Summary
A key limitation to the use of CRISPR-Cas9 proteins for genome editing and other applications is the requirement that a protospacer adjacent motif (PAM) be present at the target site. For the most commonly used Cas9 from Streptococcus pyogenes (SpCas9), this PAM requirement is NGG. No natural or engineered Cas9 variants shown to function efficiently in mammalian cells offer a PAM less restrictive than NGG. Here we used phage-assisted continuous evolution (PACE) to evolve an expanded PAM SpCas9 variant (xCas9) that can recognize a broad range of PAM sequences including NG, GAA, and GAT. The PAM compatibility of xCas9 is the broadest reported to date among Cas9s active in mammalian cells, and supports applications in human cells including targeted transcriptional activation, nuclease-mediated gene disruption, and both cytidine and adenine base editing. Remarkably, despite its broadened PAM compatibility, xCas9 has much greater DNA specificity than SpCas9, with substantially lower genome-wide off-target activity at all NGG target sites tested, as well as minimal off-target activity when targeting genomic sites with non-NGG PAMs. These findings expand the DNA targeting scope of CRISPR systems and establish that there is no necessary trade-off between Cas9 editing efficiency, PAM compatibility, and DNA specificity.
The clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) system has facilitated widely used genome manipulation capabilities including targeted gene disruption1,2, transcriptional activation and repression3, epigenetic modification3, and direct conversion of a target base pair to a different base pair4,5 in a broad range of organisms and cell types6. CRISPR-Cas9 targets DNA in a manner that is programmed by an RNA (typically a single-guide RNA, or sgRNA7) that contains a “spacer” sequence complementary to the target DNA site, the “protospacer”. In addition to a protospacer that complements the sgRNA, a Cas9 target site must also contain a protospacer adjacent motif (PAM) sequence to support recognition by Cas9. The NGG PAM requirement of canonical SpCas9, which occurs on average only once in every ~16 randomly chosen genomic loci, greatly limits the targeting scope of Cas9 especially for applications that require precise Cas9 positioning, such as base editing, which requires a PAM ~15±2 nucleotides from the target base4,5, and some forms of homology-directed repair, which are most efficient when DNA cleavage occurs ~10–20 base pairs away from a desired alteration8,9. These requirements limit the fraction of genomic DNA that can be targeted with CRISPR systems and highlight the need for more general genome editing tools.
To address this limitation, researchers have harnessed natural CRISPR nucleases with different PAM requirements and engineered existing systems to accept variants of naturally recognized PAMs. Other natural CRISPR nucleases shown to function efficiently in mammalian cells include Staphylococcus aureus Cas9 (SaCas9)10, Acidaminococcus sp. Cpf111, Lachnospiraceae bacterium Cpf111, Campylobacter jejuni Cas912, Streptococcus thermophilus Cas913, and Neisseria meningitides Cas914. None of these mammalian cell-compatible CRISPR nucleases, however, offer a PAM that occurs as frequently as that of SpCas9. While CRISPR nucleases engineered to accept additional PAM sequences15,16 also expand the scope of genomic targets available for Cas9-mediated manipulation, many target sequences remain inaccessible.
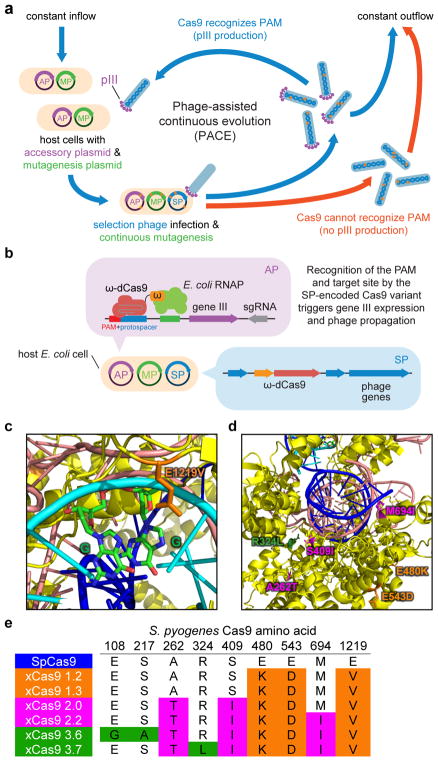
Here we used phage-assisted continuous evolution (PACE) to rapidly generate Cas9 variants that accept an expanded range of PAM sequences. During PACE, host E. coli cells continuously dilute an evolving population of bacteriophages (selection phage, SP). Since dilution occurs faster than cell division but slower than phage replication, only the SP, and not the host cells, can accumulate mutations17. Each SP carries a gene to be evolved instead of a phage gene (gene III) that is required for the production of infectious progeny phage. SP containing desired gene variants trigger host-cell gene III expression from the accessory plasmid (AP) and the production of infectious SP that propagate the desired variants. Phage encoding inactive variants do not generate infectious progeny and are rapidly diluted out of the culture vessel (Fig. 1a). As phage replication can occur in as little as 10 minutes, PACE enables hundreds of generations of directed evolution to occur per week without researcher intervention17–22.
Figure 1. Phage-assisted continuous evolution (PACE) of Cas9 variants with broadened PAM compatibility.
a, PACE takes place in a fixed-volume “lagoon” that is continuously diluted with fresh host E. coli cells. Upon infection, each selection phage (SP) that encodes a Cas9 variant capable of binding the target PAM and protospacer on the accessory plasmid (AP) induces expression of gene III, resulting in infectious progeny phage that propagate the active Cas9 variant in subsequent host cells. b, Anatomy of a phage-infected host cell during PACE. The host cell carries the AP, which links Cas9 target DNA binding to phage propagation, and the mutagenesis plasmid (MP), which elevates mutagenesis during PACE. c, d, The crystal structure of SpCas9 with the location of xCas9 mutations shown. e, Genotypes of some evolved xCas9 variants, colored by evolution stage. See Supplementary Table 5 for 95 xCas9 variant genotypes.
Results
Evolution of Cas9 variants with expanded PAM compatibility
To link Cas9 DNA recognition to phage propagation during PACE, we developed a bacterial one-hybrid selection20,23,24 in which the SP encodes a catalytically dead SpCas9 (dCas9) fused to the ω subunit of bacterial RNA polymerase. When this fusion binds an AP-encoded sgRNA and a PAM and protospacer upstream of gene III in the AP, RNA polymerase recruitment causes gene III expression and phage propagation (Fig. 1b). We envisioned installing a library of all 64 possible NNN PAM sequences at the target protospacer in the AP, so that SP encoding Cas9 variants with broader PAM compatibility would replicate in a larger fraction of host cells and thus experience a fitness advantage.
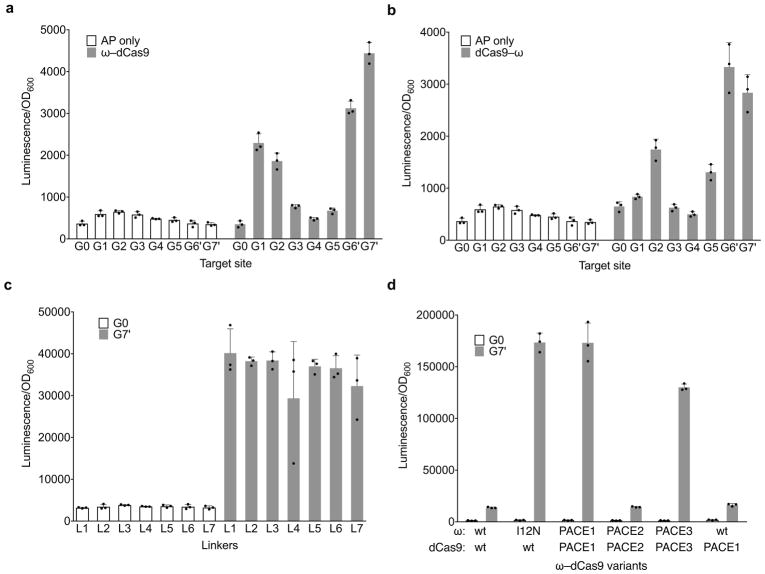
We optimized the relationship between Cas9 DNA binding and gene expression (Extended Data Fig. 1a–c). These studies revealed that (i) a fusion of the orientation N–ω–dCas9–C, (ii) a simple Ala–Ala fusion linker, and (iii) placement of the protospacer on the reverse complement strand 45 bp upstream of the −35 box together resulted in the strongest guide RNA-dependent gene expression activation of 13-fold (Extended Data Fig. 1a–c). Together, these results establish a linkage between Cas9 DNA binding activity and gene expression in a selection system suitable for PACE.
Using this selection, we first allowed an SP encoding the ω–dCas9 fusion to self-optimize on host cells containing an AP with a canonical NGG PAM, resulting in enrichment of an I12N mutation in the ω subunit. Adding this single mutation to ω–dCas9 boosted activation from 13-fold to over 100-fold (Extended Data Fig. 1d), representing early-stage optimization of ω–dCas9 during PACE prior to evolution for broadened PAM compatibility.
To evolve Cas9 variants with expanded PAM compatibility we generated three AP libraries, each containing a different sgRNA (Supplementary Table 4) and corresponding protospacer upstream of an NNN PAM library, where N is an equimolar mixture of all four DNA bases. This design imposes selection pressure to recognize many different PAM sequences, as well as to maintain compatibility with different target DNA sequences. All three AP libraries were introduced into host E. coli cells harboring the mutagenesis plasmid MP617. The resulting host cells were incubated overnight with SP containing ω(I12N)–dCas9. This phage-assisted non-continuous evolution (PANCE) system19,21 preferentially replicates Cas9 variants that bind a greater variety of PAM sequences, similar to PACE, but with lower stringency since there is no outflow of phage.
After 24 days of serial overnight propagation and 1:1000 dilution, we isolated five Cas9 clones for sequencing and characterization (xCas9 1.0–1.4). Notable recurring mutations include E480K, E543D, and E1219V (Fig. 1e and Supplementary Table 5). E1219 is close in the SpCas9 crystal structure to R1333 and R1335 (Fig. 1c), two residues known to play a critical role in PAM recognition25. The mixture of phage from the final PANCE pool were further evolved for 72 h in PACE on host cells containing the same AP libraries harboring NNN PAM sequences. Among individual Cas9 clones emerging from PACE (xCas9 2.0–2.6), E480K, E543D, and E1219V were present in all sequenced phage, along with additional mutations seen in multiple clones, including A262T, K294R, S409I, and M694I (Fig. 1e and Supplementary Table 5). Finally, the resulting phage were continuously evolved in PACE for an additional 72 h on host cells containing three protospacer-sgRNA pairs and HHH PAM libraries, where H is A, C, or T, to favor Cas9 variants with activity on non-NGG PAMs.
Fourteen resulting evolved Cas9 variants (xCas9 3.0–3.13) containing consensus mutations (Fig. 1e and Supplementary Table 5) emerged from two apparent evolution trajectories. While all phage shared E480K, E543D, and E1219V core mutations, xCas9 3.0–3.5 also all contained K294R and Q1256K mutations while xCas9 3.6–3.13 all contained A262T, S409I, and M694I mutations, with some of the latter also containing R324L. The Cas9 crystal structure predicts that R324L, S409I, and M694I lie near the DNA-sgRNA interface (Fig. 1d) and could play a role in mediating DNA sequence recognition and the switching of Cas9 from the open to the closed conformation upon target recognition26,27. Since the entire Cas9 gene was subject to mutagenesis during PACE, the mechanism of xCas9 may differ from that of engineered Cas9 variants that primarily mutated DNA-contacting residues15,16,28.
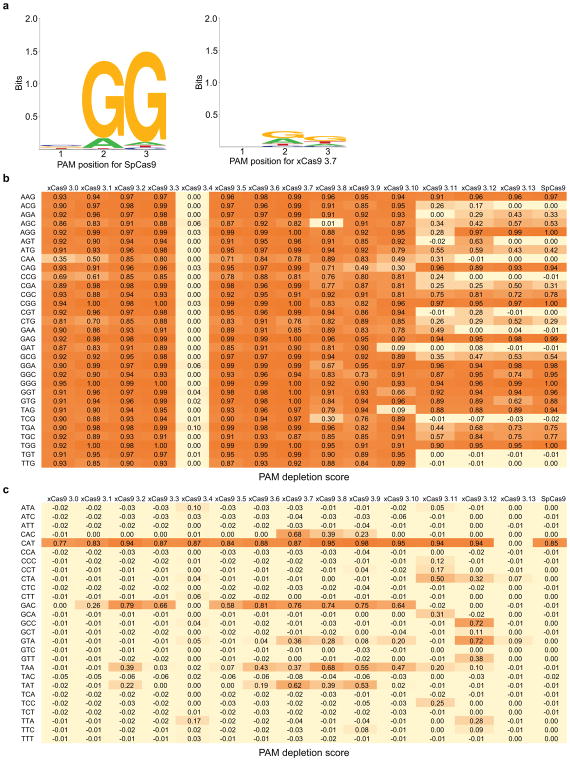
We characterized evolved xCas9 variants in several contexts. We first restored the catalytic residues Asp 10 and His 840 to test if xCas9 nucleases can cleave DNA even though they were evolved only for DNA binding. The xCas9 3.0–3.13 clones were tested in a PAM depletion assay15,16 in which they were given the opportunity to cleave a library of plasmids containing a protospacer and all possible NNN PAM sequences in an antibiotic resistance gene in bacterial cells. Plasmid cleavage results in the loss of spectinomycin resistance. This PAM depletion assay revealed that xCas9 3.0–3.3 and 3.5–3.9 cleave DNA site with NG, NNG, GAA, GAT, and CAA PAMs (Extended Data Fig. 2). The clone with the highest PAM depletion score, xCas9 3.7, depleted NG, NNG, GAA, GAT, and CAA PAMs ≥ 100-fold compared to the starting library, while xCas9 3.6 showed the second highest average PAM depletion score.
Characterization of xCas9 transcriptional activators and nucleases in human cells
To test if mutations evolved during PACE in bacteria are compatible with xCas9 function in mammalian cells, we characterized xCas9 variants for their activity and PAM compatibility in human cells in four contexts: transcriptional activation, genomic DNA cutting, cytidine base editing, and adenine base editing. To test for transcriptional activation, catalytically dead versions of xCas9 were fused to the transcriptional activator VP64–p65–Rta (dxCas9–VPR)29. Plasmids encoding dxCas9–VPR, a GFP reporter downstream of a target protospacer, and a corresponding sgRNA were co-transfected into HEK293T cells29. Target gene transcriptional activation was measured by cellular GFP fluorescence after three days. Three different target site PAM sets were tested: a single reporter with an NGG PAM, a reporter library containing a NNN PAM library, and a reporter library containing a NNNNN PAM library. In addition, two different protospacer sequences, reporter 1 and reporter 2, were tested with their corresponding sgRNAs.
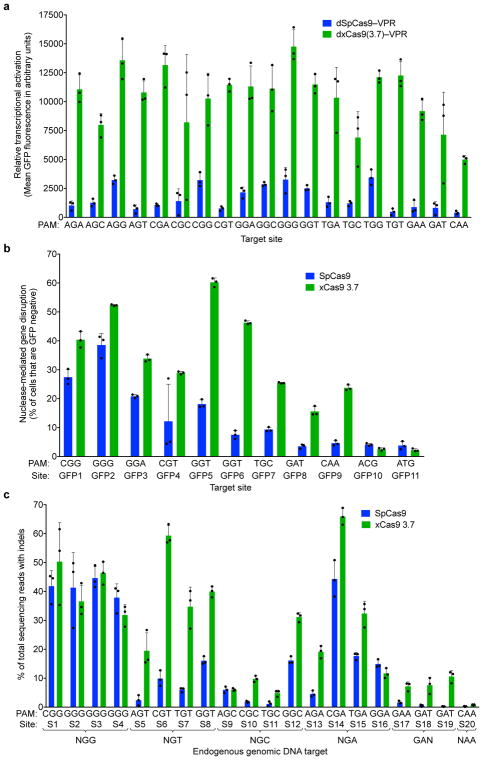
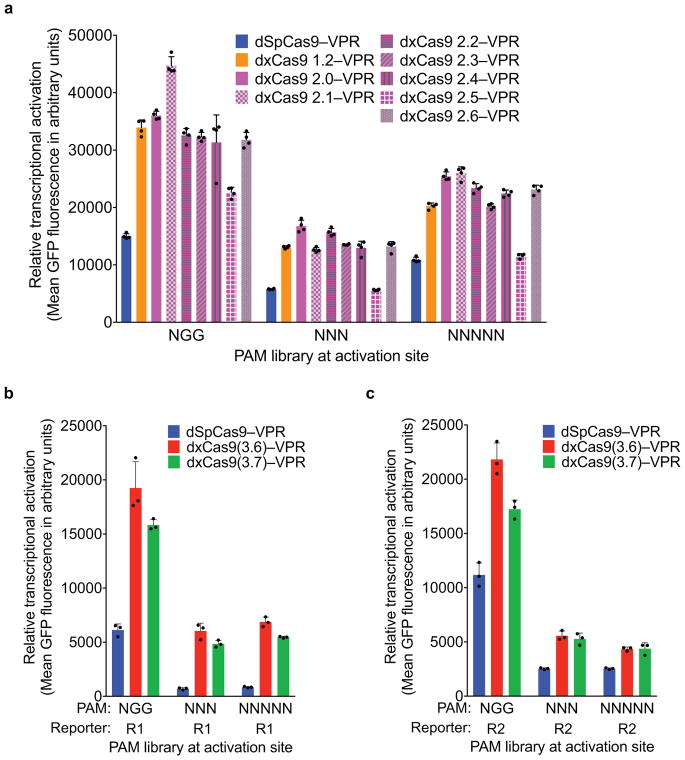
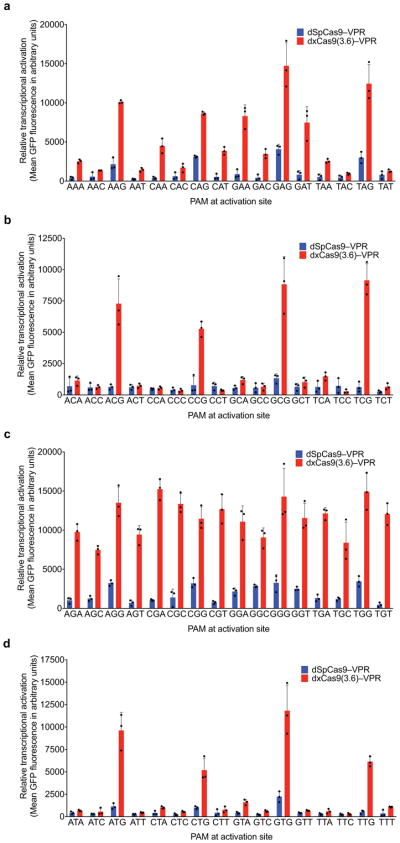
Most early-stage xCas9 variants outperformed wild-type SpCas9 on sites with NGG PAMs, as well as with NNN and NNNNN PAM libraries (Extended Data Fig. 3a). For reporter 1 and reporter 2, respectively, xCas9 3.7 achieved 2.8- and 1.5-fold higher mean fluorescence for the NGG PAM, 7.9- and 2.1-fold higher mean fluorescence for the NNN PAM library, and 5.2- and 1.7-fold higher mean fluorescence for the NNNNN PAM library compared with SpCas9 (Extended Data Fig. 3b, c). The similar performances on NNN and NNNNN PAM libraries suggest that xCas9 3.7 did not evolve strong sequence preferences at nucleotides immediately downstream of the NNN PAM. The xCas9 3.6 variant showed similar results to those of xCas9 3.7 in this assay (Extended Data Fig. 3b, c).
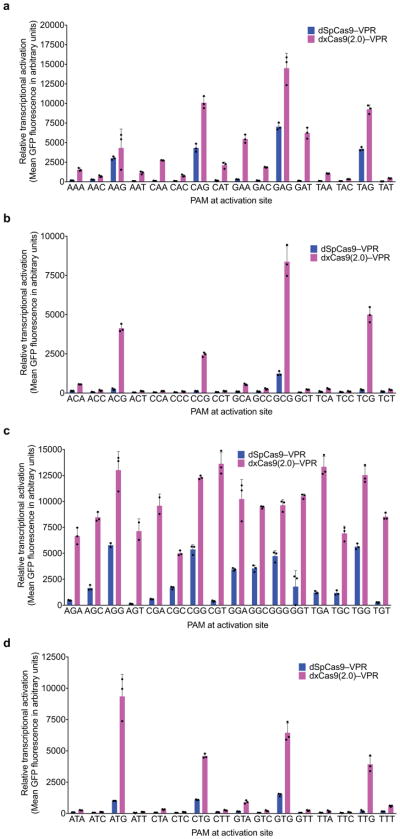
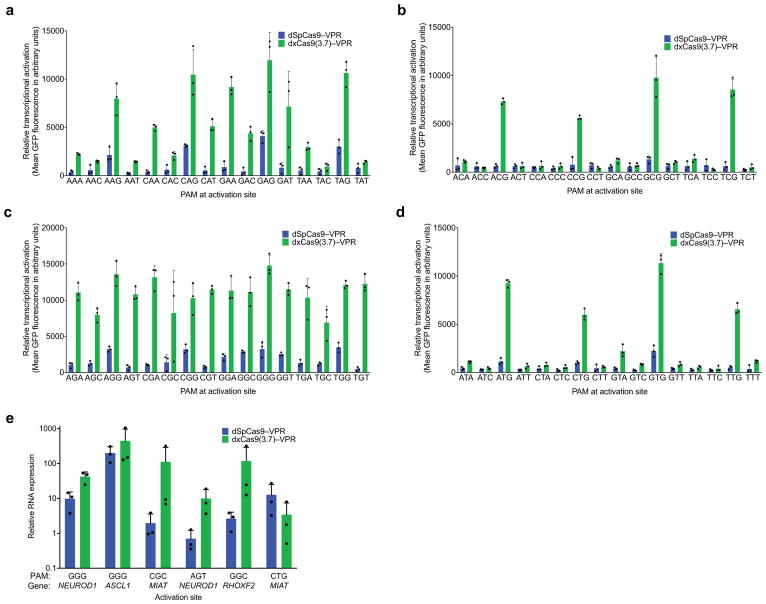
To dissect activity on individual PAM sequences, we tested dxCas9–VPR transcriptional activators on individual target sites containing each of the 64 possible three-nucleotide PAM sequences (Fig. 2a and Extended Data Figs 4–6) in HEK293T cells. Consistent with the PAM library results, dxCas9(3.7)–VPR showed broad improvements in transcriptional activity relative to dSpCas9–VPR across many individual non-NGG PAMs. Transcriptional activation by dxCas9(3.7)–VPR at sites containing NGT, NGA, NGC, NNG, GAA, and GAT PAMs averaged 56–91% of the average activity of dxCas9(3.7)–VPR on the four NGG PAM sites (Fig. 2a and Extended Data Fig. 5). The performance of xCas9 3.6 transcriptional activators was similar to that of xCas9 3.7 (Extended Data Fig. 6). To test if broadened transcriptional activation by xCas9 is limited to reporter plasmids that may be only partially chromatinized, we also tested the ability of dxCas9(3.7)–VPR to activate transcription of six endogenous genomic loci in human cells and observed 3.3-fold average improved activation of the two NGG PAM sites and 39-fold average improved activation among the three NGN PAM sites, but no improvement on the tested NNG site, relative to dSpCas9–VPR (Extended Data Fig. 5e). Overall, these results establish that xCas9 is compatible with the dCas9–VPR architecture and can serve as potent transcriptional activators in human cells at a substantially expanded set of PAMs. Based on their strong performance in the PAM depletion assay and as transcriptional activators, we chose xCas9 3.7 and xCas9 3.6 for further characterization.
Figure 2. Transcriptional activation and genomic DNA cleavage by evolved xCas9 3.7 in human cells.
a, Transcriptional activation by dSpCas9–VPR and dxCas9(3.7)–VPR targeting GFP reporter plasmids containing the same protospacer but different PAM sites in HEK293T cells. b, Genomic DNA cleavage in HEK293-GFP cells containing a genomically integrated GFP reporter gene by SpCas9 or xCas9 3.7. After 5 days, the cells were analyzed for loss of GFP fluorescence by flow cytometry. c, DNA cleavage of endogenous genomic DNA sites with NGG and non-NGG PAMs by SpCas9 and xCas9 3.7 in HEK293T cells. Indel rates were measured by HTS 5 days after plasmid transfection. Values and error bars reflect the mean and s.d. of n=3 biologically independent samples. Target sites are in Supplementary Tables 8, 11, and 12.
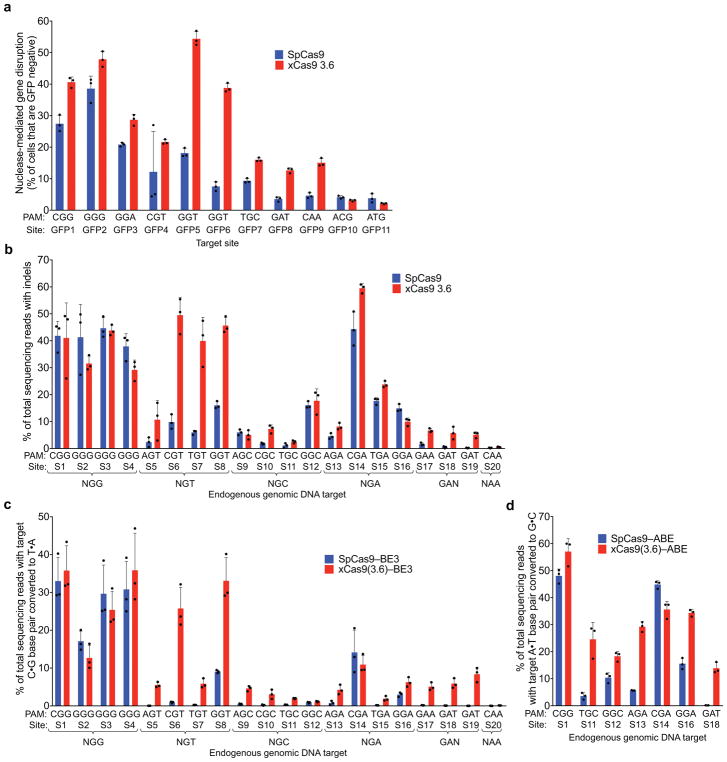
To test targeted genomic DNA cleavage in human cells we expressed xCas9 3.7 and 3.6 nuclease in a HEK293T cell line with a genomically integrated GFP gene and measured the loss of GFP fluorescence reflecting DNA cleavage and indel-mediated disruption of the target site. Two NGG PAM sites, three NGT sites, two NGC sites, and one NGA, GAT, NCG, and NTG site are present in the GFP sequence; all were tested with SpCas9, xCas9 3.7, and xCas9 3.6. For NGG PAM sites, xCas9 3.7 modestly outperformed SpCas9, resulting in 46±2.0% compared to 33±3.4% GFP disruption, respectively (mean ± SD of three independent replicates, Fig. 2b). For all tested non-NGG PAM sites, xCas9 3.7 showed substantially higher (1.6- to 6.4-fold) average apparent cleavage activity than SpCas9 (Fig. 2b). At all tested sites, xCas9 3.6 showed comparable or slightly lower GFP disruption percentages than xCas9 3.7 (Extended Data Fig. 7a). Neither xCas9 3.7 nor 3.6 increased GFP loss relative to SpCas9 for either NNG PAM site tested, suggesting that the transcriptional activation observed at some NNG PAM sites by dxCas9–VPR activators, and the strong NNG PAM signal in the bacterial PAM depletion assay, do not necessarily translate to DNA cleavage at all NNG sites in mammalian cells. Target site dependence is also well-known for SpCas930.
To further characterize DNA cleavage in human cells by xCas9 variants, we targeted endogenous genomic sites in HEK293T cells and measured indel formation by high-throughput sequencing (HTS). Twenty endogenous sites were tested covering four NGG PAM sites, all 12 possible NGT, NGC, and NGA PAMs, and GAT, GAA, and CAA PAMs (Fig. 2c). On the four NGG PAM sites tested, xCas9 3.7 showed comparable activity to SpCas9, averaging 41±6.4% indels compared to 41±6.0% for SpCas9 (Fig. 2c). All four NGT PAM sites showed much higher indel formation with xCas9 3.7 than with SpCas9, averaging 38±4.1% indels compared to 8.6±1.5% for SpCas9, a 4.5-fold increase. The four NGA PAM sites averaged 32±2.4% indels for xCas9 and 20±2.7% for SpCas9, a 1.6-fold increase, consistent with previous reports that NGA can serve as a secondary PAM for SpCas931. While indel frequencies at the endogenous NGC PAM sites were more variable, ranging from 4.8–31%, xCas9 3.7 averaged 13±0.90% indels compared to 6.3±0.77% for SpCas9, a 2.1-fold increase. Among the three GAA and GAT sites tested, SpCas9 showed virtually no activity, averaging 1.4±1.3% indel formation, while xCas9 3.7 averaged 7.2±2.8% indel formation, a 5.2-fold increase. The xCas9 3.6 variant showed comparable or slightly lower indel frequencies for all sites tested (Extended Data Fig. 7b). Negative control experiments lacking sgRNA plasmid resulted in no indels above background (Extended Data Fig. 8). Taken together, these results indicate that xCas9 3.7 nuclease mediates target gene disruption at NGG PAM sites with comparable efficiencies as wild-type SpCas9, but cleaves NG, GAA, and GAT PAM sites with substantially higher efficiencies than SpCas9. The greater PAM-dependent and protospacer-dependent variability of xCas9 nuclease-mediated gene disruption relative to transcriptional activation (Fig. 2 and Extended Data Figs 5–7) may reflect more extensive requirements for DNA cleavage than DNA binding27,32, or differences in the chromatin state of plasmid (Fig. 2a and Extended Data Figs 4, 5a–5d, 6) versus genomic targets (Fig. 2b, c, and Extended Data Figs 5e, 7).
Base editing by xCas9 variants in human cells
Base editing is a newer genome editing approach that uses a catalytically impaired Cas9 fused to a natural or laboratory-evolved nucleobase deaminase enzyme and, in some cases, a DNA glycosylase inhibitor to directly convert a target C•G to T•A, or a target A•T to G•C, without introducing double-stranded DNA breaks or requiring homology-directed repair4,5,33. The suitability of a target site for base editing is highly dependent on the presence of a suitably positioned PAM, which must exist within a narrow window downstream of the target base pair (typically 15±2 nucleotides). The broad PAM compatibility of xCas9 variants thus has the potential to expand the DNA targeting scope of base editors (Fig. 3c).
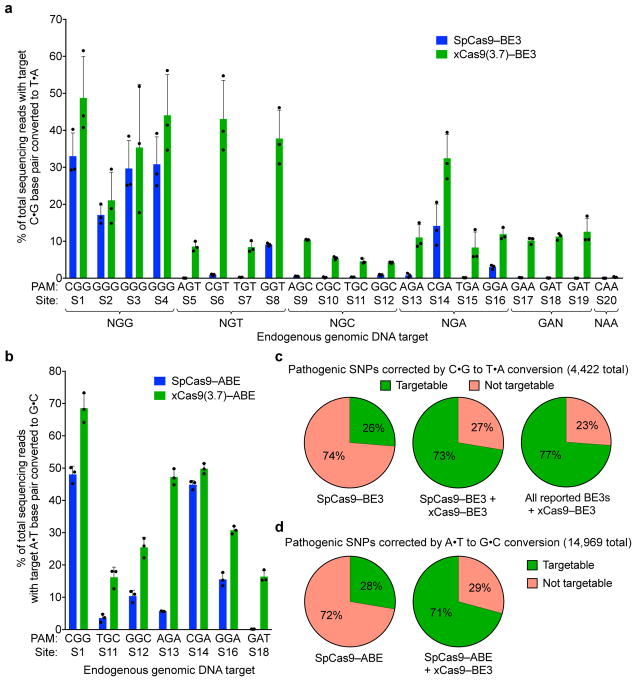
Figure 3. Cytidine and adenine base editing by xCas9.
a, The C•G to T•A conversion frequencies for 20 endogenous genomic loci in HEK293T cells at the most efficiently edited base 3 days after plasmid transfection are shown. b, The A•T to G•C conversion frequencies for seven endogenous genomic loci in HEK293T cells at the most efficiently edited base 5 days after plasmid transfection are shown. Values and error bars in (a) and (b) reflect the mean and s.d. of n=3 biologically independent samples. See Supplementary Table 14 for complete HTS results across the protospacer. c, Fraction of T•A to C•G pathogenic SNPs in ClinVar38 that, in principle, can be corrected by SpCas9–BE3 (left), SpCas9–BE3 + xCas9(3.7)–BE3 (middle), or xCas9(3.7)–BE3 + all BE3 variants reported to date (right). d, Fraction of G•C to A•T pathogenic SNPs in ClinVar that, in principle, can be corrected by SpCas9–ABE (left) or SpCas9–ABE + xCas9(3.7)–ABE (right).
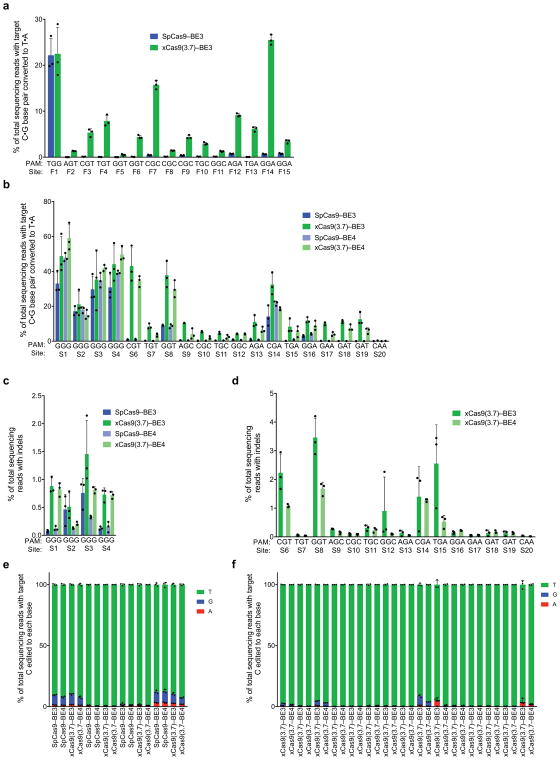
To evaluate C•G-to-T•A base editing activity of xCas9 variants, we substituted SpCas9 with xCas9 3.7 and 3.6 in the third-generation (BE3) base editor architecture4. Both xCas9–BE3s and SpCas9–BE3 were separately transfected into mammalian cells to compare editing efficiency on all 20 sites tested above for endogenous genomic DNA cleavage (Fig. 3a). At the four NGG PAM target sites tested, xCas9(3.7)–BE3 averaged 37±10% C•G to T•A conversion, while SpCas9–BE3 averaged 28±5.2% (Fig. 3a). At NG, GAA, and GAT PAM sites tested, xCas9(3.7)–BE3 resulted in substantially improved base editing, averaging 24±5.4% editing at NGT PAM sites, an 9.5-fold increase over that of SpCas9; 16±3.5% editing at NGA sites, a 3.5-fold increase over SpCas9; 6.2±0.34% editing at NGC sites, a 13-fold increase over SpCas9; 10±0.75% editing on the GAA PAM site, a > 50-fold increase over SpCas9; and 12±1.5% editing on the GAT sites, a > 100-fold increase over SpCas9 (Fig. 3a). The base editing efficiencies of xCas9(3.6)–BE3 were comparable to, or slightly worse than, those of xCas9(3.7)–BE3 (Extended Data Fig. 7c). We also tested cytosine base editing at an additional 15 endogenous genomic sites within the FANCF gene and observed similar large improvements for xCas9(3.7)–BE3 over SpCas9–BE3 (Extended Data Fig. 9a). Overall, these results indicate that xCas9 variants are compatible with the BE3 architecture, and enable cytidine base editing of target sites that cannot be accessed by SpCas9–BE3 or, with the exception of NGA PAM sites33, by any other previously reported base editors. We also tested xCas9 3.7 in the BE4 architecture designed to reduce undesired byproducts34, and observed fewer indels and higher product purities, although with slightly lower editing efficiencies (Extended Data Fig. 9b–d).
The recent development of an adenine base editor (ABE) enables programmable installation of A•T to G•C mutations5. No ABEs have been reported yet that can target non-NGG PAM sites, limiting its targeting scope. We replaced SpCas9 in ABE 7.105 with xCas9 3.7 and 3.6, and assayed the resulting xCas9–ABEs in HEK293T cells at the seven endogenous genomic sites tested above that contain an A in the targeting window of ABE (positions 4–8, counting the PAM as positions 21–23). At all seven of these sites, xCas9(3.7)–ABE resulted in higher base editing efficiencies than the original SpCas9–ABE (Fig. 3b). Average base editing efficiency at the NGG PAM site tested increased from 48±2.1% to 69±3.7%. At the GAT PAM site tested, xCas9(3.7)–ABE resulted in 16±1.5% base editing, while SpCas9–ABE yielded no detectable editing (≤ 0.1%), representing a > 100-fold increase. On the two NGC and three NGA sites tested, xCas9(3.7)–ABE averaged 21±2.5% and 43±1.5% base editing, respectively, while SpCas9–ABE averaged 7.0±1.3% and 22±1.2%, respectively. Base editing by xCas9(3.7)–ABE was comparable to or higher than that of xCas9(3.6)–ABE (Extended Data Fig. 7d). Collectively, these results establish that xCas9–ABE mediates adenine base editing at sites that cannot currently be accessed otherwise.
Improved DNA specificity of xCas9 variants in human cells
Because PAM recognition is a crucial component of Cas9 DNA specificity27, the substantially broadened PAM compatibility of xCas9s would be expected to increase their off-target activity32,35. Indeed, the engineered S. aureus KKH-SaCas9, which accepts an NNNRRT PAM instead of the native NNGRRT SaCas9 PAM, exhibits comparable or higher off-target editing than SaCas915. Most of the xCas9 mutations are close to the PAM or to the DNA:sgRNA interface (Fig. 1), raising the possibility that these mutations might alter the degree to which mismatches between the spacer and protospacer impede productive DNA binding or editing. Previous studies have demonstrated that some mutations near the Cas9:DNA interface can reduce off-target activity, in some cases without sacrificing on-target modification efficiency26,36,37.
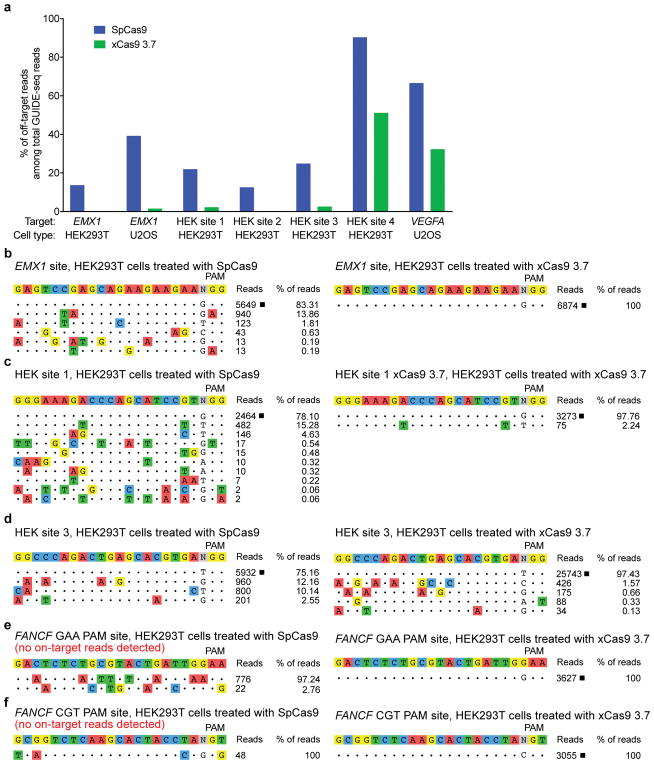
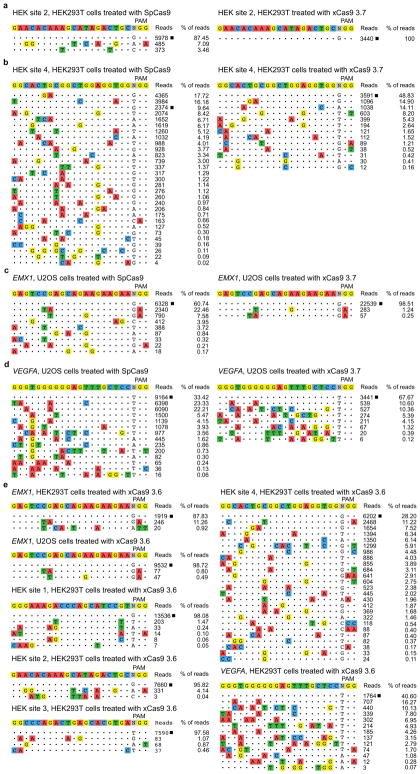
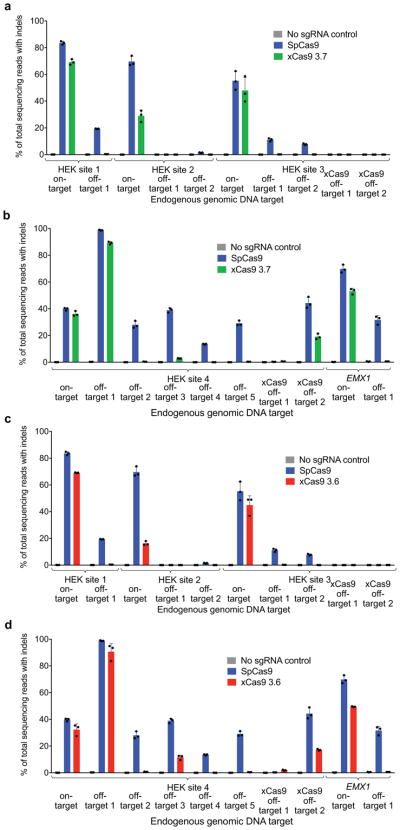
In order to test the off-target activity of xCas9 variants, we performed GUIDE-seq, an unbiased genome-wide off-target analysis32, on xCas9 3.7, xCas9 3.6, and SpCas9 in HEK293T and U2OS cells. Remarkably, for all five endogenous genomic NGG PAM sites tested in HEK293T cells and for both NGG PAM sites tested in U2OS cells, GUIDE-seq analysis revealed that xCas9 3.7 and 3.6 resulted in much lower off-target activity than SpCas9, as reflected by both the number of detected off-target sites, as well as the total modification frequency at each detected off-target site (Fig. 4 and Extended Data Fig. 10). For example, at the EMX1 target site in HEK293T cells, SpCas9 showed 5,649 on-target reads and a total of 1,132 off-target reads, while xCas9 3.7 showed 6,874 on-target reads but zero off-target reads (Fig. 4b). In U2OS cells, the off-target:on-target ratio for the same site was 0.65 for SpCas9 with 6,328 on-target reads, and 0.015 for xCas9 3.7 with 22,539 on-target reads, representing a 43-fold reduction in off-target modification (Extended Data Fig. 10c). Likewise, for HEK sites 1, 2, and 3, xCas9 3.7 resulted in ≥ 100-fold lower off-target to on-target modification ratios (0.023, < 0.001, and 0.028, respectively) compared to those of SpCas9 (0.28, 0.14, and 0.33, respectively) (Fig. 4 and Extended Data Fig. 10). For the known highly promiscuous target sites HEK site 4 and VEGFA32, the off-target:on-target ratios were 9.4 and 2.0, respectively, for SpCas9, but only 1.0 and 0.48 for xCas9 3.7, a 4.2- to 9.4-fold improvement (Extended Data Fig. 10). We observed these large improvements in DNA specificity for xCas9 3.7 and 3.6 even though, consistent with the above findings, xCas9 variants showed a much broader range of PAM sequences among detected off-target sites (Fig. 4 and Extended Data Fig. 10). These GUIDE-seq results were verified by HTS of many individual on-target and off-target sites from the genomic DNA of treated cells (Extended Data Fig. 11).
Figure 4. Off-target editing analysis of xCas9.
a, GUIDE-seq32 was performed on SpCas9 and xCas9 3.7 nucleases. Six endogenous genomic sites with NGG PAMs were tested in HEK293T or U2OS cells. The percentage of off-target sequencing reads relative to total reads are shown. b–d, All GUIDE-seq on-target reads (□) and off-target reads for three sites in HEK293T cells are shown for SpCas9 and xCas9 3.7. See Extended Data Fig. 10 for additional GUIDE-seq results. e, f, GUIDE-seq results for two endogenous genomic non-NGG PAM sites in HEK293T cells. No on-target GUIDE-seq reads (□) were detected for SpCas9 at either of these non-NGG sites. See Extended Data Fig. 10 for GUIDE-seq analysis of xCas9 3.6 and Extended Data Fig. 11 for HTS validation of GUIDE-seq results. Target sequences are listed in Supplementary Table 15.
We also evaluated the off-target DNA specificity of xCas9 3.7 at two non-NGG PAM (GAA and CGT) sites with both SpCas9 and xCas9 3.7 in HEK293T cells. As expected, GUIDE-seq did not yield any on-target reads for SpCas9 at either of these non-NGG PAM sites, while xCas9 3.7 had 3,627 on-target reads for the GAA PAM site and 3,055 on-target reads for the CGT PAM site (Fig. 4e, f). Importantly, neither site was accompanied by any detected off-target GUIDE-seq reads for xCas9 3.7, although potential off-target reads were detected for SpCas9 (Fig. 4e, f). Collectively, these findings reveal that xCas9 3.7 and 3.6 offer greatly reduced off-target activity compared with wild-type SpCas9, despite their broader PAM compatibility. These results also establish that there is no necessary trade-off between Cas9-mediated editing efficiency, PAM compatibility, and DNA specificity, a key finding as natural and engineered genome editing agents advance into widespread applications including human clinical trials.
These results, together with the success of multiple independent efforts to create high-fidelity Cas9 variants26,36,37, suggest that the DNA promiscuity of wild-type Cas9, which likely evolved to impede viral evasion, can readily be overcome by protein engineering or evolution. That xCas9 exhibits much higher DNA specificity than SpCas9 even though it was not explicitly selected for this property suggests that the off-target activity of wild-type SpCas9 may lie at a narrow fitness peak suitable for defending the much smaller bacterial genome but not optimal for genome editing in mammalian cells. These observations are therefore consistent with a model in which native SpCas9 is poised to become more specific, rather than less specific, upon mutation.
To our knowledge, the targeting scope of xCas9 is the broadest among Cas9 variants known to function efficiently in mammalian cells. Evolved xCas9 variants are also the first to offer improvements in targeting scope, activity, and DNA specificity in a single entity relative to wild-type SpCas9. While the efficacy of xCas9 on non-NGG PAMs varies based on application—here, transcriptional activation, DNA cleavage, or base editing—and on target site, the ability to access some NG, GAA, and GAT PAM sequences greatly expands the breadth of targets available for site-sensitive genome editing applications. Indeed, compared to SpCas9–BE3, xCas9(3.7)–BE3 increases the percentage of 4,422 pathogenic SNPs in the ClinVar database38 that in principle could be targeted by C•G to T•A base editing from 26% to 73% (Fig. 3c). Likewise, xCas9(3.7)–ABE increases the fraction of 14,969 pathogenic ClinVar SNPs that could be targeted by A•T to G•C base editing from 28% to 71% (Fig. 3d). We anticipate that xCas9 and additional CRISPR enzyme variants with broadened PAM compatibilities may also expand the scope of other forms of nucleic acid editing, CRISPR-based screens, and epigenetic modification.
Online Content
Methods, along with any additional Extended Data display items, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Methods
General methods and cloning
DNA sequences used in this work are listed in the Supplementary Information. PCR was performed using Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs) or Phusion U Green Multiplex PCR Master Mix (Thermo Fisher Scientific). PACE plasmids and phage were constructed by USER cloning or Gibson cloning (New England Biolabs). Cas9 genes and plasmid backbones for PACE were obtained from previously reported plasmids19,20 available from Addgene. Plasmids encoding dxCas9–VPR29, xCas9 nucleases, xCas9–BE34, xCas9-BE434, and xCas9-ABE5 were constructed by replacing SpCas9 using Gibson cloning. Plasmids for sgRNA expression were constructed using one-piece blunt-end ligation of a PCR product containing a variable 20-nt sequence corresponding to the desired sgRNA targeted site. Primers and templates used in the synthesis of all sgRNA plasmids used in this work are listed in Supplementary Tables 8, 9, 11, 12, 15, and 16. All guide RNAs used in this study were transcribed from a U6 promoter, and natively started with a G at the 5′ end to avoid possible losses in activity caused by a mismatched 5′ guide terminus. PCR was performed using Q5 Hot Start High-Fidelity Polymerase (New England Biolabs) with the phosphorylated primers and the plasmid pFYF1320 (EGFP sgRNA expression plasmid) as a template according to the manufacturer’s instructions. PCR products were analyzed by agarose gel electrophoresis, the band of the expected molecular weight was cut out, and the DNA was extracted using a Zymoclean Gel DNA Recovery Kit (Zymo Research) and ligated using T4 DNA Ligase (New England Biolabs) according to the manufacturer’s instructions. DNA vector amplification was carried out using Mach1 competent cells (Thermo Fisher Scientific). All mammalian ABE constructs, sgRNA plasmids and bacterial constructs were transformed and stored as glycerol stocks at −80 °C in Mach1 T1R Competent Cells (Thermo Fisher Scientific), which are recA−. Molecular biology grade Hyclone water (GE Healthcare Life Sciences) was used in all assays and PCR reactions. All vectors used in evolution experiments and mammalian cell assays were purified using ZympPURE Plasmid Midiprep (Zymo Research Corporation), which includes endotoxin removal. Antibiotics were purchased from Gold Biotechnology.
Cell culture
HEK293T (ATCC CRL-3216) and U2OS (ATCC-HTB-96) were maintained in Dulbecco’s Modified Eagle’s Medium plus GlutaMax (Thermo Fisher Scientific) supplemented with 10% (v/v) fetal bovine serum (FBS) at 37 °C with 5% CO2.
Transfections
HEK293T cells were seeded on 48-well poly-D-lysine-coated BioCoat plates (Corning) and transfected at approximately 85% confluency. For genomic DNA cutting or base editing, 750 ng of Cas9 or BE3 and 250 ng of sgRNA expression plasmids were transfected using 1.5 μL of Lipofectamine 2000 (Thermo Fisher Scientific) per well according to the manufacturer’s protocol. For GFP activation, 200 ng of dCas9–VPR plasmid, 50 ng of sgRNA expression plasmid, 60 ng of GFP reporter plasmid, and 30 ng of iRFP expression plasmid were transfected using 1.5 μL of Lipofectamine 2000 (Thermo Fisher Scientific) per well according to the manufacturer’s protocol. Endogenous gene activation was done similarly but with 200 ng of dCas9–VPR plasmid and 50 ng of sgRNA expression plasmid only.
GFP transcriptional activation assay
Transfected HEK293T cells were trypsinized and resuspended in Dulbecco’s Modified Eagle’s Medium plus GlutaMax (Thermo Fisher Scientific) supplemented with 10% (v/v) fetal bovine serum (FBS). The cells were kept on ice and flow cytometry was performed using a LSRFortessa from BD Biosciences. Events were gated for iRFP positive cells to analyze transfected cell. The percentage of GFP-positive cells and the intensity of GFP fluorescence from each cell was collected.
RNA expression quantification for endogenous transcriptional activation assay
RNA was extracted from HEK293T cells using the Quick-RNA Plus Kit (Zymo Research). cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad) and qPCR was performed on a Bio-Rad CFX96 Real-Time PCR Detection System using Q5 Polymerase (NEB) and SYBR Green (Lonza). Primers for qPCR are listed in Supplementary Table 10.
PAM depletion assay
Electrocompetent NEB 10-beta cells (New England Biolabs) were electroporated with two plasmids. The first plasmid expresses Cas9 (inducible by anhydrotetracycline, ATc), the sgRNA (inducible by arabinose), and a spectinomycin resistance gene. The second plasmid contains the target protospacer and a kanamycin resistance gene. After incubation with SOC outgrowth medium (New England Biolabs) for 1 hour the bacteria were plated on agar plates containing both spectinomycin and kanamycin along with ATc and arabinose inducers. After incubating overnight, the bacterial cells on the agar plates were scraped and the plasmids extracted using the ZymoPURE Plasmid Midiprep Kit (Zymo Research). The resulting post-selection DNA included all of the protospacer plasmids not cleaved by Cas9. The same region around the protospacer in the pre-selection library and the post-selection DNA was then amplified separately using NEBNext High-Fidelity PCR Polymerase (New England Biolabs) with flanking HTS primer pairs listed in the Supplementary Table 7. Illumina barcoding PCR reaction was assembled with NEBNext High-Fidelity PCR Polymerase. PCR products were purified by electrophoresis with a 2% agarose gel using a QIAquick Gel Extraction Kit, eluting with 15 μL of water. DNA concentration was quantified with the KAPA Library Quantification Kit-Illumina (KAPA Biosystems) and sequenced on an Illumina MiSeq instrument according to the manufacturer’s protocols.
High-throughput DNA sequencing of genomic DNA samples
Transfected cells were harvested after 3 days (BE3 and BE4) or 5 days (DNA cutting and ABE). Media was removed and cells were washed with 1× PBS solution (Thermo Fisher Scientific). Genomic DNA was extracted by addition of 100 μL freshly prepared lysis buffer (10 mM Tris-HCl, pH 7.0, 0.05% SDS, 25 μg/mL Proteinase K (Thermo Fisher Scientific) directly into each well of the tissue culture plate. The plate was incubated at 37 °C for 1 h. The genomic DNA mixture was transferred to a 96-well PCR plate and incubated at 80 °C for 15 min to denature enzymes. Genomic regions of interest were amplified by PCR with flanking HTS primer pairs listed in the Supplementary Table 13. Each 25 μL PCR reaction was assembled with Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Fisher Scientific) according to manufacturer’s instructions using 1.0 μM of each forward and reverse primer and 1 μL of genomic DNA extract. PCR reactions were carried out as follows: 95°C for 3 min, then 30 cycles of [98°C for 30 s, 60°C for 20 s, and 72°C for 1 min], followed by a final 72 °C extension for 5 min. PCR products were verified by comparison to DNA standards (1-kb Plus DNA Ladder) on a 2% agarose gel with ethidium bromide. Each 25-μL Illumina barcoding PCR reaction was assembled with Phusion DNA polymerase according to manufacturer’s instructions using 0.5 μM of each unique forward and reverse Illumina barcoding primer pair and 1 μL of unpurified genomic amplification PCR reaction mixture. The barcoding reactions were carried out as follows: 98°C for 2 min, then 8 cycles of [98°C for 12 s, 61°C for 25 s, and 72°C for 30 s], followed by a final 72 °C extension for 1.5 min. PCR products were purified by electrophoresis with a 2% agarose gel using a QIAquick Gel Extraction Kit, eluting with 15 μL of water. DNA concentration was determined with the KAPA Library Quantification Kit-Illumina (KAPA Biosystems) and sequenced on an Illumina MiSeq instrument according to the manufacturer’s protocols. Analysis was carried out using previously published Matlab code5, provided in Supplementary Notes 1 and 2.
Analysis of human disease-associated mutations in ClinVar database
Bioinformatic analysis of the ClinVar database was carried out in a manner similar to previously described analysis33. The code is provided in Supplementary Note 3.
GUIDE-Seq
HEK293T cells were transfected with 750 ng of the Cas9 plasmid, 250 ng of the gRNA plasmid, and 20 pmol of GUIDE-seq dsODN. U2OS cells were transfected with 750 ng of the Cas9 plasmid, 250 ng of the gRNA plasmid, and 100 pmol of GUIDE-seq dsODN. For both cell types, 20 μL of Solution SE (Lonza) was used along with a Lonza Nucleofector 4-D. Program CM-137 was used for HEK293T cells while program DN-100 was used for U2OS cells. Genomic DNA was extracted using the Quick-DNA Miniprep Plus Kit (Zymo Research) following the manufacturer’s protocol. The DNA was sheared to an average of 500 bp using a Covaris S220 focused ultrasonicator as previously described32. End repair, dA-tailing, adapter ligation, tag-specific PCR1, and tag-specific PCR2 were carried out using the primers and methods previously described32. DNA concentration was quantified with the KAPA Library Quantification Kit-Illumina (KAPA Biosystems) and sequenced on an Illumina MiSeq instrument according to the manufacturer’s protocols. Analysis was carried out using the previously published Python code32.
Data availability
High-throughput sequencing data have been deposited in the NCBI Sequence Read Archive database under accession code SRP130166. Plasmids encoding xCas9 3.7 and 3.6 transcriptional activators, nucleases, BE3, BE4, and ABE variants will be available from Addgene.
Extended Data
Extended Data Figure 1. Optimization of Cas9 PACE.
Luciferase expression in E. coli was used as a proxy of gene III expression during efforts to link Cas9 binding to gene expression for PACE. a, b, Seven guide RNAs targeting the luciferase reporter (G1–G7′, see Supplementary Table 1), as well as a scrambled guide RNA negative control (G0) were tested without dCas9 (white bars) and with ω–dCas9 (a) or dCas9–ω (b) fusions (grey bars). c, Tests of seven different linkers between ω and dCas9. See Supplementary Table 2 for linker sequences. d, Evolution of ω–dCas9 on an NGG PAM site in PACE yielded variants (PACE1, PACE2, and PACE3) that were tested in comparison with canonical (“wt”) ω–dCas9, ω tethered to PACE1 dCas9, and the I12N ω mutant tethered to canonical dCas9. Values and error bars reflect the mean and s.d. of n=3 biologically independent samples.
Extended Data Figure 2. PAM profiling of xCas9 variants.
a, A plasmid library containing a protospacer with all possible NNN PAM sequences and a spectinomycin resistance gene was electroporated into E. coli along with a plasmid expressing SpCas9 or the xCas9 variant shown in separate experiments. PAMs that are cleaved are depleted from the library when plated on media containing spectinomycin. HTS of the library before versus after selection enables quantification of the change in library composition, resulting in a sequence logo39 for the PAM preference of SpCas9 (left) and xCas9 3.7 (right). b, c, PAM depletion scores of Cas9 variants from spectinomycin selection in E. coli, calculated as described previously35, with 1.0 representing complete cleavage of that PAM sequence. Scores for NGN, NNG, GAA, GAT, and CAA are shown in (b) while the rest of the PAM sequences are shown in (c).
Extended Data Figure 3. Transcriptional activation of reporter site PAM libraries with xCas9.
Transcriptional activation by dSpCas9–VPR and dxCas9–VPR variants, transfected as plasmids, on GFP reporter plasmids containing different PAM sites in HEK293T cells. a, Earlier generations of xCas9 variants were tested on the R1 site with the NGG, NNN, or NNNNN PAM libraries. Values and error bars reflect the mean and s.d. of n=3 biologically independent samples. b, c, Transcriptional activators dxCas9(3.6)–VPR and dxCas9(3.7)–VPR were tested on two different protospacer reporters, R1 in (a) and R2 in (b), containing adjacent NGG, NNN, or NNNNN PAM libraries. Values and error bars reflect the mean and s.d. of n=3 biologically independent samples.
Extended Data Figure 4. Transcriptional activation with xCas9 2.0.
a–d, The transcriptional activator dxCas9(2.0)–VPR was tested on the R1 protospacer (Extended Data Fig. 3 and Supplementary Table 8) with each of the 64 possible NNN PAMs (NAN, NCN, NGN, and NTN) in HEK293T cells. Values and error bars reflect the mean and s.d. of n=3 biologically independent samples.
Extended Data Figure 5. Transcriptional activation with xCas9 3.7 on all 64 NNN PAM sites and endogenous gene activation in human cells.
a–d, The transcriptional activator dxCas9(3.7)–VPR was tested on the R1 protospacer (Extended Data Fig. 3 and Supplementary Table 8) with each of the 64 possible NNN PAMs (NAN, NCN, NGN, and NTN) in HEK293T cells. e, Endogenous gene activation was tested using both dSpCas9–VPR and dxCas9(3.7)–VPR to activate expression of the NEUROD1, ASCL1, MIAT, or RHOXF2 at six total sites. RNA expression as measured by RT-qPCR was compared to background expression levels for each gene (measured in the control with no sgRNA) and was normalized to ACTB expression. Values and error bars reflect the mean and s.d. of n=3 biologically independent samples. Target sites are in Supplementary Table 9.
Extended Data Figure 6. Transcriptional activation with xCas9 3.6 on all 64 NNN PAM sites.
a–d, The transcriptional activator dxCas9(3.6)–VPR was tested on the R1 protospacer (Extended Data Fig. 3 and Supplementary Table 8) with each of the 64 possible NNN PAMs (NAN, NCN, NGN, and NTN) in HEK293T cells. Values and error bars reflect the mean and s.d. of n=3 biologically independent samples.
Extended Data Figure 7. Genomic DNA cleavage and base editing by evolved xCas9 3.6.
a, Genomic DNA cleavage in HEK293-GFP cells containing a genomically integrated GFP gene by SpCas9 or xCas9 3.6, transfected as plasmids. After 5 days, the cells were analyzed for loss of GFP fluorescence by flow cytometry. Sequences for all target sites are listed in Supplementary Table 11. b, DNA cleavage of endogenous genomic DNA sites with a variety of NGG and non-NGG PAMs by SpCas9 and xCas9 3.6 in HEK293T cells. Indel rates were measured by HTS 5 days after plasmid transfection. Sequences for all target sites are listed in Supplementary Table 12. c, 20 sites containing NG, GAA, GAT, or CAA PAM sites were tested for C•G to T•A base editing in HEK293T cells by SpCas9–BE3 or xCas9(3.6)–BE3. The C•G to T•A conversion frequency by HTS at the most efficiently edited base 3 days after plasmid transfection is shown. d, Of the 20 sites in (c), seven contained an A in the canonical window for ABE editing5 and were tested for A•T to G•C base editing by SpCas9–ABE and xCas9(3.6)–ABE. The A•T to G•C conversion frequency by HTS at the most efficiently edited base 5 days after plasmid transfection is shown. Values and error bars reflect the mean and s.d. of n=3 biologically independent samples. Complete HTS results across the protospacer are provided in Supplementary Table 14.
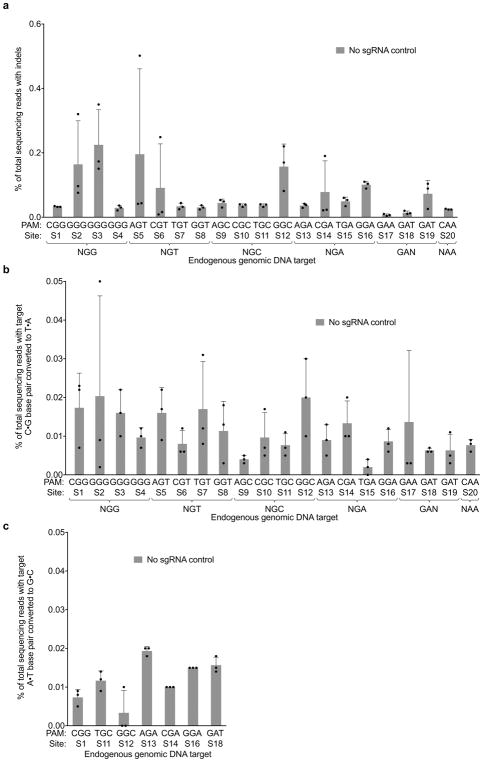
Extended Data Figure 8. Negative controls lacking guide RNA for nuclease and base editing experiments.
To verify genomic DNA cleavage and base editing results, the same sites were sequenced after treatment with SpCas9 nuclease, SpCas9-BE3, or SpCas9-ABE but without any sgRNA. a, Indel rates at endogenous target sites 5 days after treatment of HEK293T cells with SpCas9. b, Target C•G to T•A conversion 3 days after treatment of HEK293T cells with SpCas9-BE3. c, Target A•T to G•C base conversion 5 days after treatment of HEK293T cells with SpCas9-ABE. Values and error bars reflect the mean and s.d. of n=3 biologically independent samples. Complete HTS results across the protospacer are provided in Supplementary Table 14.
Extended Data Figure 9. Cytidine base editing at 15 additional genomic sites and xCas9 base editing with the BE4 architecture.
a, Base editing by SpCas9–BE3 and xCas9(3.7)–BE3 at 15 sites within the FANCF gene in HEK293T cells. The C•G to T•A conversion frequency at the most efficiently edited base 3 days after plasmid transfection is shown. b, Test of xCas9 3.7 in the BE4 architecture34 on the same sites tested in Fig. 3. The C•G to T•A conversion frequency in HEK293T cells at the most efficiently edited base 3 days after plasmid transfection is shown. c, d, Indel frequency following treatment with BE3 or BE4 variants targeting sites with NGG PAMs (c) and non-NGG PAMs (d). e, f, Product distribution among edited DNA sequence reads (reads in which the target C is mutated) following treatment with BE3 or BE4 variants targeting sites with NGG PAMs (e) and non-NGG PAMs (f). Since SpCas9 has minimal activity on non-NGG PAM sites, only xCas9(3.7)–BE3 and xCas9(3.7)–BE4 data is compared on non-NGG PAM sites. Values and error bars reflect the mean and s.d. of n=3 biologically independent samples. Target sites are in Supplementary Table 12.
Extended Data Figure 10. Additional characterization of xCas9 3.7 and xCas9 3.6 by GUIDE-seq.
In addition to the GUIDE-seq data shown in Fig. 4, two additional sites in HEK293T cells (a, b) and two sites in U2OS cells (c, d) were analyzed after treatment with SpCas9 and xCas9 3.7. e, All six GUIDE-seq sites with an NGG PAM that were tested with SpCas9 and xCas9 3.7 in HEK293T and U2OS cells were also tested with xCas9 3.6. On-target reads (□) and off-target reads for all sites are shown. Target sequences are listed in Supplementary Table 15.
Extended Data Figure 11. Validation by high-throughput sequencing of GUIDE-seq results.
The most frequent off-target sites identified by GUIDE-seq were verified by HTS of genomic DNA following treatment of HEK293T cells with SpCas9 or xCas9 3.7 (a, b), or following treatment with SpCas9 or xCas9 3.6 (c, d). New sites with non-NGG PAMs that were identified as xCas9 off-target sites were also analyzed in (a–d). Values and error bars reflect the mean and s.d. of n=3 biologically independent samples. Target sequences are in Supplementary Table 16.
Supplementary Material
Acknowledgments
This work was supported by DARPA HR0011-17-2-0049, U.S. NIH RM1 HG009490, R01 EB022376, and R35 GM118062, and HHMI. J.H.H. was supported by NDSEG and NSF graduate fellowships. S.M.M. was supported by an NSF graduate fellowship. W.T. is an HHMI Fellow of the Jane Coffin Childs Memorial Fund. L.C. was supported by the Agency for Science, Technology, and Research, Singapore. We thank Ahmed Badran, Basil Hubbard, Jonathan Levy, Tony Huang, George Church, Alejandro Chavez, Kevin Esvelt, Su Vora, and Jonathan Scheiman for helpful discussions.
Footnotes
Supplementary Information is available in the online version of the paper.
Author contributions
J.H.H. designed the research, performed PACE, characterized variants in bacteria, conducted human cell experiments, analyzed data, and performed off-target analysis, and wrote the manuscript. S.M.M. performed human cell experiments, analyzed data, and wrote the manuscript. M.H.G. performed human cell experiments and data analysis. W.T. performed human cell experiments and cloning. L.C. assisted with cloning and PACE. N.S. optimized Cas9 PACE. C.Z. assisted with cloning and PACE. X.G. assisted with off-target analysis. H.A.R. assisted with indel and base editing analyses. D.R.L designed and supervised the research and wrote the manuscript.
The authors declare competing financial interests: J.H.H. and D.R.L. have filed patent applications on this work. D.R.L. is a consultant and co-founder of Editas Medicine, Beam Therapeutics, and Pairwise Plants, companies that use genome editing technologies. The authors declare no competing non-financial interests.
References
- 1.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitsunobu H, Teramoto J, Nishida K, Kondo A. Beyond Native Cas9: Manipulating Genomic Information and Function. Trends Biotechnol. 2017;35:983–996. doi: 10.1016/j.tibtech.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaudelli NM, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim E, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller M, et al. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Ther. 2016;24:636–644. doi: 10.1038/mt.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CM, Cradick TJ, Bao G. The Neisseria meningitidis CRISPR-Cas9 System Enables Specific Genome Editing in Mammalian Cells. Mol Ther. 2016;24:645–654. doi: 10.1038/mt.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleinstiver BP, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badran AH, Liu DR. Development of potent in vivo mutagenesis plasmids with broad mutational spectra. Nat Commun. 2015;6:8425. doi: 10.1038/ncomms9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Badran AH, et al. Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature. 2016;533:58–63. doi: 10.1038/nature17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbard BP, et al. Continuous directed evolution of DNA-binding proteins to improve TALEN specificity. Nat Methods. 2015;12:939–942. doi: 10.1038/nmeth.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryson DI, et al. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat Chem Biol. 2017 doi: 10.1038/nchembio.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Packer MS, Rees HA, Liu DR. Phage-assisted continuous evolution of proteases with altered substrate specificity. Nat Commun. 2017;8:956. doi: 10.1038/s41467-017-01055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng X, Wolfe SA. Identifying DNA sequences recognized by a transcription factor using a bacterial one-hybrid system. Nat Protoc. 2006;1:30–45. doi: 10.1038/nprot.2006.6. [DOI] [PubMed] [Google Scholar]
- 24.Dove SL, Joung JK, Hochschild A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 25.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen JS, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao L, et al. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 2017;35:789–792. doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci Rep. 2014;4:5405. doi: 10.1038/srep05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim YB, et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komor AC, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv. 2017;3:eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landrum MJ, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput sequencing data have been deposited in the NCBI Sequence Read Archive database under accession code SRP130166. Plasmids encoding xCas9 3.7 and 3.6 transcriptional activators, nucleases, BE3, BE4, and ABE variants will be available from Addgene.