, CA), and PCR was performed with SsoFast SYBR Mastermix (Biorad, Hercules, CA). Fold expression was measured as ΔΔCT using ef1a[28] as a reference gene and whole kidney cDNA as a reference tissue.
Statistics
Relative fold change was done by setting the control as the standard. For triplicates, fold change per experiments were averaged and plotted with standard deviation. To discern statistical difference, data were analyzed using an unpaired two-tailed Student’s T test. * = p<0.05, ** = p<0.0001, and N.S. = no significance.
Results
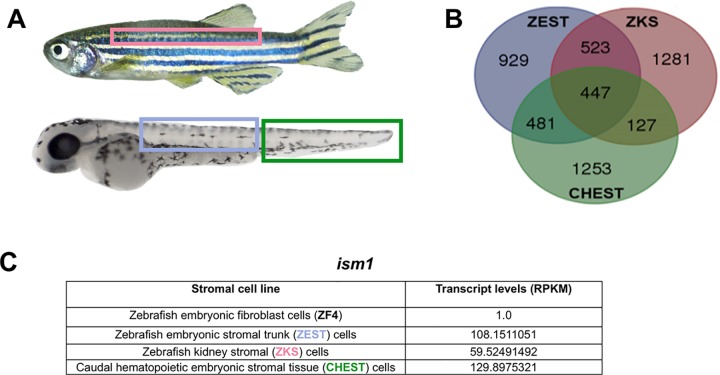
Zebrafish kidney stroma (ZKS) cells[50], zebrafish embryonic stromal trunk (ZEST) cells[48], and caudal hematopoietic embryonic stromal tissue (CHEST) cells[49] are hematopoietic-supportive stromal cell lines isolated and grown from adult and developing zebrafish. These stromal cell lines are derived from different physical locations of adult hematopoietic maintenance[64] (ZKS cells), embryonic HSC emergence[7, 8] (ZEST cells), and embryonic HSC expansion[65] (CHEST cells) at several key time points during hematopoietic development (Fig 1A) and support hematopoietic proliferation and differentiation of all blood cell types in culture[27, 48–50, 66, 67]. Therefore, we hypothesized that these cells would generate and secrete proteins required for hematopoiesis. Total RNA from each stromal cell line was isolated, sequenced, and compared to a non-supportive hematopoietic cell line (zebrafish fibroblast cells; ZF4[68]) to remove housekeeping genes from our analysis. RNA-seq of the supportive hematopoietic stromal cell lines identified 447 shared transcripts (Fig 1B; top genes involved in hematopoiesis are listed in Table 1). Transcripts were analyzed as reads per kilobase of transcript per million reads mapped (RPKM), which normalized and accounted for transcript length. From these transcripts, ism1 was chosen for further research due to its high RPKM levels across all three cell lines (Fig 1C), the fact that it is a secreted protein, and because it has a conserved ortholog in humans. To confirm that ism1 was expressed in ZKS, ZEST, and CHEST stromal cells, we performed RT-PCR for ism1, validating its expression in these tissues (S1 Fig). We also observed ism1 expression in whole kidney, the main site of hematopoiesis in the adult zebrafish[64]. To confirm that ism1 was only expressed in the stromal cells of the kidney, we fractionated cells from WKM using FACS; myeloid, lymphoid, and precursor cell populations can be easily separated based on their size and granularity[57]. RT-PCR analysis of these cells confirmed that ism1 is expressed in the supportive hematopoietic tissues, but not in hematopoietic cells themselves (S1 Fig).
Fig 1. ism1 is highly expressed in hematopoietic-supportive stromal cell lines.
(A) Each stromal cell line is derived from sites of hematopoietic emergence, expansion, and maintenance in adult and embryonic zebrafish; adult zebrafish kidney stroma (ZKS, derived from the adult kidney; pink[50]), zebrafish embryonic stromal trunk (ZEST, derived from the embryonic trunk; blue[48]), and caudal hematopoietic embryonic stromal tissue (CHEST, derived from the CHT; green[49]). (B) Each stromal cell line’s transcriptome was analyzed with RNA-seq and compared to a non-hematopoietic supportive stromal cell line (ZF4); transcripts were plotted as a Venn diagram demonstrating 447 conserved transcripts. (C) ism1 transcript levels for each cell line presented as reads per kilobase of transcript per million reads mapped (RPKM). See Table 1 for genes identified in this screen.
Table 1. Expression levels of highly expressed transcripts in hematopoietic supportive cell lines.
| Gene ID (Zv9) | Gene name | Gene description | RPKM (ZKS cells) | RPKM (ZEST cells) | RPKM (CHEST cells) |
|---|---|---|---|---|---|
| ENSDARG00000056680 | stc2a | Stanniocalcin 2a | 1171.390702 | 1231.42446 | 706.1714178 |
| ENSDARG00000015050 | calm3a | Calmodulin 3a (phosphorylase kinase, delta) | 398.2576181 | 445.6855878 | 969.0665779 |
| ENSDARG00000026726 | anxa1a | Annexin A1a | 386.1286169 | 280.2930981 | 417.0527221 |
| ENSDARG00000071498 | fhl1a | Four and a half LIM domains 1a | 392.740221 | 209.8129055 | 350.3135956 |
| ENSDARG00000055439 | adamtsl7 | ADAMTS-like 7 | 377.7311012 | 296.1991765 | 268.6541787 |
| ENSDARG00000069752 | ckba | Creatine kinase, brain a | 410.942751 | 156.7429014 | 276.7297589 |
| ENSDARG00000016691 | cd9b | CD9 antigen, b | 506.2882968 | 266.2312285 | 66.00918816 |
| ENSDARG00000093440 | tnfaip6 | Tumor necrosis factor, alpha-induced protein 6 | 197.4394341 | 109.3081398 | 491.6323329 |
| ENSDARG00000038123 | myl9a | Myosin, light chain 9a, regulatory | 179.6584981 | 96.56364658 | 511.0645904 |
| ENSDARG00000014626 | dlx3b | Distal-less homeobox gene 3b | 285.6044597 | 300.5094173 | 166.0309536 |
| ENSDARG00000087402 | tpm1 | Tropomyosin 1 (alpha) | 147.8702844 | 67.17783708 | 530.8673917 |
| ENSDARG00000029353 | serpine2 | Serine (or cysteine) proteinase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 | 155.1438879 | 185.8865435 | 362.6601572 |
| ENSDARG00000008359 | cnn3a | Calponin 3, acidic a | 200.6466151 | 175.5868376 | 322.1817671 |
| ENSDARG00000013804 | capns1b | Calpain, small subunit 1 b | 295.8800252 | 192.2972637 | 137.1191417 |
| ENSDARG00000062363 | phex | Phosphate regulating gene with homologues to endopeptidases on the X chromosome | 374.7902084 | 246.0473474 | 2.923096804 |
| ENSDARG00000007697 | fabp7a | Fatty acid binding protein 7, brain, a | 195.9918097 | 237.0929702 | 156.4171196 |
| ENSDARG00000054749 | lmo4b | LIM domain only 4b | 171.4754089 | 158.0390988 | 259.5479538 |
| ENSDARG00000040362 | ehd2 | EH-domain containing 2 | 161.7766861 | 245.0843826 | 179.9114927 |
| ENSDARG00000012066 | dcn | Decorin | 138.738941 | 110.0299673 | 294.5159175 |
| ENSDARG00000007396 | rnd3b | Rho family GTPase 3b | 230.919067 | 106.1477521 | 180.3513591 |
| ENSDARG00000075045 | cxcl-c1c | Chemokine (C-X-C motif) ligand C1c | 205.5342189 | 222.3706168 | 85.84923297 |
| ENSDARG00000030449 | crabp2b | Cellular retinoic acid binding protein 2, b | 105.0984741 | 221.8625863 | 181.7852462 |
| ENSDARG00000089187 | wfdc2 | WAP four-disulfide core domain 2 | 215.3192844 | 100.2819887 | 173.5050759 |
| ENSDARG00000061226 | timp2a | Tissue inhibitor of metalloproteinase 2a | 110.5132145 | 134.7435353 | 217.234448 |
| ENSDARG00000030694 | atp6v1e1b | ATPase, H+ transporting, lysosomal, V1 subunit E isoform 1b | 211.5850639 | 104.0432062 | 140.8175355 |
| ENSDARG00000016238 | arl6ip5b | ADP-ribosylation factor-like 6 interacting protein 5b | 180.1080903 | 105.0459892 | 159.1043514 |
| ENSDARG00000019307 | dusp5 | Dual specificity phosphatase 5 | 126.478358 | 236.8883821 | 68.27162288 |
| ENSDARG00000054823 | id3 | Inhibitor of DNA binding 3 | 123.0298414 | 119.957024 | 185.8068645 |
| ENSDARG00000096118 | ost4 | Oligosaccharyltransferase 4 homolog | 293.697141 | 56.14183101 | 47.90212635 |
| ENSDARG00000070391 | tspan4b | Tetraspanin 4b | 124.4627775 | 134.439824 | 107.2748044 |
| ENSDARG00000002507 | itga10 | Integrin, alpha 10 | 89.61611629 | 201.1502394 | 71.71587866 |
| ENSDARG00000008363 | mcl1b | Myeloid cell leukemia sequence 1b | 138.0029954 | 100.3628761 | 121.8545135 |
| ENSDARG00000078674 | hspb9 | Heat shock protein, alpha-crystallin-related, 9 | 208.1065389 | 130.9142075 | 9.6011956 |
| ENSDARG00000056627 | cxcl14 | Chemokine (C-X-C motif) ligand 14 | 30.54406211 | 119.5273691 | 197.7629774 |
| ENSDARG00000026070 | cd82b | CD82 antigen, b | 117.1867309 | 119.7501676 | 99.79403009 |
| ENSDARG00000032831 | htra1a | HtrA serine peptidase 1a | 62.04851894 | 79.54294838 | 191.2823828 |
| ENSDARG00000027088 | ptgdsb | Prostaglandin D2 synthase b | 295.7620764 | 16.83617366 | 7.299828587 |
| ENSDARG00000069983 | scinla | Scinderin like a | 229.2763242 | 77.71766924 | 12.73139596 |
| ENSDARG00000014103 | dkk1a | Dickkopf 1a | 45.36030464 | 95.82598311 | 174.3537947 |
| ENSDARG00000044001 | lgals3l | Lectin, galactoside-binding, soluble, 3 (galectin 3)-like | 151.485882 | 94.79980323 | 68.69664737 |
| ENSDARG00000094752 | rpe65b | Retinal pigment epithelium-specific protein 65b | 51.81279893 | 157.9199935 | 85.8643745 |
| ENSDARG00000006275 | irf2bp2b | Interferon regulatory factor 2 binding protein 2b | 83.23427126 | 90.32706729 | 118.4504944 |
| ENSDARG00000096594 | lratb | Lecithin retinol acyltransferase b (phosphatidylcholine—retinol O-acyltransferase b) | 54.62075299 | 126.7544822 | 101.4833434 |
| ENSDARG00000045219 | dkk1b | Dickkopf 1b | 115.2674028 | 123.3601222 | 41.36781122 |
| ENSDARG00000073891 | gdf10b | Growth differentiation factor 10b | 33.67260166 | 219.1972506 | 6.710000293 |
| ENSDARG00000063704 | gpr1 | G protein-coupled receptor 1 | 11.47967914 | 67.08867022 | 174.2527408 |
| ENSDARG00000042690 | s1pr1 | Sphingosine-1-phosphate receptor 1 | 51.05707689 | 120.3548169 | 69.24951973 |
| ENSDARG00000070420 | cyp24a1 | Cytochrome P450, family 24, subfamily A, polypeptide 1 | 18.72705544 | 177.0835255 | 44.13670934 |
| ENSDARG00000086842 | dap1b | Death associated protein 1b | 108.3112632 | 77.42847935 | 53.19516114 |
| ENSDARG00000068710 | nid1a | Nidogen 1a | 62.19315736 | 114.8440231 | 50.83336553 |
| ENSDARG00000055381 | bambia | BMP and activin membrane-bound inhibitor homolog a | 88.97243166 | 57.43071367 | 78.67391423 |
| ENSDARG00000067742 | eva1bb | Eva-1 homolog Bb | 63.44368564 | 84.2765728 | 74.97828951 |
| ENSDARG00000005789 | enpp1 | Ectonucleotide pyrophosphatase/ phosphodiesterase 1 | 64.34284892 | 137.6726653 | 19.82855613 |
| ENSDARG00000056023 | hoxb9a | Homeo box B9a | 69.19604476 | 83.45231228 | 68.31730246 |
| ENSDARG00000094857 | dio2 | Deiodinase, iodothyronine, type II | 79.95831483 | 112.2283984 | 26.87641183 |
| ENSDARG00000089078 | col23A1 | Collagen, type XXIII, alpha 1 | 63.62339938 | 147.7007832 | 2.289688031 |
| ENSDARG00000030177 | uchl3 | Ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) | 105.7558676 | 48.45357406 | 49.98198164 |
| ENSDARG00000024877 | ptgr1 | Prostaglandin reductase 1 | 103.6331577 | 54.73419065 | 37.55761808 |
| ENSDARG00000094854 | ms4a17a.8 | Membrane-spanning 4-domains, subfamily A, member 17A.8 | 116.1375676 | 55.21694763 | 23.36757024 |
| ENSDARG00000019128 | tpm4b | Tropomyosin 4b | 54.49279439 | 41.58138656 | 98.05974096 |
| ENSDARG00000058348 | scinlb | Scinderin like b | 43.90721933 | 56.88955333 | 90.22067671 |
| ENSDARG00000005541 | wif1 | Wnt inhibitory factor 1 | 93.47095208 | 67.6434638 | 28.05685047 |
| ENSDARG00000010462 | sp9 | Sp9 transcription factor | 57.92009712 | 49.7884664 | 76.65945541 |
| ENSDARG00000044803 | dhrs3b | Dehydrogenase/reductase (SDR family) member 3b | 130.0154057 | 51.68314191 | 1.624036879 |
| ENSDARG00000087263 | ssr3 | Signal sequence receptor, gamma | 91.92315352 | 47.64499378 | 39.03838439 |
| ENSDARG00000058733 | ihha | Indian hedgehog homolog a | 7.794542694 | 88.86938529 | 78.16679263 |
| ENSDARG00000015966 | yaf2 | YY1 associated factor 2 | 67.3993785 | 69.0931997 | 38.15094538 |
| ENSDARG00000090945 | clec4f | C-type lectin domain family 4, member F | 120.1210729 | 48.41536445 | 3.207217092 |
| ENSDARG00000042296 | dlx5a | Distal-less homeobox gene 5a | 126.8385026 | 31.88420836 | 12.30766798 |
| ENSDARG00000053381 | ppap2a | Phosphatidic acid phosphatase type 2A | 36.20007779 | 67.05858162 | 65.57173276 |
| ENSDARG00000069376 | tnfsf12 | Tumor necrosis factor (ligand) superfamily, member 12 | 38.06281765 | 121.2490869 | 7.954191675 |
All transcripts are presented as reads per kilobase of transcript per million reads mapped (RPKM). Each stromal cell line’s transcriptome was analyzed and compared to a non-hematopoietic supportive stromal cell line (ZF4). The transcripts below have been described previously as playing a role in hematopoiesis.
ism1 morphants exhibit reduced erythrocyte production
To elucidate ism1’s role in erythroid cell production, gata1a:DsRed transgenic zebrafish were used; Gata1 is a transcription factor expressed in erythrocytes, allowing for real time visualization of red blood cells in developing embryos. ism1 knockdown with a specific MO caused slowed circulation and caused blood to pool in gata1a:DsRed+ zebrafish (Fig 2A) when examined at 48 hpf. The control group expressed no abnormalities with respect to blood circulation, compared to 38% of the ism1 morphants. The hearts of embryos injected with ism1 MO were pumping and appeared morphologically normal. Additionally, no blood was observed leaking from blood vessels. Injection of ism1 mRNA reduced the amount of pooled blood seen in ism1 morphants and increased the speed of erythrocyte circulation (Fig 2B).
Fig 2. ism1 knockdown decreases erythrocytes.
gata1:DsRed zebrafish embryos were injected at the single-cell-stage with 7 ng ism1 MO (MO), or 7 ng ism1 MO and 17.88 ng of ism1 mRNA (Rescue); uninjected embryos served as controls. (A) 24 hpf zebrafish visualized at 5x that have normal (left), pooled blood (middle), and slowed circulation (right) phenotypes. (B) Quantitation of zebrafish shown in (A); pooled blood (yellow), slowed circulation (blue), and normal (red) phenotypes. (C) Flow cytometry results quantitating gata1:DsRed+ erythrocytes in uninjected (control; red), ism1 MO injected (MO; peach), and ism1 MO with ism1 mRNA (Rescue; pink) 72 hpf zebrafish. (D) qRT-PCR was performed comparing transcript levels of hbaa in ism1 MO injected (MO) and uninjected controls (control). Bars represent the mean, and error bars represent standard deviation. Data was normalized as fold change over control and analyzed using a two-tailed unpaired Student’s T test. * denotes p<0.05, ** denotes p<0.0001, N.S indicates no significance.
Regardless of their circulation phenotypes, we speculated that the morphant embryos also had decreased numbers of erythrocytes. To quantitate total red blood cell numbers, flow cytometry was utilized. All ism1 MO injected embryos were analyzed without grouping them based on their circulation phenotype. Three individual 72 hpf gata1a:DsRed+ ism1 MO injected zebrafish were randomly grouped together, enzymatically dissociated in a sample tube, and analyzed on a flow cytometer to quantitate gata1a:DsRed+ cells. Percentages of gata1a:DsRed+ cells were normalized to uninjected control embryos and expressed as fold change over control. ism1 knockdown caused an approximate 2-fold decrease in gata1:DsRed+ cells compared to control (Fig 2C) and the rescued group displayed erythrocyte levels similar to control groups (Fig 2C), indicating that ism1 depletion was the cause of the erythrocyte defect. Additionally, ism1 MO injected whole embryos were pooled into groups of 10, digested, and qRT-PCR was used to analyze alpha globin (hbaa) mRNA levels, a specific marker of differentiated erythrocytes. qRT-PCR demonstrated a 29-fold reduction of hbaa mRNA levels in ism1 MO injected animals (Fig 2D). Taken together, these data indicate that ism1 reduction causes a decrease in embryonic erythrocytes and a decrease in hbaa mRNA levels in the whole organism.
While intersegmental vessels (ISVs) are not involved in blood development, previous studies indicated that ism1 was necessary for proper development of ISVs in zebrafish[55]. To confirm that ism1 was not necessary for the formation of the dorsal aorta, the source of HSC generation during development, we injected ism1 MO into the vasculature-specific kdrl:EGFP zebrafish transgenic line. 24 hpf zebrafish were visualized and exhibited shortened ISVs, as reported previously (S2 Fig)[55]. However, the dorsal aorta was not negatively affected in morphants (S2 Fig), also in agreement with previous studies[55]. Although not previously described, the shortened ISVs recovered by 48 hpf without addition of exogenous ism1. In conclusion, ism1 morphants experience a reduction in ISVs that recovers by 48 hpf but have a normal dorsal aorta.
ism1 morphants exhibit reduced myeloid cells
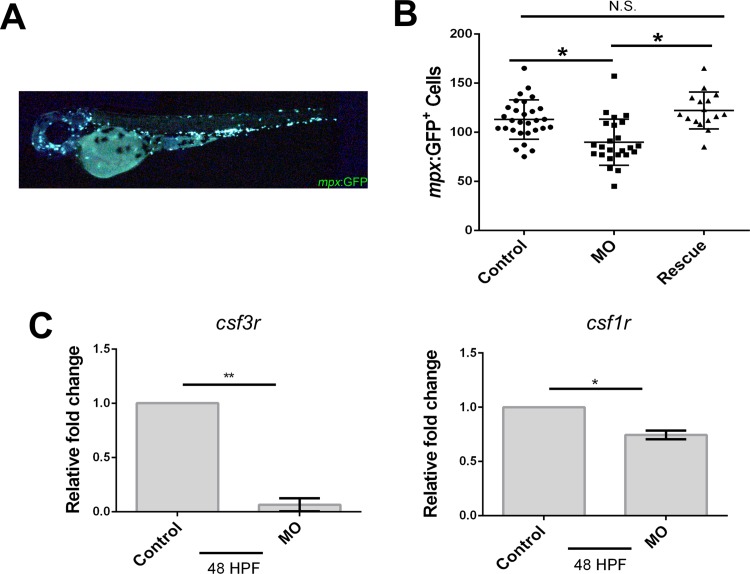
Myeloid cells such as neutrophils and macrophages are cellular components critical for innate immune responses. To investigate ism1’s role in myeloid cell biology, mpx:EGFP transgenic zebrafish were utilized. mpx:EGFP zebrafish embryos were injected with ism1 MO; all injected embryos were visualized and total mpx:EGFP+ cells were individually enumerated at 48 hpf (Fig 3A). ism1 MO injection resulted in a 20% reduction of mpx:EGFP+ cells compared to control groups (Fig 3B) and rescue with ism1 mRNA showed a recovery of mpx:EGFP+ cells compared to ism1 MO injected embryos. qRT-PCR was performed on pools of 10 injected embryos for colony stimulating factor 3 receptor (csf3r; also called granulocyte colony stimulating factor receptor), and colony stimulating factor 1 receptor (csf1r; also called macrophage colony stimulating factor receptor), two markers for neutrophils and macrophages, respectively. qRT-PCR indicated a 11-fold reduction in csf3r mRNA levels and a 30% reduction of csf1r mRNA levels in ism1 MO injected embryos (Fig 3C). Overall, these data indicate that ism1 knockdown causes a reduction in mpx:EGFP+ myeloid cells.
Fig 3. ism1 knockdown decreases neutrophils and macrophages in 48 hpf zebrafish.
mpx:GFP single-cell-stage embryos were injected with 7 ng ism1 MO (MO; squares), or 7 ng ism1 MO and 17.88 ng of ism1 mRNA (Rescue; triangles); uninjected embryos served as controls (circles). (A) Developing 48 hpf zebrafish were visualized, and individual mpx:GFP+ cells were enumerated; representative fish shown for reference. (B) Each data point represents total amount of mpx:GFP+ cells present in one zebrafish. (C) qRT-PCR was performed comparing transcript levels of csf3r (left) and csf1r (right) in ism1 MO injected (MO) and uninjected controls (control) at 48 hpf. Bars represent the mean, and error bars represent standard deviation. A two-tailed unpaired Student’s T test was performed to determine statistical significance. * denotes p<0.05, ** denotes p<0.0001, N.S indicates no significance.
HSPCs are reduced in ism1 morphants
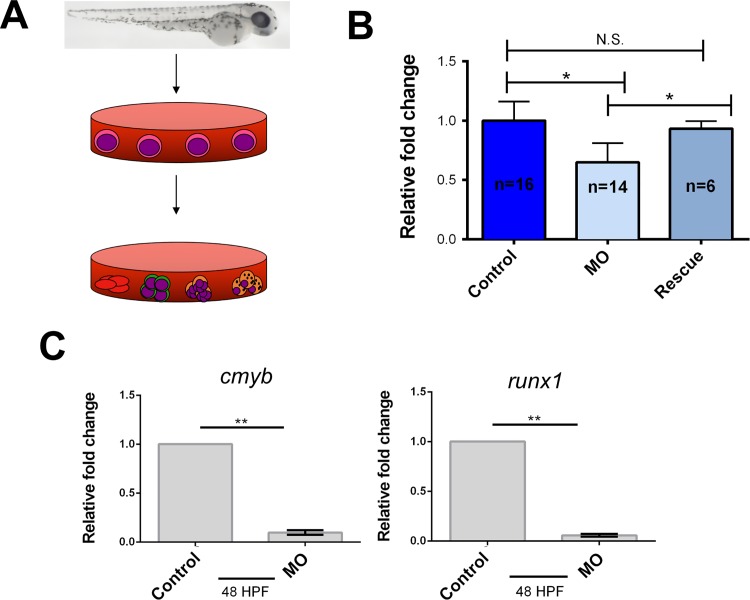
To confirm that ism1 loss was causing a reduction in HSPCs, groups of 10 MO-injected embryos at 48 hpf were placed in a tube, enzymatically digested, and plated in methylcellulose with exogenous hematopoietic-supportive growth factors[61]. After plating, the embryos were incubated for 7 days, allowing HSPCs to proliferate and differentiate (Fig 4A). Fish were not phenotypically selected before digestion; all injected fish were randomly pooled into groups of 10 and plated in methylcellulose. ism1 MO injected and rescued embryos generated hematopoietic colonies, which were enumerated. Colony forming units (CFUs) for ism1 MO injected zebrafish were reduced by 35% compared to control and rescue groups (Fig 4B). There was no statistical difference between control and rescue groups, indicating that ism1 is necessary for the generation of the proper numbers of HSPCs in developing zebrafish embryos. qRT-PCR on 48 hpf embryos was also performed to quantitate HSPC-related transcription factors. Data showed an overall 20-fold reduction in runx1, and a 10-fold reduction in cmyb mRNA levels between controls and ism1 MO injected embryos (Fig 4C). Overall, these data indicate that HSPC formation is reduced in ism1 morphants.
Fig 4. Hematopoietic stem and progenitor cells (HSPCs) are reduced in ism1 knockdown embryos.
Single-cell-stage embryos were injected with 7 ng of ism1 MO (MO), or 7 ng of ism1 MO and 17.88 ng of ism1 mRNA (Rescue); uninjected embryos served as a control. (A) Ten 48 hpf embryos from each treatment were digested, plated in methylcellulose with exogenous hematopoietic-supportive growth factors, and incubated for 7 days. (B) Colony forming units (CFUs) generated by ism1 MO (light blue) and ism1 MO with ism1 mRNA (Rescue; darker blue) injected embryos presented as fold change over control embryos (dark blue). (C) qRT-PCR was performed comparing transcript levels of cmyb and runx1 in ism1 MO injected (MO) and uninjected controls (control) at 48 hpf. Bars represent the mean, and error bars represent standard deviation. A two-tailed unpaired Student’s T test was performed to determine statistical significance. * denotes p<0.05, ** denotes p<0.0001, N.S indicates no significance.
Discussion
In this study, we sought to elucidate molecular requirements for HSPC generation, support, and expansion. Utilizing previously described zebrafish hematopoietic-supportive stromal cell lines[48–50] we identified highly expressed transcripts determined by RNA-seq. ism1 was chosen from this list due to its involvement in angiogenesis[55, 69], its presence in embryonic[52, 54, 55] and adult[53, 54] hematopoietic tissue, the fact that it is a secreted protein[52, 53, 55], and because it shares synteny with human ISM1. Our study findings indicate that ism1 is required for proper embryonic hematopoiesis.
To elucidate ism1’s role in hematopoiesis we performed loss-of-function experiments using MOs to generate ism1 morphants. Other MOs have been utilized to study ism1[55], and our MO was directed towards a similar location in the genome 8bp 3’ of Xiang et. al’s translation-blocking MO; the location we chose was suggested by modeling software to be a more effective site for blocking ism1 translation. The MO we designed binds 10bp 5’ to ism1’s start site; as two splice variants are predicted for ism1, we wanted to ensure that the MO blocked all versions of Ism1 so we would not have multiple splice isoforms potentially confounding our results. Importantly, our MO and Xiang et. al’s have the same phenotypic effect on ISV formation early in development. The dorsal aorta, the site of HSPC generation between 36–52 hpf[7, 8], was unaffected (our data and [55]). We also observed that ISVs were only affected between 24–48 hpf, after which ISV branching was normal. To observe hematopoietic defects, we injected ism1 MOs into previously characterized transgenic fish lines that have fluorescent proteins expressed in specific hematopoietic cell lineages. ism1 morphants did not express any morphological defects such as bent tails, spinal deformation, or stunted growth (data not shown). However, they did have phenotypes in their hematopoietic tissues. Coupled with the fact that these phenotypes were rescued by the addition of exogenous ism1 mRNA indicates that Ism1 is a key protein involved in normal zebrafish hematopoiesis.
It is important to note that we are utilizing zebrafish as a model system to investigate the evolution of vertebrate hematopoiesis. While mammals are derived from lobe-finned fish and zebrafish are ray-finned fish (reviewed in [70]), zebrafish have proven to be an excellent model of hematopoiesis; their molecular control of blood formation and differentiation is well conserved with mammals[24–26, 71]. Further evidence of this is that drugs identified in zebrafish are now being utilized in the clinic to treat human blood diseases[72]. One concern with using zebrafish as a model is that the MO may not be specific for only one copy of ism1. While the genome was duplicated early in teleost evolution[73, 74], there is only one described copy of ism1 in the zebrafish genome; we found no additional copies in our scanning of genomic data and RNA-seq results, in agreement with others[55]. This one copy of zebrafish ism1 shares synteny with human ISM1; JAG1 (jag1b), BTBD3 (btbd3b), and SPTLC3 (sptlc3) are 5’ to ISM1 (ism1), while TASP1 (tasp1), KIF16B (kif16bb), and FLRT3 (flrt3) are 3’. The only other genes in the zebrafish ism1 family are ism2a and ism2b. These two isthmin paralogs are located on different chromosomes (ism1 is on chromosome 13, ism2a is on 17, and ism2b is on 20) and share synteny with only the genes on the 5’ side of human ISM2. Little is known about ism2a or ism2b’s roles/functions in zebrafish. These genes share no synteny with ISM1 or ism1, indicating that ism2a and ism2b are likely different genes as opposed to duplicates of ism1. There are 9 ism1 orthologs in other fish species like cod, amazon molly, fugu, medaka, platyfish, spotted gar, stickleback, tetraodon, and tilapia, all of which only have one identified copy of ism1 in their genome. Overall, it appears that ism1 only has one identifiable copy in the zebrafish genome, is not mislabeled, and is similar to human ISM1.
To examine the role of ism1 in hematopoiesis, we first examined erythrocytes due to their abundance and importance during development; they are also the first hematopoietic cells to arise. gata1a:DsRed+ ism1 morphants exhibited pooled blood and we observed what appeared to be slowed circulation. While some “pooled blood” phenotypes have been attributed to functional cardiac defects[75], no such phenotype was visualized in our experiments. This suggested that the morphant’s vasculature might be disrupted, but our data, in agreement with prior studies[55], did not see a disruption of the dorsal aorta after ism1 knockdown. To further confirm that erythrocytes were specifically reduced, we enumerated them with flow cytometry and found a significant decrease in erythrocytes in ism1 morphants. Reduction of erythrocytes was further validated by qRT-PCR data showing reduction of the hbaa transcript, a hemoglobin gene. In conclusion, ism1 knockdown caused an overall reduction of erythrocytes and the phenotype was rescued by the addition of exogenous ism1 mRNA, indicating that ism1 is required for normal erythropoiesis.
mpx:GFP zebrafish express GFP in both neutrophils and macrophages[58, 63]; both are differentiated myeloid cell lineages. ism1 morphants had a 20% reduction in these myeloid cells, suggesting that myeloid cell lineages were not properly forming. We also saw a reduction in csf3r transcript, the signaling receptor necessary for granulocyte formation and differentiation, measured by qRT-PCR. While ism1 morphants exhibited an overall decrease in myeloid cells, these cell lineages were still generated, suggesting that ism1 isn’t required for myeloid differentiation but is necessary for the generation or proliferation of myeloid HSPCs. Together, the reduction in both myeloid and erythroid cell lineages suggested a reduction of a common upstream progenitor cell. Therefore, ism1 is likely involved in the proliferation or differentiation of a multipotent progenitor such as the CMP or HSC.
Together, decreased production of differentiated erythroid and myeloid cell lineages indicated that ism1 knockdown somehow decreased the generation or proliferation of HSPCs during development. To investigate this possibility, we developed a surrogate assay to assess HSPC numbers in morphant zebrafish embryos[61]. This assay involved plating dissociated whole zebrafish embryos and adding exogenous factors that promote only HSPC differentiation and proliferation in vitro[62, 63, 76]. Plating HSPCs generates CFUs that arise from a single progenitor cell. Therefore, the number of CFUs counted after 7 days in culture corresponds to the number of total progenitor cells present in the organism at the time they were assayed. These results suggest that ism1 is transcribed, translated, and secreted by stromal cells during a window of HSPC formation in the embryo; reducing ism1 expression during this time reduced the number of HSPCs in the developing zebrafish. This non-cell-autonomous effect of ism1 is seen with the clonal methylcellulose assay- because less Ism1 is secreted by the hematopoietic stroma during development, less HSPCs are generated in the embryo. 48 hpf was chosen for these assays due to the fact that functional HSPCs are generated by this time in development[7, 8, 27–30]. Our data, in agreement with previous studies[54], indicates that ism1 is expressed in the hematopoietic stroma, but not in hematopoietic cells themselves. These results reinforce that Ism1 is a secreted protein made in the stromal niche, and not produced by HSPCs themselves; HSPCs are present in the “precursor”[62, 63, 76, 77] and “lymphoid”[29, 57, 62, 63, 76, 77] fractions of blood cells analyzed in this study. Importantly, this was not a developmental delay; plating dissociated ism1 morphant embryos at later time points did not rescue the number of HSPC-derived colonies present (data not shown). Overall, the fact that HSPCs are reduced in ism1 morphants likely accounts for the decreased counts of differentiated erythroid and myeloid lineages, suggesting that ism1 is needed for normal levels of HSPC formation and differentiation during embryonic development.
ism1 is extremely enigmatic, and many seemingly conflicting roles for this gene exist in the literature. ism1 was originally identified as a secreted protein expressed along with fibroblast growth factor (Fgf) 8 in the Xenopus midbrain-hindbrain organizer[52], and it has recently been described in craniofacial patterning in humans[78]. Importantly, it has been identified in multiple embryonic tissues, many of which are involved in various regions of embryonic development. Of interest for blood formation, ism1 was identified in Xenopus blood islands[52] and the lateral plate mesoderm of chicks and mice[54], which is tissue that gives rise to blood. It is also found in bone marrow[54], the site of adult hematopoiesis in mammals. While ism1 was identified in the supportive hematopoietic stroma and not actual blood cells (our data and [54]), other studies indicate Ism1 is expressed in specific subsets of mouse lymphoid cells, including T and natural killer (NK) cells[53]. Our data does not correlate with this; zebrafish T and NK cells are present in the “lymphoid” population of WKM[57], and we detected no ism1 in this fraction, indicating that this may be a difference between fish and mammals. Another difference across species is the description of ism1 as an angiogenesis inhibitor; in mice exogenous ISM1 suppressed tumor angiogenesis[55, 69], but in zebrafish the knockdown of ism1 reduced the growth of intersegmental vessels[55]. While Ism1 plays a role in apoptosis, its role in this process is also complicated. It specifically reduces endothelial cell (EC) growth by inducing apoptosis[55, 69], while it has little to no effect on the proliferation or survival of glioma cells[69], NIH3T3 fibroblasts[55], Swiss 3T3 fibroblasts[55], B16 melanoma cells[55], and hepatocellular carcinoma cells[55]. This differential apoptotic capacity is not just different across cell types; it is linked to the solubility of ISM1. Soluble ISM1 causes apoptosis, while extracellular matrix (ECM)-immobilized ISM1 promotes EC adhesion, migration, and survival[79]. Unfortunately, no zebrafish-specific Ism1 antibody exists, preventing analysis of this in our experiments. It is important to note that the role of Ism1-induced apoptosis of hematopoietic cells is a possibility that could also explain a reduction in blood cells; further investigation may shed light on this subject.
Overall, these findings demonstrate a new role for ism1; its involvement in de novo hematopoietic cell formation. ism1 reduction negatively impacted the number of HSPCs formed and their ability to properly generate mature erythrocytes, neutrophils, and macrophages. This could be due to ism1 affecting a variety of signaling pathways; we speculate that ism1 is involved in molecular pathways that instruct HSPC formation and differentiation, as its knockdown decreases various mature blood cell lineages. This is of keen interest, because ism1 is secreted by our hematopoietic-supportive cell lines and affects various cell lineages, yet it has only been identified in a handful of tissues[52–55, 78] and lymphoid cells[53]. ism1 expression is present in Xenopus blood islands suggesting its role in generating primitive blood[52]. This expression also implicates its importance in definitive hematopoiesis, as primitive erythropoiesis expresses many of the same molecular factors as its definitive counterpart[80]. ism1 is also expressed at the same time and in the same location as Xenopus Fgf8 in the notochord[52]; it is also present in the zebrafish notochord[55]. FGF signaling is involved in hematopoiesis[81–87] as it is a mediator of Notch and Wnt signaling[83]. It is also modulated by BMP signaling[82]. All of these pathways are required for HSC production. Interestingly, ism1 is a Wnt target[56] that is expressed (and likely secreted from) the notochord[52, 55]; this is the first time a secreted signal from the notochord has been implicated in hematopoietic development. Overall, these previous studies and our results indicate ism1 is signaling to induce formation and functionality of hematopoietic tissues in the developing zebrafish. Further investigation of ism1’s role in this process may shed light on an important signaling pathway for blood formation and regulation.
Supporting information
RT-PCR was performed for ism1 from ZKS, ZEST, CHEST, and kidney mRNA. Myeloid, lymphoid, and precursor cell mRNA isolated from zebrafish kidney was also interrogated for ism1 transcripts.
(TIF)
flk1:GFP single-cell-stage embryos were injected with 7 ng of ism1 MO (bottom); uninjected embryos served as controls (top). 24 hpf (left column) and 48 hpf (right column) zebrafish were visualized at 40x for flk1:GFP fluorescence within the trunk area denoted by black box in brightfield image at top center. Arrows indicate shortened intersegmental vessels in ism1 morphants. Numbers in corner of images denote the number of embryos displaying the imaged phenotype.
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the California State University Program for Education and Research in Biotechnology (CSUPERB; New Investigator Grant), a NSF MRI award (proposal 1626406), and a California State University Chico Internal Research Grant (to D.L.S.).
References
- 1.Cumano A, Godin I. Ontogeny of the hematopoietic system. Annu Rev Immunol. 2007;25:745–85. doi: 10.1146/annurev.immunol.25.022106.141538 . [DOI] [PubMed] [Google Scholar]
- 2.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–44. doi: 10.1016/j.cell.2008.01.025 ; PubMed Central PMCID: PMCPMC2628169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eaves CJ. Hematopoietic stem cells: concepts, definitions, and the new reality. Blood. 2015;125(17):2605–13. doi: 10.1182/blood-2014-12-570200 ; PubMed Central PMCID: PMCPMC4440889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemischka IR. Clonal, in vivo behavior of the totipotent hematopoietic stem cell. Semin Immunol. 1991;3(6):349–55. . [PubMed] [Google Scholar]
- 5.Spangrude GJ, Smith L, Uchida N, Ikuta K, Heimfeld S, Friedman J, et al. Mouse hematopoietic stem cells. Blood. 1991;78(6):1395–402. . [PubMed] [Google Scholar]
- 6.Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464(7285):116–20. doi: 10.1038/nature08764 . [DOI] [PubMed] [Google Scholar]
- 7.Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464(7285):108–11. doi: 10.1038/nature08738 ; PubMed Central PMCID: PMCPMC2858358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464(7285):112–5. doi: 10.1038/nature08761 . [DOI] [PubMed] [Google Scholar]
- 9.Ciau-Uitz A, Walmsley M, Patient R. Distinct origins of adult and embryonic blood in Xenopus. Cell. 2000;102(6):787–96. . [DOI] [PubMed] [Google Scholar]
- 10.Oberlin E, Tavian M, Blazsek I, Peault B. Blood-forming potential of vascular endothelium in the human embryo. Development. 2002;129(17):4147–57. . [DOI] [PubMed] [Google Scholar]
- 11.Jaffredo T, Gautier R, Eichmann A, Dieterlen-Lievre F. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development. 1998;125(22):4575–83. . [DOI] [PubMed] [Google Scholar]
- 12.de Bruijn MF, Ma X, Robin C, Ottersbach K, Sanchez MJ, Dzierzak E. Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity. 2002;16(5):673–83. . [DOI] [PubMed] [Google Scholar]
- 13.North TE, de Bruijn MF, Stacy T, Talebian L, Lind E, Robin C, et al. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16(5):661–72. . [DOI] [PubMed] [Google Scholar]
- 14.Tavian M, Hallais MF, Peault B. Emergence of intraembryonic hematopoietic precursors in the pre-liver human embryo. Development. 1999;126(4):793–803. . [DOI] [PubMed] [Google Scholar]
- 15.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457(7231):887–91. doi: 10.1038/nature07619 ; PubMed Central PMCID: PMCPMC2744041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3(6):625–36. doi: 10.1016/j.stem.2008.09.018 ; PubMed Central PMCID: PMCPMC2631552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91(5):661–72. . [DOI] [PubMed] [Google Scholar]
- 18.Galy A, Travis M, Cen D, Chen B. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 1995;3(4):459–73. . [DOI] [PubMed] [Google Scholar]
- 19.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–7. doi: 10.1038/35004599 . [DOI] [PubMed] [Google Scholar]
- 20.Manz MG, Miyamoto T, Akashi K, Weissman IL. Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci U S A. 2002;99(18):11872–7. doi: 10.1073/pnas.172384399 ; PubMed Central PMCID: PMCPMC129361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulais PE, Frenette PS. Making sense of hematopoietic stem cell niches. Blood. 2015;125(17):2621–9. doi: 10.1182/blood-2014-09-570192 ; PubMed Central PMCID: PMCPMC4408288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132(4):598–611. doi: 10.1016/j.cell.2008.01.038 ; PubMed Central PMCID: PMCPMC4505728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiel MJ, Morrison SJ. Uncertainty in the niches that maintain haematopoietic stem cells. Nat Rev Immunol. 2008;8(4):290–301. doi: 10.1038/nri2279 . [DOI] [PubMed] [Google Scholar]
- 24.Gore AV, Pillay LM, Venero Galanternik M, Weinstein BM. The zebrafish: A fintastic model for hematopoietic development and disease. Wiley Interdiscip Rev Dev Biol. 2018. doi: 10.1002/wdev.312 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frame JM, Lim SE, North TE. Hematopoietic stem cell development: Using the zebrafish to identify extrinsic and intrinsic mechanisms regulating hematopoiesis. Methods Cell Biol. 2017;138:165–92. doi: 10.1016/bs.mcb.2016.08.004 . [DOI] [PubMed] [Google Scholar]
- 26.Carroll KJ, North TE. Oceans of opportunity: exploring vertebrate hematopoiesis in zebrafish. Exp Hematol. 2014;42(8):684–96. doi: 10.1016/j.exphem.2014.05.002 ; PubMed Central PMCID: PMCPMC4461861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134(23):4147–56. doi: 10.1242/dev.012385 ; PubMed Central PMCID: PMCPMC2735398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertrand JY, Kim AD, Teng S, Traver D. CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development. 2008;135(10):1853–62. doi: 10.1242/dev.015297 ; PubMed Central PMCID: PMCPMC2762343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma D, Zhang J, Lin HF, Italiano J, Handin RI. The identification and characterization of zebrafish hematopoietic stem cells. Blood. 2011;118(2):289–97. doi: 10.1182/blood-2010-12-327403 ; PubMed Central PMCID: PMCPMC3138684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam EY, Hall CJ, Crosier PS, Crosier KE, Flores MV. Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood. 2010;116(6):909–14. doi: 10.1182/blood-2010-01-264382 . [DOI] [PubMed] [Google Scholar]
- 31.Stachura DL, Traver D. Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol. 2016;133:11–53. doi: 10.1016/bs.mcb.2016.03.022 . [DOI] [PubMed] [Google Scholar]
- 32.Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. . [DOI] [PubMed] [Google Scholar]
- 33.Weinstein BM, Schier AF, Abdelilah S, Malicki J, Solnica-Krezel L, Stemple DL, et al. Hematopoietic mutations in the zebrafish. Development. 1996;123:303–9. . [DOI] [PubMed] [Google Scholar]
- 34.Ransom DG, Haffter P, Odenthal J, Brownlie A, Vogelsang E, Kelsh RN, et al. Characterization of zebrafish mutants with defects in embryonic hematopoiesis. Development. 1996;123:311–9. . [DOI] [PubMed] [Google Scholar]
- 35.Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, et al. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13(20):2713–24. ; PubMed Central PMCID: PMCPMC317115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaiano N, Amsterdam A, Kawakami K, Allende M, Becker T, Hopkins N. Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature. 1996;383(6603):829–32. doi: 10.1038/383829a0 . [DOI] [PubMed] [Google Scholar]
- 37.North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447(7147):1007–11. doi: 10.1038/nature05883 ; PubMed Central PMCID: PMCPMC2775137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paik EJ, de Jong JL, Pugach E, Opara P, Zon LI. A chemical genetic screen in zebrafish for pathways interacting with cdx4 in primitive hematopoiesis. Zebrafish. 2010;7(1):61–8. doi: 10.1089/zeb.2009.0643 ; PubMed Central PMCID: PMCPMC2897134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridges S, Heaton WL, Joshi D, Choi H, Eiring A, Batchelor L, et al. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood. 2012;119(24):5621–31. doi: 10.1182/blood-2011-12-398818 ; PubMed Central PMCID: PMCPMC3382926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeh JR, Munson KM, Elagib KE, Goldfarb AN, Sweetser DA, Peterson RT. Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat Chem Biol. 2009;5(4):236–43. doi: 10.1038/nchembio.147 ; PubMed Central PMCID: PMCPMC2658727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Astuti Y, Kramer AC, Blake AL, Blazar BR, Tolar J, Taisto ME, et al. A Functional Bioluminescent Zebrafish Screen for Enhancing Hematopoietic Cell Homing. Stem Cell Reports. 2017;8(1):177–90. doi: 10.1016/j.stemcr.2016.12.004 ; PubMed Central PMCID: PMCPMC5233450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dexter TM, Allen TD, Lajtha LG, Schofield R, Lord BI. Stimulation of differentiation and proliferation of haemopoietic cells in vitro. J Cell Physiol. 1973;82(3):461–73. doi: 10.1002/jcp.1040820315 . [DOI] [PubMed] [Google Scholar]
- 43.Dexter TM, Lajtha LG. Proliferation of haemopoietic stem cells in vitro. Br J Haematol. 1974;28(4):525–30. . [DOI] [PubMed] [Google Scholar]
- 44.Dexter TM, Testa NG. Differentiation and proliferation of hemopoietic cells in culture. Methods Cell Biol. 1976;14:387–405. . [DOI] [PubMed] [Google Scholar]
- 45.Dexter TM, Allen TD, Lajtha LG. Conditions controlling the proliferation of haemopoietic stem cells in vitro. J Cell Physiol. 1977;91(3):335–44. doi: 10.1002/jcp.1040910303 . [DOI] [PubMed] [Google Scholar]
- 46.Moore MA, Sheridan AP. Pluripotential stem cell replication in continuous human, prosimian, and murine bone marrow culture. Blood Cells. 1979;5(2):297–311. . [PubMed] [Google Scholar]
- 47.Gartner S, Kaplan HS. Long-term culture of human bone marrow cells. Proc Natl Acad Sci U S A. 1980;77(8):4756–9. ; PubMed Central PMCID: PMCPMC349925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell C, Su T, Lau RP, Shah A, Laurie PC, Avalos B, et al. Zebrafish embryonic stromal trunk (ZEST) cells support hematopoietic stem and progenitor cell (HSPC) proliferation, survival, and differentiation. Exp Hematol. 2015;43(12):1047–61. doi: 10.1016/j.exphem.2015.09.001 ; PubMed Central PMCID: PMCPMC4666736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf A, Aggio J, Campbell C, Wright F, Marquez G, Traver D, et al. Zebrafish Caudal Haematopoietic Embryonic Stromal Tissue (CHEST) Cells Support Haematopoiesis. Sci Rep. 2017;7:44644 doi: 10.1038/srep44644 ; PubMed Central PMCID: PMCPMC5353684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stachura DL, Reyes JR, Bartunek P, Paw BH, Zon LI, Traver D. Zebrafish kidney stromal cell lines support multilineage hematopoiesis. Blood. 2009;114(2):279–89. doi: 10.1182/blood-2009-02-203638 ; PubMed Central PMCID: PMCPMC2714204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Charbord P, Pouget C, Binder H, Dumont F, Stik G, Levy P, et al. A systems biology approach for defining the molecular framework of the hematopoietic stem cell niche. Cell Stem Cell. 2014;15(3):376–91. doi: 10.1016/j.stem.2014.06.005 . [DOI] [PubMed] [Google Scholar]
- 52.Pera EM, Kim JI, Martinez SL, Brechner M, Li SY, Wessely O, et al. Isthmin is a novel secreted protein expressed as part of the Fgf-8 synexpression group in the Xenopus midbrain-hindbrain organizer. Mech Dev. 2002;116(1–2):169–72. . [DOI] [PubMed] [Google Scholar]
- 53.Valle-Rios R, Maravillas-Montero JL, Burkhardt AM, Martinez C, Buhren BA, Homey B, et al. Isthmin 1 is a secreted protein expressed in skin, mucosal tissues, and NK, NKT, and th17 cells. J Interferon Cytokine Res. 2014;34(10):795–801. doi: 10.1089/jir.2013.0137 ; PubMed Central PMCID: PMCPMC4186767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Osorio L, Wu X, Zhou Z. Distinct spatiotemporal expression of ISM1 during mouse and chick development. Cell Cycle. 2014;13(10):1571–82. doi: 10.4161/cc.28494 ; PubMed Central PMCID: PMCPMC4050162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiang W, Ke Z, Zhang Y, Cheng GH, Irwan ID, Sulochana KN, et al. Isthmin is a novel secreted angiogenesis inhibitor that inhibits tumour growth in mice. J Cell Mol Med. 2011;15(2):359–74. doi: 10.1111/j.1582-4934.2009.00961.x ; PubMed Central PMCID: PMCPMC3822802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weidinger G, Thorpe CJ, Wuennenberg-Stapleton K, Ngai J, Moon RT. The Sp1-related transcription factors sp5 and sp5-like act downstream of Wnt/beta-catenin signaling in mesoderm and neuroectoderm patterning. Curr Biol. 2005;15(6):489–500. doi: 10.1016/j.cub.2005.01.041 . [DOI] [PubMed] [Google Scholar]
- 57.Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4(12):1238–46. doi: 10.1038/ni1007 . [DOI] [PubMed] [Google Scholar]
- 58.Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108(13):3976–8. doi: 10.1182/blood-2006-05-024075 . [DOI] [PubMed] [Google Scholar]
- 59.Cross LM, Cook MA, Lin S, Chen JN, Rubinstein AL. Rapid analysis of angiogenesis drugs in a live fluorescent zebrafish assay. Arterioscler Thromb Vasc Biol. 2003;23(5):911–2. doi: 10.1161/01.ATV.0000068685.72914.7E . [DOI] [PubMed] [Google Scholar]
- 60.Westerfield M . The zebrafish book A guide for the laboratory use of zebrafish (Danio rerio) 4th ed: Eugene: University of Oregon Press; 2000. [Google Scholar]
- 61.Berrun A, Stachura DL. Development of an in vitro assay to quantitate hematopoietic stem and progenitor cells (HSPCs) in Developing Zebrafish Embryos. Journal of Visualized Experiments. 2017;(129). doi: 10.3791/56836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stachura DL, Svoboda O, Lau RP, Balla KM, Zon LI, Bartunek P, et al. Clonal analysis of hematopoietic progenitor cells in the zebrafish. Blood. 2011;118(5):1274–82. doi: 10.1182/blood-2011-01-331199 ; PubMed Central PMCID: PMCPMC3152495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stachura DL, Svoboda O, Campbell CA, Espin-Palazon R, Lau RP, Zon LI, et al. The zebrafish granulocyte colony-stimulating factors (Gcsfs): 2 paralogous cytokines and their roles in hematopoietic development and maintenance. Blood. 2013;122(24):3918–28. doi: 10.1182/blood-2012-12-475392 ; PubMed Central PMCID: PMCPMC3854111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zapata A. Ultrastructural study of the teleost fish kidney. Dev Comp Immunol. 1979;3(1):55–65. . [DOI] [PubMed] [Google Scholar]
- 65.Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF, et al. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25(6):963–75. doi: 10.1016/j.immuni.2006.10.015 . [DOI] [PubMed] [Google Scholar]
- 66.Bertrand JY, Cisson JL, Stachura DL, Traver D. Notch signaling distinguishes 2 waves of definitive hematopoiesis in the zebrafish embryo. Blood. 2010;115(14):2777–83. doi: 10.1182/blood-2009-09-244590 ; PubMed Central PMCID: PMCPMC2854425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng H, Stachura DL, White RM, Gutierrez A, Zhang L, Sanda T, et al. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell. 2010;18(4):353–66. doi: 10.1016/j.ccr.2010.09.009 ; PubMed Central PMCID: PMCPMC3003429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Driever W, Rangini Z. Characterization of a cell line derived from zebrafish (Brachydanio rerio) embryos. In Vitro Cell Dev Biol Anim. 1993;29A(9):749–54. . [DOI] [PubMed] [Google Scholar]
- 69.Yuan B, Xian R, Ma J, Chen Y, Lin C, Song Y. Isthmin inhibits glioma growth through antiangiogenesis in vivo. J Neurooncol. 2012;109(2):245–52. doi: 10.1007/s11060-012-0910-8 ; PubMed Central PMCID: PMCPMC3432204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amaral DB, Schneider I. Fins into limbs: Recent insights from sarcopterygian fish. Genesis. 2018;56(1). doi: 10.1002/dvg.23052 . [DOI] [PubMed] [Google Scholar]
- 71.Ciau-Uitz A, Monteiro R, Kirmizitas A, Patient R. Developmental hematopoiesis: ontogeny, genetic programming and conservation. Exp Hematol. 2014;42(8):669–83. doi: 10.1016/j.exphem.2014.06.001 . [DOI] [PubMed] [Google Scholar]
- 72.Cutler C, Multani P, Robbins D, Kim HT, Le T, Hoggatt J, et al. Prostaglandin-modulated umbilical cord blood hematopoietic stem cell transplantation. Blood. 2013;122(17):3074–81. doi: 10.1182/blood-2013-05-503177 ; PubMed Central PMCID: PMCPMC3811179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ravi V, Venkatesh B. Rapidly evolving fish genomes and teleost diversity. Curr Opin Genet Dev. 2008;18(6):544–50. doi: 10.1016/j.gde.2008.11.001 . [DOI] [PubMed] [Google Scholar]
- 74.Glasauer SM, Neuhauss SC. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics. 2014;289(6):1045–60. doi: 10.1007/s00438-014-0889-2 . [DOI] [PubMed] [Google Scholar]
- 75.Mitchell IC, Brown TS, Terada LS, Amatruda JF, Nwariaku FE. Effect of vascular cadherin knockdown on zebrafish vasculature during development. PLoS One. 2010;5(1):e8807 doi: 10.1371/journal.pone.0008807 ; PubMed Central PMCID: PMCPMC2808391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Svoboda O, Stachura DL, Machonova O, Zon LI, Traver D, Bartunek P. Ex vivo tools for the clonal analysis of zebrafish hematopoiesis. Nat Protoc. 2016;11(5):1007–20. doi: 10.1038/nprot.2016.053 ; PubMed Central PMCID: PMCPMC5560128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Svoboda O, Stachura DL, Machonova O, Pajer P, Brynda J, Zon LI, et al. Dissection of vertebrate hematopoiesis using zebrafish thrombopoietin. Blood. 2014;124(2):220–8. doi: 10.1182/blood-2014-03-564682 ; PubMed Central PMCID: PMCPMC4093681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lansdon LA, Darbro BW, Petrin AL, Hulstrand AM, Standley JM, Brouillette RB, et al. Identification of Isthmin 1 as a Novel Clefting and Craniofacial Patterning Gene in Humans. Genetics. 2018;208(1):283–96. doi: 10.1534/genetics.117.300535 ; PubMed Central PMCID: PMCPMC5753863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y, Chen M, Venugopal S, Zhou Y, Xiang W, Li YH, et al. Isthmin exerts pro-survival and death-promoting effect on endothelial cells through alphavbeta5 integrin depending on its physical state. Cell Death Dis. 2011;2:e153 doi: 10.1038/cddis.2011.37 ; PubMed Central PMCID: PMCPMC3122116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Davidson AJ, Zon LI. The 'definitive' (and 'primitive') guide to zebrafish hematopoiesis. Oncogene. 2004;23(43):7233–46. doi: 10.1038/sj.onc.1207943 . [DOI] [PubMed] [Google Scholar]
- 81.Yamauchi H, Hotta Y, Konishi M, Miyake A, Kawahara A, Itoh N. Fgf21 is essential for haematopoiesis in zebrafish. EMBO Rep. 2006;7(6):649–54. doi: 10.1038/sj.embor.7400685 ; PubMed Central PMCID: PMCPMC1479588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pouget C, Peterkin T, Simoes FC, Lee Y, Traver D, Patient R. FGF signalling restricts haematopoietic stem cell specification via modulation of the BMP pathway. Nat Commun. 2014;5:5588 doi: 10.1038/ncomms6588 ; PubMed Central PMCID: PMCPMC4374634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee Y, Manegold JE, Kim AD, Pouget C, Stachura DL, Clements WK, et al. FGF signalling specifies haematopoietic stem cells through its regulation of somitic Notch signalling. Nat Commun. 2014;5:5583 doi: 10.1038/ncomms6583 ; PubMed Central PMCID: PMCPMC4271318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakazawa F, Nagai H, Shin M, Sheng G. Negative regulation of primitive hematopoiesis by the FGF signaling pathway. Blood. 2006;108(10):3335–43. doi: 10.1182/blood-2006-05-021386 . [DOI] [PubMed] [Google Scholar]
- 85.Berardi AC, Wang A, Abraham J, Scadden DT. Basic fibroblast growth factor mediates its effects on committed myeloid progenitors by direct action and has no effect on hematopoietic stem cells. Blood. 1995;86(6):2123–9. . [PubMed] [Google Scholar]
- 86.de Haan G, Weersing E, Dontje B, van Os R, Bystrykh LV, Vellenga E, et al. In vitro generation of long-term repopulating hematopoietic stem cells by fibroblast growth factor-1. Dev Cell. 2003;4(2):241–51. . [DOI] [PubMed] [Google Scholar]
- 87.Walmsley M, Cleaver D, Patient R. Fibroblast growth factor controls the timing of Scl, Lmo2, and Runx1 expression during embryonic blood development. Blood. 2008;111(3):1157–66. doi: 10.1182/blood-2007-03-081323 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RT-PCR was performed for ism1 from ZKS, ZEST, CHEST, and kidney mRNA. Myeloid, lymphoid, and precursor cell mRNA isolated from zebrafish kidney was also interrogated for ism1 transcripts.
(TIF)
flk1:GFP single-cell-stage embryos were injected with 7 ng of ism1 MO (bottom); uninjected embryos served as controls (top). 24 hpf (left column) and 48 hpf (right column) zebrafish were visualized at 40x for flk1:GFP fluorescence within the trunk area denoted by black box in brightfield image at top center. Arrows indicate shortened intersegmental vessels in ism1 morphants. Numbers in corner of images denote the number of embryos displaying the imaged phenotype.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.