Abstract
Whereas microglia involvement in virtually all brain diseases is well accepted their role in the control of homeostasis in the central nervous system (CNS) is mainly thought to be the maintenance of neuronal function through the formation, refinement, and monitoring of synapses in both the developing and adult brain. Although the prenatal origin as well as the neuron-centered function of cortical microglia has recently been elucidated, much less is known about a distinct amoeboid microglia population formerly described as the “fountain of microglia” that appears only postnatally in myelinated regions such as corpus callosum and cerebellum. Using large-scale transcriptional profiling, fate mapping, and genetic targeting approaches, we identified a unique molecular signature of this microglia subset that arose from a CNS endogenous microglia pool independent from circulating myeloid cells. Microglia depletion experiments revealed an essential role of postnatal microglia for the proper development and homeostasis of oligodendrocytes and their progenitors. Our data provide new cellular and molecular insights into the myelin-supporting function of microglia in the normal CNS.
Keywords: Amoeboid microglia, Myelinogenesis, Oligodendrocytes, Oligodendrocyte progenitors, Development
Introduction
In addition to non-parenchymal macrophages at tissue interfaces, microglia are the elementary endogenous tissue macrophages in the central nervous system (CNS) [51]. As such, they are critically involved in the scavenging of dying cells, pathogens, and molecules through phagocytosis and endocytosis as well as the usage of pathogen-associated molecular pattern (PAMPs) receptors [6, 56]. Moreover, microglia represent a highly plastic type of myeloid cells with highly divergent functional and genetic features depending on the environmental stimulus defined by the CNS neighboring cells [1, 15, 63] and their developmental or aging status [16, 28, 32].
It has been appreciated already for a long time that microglia have a protective role in the CNS by providing host defense that is common to all myelomonocytic cells in the body [33]. Only recently, it became clear that microglia are instrumental for CNS homeostasis as well. It has been found that they are involved in physiological synaptic pruning and remodeling during development and adulthood [45, 59, 62, 69, 72]. Furthermore, cortical microglia serve important physiological functions in learning and memory by producing brain-derived neurotrophic factor and other mediators that affect synaptic plasticity [46]. Deep sequencing of adult murine microglia has characterized the microglial “sensome”, a unique cluster of proteins that enable microglia to carry out homeostatic functions, including sensing of cytokines, chemokines, purines, inorganic substances, changes in pH, and amino acids [21].
Given the importance of microglia in the physiological brain function, it is not surprising that an increasing number of microglia-related genes have now been identified as disease triggering mechanisms. For example, mutations in the colony stimulating factor-1 receptor (CSF-1R) in patients with hereditary diffuse leukoencephalopathy with spheroids (HDLS, [55]), in the triggering receptor expressed on myeloid cell (TREM) 2 inducing Nasu–Hakola disease [44] or in the ubiquitin specific peptidase (USP) 18 causing a destructive type I interferon-triggered encephalopathy in mouse [15] or man [34]. In addition to these rare autosomal linked neuroinflammatory diseases, several microglia-associated gene polymorphisms have been described in large genome-wide association studies, such as CD33 and Trem2, in Alzheimer’s disease [12, 17, 18, 22, 41], TNFRSF1A and IRF8 in multiple sclerosis [7] and others.
Microglia function in relation to the myelinating cells of the CNS, the oligodendrocytes, was so far linked to pathophysiology. Classically activated microglia were thought to exert cytotoxic effects on oligodendrocytes in vitro, whereas alternatively activated microglia exhibit phagocytotic capacity [29]. This disease-associated phagocytic activity of microglia was found to support CNS remyelination by driving subsequent oligodendrocyte differentiation [37].
The role of microglia for physiological myelination during CNS development or for myelin homeostasis in the adult brain is currently unclear. Here, we identified highly activated amoeboid microglia subpopulations occurring in the early postnatal stages in myelinating regions of the murine brain. These phagocytic microglia subsets shared their origin from the early prenatal precursors and their independence from circulating monocytes with cortical microglia that already exhibited features of resting sessile microglia. Moreover, we identified a crucial role of early postnatal microglia for oligodendrocyte progenitor maintenance and maturation and subsequent physiological myelinogenesis. Our data additionally suggest that microglia in the adult healthy brain are essential for oligodendrocyte progenitor cell homeostasis.
Materials and methods
Mice
Cx3cr1GFP/WT, Cx3cr1CreER, C57BL/6N, Ccr2−/−, Nr4a1−/− (kindly provided by Antigoni Triantafyllopoulou, University Hospital Freiburg, Germany), NG2YFP/WT ([27] kindly provided by Hauke Werner and Sandra Goebbels, MPI of Experimental Medicine, Göttingen, Germany), Cd11cCreER (originally provided by Hans Christian Probst, University Mainz, Germany), Csf1r−/− FVB/NJ [42], Sox10-iCreERT2 × CAG-eGFP (Tg(RP 23-28O11-iCreERT2)LDxTg(CAG-eGFP) [67] and R26-tomato/R26-yfp mice were bred in-house under specific pathogen-free conditions. Ccr2RFP/WT mice were provided by Mathias Jucker (German Center for Neuro-degenerative Diseases, Tübingen, Germany). All animal experiments were revised and approved by the Society of Neuroscience and by local administration and were performed in accordance to the guidelines on the use of animals and humans in Neuroscience Research as well as the respective national, federal, and institutional regulations.
Tamoxifen induction
Tamoxifen (TAM, Sigma-Aldrich, Taufkirchen, Germany) was dissolved in corn oil (Sigma-Aldrich, Taufkirchen, Germany) to get a 20 mg/ml solution for injections. Cx3cr1CreER:Rosa26-yfp mice were intraperitoneally injected (i.p.) with 0.1 mg TAM on postnatal day (P) 1; Cd11cCreER:Rosa26-tomato mice were injected i.p. with 0.1 mg TAM on P1–P7. For adult induction of Sox10-iCreERT2 × CAG-eGFP mice, a 40 mg/ml TAM diluted in ethanol (EtOH final concentration 10%), and corn oil was prepared. 8–10-week-old animals received for three times (every second day) 10 mg TAM (total of 30 mg) by gavage.
Microglia depletion
The CSF-1R inhibitor BLZ945 (kindly provided by Novartis, Basel, Switzerland) was dissolved in 20% (2-hydroxypropyl)-β-cyclodextrin (Sigma-Aldrich, Taufkirchen, Germany). A dose of 200 mg/kg bodyweight was used. In adult (8–10 weeks old) mice, BLZ945 was applied by oral gavage for 7 consecutive days. For experiments with Sox10-iCreERT2 × CAG-GFP mice, mice received BLZ945 orally for 5 consecutive days 1 week after the last TAM application. Adult mice were killed and analyzed 1 day after the last application. Newborn mice were injected (i.p.) with BLZ945 on day P2, 4, 6, and 7 using a Hamilton syringe. Mice were analyzed at P8, P20, or P40–P42.
Proliferation rate
To label cell proliferation during the early postnatal development EdU (5-ethynyl-2′-deoxyuridin, 10 μg/g bodyweight, Thermo Fisher Scientific, Schwerte, Germany) was injected i.p. either at P0–P3 or at P4–P6. Mice were analyzed at P8, P10, and P21 by histology. Staining was performed according to manufacturer’s protocol.
Preparation of tissue samples
Mice were lethally anesthetized with ketamine (100 mg/kg bodyweight, Medistar, Germany) and xylazine (5 mg/kg body weight, Bayer, Leverkusen, Germany) followed by the perfusion with 1× PBS through the left heart chamber. For histology, the brain was transferred to 4% PFA. For immunofluorescence in Sox10-iCreERT2 mice, animals were perfused with 4% paraformaldehyde (PFA). For preparation of single cell suspension for flow cytometry, the tissue was removed, brain regions (corpus callosum, cortex, and cerebellum) were separated under the binocular, homogenized, and filtered through a 70 μm mesh. After centrifugation (10 min 1200 rpm, 4 °C), the pellet from P3–P10 old mice was directly resuspended and FACS antibody staining was performed, whereas the brain homogenate from mice older than P10 was resuspended in 37% Percoll (Sigma-Aldrich, Taufkirchen, Germany) followed by centrifugation for 30 min, 800g at 4 °C without brake. Myelin was removed from the top of the cell suspension and microglia were taken from the pellet on the bottom of the tube. To isolate OPCs from P8 brains, tissue was digested for 15 min at 37 °C using papain (Worthington, Lakewood, NJ, USA) followed by a gradient centrifugation. Cell pellet was taken for further analysis. For RNA and protein extraction, 1 mm-thick brain slices were prepared and the corpus callosum, cortex, and cerebellum were separated under the binocular. The tissue was either snap-frozen in liquid nitrogen and stored at −80 °C (protein analyses) or stored in RNAl-ater solution (Thermo Fisher Scientific, Schwerte, Germany; RNA extraction). For electron microscopy, 2 mm-thick brain slices were prepared and small tissue pieces including the corpus callosum directly transferred to 3% glutaraldehyde.
Flow cytometry
Cells were acquired on a FACSCanto II (BD Bioscience, Heidelberg, Germany) and analyzed with FlowJo software (TreeStar). Cell sorting was performed on a MoFlo Astrios (Beckman Coulter, Krefeld, Germany). The following antibodies were used for staining cells: anti-CD45 (clone 30-F11, eBioscience, San Diego, USA), anti-CD11b (clone M1/70, eBioscience, San Diego, USA), anti-Gr1 (clone RB6-8C5, Biolegend, Fell, Germany), anti-Mac3 (clone M3/84 Biolegend, Fell, Germany), and anti-CD11c (N418, eBioscience San Diego, USA). Before surface staining, dead cells were stained using the Fixable Viability Dye eFluor® 506 (eBioscience, San Diego, USA) followed by incubation with Fc receptor blocking antibody CD16/CD32 (clone 2.4G2, BD Bioscience, Heidelberg, Germany).
Histology
For immunofluorescence staining with cryosections, tissues were fixed overnight at 4 °C (for immunofluorescence in Sox10-iCreERT2 mice, tissue was only postfixed with PFA for max. 2 h), dehydrated in 30% sucrose solution and embedded in Tissue-Tek® O.C.T.TM Compound (Sakura Finetek Europ B.V., Netherlands). 12 μm or 30 μm (for reconstruction) parasagittal sections were cut on a cryotome and mounted onto microscope slides (Langenbrinck, Emmendingen, Germany) for staining. The brain slices were fixed in acetone for 20 min at −20 °C and rehydrated in PBS. Blocking was performed with 5% BSA, 0.1% Triton-X100 in PBS for 1 h. The 12 μm sections were incubated with the primary antibodies anti-GFP (1:1000, Cat. No. 600-101-215S, Rockland, Limerick, USA), anti-APC (1:400, Cat. No. OP80, Merck Chemicals, Darmstadt, Germany), anti-PDGFRa (1:300, Cat. No. 3174, Cell Signaling, Frankfurt am Main, Germany), anti-Sox9 (1:200, Cat. No. AF3075, R&D Systems, Wiesbaden-Nordenstadt, Germany), anti-NeuN (1:200, Cat. No. MAB377, Merck Chemicals, Darmstadt, Germany), Iba-1 (WAKO, Cat. No. 019-19741, Wako, Neuss, Germany), and Olig2 (1:200, Cat. No. AB9610, Merck Millipore, Darmstadt, Germany) in 1% BSA in PBS at 4 °C overnight followed by the incubation with the secondary antibodies (Alexa Fluor 555, Alexa Fluor 568, Alexa Fluor 488, Alexa Fluor 647 1:500, Life technologies, Darmstadt, Germany) in 1% BSA in PBS for 1 h at RT in the dark. Nuclei were counterstained with 4,6-diami-dino-2-phenylindole (DAPI, 1:10 000, Cat. No. 236276, Boehringer, Mannheim, Germany) for 10–15 min and the slides were mounted using Mowiol (Merck Millipore, Darmstadt, Germany). Pictures were taken with BZ-9000 Biorevo microscope (Keyence, Neu-Isenburg, Germany).
For immunofluorescence in Sox10-iCreERT2 brains, coronal sections (30 μm) were cut and stained as described [71], with the following primary antibodies: chick-GFP (1:1000, Cat. No. GFP-1020, Aves Lab, Tigard, Oregon, USA), rabbit (rb)-NG2 (1:500, Cat. No. AB5320, Merck Millipore, Darmstadt, Germany), rb-Iba1 (1:500, Cat. No. 01919741, Wako, Neuss, Germany), m-S100β (1:250, Cat. No. S2532, Sigma-Aldrich, Taufkirchen, Germany), and m-GFAP (1:500, Cat. No. G3893, Sigma-Aldrich, Taufkirchen, Germany). Fluorochrome-conjugated secondary antibodies were chosen according to the primary antibody: anti-chick A488 (1:500, Cat. No. A11039, Life Technologies, Darmstadt, Germany), anti-rb Cy3, Cy5, or DyLight 649 (1:500, Cat. No. 711-165-152, 111-175-144 or 111-495-144, respectively, Dianova, Hamburg, Germany), anti-m Cy3, Cy5 or DyLight 649 (1:500, Cat. No. 115-165-003, 115-176-072 or 115-496-072, respectively, Dianova, Hamburg, Germany). Nuclei were stained with DAPI (1:1000, Cat. No. D9564, Sigma-Aldrich, Taufkirchen, Germany). Images were collected with a Leica confocal microscope system (TCS SPE). Immunohistochemical analysis was performed on multi-channel, confocal stacks using the cell counter plug-in for Fiji (http://fiji.sc/Fiji based on ImageJ 1.48i). Analysis was performed on three sections of n = 5/6 animals per condition.
Primary OPC cultures
OPCs from the cerebral cortex of postnatal (P6–8) C57Bl/6 mice were isolated by magnetic associated cell sorting (MACS, Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions with some modifications. Shortly, cells from dissected cerebral cortex were isolated by the Adult Brain dissociation Kit (Cat. No. 130-107-677, Miltenyi Biotec, Bergisch Gladbach, Germany) in combination with the gentle MACS Octo Dissociator™ (Cat. No. 130-095-937, Miltenyi Biotec, Bergisch Gladbach, Germany). All steps were performed as described without the red cell blood removal. Subsequently cells were kept for 45 min in MACS NeuroBrew medium (Cat. No. 130-093-566, Miltenyi Biotec, Bergisch Gladbach, Germany) for re-expression of proteins. Cells were then blocked with the FcR blocking reagent mouse (Cat. No. 130-092-575, Miltenyi Biotec, Bergisch Gladbach, Germany) and labeled with anti-AN2-, -CD140a-, and -O4 microbeads. After washing OPCs were selected by a magnetic separation on a MiniMACS separator (Cat. No. 130-042-102, Miltenyi Biotec, Bergisch Gladbach, Germany). For OPC cultures, 10000 OPCs were plated on poly-ornithine (PLO) coated coverslips (10 mm diameter) and cultured with proliferation [DMEM (Gibco) containing 1:50 B27 Supplement (Gibco), 1:100 Pen/Strep (Gibco), 1:100 FBS (Gibco), 5% BSA (Sigma), 10 μg/ml Apo-Transferrin human (Sigma), 100 μM putrescine dihydrochloride (Sigma), 2 μM progesterone (Sigma), 30 μM sodium selenite (Sigma), 10 μg/ml insulin from bovine pancreas (Sigma), 500 nM L-thyroxine (Sigma), 0.5 nM 3,3′,5-triiodo-L-thyronine sodium salt (T3; Sigma), 2 mM glutamine (Gibco), 10 ng/ml PDGF-AA human recombinant (PeproTech, Hamburg, Germany), 20 ng/ml bFGF (FGF2) recombinant human protein (Gibco)] or differentiation [DMEM (Gibco) containing 1 μg/ml apo-transferrin human (Sigma), 100 μM putrescine dihydrochloride (Sigma), 200 mM progesterone (Sigma), 220 nM sodium selenite (Sigma), 10 μg/ml insulin from bovine pancreas (Sigma), 520 nM L-thyroxine (Sigma), 0.5 nM 3,3′,5-triiodo-L-thyronine sodium salt (T3; Sigma), 25 μg/ml gentamicin (Gibco), and 2% Horese serum (Gibco)] medium. After 12 h in culture vehicle or BLZ945 was added into the culture in different concentrations (0.5, 1 or 5 μM). OPCs were cultured for 3 days with every day changing the medium and adding fresh BLZ945. Cultures were then fixed for 10 min with 4% PFA. Cells were then stained with rb-Olig2 antibody (1:500, Cat. No. MABN50, Merck Millipore, Darmstadt, Germany) and counterstained with DAPI (1:1000). For counting of alive cells (DAPI+ cells) images were collected with a Keyence Biorevo BZ-9000 microscope. For merging and analyzing a VK analyzer was used, with which all DAPI+ cells within a coverslip were counted. As numbers were similar in the vehicle controls in both medium conditions, we pooled the results. The average number of cells on 2–3 coverslips per condition and experiment were used from 4 to 5 dishes (two completely independent experiments).
Three-dimensional cell reconstruction
Three-dimensional reconstruction of microglia was done using the IMARIS software (Bitplane, Zürich, Switzerland) as described previously [11]. In brief, CX3CR1+ Iba-1+ immunostained 30 μm cryosections from P7 old brains were used. Imaging was performed on an Olympus Fluoview 1000 confocal laser scanning microscope (Olympus, Hamburg, Germany) using a 20 × 0.95 NA objective. Z stacks were done with 3× zoom in 1.14 μm steps in z direction, 1024 × 1024 pixel resolution were recorded and analyzed using IMARIS software (Bitplane, Zürich, Switzerland).
Microarray analysis
FACS-sorted microglia were directly collected in cell lysis buffer, and subsequently, RNA was isolated with the Arcturus Pico Pure RNA Isolation Kit (Life technologies, Darmstadt, Germany) according to the manufacturer’s protocol. Transcriptional profiles of the microglia populations were assessed using Affymetrix® (Affymetrix, Santa Clara, USA) GeneChip Arrays (Mouse Gene 2.0 ST Arrays). Affymetrix® GeneChip array data were pre-processed using Affymetrix® Expression Console and normalized through the Robust Multi-array Average (RMA) implementation in the Expression Console. Sample processing and Affymetrix microarray hybridization were carried out at a genomics core facility: Center of Excellence for Fluorescent Bioanalytics (KFB, University of Regensburg, Germany.
Pre-processing of gene expression data
Analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team. The Robust Multi-array Average (RMA) function as implemented in BRB Tools was used to normalize the data during import of the raw Affymetrix data (CEL files). Quartile normalization was applied on a total of 18 samples. Gene probes were only kept for further analysis if their mean expression values were higher than 1.5-fold of the median of all samples in at least 15% of samples within the whole data set. Next, multiple probes for one gene locus were filtered allowing only one probe with the highest overall variance. These pre-processing steps resulted in 3099 unique genes to be present in at least one group or condition. These genes were retained for further analyses.
Identification of differentially expressed genes and hierarchical clustering
Differentially expressed (DE) genes were defined in BRB Tools by Class comparison with an adjusted P value of <0.01. Hierarchical Clustering and heatmap generation were performed using the heatmap.2 function in the R package gplots [74] in conjunction with the R hclust function [54]. Clustering was performed according to the ward.D2 implementation of the clustering method described by Ward [40, 73].
Pathway analysis by IPA
Gene Ontology (GO) enrichment analysis and pathway analysis was performed using the Ingenuity Pathway Analysis suite by Qiagen Inc.
Gene expression analysis
Brain tissue samples were collected in RNAlater solution (Thermo Fisher Scientific, Schwerte, Germany) and stored for 1–3 days at 4 °C before RNA was isolated with the Qiagen RNeasy Mini Kit (Qiagen, Germantown, USA) according to the manufacturer’s protocol. Reverse transcription and real-time PCR analysis were performed using the SuperScript Reverse Transcriptase II (Thermo Fisher Scientific, Schwerte, Germany) and Gene Expression Master Mix reagents (Applied Biosystems, Schwerte, Germany) according to the manufacturer’s recommendations. FACS-sorted microglial and oligodendrocyte progenitor cell populations were collected directly in cell extraction buffer, and subsequently, RNA was isolated with the Arcturus Pico Pure RNA Isolation Kit (Life Technologies, Darmstadt, Germany) according to the manufacturer’s protocol. Reverse transcription and real-time PCR analysis were performed using high-capacity RNA-to-cDNA-Kit and Gene Expression Master Mix reagents (Applied Biosystems, Schwerte, Germany) according to the manufacturer’s recommendations. RT-PCRs were analyzed with a LightCycler 480 (Roche, Darmstadt, Germany). For gene expression analysis, we used the following TaqMan Gene Expression Assays: Actb (Mm01205647_g1), Gapdh (Mm03302249_ g1), Mbp (Mm01266402_m1), Plp1 (Mm01297210_m1), Mog (Mm00447824_m1), Pdgfra (Mm00440701_m1), Cspg4 (Mm00507257_m1), Sox10 (Mm00569909_m1), Aif1 (Mm01132448_g1), Csf1r (Mm01266652_m1), Itgam (Mm00434455_m1), Emr1 (Mm00802529_m1), P2ry12 (Mm01950543_s1), and Gfap (Mm01253033_m1). Beta-Actin and Gapdh were used as housekeeping genes.
Western blot
Tissue samples were harvested and extracted in RIPA buffer (25 mM Tris–HCl, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, pH 7.5). Samples were separated by SDS–PAGE and immunoblotted using antibodies to MBP (1:1000, Cat. No. AB980, Merck Chemicals, Darmstadt, Germany), PLP (1:1000, Cat. No. ab28488, Abcam, Cambridge, UK), and GAPDH (1:2500, Cat. No. Mab374, Millipore, Billerica, USA).
Electron microscopy
Following standard EM protocols, samples were first fixed overnight in 3% glutaraldehyde at 4 °C, washed with Sörensen buffer and then transferred to 1% osmium tetroxide for 2 h at RT. After washing samples with aqua bidest, they were dehydrated by a graded series of ethanol (30–100%) followed by 100% propylene oxide (PO), resin/PO [1:2 (v/v)], and resin/PO [2:1 (v/v)]. Samples were embedded in resin via polymerization for 24 h at 75 °C. 700 nm semi-thin sections were cut and stained with 2% toluidine blue to define the region of interest for further preparation of 70 nm ultra-thin sections using an ultramicrotome (Leica Reichert Ultrachut S). After contrasting the sections with uranyl acetate and lead citrate (Leica Reichert Ultrostainer) following standard protocols, images were taken at 7900× magnification using CM100 Electron Microscope (Philips). G ratio and number of axons were analyzed using iTEM Software (Olympus).
Statistical analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software, Version 6.0, La Jolla, USA). All data were tested for normality applying the F test to compare variance. If normality was given, an unpaired t test was applied. If the data did not meet the criteria of normality, the Mann–Whitney U test was applied. Differences were considered significant when P value <0.05.
Results
The postnatal “fountain of microglia” population in the white matter has a distinct gene expression profile
To determine the spatiotemporal features of a special microglia population that selectively appears during post-natal days in periventricular sites and has been described as the “fountain of microglia” [25, 30, 58], we analyzed different postnatal stages in Cx3Cr1GFP/WT mice, which have a GFP knock-in in one allele of the Cx3Cr1 gene [26]. In the CNS, CX3CR1 is expressed in microglia and other CNS macrophages and their precursors starting from the early stages of prenatal development [9, 14, 20, 28, 39, 51, 53].
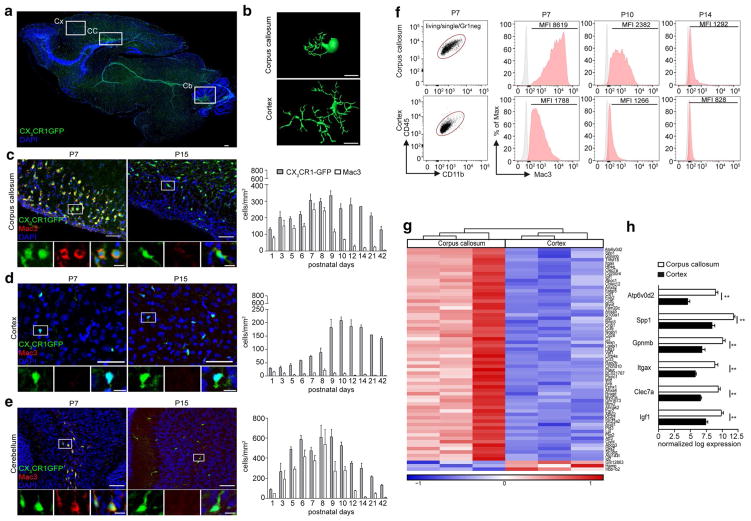
First, we compared by a comprehensive high-resolution immunofluorescence analysis, the kinetics of microglia in distinct CNS cortical and subcortical regions immediately after birth, especially in the white matter regions of the corpus callosum and cerebellum and the frontal cortex (Fig. 1a–e). The number of developing CX3CR1+ microglia steadily increased from postnatal day (P) 1 onwards in all regions examined (Fig. 1c–e). The microglia increase was accompanied by a high proliferation rate measured by 5-ethynyl-2′-deoxyuridin (EdU) incorporation in all three brain regions (Figure S1). Importantly, whereas CX3CR1+ microglia in the frontal cortex showed already at P7 several delineated processes resembling the adult microglial phenotype, cells in the developing white matter of the corpus callosum and cerebellum displayed a marked amoeboid morphology with concomitant high expression of the activation marker Mac3 (LAMP2, CD107b; Fig. 1b–e). Remarkably, the expression of Mac3 was restricted to white matter microglia from P1 to P8 but absent from cortical microglia at the same time points. Even though the number of subcortical CX3CR1+ microglia remained relatively stable over time, the expression of Mac3 in subcortical microglia dropped dramatically within 1 day (at P9). At P14, almost no microglia in the corpus callosum and cerebellum expressed Mac3 anymore (Fig. 1c–e). Elevated Mac3 expression in these clusters of activated subcortical microglia could additionally be measured by flow cytometry (Fig. 1f). Compared to the cortex, microglia in the developing corpus callosum showed a high Mac3 expression at P7 which drastically decreased already at P10 and even further at P14.
Fig. 1.
Amoeboid microglia population with distinct expression profile transiently accumulates in white matter regions shortly after birth. a Representative sagittal section of a postnatal (P) P7 Cx3cr1GFP/Wt mouse brain depicting amoeboid parenchymal CX3CR1+ microglia (green) specifically in white matter regions. 4′,6-diamidino-2-phenylindole (DAPI) in blue. White squares indicate the regions analyzed in b–e (Cx cortex, CC corpus callosum, Cb cerebellum). Scale bar 50 μm. b Imaris-based three-dimensional reconstruction of a typical (CX3CR1+Iba-1+) amoeboid microglia in the corpus callosum and ramified microglia in the cortex at P7. Scale bar 10 μm. c, d Left representative immunofluorescence images of the corpus callosum (c), cortex (d), and cerebellum (e) at P7 and P15. CX3CR1 (green), Mac3 (red), and DAPI (blue). Scale bar 50 μm. Inset: scale bar 10 μm. Right kinetics of CX3CR1+ microglia (grey bars) and the activation marker Mac3+ (white bars) at the indicated time points. Bars indicate the mean ± SEM. At least three mice per time point were analyzed. f Flow cytometric analysis of microglia from P7, P10, and P14 old Cx3cr1GFP/Wt mice. CD45+CD11b+ microglia were pre-gated on living cells, single cells, Gr1− cells. The mean fluorescent intensity (MFI) of the Mac3 expression is depicted (n = 3). g Hierarchical clustering created on the most significantly differentially expressed (DE) genes (cut off adjusted to P < 0.01) between microglia from the corpus callosum and cortex at postnatal day 7. Data are sorted by most significant changes from top to bottom. Heat map displays row z-score values from red to blue via white. h Display of genes identified in g known to be related to priming and phagocytic function of microglia. Data presented as normalized log expression. Bars indicate the mean ± SEM. Significant differences were determined by an unpaired t test: *P < 0.05, **P < 0.01
We next performed a whole-genome expression analysis using the Affymetrix Mouse Gene 2.0 ST Array from P7 microglia from subcortical and cortical regions (Fig. 1g). Subsequent investigation of the genes significantly differentially regulated (cut off adjusted to P < 0.01) between microglia from the P7 corpus callosum and cortex revealed a highly activated phenotype of the microglia in the corpus callosum. Genes such as secreted phosphoprotein 1 (Spp1), glycoprotein non-metastatic melanoma protein B (Gpnmb), integrin subunit alpha X (Itgax, also known as Cd11c), c-type lectin domain family 7 member A (Clec7a), and insulin-like growth factor 1 (Igf1) known to be related to primed and/or activated microglia were significantly upregulated in the corpus callosum compared to the cortex (Fig. 1h). Moreover, ATPase H+ transporting V0 Subunit D2 (Atp6v0d2) reported to be an important factor for mediating fusion between phagosomes and lysosomes during phagocytosis was the most upregulated gene in P7 corpus callosum microglia [48]. The specific expression of Itgax in the “fountain of microglia” was additionally confirmed on protein level using Cd11cCreER:Rosa26-tomato mice (Figure S2c).
These data indicate that different microglia populations exist in the early postnatal mouse brain that exhibit marked differences in their localization, activation pattern, and distinct genetic profiles.
Subcortical microglia cluster arises from local microglia precursors and develop in the absence of blood monocytes
The current concept that the cells of the “fountain of microglia” are in contrast to other microglia in the brain monocyte-derived largely depends on the previous work using carbon or fluosphere labeling methods in rodents that have suggested circulating blood cells as the origin of subcortical microglia after birth [24, 25, 30].
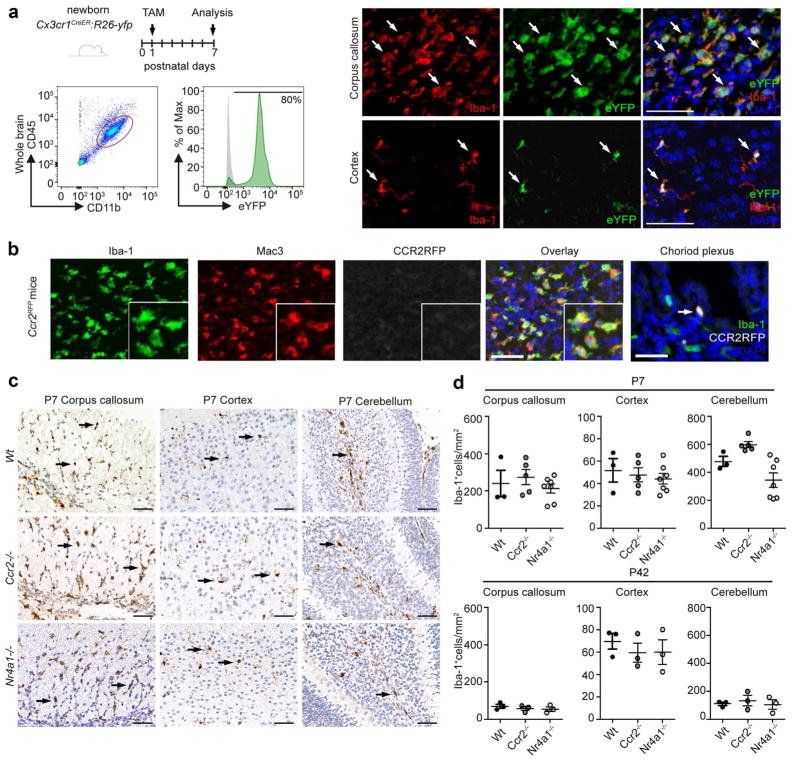
Tissue macrophages can arise either from precursors that seed tissues before birth, like cortical microglia and non-parenchymal CNS macrophages, or alternatively from circulating monocytes after birth such as macrophages in the intestine or heart [39, 51, 52]. To test whether the early progenitors contribute to the pool of subcortical amoeboid microglia to the same extent as microglia in the frontal cortex, we adapted the Cx3Cr1CreER mouse system that allows fate mapping studies by tamoxifen-induced recombination in CX3CR1-expressing cells at defined time points [9, 14, 20]. Considering that cortical microglia are entirely of prenatal origin [13, 64], we asked whether the amoeboid microglia clusters in white matter regions share this source. To target the early postnatal microglia population that is already present prior to the appearance of the “fountain of microglia,” we treated newborn Cx3Cr1CreER: Rosa26-YFP mice with tamoxifen at P1 and assessed YFP labeling of corpus callosum-associated foamy macrophages in comparison to cortical microglia at P7 (Fig. 2a). Flow cytometry measurements showed strong expression (approx. 80%) of YFP in CD45loCD11b+ microglia at P7 indicating high labeling efficacy of microglia. Subsequent examination of histological slices revealed a robust expression of the reporter gene in the “fountain of microglia” cluster, comparable to that observed in cortical microglia. These data indicate that both cortical microglia with an arborized morphology and subcortical amoeboid microglia share the same origin from a pool of CX3CR1-expressing tissue progenitors in the CNS.
Fig. 2.
Amoeboid microglia clusters are derived from local microglia precursors and develop normally in the absence of blood monocytes. a Scheme of experimental setup. Cx3cr1CreER:R26-yfp mice were injected intraperitoneally (i.p.) with tamoxifen at P1. Analysis was performed at P7. Representative blot of flow cytometric analysis of whole brain microglia (CD45lo, CD11b+) and subsequent immunofluorescent analysis of brain slices, Iba-1 (red), CX3CR1 (eYFP), and DAPI (blue). Arrows show triple positive cells. Scale bar 50 μm. b Representative immunofluorescent images of the developing corpus callosum of P7 Ccr2RFP mice. The P7 choroid plexus was used as a CCR2-RFP positive control. Iba-1 (green), Mac3 (red), CCR2-RFP (white), and DAPI (blue). Arrow shows a triple positive cell next to an Iba-1+ macrophage (asterisk). Scale bar 50 μm. c Representative images of Iba-1 immunohistochemistry in brain slices of P7 Wildtype (Wt), Ccr2−/−, and Nr4a1−/− mice. Scale bar 50 μm. d Quantification of Iba-1+ microglia in the corpus callosum, cortex, and cerebellum of P7 (upper lane) and P42 (lower lane) Wildtype (Wt), Ccr2−/−, and Nr4a1−/− mice (n = 3–7). Each symbol represents one mouse. Means with SEM are represented. Samples were collected from two independent experiments
CCR2+Ly-6Chi monocytes have previously been shown to be recruited to the adult CNS especially following damage [35, 36]. To further test whether circulating CCR2+Ly-6Chi monocytes physiologically colonize the developing brain shortly after birth, we used Ccr2RFP reporter mice. CCR2 is known to be expressed exclusively by blood monocytes but never by microglia during development or disease [38, 60]. Upon thorough histopathological examination, we were unable to identify any RFP+ microglia either within the corpus callosum or other parenchymal regions but only in the blood vessels of the choroid plexus at P7 (Fig. 2b). Furthermore, we found neither Ly-6Chi nor Ly-6Clo monocytes being essential for the establishment of amoeboid microglia clusters in the subcortical regions, as mice lacking CCR2 or the transcription factor Nr4a1 presented normal proportions of these macrophage subsets (Fig. 2c, d). In summary, our data suggest that distinct macrophage populations found in the postnatal subcortical (amoeboid shaped) and cortical regions (with cellular processes) share their early origin, are derived from the same CX3CR1+-derived CNS endogenous precursors, and have no contribution from circulating blood monocytes.
Early postnatal microglia vitally regulate myelinogenesis on the progenitor level
The precise functional role of early postnatal microglia is poorly understood. It has been shown that during P7–P18 hippocampal microglia fundamentally shape the maturating neuronal network to achieve full synaptic functionality [45] and to support juvenile neurogenesis [66]. However, the impact of the transiently activated microglia population in the first postnatal week in white matter regions is currently unknown.
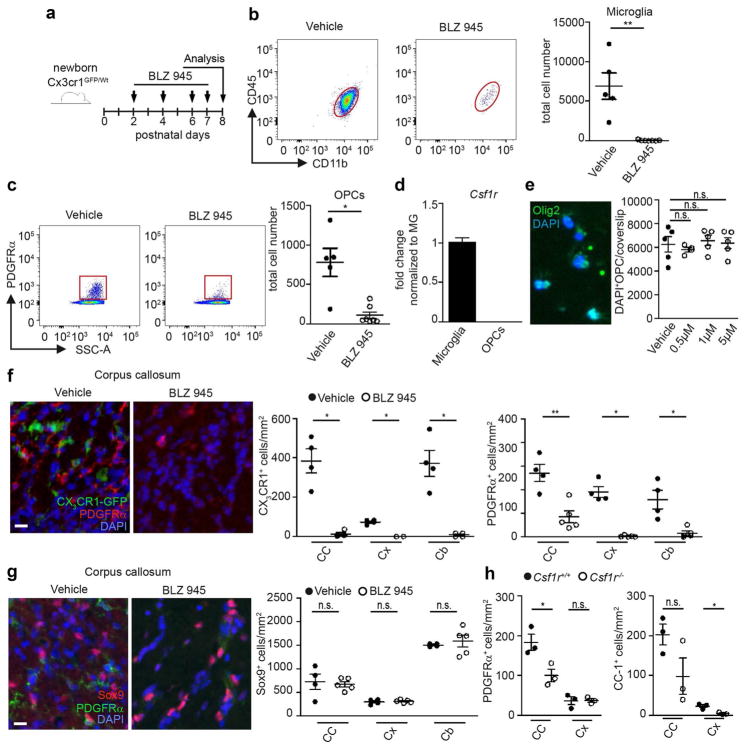
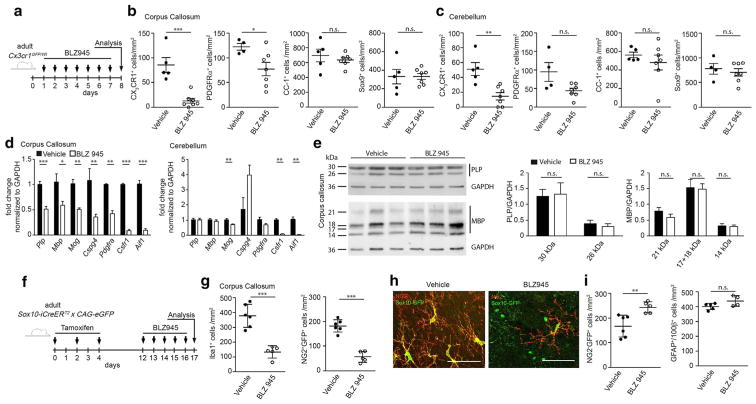
Because the early postnatal white matter region is highly active due to the ongoing maturation, including myelination of the central axonal tracts [57], we asked whether the local amoeboid microglia population might be involved in the process of physiological myelination. To do so, we applied the potent and selective CSF-1R inhibitor BLZ945, which efficiently depletes microglia [8] (Fig. 3a). Intra-peritoneal injection of vehicle or BLZ945 in neonatal Cx3cr1GFP/Wt mice was performed at P2, P4, P6, and P7. One day after the last injection, microglia were virtually completely depleted in the whole brain of Cx3cr1GFP/Wt mice as analyzed by flow cytometry (Fig. 3b) in samples that included both the white matter regions of the corpus callosum and the cerebellum, as well as the frontal cortex. We also confirmed these data by quantification of immunohistochemically detected CX3CR1+ microglia (Fig. 3f). Surprisingly, platelet derived growth factor receptor (PDGFR)α+ oligodendrocyte progenitor cells (OPCs) were strongly reduced in whole brain preparations as shown by flow cytometry (Fig. 3c). To ensure cell-specificity of BLZ945 treatment, we measured Csf1r expression on isolated OPCs and microglia by quantitative RT-PCR using NG2YFP/Wt reporter mice (Fig. 3d). The purity of isolated microglia and OPCs was demonstrated by the presence of the microglia-specific genes allograft inflammatory factor 1 (Aif1), integrin subunit alpha M (Itgam), Emr1 and P2ry12 as well as the OPC specific genes chondroitin sulfate proteoglycan 4 (Cspg4, also known as NG2), platelet derived growth factor receptor alpha (Pdgfr) and SRY-Box 10 (Sox10) (Figure S3). Csf1r mRNA was found to be highly expressed on isolated microglia, whereas no mRNA could be detected on NG2/PDGFRα double positive OPCs at P8. Moreover, cultivation of primary postnatal OPCs in the presence of 0.5, 1, or 5 μM BLZ945 did not influence cell survival of OPCs (Fig. 3e), whereas primary microglia show a reduced cell number already at a dose of 1 μM (data not shown). In addition to flow cytometry, a concomitant PDGFRα+ OPC reduction could be visualized in different CNS regions on tissue sections (Fig. 3f, g). This was further confirmed using Olig2 as a second marker for the oligodendrocyte lineage (Figure S4). Interestingly, CX3CR1+ white matter microglia were found in close vicinity to PDGFRα+ OPCs but had no detectable physical contacts. Importantly, application of BLZ945 did not lead to generalized effects on other brain cells as the number of astrocytes, visualized by the transcription factor Sox9, was not altered in all CNS regions examined (Fig. 3g). In addition, in mice in which microglia have been genetically depleted by the lack of Csf1r, PDGFRα+ OPCs were significantly reduced within the corpus callosum and the number of CC-1+ oligodendrocytes was diminished at P20 (Fig. 3h). Interestingly, oligodendrocytes in the cortex of P20 Csf1r−/− mice were reduced, whereas OPCs were not affected, indicating regional differences in the regulation of oligodendrocyte maintenance. In summary, these data clearly indicate that microglia-specific deletion has detrimental effects on OPC number.
Fig. 3.
Essential role of microglia for adequate induction of oligodendrocyte progenitors. a Scheme of experimental setup. Cx3cr1GFP/Wt mice were injected intraperitoneally (i.p.) with BLZ945 at P2, P4, P6, and P7. Analysis was performed at P8. b, c Representative blots and quantification of flow cytometric analysis of CD45+CD11b+ microglia (b) and PDGFRα+ oligodendrocyte progenitors (OPCs; c) pre-gated on living cells, single cells, Gr1− cells of mice treated as depicted in a (n = 5–6, samples are from four independent experiments). Each symbol represents one mouse. Means with SEM are represented. Significant differences were determined by an unpaired t test: *P < 0.05, **P < 0.01. d Quantitative RT-PCR of Csf1r of sorted CD45+CD11b+ microglia and NG2+PDGFRα+ OPCs from P8 NG2YFP/Wt mice (n = 5). Data are normalized to Gapdh and β-Actin and presented as fold change to microglia levels. e Representative image of a primary postnatal OPC culture showing overlapping Olig2+/DAPI+ cells (left). OPC cultures were treated for 3 days with vehicle or different concentrations of BLZ945 (0.5, 1, and 5 μM). The number of DAPI+OPCs/coverslip did not change after treatment compared to the vehicle controls. f Representative immunofluorescent images (left) of the corpus callosum showing CX3CR1+ (green) microglia and PDGFRα+ (red) OPCs in vehicle or BLZ945-treated mice. Right quantification of these cells in the corpus callosum (CC), cortex (Cx), and cerebellum (Cb). Each symbol represents one mouse. Means with SEM are represented. n = 4; samples from two independent experiments. Scale bar 40 μm. Significant differences were examined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01). g Representative immunofluorescent images (left) of the corpus callosum showing Sox9+ (red) astrocytes and PDGFRα+ (green) OPCs in vehicle or BLZ945-treated mice. Right quantification of Sox9+/PDGFRα− astrocytes in the corpus callosum (CC), cortex (Cx), and cerebellum (Cb). n = 4–5; samples from two independent experiments. Scale bar 40 μm. Significant differences were examined by an unpaired t test. n.s. not significant. h Quantification of PDGFRα+ OPCs and CC1+ oligodendrocytes in the corpus callosum (CC) and cortex (Cx) of P20 Csf1r+/+ and Csf1r−/− mice. Significant differences were examined by an unpaired t test and marked with asterisks (*P < 0.05). n.s. not significant
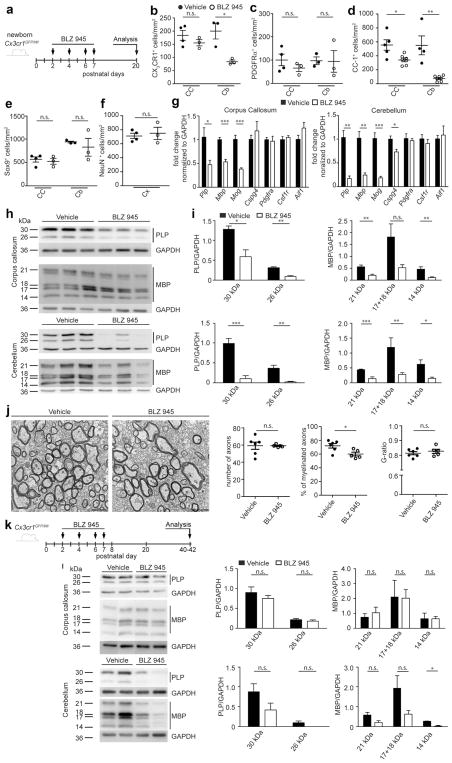
To decipher the functional role of the “fountain of microglia” for the white matter regions, we examined potential functional consequences of a disrupted early postnatal microglia–OPC axis on the subsequent process of myelination. Therefore, after depletion of microglia by BLZ945 application at P2, P4, P6, and P7, mice were investigated at P20 (Fig. 4a). As changes of oligodendrocytes should mainly affect white matter regions, we focused on the corpus callosum as well as the white matter of the cerebellum. Microglia numbers were normal in the corpus callosum but were still reduced in the white matter of the cerebellum (Fig. 4b) indicating a high plasticity of these cells with regional differences, as previously described [3, 8]. Whereas no changes in the number of PDGFRα+ OPCs could be detected in either the corpus callosum or the white matter of the cerebellum, the number of CC1+ mature oligodendrocytes was significantly reduced in both analyzed areas (Fig. 4c, d). Notably, no changes in the number of Sox9+/PDGFRα− astrocytes in the corpus callosum and cerebellum or NeuN+ neurons in the cortex could be detected, again confirming that the consequences of postnatal microglia depletion are restricted to oligodendrocytes (Fig. 4e, f).
Fig. 4.
Reduced postnatal myelinogenesis in microglia-depleted mice. a Scheme of experimental setup. Cx3cr1GFP/Wt mice were injected intraperitoneally (i.p.) with BLZ945 at P2, P4, P6, and P7. Analysis was done at P20. b–f Quantification of CX3CR1+ microglia (b), PDGFRα+ OPCs (c), CC1+ oligodendrocytes (d), Sox9+/PDGFRα− astrocytes (e), and NeuN+ neurons (f) in the indicated brain regions using immunohistochemically stained brain slices. Significant differences were examined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01). Each symbol represents one mouse. Bars represent mean ± SEM. g Quantitative RT-PCR of the genes proteolipid protein (Plp), myelin basic protein (Mbp), myelin oligodendrocyte glycoprotein (Mog), chondroitin sulfate proteoglycan 4 (Cspg4 = NG2), platelet derived growth factor receptor alpha (Pdgfrα), colony stimulating 1 receptor (Csf1r), and allograft inflammatory factor 1 (Aif1) in tissue isolated from the corpus callosum and cerebellum at P20 (n = 4; samples from two independent experiments). Data were normalized to Gapdh and β-actin and are presented as fold change normed to vehicle. Significant differences were examined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001). h, i Western blot analysis for the myelin proteins proteolipid protein (PLP) and myelin basic protein (MBP) in the corpus callosum and cerebellum at P20. GAPDH was used as a loading control. Quantification (right) was performed by normalization of the myelin proteins to GAPDH. Significant differences were examined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001). Bars indicate mean ± SEM. j Representative electron microscope images from the corpus callosum treated with vehicle or BLZ945 at P20 (left, scale bar 2 μm) and quantification thereof (right). The total number of axons (myelinated and non-myelinated cut off ≥0.5 μm), the percentage of myelinated axons of all axons, and the G-ratio (axon diameter/filament diameter) were quantified (n = 5–6; samples from two independent experiments). Each symbol represents one mouse. Mean ± SEM are presented. Significant differences were examined by an unpaired t test and marked with asterisks (*P < 0.05). k Scheme of experimental setup. Cx3cr1GFP/Wt mice were injected i.p. with BLZ945 at P2, P4, P6, and P7. Analysis was done at P40–42. l Western blot analysis for the myelin proteins PLP and MBP in the corpus callosum and cerebellum at P40–42. GAPDH was used as a loading control. Quantification (right) was performed by normalization of the myelin proteins to GAPDH. Significant differences were examined by an unpaired t test and marked with asterisks (*P < 0.05). Bars indicate mean ± SEM
To specifically see if the loss of mature oligodendrocytes also leads to lack of myelin formation, we analyzed the expression levels of the myelin-specific genes proteolipid protein (Plp), myelin basic protein (Mbp) and myelin oligodendrocyte glycoprotein (Mog) at P20 (Fig. 4g). All these myelin-specific genes were significantly reduced in both the corpus callosum and cerebellum while genes for microglia (Csf1r, Aif1) and OPCs (Cspg4, Pdgfra) were mostly unaltered at this time point. Only Cspg4 was still reduced in the cerebellum. To further confirm that these effects lead to disturbances of myelin on protein level, the presence of PLP and MBP was measured by Western blot analysis (Fig. 4h, i). A robust reduction of PLP and MBP was detectable in the corpus callosum and cerebellum. To discover the consequences of the reduced myelin protein amount after microglia depletion, the corpus callosum was further analyzed by electron microscopy (Fig. 4j). Notably, the percentage of myelinated axons in microglia-depleted mice was significantly reduced, whereas the total number of axons (myelinated and non-myelinated) was unchanged (Fig. 4j). No change in the G-ratio (relation of axon diameter to the myelinated fiber diameter) was observed. The influence of microglia on oligodendrocytes and myelination was further supported by transcriptome analyses of genes related to the GO-terms myelination and axogenesis. Compared to adult (P42) microglia, most of these genes were highly upregulated at the early postnatal time point irrespective of the brain region (Figure S2a). However, focusing only on the differences between microglia of the P7 corpus callosum and cortex, we found that a subgroup of the genes involved in oligodendrocyte maturation and myelination as Igf1, C-X-C chemokine receptor type 4 (Cxcr4) and myosin ID (Myo Id) was highly expressed specifically in microglia of the P7 corpus callosum (Figure S2b). This is of interest as Igfr1 has been shown to be important for oligodendrocyte development and myelination [75], whereas Cxcr4 is essential for OPC differentiation and migration [2, 47]. These data clearly demonstrate that microglia and oligodendrocytes in white matter regions are functionally connected to each other. Early postnatal microglia localized in the “fountain of microglia” shape the number of oligodendrocyte progenitor cells, with consequences for subsequent myelinogenesis. To further elucidate if the effect on myelinogenesis is long lasting or if the fast recovery of microglia is able to compensate the phenotype, BLZ945 was applied at P2, P4, P6, and P7 and myelin proteins were assessed at P40–P42 (Fig. 4k). Surprisingly, at this time point, no significant differences in the amount of both PLP and MBP could be seen in the corpus callosum, whereas a distinct reduction was still present in the cerebellum (Fig. 4l). These data show that the developing brain is highly plastic and has substantial compensatory ability with distinct regional differences.
Microglia maintain oligodendrocyte progenitor numbers in the adult healthy brain
After having shown that the early postnatal microglia vitally regulate primary formation of the myelin compartment, we wondered whether there might be an additional functional role of microglia in oligodendrocyte and oligodendrocyte progenitor homeostasis in the adult brain. We, therefore, again depleted microglia in adult Cx3cr1GFP/Wt mice with the CSF-1R inhibitor BLZ945 for 7 days and assessed the brain thereafter (Fig. 5a). Quantification of CX3CR1+ cells in brain sections revealed a strong reduction of parenchymal microglia in both the corpus callosum (Fig. 5b) and the white matter of the cerebellum (Fig. 5c) and a significantly reduced number of PDGFRα+ OPCs in the corpus callosum (Fig. 5b). The number of CC-1+ oligodendrocytes in the corpus callosum and cerebellum was not significantly changed at this time point (Fig. 5b, c). Despite significantly reduced mRNA levels of Plp, Mbp, and Mog, no changes on protein levels of PLP and MBP were visible in the corpus callosum (Fig. 5d, e). No treatment effect on astrocytes was detectable, as the number of Sox9+ astrocytes was unaltered upon immunohistochemical quantification (Fig. 5b, c). To trace the oligodendrocyte lineage by an independent reporter protein, we used adult Sox10-iCre-ERT2 × CAG-eGFP mice that label after induction with tamoxifen OPCs and oligodendrocytes with the enhanced green fluorescent protein (eGFP). One week after induction with TAM, BLZ945 was applied orally for 5 consecutive days (Fig. 5f). Quantification of Iba-1+ cells on day 6 confirmed a strong reduction of microglia in the corpus callosum and a significant reduction of GFP+/NG2+ OPCs (Fig. 5g). These data additionally show that microglia are required to maintain the OPC pool in the adult brain. Interestingly, although the absolute number of recombined (GFP+) cells was decreased (from 349 ± 13 to 299 ± 13 GFP+ cells/mm2) upon microglia depletion, the number of GFP+NG2-negative cells was increased (Fig. 5i). However, as the GFP signal specifically in these GFP+/NG2− cells is very weak (Fig. 5h), it needs to be further clarified if these cells represent premature not myelinating oligodendrocytes or dying/unhealthy cells. Nevertheless, these results underlined again that microglia are necessary for proper regulation of OPC differentiation even in adult stages. As expected by the previous results, no treatment effect on astrocytes could be observed by quantification of GFAP+/S100β+ astrocytes in the corpus callosum (Fig. 5i).
Fig. 5.
Microglia are necessary for myelin homeostasis during adulthood. a Scheme of experimental setup. 6–10-week-old Cx3cr1GFP/Wt mice were treated with BLZ945 for 7 consecutive days by oral gavage. Analysis was performed 1 day after the last application. b, c Quantification of CX3CR1+ microglia, PDGFRα+ OPCs, CC-1+ oligodendrocytes, Sox9+ astrocytes (b, d) in the corpus callosum (b) and cerebellum (c) using immunohistochemically stained brain sections at day 8. n = 4–7; samples from two independent experiments. Significant differences were examined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001; n.s not significant.) Each symbol represents one mouse. Mean ± SEM are depicted. d Quantitative RT-PCR of the genes proteolipid protein (Plp), myelin basic protein (Mbp), myelin oligodendrocyte glycoprotein (Mog), chondroitin sulfate proteoglycan 4 (Cspg4), platelet derived growth factor receptor alpha (Pdgfrα), colony stimulating 1 receptor (Csf1r), and allograft inflammatory factor 1 (Aif1) in tissue isolated from the corpus callosum and cerebellum at day 8 (n = 4–7; samples from two independent experiments). Data were normalized to Gapdh and β-actin and are presented as fold change normed to vehicle. Significant differences were examined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001). Bars represent mean ± SEM. e Western blot analysis for proteolipid protein (PLP) and myelin basic protein (MBP) in the corpus callosum at P8, 1 day after the last BLZ945 application. GAPDH was used as a loading control. Quantification (right) was performed by normalization of the myelin proteins to GAPDH. Bars indicate mean ± SEM. Significant differences were examined by an unpaired t test (n.s not significant). f Scheme of experimental setup. 8–10-week-old Sox10-iCreERT2 × CAG-eGFP mice were treated with BLZ945 for 5 consecutive days by oral gavage. Analysis was performed 1 day after the last application. g, i Quantification of Iba-1+ microglia, Sox10-GFP+/NG2+ OPCs (g), Sox10-GFP+/NG2− oligodendrocytes and GFAP+/S100β+ astrocytes (i) in the corpus callosum using immunohistochemically stained brain sections at day 6. n = 5–6, samples from two independent experiments. Significant differences were examined by an unpaired t test and marked with asterisks (**P < 0.01, ***P < 0.001; n.s not significant.) Each symbol represents one mouse. Mean ± SEM are depicted. h Representative confocal images of a NG2-immunofluorescence in the corpus callosum of a mouse treated with vehicle (left) or BLZ945 for 5 consecutive days. Scale bar 50 μm
These data collectively suggest that microglia have an important role in the maintenance of OPC numbers already after a very short timeframe of microglia removal in the healthy adult mouse brain without affecting oligodendrocytes and myelin homeostasis yet.
Discussion
Here, we have described in detail the dynamics and features of a microglia subset that is specifically present in the normal postnatal mouse brain only in the developing white matter regions. We have furthermore identified a unique transcriptional profile of this distinct microglia population compared to cortical microglia and have provided evidence that these cells have no contribution from circulating blood monocytes and share their tissue endogenous precursors with cortical microglia. The “fountain of microglia” showed a distinct transcriptomic signature with some genes such as Clec7a, Spp1, Igf1, Anxa5, Itgax, and Gpnmb that have been mainly described for microglia during aging or in disease models [21, 23]. In addition, the presence of early postnatal microglia was required for the proper induction of oligodendrocyte progenitors and subsequent myelination. Furthermore, OPC homeostasis in the adult brain was also found to be dependent on the presence of functional microglia. These data also suggest direct effects of microglia on the oligodendrocyte lineage as we found CSF-1R mRNA expression on FACS-sorted microglia only and not on oligodendrocyte progenitors, consistent with the reported absence of CSF-1R expression on oligodendrocytes [42].
These data are of great significance, because they provide detailed in vivo evidence for an active interaction between microglia and OPCs during normal development and in the steady state CNS. Especially during development, this interaction seems to be required for proper myelinogenesis. Despite the fact that the early postnatal microglia are known to be densely populated in neurogenic niches and in the subventricular zone their role for oligodendrogenesis was only hypothesized thus far [65]. Earlier in vitro studies proposed that microglia might be able to differentiate neural precursor cells into oligodendrocytes especially in the presence of interleukin-4 [4].
Myelin is formed by oligodendrocytes as a multilamellar structure that encloses segments of axons in the CNS of vertebrates [43]. Subtle changes in myelin protein levels have an impact on neurodegeneration and neuropsychiatric diseases, which makes it even more important to understand the underlying mechanisms [19, 49]. A mutual interaction of microglia and oligodendrocytes in the adult rodent brain was described recently when a key role of microglia clearance has been shown for the process of age-dependent myelin fragmentation [61]. In this scenario, blockade of microglial-specific phagocytosis through inhibition of Rab7 resulted in enhanced accumulation of myelin products in microglial endosomes. It is furthermore widely accepted that microglia are involved in myelin debris clearance during disease conditions, such as cuprizone-induced demyelination [50, 68].
What are the microglia-derived molecules that induce survival of OPCs, their differentiation to oligodendrocytes and myelinogenesis? There are some candidate molecules that are produced by early postnatal microglia such as insulin-like growth factor-1 (IGF-1). IGF-1 not only plays a role in regulating the survival of layer V neurons during postnatal development, but it has been further shown to inhibit oligodendrocyte apoptosis during cuprizone-induced demyelination and thereby promotes remyelination [31, 70]. It also directly acts on oligodendrocytes and myelination, as demonstrated in oligodendrocyte lineage-specific knockout mice of the IGF-1R [75]. In contrast, another study excluded IGF-1 as a main microglia-linked factor, because IGF-1 expression was not changed upon minocycline treatment that is thought to decrease microglia activation [65]. Therefore, to identify the underlying molecular mechanism will be of particular interest in future studies. Preliminary studies in mice have shown that although mature oligodendrocytes do not express the CSF-1R, in CSF-1R deficiency, they are significantly reduced in cortical regions, including the corpus callosum. In addition, cultured postnatal cortical neural progenitors could only exhibit increased oligodendrocyte differentiation in response to CSF-1 if microglia were present [5, 42].
Our unexpected finding of microglia involvement in the maintenance of OPC numbers as well as the consequences of a reduced OPC number on myelin development is also mirrored by the fact that several classical “microgliopathies” described in patients display severe white matter changes. These include sclerosing leukencephalopathy in Nasu–Hakola disease [44], diffuse leukencephalopathy in HDLS [55], or in USP18 deficiency [34]. These changes were so far considered to be secondary to neuronal damage upon microglia dysfunction but might now also be triggered by a primary oligodendrocyte loss in patients with mutated microglia. Interestingly, CSF-1R mutant mice display severe destruction of the cerebral cortex that is accompanied by a significant reduction of NogoA+ oligodendrocytes and reactive astrogliosis but unaltered number of cortical neurons [10]. Moreover, our data show a reduced number of OPCs as well as a diminished number of CC1+ oligodendrocytes in the corpus callosum of CSF-1R mutant mice. Earlier pharmacological inhibition of the CSF-1R, however, failed to detect a reduction of the myelin compartment that is possibly due to the fact that only one myelin gene was investigated [8].
In summary, our findings substantially increase the understanding of microglia functions in the healthy brain to include the homeostatic regulation of oligodendrocyte progenitors and the myelin compartment of the CNS. Further experiments are required to clarify the role of the precise signals derived from the “fountain of microglia” population. In addition, potential differences in the influence of these cells on the oligodendrocyte lineage between developmental myelinogenesis and myelin turnover and maintenance in the adult brain need to be further characterized. These will pinpoint the precise interaction partners of the microglia–oligodendrocyte lineage axis that could inform new therapeutic approaches.
Supplementary Material
Acknowledgments
We thank Margarethe Ditter, Maria Oberle, Dr. Alexandra Müller, and Katrin Seidel for excellent technical assistance, Stefan Bohlen for the Imaris reconstructions, Antigoni Trian-tafyllopoulou for providing Nr4a1−/− mice, Hauke Werner and Sandra Goebbels for providing NG2YFP/WT mice, Peter Wieghofer for establishing microglia depleting experiments using BLZ945, Hans Christian Probst for providing Cd11cCreER mice, Mathias Jucker for the Ccr2RFP/WT mice, and Jaclyn Wamsteeker Cusulin for critically reading and editing the manuscript. We apologize to colleagues whose work could not be cited because of space constraints. M.P. is supported by the BMBF-funded competence network of multiple sclerosis (KKNMS), the Sobek-Stiftung, the DFG (SFB 992, SFB1140, SFB/TRR167, Reinhart-Koselleck-Grant), the ERA-Net NEURON initiative “NEURO-IFN”, and the Sonderlinie Hochschulmedizin, project “neuroinflammation in neurodegeneration”. ESP and ERS are supported by NIH grant R01 NS091519.
Footnotes
Geo accession numbers The main GEO accession number for microarray: GSE96623.
Author contribution
NH designed and conducted the experiments. KH performed immunofluorescent analysis related to the cell number during development and proliferation. MAA performed analysis of electron microscope data. NU performed experiments with Sox10-iCreERT2 × CAG-eGFP mice and OPC cultures. ESP provided Csf1r−/− material. LD and ERS provided scientific input and edited the manuscript. OS analyzed the microarray data. NH and MP supervised the project and wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material The online version of this article (doi:10.1007/s00401-017-1747-1) contains supplementary material, which is available to authorized users.
References
- 1.Amit I, Winter DR, Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol. 2016;17:18–25. doi: 10.1038/ni.3325. [DOI] [PubMed] [Google Scholar]
- 2.Banisadr G, Frederick TJ, Freitag C, Ren D, Jung H, Miller SD, Miller RJ. The role of CXCR4 signaling in the migration of transplanted oligodendrocyte progenitors into the cerebral white matter. Neurobiol Dis. 2011;44:19–27. doi: 10.1016/j.nbd.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruttger J, Karram K, Wörtge S, Regen T, Marini F, Hoppmann N, Klein M, Blank T, Yona S, Wolf Y, Mack M, Pinteaux E, Müller W, Zipp F, Binder H, Bopp T, Prinz M, Jung S, Waisman A. Genetic cell ablation reveals clusters of local self-renewing microglia in the mammalian central nervous system. Immunity. 2015;43:92–106. doi: 10.1016/j.immuni.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Butovsky O, Ziv Y, Schwartz A, Landa G, Talpalar AE, Pluchino S, Martino G, Schwartz M. Microglia activated by IL-4 or IFN-γ differentially induce neurogenesis and oligodendrogenesis from adult stem/progenitor cells. Mol Cell Neurosci. 2006;31:149–160. doi: 10.1016/j.mcn.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Chitu V, Gokhan S, Nandi S, Mehler MF, Stanley ER. Emerging roles for CSF-1 receptor and its ligands in the nervous system. Trends Neurosci. 2016;39:378–393. doi: 10.1016/j.tins.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colonna M, Butovsky O. Microglia function in the central nervous system during health and neurodegeneration. Annu Rev Immunol. 2016 doi: 10.1146/annurev-immunol-051116-052358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Jager PL, Jia X, Wang J, de Bakker PIW, Ottoboni L, Aggarwal NT, Piccio L, Raychaudhuri S, Tran D, Aubin C, Briskin R, Romano S, Baranzini SE, McCauley JL, Pericak-Vance MA, Haines JL, Gibson RA, Naeglin Y, Uitdehaag B, Matthews PM, Kappos L, Polman C, McArdle WL, Strachan DP, Evans D, Cross AH, Daly MJ, Compston A, Sawcer SJ, Weiner HL, Hauser SL, Hafler DA, Oksenberg JR. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elmore MRP, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, Kitazawa M, Matusow B, Nguyen H, West BL, Green KN. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–397. doi: 10.1016/j.neuron.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ensan S, Li A, Besla R, Degousee N, Cosme J, Roufaiel M, Shikatani EA, El-Maklizi M, Williams JW, Robins L, Li C, Lewis B, Yun TJ, Lee JS, Wieghofer P, Khattar R, Farrokhi K, Byrne J, Ouzounian M, Zavitz CCJ, Levy GA, Bauer CMT, Libby P, Husain M, Swirski FK, Cheong C, Prinz M, Hilgendorf I, Randolph GJ, Epelman S, Gramolini AO, Cybulsky MI, Rubin BB, Robbins CS. Self-renewing resident arterial macrophages arise from embryonic CX3CR1+ precursors and circulating monocytes immediately after birth. Nat Immunol. 2016;17:159–168. doi: 10.1038/ni.3343. [DOI] [PubMed] [Google Scholar]
- 10.Erblich B, Zhu L, Etgen AM, Dobrenis K, Pollard JW. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One. 2011;6:e26317. doi: 10.1371/journal.pone.0026317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, Schwierzeck V, Utermohlen O, Chun E, Garrett WS, McCoy KD, Diefenbach A, Staeheli P, Stecher B, Amit I, Prinz M. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank S, Burbach GJ, Bonin M, Walter M, Streit W, Bechmann I, Deller T. TREM2 is upregulated in amyloid plaque-associated microglia in aged APP23 transgenic mice. Glia. 2008;56:1438–1447. doi: 10.1002/glia.20710. [DOI] [PubMed] [Google Scholar]
- 13.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldmann T, Wieghofer P, Jordao MJC, Prutek F, Hagemeyer N, Frenzel K, Amann L, Staszewski O, Kierdorf K, Krueger M, Locatelli G, Hochgerner H, Zeiser R, Epelman S, Geissmann F, Priller J, Rossi FMV, Bechmann I, Kerschensteiner M, Linnarsson S, Jung S, Prinz M. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol. 2016;17:797–805. doi: 10.1038/ni.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldmann T, Zeller N, Raasch J, Kierdorf K, Frenzel K, Ketscher L, Basters A, Staszewski O, Brendecke SM, Spiess A, Tay TL, Kreutz C, Timmer J, Mancini GM, Blank T, Fritz G, Biber K, Lang R, Malo D, Merkler D, Heikenwälder M, Knobeloch K-P, Prinz M. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J. 2015;34:1612–1629. doi: 10.15252/embj.201490791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK, Stevens MP, Freeman TC, Summers KM, McColl BW. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci. 2016;19:504–516. doi: 10.1038/nn.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, Hooli B, Choi SH, Hyman BT, Tanzi RE. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 2013;78:631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JSK, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert J-C, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J. TREM2 Variants in Alzheimer’s Disease. N Engl J Med. 2012;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemeyer N, Goebbels S, Papiol S, Kästner A, Hofer S, Begemann M, Gerwig UC, Boretius S, Wieser GL, Ronnenberg A, Gurvich A, Heckers SH, Frahm J, Nave K-A, Ehrenreich H. A myelin gene causative of a catatonia-depression syndrome upon aging. EMBO Mol Med. 2012;4:528–539. doi: 10.1002/emmm.201200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagemeyer N, Kierdorf K, Frenzel K, Xue J, Ringelhan M, Abdullah Z, Godin I, Wieghofer P, Costa Jordão MJ, Ulas T, Yorgancioglu G, Rosenbauer F, Knolle PA, Heikenwalder M, Schultze JL, Prinz M. Transcriptome-based profiling of yolk sac-derived macrophages reveals a role for Irf8 in macrophage maturation. EMBO J. 2016;35:1730–1744. doi: 10.15252/embj.201693801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickman SE, Kingery ND, Ohsumi T, Borowsky M, Wang L, Means TK, Khoury JE. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holtman IR, Raj DD, Miller JA, Schaafsma W, Yin Z, Brouwer N, Wes PD, Möller T, Orre M, Kamphuis W, Hol EM, Boddeke EWGM, Eggen BJL. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: a co-expression meta-analysis. Acta Neuropathol Commun. 2015;3:31. doi: 10.1186/s40478-015-0203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hristova M, Cuthill D, Zbarsky V, Acosta-Saltos A, Wallace A, Blight K, Buckley SMK, Peebles D, Heuer H, Waddington SN, Raivich G. Activation and deactivation of periventricular white matter phagocytes during postnatal mouse development. Glia. 2010;58:11–28. doi: 10.1002/glia.20896. [DOI] [PubMed] [Google Scholar]
- 25.Imamoto K, Leblond CP. Radioautographic investigation of gliogenesis in the corpus callosum of young rats II. Origin of microglial cells. J Comp Neurol. 1978;180:139–163. doi: 10.1002/cne.901800109. [DOI] [PubMed] [Google Scholar]
- 26.Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karram K, Goebbels S, Schwab M, Jennissen K, Seifert G, Steinhäuser C, Nave K-A, Trotter J. NG2-expressing cells in the nervous system revealed by the NG2-EYFP-knockin mouse. Genesis. 2008;46:743–757. doi: 10.1002/dvg.20440. [DOI] [PubMed] [Google Scholar]
- 28.Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Holscher C, Muller DN, Luckow B, Brocker T, Debowski K, Fritz G, Opdenakker G, Diefenbach A, Biber K, Heikenwalder M, Geissmann F, Rosenbauer F, Prinz M. Microglia emerge from erythro-myeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 29.Kigerl KA, Gensel JC, Ankeny DP, Alexander JK, Donnelly DJ, Popovich PG. Identification of two distinct macrophage subsets with divergent effects causing either neurotoxicity or regeneration in the injured mouse spinal cord. J Neurosci. 2009;29:13435. doi: 10.1523/JNEUROSCI.3257-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ling EA. Transformation of monocytes into amoeboid microglia in the corpus callosum of postnatal rats, as shown by labelling monocytes by carbon particles. J Anat. 1979;128:847–858. [PMC free article] [PubMed] [Google Scholar]
- 31.Mason JL, Jones JJ, Taniike M, Morell P, Suzuki K, Matsushima GK. Mature oligodendrocyte apoptosis precedes IGF-1 production and oligodendrocyte progenitor accumulation and differentiation during demyelination/remyelination. J Neurosci Res. 2000;61:251–262. doi: 10.1002/1097-4547(20000801)61:3<251:AID-JNR3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 32.Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada González F, Perrin P, Keren-Shaul H, Gury M, Lara-Astaiso D, Thaiss CA, Cohen M, Bahar Halpern K, Baruch K, Deczkowska A, Lorenzo-Vivas E, Itzkovitz S, Elinav E, Sieweke MH, Schwartz M, Amit I. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016 doi: 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- 33.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 34.Meuwissen MEC, Schot R, Buta S, Oudesluijs G, Tinschert S, Speer SD, Li Z, van Unen L, Heijsman D, Goldmann T, Lequin MH, Kros JM, Stam W, Hermann M, Willemsen R, Brouwer RWW, Van Ijcken WFJ, Martin-Fernandez M, de Coo I, Dudink J, de Vries FAT, Bertoli Avella A, Prinz M, Crow YJ, Verheijen FW, Pellegrini S, Bogunovic D, Mancini GMS. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J Exp Med. 2016;213:1163. doi: 10.1084/jem.20151529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mildner A, Mack M, Schmidt H, Brück W, Djukic M, Zabel MD, Hille A, Priller J, Prinz M. CCR2+ Ly-6Chi monocytes are crucial for the effector phase of autoimmunity in the central nervous system. Brain. 2009;132:2487–2500. doi: 10.1093/brain/awp144. [DOI] [PubMed] [Google Scholar]
- 36.Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch U-K, Mack M, Heikenwalder M, Bruck W, Priller J, Prinz M. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 37.Miron VE, Boyd A, Zhao J-W, Yuen TJ, Ruckh JM, Shadrach JL, van Wijngaarden P, Wagers AJ, Williams A, Franklin RJ, ffrench-Constant C. M2 microglia/macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat Neurosci. 2013;16:1211–1218. doi: 10.1038/nn.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizutani M, Pino PA, Saederup N, Charo IF, Ransohoff RM, Cardona AE. The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood. J Immunol. 2011;188:29. doi: 10.4049/jimmunol.1100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molawi K, Wolf Y, Kandalla PK, Favret J, Hagemeyer N, Frenzel K, Pinto AR, Klapproth K, Henri S, Malissen B, Rodewald H-R, Rosenthal NA, Bajenoff M, Prinz M, Jung S, Sieweke MH. Progressive replacement of embryo-derived cardiac macrophages with age. J Exp Med. 2014;211:2151–2158. doi: 10.1084/jem.20140639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murtagh F, Legendre P. Ward’s hierarchical agglomerative clustering method: which algorithms implement ward’s criterion? J Classif. 2014;31:274–295. doi: 10.1007/s00357-014-9161-z. [DOI] [Google Scholar]
- 41.Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nandi S, Gokhan S, Dai X-M, Wei S, Enikolopov G, Lin H, Mehler MF, Stanley ER. The CSF-1 receptor ligands IL-34 and CSF-1 exhibit distinct developmental brain expression patterns and regulate neural progenitor cell maintenance and maturation. Dev Biol. 2012;367:100–113. doi: 10.1016/j.ydbio.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nave K-A, Werner HB. Myelination of the nervous system: mechanisms and functions. Annu Rev Cell Dev Biol. 2014;30:503–533. doi: 10.1146/annurev-cellbio-100913-013101. [DOI] [PubMed] [Google Scholar]
- 44.Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, Bianchin M, Bird T, Miranda R, Salmaggi A, Tranebjærg L, Konttinen Y, Peltonen L. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71:656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, Ragozzino D, Gross CT. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 46.Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, III, Lafaille JJ, Hempstead BL, Littman DR, Gan W-B. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel JR, McCandless EE, Dorsey D, Klein RS. CXCR4 promotes differentiation of oligodendrocyte progenitors and remyelination. Proc Natl Acad Sci USA. 2010;107:11062–11067. doi: 10.1073/pnas.1006301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peri F, Nüsslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133:916–927. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- 49.Poggi G, Boretius S, Möbius W, Moschny N, Baudewig J, Ruhwedel T, Hassouna I, Wieser GL, Werner HB, Goebbels S, Nave K, Ehrenreich H. Cortical network dysfunction caused by a subtle defect of myelination. Glia. 2016;64:2025–2040. doi: 10.1002/glia.23039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poliani PL, Wang Y, Fontana E, Robinette ML, Yamanishi Y, Gilfillan S, Colonna M. TREM2 sustains microglial expansion during aging and response to demyelination. J Clin Invest. 2015;125:2161–2170. doi: 10.1172/JCI77983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prinz M, Erny D, Hagemeyer N. Ontogeny and homeostasis of CNS myeloid cells. Nat Immunol. 2017;18:385. doi: 10.1038/ni.3703. [DOI] [PubMed] [Google Scholar]
- 52.Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15:300–312. doi: 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- 53.Prinz M, Priller J. The role of peripheral immune cells in the CNS in steady state and disease. Nat Neurosci. 2017;20:136–144. doi: 10.1038/nn.4475. [DOI] [PubMed] [Google Scholar]
- 54.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: [Google Scholar]
- 55.Rademakers R, Baker M, Nicholson AM, Rutherford NJ, Finch N, Soto-Ortolaza A, Lash J, Wider C, Wojtas A, DeJesus-Hernandez M, Adamson J, Kouri N, Sundal C, Shuster EA, Aasly J, MacKenzie J, Roeber S, Kretzschmar HA, Boeve BF, Knopman DS, Petersen RC, Cairns NJ, Ghetti B, Spina S, Garbern J, Tselis AC, Uitti R, Das P, Van Gerpen JA, Meschia JF, Levy S, Broderick DF, Graff-Radford N, Ross OA, Miller BB, Swerdlow RH, Dickson DW, Wszolek ZK. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat Genet. 2012;44:200–205. doi: 10.1038/ng.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ransohoff RM, Perry Hugh H. Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- 57.Rezaie P, Dean A. Periventricular leukomalacia, inflammation and white matter lesions within the developing nervous system. Neuropathology. 2002;22:106–132. doi: 10.1046/j.1440-1789.2002.00438.x. [DOI] [PubMed] [Google Scholar]
- 58.Rio-Hortega P. Microglia. Cytol Cell Pathol Nerv Syst. 1932;2:482–534. [Google Scholar]
- 59.Roumier A, Béchade C, Poncer J-C, Smalla K-H, Tomasello E, Vivier E, Gundelfinger ED, Triller A, Bessis A. Impaired synaptic function in the microglial KARAP/DAP12-deficient mouse. J Neurosci. 2004;24:11421. doi: 10.1523/JNEUROSCI.2251-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saederup N, Cardona AE, Croft K, Mizutani M, Cotleur AC, Tsou C-L, Ransohoff RM, Charo IF. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS One. 2010;5:e13693. doi: 10.1371/journal.pone.0013693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Safaiyan S, Kannaiyan N, Snaidero N, Brioschi S, Biber K, Yona S, Edinger AL, Jung S, Rossner MJ, Simons M. Age-related myelin degradation burdens the clearance function of microglia during aging. Nat Neurosci. 2016;19:995–998. doi: 10.1038/nn.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheffel J, Regen T, Van Rossum D, Seifert S, Ribes S, Nau R, Parsa R, Harris RA, Boddeke HWGM, Chuang H-N, Pukrop T, Wessels JT, Jürgens T, Merkler D, Brück W, Schnaars M, Simons M, Kettenmann H, Hanisch U-K. Toll-like receptor activation reveals developmental reorganization and unmasks responder subsets of microglia. Glia. 2012;60:1930–1943. doi: 10.1002/glia.22409. [DOI] [PubMed] [Google Scholar]
- 64.Schulz C, Perdiguero EG, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SEW, Pollard JW, Frampton J, Liu KJ, Geissmann F. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 65.Shigemoto-Mogami Y, Hoshikawa K, Goldman JE, Sekino Y, Sato K. Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J Neurosci. 2014;34:2231–2243. doi: 10.1523/JNEUROSCI.1619-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sierra A, Encinas JM, Deudero JJP, Chancey JH, Enikolopov G, Overstreet-Wadiche LS, Tsirka SE, Maletic-Savatic M. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell. 2010;7:483–495. doi: 10.1016/j.stem.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simon C, Lickert H, Götz M, Dimou L. Sox10-iCreERT2: a mouse line to inducibly trace the neural crest and oligodendrocyte lineage. Genesis. 2012;50:506–515. doi: 10.1002/dvg.22003. [DOI] [PubMed] [Google Scholar]
- 68.Skripuletz T, Miller E, Moharregh-Khiabani D, Blank A, Pul R, Gudi V, Trebst C, Stangel M. Beneficial effects of minocycline on cuprizone induced cortical demyelination. Neurochem Res. 2010;35:1422–1433. doi: 10.1007/s11064-010-0202-7. [DOI] [PubMed] [Google Scholar]
- 69.Tremblay M-È, Lowery RL, Majewska AK. Microglial interactions with synapses are modulated by visual experience. PLoS Biol. 2010;8:e1000527. doi: 10.1371/journal.pbio.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, Yamashita T. Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci. 2013;16:543–551. doi: 10.1038/nn.3358. [DOI] [PubMed] [Google Scholar]
- 71.Viganò F, Schneider S, Cimino M, Bonfanti E, Gelosa P, Sironi L, Abbracchio MP, Dimou L. GPR17 expressing NG2-Glia: oligodendrocyte progenitors serving as a reserve pool after injury. Glia. 2016;64:287–299. doi: 10.1002/glia.22929. [DOI] [PubMed] [Google Scholar]
- 72.Wake H, Moorhouse AJ, Jinno S, Kohsaka S, Nabekura J. Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J Neurosci. 2009;29:3974. doi: 10.1523/JNEUROSCI.4363-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ward JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;69:236–244. [Google Scholar]
- 74.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B. R package version 3.0.1. 2016. gplots: various R programming tools for plotting data. [Google Scholar]
- 75.Zeger M, Popken G, Zhang J, Xuan S, Lu QR, Schwab MH, Nave K-A, Rowitch D, D’Ercole AJ, Ye P. Insulin-like growth factor type 1 receptor signaling in the cells of oligodendrocyte lineage is required for normal in vivo oligodendrocyte development and myelination. Glia. 2007;55:400–411. doi: 10.1002/glia.20469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.