Abstract
The transient receptor potential vanilloid 5 (TRPV5) channel is a member of the transient receptor potential (TRP) channel family, which is highly selective for Ca2+, that is present primarily at the apical membrane of distal tubule epithelial cells in the kidney and plays a key role in Ca2+ reabsorption. Here we present the structure of the full-length rabbit TRPV5 channel as determined using cryo-EM in complex with its inhibitor econazole. This structure reveals that econazole resides in a hydrophobic pocket analogous to that occupied by phosphatidylinositides and vanilloids in TRPV1, thus suggesting conserved mechanisms for ligand recognition and lipid binding among TRPV channels. The econazole-bound TRPV5 structure adopts a closed conformation with a distinct lower gate that occludes Ca2+ permeation through the channel. Structural comparisons between TRPV5 and other TRPV channels, complemented with molecular dynamics (MD) simulations of the econazole-bound TRPV5 structure, allowed us to gain mechanistic insight into TRPV5 channel inhibition by small molecules.
Nephrolithiasis, commonly known as kidney stones, is a common disease that affects approximately 30 million Americans1. The occurrence of kidney stones in the United States has been rising over the last decade1. One of the most critical risk factors is hypercalciuria, or high levels of Ca2+ in urine2. Around 80% of all kidney stones are comprised primarily of Ca2+ salts2. TRPV5 is a highly Ca2+-selective kidney-specific TRP channel that plays a major role in systemic calcium homeostasis3. TRPV5 is found in the apical membrane of the nephron epithelium and allows for Ca2+ reabsorption from the urine along its concentration gradient3. Loss or dysfunction of TRPV5 has been shown to severely increase urine Ca2+ levels and the occurrence of kidney stones3,4.
TRPV5 belongs to the TRPV subfamily of TRP channels, which contains six members (TRPV1–TRPV6)5. In the TRPV subfamily, TRPV5 and TRPV6 are the only Ca2+-selective channels and share high sequence identity3,6. In the absence of modulators, TRPV5 and TRPV6 are constitutively active channels3. Several endogenous modulators such as calmodulin and phosphatidylinositol 4,5-bisphosphate (PIP2) have been found to stabilize these channels in the closed or open conformation, respectively7,8. Additionally, the small-molecule antifungals econazole and miconazole, which are commonly used topically to treat skin infections, have been shown to effectively inhibit both TRPV5 and TRPV6 (refs. 3,9). Whereas TRPV6 has a wide tissue distribution6,10, TRPV5 expression is limited to the kidney3, where it plays a key role in Ca2+ reabsorption in the human body3.
No atomic structures for TRPV subfamily members were available until the recent revolution in the cryo-EM field enabled the first atomic structure determinations for TRPV1 and TRPV2 (refs. 11–13). Subsequently, the structures of other TRP channels have been determined using cryo-EM, revealing structural diversity among family members14–18. The X-ray structures of modified and truncated TRPV6 constructs have also been solved19,20, and these structures indicated a mechanism of Ca2+ permeation but did not provide information regarding ligand binding, modulation and channel gating. Prior to our work, the atomic structure of full-length TRPV5 and how small molecules modulate TRPV5 at the molecular level had remained unknown.
Here, we report a cryo-EM structure of the full-length TRPV5 channel in the presence of econazole at a resolution of 4.8 Å, with the local resolution at the transmembrane (TM) region of the channel resolved to 3.5–4.0 Å. This full-length TRPV5 structure reveals that, like other TRPV subfamily members11–13, the channel adopts a domain-swapped architecture that is common to many voltage-gated ion channels21. The structure depicts the inhibited state of the TRPV5 channel and allows for the identification of a putative binding site for econazole. Based on currently available structural information for TRPV channels combined with energy refinements of the econazole-bound TRPV5 structure in a membrane-mimetic environment by molecular dynamics (MD) simulations, we gained the first insight into the mechanism of TRPV5 inhibition.
Results
Determination of the inhibitor-bound TRPV5 structure
The full-length rabbit TRPV5 channel was heterologously expressed using Saccharomyces cerevisiae and purified using established and previously published protocols11,22–24 (Supplementary Fig. 1a). Rabbit TRPV5 has high sequence homology to human TRPV5 (> 80%), particularly in the pore region, and was favorably expressed in our system (Supplementary Fig. 1a). To investigate the structure of full-length rabbit TRPV5 in the presence of econazole, we used cryo-EM, which allowed us to image TRPV5 in an aqueous environment and avoid potential artifacts of protein crystallization, such as crystal packing that may not be biologically relevant.
We determined that the econazole half-maximum inhibition (IC50) was 2 µM for rabbit TRPV5 in HEK293 cells (Supplementary Fig. 1b,c), which is similar to that reported previously (IC50 1.3 µM)9. Detergent-solubilized rabbit TRPV5 was frozen in vitreous ice in the presence of 6 µM econazole. This concentration of econazole is three to four times higher than the IC50 values reported by our group (Supplementary Fig. 1c) and by others in mammalian cells9. This was the highest concentration we could achieve while minimizing the effect of DMSO on freezing conditions and taking into account the limit of econazole solubility.
The overall resolution of the cryo-EM TRPV5 density map (TRPV5ECN) is 4.8 Å, with the local resolution in the TM region between 3.5 and 4 Å. This structure was reconstructed from one 3D class that contained 50,566 particles using RELION 2.0 (refs. 25,26) (Table 1, Fig. 1a and Supplementary Figs. 1d,e and 2). The number of particles that went into the final reconstruction is in line with recently published TRPV1 channel structures with various ligands27,28, and we believe that the other 3D classes from this data-set represent poor-quality TRPV5 particles. The quality of the cryo-EM densities in the TM region allowed us to confidently build an atomic model of this portion of the protein (Supplementary Figs. 2 and 3). The N-terminal ankyrin-repeat domain (ARD) and C terminus were resolved in the range of 4–5.5 Å, which allowed model building in this region and the fitting of a TRPV5 ARD homology model based on the TRPV6 ARD structure29 (Supplementary Figs. 2 and 3). The final TRPV5 structural model (TRPV5ECN) is presented in Fig. 1b.
Table 1.
Cryo-EM data collection, refinement, and validation
| TRPV5ECN (EMD-7058, PDB 6B5V) |
|
|---|---|
| Data collection and processing | |
| Magnification | 45,455 |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å 2) | 44 |
| Defocus range (µm) | 1.5–4.0 |
| Pixel size (Å) | 0.55 |
| Symmetry imposed | C4 |
| Initial particle images (no.) | 925,927 |
| Final particle images (no.) | 50,566 |
| Map resolution (Å) | 4.8 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 3.5–5.5 |
| Refinement | |
| Model resolution cutoff (Å) | 4.8 |
| FSC threshold | 0.143 |
| Map sharpening B factor (Å2) | −338 |
| Model composition | |
| Nonhydrogen atoms | 1 |
| Protein residues | 2,352 |
| Ligands | 8 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.03 |
| Bond angles (°) | 0.791 |
| Validation | |
| MolProbity score | 2.14 |
| Clashscore | 8.79 |
| Poor rotamers (%) | 2.37 |
| Ramachandran plot | |
| Favored (%) | 94.36 |
| Allowed (%) | 5.64 |
| Disallowed (%) | 0.00 |
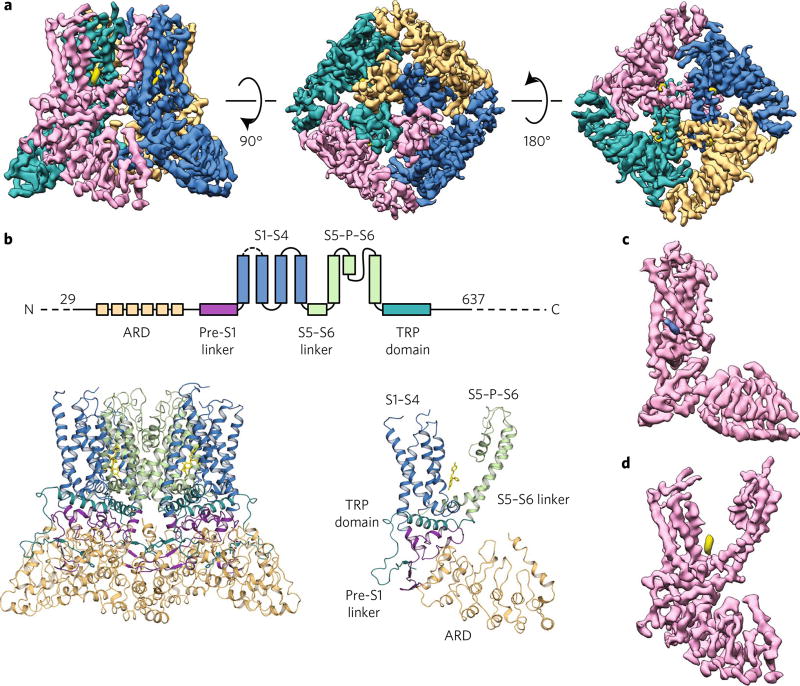
Fig. 1. Econazole-bound TRPV5 (TRPV5ECN) structure as determined by cryo-EM.
a, Density map of TRPV5ECN at 4.8-Å resolution. Each monomer is depicted in a different color (orange, blue, pink, and teal), and the density attributed to econazole is shown in yellow. The threshold of the density map was adjusted so that the putative calcium density is not visible for better visualization of the pore in this figure. b, A schematic representation of the domains present in TRPV5. Dashed lines indicate regions for which a model could not be built. Below, the TRPV5ECN model in tetrameric (left) and monomeric (right) form. c,d, A density map of a single monomer of TRPV5ECN (pink) with densities ascribed to phosphatidylinositol (blue) (c) and econazole (yellow) (d).
Overall TRPV5 channel architecture
All recently determined cryo-EM structures of TRP channels have revealed a domain-swapped architecture that is shared by many voltage-gated ion channels11–18. As expected, the full-length TRPV5 structure is similar to other TRPV subfamily structures11–13 (Fig. 1a,b). Specifically, it is a tetrameric ion channel comprised of classic TRPV channel domains, such as transmembrane helices (S1–S6), an N-terminal ARD, a linker domain, and the C-terminal TRP domain (Fig. 1b). While our construct is a full-length TRPV5, we only were able to resolve ~85% of the total protein (Trp29–His637), owing to the N-terminal and C-terminal flexibility (Fig. 1b). The loop region between S1 and S2 (Arg355–Ala376) is also not resolved (Fig. 1b). The loop between S2 and S3, which is not resolved in the structures of the close homolog TRPV6 (refs. 19,20), is resolved in our TRPV5 structure. This region resembles that of TRPV1 and TRPV2.
Additionally, two nonprotein densities adjacent to each TM domain of the channel were observed in our structure (Fig. 1c,d). One was clearly visible between the S1 and S2 helices and was assigned to a tightly bound lipid molecule (Fig. 1c and Supplementary Fig. 4a). This density is in an analogous position to the lipid densities that have been found in previously published TRPV1 and TRPV2 structures resolved in amphipol and nanodiscs13,28 (Supplementary Fig. 4a). TRPV5 was imaged in the presence of decyl maltose neo-pentyl glycol (DMNG) detergent, which suggests that this tightly bound lipid is preserved independently of the sample preparation method. On the basis of the shape of this density and the densities in published results for other TRPV channels, we assigned this density to phosphatidylcholine (Fig. 1c and Supplementary Fig. 4a).
The second density was found positioned between S4 and S5 of each subunit and S6 of the neighboring subunit and was putatively assigned to econazole (Fig. 1d). The TRPV1 structure with capsazepine is the only other TRPV subfamily member cryo-EM structure that was solved in the presence of an inhibitor28 (Supplementary Fig. 4b). We compared the position of the econazole density in our TRPV5 structure to that of the density assigned to capsazepine in the TRPV1 structure (Supplementary Fig. 4b). In both structures, these inhibitors occupy similar positions, suggesting that this ligand-binding pocket may be conserved among TRPV channels (Supplementary Fig. 4b).
Econazole-binding pocket
Econazole is an antifungal drug that has been shown to inhibit both TRPV5 and TRPV6 channels9,30. In our experiments, rabbit TRPV5 was inhibited in the presence of 2 µM econazole when overexpressed in HEK293 cells (Supplementary Fig. 1b,c). Purified TRPV5 was frozen in the presence of 6 µM econazole, which is in the upper concentration range of the published econazole IC50 (1.3 µM)9 and the IC50 of 2 µ M determined by our group for TRPV5 expressed in the HEK293 cell line (Supplementary Fig. 1b,c). An econazole molecule (either the R or S enantiomer) can be fit reasonably well into the assigned cryo-EM density by manual docking (Supplementary Fig. 5a – c). Although the low resolution of the structure does not allow us to unambiguously determine by cryo-EM which enantiomeric form of the drug is bound to the protein, it seems that the chlorophenyl moiety of (R)-econazole fits better in the electron density (Supplementary Fig. 5a – c). Whereas the chlorophenyl and dichlorophenyl moieties of econazole can be docked successfully into the density (Supplementary Fig. 5a – c), the density for the imidazole ring was not resolved. However, this is probably due to the flexible nature of this moiety compared to the rest of the compound and the current resolution of the cryo-EM structure in this region.
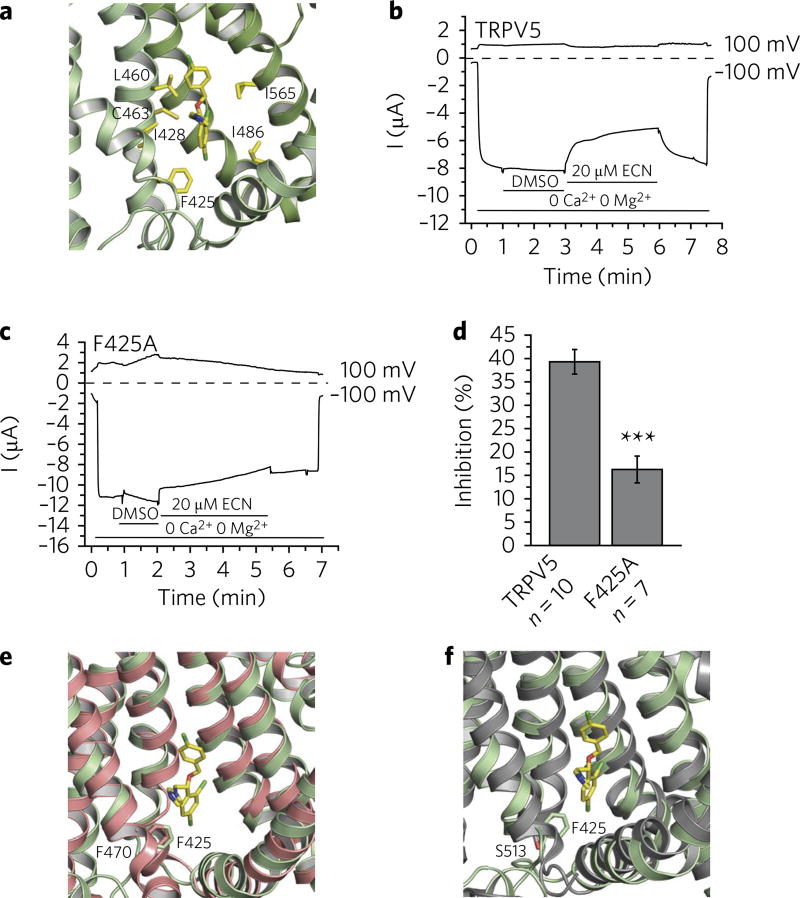
Several residues surrounding the density assigned to econazole were observed to be within interaction distance of the econazole molecule (minimum distance within 4.0 Å between heavy atoms) and, therefore, could be involved in econazole binding (Supplementary Table 1). However, it is of note that not all residues within interaction distance remained so during MD simulations, suggesting that some residues may play a less significant or transient role in econazole binding. Residues that stayed within 4.0 Å from at least three of the four econazole molecules during the MD simulations with either enantiomeric form (shown in boldface in Supplementary Table 1) were assigned as the putative econazole-binding pocket and include Phe425 and Ile428 located on the S3 helix, Leu460 and Cys463 located on the S4 helix, Ile486 from the S4–S5 linker region, and Ile565 located on the S6 helix of an adjacent monomer (Fig. 2a). Although the relative contribution of these residues to the free energy of econazole binding cannot be inferred by simple energy relaxation, the MD data support the hypothesis that this mainly hydrophobic pocket may recognize and bind small molecules like econazole and miconazole that inhibit TRPV5 and TRPV6 (Supplementary Fig. 5d,e).
Fig. 2. Econazole inhibition of TRPV5.
a, Zoomed-in view of the econazole-binding pocket. (R)-econazole is represented as yellow sticks. Residues that stayed within 4.0 Å from at least three of the four econazole molecules during the MD simulations with either enantiomeric form are labeled and shown in yellow. b, A representative TEVC trace of wild-type rabbit TRPV5 in Xenopus oocytes (n = 10 individual traces) at 100 mV and −100 mV in the presence of 2.2% DMSO (solvent) and 20 µM econazole (ECN), indicated by horizontal lines. TRPV5 currents under basal conditions are largely blocked by the 1 mM Mg2+ and trace amounts of Ca2+ in the bath solution. Monovalent currents are initiated by removing Mg2+ and chelating Ca2+ with 1 mM EGTA. c, A representative TEVC trace of the F425A mutant rabbit TRPV5 in Xenopus oocytes (n = 7 individual traces) at 100 and −100 mV in the presence of 2.2% DMSO (solvent) and 20 µM econazole (ECN), indicated by horizontal lines. TRPV5 currents under basal conditions are largely blocked by the 1 mM Mg2+ and trace amounts of Ca2+ in the bath solution. Monovalent currents are initiated by removing Mg2+ and chelating Ca2+ with 1 mM EGTA. d, Statistical summary (% inhibition) at −100 mV, ***P < 0.001, two-sample t test. Data shown are mean ± s.e.m. e,f, The econazole binding-pocket aligned to the ligand-binding pockets of TRPV2 (PDB 5AN8) (e) and TRPV1 (PDB 3J5R) (f). Residues of interest are labeled and shown as sticks.
In order to provide additional evidence that this density is indeed econazole and that these residues constitute the econazole-binding pocket, we used the relatively high-throughput Xenopus oocyte two-electrode voltage clamp (TEVC) technique and tested the effect of mutating one residue on each region of the proposed putative binding pocket. Specifically, we tested the effect of Phe425Ala, Leu460Ala, Ile486Ala, and Ile565Ala mutations on TRPV5 inhibition by econazole (Supplementary Fig. 6). Consistent with previous observations30, higher concentrations of econazole were required to achieve rabbit TRPV5 inhibition in Xenopus oocytes compared to the HEK293 expression system. In Xenopus oocytes, we used 20 µM econazole, which resulted in ~40% inhibition of the wild-type channel (Fig. 2b,d). Among the mutants tested, those with Phe425Ala showed a significantly reduced inhibition by econazole (Fig. 2c,d), implicating the involvement of this residue in econazole binding. The other tested alanine mutations (Leu460Ala, Ile486Ala, and Ile565Ala) did not reduce the inhibitory effect of econazole on TRPV5 (Supplementary Fig. 6). Ile486Ala and Ile565Ala were inhibited more than the wild-type TRPV5, but those mutants also displayed some inhibition by DMSO at the concentration necessary to dissolve econazole (Supplementary Fig. 6), implying that processes other than econazole binding may be involved. Overall, it is likely that the Leu460, Ile486, and Ile565 residues do not contribute to econazole inhibition to the same extent as Phe425.
The econazole-binding pocket in TRPV5 resembles the ‘vanilloid’-binding pocket that was identified in TRPV1 by cryo-EM28 (Supplementary Fig. 7). The TRPV1 vanilloid-binding pocket can accommodate activators (resiniferatoxin and capsaicin), as well as inhibitors, such as capsazepine, and modulators of channel activity, such as phosphatidylinositol lipids28 (Supplementary Fig. 7). It has been proposed that the exchange between phosphoinositides and the TRPV1 activators and inhibitors in this vanilloid-binding pocket allosterically regulates TRPV1 (ref. 28). Moreover, the vanilloid-insensitive TRPV2 channel can be transformed into the vanilloid-sensitive channel by modification of four critical residues in this vanilloid-binding pocket31,32. One of these residues is located on the S3 helix: for mouse TRPV1, it is a polar Ser513, and for mouse TRPV2, it is a hydrophobic Phe467 (Fig. 2e,f). Changing TRPV2 Phe467 to Ser467 facilitates capsaicin entering into the vanilloid-binding pocket of TRPV2 (refs. 31,32). Changing TRPV1 Ser513 to Tyr513, a residue that is equally as bulky as Phe467 in TRPV2, diminishes TRPV1 activation by capsaicin31,32. The S3 helix of rabbit TRPV5 harbors Phe425 at the same position as mouse TRPV2 Phe467 (or rabbit TRPV2 Phe470) (Fig. 2e) and mouse TRPV1 Ser513 (Fig. 2f). Intriguingly, like TRPV2, TRPV5 does not respond to capsaicin or capsazepine9, suggesting that the presence of the hydrophobic phenylalanine at this position may prevent these TRPV1-specific modulators from entering the TRPV5 ligand-binding pocket, but could allow for the interaction with its specific modulators via a mechanism similar to that seen for TRPV2 vanilloid insensitivity31,32
Additionally, our MD simulations of the econazole-bound TRPV5 tetramer also indicated high flexibility of the drug within the binding pocket of the TRPV5 subunits (r.m.s. deviations of 2.0–6.4 Å for (R)-econazole and 1.9–7.2 Å for (S)-econazole; Supplementary Fig. 8), whereas the TRPV5 tetramer kept its overall conformation during 25-ns MD simulation (average Cα r.m.s. deviations of the TM helices is 2.3 ± 0.1 Å for the protein bound to (R)-econazole and 1.9 ± 0.1 Å for the protein bound to (S)-econazole with respect to the initial structure derived from the electron density map; Supplementary Fig. 9a,c). As also seen in simulations of TRPV1 bound to antagonists or agonists33, the four monomers in the TRPV5 tetramer show different dynamic behavior, as evident from comparison of their Cα r.m.s. fluctuations (Supplementary Fig. 9b,d), in which peaks correspond to loop regions. The initial identical poses of econazole molecules (named ECN_1–ECN_4) in the four monomers of the TRPV5 tetramer (monomers A–D) differed during dynamics, with ECN_1 and ECN_2 molecules of both (R)-econazole and (S)-econazole fluctuating more than ECN_3 and ECN_4 during the production run (Supplementary Figs. 8 and 10). This differential flexibility of the econazole molecules within the binding pockets may be dependent on the surrounding environment. Interestingly, the number of 1-palmitoyl-2-oleoyl phosphatidyl choline (POPC) molecules in contact with each of the three ring moieties of econazole (chlorophenyl, dichlorophenyl, and imidazole) showed moderate-to-strong correlation with the r.m.s. deviation of the most-fluctuating econazole molecules during the production run (correlation coefficients are 0.53 and 0.46 for the ECN_1 and ECN_2 (R)-molecules, respectively, and 0.7 and 0.68 for the corresponding (S)-molecules) but much lower correlation with the two econazole molecules that moved less (ECN_3 and ECN_4), suggesting a direct effect of lipid contacts on the flexibility of the econazole molecules.
TRPV5 ion-permeation pore in the presence of econazole
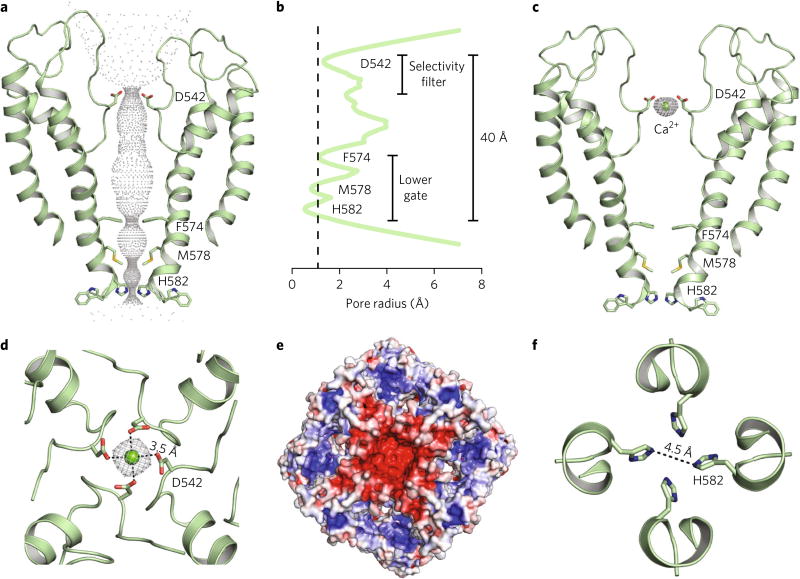
TRPV5 and TRPV6 are Ca2+-selective non-voltage-gated ion channels that belong to the TRP family of proteins5. The ion-permeation pore of the econazole-bound TRPV5 is comparable to that of other TRPV channels, which is formed between the S5 helix, the P-helix loop, and the S6 helix (S5–P–S6) (Fig. 3a,b). Nevertheless, TRPV5 is a Ca2+-selective channel, and unique pore features are noted in the structure. Four aspartate residues (Asp542), one from each monomer, line the selectivity filter of the channel and coordinate Ca2+ ions. In fact, a density possibly corresponding to a Ca2+ ion along the central-pore axis was resolved in our econazole-bound TRPV5 structure (Fig. 3c). The carboxylate groups of Asp542 are not clearly resolved in the EM map, owing to radiation damage (Supplementary Fig. 3a); however, the quality of the cryo-EM densities in this region allowed backbone assignment. The position of the Asp542 side chain in the TRPV5 pore was selected based on the best-allowed rotameric position (Supplementary Fig. 3a).
Fig. 3. Ion pore of TRPV5ECN.
a, Dimer cartoon representation and pore profile of TRPV5ECN generated with HOLE. Constricting residues are labeled and represented as sticks. b, Graphic representation of the radius of the TRPV5ECN pore profile. The dotted line represents the radius of a nonhydrated calcium ion. Residues involved in pore constriction are labeled. c, Dimer cartoon representation of the TRPV5ECN pore. Density attributed to Ca2+ is shown in gray mesh. d, Top-down (extracellular) view of the selectivity filter of the tetrameric TRPV5ECN. The density attributed to Ca2+ is shown in gray mesh. Dashed lines indicate the length from the center of the oxygen atom of Asp542 to the center of the Ca2+ atom. e, Electrostatic map of the extracellular view of TRPV5ECN. Blue indicates positive and partial-positive regions. Red indicates negative and partial-negative regions. f, Top-down view of the tightest constriction point of the TRPV5ECN pore at His582. Dashed line indicates distance between the centers of each atom.
The ionic diameter of Ca2+ is 2.28 Å and ~10–12 Å when fully hydrated34,35. The distance between the oxygen atoms of two opposing Asp542 carboxylates in the selectivity filter, as measured from our model, is ~7 Å, which may require water molecules to facilitate Ca2+ ion interaction with the filter35. The distance between the putative Ca2+ ion and carboxylate oxygen atoms of the Asp542 residues in our model (Fig. 3d, dash lines) is 3.5 Å, suggesting that the density represents a partially hydrated Ca2+ ion coordinated by the selectivity filter (Fig. 3c,d). Moreover, the extracellular vestibule of the TRPV5 pore is predominantly electronegative (Fig. 3e), suggesting that it directs Ca2+ ions toward the selectivity filter. In our MD simulations, a Ca2+ ion remains stably coordinated by the four Asp542 residues along with one (in the (R)-econazole-bound TRPV5 complex) or up to two (in the (S)-econazole-bound TRPV5) water molecules, thus maintaining a coordination number of ~7 throughout the simulation. Whereas all four oxygen atoms of the Asp542 carboxylate moieties on monomers B and C maintain an average minimum distance of 2.9 ± 0.2 Å from each other in the TRPV5 complex bound to (R)-econazole and an average distance of ~2.2 Å from the Ca2+ ion, only two out of four oxygen atoms of the Asp542 residues on monomers A and D contribute to the Ca2+ coordination through an average minimum distance of 4.1 ± 0.3 Å from each other and an average distance of ~2.2 Å from the Ca2+ ion. The simulation of TRPV5 bound to (S)-econazole behaves in a similar manner.
The lower gate of econazole-bound TRPV5 is tightly closed, as expected, owing to channel inhibition by this small molecule (Fig. 3a,b). The inner helical bundle at the Phe574, Met578, and His582 positions constrict the pore to a diameter of ~5.9 Å at Phe574, ~5.3 Å at Met578, and ~4.5 Å at His582 (Fig. 3b,f). As previously noted in the TRPV6 structure, Met577 is the key residue involved in channel closure19,20 (Fig. 4a). In our current TRPV5 model, His582 is the narrowest point in the pore (Fig. 3f and Fig. 4b). However, our 25-ns MD simulations, during which these residues moved to average minimum distances > 9 Å, did not support this assessment. Whereas both Phe574 and Met578 residues on monomers A and D maintained a 4.0 ± 0.4 and 4.1 ± 0.4 Å average minimum distance, respectively, between side chain heavy atoms of TRPV5 bound to (R)-econazole during MD simulations, the two residues on monomers B and C assumed an average minimum distance of 7.8 ±0.4 and 7.2 ± 0.5 Å, respectively. Taken together, our data suggest that the pore of the econazole-bound TRPV5 is nonconducting, as both Phe574 and Met578 residues might obstruct Ca2+ flow A similar dynamic behavior was observed in the TRPV5 complex bound to (S)-econazole.
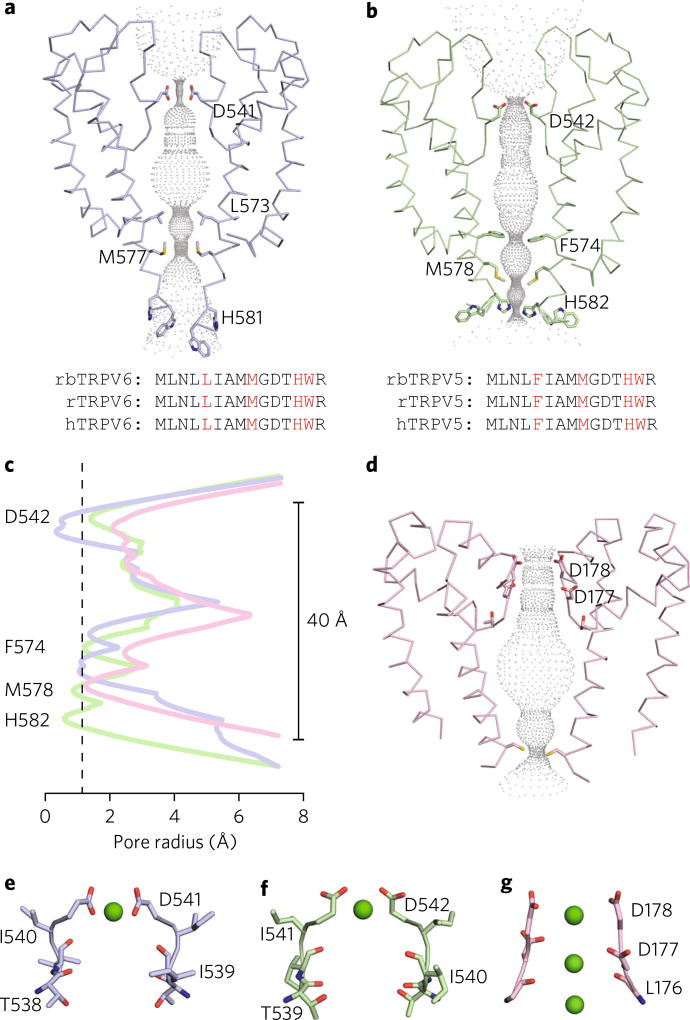
Fig. 4. Pore comparison of select Ca2+-selective ion channels.
a,b, Dimer ribbon representation and pore profile of TRPV6* (blue, PDB 5WO7 (ref. 20)) (a) and TRPV5ECN (green) b. Pore profiles were generated using HOLE. Constricting residues for each are labeled and represented as sticks. Corresponding sequence alignments of rabbit (rb), rat (r), and human (h) homologs of TRPV5 and TRPV6 are shown below. Constricting residues are in red. c, Graphic representation of the radii of the TRPV6* (blue), TRPV5ECN (green), and CavAB (pink, PDB 4MVM) pore profiles. The dotted vertical line represents the radius of a nonhydrated calcium ion. TRPV5ECN residues involved in pore constriction are labeled. d, Dimer ribbon representation and pore profile of CavAB generated with HOLE. Constricting residues are labeled and represented as sticks. e–g, Dimer ribbon representation of the selectivity filters for TRPV6* (blue) (e), TRPV5ECN (green) (f) and CavAB (pink) (g). Residues expected to be involved in Ca2+ coordination are labeled and represented as sticks. Calcium ions are shown as green spheres.
Although TRPV5 and TRPV6 share high levels of sequence homology, we noted several differences in the pore architecture between full-length rabbit TRPV5 and the mutated and truncated rat TRPV6 construct (TRPV6*) employed in its structural studies20 (Fig. 4a,b). Specifically, the first residue that constricts the TRPV5 pore is Phe574, which is conserved between rabbit, rat, and human TRPV5, but diverges to Leu573 in rabbit, rat, and human TRPV6 (Fig. 4a,b). While both side chains are hydrophobic, the observed difference in the level of pore obstruction at this amino acid position is due to the size difference between phenylalanine and leucine (Fig. 4a – c). Another notable difference is at the position of Met578 in rabbit TRPV5 versus that of Met577 in rat TRPV6* (Fig. 4a,b). In both channel structures, the methionine residues create a constricted region in the pore (Fig. 4a,b), but the position of the methionine side chains are distinct for each structure (Fig. 4a,b).
The major difference noted between the full-length rabbit TRPV5 and the rat TRPV6* structures is at the selectivity filter (Fig. 4a,b). The side chain of Asp541 in rat TRPV6* projects toward the pore (Fig. 4a), whereas the position of the analogous Asp542 side chain in the rabbit TRPV5 structure resembles that of voltage-gated Ca2+ channels34,35 (Fig. 4b – d). The distance between the center of the oxygen atoms from two opposing Asp541 in the rat TRPV6*, which coordinate Ca2+ at the selectivity filter, is 4.6 Å, suggesting that an aspartate from each subunit binds dehydrated Ca2+ (ref. 19) (Fig. 4e). Although we could not completely resolve the side chain of Asp542 in rabbit TRPV5, our model and MD data suggest that the aspartate side chain positions in the pore could coordinate partially hydrated Ca2+ (Fig. 4f), as has been proposed for CavAb and Cav1.1 channels34–36 (Fig. 4g).
Insight into the mechanism of econazole inhibition of TRPV5
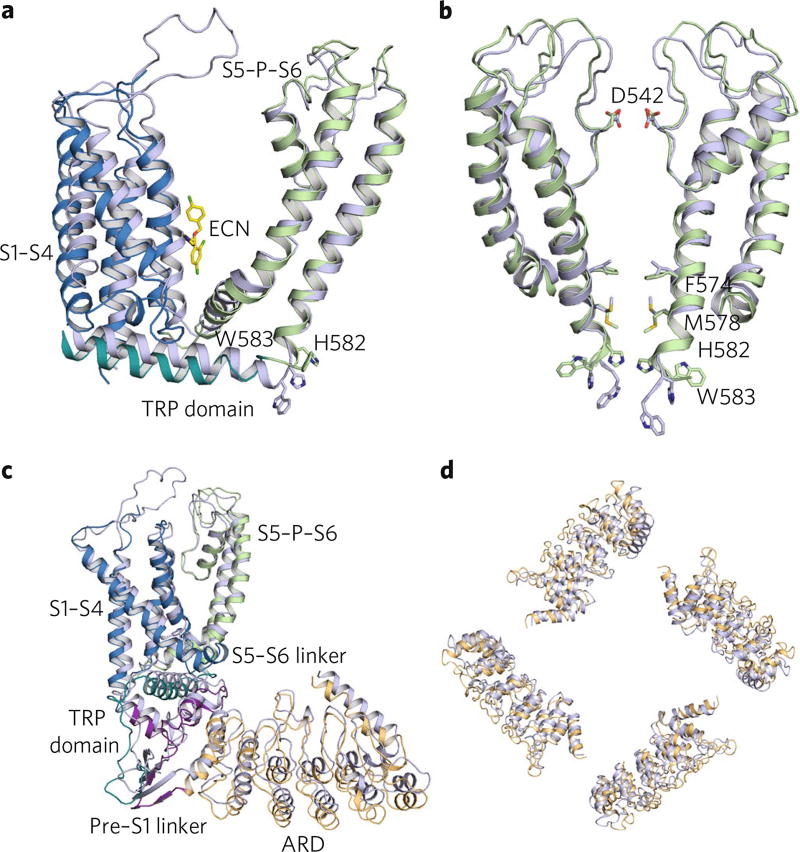
Econazole is a potent inhibitor of TRPV5 and TRPV6 channels, which, upon binding, critically affects ion conduction through the channel pore9,30. The high level of sequence homology between these two TRPV subfamily members, as well as their comparable functional properties, has allowed us to propose a mechanism of econazole inhibition of TRPV5. The structure of TRPV6*(ref. 20) was solved via X-ray crystallography in the absence of any organic small-molecule inhibitor, and the rabbit TRPV5 structure that is presented here is econazole bound. Superimposition of these two nonconducting structures revealed several conformational differences between the unbound TRPV6* (ref. 20) and inhibited TRPV5 structures independent of sequence conservation (Fig. 5). Econazole binding to the hydrophobic pocket between the S3–S4 helices, the S4–S5 linker, and the S6 helix of the adjacent subunit appeared to have produced a movement of the S1–S4 helical bundle together with the S4–S5 linker away from the pore axis and to have enabled a conformational change in the loop that connects the S6 helix to the TRP domain (Fig. 5a). This loop movement caused constriction of the lower gate, specifically at conserved residues Phe574, Met578, and His582, thus obstructing the pore (Fig. 5b). Moreover, this loop movement produced additional conformational changes in the TRP domain, leading to large vertical and rotational motions of the ARDs of the channel (Fig. 5c,d). Our structural observation is consistent with a recent mutagenesis study7 that suggested a similar gating mechanism for the TRPV5 channel and implicated His582 in this mechanism.
Fig. 5. Structural comparison of TRPV5ECN to TRPV6*.
a, Overlaid TM domains of TRPV5ECN (multicolored) and TRPV6* (blue). The regions of TRPV5ECN are labeled and colored based on the diagram in Fig. 1b. The histidine and tryptophan of interest in each pore are represented as sticks. The econazole molecule (ECN, yellow) from the TRPV5ECN structure is also presented as sticks. b, Aligned TRPV5ECN (green) and TRPV6* (blue) dimer pores. Residues of interest are labeled and shown as sticks. c, Aligned TRPV5ECN (multicolored) and TRPV6* (blue) monomers. The regions of TRPV5ECN are labeled and colored based on the diagram in Fig. 1b. d, Superimposed ARDs of TRPV5ECN (orange) and TRPV6* (blue).
Intriguingly, these econazole-induced conformational changes did not significantly affect the conformation of the outer pore region of the TRPV5 channel or its selectivity filter (Fig. 5b). This finding suggests that whereas the gating of some TRPV subfamily members involves the outer pore region, called the turret region in the case of TRPV1 (ref. 27) and TRPV2 (ref. 11), TRPV5 and TRPV6 do not require large conformational changes in this region for channel gating.
Discussion
Our cryo-EM structure of the econazole-bound full-length TRPV5 channel exhibits a domain-swapped architecture, which is a hallmark of all currently determined TRP channels at atomic resolution11–18 and was recently confirmed to be present in TRPV6 (refs. 19,20). Our data revealed that overall, TRPV5 and TRPV6 are similar, but the principal structural differences between them could be used to further understand the TRPV subfamily of the channels. The TRPV5 pore architecture suggests that like other Ca2+-selective ion channels34–36, it selects Ca2+ ions through interaction with their hydration shell35 and may not interact with Ca2+ ions directly19,20. Nevertheless, further studies will be required to better understand the mechanism of Ca2+ ion permeation through these Ca2+-selective non-voltage-gated channels.
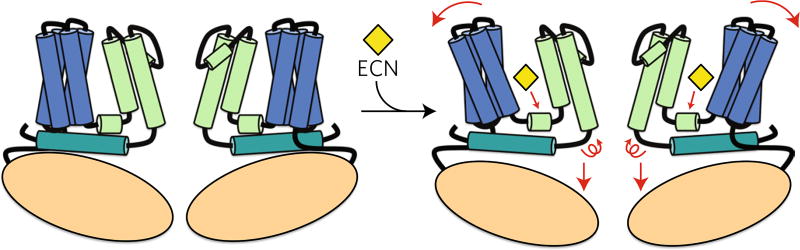
On the basis of comparisons between the TRPV5 and TRPV6 structures, we were able to make initial inferences about the mechanism of channel inhibition by econazole (Fig. 6). First, we demonstrated that inhibitors of TRPVs interact with the channels through a conserved hydrophobic ligand-binding pocket (Fig. 6). Next, we showed that inhibitor binding to the TRPV5 channel is associated with movements in the S1–S4 helices bundle and S4–S5 linker, which are accompanied by a conformational change in the lower gate of the channel and may lead to its closure (Fig. 6). Finally, consistent with the recent mutagenesis and structural studies7, we determined that in addition to the Met578, the rotation of the Phe574 and possibly His582 into the pore may further block Ca2+ ion flow through TRPV5 (Fig. 6).
Fig. 6. Schematic representation of the proposed TRPV5 inhibition mechanism by econazole.
The regions of TRPV5 are colored based on the diagram in Fig. 1b. Red arrows denote proposed movement between the uninhibited and inhibited states of TRPV5.
Here we presented a structure of the TRPV5 channel in an inhibited state. The determination of full-length TRPV5 in ligand-free and ligand-activated states will be essential to understanding the structural diversity among TRPV channel gating mechanisms and their physiological functions. This study in conjunction with future findings will potentially pave the way for novel targeted therapies for hypercalciuria and nephrolithiasis.
Methods
Protein expression and purification
Full-length wild-type rabbit TRPV5 was expressed with a C-terminal 1D4 epitope (TETSQVAPA) in the BJ5457 S. cerevisiae strain as previously reported11. In order to maintain protein stability, all purification steps were conducted at 4 °C. Cells were lysed via microfluidizer (M-110Y, Microfluidics), and plasma membranes were isolated using multiple rounds of centrifugation (3,000g for 10 min, 14,000g for 35 min, 100,000g for 45 min). Plasma membranes were stored at −80 °C until needed. To purify the channel, the plasma membranes were solubilized in a buffer containing 20 mM HEPES, pH 8.0, 150 mM NaCl, 0.87 mM LMNG, 5% glycerol, 2 mM TCEP, and 1 mM PMSF for 1 h. The sample was then centrifuged at 100,000g to remove any insoluble material. The protein was then bound to an affinity-chromatography column containing 1D4 antibody conjugated CnBr-activated Sepharose 4B beads. The column was washed with a buffer containing 20 mM HEPES, pH 8.0, 150 mM NaCl, 0.064 mM DMNG, and 2 mM TCEP, and TRPV5 was eluted using buffer containing 20 mM HEPES, pH 8.0, 150 mM NaCl, 0.064 mM DMNG, 2 mM TCEP and 3 mg/mL 1D4 peptide (NH2-TETSQVAPA-CO2H). Eluted protein was concentrated and further purified by size-exclusion chromatography (Superose 6, GE Healthcare) in 20 mM HEPES, pH 8.0, 150 mM NaCl, 0.064 mM DMNG, and 2 mM TCEP. The peak fraction was centrifugally concentrated and used for subsequent cryo-EM studies. No Ca2+ chelators were included in any of the purification buffers.
Electrophysiological recordings
Two electrode voltage clamp (TEVC) experiments in Xenopus oocytes were performed as described previously37. Briefly, ovarian sacs were removed from female Xenopus laevis frogs, then oocytes were digested using 0.2 mg/mL collagenase (Sigma) in a solution containing 82.5 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM HEPES, pH 7.4 (OR2 solution) overnight for ~16 h at 18 °C in a temperature-controlled incubator. Defolliculated oocytes were selected and kept at 18 °C in OR2 solution supplemented with 1% penicillin/streptomycin (Mediatech) and 1.8 mM CaCl2. cRNA was generated from the linearized rabbit TRPV5 and its mutants in the pGEMSH oocyte vector, using the mMessage mMachine kit (Ambion). 5 ng RNA was microinjected into each oocyte using a nanoliter injector system (World Precision Instruments). The experiments were performed 48–72 h after injection. TEVC measurements were performed as described previously. Briefly, oocytes were placed in a solution containing 97 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM HEPES, pH 7.4, in which TRPV5 currents are largely blocked by Mg2+ and trace amounts of Ca2+ in the medium. Monovalent currents were initiated by a solution containing 96 mM LiCl, 1 mM EGTA, and 5 mM HEPES, pH 7.4. Currents were recorded with thin-wall inner filament-containing glass pipettes (World Precision Instruments) filled with 3 M KCl in 1% agarose. Currents were measured with a ramp protocol from −100 mV to 100 mV applied every 500 ms, preceded by a step to −100 mV for 50 ms. Econazole was dissolved in DMSO as a stock solution at 0.4 mg/mL (0.9 mM). The stock was added directly to the aqueous buffer solutions for measurements in oocytes. To apply 20 µM of econazole in the oocyte measurements, the final concentration of DMSO was 2.2%. Higher concentrations of DMSO inhibited wild-type TRPV5 currents, thus we could not apply higher concentrations of econazole.
Whole-cell patch clamp was performed similarly to that described for TRPV6 (ref. 37), with some modifications. HEK293 cells were cultured in MEM supplemented with 10% FBS and kept in a humidified incubator with 5% CO2 at 37 °C. Cells were transfected using the Effectene reagent (Qiagen), and measurements were performed 48–72 h after transfection. Currents were measured with an Axpatch 200B amplifier using a ramp protocol from −100 mV to 100 mV applied every second. The extracellular solution contained 142 mM LiCl, 1 mM MgCl2, 10 mM HEPES, and 10 mM glucose, pH 7.4. Monovalent currents were initiated by the same solution containing 1 mM EGTA but no MgCl2. The intracellular pipette solution contained 140 mM K-gluconate, 10 mM HEPES, 5 mM EGTA, 2 mM MgCl, and 2 mM ATP, pH 7.3. For both oocytes and HEK cells, we used a LiCl-based extracellular solution, because in this solution, removal of extracellular Mg2+ and Ca2+ does not induce endogenous currents in nontransfected cells, unlike in Na+ or K+ based solutions. The econazole stock as described above was added directly to the aqueous buffer solutions for measurements in HEK cells.
Cryo-electron microscopy data collection
Filtered, concentrated TRPV5 (~2.5 mg/mL) was incubated with 5.6 µM econazole and 3 mM fluorinated Fos-Choline-8, which improved particle distribution. The sample was double blotted (3.5 µL per blot) onto 200 mesh Quantifoil 1.2/1.3 grids (Quantifoil Micro Tools); the second blot was incubated for 30 s, and the grid was plunge frozen using a Vitrobot (FEI). The grids were imaged using a 300 kV FEI Titan Krios microscope equipped with a Gatan K2 Summit direct detector camera. 50-frame movies were collected at a nominal magnification of 45,455× in super-resolution mode, resulting in a pixel size of 0.55 Å at sample level and a dose rate of ~8 e−/pixel/s in the detector. Total exposure time for each movie was 10 s, with one frame collected every 0.2 s. Images were collected 1.5–4.0 µm under focus, as per the automated imaging software Leginon38.
Image processing
Movies were motion corrected to compensate for beam-induced motion using MotionCor2, resulting in summed and dose-weighted micrographs39. The preprocessing procedures, CTF estimation, and autopicking procedures were performed on the summed micrographs, and the 2D classification, 3D classification, and 3D autorefinement were performed with the dose-weighted micrographs. The defocus values of the motion-corrected micrographs were estimated using Gctf software40. All subsequent data processing was conducted in RELION 2.0 (refs. 25,26). Approximately 2,000 particles were manually picked from the 3,313 micrographs and sorted into 2D classes. The best of these classes were used as templates for autopicking. A loose autopicking threshold was selected to ensure that no good particles were missed. This resulted in ~900,000 autopicked particles that were subjected to multiple rounds of 2D classification to remove false positives and suboptimal particles. The best 137,679 particles were then subjected to 3D autorefinement, followed by 3D classification into four classes. A published structure of TRPV2 low-pass filtered to 60 Å was used as an initial model for the first 3D autorefinement11. Subsequent classifications and refinements used the TRPV5 map from the first 3D autorefinement low-pass filtered to 60 Å as the initial model. The mask used for processing was created from the TRPV5 map from the first 3D autorefinement adjusted to a threshold of 0.005 and low-pass filtered to 15 Å. The best 3D class, containing 70,059 particles, was subjected to an additional round of 3D autorefinement and 3D reclassification. From this, the best 50,566 particles were used in the final 3D autorefinement and post-processing to yield a TRPV5 structure at an overall resolution of 4.8 Å based on the 0.143 cutoff criterion. Local resolutions were estimated using the RESMAP software41.
TRPV5 model building
An initial homology model was generated with the I-TASSER42 server employing all previous structures of TRPV1 (apo-TRPV1, PDB 3J5P; capsaicin–TRPV1, PDB 3J5R; RTX/DkTx–TRPV1, PDB 3J5Q), TRPV6 (apo-TRPV6, PDB 5IWK) and our previous structure of full-length TRPV2 (PDB 5HI9). This initial model was aligned to our TRPV5 cryo-EM map calculated with RELION 2.0 and was manually adjusted to fit the electron density in COOT43. After this initial model fitting, we refined this model against the EM-derived maps using the phenix.real_space_refinement tool from the PHENIX software package44, employing rigid-body refinement, gradient minimization, local grid search and ADP refinement with secondary structure restraints and NCS constraints. The structures of TRPV1 (PDB 3J5P, PDB 3J5R, and PDB 3J5Q), rabbit TRPV2 (PDB 5AN8), and the new TRPV6* structure20 (generously provided by A. I. Sobolevsky (Columbia University)) were used as references during manual building and structure adjustment and assisted in assignment of topology and improved connectivity in the model. The model was subjected to additional rounds of manual model fitting and real space refinement (RSR), resulting in a final model to map cross-correlation coefficient of 0.722.
Molecular docking and MD simulations
Both enantiomers of econazole were manually docked to fit the assigned cryo-EM density within the four TRPV5 monomers. The atomic coordinates of the (R)-econazole-bound and (S)-econazole-bound TRPV5 tetramer embedded in a 1-palmitoyl-2-oleoyl phosphatidyl choline (POPC) bilayer solvated with TIP3P water, 0.15 M NaCl, and additional neutralizing ions were used for MD simulations. Missing residues of the loop between TM helices S1 and S2 (residues 356–375) were added with MODELLER45 using the corresponding region of the TRPV6 crystal structure (PDB 5WIP) as a template for homology modeling. A total of 50 loop models were generated, and the structure with the top discrete optimized protein energy (DOPE)46 score was selected as an initial conformation for the loop. The starting system for MD simulations was assembled with the Membrane Builder functionality of the CHARMM-GUI webserver47,48. This system (box dimension: 143 Å × 143 Å × 147 Å) consisted of the TRPV5 tetramer, (R)- or (S)-econazole molecules bound to each tetramer subunit, 450 POPC molecules, 58,725–59,185 water molecules, 213–215 sodium ions, 183–185 chloride ions, and a calcium ion in the outer pore region, totaling 276,224 atoms in the case of TRPV5 bound to (R)-econazole and 277,608 atoms for TRPV5 bound to (S)-econazole.
MD simulations were run using Gromacs 2016.3 (ref. 49) with a time step of 2 fs for computation and CHARMM36 force field50–52. Initial parameters for either (R)-or (S)-econazole were obtained using the CHARMM General force field via the ParamChem webserver (https://cgenff.paramchem.org) and were further validated and optimized following published guidelines53. The system was first minimized using steepest descent for 5,000 steps, then 50 ps of NVT and 50 ns of NPT equilibration. The NPT equilibration was performed in 18 steps, with the first 17 steps consisting of gradually decreasing positional restraints on the heavy atoms of lipids first, then protein side chains, protein backbone and ligand ring atoms, and lastly, the remaining ligand atoms. The last step consisted of a 1 ns unrestrained NPT equilibration, which was followed by a 25-ns production run. During the equilibration, system temperature and pressure were maintained at 300 K and 1 bar, respectively, using velocity rescale54 for temperature coupling and a Parrinello– Rahman barostat for pressure coupling. Semi-isotropic pressure coupling was applied during simulation. All bonds involving hydrogen atoms were constrained using the LINCS algorithm. Short-range nonbonded interactions were cut at 12 Å. Long-range electrostatic interactions were computed using the particle mesh Ewald (PME) summation with a Fourier grid spacing of 1.6 Å. The production run was performed with the same parameters as described above but with the thermostat switched to Nose-Hoover55.
The resulting production trajectory was analyzed using PyEMMA 2 (ref. 56) and VMD57. PyMOL (Version 1.8 Schrödinger, LLC; http://www.pymol.org) was used to create images. R.m.s. deviation of the protein Cα atoms and econazole heavy atoms were calculated following superposition of residues of TM helices S1–S6 in the trajectory frames onto the coordinates of the starting cryo-EM structure. Minimum heavy atom distance between POPC molecules and each of the three ring moieties (chlorophenyl, dichlorophenyl, and imidazole) of econazole were calculated for each trajectory frame using PyEMMA 2, and the number of POPC molecules in contact with each of the rings of econazole was computed with a minimum heavy atom distance cutoff of 4.5 Å.
All presented MD values in the main text are means calculated from n =2,500 frames per simulation, and presented error values represent the s.d. of the 2,500 frames.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
The cryo-EM density map and the atomic coordinates of the econazole-bound full-length TRPV5 channel are deposited into the Electron Microscopy Data Bank and Protein Data Bank under access codes EMD-7058 and PDB 6B5V. All other data are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank S. Molugu for support and training of future cryo-EM microscopists at Cleveland Center for Membrane and Structural Biology. We thank D. Major for assistance with hybridoma and cell culture at Department of Ophthalmology and Visual Sciences (supported by the National Institutes of Health Core Grant P30EY11373). We thank A. I. Sobolevsky (Columbia University) for generously providing the structure of TRPV6* before its release. MD simulations were run on resources available through the Scientific Computing Facility at the Icahn School of Medicine at Mount Sinai and the Extreme Science and Engineering Discovery Environment under MCB080077 (to M.F.), which is supported by National Science Foundation grant number ACI-1053575. We acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by the NIH (1S10RR23057 and 1S10OD018111), NSF (DBI-1338135) and CNSI at UCLA. This work was supported by grants from the National Institute of Health (R01GM103899 to V.Y.M.-B., R01GM093290 to T.R., and U24 GM116792 to Z.H.Z and V.Y.M.-B).
Footnotes
Author contributions
T.E.T.H. conducted all biochemical and cryo-EM studies, including protein purification, sample preparation, imaging, data analysis, and interpretation; D.T.L. built and refined the atomic model; K.W.H. assisted T.E.T.H. in cryo-EM data collection and analysis; A.Y., J.D.R., and T.R. performed and interpreted electrophysiological data; A.K. performed MD simulations, and M.F. helped interpret the data; A.S. trained T.E.T.H. in cryo-EM sample preparation; X.H. participated in the initial stage of this project; S.B. and S.C. trained T.E.T.H. in data analysis; Z.H.Z. supervised cryo-EM data collection and analysis; S.H. assisted with data analysis; V.Y.M.-B. designed and supervised the execution of all experiments in this manuscript; T.E.T.H. and V.Y.M.-B. drafted the manuscript; all authors reviewed the final manuscript.
Competing interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41594-017-0009-1.
References
- 1.Scales CD, Smith AC, Hanley JM, Saigal CS, Urologic Diseases in America Project Prevalence of kidney stones in the United States. Eur. Urol. 2012;62:160–165. doi: 10.1016/j.eururo.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nie M, et al. Mucin-1 increases renal TRPV5 activity in vitro, and urinary level associates with calcium nephrolithiasis in patients. J. Am. Soc. Nephrol. 2016;27:3447–3458. doi: 10.1681/ASN.2015101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Na T, Peng JB. TRPV5: a Ca(2+) channel for the fine-tuning of Ca(2+) reabsorption. Handb. Exp. Pharmacol. 2014;222:321–357. doi: 10.1007/978-3-642-54215-2_13. [DOI] [PubMed] [Google Scholar]
- 4.Renkema KY, et al. Hypervitaminosis D mediates compensatory Ca2+ hyperabsorption in TRPV5 knockout mice. J. Am. Soc. Nephrol. 2005;16:3188–3195. doi: 10.1681/ASN.2005060632. [DOI] [PubMed] [Google Scholar]
- 5.Venkatachalam K, Montell C. TRP channels. Annu. Rev. Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoenderop JG, et al. Function and expression of the epithelial Ca(2+) channel family: comparison of mammalian ECaC1 and 2. J. Physiol. (Lond.) 2001;537:747–761. doi: 10.1111/j.1469-7793.2001.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Wijst J, et al. A gate hinge controls the epithelial calcium channel TRPV5. Sci. Rep. 2017;7:45489. doi: 10.1038/srep45489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Goor MKC, Hoenderop JGJ, van der Wijst J. TRP channels in calcium homeostasis: from hormonal control to structure-function relationship of TRPV5 and TRPV6. Biochim. Biophys. Acta. 2017;1864:883–893. doi: 10.1016/j.bbamcr.2016.11.027. [DOI] [PubMed] [Google Scholar]
- 9.Nilius B, et al. Pharmacological modulation of monovalent cation currents through the epithelial Ca2+ channel ECaC1. Br. J. Pharmacol. 2001;134:453–462. doi: 10.1038/sj.bjp.0704272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fecher-Trost C, Wissenbach U, Weissgerber P. TRPV6: from identification to function. Cell Calcium. 2017;67:116–122. doi: 10.1016/j.ceca.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Huynh KW, et al. Structure of the full-length TRPV2 channel by cryo-EM. Nat. Commun. 2016;7:11130. doi: 10.1038/ncomms11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zubcevic L, et al. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat. Struct. Mol. Biol. 2016;23:180–186. doi: 10.1038/nsmb.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulsen CE, Armache JP, Gao Y, Cheng Y, Julius D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 2015;520:511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grieben M, et al. Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2) Nat. Struct. Mol. Biol. 2017;24:114–122. doi: 10.1038/nsmb.3343. [DOI] [PubMed] [Google Scholar]
- 16.Wilkes M, et al. Molecular insights into lipid-assisted Ca2+ regulation of the TRP channel Polycystin-2. Nat. Struct. Mol. Biol. 2017;24:123–130. doi: 10.1038/nsmb.3357. [DOI] [PubMed] [Google Scholar]
- 17.Shen PS, et al. The structure of the polycystic kidney disease channel PKD2 in lipid nanodiscs. Cell. 2016;167:763–773. doi: 10.1016/j.cell.2016.09.048. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, et al. Structural basis of dual Ca2+/pH regulation of the endolysosomal TRPML1 channel. Nat. Struct. Mol. Biol. 2017;24:205–213. doi: 10.1038/nsmb.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saotome K, Singh AK, Yelshanskaya MV, Sobolevsky AI. Crystal structure of the epithelial calcium channel TRPV6. Nature. 2016;534:506–511. doi: 10.1038/nature17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh AK, Saotome K, Sobolevsky AI. Swapping of transmembrane domains in the epithelial calcium channel TRPV6. Sci. Rep. 2017;7:10669. doi: 10.1038/s41598-017-10993-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Catterall WA. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron. 2010;67:915–928. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huynh KW, et al. Structural insight into the assembly of TRPV channels. Structure. 2014;22:260–268. doi: 10.1016/j.str.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cvetkov TL, Huynh KW, Cohen MR, Moiseenkova-Bell VY. Molecular architecture and subunit organization of TRPA1 ion channel revealed by electron microscopy. J. Biol. Chem. 2011;286:38168–38176. doi: 10.1074/jbc.M111.288993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moiseenkova-Bell VY, Stanciu LA, Serysheva II, Tobe BJ, Wensel TG. Structure of TRPV1 channel revealed by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA. 2008;105:7451–7455. doi: 10.1073/pnas.0711835105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheres SH. Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 2016;579:125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 26.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phelps CB, Huang RJ, Lishko PV, Wang RR, Gaudet R. Structural analyses of the ankyrin repeat domain of TRPV6 and related TRPV ion channels. Biochemistry. 2008;47:2476–2484. doi: 10.1021/bi702109w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landowski CP, Bolanz KA, Suzuki Y, Hediger MA. Chemical inhibitors of the calcium entry channel TRPV6. Pharm. Res. 2011;28:322–330. doi: 10.1007/s11095-010-0249-9. [DOI] [PubMed] [Google Scholar]
- 31.Yang F, Vu S, Yarov-Yarovoy V, Zheng J. Rational design and validation of a vanilloid-sensitive TRPV2 ion channel. Proc. Natl. Acad. Sci. USA. 2016;113:E3657–E3666. doi: 10.1073/pnas.1604180113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F, et al. Engineering vanilloid-sensitivity into the rat TRPV2 channel. eLife. 2016;5:e16409. doi: 10.7554/eLife.16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Z, et al. Structural insight into tetrameric hTRPV1 from homology modeling, molecular docking, molecular dynamics simulation, virtual screening, and bioassay validations. J. Chem. Inf. Model. 2015;55:572–588. doi: 10.1021/ci5007189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang L, et al. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature. 2014;505:56–61. doi: 10.1038/nature12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Catterall WA, Wisedchaisri G, Zheng N. The chemical basis for electrical signaling. Nat. Chem. Biol. 2017;13:455–463. doi: 10.1038/nchembio.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J, et al. Structure of the voltage-gated calcium channel Cav1.1 at 3.6 Å resolution. Nature. 2016;537:191–196. doi: 10.1038/nature19321. [DOI] [PubMed] [Google Scholar]
References
- 37.Velisetty P, et al. A molecular determinant of phosphoinositide affinity in mammalian TRPV channels. Sci. Rep. 2016;6:27652. doi: 10.1038/srep27652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potter CS, et al. Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy. 1999;77:153–161. doi: 10.1016/s0304-3991(99)00043-1. [DOI] [PubMed] [Google Scholar]
- 39.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Virtanen J, Xue Z, Zhang Y. I-TASSER-MR: automated molecular replacement for distant-homology proteins using iterative fragment assembly and progressive sequence truncation. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx349. https://doi.org/10.1093/nar/gkx349. [DOI] [PMC free article] [PubMed]
- 43.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 45.Eswar N, et al. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2007;50:2.9.1–2.9.31. doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 46.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 48.Lee J, et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Teory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Der Spoel D, et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 50.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 51.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Teory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanommeslaeghe K, et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. https://doi.org/10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 55.Hoover WG. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 56.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 57.rer MK, et al. PyEMMA 2: a software package for estimation, validation, and analysis of Markov models. J. Chem. Theory Comput. 2015;11:5525–5542. doi: 10.1021/acs.jctc.5b00743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density map and the atomic coordinates of the econazole-bound full-length TRPV5 channel are deposited into the Electron Microscopy Data Bank and Protein Data Bank under access codes EMD-7058 and PDB 6B5V. All other data are available from the corresponding author upon reasonable request.