Abstract
Cryptococcus neoformans is an opportunistic fungal pathogen that is ubiquitous in the environment. It causes a deadly meningitis that is responsible for over 180,000 deaths worldwide each year, including 15% of all AIDS-related deaths. The high mortality rates for this infection, even with treatment, suggest a need for improved therapy. Unique characteristics of C. neoformans may suggest directions for drug discovery. These include features of three structures that surround the cell: the plasma membrane, the cell wall around it, and the outermost polysaccharide capsule. We review current knowledge of the fundamental biology of these fascinating structures and highlight open questions in the field, with the goal of stimulating further investigation that will advance basic knowledge and human health.
Key words: Cryptococcus neoformans, polysaccharide capsule, cell wall, plasma membrane
Cryptococcus neoformans is an opportunistic fungal pathogen that causes severe infection of the central nervous system. Inhalation of this microbe, as either a spore or desiccated yeast cell (Giles et al. 2009), causes a pulmonary infection that in immunocompetent individuals is minimally symptomatic, although it may remain latent for extended periods of time (Kwon-Chung et al. 2014, Ballou and Johnston 2017). In severely immunocompromised individuals, however, C. neoformans can disseminate from the lungs and cross the blood-brain barrier (Santiago-Tirado et al. 2017), causing an often-lethal meningoencephalitis. Close to 220,000 cases of cryptococcal meningitis are reported annually. These result in over 180,000 deaths worldwide, including 15% of all AIDS-related deaths (Rajasingham et al. 2017). Mortality rates range from 10 to 75% (Day et al. 2013, Jarvis et al. 2014), even with carefully developed treatment regimens (Perfect et al. 2010), due to challenges that include drug toxicity, efficacy, cost, and availability. Increasing drug resistance has also been reported (Sionov et al. 2013, Chen et al. 2015, Smith et al. 2015). Clearly, improved therapies are needed to combat this infection.
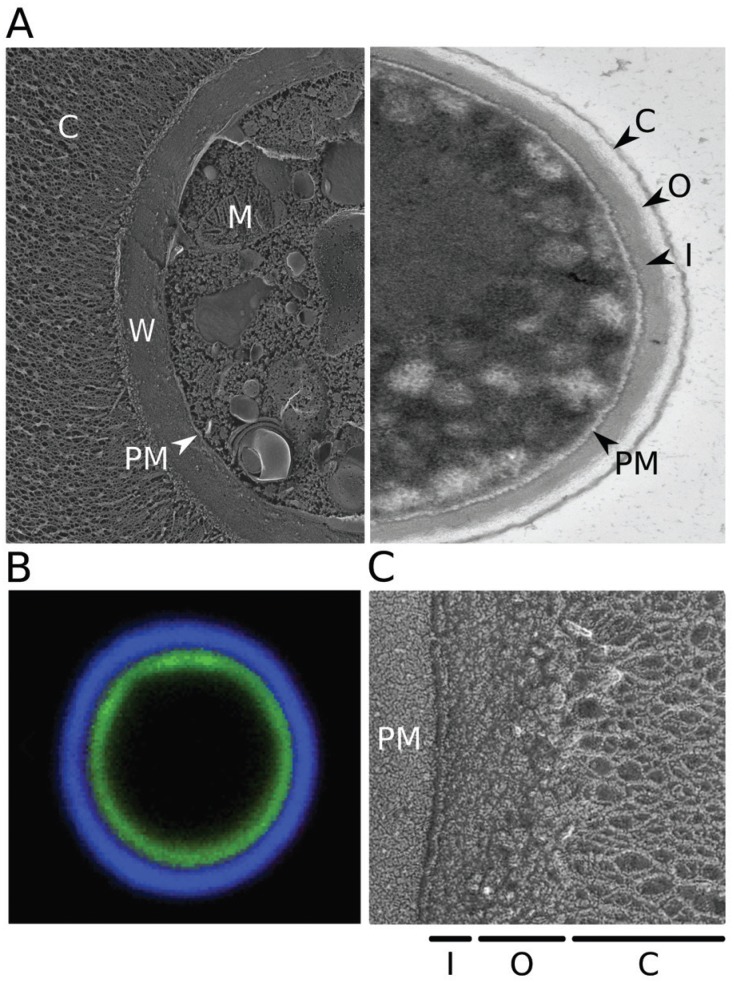
The outer layers of C. neoformans have unique features that may offer directions for drug discovery. These consist of three concentric structures: the capsule, cell wall, and plasma membrane (Fig. 1A, left). The outermost layer, the polysaccharide capsule, is the hallmark of this organism and is required for virulence. This highly dynamic structure modulates fungal interactions with immune cells. Capsule components are also shed into the environment, where they further influence the host response and may be exploited for diagnosis and monitoring of cryptococcal infection (Alspaugh 2015). Both capsule thickness and shedding are tightly regulated in response to environmental conditions (Kumar et al. 2011, Maier et al. 2015).
(A) Electron micrographs of Cryptococcus neoformans. Left, quick-freeze deep-etch image of cells grown in capsule-inducing conditions (as in Haynes et al. 2011); right, transmission image of cells grown in rich medium (YPD), which yields only thin capsules. C: capsule; W: cell wall; PM: plasma membrane; M: mitochondrion; O: outer cell wall layer; I: inner cell wall layer. (B) Fluorescent micrograph highlighting the cell wall and capsule. Cryptococcal cells were induced to form capsule and stained with florescein (green) to label the cell wall and monoclonal antibody 2H1 (blue, generously provided by Arturo Casadevall) to label the capsule, as in Pierini and Doering (2001). (C) Quick-freeze deep-etch image highlighting the two layers of the cell wall.
Below the capsule, and anchoring it, lies the fungal cell wall. This complex structure surrounds the plasma membrane and helps the cell withstand environmental challenges such as osmotic and mechanical stress. It is composed of glucans, chitin, chitosan, and glycosylated proteins (Gow et al. 2017). Melanin pigment associated with the cell wall further helps C. neoformans resist environmental stress and antifungal drug toxicity.
The innermost of the three layers we will consider is the plasma membrane. Unique features of fungal membranes are already the targets of several important classes of antifungal drugs, the polyenes and azoles. The plasma membrane is also the site of multiple proteins implicated in fungal virulence and has been implicated in the production of extracellular vesicles with potential roles in virulence (Rodrigues et al. 2014, Brown et al. 2015, Joffe et al. 2016, Rella et al. 2016).
The unique features of the cryptococcal capsule, cell wall, and plasma membrane demonstrate fascinating fundamental biology that may potentially be exploited for therapy. Below we review current knowledge of these structures and their synthesis, and highlight some open questions in the field. We do not address the regulation of these structures in this short article, but refer interested readers to recent reviews that address this important topic (Doering 2009, Gilbert et al. 2011, O’Meara and Alspaugh 2012, Bahn and Jung 2013, Kwon-Chung et al. 2014, Srikanta et al. 2014, Alspaugh 2015, Rella et al. 2016, Gow et al. 2017).
The capsule - The polysaccharide capsule is a major cryptococcal virulence factor, which impedes the host immune response and is required for fungal survival within the host. This structure is composed of two polymers, glucuronoxylomannan (GXM) and glucuronoxylomannogalactan (GXMGal), along with trace mannoproteins (Cherniak et al. 1998, Doering 2009). GXM, which typically has a molecular weight in the millions, consists of an a-1,3-linked mannose backbone substituted with glucuronic acid and xylose (Cherniak et al. 1988, Cherniak et al. 1998, Heiss et al. 2009). GXMGal, roughly an order of magnitude smaller, is a galactan with galactomannan side chains bearing a variable number of xylose residues; the galactose backbone also bears galactofuranose (all sugars are pyranose where not specified) (Heiss et al. 2013, Previato et al. 2017). Mannose residues of both polymers may also be O-acetylated (Gates-Hollingsworth and Kozel 2009, Previato et al. 2017). Based on analysis of shed capsule, GXM constitutes ~90% of the capsule mass and GXMGal the remainder (Cherniak and Sundstrom 1994), although this may differ for surface-associated capsule.
The reactions that incorporate individual monosaccharides into complex polysaccharides like those of the capsule or cell wall use activated sugar molecules, usually in the form of nucleotide sugars, as donors. These compounds are mainly synthesized in the cytosol, although most glycan biosynthesis, including the formation of capsule polysaccharides (Yoneda and Doering 2006), occurs in the lumen of the secretory pathway. For this to occur, the precursors must be moved into the synthetic compartment by specific nucleotide sugar transporters (NSTs) (Caffaro and Hirschberg 2006, Hadley et al. 2014). C. neoformans transporters have been identified for all of the predicted capsule precursors: GDP-mannose (Cottrell et al. 2007, Wang et al. 2014), UDP-galactose (Cottrell et al. 2007, Moyrand et al. 2007, Wang et al. 2014, Li et al. 2017), UDP-galactofuranose (Li et al. 2018a), UDP-glucuronic acid (Li et al. 2018b), and UDP-xylose (Li et al. 2018a).
Once nucleotide sugars are transported into the lumen of secretory organelles, they serve as substrates for specific glycosyltransferases, which mediate the synthetic reactions that form capsule polysaccharides. The complex structures of GXM and GXMGal suggest the involvement of multiple such enzymes. However, only a single glycosyltransferase, Cxt1, has been directly implicated in capsule synthesis. This β-1,2-xylosyltransferase is required to produce fully xylosylated GXM and GXMGal (Klutts et al. 2007, Klutts and Doering 2008). Two other cryptococcal glycosyltransferases have been biochemically characterized (Sommer et al. 2003, Reilly et al. 2009, Reilly et al. 2011), but only one of them has specificity appropriate for a role in capsule production, and deletion of the corresponding gene does not alter capsule composition (Sommer et al. 2003). It remains to be determined whether this reflects the presence of compensating activities or indicates that this enzyme does not participate in capsule synthesis. The many other activities required for capsule synthesis (Bose et al. 2003, Klutts et al. 2006, Doering 2009) are likely encoded by some of the ~70 putative glycosyltransferase genes observed in the C. neoformans genome (Cantarel et al. 2009, Lombard et al. 2014); future work will be required to identify them.
Most glycan synthetic machinery is localized to the Golgi, where elaboration of core glycans on lipids and proteins occurs and capsule is synthesized (Yoneda and Doering 2006); Cxt1 also resides in this compartment (Klutts et al., unpublished observations). From here, the classical secretory pathway transports capsule material to the cell surface (Yoneda and Doering 2006). There are several models for the subsequent incorporation of newly synthesized polysaccharides into the existing capsule, which propose that this material is incorporated either at its inner face, near the cell wall (Pierini and Doering 2001, Cordero et al. 2013), or at the outer edge of the structure (Zaragoza et al. 2006).
Although capsule polysaccharides are generally described as linear polymers, they may also contain branches, as suggested by their viscosity and shape factor (Cordero et al. 2011). Increased branching has been reported to confer increased resistance to oxidative stress, inhibit nitric oxide production by macrophages, and enhance fungal survival in serum (Cordero et al. 2011). Interestingly, Cryptococcus liquefaciens, which has a capsule that is chemically identical to that of C. neoformans although with no evidence of branching, is not as adept at resisting predation by amoeba, a potential environmental host of Cryptococci (Araújo et al. 2012). However, this difference in intracellular survival does not hold in mammalian macrophages, suggesting that the biological activity of capsular polysaccharide may be context-dependent (Araújo et al. 2017) or other factors may be involved. The details of branch formation and its role in host-pathogen interactions are still unclear, but promise to be exciting areas of study.
Mannoproteins comprise a small fraction of the capsule mass (Cherniak and Sundstrom 1994). These polypeptides appear mainly in the inner region of the capsule, close to the cell wall (Jesus et al. 2010); they may represent an integral part of the capsule or perhaps secreted proteins that are in transit through it. These predominantly cell wall proteins are discussed further below.
The cell wall - Moving inward from the capsule, the next protective barrier that surrounds C. neoformans is the cell wall (Fig. 1B). This structure is essential for cell viability, because it protects the fungus from osmotic and other environmental stresses; it also serves as a scaffold for the capsule. The cell wall is composed of a- and β-linked glucans (glucose polymers), chitin (a polymer of β-1,4-N-acetyl-glucosamine), chitosan (deacetylated chitin), glycoproteins, and, in the presence of appropriate precursors, the pigment melanin (Agustinho and Nosanchuk 2017). Because the wall contains multiple components that are not shared with mammalian hosts, it has been the focus of much research for the development of antifungal compounds.
The cryptococcal cell wall is arranged in two layers. In quick-freeze deep-etch electron micrographs the inner layer appears more striated and the outer one more particulate (Fig. 1C); they also differ in electron density on thin section electron micrographs (Fig. 1A, right). The inner layer is composed of β-glucans and chitin; mannoproteins and melanin are also most abundant in this layer although they occur throughout the cell wall (Vartivarian et al. 1989, Wang et al. 1995). The outer layer mainly contains a- and β-glucans (Reese et al. 2007).
Each of the wall components has a specific role in maintaining cell wall structure and function. β-1,3-glucan underlies the cell wall framework. It occurs as long polymers (James et al. 1990), with short β-1,6-glucan branches that crosslink the polymers to each other, as well as to chitin and glycoproteins (Reilly and Doering 2009, Gilbert et al. 2011, Gow et al. 2017). Chitin and chitosan contribute to maintaining the integrity and flexibility of the wall structure (Banks et al. 2005), while chitin and chitooligomers have been implicated in capsule architecture (Banks et al. 2005, Rodrigues et al. 2008, Fonseca et al. 2009). a-1,3- glucans are required for tethering the polysaccharide capsule to the cell (Reese and Doering 2003, Reese et al. 2007).
Unlike capsule components, cell wall polysaccharides are made at the plasma membrane and extruded through it. Once outside the cell, they associate with each other and with cell wall proteins that have exited via the secretory pathway. Some of these interactions are directed by branching or cross-linking enzymes, which together establish the complex wall structure (Gilbert et al. 2011, Free 2013, Gow et al. 2017).
β-1,3-glucan is synthesized at the plasma membrane from UDP-glucose by Fks1. The antifungal drug caspofungin (Kartsonis et al. 2003) targets Fks1 in other yeasts, but notably is not effective against C. neoformans, even though it inhibits the cryptococcal enzyme in vitro (Maligie and Selitrennikoff 2005). The synthases Skn1 and Kre6 participate in formation of β-1,6-glucan, although their specific biochemical roles are not known; deletion of the corresponding genes also perturbs capsular architecture, likely due to disorganization of the underlying cell wall (Gilbert et al. 2010). Finally, a membrane-associated alpha glucan synthase [Ags1; (Reese and Doering 2003)] forms a-1-3-glucan.
Chitin is made at the plasma membrane by a family of chitin synthases. Although no individual family member is essential for C. neoformans viability, the deletion of genes encoding chitin synthase 3 (Chs3) and a chitin synthase regulator (Csr2) drastically impairs cell wall integrity (Banks et al. 2005). Unlike in other fungi, most of the chitin in C. neoformans is deacetylated to form chitosan, a polymer that confers extra flexibility on the cell wall. Cells from strains lacking all three chitin deacetylases (Cda1, Cda2, and Cda3) have no cell wall chitosan and exhibit defects in cell integrity (Baker et al. 2007). Chitosan is also essential for virulence (Baker et al. 2011), a feature that has been successfully exploited in using a chitosan-deficient strain of C. neoformans to induce robust protective immunity in a murine model of infection (Upadhya et al. 2016).
Another component of the cell wall that has been implicated in virulence is melanin, a negatively-charged polymeric and hydrophobic pigment made from phenolic or indolic precursors (Nosanchuk and Casadevall 2006). This material is associated with the cryptococcal cell wall in a chitin-dependent manner (Baker et al. 2007, Camacho et al. 2017). Melanization enhances cryptococcal survival within natural predators, such as amoebae (Steenbergen et al. 2001) or nematodes (Mylonakis et al. 2002). Disruption of its synthesis during infection reduces cryptococcal dissemination (Noverr et al. 2004) and virulence (Salas et al. 1996), possibly due to melanin-mediated inhibition of phagocytosis and modulation of host cell cytokine responses (Huffnagle et al. 1995, Mednick et al. 2005). Melanization also increases resistance to antifungal compounds such as amphotericin B and caspofungin (Wang and Casadevall 1994a, Martinez and Casadevall 2006), and to environmental stresses, including host oxidative and nitrosative responses (Wang and Casadevall 1994b).
Many proteins are present in the cell wall, most of them heavily modified with N- and O-linked glycans (Klutts et al. 2006, Levitz and Specht 2006, Doering 2009, Reilly et al. 2011). The majority of these originate as plasma membrane localized glycosylphosphatidylinositol (GPI)-linked polypeptides, which are transferred, along with part of their anchor glycan, to covalent linkage with cell wall β-1,6-glycans (Orlean and Menon 2007, Muniz and Zurzolo 2014). Computational analysis predicts over 50 GPI-linked proteins encoded by the cryptococcal genome (de Groot et al. 2005, Loftus et al. 2005, Levitz and Specht 2006); some of these have been confirmed in studies of the C. neoformans secreted proteome (Eigenheer et al. 2007). Other proteins associate with the cell wall via non-covalent interactions, linkage to β-1,3-glucan (Yin et al. 2008, Karkowska-Kuleta and Kozik 2015, Gow et al. 2017), or disulfide bonds to polypeptides that are themselves covalently bound to structural glycans (de Nobel and Lipke 1994, Jaafar et al. 2003); these processes are less well studied.
Multiple cell wall proteins have key functions in cryp-tococcal biology. Some have been implicated in the dynamic responses of the wall to environmental conditions, such as the GPI-linked β-glucanase Gas1 that acts in β-1,3-glucan remodeling (Levitz and Specht 2006, Eigenheer et al. 2007). Another important GPI-linked protein is phospholipase B1 (Plb1), which has been implicated in C. neoformans virulence (Siafakas et al. 2007, Maruvada et al. 2012). Cell wall mannoproteins, small amounts of which can be found in the capsule (see above), are often 80-90% mannose by mass (Mansour and Levitz 2003). They are highly immunogenic (Levitz and Specht 2006, Wozniak and Levitz 2009) due to their activation of the mannose receptor on dendritic cells and consequent activation of T-cells, which leads to a protective immune response against C. neoformans (Specht et al. 2007, Dan et al. 2008a, Dan et al. 2008b). These proteins are being explored as an adjuvant component of a vaccine for cryptococcosis (Chow and Casadevall 2011, Levitz et al. 2015).
Several cryptococcal mannoproteins have been studied in depth. The first one described, MP98, was shown to be involved in T-cell activation (Levitz et al. 2001) and turns out to be the same protein as the chitin deacetylase Cda2 discussed above (Gilbert et al. 2012). This protein, despite originating as a GPI-anchored membrane protein, associates with the cell wall in a manner that is independent of its GPI structure and β-1,6-glucans; the enzymatic activity is associated with the membrane form (Gilbert et al. 2012). Other mannoproteins that have been studied include MP88, involved in T-cell activation (Huang et al. 2002), and others (MP84 and MP115) that have homology to chitin deacetylase and carboxylesterase proteins (Biondo et al. 2005). MP84 has been reported to mediate adhesion of C. neoformans yeasts to lung epithelial cells, suggesting a role early in infection (Teixeira et al. 2014). Finally, the mannoprotein Cig1 participates in iron uptake and contributes to virulence in a mouse model (Jung et al. 2006, Cadieux et al. 2013). Future studies of these and other cell wall proteins will further illuminate the synthesis and function of this complex structure, and may also advance efforts to develop vaccines or therapies.
Plasma membrane - Beneath the polysaccharide capsule and cell wall is the plasma membrane (Fig. 1), which serves as a barrier to the passage of hydrophilic molecules (van der Rest et al. 1995). Fungal membranes are composed of sterols, glycerophospholipids, and sphingolipids (Ejsing et al. 2009, Singh et al. 2017), although these differ in many respects from their mammalian counterparts. The plasma membrane also contains proteins, which maintain their association with the membrane via transmembrane domains, GPI anchors, or various lipid modifications (Santiago-Tirado and Doering 2016). Membrane structure and composition are dynamic, and vary with the fungal species analyzed and the environmental conditions (Singh and Prasad 2011, Xia et al. 2011).
Fungal membranes differ from those of mammals in containing ergosterol in place of cholesterol. This feature has been exploited by two major classes of antifungal drugs, Amphotericin B, which binds ergosterol, and the azoles, which inhibit its synthesis. C. neoformans also produces glycosylated ergosterols, termed sterylglycosides (SGs) (Weete et al. 2010, Rella et al. 2016). Although the enzyme(s) responsible for SG synthesis has not been identified (Warnecke et al. 1999), a glucosidase involved in SG degradation (termed EGCrP2 or Sgl1) is known. Deletion of the corresponding gene yields growth arrest, abnormal budding, and abnormal vacuole morphology (Watanabe et al. 2015); it also eliminates virulence in a murine model (Rella et al. 2015). Interestingly, inoculation with this mutant protected mice against subsequent lethal doses of C. neoformans H99 and Cryptococcus gattii R265 (Rella et al. 2015).
Yeast glycerophospholipids resemble those of higher eukaryotes, although their fatty acid composition may vary, while glycosphingolipids are more distinct. One of the latter that has been studied in detail in C. neoformans is glucosylceramide (GlcCer) (Nimrichter and Rodrigues 2011), which influences cryptococcal pathogenicity and is required for normal growth, resistance to alkaline conditions, spore production, and germination (Del Poeta et al. 2014). GlcCer is formed by the enzyme glucosyl-ceramide synthase 1, which transfers glucose from the nucleotide sugar UDP-glucose to a ceramide backbone (Rittershaus et al. 2006, Rella et al. 2016); disruption of the corresponding gene abrogates growth and virulence (Rittershaus et al. 2006). Notably, fungal GlcCer differs from that of other eukaryotes by the presence of a methyl group in the sphingoid base (Rodrigues et al. 2000). Eliminating this methylation alters membrane integrity and reduces virulence (Singh et al. 2012, Raj et al. 2017). Little is known about GlcCer catabolism, but a cryptococcal glucosylceramidase (EGCrP1) may act in GlcCer quality control (Ishibashi et al. 2012).
Within the plasma membrane of eukaryotes, distinct microdomains are enriched in ergosterol, GlcCer, other sphingolipids, and GPI-proteins. Such ‘lipid rafts’ are also found in C. neoformans, although they contain more saturated fatty acids (e.g. palmitic and stearic acid), fewer unsaturated fatty acids (e.g. oleic and linoleic acid), and none of the very long chain fatty acids (> 20 carbons) found in lipid rafts from mammalian cells and S. cerevisiae (Siafakas et al. 2006). Several virulence factors cluster in these domains, including the phospholipase Plb1 (Maruvada et al. 2012), the antioxidant Cu/ Zn superoxide dismutase (Siafakas et al. 2006), and the plasma membrane ATPase (Pma1) (Farnoud et al. 2014).
Final thoughts - The AIDS epidemic allowed the explosive emergence of opportunistic pathogens such as C. neoformans. The death toll caused by this fungus continues to be an enormous burden, especially in regions with limited health care resources. This impact, coupled with the challenges of drug cost, availability, toxic side effects, lengthy treatment regimens, and resistance, creates an urgent need for improved therapies.
C. neoformans is protected by concentric surface structures, each of which influences multiple aspects of pathogenesis. Unique features of these structures may offer targets for new antifungal agents, but many of them remain poorly defined. For capsule, we still do not know how GXM and GXMGal are arranged and associate with the cell, or most of the glycosyltransferases required for their synthesis. The mechanisms of capsule branching, shedding, and degradation also remain unexplored. Fungal cell wall synthesis is fairly well understood and has been successfully exploited for antifungal therapy by glucan synthase inhibitors, but this class of drugs is not effective against cryptococcal infection. Continued exploration of wall synthesis and regulation, and of cell wall proteins or strains defective in cell wall components that may act as vaccines, may help compensate for this gap in efficacy. Finally, the plasma membrane is the site of unique glycolipids whose synthesis and catabolism is yet to be fully elucidated; these and novel membrane proteins may offer targets for drug development or potential for diagnostics. Addressing these many fascinating questions, as we peel away the layers of this fascinating yeast, should lead to advances in fundamental biology and point the way to new ways to combat a formidable pathogen.
ACKNOWLEDGEMENTS
To our colleagues in the cryptococcal community for stimulating interactions, exemplified by the excellent 2017 conference in Foz do Iguaçu, Brazil. We thank Felipe Santiago-Tirado, Lynda Pierini, John Heuser, and Robyn Roth for micrographs shown in Fig. 1.
Footnotes
Financial support: NIH (grants AI049173, GM071007, AI109623).
REFERENCES
- Agustinho DP, Nosanchuk JD. Functions of fungal melanins. In: Reference module in life sciences. 2017. Available from: http://www.sciencedirect.com/science/article/pii/B9780128096338120916.
- Alspaugh JA. Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet Biol. 2015;78:55–58. doi: 10.1016/j.fgb.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo GRS, Freitas GJC, Fonseca FL, Leite PEC, Rocha GM, de Souza W, et al. The environmental yeast Cryptococcus liquefaciens produces capsular and secreted polysaccharides with similar pathogenic properties to those of C. neoformans . Sci Rep. 2017;4:67–68. doi: 10.1038/srep46768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo GS, Fonseca FL, Pontes B, Torres A, Cordero RJB, Zancop-Oliveira RM, et al. Capsules from pathogenic and non-pathogenic Cryptococcus spp. manifest significant differences in structure and ability to protect against phagocytic cells. PLoS ONE. 2012;7(1):e29561. doi: 10.1371/journal.pone.0029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, Jung KW. Stress signaling pathways for the pathogenicity of Cryptococcus . Eukaryot Cell. 2013;12(12):1564–1577. doi: 10.1128/EC.00218-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LG, Specht CA, Donlin MJ, Lodge JK. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans . Eukaryot Cell. 2007;6(5):855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LG, Specht CA, Lodge JK. Cell wall chitosan is necessary for virulence in the opportunistic pathogen Cryptococcus neoformans . Eukaryot Cell. 2011;10(9):1264–1268. doi: 10.1128/EC.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou ER, Johnston SA. The cause and effect of Cryptococcus interactions with the host. Curr Opin Microbiol. 2017;40:88–94. doi: 10.1016/j.mib.2017.10.012. [DOI] [PubMed] [Google Scholar]
- Banks IR, Specht CA, Donlin MJ, Gerik KJ, Levitz SM, Lodge JK. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans . Eukaryot Cell. 2005;4(11):1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondo C, Messina L, Bombaci M, Mancuso G, Midiri A, Beninati C, et al. Characterization of two novel cryptococcal mannoproteins recognized by immune sera. Infect Immun. 2005;73(11):7348–7355. doi: 10.1128/IAI.73.11.7348-7355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose I, Reese AJ, Ory JJ, Janbon G, Doering TL. A yeast under cover: the capsule of Cryptococcus neoformans . Eukaryot Cell. 2003;2(4):655–663. doi: 10.1128/EC.2.4.655-663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol. 2015;13(10):620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieux B, Lian T, Hu G, Wang J, Biondo C, Teti G, et al. The mannoprotein Cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans . J Infect Dis. 2013;207(8):1339–1347. doi: 10.1093/infdis/jit029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffaro CE, Hirschberg CB. Nucleotide sugar transporters of the Golgi apparatus: from basic science to diseases. Acc Chem Res. 2006;39(11):805–812. doi: 10.1021/ar0400239. [DOI] [PubMed] [Google Scholar]
- Camacho E, Chrissian C, Cordero RJB, Liporagi-Lopes L, Stark RE, Casadevall A. N-acetylglucosamine affects Cryptococcus neoformans cell-wall composition and melanin architecture. Microbiology. 2017;163(11):1540–1556. doi: 10.1099/mic.0.000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active enZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Chang TY, Liu JW, Chen FJ, Chien CC, Lee CH, et al. Increasing trend of fluconazole-non-susceptible Cryptococcus neoformans in patients with invasive cryptococcosis: a 12-year longitudinal study. BMC Infect Dis. 2015;15:277–277. doi: 10.1186/s12879-015-1023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherniak R, Jones RG, Reiss E. Structure determination of Cryptococcus neoformans serotype A-variant glucuronoxylomannan by 13C-n.m.r. spectroscopy. Carbohydr Res. 1988;172(1):113–138. doi: 10.1016/s0008-6215(00)90846-2. [DOI] [PubMed] [Google Scholar]
- Cherniak R, Sundstrom JB. Polysaccharide antigens of the capsule of Cryptococcus neoformans . Infect Immun. 1994;62(5):1507–1512. doi: 10.1128/iai.62.5.1507-1512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherniak R, Valafar H, Morris LC, Valafar F. Cryptococcus neoformans chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronoxylomannans with a computer-simulated artificial neural network. Clin Diagn Lab Immunol. 1998;5(2):146–159. doi: 10.1128/cdli.5.2.146-159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow SK, Casadevall A. Evaluation of Cryptococcus neoformans galactoxylomannan-protein conjugate as vaccine candidate against murine cryptococcosis. Vaccine. 2011;29(10):1891–1898. doi: 10.1016/j.vaccine.2010.12.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero RJ, Bergman A, Casadevall A. Temporal behavior of capsule enlargement by Cryptococcus neoformans . Eukaryot Cell. 2013;12(10):1383–1388. doi: 10.1128/EC.00163-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero RJ, Frases S, Guimaraes AJ, Rivera J, Casadevall A. Evidence for branching in cryptococcal capsular polysaccharides and consequences on its biological activity. Mol Microbiol. 2011;79(4):1101–1117. doi: 10.1111/j.1365-2958.2010.07511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell TR, Griffith CL, Liu H, Nenninger AA, Doering TL. The pathogenic fungus Cryptococcus neoformans expresses two functional GDP-mannose transporters with distinct expression patterns and roles in capsule synthesis. Eukaryot Cell. 2007;6(5):776–785. doi: 10.1128/EC.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan JM, Kelly RM, Lee CK, Levitz SM. Role of the mannose receptor in a murine model of Cryptococcus neoformans infection. Infect Immun. 2008a;76(6):2362–2367. doi: 10.1128/IAI.00095-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan JM, Wang JP, Lee CK, Levitz SM. Cooperative stimulation of dendritic cells by Cryptococcus neoformans mannoproteins and CpG oligodeoxynucleotides. PLoS One. 2008b;3(4):e2046. doi: 10.1371/journal.pone.0002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JN, Chau TT, Lalloo DG. Combination antifungal therapy for cryptococcal meningitis. N Engl J Med. 2013;368(26):2522–2523. doi: 10.1056/NEJMc1305981. [DOI] [PubMed] [Google Scholar]
- de Groot PW, Ram AF, Klis FM. Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet Biol. 2005;42(8):657–675. doi: 10.1016/j.fgb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- de Nobel H, Lipke PN. Is there a role for GPIs in yeast cell-wall assembly? Trends Cell Biol. 1994;4(2):42–45. doi: 10.1016/0962-8924(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Del Poeta M, Nimrichter L, Rodrigues ML, Luberto C. Synthesis and biological properties of fungal glucosylceramide. PLoS Pathog. 2014;10(1):e1003832. doi: 10.1371/journal.ppat.1003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering TL. How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans . Annu Rev Microbiol. 2009;63:223–247. doi: 10.1146/annurev.micro.62.081307.162753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenheer RA, Jin Lee Y, Blumwald E, Phinney BS, Gelli A. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans . FEMS Yeast Res. 2007;7(4):499–510. doi: 10.1111/j.1567-1364.2006.00198.x. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA. 2009;106(7):2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnoud AM, Mor V, Singh A, del Poeta M. Inositol phosphosphingolipid phospholipase C1 regulates plasma membrane ATPase (Pma1) stability in Cryptococcus neoformans . FEBS Lett. 2014;588(21):3932–3938. doi: 10.1016/j.febslet.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca FL, Nimrichter L, Cordero RJ, Frases S, Rodrigues J, Goldman DL, et al. Role for chitin and chitooligomers in the capsular architecture of Cryptococcus neoformans . Eukaryot Cell. 2009;8(10):1543–1553. doi: 10.1128/EC.00142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free SJ. Fungal cell wall organization and biosynthesis. Adv Genet. 2013;81:33–82. doi: 10.1016/B978-0-12-407677-8.00002-6. [DOI] [PubMed] [Google Scholar]
- Gates-Hollingsworth MA, Kozel TR. Phenotypic heterogeneity in expression of epitopes in the Cryptococcus neoformans capsule. Mol Microbiol. 2009;74(1):126–138. doi: 10.1111/j.1365-2958.2009.06855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert NM, Baker LG, Specht CA, Lodge JK. A glycosylphosphatidylinositol anchor is required for membrane localization but dispensable for cell wall association of chitin deacetylase 2 in Cryptococcus neoformans . MBio. 2012;3(1):e00007–e00012. doi: 10.1128/mBio.00007-12. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert NM, Donlin MJ, Gerik KJ, Specht CA, Djordjevic JT, Wilson CF, et al. KRE genes are required for beta-1,6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans . Mol Microbiol. 2010;76(2):517–534. doi: 10.1111/j.1365-2958.2010.07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert NM, Lodge JK, Specht CA. The cell wall of Cryptococcus. In: Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadeval A, editors. Cryptococcus: from human pathogen to model yeast. Washington (DC): American Society of Microbiology; 2011. pp. 67–79. [Google Scholar]
- Giles SS, Dagenais TR, Botts MR, Keller NP, Hull CM. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans . Infect Immun. 2009;77(8):3491–3500. doi: 10.1128/IAI.00334-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NAR, Latge JP, Munro CA. The fungal cell wall: structure, biosynthesis, and function. Microbiol Spectr. 2017;5(3) doi: 10.1128/microbiolspec.FUNK-0035-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadley B, Maggioni A, Ashikov A, Day CJ, Haselhorst T, Tiralongo J. Structure and function of nucleotide sugar transporters: current progress. Comput Struct Biotechnol J. 2014;10(16):23–32. doi: 10.1016/j.csbj.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BC, Skowyra ML, Spencer SJ, Gish SR, Williams M, Held EP, et al. Toward an integrated model of capsule regulation in Cryptococcus neoformans . PLoS Pathog. 2011;7(12):e1002411. doi: 10.1371/journal.ppat.1002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss C, Klutts JS, Wang Z, Doering TL, Azadi P. The structure of Cryptococcus neoformans galactoxylomannan contains beta-D-glucuronic acid. Carbohydr Res. 2009;344(7):915–920. doi: 10.1016/j.carres.2009.03.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss C, Skowyra ML, Liu H, Klutts JS, Wang Z, Williams M, et al. Unusual galactofuranose modification of a capsule polysaccharide in the pathogenic yeast Cryptococcus neoformans . J Biol Chem. 2013;288(16):10994–11003. doi: 10.1074/jbc.M112.441998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Nong SH, Mansour MK, Specht CA, Levitz SM. Purification and characterization of a second immunoreactive mannoprotein from Cryptococcus neoformans that stimulates T-Cell responses. Infect Immun. 2002;70(10):5485–5493. doi: 10.1128/IAI.70.10.5485-5493.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle GB, Chen GH, Curtis JL, McDonald RA, Strieter RM, Toews GB. Down-regulation of the afferent phase of T cell-mediated pulmonary inflammation and immunity by a high melanin-producing strain of Cryptococcus neoformans . J Immunol. 1995;155(7):3507–3516. [PubMed] [Google Scholar]
- Ishibashi Y, Ikeda K, Sakaguchi K, Okino N, Taguchi R, Ito M. Quality control of fungus-specific glucosylceramide in Cryptococcus neoformans by endoglycoceramidase-related protein 1 (EGCrP1) J Biol Chem. 2012;287(1):368–381. doi: 10.1074/jbc.M111.311340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafar L, Moukadiri I, Zueco J. Characterization of a disulphidebound Pir-cell wall protein (Pir-CWP) of Yarrowia lipolytica . Yeast. 2003;20(5):417–426. doi: 10.1002/yea.973. [DOI] [PubMed] [Google Scholar]
- James PG, Cherniak R, Jones RG, Stortz CA, Reiss E. Cell-wall glucans of Cryptococcus neoformans Cap 67. Carbohydr Res. 1990;198(1):23–38. doi: 10.1016/0008-6215(90)84273-w. [DOI] [PubMed] [Google Scholar]
- Jarvis JN, Bicanic T, Loyse A, Namarika D, Jackson A, Nussbaum JC, et al. Determinants of mortality in a combined cohort of 501 patients with HIV-associated Cryptococcal meningitis: implications for improving outcomes. Clin Infect Dis. 2014;58(5):736–745. doi: 10.1093/cid/cit794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesus MD, Nicola AM, Chow SK, Lee IR, Nong S, Specht CA, et al. Glucuronoxylomannan, galactoxylomannan, and mannoprotein occupy spatially separate and discrete regions in the capsule of Cryptococcus neoformans . Virulence. 2010;1(6):500–508. doi: 10.4161/viru.1.6.13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffe LS, Nimrichter L, Rodrigues ML, Del Poeta M. Potential roles of fungal extracellular vesicles during infection. mSphere. 2016;1(4):pii–pii.:e00099-16. doi: 10.1128/mSphere.00099-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, White R, Kronstad JW. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans . PLoS Biol. 2006;4(12):e410–e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkowska-Kuleta J, Kozik A. Cell wall proteome of pathogenic fungi. Acta Biochim Pol. 2015;62(3):339–351. doi: 10.18388/abp.2015_1032. [DOI] [PubMed] [Google Scholar]
- Kartsonis NA, Nielsen J, Douglas CM. Caspofungin: the first in a new class of antifungal agents. Drug Resist Updat. 2003;6(4):197–218. doi: 10.1016/s1368-7646(03)00064-5. [DOI] [PubMed] [Google Scholar]
- Klutts JS, Doering TL. Cryptococcal xylosyltransferase 1 (Cxt1p) from Cryptococcus neoformans plays a direct role in the synthesis of capsule polysaccharides. J Biol Chem. 2008;283(21):14327–34. doi: 10.1074/jbc.M708927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klutts JS, Levery SB, Doering TL. A beta-1,2-xylosyltransferase from Cryptococcus neoformans defines a new family of glycosyltransferases. J Biol Chem. 2007;282(24):17890–9. doi: 10.1074/jbc.M701941200. [DOI] [PubMed] [Google Scholar]
- Klutts JS, Yoneda A, Reilly MC, Bose I, Doering TL. Glycosyltransferases and their products: cryptococcal variations on fungal themes. FEMS Yeast Res. 2006;6(4):499–512. doi: 10.1111/j.1567-1364.2006.00054.x. [DOI] [PubMed] [Google Scholar]
- Kumar P, Yang M, Haynes BC, Skowyra ML, Doering TL. Emerging themes in cryptococcal capsule synthesis. Curr Opin Struct Biol. 2011;21(5):597–602. doi: 10.1016/j.sbi.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Fraser JA, Doering TL, Wang Z, Janbon G, Idnurm A, et al. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med. 2014;4(7):a019760. doi: 10.1101/cshperspect.a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz SM, Huang H, Ostroff GR, Specht CA. Exploiting fungal cell wall components in vaccines. Semin Immunopathol. 2015;37(2):199–207. doi: 10.1007/s00281-014-0460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz SM, Nong S, Mansour MK, Huang C, Specht CA. Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T cell responses to Cryptococcus neoformans . Proc Natl Acad Sci USA. 2001;98(18):10422–17. doi: 10.1073/pnas.181331398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz SM, Specht CA. The molecular basis for the immunogenicity of Cryptococcus neoformans mannoproteins. FEMS Yeast Res. 2006;6(4):513–524. doi: 10.1111/j.1567-1364.2006.00071.x. [DOI] [PubMed] [Google Scholar]
- Li LX, Ashikov A, Liu H, Griffith CL, Bakker H, Doering TL. Cryptococcus neoformans UGT1 encodes a UDP-Galactose/UDPGalNAc transporter. Glycobiology. 2017;27(1):87–98. doi: 10.1093/glycob/cww078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LX, Rautengarten C, Heazlewood JL, Doering TL. Xylose donor transport is critical for fungal virulence. PLoS Pathog. 2018a;14(1):e1006765. doi: 10.1371/journal.ppat.1006765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LX, Rautengarten C, Heazlewood JL, Doering TL. UDP-Glucuronic acid transport is required for virulence of Cryptococcus neoformans . MBio. 2018b;9(1):e02319-17. doi: 10.1128/mBio.02319-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans . Science. 2005;307(5713):1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier EJ, Haynes BC, Gish SR, Wang ZA, Skowyra ML, Marulli AL, et al. Model-driven mapping of transcriptional networks reveals the circuitry and dynamics of virulence regulation. Genome Res. 2015;25(5):690–700. doi: 10.1101/gr.184101.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maligie MA, Selitrennikoff CP. Cryptococcus neoformans resistance to echinocandins: (1,3)beta-glucan synthase activity is sensitive to echinocandins. Antimicrob Agents Chemother. 2005;49(7):2851–2856. doi: 10.1128/AAC.49.7.2851-2856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MK, Levitz SM. Fungal mannoproteins: the sweet path to immunodominance-mannose residues on glycoproteins trigger key host immune response that, if better harnessed, could protect against damage and disease. ASM News. 2003;69(12):595–600. [Google Scholar]
- Martínez LR, Casadevall A. Susceptibility of Cryptococcus neoformans biofilms to antifungal agents in vitro . Antimicrob Agents Chemother. 2006;50(3):1021–1033. doi: 10.1128/AAC.50.3.1021-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruvada R, Zhu L, Pearce D, Zheng Y, Perfect J, Kwon-Chung KJ, et al. Cryptococcus neoformans phospholipase B1 activates host cell Rac1 for traversal across the blood-brain barrier. Cell Microbiol. 2012;14(10):1544–1553. doi: 10.1111/j.1462-5822.2012.01819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mednick AJ, Nosanchuk JD, Casadevall A. Melanization of Cryptococcus neoformans affects lung inflammatory responses during cryptococcal infection. Infect Immun. 2005;73(4):2012–2019. doi: 10.1128/IAI.73.4.2012-2019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyrand F, Fontaine T, Janbon G. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol Microbiol. 2007;64(3):771–781. doi: 10.1111/j.1365-2958.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- Muniz M, Zurzolo C. Sorting of GPI-anchored proteins from yeast to mammals - common pathways at different sites? J Cell Sci. 2014;127(Pt 13):2793–2801. doi: 10.1242/jcs.148056. [DOI] [PubMed] [Google Scholar]
- Mylonakis E, Ausubel FM, Perfect JR, Heitman J, Calderwood SB. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc Natl Acad Sci USA. 2002;99(24):15675–80. doi: 10.1073/pnas.232568599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimrichter L, Rodrigues ML. Fungal glucosylceramides: from structural components to biologically active targets of new antimicrobials. Front Microbiol. 2011;2:212–212. doi: 10.3389/fmicb.2011.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosanchuk JD, Casadevall A. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob Agents Chemother. 2006;50(11):3519–3528. doi: 10.1128/AAC.00545-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr MC, Williamson PR, Fajardo RS, Huffnagle GB. CNLAC1 is required for extrapulmonary dissemination of Cryptococcus neoformans but not pulmonary persistence. Infect Immun. 2004;72(3):1693–1699. doi: 10.1128/IAI.72.3.1693-1699.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Alspaugh JA. The Cryptococcus neoformans capsule: a sword and a shield. Clin Microbiol Rev. 2012;25(3):387–408. doi: 10.1128/CMR.00001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P, Menon AK. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res. 2007;48(5):993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ, et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clin Infect Dis. 2010;50(3):291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierini LM, Doering TL. Spatial and temporal sequence of capsule construction in Cryptococcus neoformans . Mol Microbiol. 2001;41(1):105–115. doi: 10.1046/j.1365-2958.2001.02504.x. [DOI] [PubMed] [Google Scholar]
- Previato JO, Vinogradov E, Maes E, Fonseca LM, Guerardel Y, Oliveira PAV, et al. Distribution of the O-acetyl groups and beta-galactofuranose units in galactoxylomannans of the opportunistic fungus Cryptococcus neoformans . Glycobiology. 2017;27(6):582–592. doi: 10.1093/glycob/cww127. [DOI] [PubMed] [Google Scholar]
- Raj S, Nazemidashtarjandi S, Kim J, Joffe L, Zhang X, Singh A, et al. Changes in glucosylceramide structure affect virulence and membrane biophysical properties of Cryptococcus neoformans . Biochim Biophys Acta. 2017;1859(11):2224–2233. doi: 10.1016/j.bbamem.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 2017;17(8):873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese AJ, Doering TL. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol Microbiol. 2003;50(4):1401–1409. doi: 10.1046/j.1365-2958.2003.03780.x. [DOI] [PubMed] [Google Scholar]
- Reese AJ, Yoneda A, Breger JA, Beauvais A, Liu H, Griffith CL, et al. Loss of cell wall alpha(1-3) glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol Microbiol. 2007;63(5):1385–1398. doi: 10.1111/j.1365-2958.2006.05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MC, Aoki K, Wang ZA, Skowyra ML, Williams M, Tiemeyer M, et al. A xylosylphosphotransferase of Cryptococcus neoformans acts in protein O-glycan synthesis. J Biol Chem. 2011;286(30):26888–99. doi: 10.1074/jbc.M111.262162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MC, Doering TL. Biosynthesis of fungal and yeast glycans. In: Moran AP, editor. Microbial Glycobiology: structures, relevance and applications. London: Academic; 2009. [Google Scholar]
- Reilly MC, Levery SB, Castle SA, Klutts JS, Doering TL. A novel xylosylphosphotransferase activity discovered in Cryptococcus neoformans . J Biol Chem. 2009;284(52):36118–27. doi: 10.1074/jbc.M109.056226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rella A, Farnoud AM, Del Poeta M. Plasma membrane lipids and their role in fungal virulence. Prog Lipid Res. 2016;61:63–72. doi: 10.1016/j.plipres.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rella A, Mor V, Farnoud AM, Singh A, Shamseddine AA, Ivanova E, et al. Role of Sterylglucosidase 1 (Sgl1) on the pathogenicity of Cryptococcus neoformans: potential applications for vaccine development. Front Microbiol. 2015;6:836–836. doi: 10.3389/fmicb.2015.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittershaus PC, Kechichian TB, Allegood JC, Merrill AH, Jr, Hennig M, Luberto C, et al. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans . J Clin Invest. 2006;116(6):1651–1659. doi: 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Alvarez M, Fonseca FL, Casadevall A. Binding of the wheat germ lectin to Cryptococcus neoformans suggests an association of chitinlike structures with yeast budding and capsular glucuronoxylomannan. Eukaryot Cell. 2008;7(4):602–609. doi: 10.1128/EC.00307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nakayasu ES, Almeida IC, Nimrichter L. The impact of proteomics on the understanding of functions and biogenesis of fungal extracellular vesicles. J Proteomics. 2014;97:177–186. doi: 10.1016/j.jprot.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Travassos LR, Miranda KR, Franzen AJ, Rozental S, de Souza W, et al. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth. Infect Immun. 2000;68(12):7049–7060. doi: 10.1128/iai.68.12.7049-7060.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas SD, Bennett JE, Kwon-Chung KJ, Perfect JR, Williamson PR. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans . J Exp Med. 1996;184(2):377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Tirado FH, Doering TL. All about that fat: lipid modification of proteins in Cryptococcus neoformans . J Microbiol. 2016;54(3):212–222. doi: 10.1007/s12275-016-5626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Tirado FH, Onken MD, Cooper JA, Klein RS, Doering TL. Trojan horse transit contributes to blood-brain barrier crossing of a eukaryotic pathogen. MBio. 2017;8(1):e02183-16. doi: 10.1128/mBio.02183-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siafakas AR, Sorrell TC, Wright LC, Wilson C, Larsen M, Boadle R, et al. Cell wall-linked cryptococcal phospholipase B1 is a source of secreted enzyme and a determinant of cell wall integrity. J Biol Chem. 2007;282(52):37508–14. doi: 10.1074/jbc.M707913200. [DOI] [PubMed] [Google Scholar]
- Siafakas AR, Wright LC, Sorrell TC, Djordjevic JT. Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase. Eukaryotic cell. 2006;5(3):488–498. doi: 10.1128/EC.5.3.488-498.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, MacKenzie A, Girnun G, Del Poeta M. Analysis of sphingolipids, sterols, and phospholipids in human pathogenic Cryptococcus strains. J Lipid Res. 2017;58(10):2017–2036. doi: 10.1194/jlr.M078600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Prasad R. Comparative lipidomics of azole sensitive and resistant clinical isolates of Candida albicans reveals unexpected diversity in molecular lipid imprints. PLoS One. 2011;6(4):e19266. doi: 10.1371/journal.pone.0019266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Wang H, Silva LC, Na C, Prieto M, Futerman AH, et al. Methylation of glycosylated sphingolipid modulates membrane lipid topography and pathogenicity of Cryptococcus neoformans . Cell Microbiol. 2012;14(4):500–516. doi: 10.1111/j.1462-5822.2011.01735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionov E, Chang YC, Kwon-Chung KJ. Azole heteroresistance in Cryptococcus neoformans: emergence of resistant clones with chromosomal disomy in the mouse brain during fluconazole treatment. Antimicrob Agents Chemother. 2013;57(10):5127–5130. doi: 10.1128/AAC.00694-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Achan B, Hullsiek KH, McDonald TR, Okagaki LH, Alhadab AA, et al. Increased antifungal drug resistance in clinical isolates of Cryptococcus neoformans in Uganda. Antimicrob Agents Chemother. 2015;59(12):7197–7204. doi: 10.1128/AAC.01299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer U, Liu H, Doering TL. An alpha-1,3-mannosyltransferase of Cryptococcus neoformans . J Biol Chem. 2003;278(48):47724–30. doi: 10.1074/jbc.M307223200. [DOI] [PubMed] [Google Scholar]
- Specht CA, Nong S, Dan JM, Lee CK, Levitz SM. Contribution of glycosylation to T cell responses stimulated by recombinant Cryptococcus neoformans mannoprotein. J Infect Dis. 2007;196(5):796–800. doi: 10.1086/520536. [DOI] [PubMed] [Google Scholar]
- Srikanta D, Santiago-Tirado FH, Doering TL. Cryptococcus neoformans: historical curiosity to modern pathogen. Yeast. 2014;31(2):47–60. doi: 10.1002/yea.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen JN, Shuman HA, Casadevall A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci USA. 2001;98(26):15245–50. doi: 10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira PA, Penha LL, Mendonca-Previato L, Previato JO. Manno-protein MP84 mediates the adhesion of Cryptococcus neoformans to epithelial lung cells. Front Cell Infect Microbiol. 2014;4:106–106. doi: 10.3389/fcimb.2014.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya R, Lam WC, Maybruck B, Specht CA, Levitz SM, Lodge JK. Induction of protective immunity to cryptococcal infection in mice by a heat-killed, chitosan-deficient strain of Cryptococcus neoformans . MBio. 2016;7(3):pii–pii.:e00547-16. doi: 10.1128/mBio.00547-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Rest ME, Kamminga AH, Nakano A, Anraku Y, Poolman B, Konings WN. The plasma membrane of Saccharomyces cerevisiae: structure, function, and biogenesis. Microbiol Rev. 1995;59(2):304–322. doi: 10.1128/mr.59.2.304-322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartivarian SE, Reyes GH, Jacobson ES, James PG, Cherniak R, Mumaw VR, et al. Localization of mannoprotein in Cryptococcus neoformans . J Bacteriol. 1989;171(12):6850–6852. doi: 10.1128/jb.171.12.6850-6852.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Aisen P, Casadevall A. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun. 1995;63(8):3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Casadevall A. Growth of Cryptococcus neoformans in presence of L-dopa decreases its susceptibility to amphotericin B. Antimicrob Agents Chemother. 1994a;38(11):2648–2650. doi: 10.1128/aac.38.11.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Casadevall A. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect Immun. 1994b;62(7):3004–3007. doi: 10.1128/iai.62.7.3004-3007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZA, Griffith CL, Skowyra ML, Salinas N, Williams M, Maier EJ, et al. Cryptococcus neoformans dual GDP-mannose transporters and their role in biology and virulence. Eukaryot Cell. 2014;13(6):832–842. doi: 10.1128/EC.00054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke D, Erdmann R, Fahl A, Hube B, Muller F, Zank T, et al. Cloning and functional expression of UGT genes encoding sterol glucosyltransferases from Saccharomyces cerevisiae, Candida albicans, Pichia pastoris, and Dictyostelium discoideum . J Biol Chem. 1999;274(19):13048–59. doi: 10.1074/jbc.274.19.13048. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Ito T, Goda HM, Ishibashi Y, Miyamoto T, Ikeda K, et al. Sterylglucoside catabolism in Cryptococcus neoformans with endoglycoceramidase-related protein 2 (EGCrP2), the first steryl-beta-glucosidase identified in fungi. J Biol Chem. 2015;290(2):1005–1019. doi: 10.1074/jbc.M114.616300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weete JD, Abril M, Blackwell M. Phylogenetic distribution of fungal sterols. PLoS One. 2010;5(5):e10899. doi: 10.1371/journal.pone.0010899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak KL, Levitz SM. Isolation and purification of antigenic components of Cryptococcus . Methods Mol Biol. 2009;470:71–83. doi: 10.1007/978-1-59745-204-5_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Jones AD, Lau MW, Yuan YJ, Dale BE, Balan V. Comparative lipidomic profiling of xylose-metabolizing S. cerevisiae and its parental strain in different media reveals correlations between membrane lipids and fermentation capacity. Biotechnol Bioeng. 2011;108(1):12–21. doi: 10.1002/bit.22910. [DOI] [PubMed] [Google Scholar]
- Yin QY, de Groot PW, de Koster CG, Klis FM. Mass spectrometry-based proteomics of fungal wall glycoproteins. Trends Microbiol. 2008;16(1):20–26. doi: 10.1016/j.tim.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Yoneda A, Doering TL. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol Biol Cell. 2006;17(12):5131–5140. doi: 10.1091/mbc.E06-08-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Telzak A, Bryan RA, Dadachova E, Casadevall A. The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Mol Microbiol. 2006;59(1):67–83. doi: 10.1111/j.1365-2958.2005.04928.x. [DOI] [PubMed] [Google Scholar]