Summary
Mitoribosomes are specialized for the synthesis of hydrophobic membrane proteins encoded by mitochondrial DNA, all essential for oxidative phosphorylation. Despite their linkage to human mitochondrial diseases and the recent cryo-EM reconstruction of yeast and mammalian mitoribosomes, how they are assembled remains obscure. Here, we dissected the yeast mitoribosome large subunit (mtLSU) assembly process by systematic genomic deletion of 44 mtLSU proteins (MRPs). Analysis of the strain collection unveiled 37 proteins essential for functional mtLSU assembly, three of which are critical for mtLSU 21S rRNA stability. Hierarchical cluster analysis of mtLSU subassemblies accumulated in mutant strains revealed cooperative assembly of protein sets forming structural clusters and preassembled modules. It also indicated crucial roles for mitochondrion-specific membrane-binding MRPs in anchoring newly transcribed 21S rRNA to the inner membrane, where assembly proceeds. Our results define the yeast mtLSU assembly landscape in vivo and provide a foundation for studies of mitoribosome assembly across evolution.
Keywords: Mitochondrial ribosome, Mitoribosome Assembly, OXPHOS
Introduction
Mitochondria are complex eukaryotic organelles of endosymbiotic origin. They have retained only a vestige of their original α-proteobacterium ancestor genome, the mitochondrial DNA (mtDNA), while in the course of evolution, most genetic information has been transferred to the nuclear genome (Kurland and Andersson, 2000). Mitochondria contain ribosomes (mitoribosomes) specialized for the synthesis of a small set of hydrophobic proteins (8 in Saccharomyces cerevisiae, 13 in human cells), encoded by mtDNA (O'Brien, 1971; Ott et al., 2016). These proteins are core components of enzymatic complexes essential for the aerobic conversion of energy stored in nutrients into adenosine 5′-triphosphate (ATP), by oxidative phosphorylation (OXPHOS). Owing to their eubacterial ancestry, mitoribosomes are similar to bacterial ribosomes as reflected by the conservation of proteins and rRNA helices and their sensitivity to similar antibiotics. However, as a consequence of their specialization, mitoribosomes also differ from bacterial and cytoplasmic ribosomes regarding their composition, structure and function (Amunts et al., 2014; Greber and Ban, 2016; Ott et al., 2016). Their biomedical relevance stems from the fact that mutations in mitoribosome proteins (MRPs), ribosomal RNAs (rRNAs) and translation factors are responsible for a heterogeneous group of human multisystemic OXPHOS disorders, such as Leigh's syndrome, sensorineural hearing loss, encephalomyopathy and hypertrophic cardiomyopathy (De Silva et al., 2015; Rotig, 2011). Furthermore, several clinically useful antibiotics used as frontline therapy against microbial infectious diseases can unintentionally target mitoribosomes, which can result in mild to severe side effects in physiological and pathological conditions (Singh et al., 2014). Moreover, mitochondrial ribosomes and their components are emerging as both cancer biomarkers and targets for cancer therapy (Kim et al., 2017). For these reasons, understanding mitoribosome biogenesis is timely.
Ribosome assembly involves the coordinated processing and modification of rRNAs with the association of ribosomal proteins. Bacterial ribosome assembly has been reconstituted in vitro, and abundant information exists regarding the assembly pathway of bacterial and cytoplasmic ribosomes (Chen and Williamson, 2013; Davis et al., 2016; Shajani et al., 2011). In contrast, although high-resolution cryoelectron microscopic structures of yeast and mammalian mitoribosomes are available (Amunts et al., 2015; Desai et al., 2017; Greber et al., 2014), as well as structures of these mitoribosomes studied in situ by cryoelectron tomography (Englmeier et al., 2017; Pfeffer et al., 2015), the mitoribosome assembly pathway and the specific factors involved remain largely uncharacterized. The facultative aerobe/anaerobe yeast Saccharomyces cerevisiae is an amenable system for studying mitochondrial biogenesis. S. cerevisiae has a 74S mitoribosome consisting of a small 37S subunit (mtSSU) formed by a 15S ribosomal RNA (rRNA) and 34 MRPs (Desai et al., 2017), and a large 54S subunit (mtLSU) containing a 21S rRNA and 46 MRPs (Amunts et al., 2014). The two rRNAs and an mtSSU protein component, Var1, are encoded in the mtDNA, whereas all other proteins are encoded in the nuclear genome. The cryo-EM reconstructions of the yeast mtLSU confirmed that its single structural RNA is the 21S rRNA whereas the area occupied by the 5S rRNA in bacteria has been substituted by proteins.
Here, to gain insight into mitoribosome biogenesis, we have dissected the yeast 54S mtLSU assembly pathway by creating and evaluating a panel of mtLSU gene deletion strains for mitoribosome assembly and function. This analysis revealed the essentiality of most, but not all, mitoribosome proteins, for building a functional ribonucleoprotein particle, disclosed an assembly pathway based on hierarchical incorporation of protein clusters and modules, and identified the use of the mitochondrial inner membrane as an mtLSU assembly platform.
Results and Discussion
Generation of a collection of mtLSU MRP mutant strains
To study the mtLSU assembly pathway in S. cerevisiae and the importance of its protein components, we performed a systematic genome deletion of 44 genes coding for mtLSU MRPs in a favorable modified genetic background (see STAR methods, Key Resources Table 1 and Table S1).
Key Resources Table.
Antibodies used. The list of antibodies against S. cerevisiae mitoribosomal proteins includes double nomenclature to avoid confusion.
Oligonucleotides listed were used for creation of MRP mutant strains. All sequences are in 5′-3′ direction. F, forward; R, reverse.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Aco1 | Gift of Dr. Xin Jie Chen | (Chen et al., 2005) |
| Coa3 | Previously generated by our group | (Fontanesi et al., 2011) |
| Cox1 | Anti-MTCO1 antibody [11D8B7] from Abcam | Cat#ab110270 |
| Cyt b | Gift from Dr. Alexander Tzagoloff | (Dieckmann and Tzagoloff, 1985) |
| Mba1 | Gift from Dr. Martin Ott and Dr. Johannes M. Herrmann | (Ott et al. 2006) |
| Mdm38 | Gift from Dr. Johannes M. Herrmann and Peter Rehling | (Bauerschmitt et al. 2010) |
| Oxa1 | Gift from Dr. Martin Ott and Dr. Johannes M. Herrmann | (Ott et al. 2006) |
| Porin | Anti-VDAC1/Porin antibody [16G9E6BC4] - Mitochondrial Loading Control from Abcam | Cat#ab110326 |
| uL2 (Rml2) | This study | Peptide aa379- 392; CRMKVKDRPRGKDAR |
| uL3 (Mrpl9) | This study | Peptide aa207- 220; QNQDPGRVLPGRKM |
| uL6 (Mrpl6) | This study | Purified recombinant protein |
| uL16 (Mrpl16) | Generated by our group | Peptide aa161-174; CAREAFRKAGTKLPG; (De Silva et al., 2017) |
| uL23 (Mrp20) | Gift from Dr. Martin Ott and Dr. Rosemary Stuart | (Gruschke et al., 2010), (Kaur et al., 2011) |
| uL24 (Mrpl40) | Gift from Dr. Rosemary Stuart | (Kaur et al. 2011) |
| bL28 (Mrpl24) | This study | Peptide aa42- 55; DAKPLHMPKERKKFC |
| uL29 (Mrpl4) | Gift from Dr. Martin Ott | (Gruschke et al., 2010) |
| bL31 (Mrpl36) | Gift from Dr. Martin Ott | (Gruschke et al., 2010) |
| bL32 (Mrpl32) | Gift from Dr. Rosemary Stuart | (Kaur et al., 2011) |
| bL33 (Mrpl39) | Generated by our group | Peptide aa57-70; CVAERKPLDFLRTAK; (De Silva et al., 2017) |
| mL50 (Mrpl13) | Gift from Dr. Alexander Tzagoloff | (Jin et al., 1997) |
| bS1 (Mrp51) | Gift from Dr. Martin Ott | (Gruschke et al., 2010) |
| mS37 (Mrp10) | Gift from Dr. Alexander Tzagoloff | (Jin et al., 1997) |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Peptides used to generate antibodies are listed in the antibodies section. All peptides were generated by Genscript. | ||
| Critical Commercial Assays | ||
| SuperSignal West Femto Maximum Sensitivity Substrate | Thermo Scientifc | Cat#34095 |
| Deposited Data | ||
| Mass spectrometry performed with sucrose fractions from mutant strain mitochondrial extracts | This paper | Figure 3 |
| Experimental Models: Organisms/Strains | ||
| αW303-I0 | (Zambrano et al., 2007) | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, ρ+ I0 |
| αW303 / ρ0 | (Zambrano et al., 2007) | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, ρ0 |
| a/αW303-I0 + VAR1 + RNR1 | This study | Diploid, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ρ+ I0 |
| a/αW303-I0 + VAR1 + YMC2 | This study | Diploid, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-YMC2, ρ+ I0 |
| CB11 | (Myers et al., 1985) | MATa, ade1, ρ0 |
| αW303-I0 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ρ+ I0 |
| αW303-I0 + VAR1 + YMC2 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-YMC2, ρ+ I0 |
| αW303 / ρ0+ VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ρ0 |
| αW303-I0 ΔuL1 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL1∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL2 + VAR1 + YMC2 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-YMC2, ΔuL2∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL3 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL3∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL4 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL4∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL5 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL5∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL6 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL6∷KanMX, ρ + I0 |
| αW303-I0 ΔbL9 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔbL9∷KanMX, ρ + I0 |
| αW303-I0 ΔuL10 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL10∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL11 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL11∷KanMX, ρ+ I0 |
| αW303-I0 ΔbL12 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔbL12∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL13 + VAR1 + YMC2 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-YMC2, ΔuL13∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL14 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL14∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL15 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL15∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL17 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL17∷KanMX, ρ+ I0 |
| αW303-I0 ΔbL19 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔbL19∷KanMX, ρ+ I0 |
| αW303-I0 ΔbL21 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔbL21∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL22 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL22∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL23 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL23∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL24 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL24∷KanMX, ρ+ I0 |
| αW303-I0 ΔbL27 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔbL27∷KanMX, ρ+ I0 |
| αW303 / I0 ΔbL28 + VAR1 + YMC2 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-YMC2, ΔbL28∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL29 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL29∷KanMX, ρ+ I0 |
| αW303-I0 ΔuL30 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔuL30∷KanMX, ρ+ I0 |
| αW303-I0 ΔbL31 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔbL31∷KanMX, ρ+ I0 |
| αW303-I0 ΔbL32 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔbL32∷KanMX, ρ+ I0 |
| αW303-I0 ΔbL34 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔbL34∷KanMX, ρ+ I0 |
| αW303-I0 ΔbL35 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔbL35∷KanMX, ρ+ I0 |
| αW303-I0 ΔbL36 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔbL36∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL38 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL38∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL40 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL40∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL41 + VAR1 + YMC2 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-YMC2, ΔmL41∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL43 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL43∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL44 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL44∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL46 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL46∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL49 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL49∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL50 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL50∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL53 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL53∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL54 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL54∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL57 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL57∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL58 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL58∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL59 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL59∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL60 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL60∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL61 + VAR1+ RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, ΔmL61∷KanMX, ρ+ I0 |
| αW303-I0 Δmhr1 + VAR1 + RNR1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, Δmhr1∷KanMX, ρ+ I0 |
| aW303 + YEplac181 | This study | MATa, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, LEU2∷YEplac181, ρ+ |
| aW303 + YMC2 | This study | MATa, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, LEU2∷YEplac181-YMC2, ρ+ |
| aW303 + RNR1 | This study | MATa, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, LEU2∷YEplac181-RNR1, ρ+ |
| aW303-I0 + YEplac181 | This study | MATa, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, LEU2∷YEplac181, ρ+ I0 |
| aW303-I0 + YMC2 | This study | MATa, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, LEU2∷YEplac181-YMC2, ρ+ I0 |
| aW303-I0 + RNR1 | This study | MATa, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, LEU2∷YEplac181-RNR1, ρ+ I0 |
| HHY294 (Δmba1, Δmdm38, oxa1ΔC) | (Bauerschmitt et al., 2010) | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, oxa1-K332*, Δmba1∷HIS5MX6, Δmdm38∷TRP1 |
| αW303-I0 + VAR1 + RNR1 +YEplac112 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, TRP1:YEplac112, ρ+ I0 |
| αW303-I0 ΔmL57 + VAR1 + RNR1+YEplac112 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, TRP1:YEplac112, ΔmL57∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL57 + VAR1 + RNR1+YEplac112-MBA1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, TRP1:YEplac112-MBA1, ΔmL57∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL57 + VAR1 + RNR1+YEplac112-MDM38 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, TRP1:YEplac112-MDM38, ΔmL57∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL57 + VAR1 + RNR1+YEplac112-OXA1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, TRP1:YEplac112-OXA1, ΔmL57∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL58 + VAR1 + RNR1+YEplac112 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, TRP1:YEplac112, ΔmL58∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL58 + VAR1 + RNR1+YEplac112-MBA1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, TRP1:YEplac112-MBA1, ΔmL58∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL58 + VAR1 + RNR1+YEplac112-MDM38 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, TRP1:YEplac112-MDM38, ΔmL58∷KanMX, ρ+ I0 |
| αW303-I0 ΔmL58 + VAR1 + RNR1+YEplac112-OXA1 | This study | MATα, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1, URA3∷pRS316-VAR1, LEU2∷YEplac181-RNR1, TRP1:YEplac112-OXA1, ΔmL58∷KanMX, ρ+ I0 |
| Oligonucleotides | ||
| See Supplemental Table S2 | ||
| Recombinant DNA | ||
| pRS316 (URA3) To overexpress ectopic recoded VAR1 | ATCC | Cat#77145 |
| YEplac181 (LEU2) To overexpress either RNR1 or YMC2 | ATCC | Cat# 87588 |
| YEplac112 (TRP1) To overexpress MBA1, MDM38 or OXA1 | ATCC | Cat# 87590 |
| Software and Algorithms | ||
| Mascot algorithm | (Hirosawa et al., 1993) | Mascot Distiller program http://www.matrixscience.com/home.html |
| R studio software | RStudio Team (2015). RStudio: Integrated Development for R. RStudio, Inc., Boston, MA. | URL http://www.rstudio.com/ |
Despite the power of yeast genetics, studies of mitoribosomal proteins (MRPs) and their assembly pathway have been hampered by three facts: that the presence of an intron in the 21S rRNA gene may confound splicing defects with ribosome assembly defects, that in strains unable to undergo mitochondrial protein synthesis the Var1 protein will not be synthesized thus compromising mtSSU assembly, and that deletions of MRP genes frequently result in the loss of mitochondrial DNA (mtDNA). Here, to study mitoribosome assembly without interference of splicing defects, we used W303 strains carrying intronless mtDNA (W303-I0) in all our experiments. To avoid mtSSU assembly defects secondary to mitochondrial translation defects, we have relocated a recoded and functional version of the mtDNA gene VAR1 to the nucleus as reported (Sanchirico et al., 1995). Additionally, to stabilize the mtDNA in our 44 mutant strains, we overexpressed a dNTP checkpoint enzyme, RNR1, encoding the large subunit of ribonucleotide reductase, known to promote mtDNA stability of mtDNA polymerase gamma mutant strains (Baruffini et al., 2006). For some MRP mutant strains in which RNR1 overexpression did not suppress the loss of mtDNA, we effectively used an alternative gene, YMC2 (Table S1), coding for an uncharacterized mitochondrial inner membrane transporter (Van Dyck et al., 1994). YMC2 was previously reported to suppress the mtDNA loss in mutants of mrh4, coding for a DEAD-box helicase essential for mtLSU biogenesis (De Silva et al., 2013). Although the precise mechanism of mtDNA stabilization by Rnr1 or Ymc2 is not known, the success of this approach allowed us to study mitoribosome assembly with a large percentage of mtDNA-containing cells (rho+) (Table S1). Importantly, overexpression of YMC2 or RNR1, does not produce any apparent effect of 21S rRNA levels, mitochondrial protein synthesis or respiratory growth (Fig. S1A-C), and therefore, the strains constructed here are optimal for studies of mitoribosome biogenesis.
Seven mtLSU proteins are not essential for mitoribosome assembly and function
None of the deletions affected viability in medium containing fermentable carbon sources. As expected, however, most deletion strains were respiratory deficient. Thirty-seven out of the 44 mutant strains were unable to grow in YPEG media containing non-fermentable carbon sources as they had lost their oxygen-consumption capacity (Fig. 1A-B). Consistently, these 37 mutant strains were unable to perform mitochondrial protein synthesis (Fig. 1C). In contrast, 7 mutant strains (ΔuL1, ΔbL9, ΔbL12, ΔbL35, ΔbL36, ΔmL53, and ΔmL61) retained some respiratory capacity, albeit variable (Fig. 1A-B), consistent with the steady-state levels of key mtDNA-encoded OXPHOS enzyme subunits (Fig. 1D) that were synthesized by the incomplete ribosomes (Fig. 1C). As in yeast, E. coli proteins L1, L9, L35 and L36 are non-essential for protein synthesis (Dabbs et al., 1983; Ikegami et al., 2005; Subramanian and Dabbs, 1980), and their absence produce effects on protein synthesis capacity similar to those observed in the corresponding yeast mutant strains (Fig. S1D). The absence of E. coli L1, involved in the interaction between ribosomes and peptidyl-tRNA, results in 40 to 60% reduced protein synthesis capacity as we see in yeast (Sander, 1983), whereas the lack of L9, which may act as a translation fidelity factor, apparently does not affect the protein synthesis capacity (Herbst et al., 1994). On the contrary, E. coli L7/L12 is critical for association and activation of translation initiation, elongation, and termination factors with the 70S ribosome (Carlson et al., 2017). The poor performance of bL12-less yeast mitoribosomes is in accordance with such an important function. Also in humans, mutations in bL12 result in growth retardation, neurological deterioration and mitochondrial translation deficiency, predominantly affecting synthesis of COX subunits (Serre et al., 2013), a selectivity that cannot be appreciated in the ΔbL12 strain. Proteins bL9, bL35 and bL36 locate on the mtLSU surface (Fig. 1E), and mL53 and mL61 were absent from the mt-LSU structure (Amunts et al., 2014), suggesting a peripheral location as well (see below). These non-essential proteins might play some regulatory role in mitoribosome assembly and translation.
Fig 1. Most, but not all mtLSU proteins are essential for mitochondrial protein synthesis and respiration. See also Figure S1.
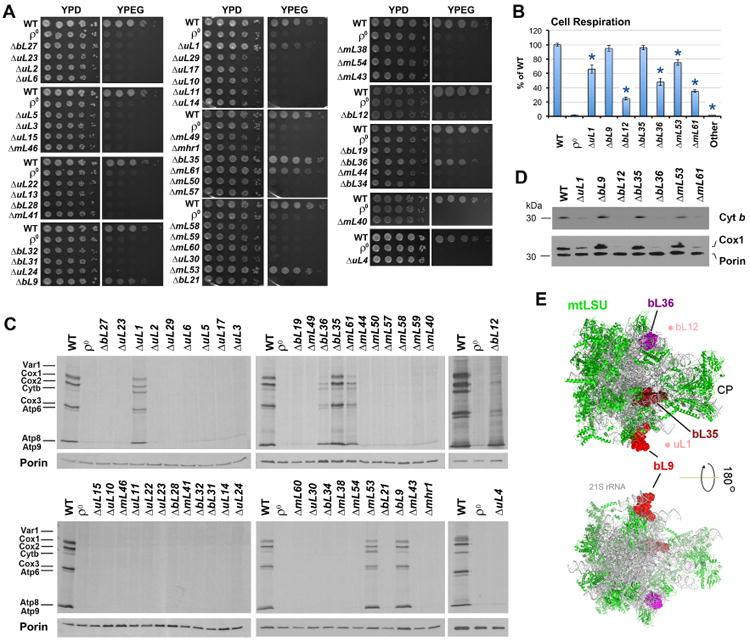
A. Serial dilution growth test of WT (W303I0) and MRP deletion mutant strains in fermentable solid media containing glucose (YPD) and non-fermentable, respiratory media containing ethanol and glycerol (YPEG).
B. Endogenous cell respiration for WT and MRP mutant strains measured polarographically in cells growing in synthetic media containing 2% galactose and 0.5% glucose. “Other” refers to the rest strains; full data is presented in Supplemental Table S2. n=3 biological replicates. Data are mean ± SD. *p<0.05.
C. Metabolic labeling with 35S-methionine of newly synthesized mitochondrial translation products in the presence of cycloheximide to inhibit cytoplasmic protein synthesis. Immunoblotting for Porin was used as a loading control.
D. Immunoblot analyses of Cox1 and cytochrome b (Cyt b) steady-state levels in the indicated strains, using Porin as a loading control.
E. Location of non-essential proteins in the mtLSU are shown on structures of the S. cerevisiae mitochondrial ribosome (PDB 5mrc) (Desai et al., 2017). Solvent-facing (upper panel) and interface-facing (lower panel) views are presented. Proteins bL36 (magenta), bL35 (firebrick) and bL9 (red) are shown as surface and the rest of proteins (green) are shown as ribbon diagrams. The 21S rRNA is shown in grey. The densities for uL1 and bL12 were not identified in the structure but the area where they localize (based on the structure of bacterial ribosomes) is indicated by light reddish dots for orientation purposes. CP, central protuberance.
Variable stability of 21S rRNA and mitoriboproteins in MRP mutant strains
We further analyzed the steady-state levels of mtLSU components (MRPs and 21S rRNA) in all mutant strains (Fig. 2A-B). Immunoblot analyses of the levels of MRPs showed that the two mtSSU proteins analyzed (bS1 and mS37) were either unaffected or increased in some mutant strains compared to the wild-type (WT), which might be due to some compensatory effects. Instead, levels of the twelve mtLSU proteins probed were decreased in most strains, likely due to degradation when mtLSU assembly is impaired (Fig. 2A and S2). Among the mtLSU proteins examined, the levels of uL6, uL16, and bL33 were decreased most markedly across the collection of mutant strains, indicating that these may be late-stage mtLSU assembly proteins. In contrast, the levels of bL31 and bL32 were elevated in numerous mutant strains (Fig. 2A and S2). The levels of these proteins may be specifically regulated since bL31 can accumulate in an extraribosomal soluble pool in the mitochondrial matrix (Prestele et al., 2009), and a precursor form of bL32 forms a complex with the m-AAA protease until it is processed to release mature bL32 that associates with preassembled ribosomal particles (Bonn et al., 2011). Deletion of uL13, uL23, mL44, mL50, mL57, and mL58 led to little or no accumulation of most mtLSU proteins, which suggests an early mtLSU assembly defect in the corresponding mutant strains. Instead, MRP levels were not affected upon deletion of uL2, uL6, uL10 and uL14, or even increased in the seven strains missing each of the non-essential mitoribosome proteins (ΔuL1, ΔbL9, ΔbL12, ΔbL35, ΔbL36, ΔmL53, and ΔmL61) (Fig. 2A and S2), indicating that mLSU assembly in these mutants was impaired at a late stage and most MRPs formed protease-protected subassemblies.
Fig 2. Altered accumulation of mtLSU proteins, 21S rRNA and subassemblies in MRP mutant strains. See also Figure S2.
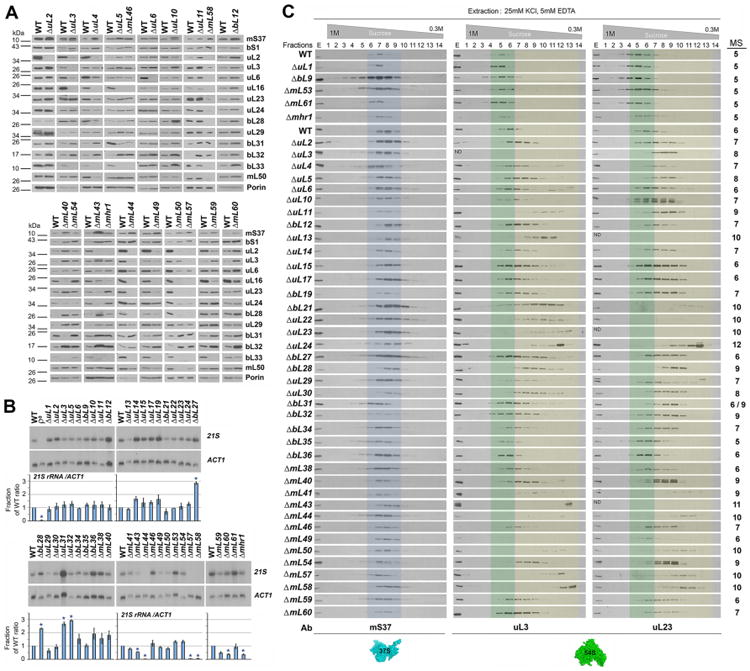
A. Immunoblot analyses of the steady-state levels of the indicated MRP proteins in WT and a selection of MRP mutant strains. Porin was used as loading control.
B. Steady-state level of 21S rRNA analyzed by northern blotting using total cellular RNA. The lower panel shows the densitometry values normalized by the signal of ACT1 and expressed relative to the WT. n=3 biological replicates. Data are mean ± SD. *p<0.05.
C. Sucrose gradient sedimentation analyses of mt-SSU (mS37) and mt-LSU proteins (uL3 and uL23) in mitochondrial extracts from the indicated strains, prepared in the presence of 25 mM KCl, 5 mM EDTA and 0.8% Triton X-100. Transparent blue and green colors mark the fractions where the WT mtSSU and mtLSU sediment, respectively. Transparent yellow color labels the fractions where mtLSU subassemblies accumulate. The fractions from these gradients that were subsequently used for mass spectrometry (MS) analyses of mitoribosome subunits and assembly factors are indicated on the right side of the figure. Ab, antibodies used.
The stability of the 21S rRNA is also expected to provide information regarding the stage of assembly of each MRP. Out of the 44 mutant strains, only 10 had abnormal 21S rRNA steady-state levels (Fig. 2B). No 21S rRNA was detected in the absence of the mitochondrion-specific proteins mL44, mL57 or mL58, consistent with previous structural findings showing that they stabilize the 21S rRNA helix 0-expansion segment (Amunts et al., 2014), and pointing toward the early assembly of these proteins. Similarly, 21S rRNA levels were also decreased in the absence of mL43, mL60 and Mhr1. Structural studies have shown that all of the 13 S. cerevisiae mitochondrion-specific mtLSU MRPs interact with rRNA, all of which, except mL43 and mL49, interact with rRNA mitochondrion-specific expansion segments (Amunts et al., 2014). Thus, whereas both protein and RNA moieties in S. cerevisiae mtLSU are substantially larger than their bacterial counterparts, the stabilization of rRNA expansions by mitochondrion-specific proteins is critical for the stability and assembly of the overall mtLSU. In contrast, 21S rRNA levels were enhanced by more than two-fold in the absence of conserved proteins whose E. coli homologs are incorporated into the LSU during late assembly stages, namely bL27, bL28, bL31 and bL32. Enhanced 21S rRNA levels correlate with increased abundance of some MRPs in the corresponding mutant strains, thus augmenting accumulation of subassembly particles.
MRP mutant mitochondria accumulate an array of 54S mtLSU subassemblies
Analysis of in vivo assembly intermediates upon depletion of various MRPs could provide insights regarding the involvement of these proteins in mitoribosome assembly. Therefore, we examined the sedimentation properties in sucrose gradients of MRPs extracted from mitochondria isolated from each of the 44 mutant strains. In all strains, the sedimentation pattern of a 37S mtSSU marker, mS37, was the same as in WT (Fig. 2C). However, the sedimentation pattern of two 54S mtLSU markers, uL3 and uL23, revealed accumulation in most strains of particles with a density lower than the WT 54S particles (Fig. 2C). Expected exceptions were the mutant strains that retain either full or some residual respiratory capacity and mitochondrial protein synthesis (Fig. 1), in which mtLSUs exhibit WT sedimentation properties (Fig. 2C).
Next, the protein composition of the 54S subassemblies, detected in the sucrose gradient fractions in each of the mutant strains (Fig. 2C), was determined by mass spectrometry analysis (LC MS/MS). All mtLSU MRPs were identified in the WT 54S mtLSU. We used Mascot scores (Log2 (mutant/WT)) to compare the composition of subassembly particles across strains. Hierarchical clustering of mtLSU protein ratios in the 54S subassemblies from each mutant strain identified five clusters containing mtLSU proteins with similar stabilities across mutant strains (Fig. 3A). Mapping of these clusters to the architecture of the yeast mtLSU (Amunts et al., 2014) revealed distinct structural modules (Fig. 3B). For example, one cluster contains proteins in the central protuberance (CP) (dark blue color in Fig. 3B), which is extensively remodeled from the bacterial CP because it lacks 5S rRNA and the 5S RNA-binding proteins L18 and L25. The 54S CP consists of mitochondrion-specific proteins (mL38, mL40, and mL46), several RNA expansion clusters, and the extensions of the five conserved proteins (uL5, uL16, bL27, bL31, and bL33) (Amunts et al., 2014). The levels of these CP MRPs were stable in the strains lacking 21S rRNA (ΔmL44, ΔmL57, and ΔmL58; Fig. 2B), which suggests that they may form an MRP subcomplex primed to be assembled into the mtLSU. The structural model previously reported did not include surface-exposed regions for which only partial density was visible, such as the L1 and L7/L12 stalks (Amunts et al., 2014). Our data, however, clustered the four known components of the L7/L12 stalk: uL10, uL11, bL12 (Kavran and Steitz, 2007) and mL53 identified in mammalian mitochondria (Brown et al., 2014). The cluster additionally included mL54 (Fig. 3A-B). Considering that mL54 and uL10 were undetectable in ΔuL11 mitochondria, our data suggest that uL10, uL11, bL12, mL53, and mL54 compose the yeast 54S L7/L12 stalk. Regarding the L1 stalk, because deletion of uL1 results in predominant attenuation of mL61 levels (Fig. 3A), these two proteins may be adjacent to each other in the mtLSU structure.
Fig 3. Mitoribosome mtLSU protein stability correlates with structural modules and defines an assembly pathway. See also Figure S3.
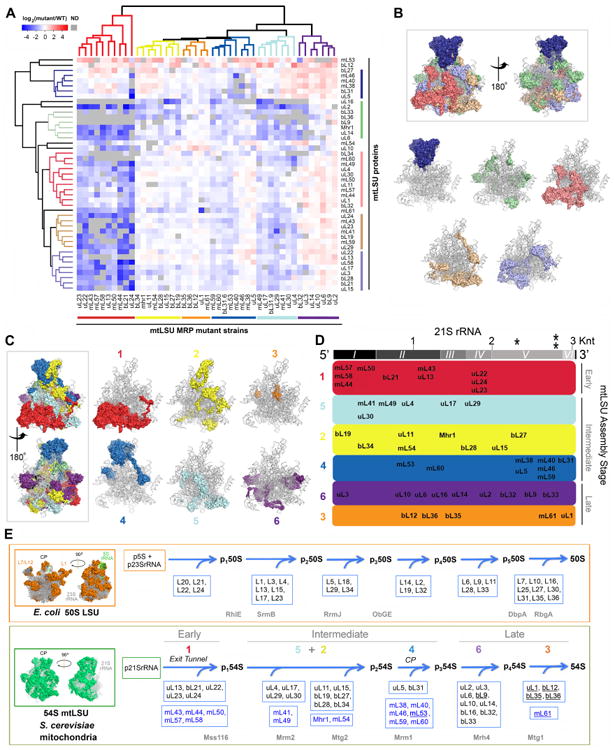
A. Following analysis by mass spectrometry of the mtLSU intermediates that accumulate in the mtLSU mutant strains (fractions indicated in Fig. 2C), the data obtained on the abundance of mtLSU subunits accumulated in mtLSU subassemblies was used for cluster analysis of MRP mutant strains (see STAR Methods). The several clusters identified are color-coded. The results presented are representative of two independent biological replicates. ND, not detected.
B. Clusters of mtLSU proteins defined in panel A (vertical clustering), mapped to the mtLSU structure (PDB-3J6B (Amunts et al., 2014)) with 21S rRNA shown in gray. The top portion presents two views of the mtLSU with all the clusters color-coded, and the lower portion presents the location of individual clusters on the 21S rRNA backbone.
C. mtLSU MRP mutant strains clusters defined in panel A (horizontal clustering) mapped to the mtLSU structure with 21S rRNA shown in gray. The left portion presents two views of the mtLSU with all the clusters color-coded, and the right portion presents the location of individual clusters (specified as 1 to 6) on the 21S rRNA backbone.
D. mtLSU assembly map following the modules defined in panel C. The proteins are distributed along the length of the 21S rRNA (grey bar on top), based on the protein-RNA interactions defined in the mtLSU structure (Amunts et al., 2014). The secondary structure domains of 21S rRNA are indicated as I to VI. * and ** indicate the two methylation sites in the 21S rRNA (Gm2270 and Um2791). Knt, kilonucleotides. The clusters are ordered following their incorporation during early, intermediate or late stages of mtLSU assembly.
E. Comparison of the mtLSU assembly map proposed here with the known assembly map of bacterial (E. coli) LSU (Chen and Williamson, 2013). On the left, the structure cartoons of the bacterial and yeast mitochondria LSU are presented. In the proposed yeast mtLSU assembly pathway, in each cluster, proteins conserved in bacteria are boxed and marked in black, and mitochondria-specific proteins are boxed and labeled in blue. Names of non-essential proteins are underlined. The clusters proposed to assemble during intermediate stages are separated into 5+2 and 4 (corresponding to the central protuberance (CP)), given the reported independent existence of a stable CP module (Box et al., 2017). At the bottom of the yeast 54S mtLSU assembly pathway are listed in grey the 6 assembly factors so far described. Proposed bacterial orthologs and paralogs of these assembly factors are listed in grey at the bottom of the 50S E. coli LSU assembly pathway.
mtLSU assembly involves hierarchical incorporation of protein clusters and modules
Hierarchical clustering of all mutant strains based on their mtLSU subassembly MRP profile revealed six clusters (Fig. 3A) that we mapped to the yeast 54S structure (Fig. 3C). Subassemblies from the mutant strains that clustered had similar MRP compositions, which we interpret as an indication that the deleted proteins assemble concurrently or at a similar stage of 54S assembly. Remarkably, these six clusters also represent distinct structural modules that we have color-coded and termed 1-6 based only on the dendrogram distribution (Fig. 3C).
Mutant strains in cluster 1 contained subassemblies with decreased levels or absence of most mtLSU MRPs. The cluster includes ΔmL44, ΔmL57, and ΔmL58 in which 21S rRNA is undetectable. This suggests that MRPs deleted in cluster 1, all of which are RNA-binding proteins, interact with the 5′-end of the 21S rRNA and are the first to be incorporated during mtLSU assembly (Fig. 3D). Some of these proteins form the polypeptide exit tunnel, which follows a path that differs from the bacterial one (Amunts et al., 2014). The boundaries of the tunnel are defined by extensions of uL22, uL23, and uL24, and several rRNA extensions and the exit is surrounded by two additional exclusive proteins, mL44 and mL50, which also form the membrane-facing protuberance (Amunts et al., 2014) that orients the ribosome for co-translational membrane insertion of newly synthesized polypeptides. The information gained through the study of yeast mitoribosomes can be highly valuable in the analysis of conserved features, particularly those regarding mitochondrion-specific MRPs. For example, mutations in mL44 were identified in a few cases of variably progressive multisystem disease with childhood-onset hypertrophic cardiomyopathy (Carroll et al., 2013; Distelmaier et al., 2015). mL44 mutant fibroblasts have lowered fully-assembled mtLSU and 16S rRNA levels, without accumulation of subassembly particles at least containing the few subunits tested (uL3, bL12, mL44 and uL13)(Carroll et al., 2013), thus suggesting an early interaction of human mL44 with the rRNA as seen in yeast.
Cluster 2 and cluster 5 included deleted proteins with separated locations on the LSU structure (Fig. 3C). These proteins may serve as the connections among other clusters and are proposed to assemble during the intermediate stages of 54S biogenesis (Fig. 3D).
Proteins deleted in cluster 3 are from the strains that retain complete or residual protein synthesis capacity and locate on the surface of the mtLSU structure (Fig. 3C), which points towards their incorporation during the terminal assembly stage (Fig. 3D). Proteins deleted in cluster 4, which mainly belong to the CP area, are proposed to form an RNA-independent assembly module (Box et al., 2017), as discussed earlier.
Proteins deleted in cluster 6 are on the surface of the mtLSU structure (Fig. 3C) and the intermediates accumulated in their absence contain WT or increased levels of most detected MRPs (Fig. 3A), indicating their late-stage assembly identity (Fig. 3D).
The distribution of the proteins forming each cluster along the linear sequence of the 21S rRNA, according to the interactions reported in the 54S structure (Amunts et al., 2014), is presented in Figure 3D. According to the bacterial 23S rRNA and the yeast 21S rRNA secondary structures, the rRNA is divided into six domains (I to VI). These domains do not, however, match well to the structural tertiary domains (Williamson, 2003) (Fig. S3A). This suggests that while anchoring of the 21S rRNA 5′ to the membrane-facing protuberance may occur in a co-transcriptional manner, the subsequent assembly steps may not follow the 5′ to 3′ transcription direction, and therefore, are not necessarily co-transcriptional. Our data indicates that proteins in each assembly cluster locate over several 21S rRNA domains, while those in cluster 4 mostly interact with helices in 21S rRNA domain V (Fig. 3D).
At least two of our protein clusters have been previously found to form rRNA-independent protein modules. First, a non-assembled MRP module present in ρ0 cells and in cells that accumulate a 54S subcomplex due to the expression of a truncated form of uL23 is composed of tunnel-exit-site and other proteins in our cluster 1 (De Silva et al., 2013; Kaur and Stuart, 2011). Second, mL38 immunoprecipation assays have allowed to disclose the existence of a stable module containing central protuberance (CP) proteins (mL38, uL5, bL27, bL31, mL46 and mL40) (Box et al., 2017). These results agree with our data showing that the proteins forming the CP (cluster 4) are largely stable in most mutants not affecting directly this structure. Hence, different modules of the 54S ribosome could assemble independently before proceeding to their hierarchical incorporation into the assembly line, in combination with other protein clusters to fulfill mtLSU biogenesis.
Early in vitro reconstitution of E. coli ribosome LSU assembly indicated that five ribosomal proteins (L4, L13, L20, L22, and L24) located at the 23S rRNA 5′ end and protein L3 that interact with the 3′ segment, are essential for the formation of a first intermediate (Nierhaus and Dohme, 1974). The binding of proteins L3 (23S rRNA domain IV) and L24 (domain I) are especially important for proper assembly since during 23S rRNA folding, they can initiate the formation of two main and independent assembly centers (Ostergaard et al., 1998). Similar results were observed in vivo ((Chen and Williamson, 2013) and Fig. 3E). Our data shows that for yeast 54S mtLSU assembly, binding of mitochondrial uL13, bL21, uL22, uL23 and uL24 to the 21S rRNA domain I in the 5′ end also occurs early, most probably during or immediately after binding to the mitochondrion-specific mL43, mL44, mL50, mL57 and mL58 membrane-facing proteins (Fig. 3E and S3A). On the contrary, L3 is required for stability of proteins in cluster 6, which assembles once the central protuberance is built. The difference might stem from the large CP remodeling that occurred from bacterial to mitochondrial ribosomes, or from the above-mentioned membrane-compartmentalized nature of the mitoribosome assembly pathway. As a note, uL3 was found mutated in cases of neonatal cardiomyopathy and psychomotor retardation associated with impaired mitoribosome function (Galmiche et al., 2011), thus highlighting the importance of this protein. Overall, the hierarchical incorporation of the protein clusters and modules identified here for yeast 54S mtLSU aligns with how the process occurs for bacterial 50S LSU assembly.
Incorporation of mitoribosome biogenetic factors to the assembly pathway
Although bacterial 50S assembly was reconstituted in vitro using purified rRNAs and RPs (Nierhaus and Dohme, 1974), the process in vivo relies on multiple ribosome assembly factors. These include rRNA processing and modifying enzymes, RNA helicases, GTPases and chaperones that help resolving kinetic traps during rRNA folding and ribonucleoprotein complex formation (Shajani et al., 2011). The factors described up to now on the biogenesis of the yeast 54S mtLSU (reviewed in (De Silva et al., 2015)) include the 21S rRNA methyltransferases Mrm1 and Mrm2, the conserved GTPases Mtg1 and Mtg2, and the helicases Mss116 and Mrh4. When we included the presence of these assembly factors in 54S subassemblies in our cluster analyses (Fig. S3B), we resolved that all of them join the assembly line following the binding and stabilization of the 21S rRNA by the membrane-bound exit tunnel MRP module (Fig. 3E). Mss116 and Mrm2 are incorporated early, followed by Mtg2 and Mrm1 during the joining of the CP MRP module, then by Mrh4 and finally by Mtg1. Orthologs or paralogs of the yeast mtLSU factors discussed here have been recognized as acting in similar stages of assembly of bacterial 50S LSU (Fig. 3E). For example, SrmB, one of the five E. coli DEAD-box proteins, participates in the assembly of the large ribosomal subunit notably by facilitating the incorporation of L13, one of the ribosomal proteins that bind 23S rRNA earliest (Proux et al., 2011). The function of E. coli SrmB during ribosome assembly is regulated by another RNA helicase, RhlE (Jain, 2008). The role of these helicases could be similar to that of yeast Mss116, a helicase that participates in early stages or mtLSU assembly (De Silva et al., 2017). Yeast Mrm2 is required for methylating U(2791) of 21S rRNA, which is important for ribosome assembly and function, possibly by inducing conformational rearrangement of the peptidyl transferase center (Pintard et al., 2002). The homolog of Mrm2 in E. coli is RrmJ, which modifies the U2552 of 23rRNA. Deletion of this protein causes destabilization and premature dissociation of the 50S LSU, which forms a 40S particle. Yeast Mtg2 belongs to the conserved Obg family of GTPases and was originally identified as a suppressor of mutant mrm2 (Datta et al., 2005). ObgE deletion in E. coli leads to the accumulation of LSU assembly intermediates containing reduced levels of L16, L33 and L34, linking Obg proteins to roles in LSU biogenesis (Sato et al., 2005). In S. cerevisiae, the rRNA Gm2270 methyltransferase, Mrm1, has an essential role in mtLSU maturation that is independent of its methyltransferase activity (Sirum-Connolly and Mason, 1993). The E. coli Mrm1 ortholog, RlmB, is indeed essential for the formation of Gm in position 2251 of 23S rRNA. However, a null rlmB mutant did not show any ribosome assembly or function defects (Lovgren and Wikstrom, 2001). In agreement with the data presented here, the yeast DEAD-box RNA helicase Mrh4 catalyzes a late assembly step required for the incorporation of uL16 and bL33 (De Silva et al., 2013). The bacterial helicase DbpA binds to the 23S rRNA helix 92, located within the PTC (Diges and Uhlenbeck, 2001) and could be performing a similar role. Overexpression of a dominant-negative dbpa mutant induced a deficit in 50S subunits and gave rise to a 45S particle containing reduced levels of L16 and L33 and surrounding proteins L25, L27, L28, L34, and L35 (Sharpe Elles et al., 2009). Finally, the yeast GTPase Mtg1 belongs to the RbgA/YlqF GTPase subfamily (Britton, 2009). Absent in E. coli, the most studied Bacillus subtilis RbgA binds to the 23S rRNA (Goto et al., 2013) and is required for late incorporation of several proteins such as L16 and L27 (Jomaa et al., 2014; Matsuo et al., 2007). In yeast mitochondria, Mtg1 interacts with the mtLSU and fully assembled mitoribosomes (De Silva et al., 2013). A Δmtg1 mutant strain is unable to perform mitochondrial protein synthesis, a phenotype that is partially suppressed by spontaneous mutations in the 21S rRNA conferring resistance to erythromycin (Barrientos et al., 2003), thus further linking the role of Mtg1 to mtLSU biogenesis.
The initial model of mtLSU assembly proposed in this manuscript is expected to serve as a framework for extended studies on mitoribosome assembly factors and the assembly steps they catalyze.
mtLSU assembly occurs on the inner membrane platform
Our data indicate that early 21S rRNA membrane anchoring is essential for its stability and 54S mtLSU ribogenesis (Fig. 2 and 3), and suggest that the 54S biogenetic process proceeds in contact with the inner membrane. This idea is compatible with the previous identification of membrane-bound pre-54S subassemblies, including the exit tunnel protein module (De Silva et al., 2013; Kaur and Stuart, 2011). Structures of yeast mitoribosomes studied in situ by cryoelectron tomography (Englmeier et al., 2017; Pfeffer et al., 2015), have identified two contact sites of the mtLSU with the inner mitochondrial membrane (Fig. S4A). One of them is formed by the 21S rRNA expansion 96-ES1. The second anchoring site involves the mitoriboproteins forming the membrane-facing protuberance (cluster 1 proteins) in contact with the ribosome receptor Mba1, a protein previously found to interact with the transmembrane Mdm38 and the Oxa1 insertase machinery (Bauerschmitt et al., 2010; Lupo et al., 2011; Ott et al., 2006; Preuss et al., 2005). To examine the dependence of mitoribosome association with the membrane on membrane anchoring proteins Mba1, Mdm38 and Oxa1, we have obtained a triple mutant of these three proteins (3×Δ or Δmba1Δmdm38, oxa1ΔC). The strain carries deletion mutations for mba1 and mdm38 and expresses a truncated version of Oxa1, missing the C-terminal domain that helps tethering the mitoribosome to the insertion complex to facilitate efficient membrane insertion of nascent chains (Bauerschmitt et al., 2010b). Although the strain is respiratory deficient (Fig. S4B), its mitochondrial protein synthesis capacity is only slightly lowered (Fig. S4C), indicating the presence of functional mitoribosomes. In fact, the steady state levels of mitoribosomal proteins are unchanged or increased compared with WT (Fig. S4D). Importantly, flotation assays have indicated that these mitoribosomes are attached to the membrane (Fig. S4E) and have a normal sedimentation profile in a sucrose gradient (Fig. S4F). Furthermore, we have overexpressed each of these proteins in several cluster-1 mutant strains, in most of which the levels of Oxa1, Mba1, Mdm38 in the mtLSU subcomplexes are increased (Fig. S4G-H), without observing any effect of 21S rRNA stabilization and mitoribosome function (Fig. S6I-K). These results suggest that Mba1, Mdm38 and Oxa1 are not essential for mitoribosome assembly.
To investigate the association of preassembly intermediates with the membrane, we have used flotation gradients. The data presented in Fig. 4A, shows that markers of these intermediates are associated with the membrane in mutants of all MRP clusters identified in Fig. 3, with some exceptions of mutants in cluster 1. In cluster-1 mutants where the 21S rRNA is absent, the stable markers are soluble (e.g. ΔmL57). In cluster-1 mutants where the 21S rRNA is present but in lowered amounts, only a portion (∼50%) of the stable markers are associated with the membrane (e.g. ΔmL50 orΔuL22). These data demonstrate that the inner membrane is used as an assembly platform for mtLSU biogenesis
Fig 4. Mitoribosomal LSU subunit assembly follows a hierarchical and module-based pathway with the initial stage on the mitochondrial membrane. See also Figure S4.
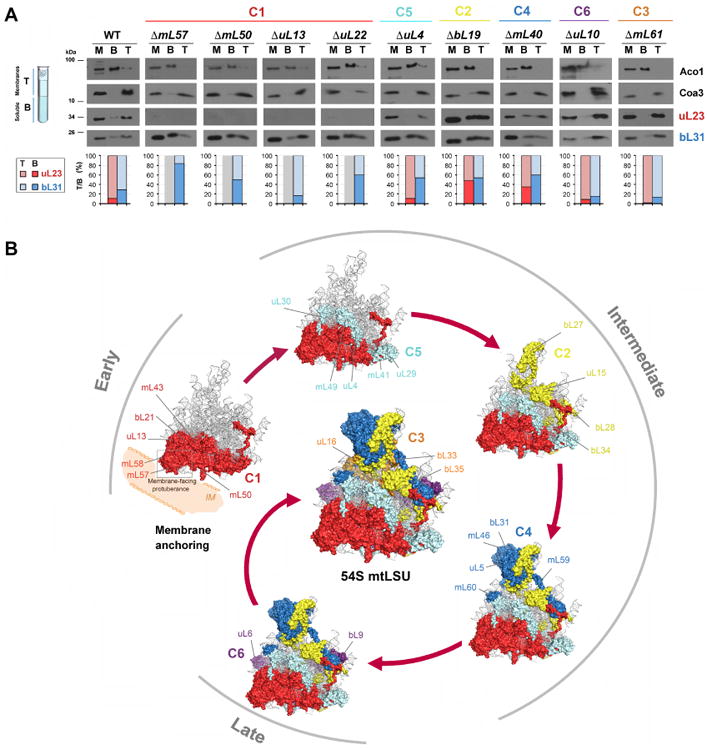
A. Mitochondria of the indicated strains were fractionated into membrane (half top of the gradient –T) and soluble (half bottom of the gradient -B) by sonication with freeze-thawing and flotation. M: total mitochondria extract. Proteins of these fractions were analyzed by immunoblotting using antibodies against uL23 and bL31. Markers of soluble (Aco1) and integral membrane proteins (Coa3) were used as controls. To quantify the proportion of soluble and membrane bound mtLSU markers, the images were digitalized, and densitometric analysis of the bands was performed using the histogram function of the Adobe Photoshop program. The data is presented as T/B ratio for uL23 (red) and bL31 (blue). The results from two independent experiments did not differ by more than 5%. C1 to C6 indicate the protein clusters identified in the analysis presented in Figure 3.
B. Model of 54S biogenesis depicting a hierarchical and module-based 54S assembly pathway. The process starts by the formation of the membrane-facing protuberance, which will anchor the growing pre-54S particle to the mitochondrial inner membrane, allowing the incorporation of the polypeptide exit tunnel-forming proteins. It then follows by the hierarchical incorporation of intermediate-stage clusters of subunits and the pre-assembled central protuberance (CP) module. The assembly would end by incorporation of a cluster of peripherally associated proteins, including those non-essentials for mitoribosome assembly (clusters C6 and C3). C1 to C6 refers to the protein clusters identified in the analysis presented in Figure 3. For each cluster, the localization in the 54S structure (PDB-3J6B) (Amunts et al., 2014) of some of the protein components is indicated.
Conclusion
Overall, the data presented here has allowed us to elaborate a model for 54S assembly (Fig. 4B) that exhibits multiple similarities with the process leading to E. coli 50S assembly (Chen and Williamson, 2013). The model also shows notable differences owing to the variances in rRNA and protein composition, the membrane-anchored nature of the mitoribosome and the use of the inner membrane as a platform for 54S mtLSU assembly. Our data showed the hierarchical protein cluster and module-based assembly of yeast mitoribosome, which is consistent with the modular assembly recently reported for the bacterial LSU, in which blocks of structured rRNA and proteins were found to assemble cooperatively following different parallel pathways (Davis et al., 2016). By defining the impact of individual mtLSU protein deletions, our work reveals the landscape of yeast mtLSU 54S biogenesis in vivo and provides a foundation to identify possible assembly factors involved during the process and to help understand mitoribosome assembly in higher eukaryotes.
Limitations of Study
The global survey of mitoribosome mutant yeast strains presented here provides an initial model for mtLSU assembly that will serve as a framework for further pathway refinement studies. Our approach to study in vivo ribosomal biogenesis is limited by the inherent complexity of the mitoribosome assembly process and the potential heterogeneity of the subassemblies analyzed. In fact, our approach does not distinguish among “on pathway” assembled intermediates, “dead end” aberrant subassemblies and potential artifact subassemblies resulting from fragmentation of larger subassemblies during experimental manipulation. In bacteria, multiple parallel pathways are operative for assembly, generating a variety of intermediates containing slightly different riboprotein sets. Our data suggest that if several parallel pathways also operate during mtLSU assembly, they would mostly vary in intermediate assembly stages. Future studies based on a combination of quantitative mass spectrometry and cryo-EM reconstruction of assembly intermediates separated by sucrose sedimentation will facilitate the identification of sub-populations of mitoribosome subcomplexes within a single sample. Finally, whereas the studies presented in this study have been performed in mutant strains, additional studies in wild-type cells will be required to refine the proposed model and clarify whether all the subassemblies identified here are true “on pathway” mitoribosome assembly intermediates.
Star Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Antoni Barrientos, Ph.D. (abarrientos@med.miami.edu).
Experimental Model and Subject Detail
All S. cerevisiae strains used are listed in the Key Resources Table. To generate all mutant strains, we obtained from GE Dharmacon (Lafayette, CO), a collection of deleted strains in the BY4741/BY4742 background, except ΔbL12, with the target MRP gene replaced by the KANMX cassette. BY4741/4742 strains could not be used directly for our studies for several reasons. These strains are known to carry a mutation affecting Hap1, a heme-dependent regulator of a number of genes involved in electron-transfer reactions. This contributes to their reduced ability to respire compared with strains carrying a WT HAP1 gene such as W303 (Ocampo et al., 2012), although other genetic differences between the strains may also contribute. Although most of the MRP gene deletion strains had lost their mtDNA, we could easily profit from their deletion cassettes. The KANMX cassette and ∼500bp flanking sequences of each corresponding MRP gene were PCR amplified using primers shown in the Key Resources Table. The KANMX cassette was then transformed into a diploid W303-I0+VAR1+RNR1 or W303-I0+VAR1+YMC2 strain. Then, haploid mutant strains with VAR1 and RNR1 or YMC2 were obtained through sporulation and tetrad dissection. In each case, strains were obtained in both mating types. Following tests of mitochondrial protein synthesis and sucrose gradient sedimentation analysis of mitoribosome subassemblies with several mutant strains that discarded any influence of the mating type on the phenotypes observed, we chose to carry all our studies using strains in the alpha mating type.
To generate the W303-I0ΔbL12 strain, the KANMX cassette from one of the MRP mutant strains was PCR amplified using primer set 1, which contains bL12 flanking sequences. Then the PCR product obtained was used as a template for a second PCR to extend the bL12 flanking sequence with primer set 2 (Key Resources Table). Then the KANMX cassette was transformed into the diploid W303-I0 strain as done for the other MRP mutant strains.
Yeast cells were grown at 30°C in the following standard media: YPD (2% glucose, 1% yeast extract, 2% peptone), YPEG (1% yeast extract, 2% peptone, 2% ethanol, 3% glycerol), WO-Gal+0.5%Glu (0.67% yeast nitrogen base, 2% galactose, 0.5% glucose).
Method Details
Quantification of mtDNA level in yeast strains
Freshly-purified yeast colonies containing mtDNA (rho+ or ρ+ colonies), carrying plasmids to overexpress VAR1 (URA3 marker) and either RNR1 or YMC2 (LEU2 marker) were grown at 30°C in liquid WO-Gal+0.5%Glu media (0.67% yeast nitrogen base, 2% galactose, 0.5% glucose) supplemented with auxotrophic requirements (adenine, histidine and tryptophan) for 20 generations. Cells were plated on fermentable YPD-agar to obtain 5 plates × ∼100 single clones. Since all our studies were made with MATα strains, in each case, the ∼100 clones were individually crossed with MATa, CB11 strains devoid of mtDNA (ρ0). Then, the percentage of rho+ cells was calculated as: (number of diploids that can grow on respiratory YPEG media after crossing with CB11)/ (total cell number). The data presented is the average of five individual plates (Table S1).
RNA isolation and analysis
Total RNA was isolated from whole cells by the hot-acidic phenol method and used for Northern-blot analyses. For Northern-blot analyses, the 100 ng RNA extracts were separated on a denaturing 1.8% agarose gel containing 2 M formaldehyde. The quality of the RNA was assessed by GelRed nucleic acid stain (Phenix, Candler, NC) prior to transferring the RNA onto a nylon membrane (Nytran®, SuPerCharge, Schleicher and Schuell, Keene, NH). The RNA bound to the nylon membrane was cross-linked with UV light and pre-hybridized at 65 °C for 1 h. Subsequently, the specific probes, in a solution containing 7% SDS, 1 μM EDTA, and 0.5 M Na2HPO4/NaH2PO4, were added to the membrane and were allowed to hybridize overnight at 65°C. After overnight incubation with probe, membrane was washed at 65°C with washing buffer containing 1% SDS, 0.1 μM EDTA, and 40 μM Na2HPO4/NaH2PO4, and exposed to Kodak X-OMAT X-ray film at -80°C. The probes for 21S rRNA and ACT1 were created by PCR on genomic DNA, column-purified (Promega, Madison, WI) and labeled with [α-32P]dCTP by random priming. The probe for ACT1 was used as a loading control. Primers used to amplify the probes were:
21S rRNA:
5′-CAGCAAAAGTATCTGAATAAGTCC-3′ and 5′-GGTTGATTCATTATGGTCCTTGC-3′
ACT1:
5′-GTGCTGTCTTCCCATCTATC-3′ and 5′-GCTTCTGGGGCTCTGAATC-3′
Characterization of the mitochondrial respiratory chain
Endogenous potassium cyanide (KCN)-sensitive cell respiration was assayed in whole cells in the presence of galactose using a Clark-type polarographic oxygen electrode from Hansatech Instruments (Norfolk, UK) at 24 °C as described (Barrientos et al., 2002).
In vivo Mitochondrial Protein Synthesis
Mitochondrial gene products were labeled with [35S]-methionine (7 mCi/mmol, Perkin Elmer) in whole cells at 30 °C for 15 min in the presence of 0.2 mg/ml cycloheximide to inhibit cytoplasmic protein synthesis (Barrientos et al., 2002). Equivalent amounts of total cellular proteins were separated by SDS-PAGE on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane and exposed to Kodak X-OMAT X-ray film.
Sucrose Gradients
The sedimentation profile of mitoribosomal proteins in WT and each MRP mutant strain from total mitochondrial extracts was analyzed essentially as described (23). Mitochondria were prepared by the method of Herrmann et al. (Herrmann et al., 1994). Four mg of protein from WT and MRP mutant strains mitochondria were solubilized in 400 μl of extraction buffer (20 mM HEPES, pH 7.4, 25 mM KCl, 5 mM EDTA, 0.5 mM PMSF and 0.8% Triton X-100) on ice for 5 min. These extraction conditions were previously used successfully to analyze an on-pathway mtLSU late assembly intermediate that accumulates in the absence of the assembly factor Mrh4 (De Silva et al., 2013). Then samples were centrifuged for 15 min at 4°C at 21,000 rpm. The supernatant was loaded onto a 5-ml linear 0.3 M–1.0 M sucrose gradient containing 20 mM HEPES, pH 7.4, 25 mM KCl, 5 mM EDTA, 0.5 mM PMSF and 0.1% Triton X-100. After centrifugation for 3 h and 10 min at 40,000 r.p.m., using a Beckman 55Ti rotor, the gradients were collected in 14 equal fractions. Forty μl from each fraction was used to determine the distributions of LSU and SSU proteins by immunoblot blot analysis. For standard Immunoblot signal detection was performed by chemiluminescence using an enhanced chemiluminescent (ECL) horseradish peroxidase (HRP) substrate that enables picogram-level protein detection. However, to detect some very low abundance proteins in the mutant strains, we used the SuperSignal West Femto Maximum Sensitivity Substrate, which is an ultra-sensitive ECL substrate for low-femtogram-level detection by immunoblot analysis (Thermo Scientific). All the antibodies used in this study are presented in the Key Resources Table. All of the gradients were performed at least in triplicate using independent mitochondrial preparations. The gradients reported are representative of each strain because the patterns observed were reproducible.
Membrane Flotation Assay
Experiment was performed using method previously described (Bauerschmitt et al., 2010). In detail, mitochondria (400 μg) were dissolved in 50 mM KCl, 10 mM MgCl2, 20 mM Tris/HCl, pH7.4, 1mM phenylmethylsulphonyl fluoride (PMSF) and disrupted by 3× 30s sonication in an ultrasonic cleaner bath (Sper Scientific 100004), with cooling intervals, followed by three freeze-thawing events. Then, the sample was adjusted to final 1.6 M sucrose in 300 μl, loaded into a centrifugation tube and layered with 250 μl 1.4 M sucrose and 100 μl 0.25 M sucrose on top. Sucrose solutions were prepared in the same buffer used to resuspend mitochondria. The samples were centrifuged in a Beckman SW55Ti rotor for 4.5 h at 255,000 g at 4 °C, and the gradient was split into a membrane (half top, T) and soluble (half bottom, B) fractions. Equal volume of each fraction was loaded on a SDS-PAGE gel and analyzed by immunoblotting.
Quantification and Statistical Analysis
Mass Spectrometry Analysis and Data Hierarchical Clustering
Proteins in sucrose gradient fractions where subassemblies sedimented in each mutant strain were precipitated using the methanol/chloroform method and analyzed by LC-MS/MS (Tandem mass spectrometry) using the services provided by the W.M. Keck Foundation proteomics facility at Yale University. All MS/MS spectra were searched in-house at the facility using the Mascot algorithm (Hirosawa et al., 1993) for un-interpreted MS/MS spectra after using the Mascot Distiller program to generate Mascot compatible files (see http://www.matrixscience.com/home.html for details). For each protein, we obtained the Mascot score ratio between each mutant strain subassembly and WT 54S particles. The log2 ratio values of each protein were processed in R studio software and the heatmap.2 program was used for the hierarchical clustering analysis.
Statistical Analysis
All of the experiments were done at least in triplicate, otherwise indicated. The data are presented as the means ± S.D. of absolute values or percentages of control. The values obtained for WT and MRP mutant strains for the different parameters studied were compared by Student's t-test. p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Dr. M. Deutscher and Dr. F. Fontanesi for scientific discussions and comments on the manuscript. We thank Dr. T. Fox, Dr. M. Ott, Dr. R. Stuart, Dr. J. Herrmann, Dr P. Rehling and Dr. A. Tzagoloff for providing reagents. This research was supported by NIGMS-R35 grant GM118141 (to AB), MDA Grant MDA-381828 (to AB), and an American Heart Association predoctoral fellowship (to R.Z).
Footnotes
Supplemental Material: Supplemental Information includes 4 figures and 1 table. Figure S1, related to Fig. 1, presents the effect of genetic background on the phenotype of yeast mutant strains and quantification of the overall protein synthesis capacity in the non-essential yeast mutant strains. Figure S2, related to Fig. 2, shows the immunoblot analyses of the steady-state levels of mitoribosome proteins in WT and MRP mutant strains. Figure S3, related to Fig. 3, presents the mapping of protein clusters on the 21S rRNA structure and presence of assembly factors in mitoribosome subassemblies. Figure S4, related to Fig. 4, includes multiple panels to demonstrate that Mba1, Mdm38 and Oxa1 are not essential for mitoribosome biogenesis. Table S1 lists all S. cerevisiae mitoribosomal LSU proteins to clarify their old and new nomenclatures and for each mutant strain, indicates their quantified respiratory capacity and their percentage of mtDNA retention following 20 generations. Table S2 presents the list of oligonucleotides used to amplify the cassettes used to generate the MRP deletion strains.
Declaration of Interests: The authors declare no competing interests.
Author Contributions: R.Z. and A.B. designed the study, designed and performed experiments, analyzed results, and wrote the paper. E.S. performed experiments.
References
- Amunts A, Brown A, Bai X, Llácer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SHW, Ramakrishnan V. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A, Brown A, Toots J, Scheres SHW, Ramakrishnan V. The structure of the human mitochondrial ribosome. Science. 2015;348:95–98. doi: 10.1126/science.aaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Korr D, Barwell KJ, Sjulsen C, Gajewski CD, Manfredi G, Ackerman S, Tzagoloff A. MTG1 codes for a conserved protein required for mitochondrial translation. Mol Biol Cell. 2003;14:2292–2302. doi: 10.1091/mbc.E02-10-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Korr D, Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 2002;21:43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruffini E, Lodi T, Dallabona C, Puglisi A, Zeviani M, Ferrero I. Genetic and chemical rescue of the Saccharomyces cerevisiae phenotype induced by mitochondrial DNA polymerase mutations associated with progressive external ophthalmoplegia in humans. Hum Mol Genet. 2006;15:2846–2855. doi: 10.1093/hmg/ddl219. [DOI] [PubMed] [Google Scholar]
- Bauerschmitt H, Mick DU, Deckers M, Vollmer C, Funes S, Kehrein K, Ott M, Rehling P, Herrmann JM. Ribosome-binding proteins Mdm38 and Mba1 display overlapping functions for regulation of mitochondrial translation. Mol Biol Cell. 2010;21:1937–1944. doi: 10.1091/mbc.E10-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn F, Tatsuta T, Petrungaro C, Riemer J, Langer T. Presequence-dependent folding ensures MrpL32 processing by the m-AAA protease in mitochondria. EMBO J. 2011;30:2545–2556. doi: 10.1038/emboj.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box JM, Kaur J, Stuart RA. MrpL35, a mitospecific component of mitoribosomes, plays a key role in cytochrome c oxidase assembly. Mol Biol Cell. 2017;28:3489–3499. doi: 10.1091/mbc.E17-04-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA. Role of GTPases in bacterial ribosome assembly. Annu Rev Microbiol. 2009;63:155–176. doi: 10.1146/annurev.micro.091208.073225. [DOI] [PubMed] [Google Scholar]
- Brown A, Amunts A, Bai XC, Sugimoto Y, Edwards PC, Murshudov G, Scheres SH, Ramakrishnan V. Structure of the large ribosomal subunit from human mitochondria. Science. 2014;2:1258026. doi: 10.1126/science.1258026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson MA, Haddad BG, Weis AJ, Blackwood CS, Shelton CD, Wuerth ME, Walter JD, Spiegel PC., Jr Ribosomal protein L7/L12 is required for GTPase translation factors EF-G, RF3, and IF2 to bind in their GTP state to 70S ribosomes. FEBS J. 2017;284:1631–1643. doi: 10.1111/febs.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CJ, Isohanni P, Poyhonen R, Euro L, Richter U, Brilhante V, Gotz A, Lahtinen T, Paetau A, Pihko H, et al. Whole-exome sequencing identifies a mutation in the mitochondrial ribosome protein MRPL44 to underlie mitochondrial infantile cardiomyopathy. J Med Genet. 2013;50:151–159. doi: 10.1136/jmedgenet-2012-101375. [DOI] [PubMed] [Google Scholar]
- Chen SS, Williamson JR. Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J Mol Biol. 2013;425:767–779. doi: 10.1016/j.jmb.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbs ER, Hasenbank R, Kastner B, Rak KH, Wartusch B, Stoffler G. Immunological studies of Escherichia coli mutants lacking one or two ribosomal proteins. Mol Gen Genet. 1983;192:301–308. doi: 10.1007/BF00392166. [DOI] [PubMed] [Google Scholar]
- Datta K, Fuentes JL, Maddock JR. The yeast GTPase Mtg2p is required for mitochondrial translation and partially suppresses an rRNA methyltransferase mutant, mrm2. Mol Biol Cell. 2005;16:954–963. doi: 10.1091/mbc.E04-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JH, Tan YZ, Carragher B, Potter CS, Lyumkis D, Williamson JR. Modular Assembly of the Bacterial Large Ribosomal Subunit. Cell. 2016;167:1610–1622. doi: 10.1016/j.cell.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva D, Fontanesi F, Barrientos A. The DEAD-Box protein Mrh4 functions in the assembly of the mitochondrial large ribosomal subunit. Cell Metab. 2013;18:712–725. doi: 10.1016/j.cmet.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva D, Poliquin S, Zeng R, Zamudio-Ochoa A, Marrero N, Perez-Martinez X, Fontanesi F, Barrientos A. The DEAD-box helicase Mss116 plays distinct roles in mitochondrial ribogenesis and mRNA-specific translation. Nucleic Acids Res. 2017;45:6628–6643. doi: 10.1093/nar/gkx426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva D, Tu YT, Amunts A, Fontanesi F, Barrientos A. Mitochondrial ribosome assembly in health and disease. Cell Cycle. 2015;14:2226–2250. doi: 10.1080/15384101.2015.1053672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N, Brown A, Amunts A, Ramakrishnan V. The structure of the yeast mitochondrial ribosome. Science. 2017;355:528–531. doi: 10.1126/science.aal2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diges CM, Uhlenbeck OC. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelmaier F, Haack TB, Catarino CB, Gallenmuller C, Rodenburg RJ, Strom TM, Baertling F, Meitinger T, Mayatepek E, Prokisch H, et al. MRPL44 mutations cause a slowly progressive multisystem disease with childhood-onset hypertrophic cardiomyopathy. Neurogenetics. 2015;24 doi: 10.1007/s10048-015-0444-2. [DOI] [PubMed] [Google Scholar]
- Englmeier R, Pfeffer S, Forster F. Structure of the human mitochondrial ribosome studied in situ by cryoelectron tomography. Structure. 2017;25:1574–1581. doi: 10.1016/j.str.2017.07.011. [DOI] [PubMed] [Google Scholar]
- Galmiche L, Serre V, Beinat M, Assouline Z, Lebre AS, Chretien D, Nietschke P, Benes V, Boddaert N, Sidi D, et al. Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum Mutat. 2011;32:1225–1231. doi: 10.1002/humu.21562. [DOI] [PubMed] [Google Scholar]
- Goto S, Muto A, Himeno H. GTPases involved in bacterial ribosome maturation. J Biochem. 2013;153:403–414. doi: 10.1093/jb/mvt022. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Ban N. Structure and function of the mitochondrial ribosome. Annu Rev Biochem. 2016;85:103–132. doi: 10.1146/annurev-biochem-060815-014343. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, Aebersold R, Ban N. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;515:283–286. doi: 10.1038/nature13895. [DOI] [PubMed] [Google Scholar]
- Herbst KL, Nichols LM, Gesteland RF, Weiss RB. A mutation in ribosomal protein L9 affects ribosomal hopping during translation of gene 60 from bacteriophage T4. Proc Natl Acad Sci U S A. 1994;91:12525–12529. doi: 10.1073/pnas.91.26.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Stuart RA, Craig EA, Neupert W. Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J Cell Biol. 1994;127:893–902. doi: 10.1083/jcb.127.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirosawa M, Hoshida M, Ishikawa M, Toya T. MASCOT: multiple alignment system for protein sequences based on three-way dynamic programming. Comput Appl Biosci. 1993;9:161–167. doi: 10.1093/bioinformatics/9.2.161. [DOI] [PubMed] [Google Scholar]
- Ikegami A, Nishiyama K, Matsuyama S, Tokuda H. Disruption of rpmJ encoding ribosomal protein L36 decreases the expression of secY upstream of the spc operon and inhibits protein translocation in Escherichia coli. Biosci Biotechnol Biochem. 2005;69:1595–1602. doi: 10.1271/bbb.69.1595. [DOI] [PubMed] [Google Scholar]
- Jain C. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA. 2008;14:381–389. doi: 10.1261/rna.800308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomaa A, Jain N, Davis JH, Williamson JR, Britton RA, Ortega J. Functional domains of the 50S subunit mature late in the assembly process. Nucleic Acids Res. 2014;42:3419–3435. doi: 10.1093/nar/gkt1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Stuart RA. Truncation of the Mrp20 protein reveals new ribosome-assembly subcomplex in mitochondria. EMBO Rep. 2011;12:950–955. doi: 10.1038/embor.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavran JM, Steitz TA. Structure of the base of the L7/L12 stalk of the Haloarcula marismortui large ribosomal subunit: analysis of L11 movements. J Mol Biol. 2007;371:1047–1059. doi: 10.1016/j.jmb.2007.05.091. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Maiti P, Barrientos A. Mitochondrial ribosomes in cancer. Semin Cancer Biol. 2017 doi: 10.1016/j.semcancer.2017.1004.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Andersson SG. Origin and evolution of the mitochondrial proteome. Microbiol Mol Biol Rev. 2000;64:786–820. doi: 10.1128/mmbr.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovgren JM, Wikstrom PM. The rlmB gene is essential for formation of Gm2251 in 23S rRNA but not for ribosome maturation in Escherichia coli. J Bacteriol. 2001;183:6957–6960. doi: 10.1128/JB.183.23.6957-6960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo D, Vollmer C, Deckers M, Mick DU, Tews I, Sinning I, Rehling P. Mdm38 is a 14-3-3-like receptor and associates with the protein synthesis machinery at the inner mitochondrial membrane. Traffic. 2011;12:1457–1466. doi: 10.1111/j.1600-0854.2011.01239.x. [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Oshima T, Loh PC, Morimoto T, Ogasawara N. Isolation and characterization of a dominant negative mutant of Bacillus subtilis GTP-binding protein, YlqF, essential for biogenesis and maintenance of the 50 S ribosomal subunit. J Biol Chem. 2007;282:25270–25277. doi: 10.1074/jbc.M703894200. [DOI] [PubMed] [Google Scholar]
- Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc Natl Acad Sci USA. 1974;71:4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien TW. The general occurrence of 55 S ribosomes in mammalian liver mitochondria. J Biol Chem. 1971;246:3409–3417. [PubMed] [Google Scholar]
- Ocampo A, Liu J, Schroeder EA, Shadel GS, Barrientos A. Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab. 2012;16:55–67. doi: 10.1016/j.cmet.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard P, Phan H, Johansen LB, Egebjerg J, Ostergaard L, Porse BT, Garrett RA. Assembly of proteins and 5 S rRNA to transcripts of the major structural domains of 23 S rRNA. J Mol Biol. 1998;284:227–240. doi: 10.1006/jmbi.1998.2185. [DOI] [PubMed] [Google Scholar]
- Ott M, Amunts A, Brown A. Organization and regulation of mitochondrial protein synthesis. Annu Rev Biochem. 2016;85:77–101. doi: 10.1146/annurev-biochem-060815-014334. [DOI] [PubMed] [Google Scholar]
- Ott M, Prestele M, Bauerschmitt H, Funes S, Bonnefoy N, Herrmann JM. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 2006;25:1603–1610. doi: 10.1038/sj.emboj.7601070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Woellhaf MW, Herrmann JM, Forster F. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nature communications. 2015;6:6019. doi: 10.1038/ncomms7019. [DOI] [PubMed] [Google Scholar]
- Pintard L, Bujnicki JM, Lapeyre B, Bonnerot C. MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J. 2002;21:1139–1147. doi: 10.1093/emboj/21.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestele M, Vogel F, Reichert AS, Herrmann JM, Ott M. Mrpl36 is important for generation of assembly competent proteins during mitochondrial translation. Mol Biol Cell. 2009;20:2615–2625. doi: 10.1091/mbc.E08-12-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss M, Ott M, Funes S, Luirink J, Herrmann JM. Evolution of mitochondrial oxa proteins from bacterial YidC. Inherited and acquired functions of a conserved protein insertion machinery. J Biol Chem. 2005;280:13004–13011. doi: 10.1074/jbc.M414093200. [DOI] [PubMed] [Google Scholar]
- Proux F, Dreyfus M, Iost I. Identification of the sites of action of SrmB, a DEAD-box RNA helicase involved in Escherichia coli ribosome assembly. Mol Microbiol. 2011;82:300–311. doi: 10.1111/j.1365-2958.2011.07779.x. [DOI] [PubMed] [Google Scholar]
- Rotig A. Human diseases with impaired mitochondrial protein synthesis. Biochim Biophys Acta. 2011;1807:1198–1205. doi: 10.1016/j.bbabio.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Sanchirico M, Tzellas A, Fox TD, Conrad-Webb H, Periman PS, Mason TL. Relocation of the unusual VAR1 gene from the mitochondrion to the nucleus. Biochem Cell Biol. 1995;73:987–995. doi: 10.1139/o95-106. [DOI] [PubMed] [Google Scholar]
- Sander G. Ribosomal protein L1 from Escherichia coli. Its role in the binding of tRNA to the ribosome and in elongation factor g-dependent gtp hydrolysis. J Biol Chem. 1983;258:10098–10103. [PubMed] [Google Scholar]
- Sato A, Kobayashi G, Hayashi H, Yoshida H, Wada A, Maeda M, Hiraga S, Takeyasu K, Wada C. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells. 2005;10:393–408. doi: 10.1111/j.1365-2443.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- Serre V, Rozanska A, Beinat M, Chretien D, Boddaert N, Munnich A, Rotig A, Chrzanowska-Lightowlers ZM. Mutations in mitochondrial ribosomal protein MRPL12 leads to growth retardation, neurological deterioration and mitochondrial translation deficiency. Biochim Biophys Acta. 2013;1832:1304–1312. doi: 10.1016/j.bbadis.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- Sharpe Elles LM, Sykes MT, Williamson JR, Uhlenbeck OC. A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res. 2009;37:6503–6514. doi: 10.1093/nar/gkp711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Sripada L, Singh R. Side effects of antibiotics during bacterial infection: mitochondria, the main target in host cell. Mitochondrion. 2014;16:50–54. doi: 10.1016/j.mito.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Sirum-Connolly K, Mason TL. Functional requirement of a site-specific ribose methylation in ribosomal RNA. Science. 1993;262:1886–1889. doi: 10.1126/science.8266080. [DOI] [PubMed] [Google Scholar]
- Subramanian AR, Dabbs ER. Functional studies on ribosomes lacking protein L1 from mutant Escherichia coli. Eur J Biochem. 1980;112:425–430. doi: 10.1111/j.1432-1033.1980.tb07222.x. [DOI] [PubMed] [Google Scholar]
- Van Dyck L, Pearce DA, Sherman F. PIM1 encodes a mitochondrial ATP-dependent protease that is required for mitochondrial function in the yeast Saccharomyces cerevisiae. J Biol Chem. 1994;269:238–242. [PubMed] [Google Scholar]
- Williamson JR. After the ribosome structures: how are the subunits assembled? RNA. 2003;9:165–167. doi: 10.1261/rna.2164903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


