Abstract
Sebaceous gland carcinoma (SGC) is a rare, but life-threatening condition with a predilection for the periocular region. Eyelid SGC can be broadly categorised into two subtypes, namely either nodular or pagetoid with the latter being more aggressive and requiring radical excision to save life. We have identified key altered microRNAs (miRNA) involved in SGC shared by both subtypes, hsa-miR-34a-5p and hsa-miR-16-5p. However, their gene targets BCL2 and MYC were differentially expressed with both overexpressed in pagetoid but unchanged in nodular suggesting different modes of action of these two miRNAs on BCL/MYC expression. Hsa-miR-150p is nodular-specifically overexpressed, and its target ZEB1 was significantly downregulated in nodular SGC suggesting a tumour suppressor role. Invasive pagetoid subtype demonstrated specific overexpression of hsa-miR-205 and downregulation of hsa-miR-199a. Correspondingly, miRNA gene targets, EZH2 (by hsa-miR-205) and CD44 (by hsa-miR-199a), were both overexpressed in pagetoid SGC. CD44 has been identified as a potential cancer stem cell marker in head and neck squamous cell carcinoma and its overexpression in pagetoid cells represents a novel treatment target. Aberrant miRNAs and their gene targets have been identified in both SGC subtypes, paving the way for better molecular understanding of these tumours and identifying new treatment targets.
Introduction
Sebaceous gland carcinoma (SGC) is a rare, aggressive cancer that has a predilection for the periorbital region, perhaps due to the multitude of glands surrounding the globe, but can come from extraocular sites, albeit mainly within the head region1. Geographical variation is significant with the incidence around 0.65 per 100,000 in Canada versus China where it represents almost a third of the malignant eyelid workload and second to BCC in frequency2,3. In Japan, the rate of SGC equalled that of BCC in one study4. Risk factors for developing SGC include radiation exposure; for example external beam radiotherapy which used to be the mainstay of treatment for retinoblastoma5. More aggressive features include vascular and lymphatic invasion, orbital invasion, involvement of both upper and lower eyelids, poor differentiation, multicentric origin, diameter greater than 10 mm, infiltrative growth pattern and pagetoid invasion of the adjacent epithelium6. SGC can be broadly divided in to two subtypes with highly contrasting prognosis, namely: nodular and pagetoid7. The nodular form often presents as a discrete mass on the eyelid and is more amenable to curative surgical resection whereas the pagetoid (intraepitheial spread on histology) subtype presents as non-specific thickening or redness of the eye and often contains skip lesions that requires more mutilating surgery to achieve complete clearance8. Surgical excision is currently the only available treatment and this may require removal of a normal seeing eye (exenteration) to protect life, thus a better understanding of its molecular biology along with identification of alternative treatment modalities is needed9,10.
Little is known about the molecular factors involved in SGC tumorigenesis although recent targeted whole exome sequencing of periocular SGC has revealed mutations in TP53 and RB1 genes11. Four differentially expressed microRNA (miRNA) have been identified in SGC (hsa-miR-486-5p and -184 upregulated; hsa-miR-211 and -518d downregulated), however, these miRNAs were compared to sebaceous adenoma rather than normal tissue and no histological subtype analysis was made12. Furthermore, progression from sebaceous adenoma to carcinoma is unknown, unlike the transition from colonic adenoma to adenocarcinoma, confounding the discovery of real SGC tumorigenesis markers.
SGC prognosis is dependent on the subtype, with pagetoid conferring a poorer prognosis and nodular a better prognosis8. In this study we assess periocular SGC whole-miRNA profile normalised to eyelid tarsal plate, the presumed tissue of origin. We have identified aberrantly expressed miRNAs unique to pagetoid and nodular SGC along with shared aberrant miRNAs. Subsequently, a combination of in-silico miRNA target search and transcriptome profiling, as well as miRNA-target gene network analysis, was performed to highlight miRNA targets in both subtypes.
Results and Discussion
Sebaceous gland carcinoma is a rare, but aggressive cancer with surgical excision as the only option for definitive treatment. In order to carry out complete excision a margin of normal tissue needs to be removed and verified using histological examination. Certain subtypes of SGC present with a local mass, termed nodular SGC and is more amenable to local resection (see Supplementary Fig. S1A and S1C), as a result confer a better prognosis. In contrast, diffuse lid swelling which demonstrates intraepithelial spread on histological and sometimes skip lesions do not demonstrate clear margins for wide local excision, thus more aggressive excision of the orbital content is required resulting in a poorer prognosis (see Supplementary Fig. S1B and S1D). However, little is known about the molecular biology of these tumours and the relationship between SGC miRNA profile, transcriptome and its cancer behaviour including the differences exhibited by the nodular and pagetoid subtypes.
Common SGC miRNA in both nodular and pagetoid subtype with target gene expression
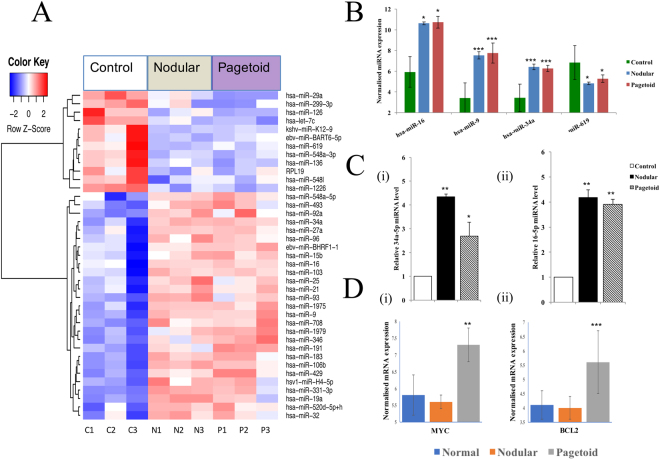
Thirty-nine differentially expressed (DE) miRNA were common to both subtypes with the majority being upregulated (Fig. 1A). The top 4 DE miRNAs are shown in Fig. 1B (see Supplementary Table S1).
Figure 1.
Shared microRNAs in nodular and pagetoid sebaceous gland carcinoma (SGC). (A) Thirty-nine significant differentially expressed microRNA shared between both subtypes using a p < 0.05 threshold. (B) Top 4 differentially expressed miRNAs present in both nodular and pagetoid SGC when compared to tarsal plate control. (C) MicroRNA verification of expression using Taqman RT-qPCR in SGC. Relative expression levels were determined for nodular and pagetoid SGC using Taqman RT-qPCR against normal eyelid tissue for miRNA (i) 34a-5p and (ii) 16-5p. (D) Expression of target genes in both nodular and pagetoid subtypes. Significance levels are shown as *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent mean +/− s.d.
Hsa-miR-34a was significantly overexpressed in both subtypes compared to the control (Fig. 1B,C). It forms part of the TP53 suppressor network by modulating TP53 targets and forms a positive feedback loop via SIRT113. Despite the common overexpression of hsa-miR-34a in both subtypes, the action of the miRNA on its target gene is different. For example, the target gene MYC was significantly overexpressed in the pagetoid subtype, but remaining unchanged in nodular (Fig. 1D). Interestingly in MYC-driven tumours, hsa-miR-34a improves cell survival based on its ability to reduce TP53 levels, in a MYC mediated and dependent fashion, hence helps the cancer cells to survive14. As MYC is over-expressed in the pagetoid subtype, our results suggest that hsa-miR-34a may play a tumour activating and enhancing role, but not the case in nodular SGC where the expression of MYC was unchanged compared to normal control (Fig. 1D). Furthermore, there is a known synergistic action between BCL2, also a target of hsa-miR-34a, and MYC in oncogenesis and this appears to be occurring in the pagetoid form only where both were significantly overexpressed (Fig. 1D)15. Hsa-miR-16 also targets BCL2 and was similarly overexpressed in both subtypes (Fig. 1B,C). Moreover, hsa-miR-16 has been shown to suppress both BCL2 and FOXO1, the latter being under-expressed in nodular SGC only16. It may therefore be acting in a similar opposing fashion as hsa-miR-34 and behave with a different mode of action according to the MYC and BCL2 status of the two subtypes. Hsa-mir-9, a known tumour suppressor, is overexpressed in both subtypes, and together with hsa-miR-619 (downregulated in both subtypes), targets MDGA2, a tumour suppressor gene which is highlighted in our common SGC miRNA-target gene network shown in Supplementary Fig. S217,18. Both hsa-miR-126 and -9 target chemokine receptor CXCR4 which plays a role in metastatic spread19,20.
Nodular SGC specific microRNA with target gene expression
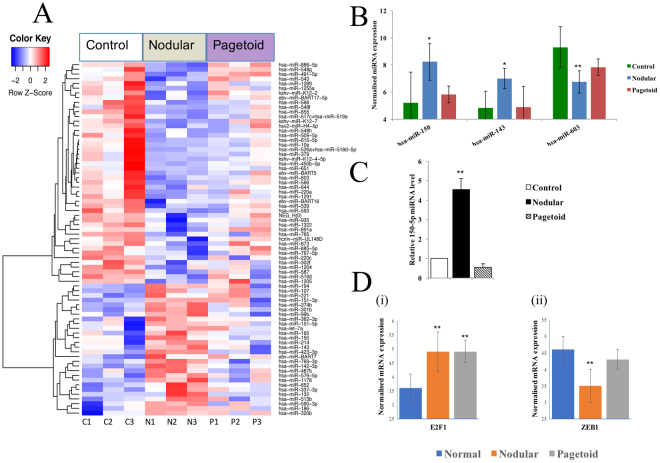
Seventy-five DE genes were found to be unique to nodular SGC compared to control and pagetoid (Fig. 2A). The majority of these (63%) were downregulated and the top 3 nodular specific miRNA are shown in Fig. 2B (see Supplementary Table S1).
Figure 2.
Nodular sebaceous gland carcinoma specific microRNAs. (A) Seventy-five significant differentially expressed microRNA unique to nodular SGC using a p < 0.05 threshold. (B) Top 3 differentially expressed miRNAs present in nodular SGC only when compared to tarsal plate control. Pagetoid expression shown to demonstrate non-significant change within the subtype. (C) MicroRNA verification of 150-5p expression using Taqman RT-qPCR in SGC. Relative expression levels were determined for nodular and pagetoid SGC using Taqman RT-qPCR against normal eyelid tissue for miRNA 150-5p. (D) Expression of target genes in nodular and pagetoid subtype for hsa-miR-150-5p. Significance levels are shown as *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent mean +/− s.d.
Hsa-miR-150 was the most overexpressed miRNA unique to nodular SGC, with more than an 8-fold change (Fig. 2B,C) compared to control. Tumour suppressor action of hsa-miR-150 has been noted in epithelial ovarian cancer and shown to reduce invasion and metastasis by suppressing the transcriptional repressor ZEB1 which was significantly downregulated in nodular SGC (Fig. 2D)21. Hsa-miR-143 was also found significantly upregulated specifically to the nodular subtype (Fig. 2B) and known for its tumour suppressive action22. It is likely to be acting through suppression of BCL2, which is also targeted by the aforementioned hsa-miR-16 and -34a, helping to prevent cancer progression. In contrast, BCL2 is significantly overexpressed in the pagetoid subtype and as an important inhibitor of apoptosis, confers an advantage to the pagetoid subtype, however, BCL2 also represents a potential novel treatment target.23 Oncomir hsa-miR-603 was found to be much more downregulated in nodular than in pagetoid subtype (Fig. 2B) and we propose it is attempting to regulate of E2F1 in a negatively correlated fashion which may contribute to nodular SGC less aggressive behaviour (Supplementary Fig. S3)24,25. A miRNA-target gene network of the proposed regulatory interactions seen in nodular SGC is shown in Supplementary Fig. S3. The MAPK/ERK pathway is intimately linked to the altered miRNAs seen in nodular SGC and it is possible that these miRNAs are acting as tumour suppressors to inhibit this pathway26,27. Moreover, synthetic miR-143 has been shown to silence KRAS signalling including its effector signalling molecules AKT and ERK (MAPK1).28 Furthermore, high expression of hsa-miR-150 specifically occurs in cancer patients and has the potential to be a biomarker of disease29.
Pagetoid specific microRNA with target gene expression
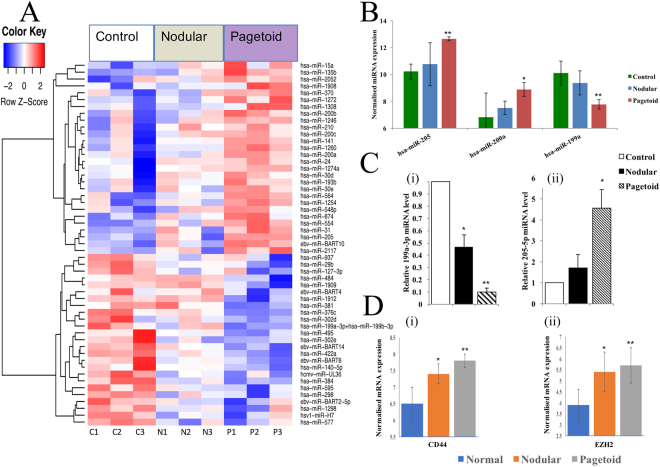
Pagetoid SGC expressed 53 specific, significantly DE genes (Fig. 3A). 47% of them were downregulated and the top 3 pagetoid specific genes are shown in Fig. 3B (see Supplementary Table S1).
Figure 3.
Pagetoid sebaceous gland carcinoma specific microRNAs. (A) Fifty-three significant differentially expressed microRNAs unique to nodular SGC using a p < 0.05 threshold. (B) Top 3 differentially expressed genes present in pagetoid SGC only when compared to tarsal plate. Nodular expression shown to demonstrate non-significant change within the subtype. (C) MicroRNA expression using Taqman RT-qPCR in SGC. Relative expression levels were determined for nodular and pagetoid SGC using Taqman RT-qPCR against normal eyelid tissue for miRNA (i) 199a-3p, (ii) 205-5p. (D) Expression of target genes in nodular and pagetoid subtype for hsa-miR-199a-3p and hsa-miR-205-5p. Significance levels are shown as *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent mean +/− s.d.
Significantly increased level of hsa-miR-205 was found to be unique to pagetoid SGC and overexpression promotes VEGF invasion of ovarian cancer cells by targeting EZR and LMNA (Fig. 3B,C)30. Higher levels of hsa-miR-205 are also associated with adverse clinical outcome in bladder and biliary cancer through its action on ZEB1 by maintaining an epithelial phenotype31,32. Upregulation of hsa-miR-205 also occurs in non-squamous cell lung cancer and directly represses PTEN (targeted by multiple pagetoid specific differentially expressed miRNAs, including hsa-miR-205 and hsa-miR-200a – see Supplementary Fig. S4) and PHLPP2 expression, which in turn activates the AKT/FOXO3a and AKT/mTOR pathways respectively33. Moreover, an interaction between hsa-miR-205 and overexpressed EZH2 occurs in renal cell carcinoma to control b-catenin, and indeed EZH2 was found to be significantly overexpressed in pagetoid SGC (Fig. 3D)34.
Hsa-miR-200a, also a pagetoid specific upregulated miRNA, is involved in EMT too, but works together with hsa-miR-141 (upregulated in pagetoid only) to target MAPK14 to enhance the oxidative stress tumour growth response in ovarian cancer and effectively behaving as oncomiRs (Fig. 3B and see Supplementary Fig. S4).35
Hsa-miR-199 acts as a tumour suppressor gene in several cancers including colorectal and thyroid cancer36,37. Indeed, it was found to be the most downregulated miRNA unique to pagetoid SGC (Fig. 3B,C). Hsa-miR-199 has been shown to target CD44 glycoprotein, an oncoprotein that aids in cell adhesion and migration, with increased CD44 expression contributing to the aggressive nature of tumours and represents a therapeutic target plus possible stem cell biomarker38,39. This inverse association has also been noted in hepatocellular carcinoma40. CD44 was found to be significantly overexpressed in pagetoid subtype, which supports the suppressing role of hsa-miR-199 on CD44 seen in this subtype (Fig. 3). A summary of a pagetoid SGC specific miRNA-target gene network is shown in Supplementary Fig. S4, with common aforementioned gene targets highlighted along with the p53 feedback pathway (MAPK and PTEN).
MiRNA-target gene networks and integrative analysis of sebaceous gland carcinoma
Integrative genomic analysis has been shown to be a powerful tool in identifying cancer subtype-specific highly connected network genes and signatures, which could offer potential subtype-specific drug targets41–43. These kind of studies often benefit from the availability of multi-omics data sets, especially on the same patient samples, such as TCGA studies: for example, in ovary44, lung45 and oesophageal46. The integration of different types of molecular data along with known human signalling plus protein-protein interaction, miRNA-target gene networks can further point us to a set of genes or pathways frequently targeted by various types of alterations and the interplay between them. To further identify robust SGC subtype specific signatures, we will need to generate and curate more multi-omic data sets (such as whole exome sequencing, RNA sequencing, copy number aberration and methylation), however SGC samples are rare to obtain, making it difficult to generate such datasets. More genomic studies involving multiple national and international centres are therefore much desired.
Development of computational models for SGC related miRNA prediction
To further validate the functional significance and disease causal potential for our identified shared and specific miRNAs, curated, known miRNA-disease associations were queried for these miRNAs using HMDD v2.047, MiRCancer48 and MiR2Disease49. These databases collect and record miRNA-disease associations through text mining in literature, manual confirmation, and experimentally supported evidence. Out of all 167 significantly differentially expressed miRNAs in SGC compared to control, ~100 (60%) were reported to be associated with at least one human disease with experiment-supported evidence based on HMDDv2.0. Similar percentages of miRNAs were also reported to have human disease and/or cancer associations based on MiRCancer and MiR2Disease, 60% and 48%, respectively. Thus, more than half of our SGC related miRNAs have been approved experimentally to be involved in other diseases and cancers.
Currently, there are little known, validated or predicted associations between miRNAs and SGC. Thus, it would be interesting to develop a computational framework for their association prediction, in addition to our Nanostring experimental data. This framework often requires first the prediction of miRNA functional similarity, disease semantic similarity, and the validated miRNA-disease associations. Similarity matrices are then integrated with Gaussian kernel similarity to produce the integrated similarity matrices for both miRNAs and diseases. Based on the assumption that functionally similar miRNAs tend to be associated with similar diseases, we can then identity and prioritise potential miRNA-SGC associations using graph-based learning (such as LRSSLMDA50 and PBMDA51), singular value thresholding algorithm (for example MCMDA52), or a within and between score model (for example WBSMDA53). Nevertheless, more studies are still needed to understand this rare and aggressive disease to improve the disease similarity prediction within this computational framework.
Conclusion
We have identified novel aberrant miRNAs in SGC and related them to specific subtypes of SGC with different tumour behaviour to infer their role in carcinogenesis. Concomitantly, we assessed the transcriptome to explore the differential expression of predicted key target genes by these aberrant miRNAs, with the integration of miRNA-target gene networks associated with SGC and its subtypes. Further work is needed to understand the interplay of these novel miRNA in SGC tumour progression and their potential as treatment targets in the future.
Methods
Patients and tumours
This study included eight SGC samples from archival tissue housed at Moorfields Biobank and obtained both institutional (local) review board approval and national research ethics committee approval: Moorfields Eye Hospital Biobank internal research ethics committee agreement (reference: 10/H0106/57-2012ETR28), sponsor approval by Queen Mary University of London (number: 008621GM) and overall approval from the heath research authority, national research ethics service, Committee North West - Greater Manchester South (national research ethics committee number 14/NW/1080). As a result, all methods were carried out in accordance with relevant guidelines and regulations. In addition, all experimental protocols were approved by the aforementioned institutional and national ethical licensing committees. Four samples were pagetoid, 4 were nodular and compared to 4 normal tarsal plate as controls (see Supplementary Table S2). Six SGC were analysed for miRNA and 8 for messenger (mRNA) whilst being compared to tarsal plate control. Informed consent was taken for each patient for study publication and publication of identifying images in an online open access publication.
Micro and messenger RNA extraction
RNeasy® FFPE kit from Qiagen® was used to extract both messenger and microRNA.
A Qubit® 2.0 Fluorometer (invitrogen™ by Life technologies™) and an Agilent 2100 bioanalyzer instrument (Agilent technologies, Inc) was used to determine concentration and integrity of messenger RNA (mRNA).
Nanostring® nCounter®v2 microRNA expression assay and bioinformatics pipeline
Hybridisation of 800 unique oligonucleotide miRNA tags based on the miRBase version 18 reaction was performed. Raw miRNA expression and normalised data were first generated from Nanostring nCounter Digital Analyzer. Filtered data from the Nanostring platform were log2 transformed and further normalised using the quantile normalisation method in R (http://www.r-project.org/). Differential expression (DE) analyses were performed using the limma R package54. Pairwise comparisons of nodular versus control and pagetoid versus control were conducted to identify common and unique DE miRNA to the two subtypes using p < 0.05 (see Supplementary Table S1).
Affymetrix™GeneChip® Human Gene 2.0 messenger RNA expression array and pipeline
Twelve samples were analysed (4 pagetoid, 4 nodular and 4 control) covering 47,000 transcripts. Expression data was normalised using the GC-robust multiarray average55. DE genes were identified using the threshold of p < 0.01 with limma R (see Supplementary Table S1). Data has been deposited to the Gene Expression Omnibus under the accession of GSE101476 (www.ncbi.nlm.nih.gov/geo).
Real-time quantitative reverse transcriptase–PCR (rt-qPCR) for microRNA validation
A library of cDNA from isolated miRNA was made using TaqMan® Advanced miRNA cDNA synthesis kit. Customised oligonucleotides for micoRNA were designed (see Supplementary Table S3) and hsa-miR-26a-5p was chosen as the control (see Supplementary Table S4). A minimum of three replicates for each reaction and relative transcript expression was calculated by the 2−ΔΔCt method and miRNA levels of each gene were normalised to the geometric mean of hsa-miR-26a-5p.
In silico miRNA-target gene network
For selected top differentially expressed miRNAs, their targeted genes were identified in silico using miRNA target prediction databases (www.mirbase.org; www.mirdb.org; www.mirtarbase.mbc.nctu.edu.tw)56–58. The differential expression patterns of these target genes were then explored based on our mRNA expression data. The miRNA-target gene networks were further constructed using Cytoscape v3.3.0 (http://www.cytoscape.org/), with various shapes and colours indicating different attributes of miRNAs and genes.
Accession codes
Data has been deposited to the Gene Expression Omnibus under the accession of GSE101476 (www.ncbi.nlm.nih.gov/geo).
Electronic supplementary material
Supplementary Figure S1–4 and Table S2-S4
Acknowledgements
We gratefully acknowledge the funding for a Fight For Sight Fellowship, UK, and the support of the National Institute for Health Research (NIHR) Biomedical Research Centre based at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology. J.W. and C.C. acknowledge support from Cancer Research UK Centre of Excellence Award to Barts Cancer Centre [C16420/A18066].
Author Contributions
D.G.E. conceived the study. J.C.B., M.B. and D.G.E. collected samples. J.C.B. and C.F. performed miRNA and mRNA extraction. J.C.B. and M.M. carried out rt-qPCR. J.C.B., J.W., A.S., M.M., C.C., E.A.T., M.P.P. carried out data analysis. J.C.B., J.W., M.M., E.A.T., M.P.P. and D.G.E. contributed to the design of the study. All authors reviewed and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
John C. Bladen and Jun Wang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25900-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Deprez M, Uffer S. Clinicopathological features of eyelid skin tumors. A retrospective study of 5504 cases and review of literature. The American Journal of dermatopathology. 2009;31:256–262. doi: 10.1097/DAD.0b013e3181961861. [DOI] [PubMed] [Google Scholar]
- 2.Kuzel P, Metelitsa AI, Dover DC, Salopek TG. Epidemiology of sebaceous carcinoma in Alberta, Canada, from 1988 to 2007. Journal of cutaneous medicine and surgery. 2012;16:417–423. doi: 10.1177/120347541201600610. [DOI] [PubMed] [Google Scholar]
- 3.Xu XL, et al. Eyelid neoplasms in the Beijing Tongren Eye Centre between 1997 and 2006. Ophthalmic surgery, lasers & imaging: the official journal of the International Society for Imaging in the Eye. 2008;39:367–372. doi: 10.3928/15428877-20080901-18. [DOI] [PubMed] [Google Scholar]
- 4.Obata H, Aoki Y, Kubota S, Kanai N, Tsuru T. Incidence of benign and malignant lesions of eyelid and conjunctival tumors. Nippon Ganka Gakkai zasshi. 2005;109:573–579. [PubMed] [Google Scholar]
- 5.Kivela T, Asko-Seljavaara S, Pihkala U, Hovi L, Heikkonen J. Sebaceous carcinoma of the eyelid associated with retinoblastoma. Ophthalmology. 2001;108:1124–1128. doi: 10.1016/S0161-6420(01)00555-3. [DOI] [PubMed] [Google Scholar]
- 6.Rao NA, Hidayat AA, McLean IW, Zimmerman LE. Sebaceous carcinomas of the ocular adnexa: A clinicopathologic study of 104 cases, with five-year follow-up data. Human pathology. 1982;13:113–122. doi: 10.1016/S0046-8177(82)80115-9. [DOI] [PubMed] [Google Scholar]
- 7.Mulay K, Aggarwal E, White VA. Periocular sebaceous gland carcinoma: A comprehensive review. Saudi journal of ophthalmology: official journal of the Saudi Ophthalmological Society. 2013;27:159–165. doi: 10.1016/j.sjopt.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson, B. R., Hamlet, K. R., Gillard, M., Railan, D. & Johnson, T. M. Sebaceous carcinoma. Journal of the American Academy of Dermatology33, 1–15 quiz 16–18 (1995). [DOI] [PubMed]
- 9.Chao AN, Shields CL, Krema H, Shields JA. Outcome of patients with periocular sebaceous gland carcinoma with and without conjunctival intraepithelial invasion. Ophthalmology. 2001;108:1877–1883. doi: 10.1016/S0161-6420(01)00719-9. [DOI] [PubMed] [Google Scholar]
- 10.Dasgupta T, Wilson LD, Yu JB. A retrospective review of 1349 cases of sebaceous carcinoma. Cancer. 2009;115:158–165. doi: 10.1002/cncr.23952. [DOI] [PubMed] [Google Scholar]
- 11.Tetzlaff MT, et al. Next-generation sequencing identifies high frequency of mutations in potentially clinically actionable genes in sebaceous carcinoma. J Pathol. 2016;240:84–95. doi: 10.1002/path.4759. [DOI] [PubMed] [Google Scholar]
- 12.Tetzlaff MT, et al. Distinct Pathways in the Pathogenesis of Sebaceous Carcinomas Implicated by Differentially Expressed MicroRNAs. JAMA ophthalmology. 2015;133:1109–1116. doi: 10.1001/jamaophthalmol.2015.2310. [DOI] [PubMed] [Google Scholar]
- 13.Lai X, Wolkenhauer O, Vera J. Modeling miRNA regulation in cancer signaling systems: miR-34a regulation of the p53/Sirt1 signaling module. Methods in molecular biology (Clifton, N.J.) 2012;880:87–108. doi: 10.1007/978-1-61779-833-7_6. [DOI] [PubMed] [Google Scholar]
- 14.Sotillo E, et al. Myc overexpression brings out unexpected antiapoptotic effects of miR-34a. Oncogene. 2011;30:2587–2594. doi: 10.1038/onc.2010.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 16.Jia X, et al. miR-16 controls myoblast proliferation and apoptosis through directly suppressing Bcl2 and FOXO1 activities. Biochimica et biophysica acta. 2017;1860:674–684. doi: 10.1016/j.bbagrm.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Emmrich S, et al. miR-9 is a tumor suppressor in pediatric AML with t(8;21) Leukemia. 2014;28:1022–1032. doi: 10.1038/leu.2013.357. [DOI] [PubMed] [Google Scholar]
- 18.Wang K, et al. MDGA2 is a novel tumour suppressor cooperating with DMAP1 in gastric cancer and is associated with disease outcome. Gut. 2016;65:1619–1631. doi: 10.1136/gutjnl-2015-309276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–1767. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- 20.Furusato B, Mohamed A, Uhlen M, Rhim JS. CXCR4 and cancer. Pathology international. 2010;60:497–505. doi: 10.1111/j.1440-1827.2010.02548.x. [DOI] [PubMed] [Google Scholar]
- 21.Jin M, Yang Z, Ye W, Xu H, Hua X. MicroRNA-150 predicts a favorable prognosis in patients with epithelial ovarian cancer, and inhibits cell invasion and metastasis by suppressing transcriptional repressor ZEB1. PLoS One. 2014;9:e103965. doi: 10.1371/journal.pone.0103965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou LL, Dong JL, Huang G, Sun ZL, Wu J. MicroRNA-143 inhibits cell growth by targeting ERK5 and MAP3K7 in breast cancer. Brazilian journal of medical and biological research=Revista brasileira de pesquisas medicas e biologicas. 2017;50:e5891. doi: 10.1590/1414-431X20175891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fesler A, Liu H, Ju J. Modified miR-15a has therapeutic potential for improving treatment of advanced stage colorectal cancer through inhibition of BCL2, BMI1, YAP1 and DCLK1. Oncotarget. 2018;9:2367–2383. doi: 10.18632/oncotarget.23414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma C, Zhan C, Yuan H, Cui Y, Zhang Z. MicroRNA-603 functions as an oncogene by suppressing BRCC2 protein translation in osteosarcoma. Oncology reports. 2016;35:3257–3264. doi: 10.3892/or.2016.4718. [DOI] [PubMed] [Google Scholar]
- 25.D’Angelo D, et al. Altered microRNA expression profile in human pituitary GH adenomas: down-regulation of miRNA targeting HMGA1, HMGA2, and E2F1. The Journal of clinical endocrinology and metabolism. 2012;97:E1128–1138. doi: 10.1210/jc.2011-3482. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, et al. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget. 2014;5:5416–5427. doi: 10.18632/oncotarget.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prakhar P, et al. Ac2PIM-responsive miR-150 and miR-143 target receptor-interacting protein kinase 2 and transforming growth factor beta-activated kinase 1 to suppress NOD2-induced immunomodulators. J Biol Chem. 2015;290:26576–26586. doi: 10.1074/jbc.M115.662817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akao, Y. et al. Impairment of K-Ras signaling networks and increased efficacy of EGFR inhibitors by a novel synthetic miR-143. Cancer science, 10.1111/cas.13559 (2018). [DOI] [PMC free article] [PubMed]
- 29.Zeng XL, Zhang SY, Zheng JF, Yuan H, Wang Y. Altered miR-143 and miR-150 expressions in peripheral blood mononuclear cells for diagnosis of non-small cell lung cancer. Chinese medical journal. 2013;126:4510–4516. [PubMed] [Google Scholar]
- 30.Li J, et al. The role of miR-205 in the VEGF-mediated promotion of human ovarian cancer cell invasion. Gynecologic oncology. 2015;137:125–133. doi: 10.1016/j.ygyno.2015.01.531. [DOI] [PubMed] [Google Scholar]
- 31.Tran MN, et al. Thep63 protein isoform DeltaNp63alpha inhibits epithelial-mesenchymal transition in human bladder cancer cells: role of MIR-205. J Biol Chem. 2013;288:3275–3288. doi: 10.1074/jbc.M112.408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urbas, R. et al. Relevance of MicroRNA200 Family and MicroRNA205 for Epithelial to Mesenchymal Transition and Clinical Outcome in Biliary Tract Cancer Patients. International journal of molecular sciences17, 10.3390/ijms17122053 (2016). [DOI] [PMC free article] [PubMed]
- 33.Cai J, et al. miR-205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non-small cell lung cancer. Cancer Res. 2013;73:5402–5415. doi: 10.1158/0008-5472.CAN-13-0297. [DOI] [PubMed] [Google Scholar]
- 34.Hirata H, et al. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mateescu B, et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17:1627–1635. doi: 10.1038/nm.2512. [DOI] [PubMed] [Google Scholar]
- 36.Han Y, et al. NLK, a novel target of miR-199a-3p, functions as a tumor suppressor in colorectal cancer. Biomedicine & pharmacotherapy=Biomedecine & pharmacotherapie. 2014;68:497–505. doi: 10.1016/j.biopha.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Minna E, et al. miR-199a-3p displays tumor suppressor functions in papillary thyroid carcinoma. Oncotarget. 2014;5:2513–2528. doi: 10.18632/oncotarget.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Y, et al. CD44 is a direct target of miR-199a-3p and contributes to aggressive progression in osteosarcoma. Sci Rep. 2015;5:11365. doi: 10.1038/srep11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan Y, Zuo X, Wei D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem cells translational medicine. 2015;4:1033–1043. doi: 10.5966/sctm.2015-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henry JC, et al. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochemical and biophysical research communications. 2010;403:120–125. doi: 10.1016/j.bbrc.2010.10.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaman N, et al. Signaling network assessment of mutations and copy number variations predict breast cancer subtype-specific drug targets. Cell reports. 2013;5:216–223. doi: 10.1016/j.celrep.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 42.Patel N, et al. Integrated genomics and functional validation identifies malignant cell specific dependencies in triple negative breast cancer. Nat Commun. 2018;9:1044. doi: 10.1038/s41467-018-03283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serra-Musach J, et al. Cancer network activity associated with therapeutic response and synergism. Genome medicine. 2016;8:88. doi: 10.1186/s13073-016-0340-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Integrated genomic analyses of ovarian carcinoma. Nature474, 609–615, 10.1038/nature10166 (2011). [DOI] [PMC free article] [PubMed]
- 45.Comprehensive molecular profiling of lung adenocarcinoma. Nature511, 543–550, 10.1038/nature13385 (2014). [DOI] [PMC free article] [PubMed]
- 46.Integrated genomic characterization of oesophageal carcinoma. Nature541, 169–175, 10.1038/nature20805 (2017). [DOI] [PMC free article] [PubMed]
- 47.Li Y, et al. HMDDv2.0: a database for experimentally supported human microRNA and disease associations. Nucleic acids research. 2014;42:D1070–1074. doi: 10.1093/nar/gkt1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie B, Ding Q, Han H, Wu D. miRCancer: a microRNA-cancer association database constructed by text mining on literature. Bioinformatics (Oxford, England) 2013;29:638–644. doi: 10.1093/bioinformatics/btt014. [DOI] [PubMed] [Google Scholar]
- 49.Jiang Q, et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic acids research. 2009;37:D98–104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X, Huang L. LRSSLMDA: Laplacian Regularized Sparse Subspace Learning for MiRNA-Disease Association prediction. PLoS computational biology. 2017;13:e1005912. doi: 10.1371/journal.pcbi.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.You ZH, et al. PBMDA: A novel and effective path-based computational model for miRNA-disease association prediction. PLoS computational biology. 2017;13:e1005455. doi: 10.1371/journal.pcbi.1005455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li JQ, Rong ZH, Chen X, Yan GY, You ZH. MCMDA: Matrix completion for MiRNA-disease association prediction. Oncotarget. 2017;8:21187–21199. doi: 10.18632/oncotarget.15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X, et al. WBSMDA: Within and Between Score for MiRNA-Disease Association prediction. Sci Rep. 2016;6:21106. doi: 10.1038/srep21106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, Z., Irizarry, R. A., Gentleman, R., Murillo, F. M. & Spencer, F. A. Model-Based Background Adjustment for Oligonucleotide Expression Arrays. Journal of the American Statistical Association, 909–917 (2004).
- 56.Chou CH, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic acids research. 2016;44:D239–247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic acids research. 2015;43:D146–152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic acids research. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1–4 and Table S2-S4