ABSTRACT
Coxsackievirus A6 (CV-A6) is an emerging pathogen associated with hand, foot, and mouth disease (HFMD). Its genetic characterization and pathogenic properties are largely unknown. Here, we report 39 circulating CV-A6 strains isolated in 2013 from HFMD patients in northeast China. Three major clusters of CV-A6 were identified and related to CV-A6, mostly from Shanghai, indicating that domestic CV-A6 strains were responsible for HFMD emerging in northeast China. Four full-length CV-A6 genomes representing each cluster were sequenced and analyzed further. Bootscanning tests indicated that all four CV-A6-Changchun strains were most likely recombinants between the CV-A6 prototype Gdula and prototype CV-A4 or CV-A4-related viruses, while the recombination pattern was related to, yet distinct from, the strains isolated from other regions of China. Furthermore, different CV-A6 strains showed different capabilities of viral replication, release, and pathogenesis in a mouse model. Further analyses indicated that viral protein 2C contributed to the diverse pathogenic abilities of CV-A6 by causing autophagy and inducing cell death. To our knowledge, this study is the first to report lethal and nonlethal strains of CV-A6 associated with HFMD. The 2C protein region may play a key role in the pathogenicity of CV-A6 strains.
IMPORTANCE Hand, foot, and mouth disease (HFMD) is a major and persistent threat to infants and children. Besides the most common pathogens, such as enterovirus A71 (EV-A71) and coxsackievirus A16 (CV-A16), other enteroviruses are increasingly contributing to HFMD. The present study focused on the recently emerged CV-A6 strain. We found that CV-A6 strains isolated in Changchun City in northeast China were associated with domestic origins. These Changchun viruses were novel recombinants of the CV-A6 prototype Gdula and CV-A4. Our results imply that measures to control CV-A6 transmission are urgently needed. Further analyses revealed differing pathogenicities in strains isolated in a neonatal mouse model. One of the possible causes has been narrowed down to the viral protein 2C, using phylogenetic studies, viral sequences, and direct tests on cultured human cells. Thus, the viral 2C protein is a promising target for antiviral drugs to prevent CV-A6-induced tissue damage.
KEYWORDS: coxsackievirus A6, hand, foot, and mouth disease, lethality, mouse model, pathogenicity, recombination
INTRODUCTION
Hand, foot, and mouth disease (HFMD) is a common infectious disease among children under 5 years of age (1). Enterovirus 71 (EV-A71) and coxsackievirus A16 (CV-A16), belonging to the enterovirus subfamily of the family Picornaviridae, are most frequently associated with HFMD in Asian and Pacific countries, such as Singapore (2), Japan (3), Taiwan (4), and mainland China (5–9). In addition to the classic hand, foot, and mouth ulcerations, infection due to these viruses can also cause myocarditis, intractable shock, and poliomyelitis-like paralysis (10, 11).
In recent years, HFMD cases not associated with EV-A71 or CV-A16 have also been reported (12–17). In particular, coxsackievirus A6 (CV-A6) has been reported to be the predominant cause of HFMD in China, spreading from south to north (14, 18–26). We have reported that CV-A6-related HFMD accounted for more than 60% of all hospitalized cases in Jilin Province in northeast China in 2013 (27). In the same period, EV-A71- or CV-A16-associated HFMD accounted for only 26% of the cases.
Similar to other enteroviruses, CV-A6 has a single-stranded positive RNA genome with a length of ∼7,400 nucleotides. The genome contains a single open reading frame that encodes a polyprotein and is flanked by 5′ and 3′ untranslated regions (UTRs). The encoded polyprotein contains three major regions (P1, P2, and P3). These three regions can self-digest into four separate structural proteins (VP1, VP2, VP3, and VP4 from P1) and seven nonstructural proteins (2A, 2B, and 2C from P2; 3A, 3B, 3C, and 3D from P3). Similar to EV-A71 and CV-A16 circulating in China (7–9), partial and full-length genome analyses have revealed novel recombination patterns for these circulating CV-A6 strains. The recombinations occurred between CV-A4 and the CV-A6 prototype Gdula (16, 17).
In this study, we obtained partial VP1 sequences of 39 CV-A6 strains isolated in Changchun City in northeast China in 2013. Phylogenetic analyses indicated that these Changchun CV-A6 strains were grouped into three different subclusters but mostly derived from strains previously circulating in Shanghai. Four of the Changchun CV-A6 strains were selected to represent each subcluster and tested for pathogenicity in a widely used lethal mouse model. A one-step growth assay confirmed that the lethalities of these Changchun strains correlated with their potencies of viral replication and times of virus release. The lethalities of the four strains, along with phylogenetic analyses, divided the known CV-A6 strains into lethal and nonlethal groups. Examination of the sequence differences between the lethal and nonlethal strains suggested that the viral 2C protein might contribute to viral pathogenicity. This hypothesis was supported by the fact that exogenous expression of 2C in cultured human cells induced cell death, but not through apoptosis. Thus, our study not only provided detailed epidemiological information on CV-A6 strains circulating in northeast China, but also revealed the viral protein 2C as a pathogenesis contributor and possible target for anti-CV-A6 drug development.
RESULTS
Phylogenetic analyses of modern CV-A6 strains in northeast China.
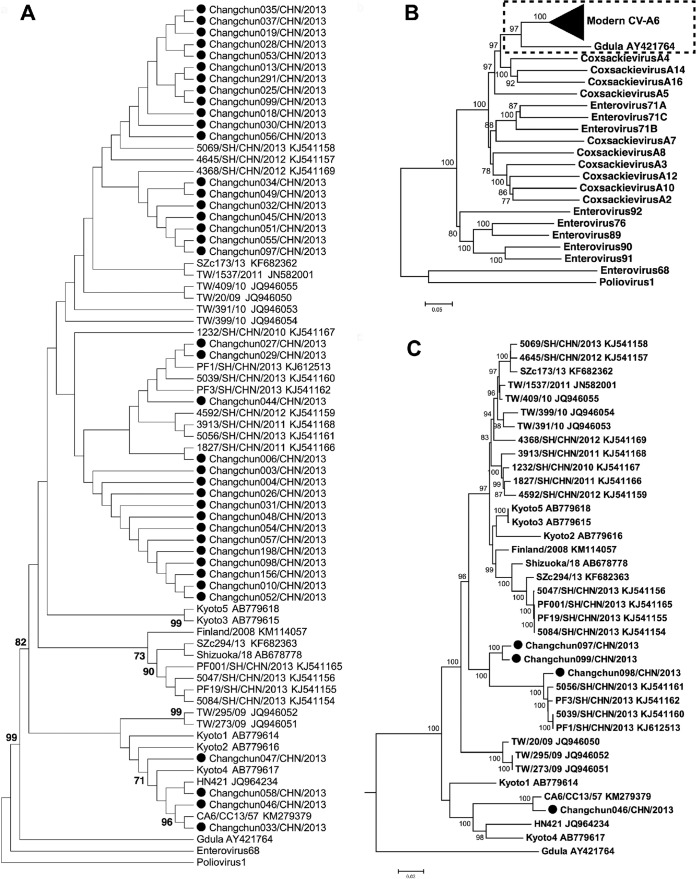
Partial VP1 sequences were obtained from 39 Changchun HFMD patient samples using CV-A6-specific primers. To determine the phylogenetic relationship between Changchun viruses and other reported CV-A6 strains, various published CV-A6 sequences were retrieved from GenBank, including the prototype Gdula, which was isolated in 1949 (28). A neighbor-joining (NJ) tree was constructed from 1,000 bootstrap replicates. Poliovirus 1 (Sabin 1; accession no. V01150) and EV-D68 (Fermon; accession no. AY426531) were used as the outgroup. Grouping of Changchun viruses with known CV-A6 strains was observed (Fig. 1A), confirming the classification of these Changchun strains as CV-A6. However, all the modern strains clustered together and were distant from the prototype Gdula. Phylogenetic analyses also identified three major clusters of CV-A6 strains as being related mostly to CV-A6 strains from Shanghai, China (Fig. 1A), indicating that multiple introductions of divergent CV-A6 strains from the same city in China were responsible for the HFMD emerging in northeast China. Some CV-A6 Changchun strains, such as Changchun 046, were more closely related to CV-A6 from Japan, while multiple CV-A6 strains persisted in Changchun for years and caused the HFMD epidemic in 2013.
FIG 1.
Phylogenetic status of modern CV-A6 strains. (A) Neighbor-joining tree based on a short region of the CV-A6 genome (partial VP1, positions 2614 to 2866, corresponding to the Gdula genome). Only topology is shown for clear demonstration of phylogenetic relationships between strains. (B and C) Neighbor-joining tree based on full-length CV-A6 genomes. Prototypes of the HEV-A group, EV-D68, and poliovirus 1 were used as references. The tree indicates that modern CV-A6 strains are phylogenetically close to the prototype Gdula (B), while the subtree indicates that strains from Changchun have different origins (C). All the trees were tested by the bootstrap method for 1,000 replicates, and values of >70 are shown at the nodes. ●, CV-A6-Changchun isolates.
To further characterize these new CV-A6 strains, we determined four full-length CV-A6 viral sequences representing the three different clusters shown in Fig. 1A, i.e., Changchun046, Changchun097, Changchun098, and Changchun099. To examine the phylogenetic relationship between modern CV-A6 strains and other members of human enterovirus A (HEV-A) groups, the sequences of 34 additional full-length CV-A6 strains and prototypic strains representing each member of the HEV-A group were retrieved from GenBank, including the CV-A6 prototype Gdula. A similar strategy successfully separated circulating CV-A16 strains from their prototype, G-10, and ultimately led to the discovery that the modern CV-A16 was in fact a recombinant form (7). Unlike CV-A16 (7), the resulting NJ tree indicated that modern CV-A6 was evolutionarily close to the CV-A6 prototype Gdula and distinct from other members of the HEV-A group (Fig. 1B). However, the four Changchun sequences were located within different subclusters (Fig. 1C). Changchun046 was closely related to another previously reported Changchun sequence, CV-A6/CC13/57, and both might share an origin in Japan. Changchun097 and Changchun099 were phylogenetically close to one another but distinct from Changchun098, whereas the last might share an ancestor with sequences from Shanghai.
Circulating CV-A6 strains produced dose-dependent morbidity and mortality in vivo.
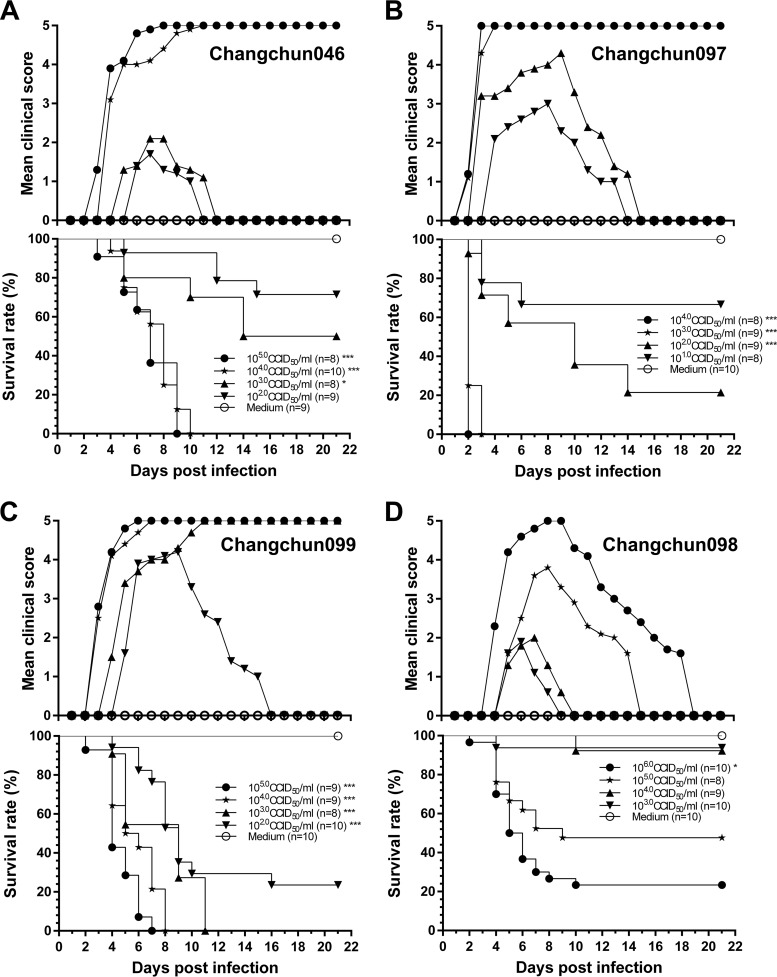
To find the pathogenic mechanism and virulence-determining region of CV-A6 strains, we used a lethal mouse model. To determine whether CV-A6-Changchun strains were lethal to neonatal mice and to measure the median lethal doses (LD50) of these viruses, 1-day-old mice were intracerebrally injected with CV-A6-Changchun. The mice were randomly divided into eight groups and injected intracerebrally (10 μl/mouse) with 10-fold serial dilutions of Changchun046 (105.0 to 102.0 50% cell culture infectious doses [CCID50]/ml), Changchun097 (104.0 to 101.0 CCID50/ml), Changchun098 (106.0 to 103.0 CCID50/ml), and Changchun099 (105.0 to 102.0 CCID50/ml), with modified Eagle's medium (MEM) as a negative control for each virus group.
The mice that were infected with Changchun046 at titers of 105.0 to 104.0 CCID50/ml started to show grade 1 symptoms (see below) on day 3 postinfection. The symptoms gradually became more severe until they reached grade 5 on days 6 and 9, respectively (Fig. 2A). The titers of 105.0 and 104.0 CCID50/ml Changchun046 caused death on days 3 and 4, with 100% mortality by days 9 and 10 postinfection (Fig. 2A). The log rank tests suggested that the survival rates in mice treated with 105.0 and 104.0 CCID50/ml of Changchun046 were statistically different from those for the control group (P < 0.001). Changchun046 at lower titers of 103.0 to 102.0 CCID50/ml caused 50 and 28.6% mortality, respectively (Fig. 2A). As expected, the control group did not experience any mortality or symptoms during the 21-day period (Fig. 2A).
FIG 2.
CV-A6-Changchun viruses cause clinical symptoms and mortality in a dose-dependent manner. One-day-old ICR mice (n = 8 to 10 per litter) were intracerebrally inoculated with 10-fold serially diluted dosages of viruses (10 μl/mouse). Control animals were mock infected with MEM instead of virus (10 μl/mouse). Clinical symptoms and mortality were monitored daily for 21 days postinfection. Neonatal mice were challenged with 102.0 to 105.0 CCID50/ml of Changchun046 virus (A), 101.0 to 104.0 CCID50/ml of Changchun097 virus (B), 102.0 to 105.0 CCID50/ml of Changchun099 virus (C), or 103.0 to 106.0 CCID50/ml of Changchun098 virus (D). The log rank test was used to compare the survival rates of newborn mice between the groups and the control group at 21 days postinfection. ***, P < 0.001; *, P < 0.05. One representative of three independent tests is shown.
For Changchun097, the challenged mice began to fall sick on day 2 at titers of 104.0 to 103.0 CCID50/ml, and all the mice in these groups quickly reached grade 5 symptoms before day 6 postinfection (Fig. 2B). One hundred percent of the mice injected with Changchun097 at titers of 104.0 to 103.0 CCID50/ml died on days 2 and 3 postinoculation (Fig. 2B). Surprisingly, even at relatively low titers of 102.0 to 101.0 CCID50/ml, Changchun097 resulted in 78.6 and 33.3% mouse mortality, respectively. In addition, the log rank tests indicated that the survival rates in newborn mice treated with 104.0 to 102.0 CCID50/ml of Changchun097 were significantly different from those of the control group (P < 0.001). No clinical symptoms or deaths were observed in the negative-control group (Fig. 2B).
The mice infected with different titers of Changchun099 started to become sick on days 3 to 5 postinfection (Fig. 2C, grade 2). After increases in the clinical grade of symptoms, the 105.0-, 104.0-, and 103.0-CCID50/ml groups reached grade 5 symptoms on days 6, 7, and 11, respectively. One hundred percent of the infected mice died on days 7, 8, and 11 at titers of 105.0 to 103.0 CCID50/ml, whereas the virus caused 76.5% mortality at a titer of 102.0 CCID50/ml. The survival rates in all four Changchun099-treated groups were significantly different from those in the control group (P < 0.001). The control group did not exhibit any mortality or clinical symptoms (Fig. 2C).
The challenged mice administered 106.0, 105.0, 104.0, and 103.0 CCID50/ml of Changchun098 began to sicken on days 4, 5, 5, and 5, respectively. These groups had survival rates of 43.8, 90, 92.3, and 93.8%, respectively, with a mean clinical score of grade 3 (Fig. 2D). However, only the mice treated with 106.0 CCID50/ml Changchun098 were different from the control group (P < 0.05), according to the log rank test results.
The above-mentioned results suggest that CV-A6-Changchun strains may be divided into lethal strains (Changchun046, Changchun097, and Changchun099) and a nonlethal strain (Changchun098), and the measured LD50s of Changchun046, Changchun097, and Changchun099 were 2.00 × 101, 3.16 × 100, and 5.01 × 100 CCID50/mouse, respectively.
Pathological analyses of CV-A6-Changchun strains in neonatal mice.
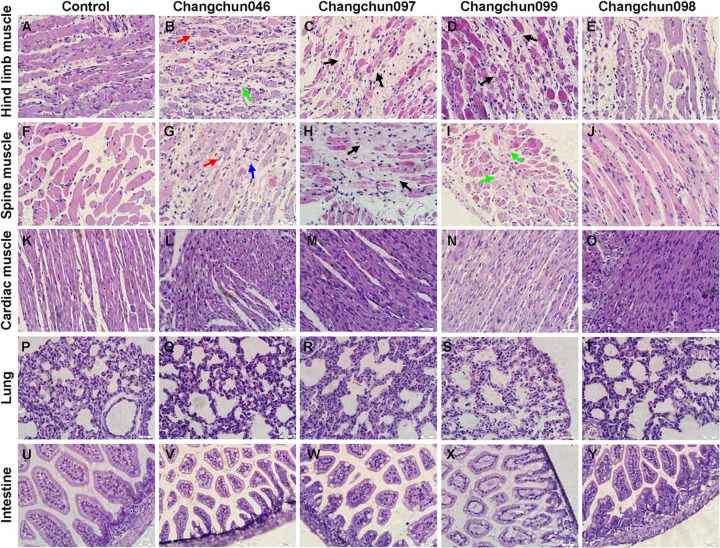
To understand the pathogenesis of CV-A6-Changchun strains that might be related to the death of the newborn mice, we performed a series of pathological analyses on nine tissues. The infected mice that were challenged with 105.0 to 104.0 CCID50/ml of Changchun046, Changchun097, Changchun098, and Changchun099 and presented grade 4 or 5 clinical scores were collected for pathological analyses. Hind limb muscle and spine muscle fibers exhibited severe necrosis, including muscle bundle fracture (black arrows), muscle fiber swelling (green arrows), nuclear dissolution (red arrows), and shrinkage (blue arrows) in Changchun046-, Changchun097-, and Changchun099-infected mice (Fig. 3B, C, D, G, H, and I) compared with those of the negative-control mice (Fig. 3A and F). However, no obvious changes were found in the hind limb muscle and spine muscle fibers of Changchun098-infected mice (Fig. 3E and J). Moreover, no detectable pathological changes were observed in the heart (Fig. 3L to O), lung (Fig. 3Q to T), or intestine (Fig. 3V to Y). No obvious pathological symptoms were found in the brain, liver, spleen, or kidney tissues of any CV-A6-Changchun-infected mice (data not shown). These results indicate that three lethal circulating CV-A6-Changchun strains have strong tropism for the muscle tissues, whereas Changchun098 is an exception. These findings correspond to the results of lethal neonatal mouse experiments (Fig. 2).
FIG 3.
Pathological analyses of CV-A6-infected newborn mice. One-day-old ICR mice were intracerebrally inoculated with Changchun046 (105.0 CCID50/ml), Changchun097 (104.0 CCID50/ml), Changchun099 (105.0 CCID50/ml), or Changchun098 (105.0 CCID50/ml) or medium (negative control). Infected mice with grade 4 or 5 that exhibited severe hind limb paralysis were dissected for H&E staining. Representative images from the hind limb muscle, spine muscle, cardiac muscle, and lung and intestine tissues after infection are shown. Hind limb muscle (B to D) and spine muscle (G to I) fibers exhibited severe necrosis, including muscle bundle fracture (black arrows), muscle fiber swelling (green arrows), nuclear dissolution (red arrows), and shrinkage (blue arrows), in Changchun046-, Changchun097-, and Changchun099-infected mice. No obvious changes were found in the hind limb muscle and spine muscle fibers of Changchun098-infected mice (E and J), and no detectable pathological changes were observed in the heart (L to O), lung (Q to T), or intestine (V to Y). Magnification, ×400 (scale bars, 20 μm). The results are representative of three independent experiments.
Viral loads in various tissues of neonatal mice infected with CV-A6-Changchun.
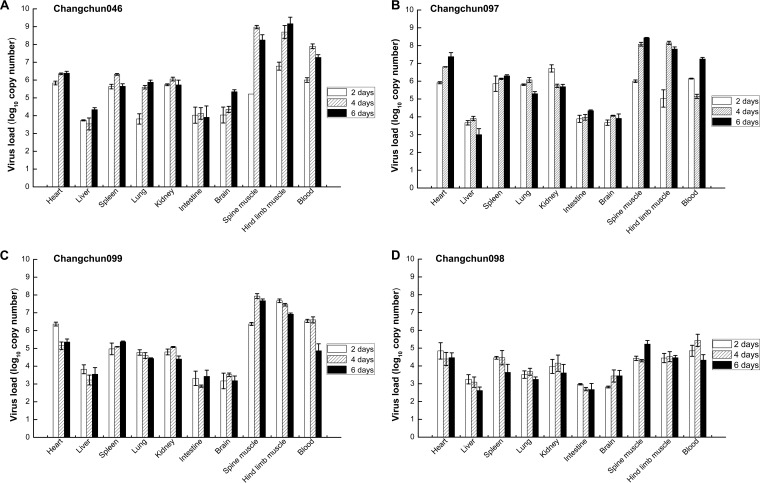
To further investigate the replication and distribution of CV-A6 strains in infected mice, we measured the viral loads in 10 samples at different time points after inoculation. Viral loads were measured on days 2, 4, and 6 in all samples from mice infected with each of the four CV-A6-Changchun strains. As shown in Fig. 4, the viruses were detected in almost all tissues at relatively low copy numbers for all CV-A6-Changchun strains on day 2 postinoculation. On day 4 postinfection, the viral loads increased steadily in spine muscle, hind limb muscle, and blood, with the copy numbers reaching 108.98 copies/mg, 108.69 copies/mg, and 107.90 copies/ml, respectively, for Changchun046; 108.08 copies/mg, 108.15 copies/mg, and 105.15 copies/ml, respectively, for Changchun097; and 107.93 copies/mg, 107.45 copies/mg, and 106.60 copies/ml, respectively, for Changchun099. However, the viral loads in Changchun098-infected mice were as low as 104.29 copies/mg, 104.52 copies/mg, and 105.43 copies/ml in the corresponding tissues on day 4. These findings suggested that the virus spread systemically and increased steadily on day 4 postinfection.
FIG 4.
Kinetics of viral load levels in various tissues of CV-A6-infected mice at different time points. One-day-old ICR mice were intracerebrally inoculated with Changchun046 (105.0 CCID50/ml) (A), Changchun097 (104.0 CCID50/ml) (B), Changchun099 (105.0 CCID50/ml) (C), and Changchun098 (105.0 CCID50/ml) (D). The viral loads were detected by qRT-PCR at days 2, 4, and 6 postinfection in 10 samples. The results represent the mean viral loads (log10 copies per milligram of tissue or log10 copies per milliliter of blood) ± SD (3 mice/group; repeated 3 times). The data shown are representative of the results of three independent experiments.
Six days after inoculation, the viruses were detected in the spine muscle, hind limb muscle, and blood at relatively high levels for Changchun046 (Fig. 4A) (108.25 copies/mg, 109.19 copies/mg, and 107.27 copies/ml, respectively), Changchun097 (Fig. 4B) (108.44 copies/mg, 107.81 copies/mg, and 107.25 copies/ml, respectively), and Changchun099 (Fig. 4C) (107.68 copies/mg, 106.92 copies/mg, and 104.87 copies/ml, respectively). Changchun098 (Fig. 4D) (105.23 copies/mg, 104.46 copies/mg, and 104.33 copies/ml, respectively) had relatively low viral loads in the three tissues. These results indicated that on day 6, the viral loads of the lethal strains were 2.4 to 3.2 log units higher in the spine muscle, 2.5 to 4.7 log units higher in the hind limb muscle, and 0.5 to 2.9 log units higher in the blood than those of the nonlethal Changchun098 strain. In the negative-control mice, no viral loads were detected in any of the 10 selected samples, indicating the specificity of our results. The above-mentioned results also suggested that CV-A6 had strong tropism to muscle tissues, which might be a major location for viral duplication. Changchun098 was slightly different than Changchun046, Changchun097, and Changchun099 in viral replication and distribution. These results were also consistent with the survival rates and clinical scores in lethally infected neonatal mice (Fig. 2) and pathological changes (Fig. 3) after infection.
One-step growth curve of CV-A6 in RD cells.
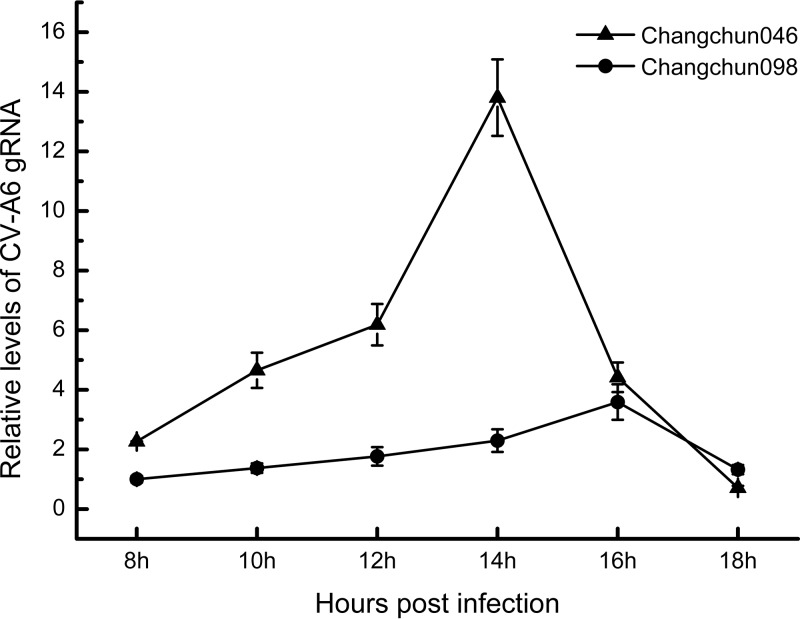
To determine whether the lethality of the Changchun CV-A6 strains was due to their potency in viral replication, a one-step growth assay with a 2-h interval was performed. Changchun046, as the weakest strain in the lethal group, and Changchun098, as the only nonlethal strain in this study, were tested in RD cells. As shown in Fig. 5, the levels of both viruses, determined by CV-A6-specific quantitative real-time reverse transcription (qRT)-PCR (data not shown), first increased in the culture medium, indicating viral replication and release, which was then followed by a decrease, most likely due to the viral adsorption to RD cells to start the next round of infection. However, even with similar viral dosages to initiate the infection, levels of the lethal strain Changchun046 were constantly higher than those of the nonlethal Changchun098 strain during the replication/release phase and showed the first peak of viral release at 14 h postadsorption, which was 2 h earlier than that detected with Changchun098. In addition, more Changchun046 than Changchun098 viruses were detected at the peak points. These results demonstrated that the viral replication and release of the lethal CV-A6 strain was much more potent than that of the nonlethal CV-A6 strain, and this finding was correlated with the lethality of associated CV-A6 strains in the mouse model (compare Fig. 5 to Fig. 2).
FIG 5.
The lethality of CV-A6 correlates with viral replication and release. Shown are one-step growth curves of CV-A6 Changchun046 and Changchun098. Similar amounts of Changchun046 and Changchun098 viruses, based on genome quantification by qRT-PCR, were used to infect RD cells. After adsorbing the virus for 6 h, the infected cells were washed, the medium was replaced with maintenance medium, and the levels of released viruses in the medium were determined through qRT-PCR at 2-h intervals, starting 8 h postadsorption. The values are shown as means ± SD.
CV-A6-Changchun strains are products of recombination between Gdula and CV-A4.
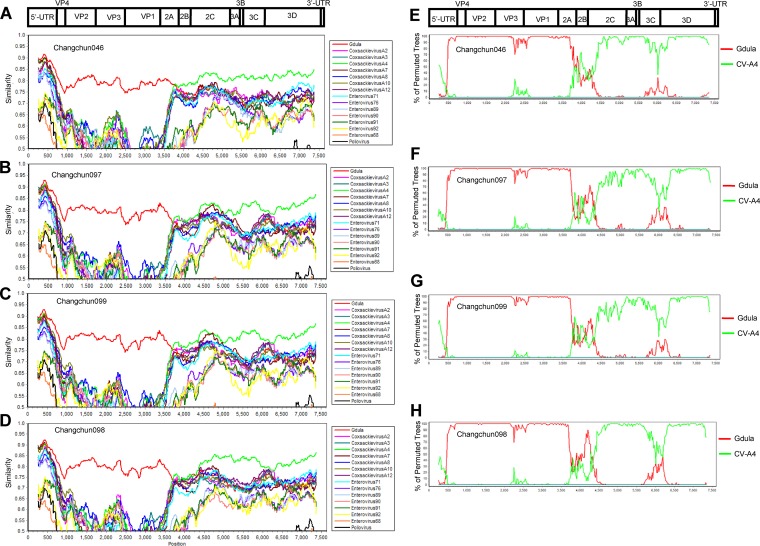
To reveal possible reasons for the differing pathogenicities observed among CV-A6-Changchun strains, we first checked whether the lethal and nonlethal strains had different recombination patterns. Similarity tests based on the complete genome were performed, with prototypic strains of members of the HEV-A group as reference sequences and each of the four full-length CV-A6-Changchun strains as the query. EV-D68 and poliovirus 1 were used as the outliers. To our surprise, despite their different potencies in pathogenesis, all four CV-A6-Changchun strains presented similar patterns of recombination, which was mostly based on the prototype of CV-A6 and CV-A4 (Fig. 6A to D). The recombination was further confirmed by full-length bootscanning analyses (Fig. 6E to H). In both tests, the P1 regions of all four CV-A6-Changchun strains were phylogenetically related to that of the Gdula prototype (Fig. 6). In addition, CV-A4 was close to the CV-A6-Changchun strains in the P2/P3 regions (Fig. 6). The recombination patterns of all CV-A6-Changchun strains were almost identical throughout the genome, with the 2C region (positions 4105 to 5091) of Changchun098 closer to that of the Gdula strain than those of the other three CV-A6-Changchun strains in both tests (Fig. 6). Indeed, several previous studies have indicated that modern CV-A6 strains are products of recombination between Gdula and other circulating enteroviruses, such as CV-A4 (16, 17). The modern CV-A6-Changchun strains are most likely recombinants between Gdula and the prototype CV-A4 or an unknown parental virus similar to CV-A4.
FIG 6.
Similarity tests and bootscanning analyses of the CV-A6-Changchun strains and the prototype Gdula and CV-A4 on the basis of full-length genomes. The similarity tests for Changchun046 (A), Changchun097 (C), Changchun099 (E), and Changchun098 (G) were performed with prototypic strains of members of the HEV-A group. The test results indicated that modern CV-A6 strains circulating in Changchun were mostly recombinants between CV-A6 and CV-A4, as previously described (16, 17). Thus, for better demonstration, only CV-A4 (accession number AY421762) and CV-A6 Gdula (accession number AY421764) were included as the reference sequences for the bootscanning tests for Changchun046 (B), Changchun097 (D), Changchun099 (F), and Changchun098 (H). EV-D68 (accession number AY426531) and poliovirus 1 (accession number V01150) were used as the outliers in both similarity tests and bootscanning analyses. All the tests were performed with full-length viral genomes. A cartoon of the enteroviral genome is shown at the top.
Phylogenetic analyses of CV-A6-Changchun strains with other CV-A6 strains from China and other countries.
To further investigate the cause of CV-A6 lethality, the four Changchun CV-A6 strains were subjected to phylogenetic analyses based on different fragments (i.e., 5′ UTR, P1, P2, and P3). To expand the search, we included 34 additional CV-A6 sequences isolated from other provinces in China and other regions of the world. The prototype CV-A6 Gdula sequence was used as the outgroup.
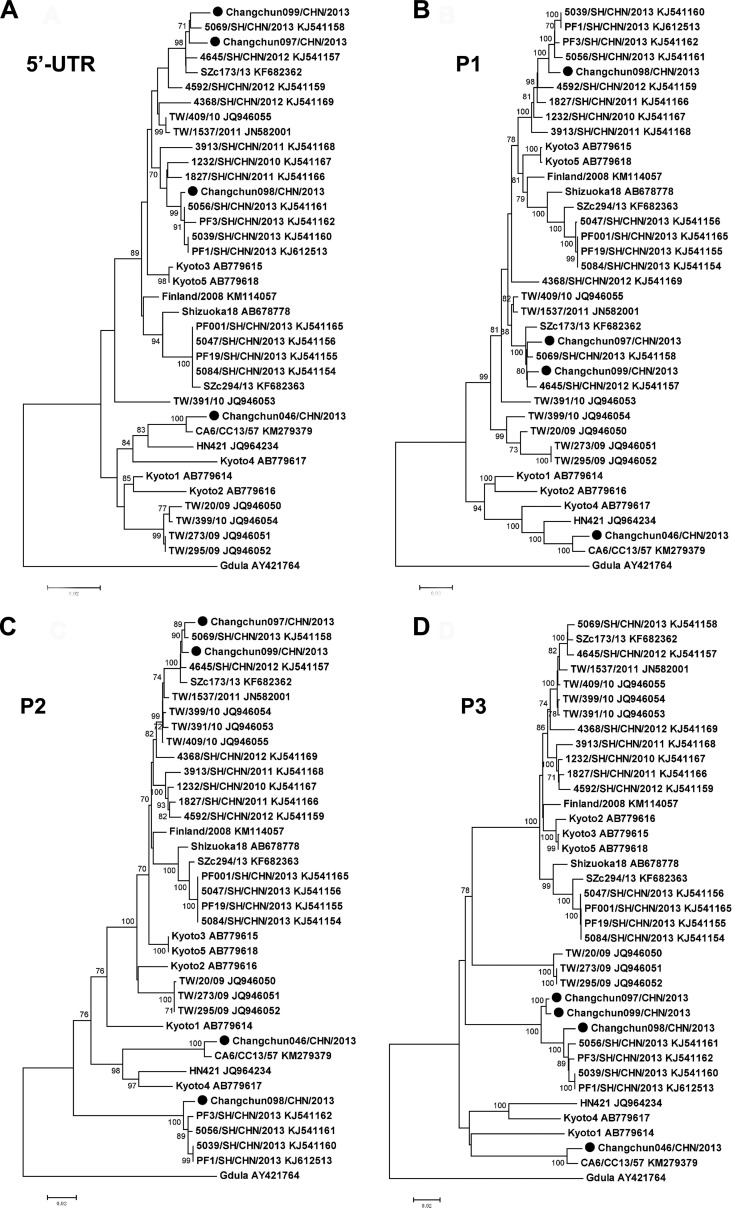
All phylogenetic analyses indicated that the Changchun098 strain was closely related to the PF3/SH/CHN/2013, 5056/SH/CHN/2013, 5039/SH/CHN/2013, and PF1/SH/CHN/2013 strains (Fig. 7A to D), which were all isolated from Shanghai, China. The NJ tree for the P2 sequences was most closely correlated with the lethality of the Changchun CV-A6 strains. The nonlethal Changchun098 was in a separate cluster from the lethal Changchun046, Changchun097, and Changchun099 (Fig. 7C). Similar clustering was not observed for other fragments, such as the 5′ UTR (Fig. 7A), P1 (Fig. 7B), and P3 (Fig. 7D). This pattern was consistent with a P2 contribution to the lethality of CV-A6-Changchun strains.
FIG 7.
Phylogenetic trees constructed for 5′ UTR, P1, P2, and P3 regions of CV-A6-Changchun strains with other CV-A6 strains retrieved from the GenBank database. Neighbor-joining trees based on the full-length 5′ UTR (A), P1 (B), P2 (C), and P3 (D) regions were generated. All the trees were tested by bootstrapping for 1,000 replicates, and values of >70 are shown at the nodes. ●, CV-A6-Changchun isolates.
Viral 2C protein is a contributor to the pathogenicity of CV-A6.
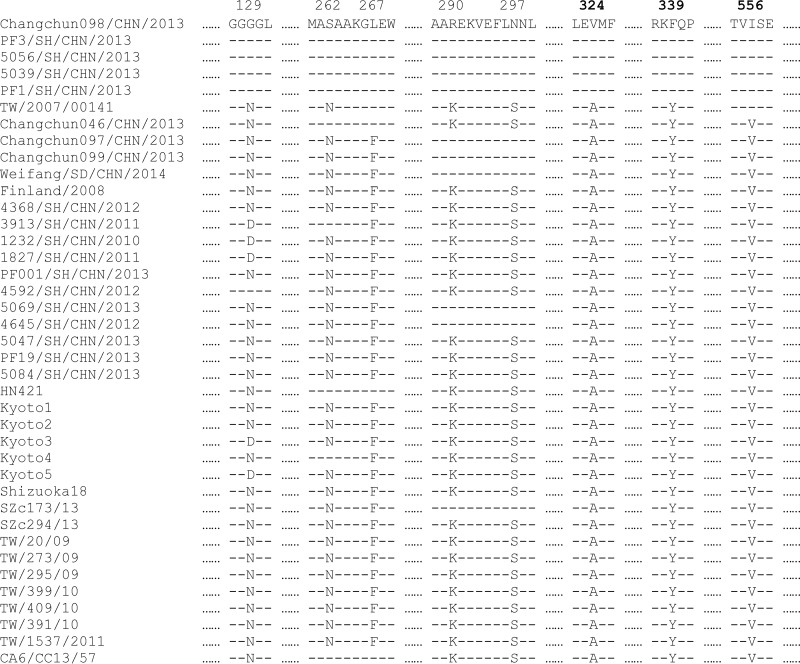
To identify residues potentially responsible for the lethality of the virus, protein sequence alignment was performed on the CV-A6 P2 region. As a result, variations between the lethal and nonlethal groups were detected at eight positions (129, 262, 267, 290, 297, 324, 339, and 556), with three of them (324, 339, and 556) being unified within each group (Fig. 8). These three positions were all located in the 2C region of CV-A6, indicating that the 2C protein might be responsible for the lethality of CV-A6.
FIG 8.
Sequence alignment of the lethal and nonlethal strains of CV-A6. The amino acid sequences of the P2 regions of CV-A6 strains from Changchun and other places in China and worldwide were aligned, and specific positions possibly associated with lethality are indicated, based on the differences between the lethal group (containing Changchun046, Changchun097, and Changchun099) and the nonlethal group (containing Changchun098). Dashes stand for amino acid residues identical to those of Changchun098.
To further confirm 2C as a contributor to the pathogenicity of CV-A6, we introduced two more 2C protein sequences from CV-A6 strains with reported lethality. Weifang/SD/CHN/2014/was effective in killing 5-day-old mice (29) and shared amino acids at positions 324, 339, and 556 with the lethal strains (Fig. 8). TW/2007/00141 had an LD50 of 1.33 × 103 CCID50/mouse (i.e., less lethal than Changchun046) (30) and had 2 residues (at positions 324 and 339) identical to those of the lethal strains, but 1 residue (position 556) was identical to that of the nonlethal strains (Fig. 8). This information supported the idea that these 3 amino acids contribute to the pathogenicity of the 2C protein and CV-A6.
CV-A6 2C protein promotes cell death by inducing autophagy.
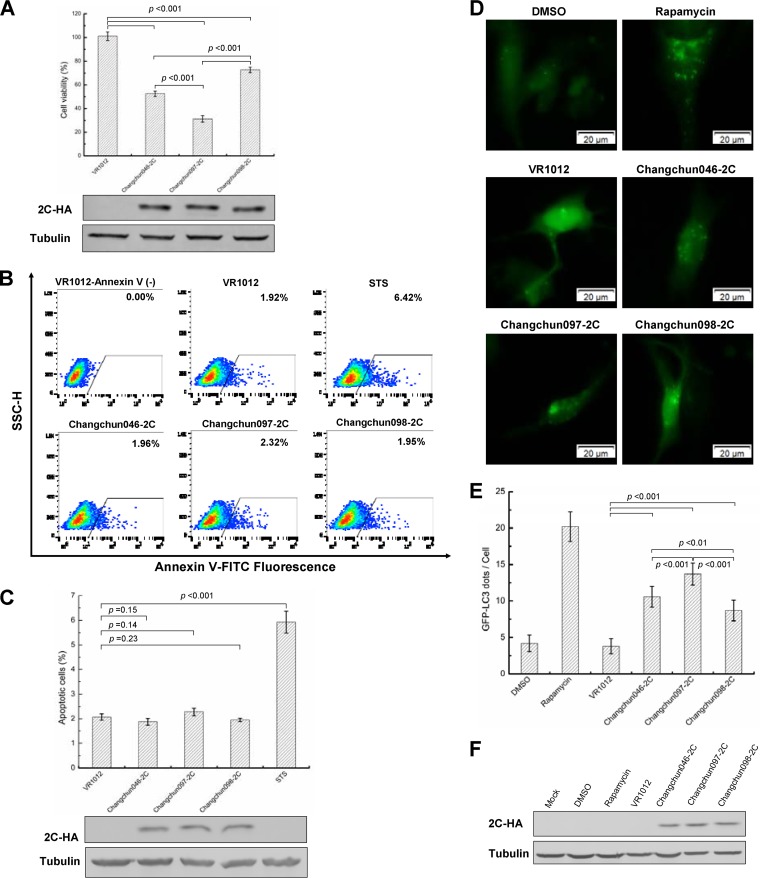
To determine the contribution of the viral 2C protein to the pathogenicity of CV-A6, we transfected Changchun046 2C-, Changchun097 2C-, and Changchun098 2C-expressing vectors into RD cells and checked the cell survival rate at 48 h posttransfection. Changchun099 2C was excluded, because parts of its amino acid sequence were identical to that of Changchun097 2C. As a result, exogenous expression of the Changchun046 2C, Changchun097 2C, and Changchun098 2C proteins resulted in 47.57, 68.89, and 27.38% rates of lethality, respectively, in transfected RD cells, which were all significantly different from that of the control group (P < 0.001) (Fig. 9A). The observed cell death rates due to the exogenous expression of 2C proteins were correlated with the lethality of associated CV-A6 strains in the mouse model (compare Fig. 9A to Fig. 2).
FIG 9.
CV-A6 2C protein induces cell death through autophagy instead of apoptosis in RD cells. (A) Cell lethality caused by different CV-A6 2C proteins in RD cells. Changchun046 2C-, Changchun097 2C-, or Changchun098 2C- expressing vectors were transfected into RD cells, and a cell proliferation assay was performed at 48 h posttransfection. Western blotting results indicating expression levels of viral 2C proteins for the cell lethality experiment are shown. (B) Representative flow charts for the apoptosis analysis. Changchun046 2C-, Changchun097 2C-, or Changchun098 2C-expressing vectors were transfected into RD cells. The RD cells were harvested at 48 h posttransfection, and cell death was evaluated by flow cytometry. The percentages of apoptotic cells corresponding to the increased FITC fluorescence intensity for each of the experimental conditions are indicated. RD cells transfected with VR1012 or treated with STS were used as negative and positive controls, respectively. (C) Bar graph of apoptosis analysis in RD cells. Shown are Western blotting results indicating the expression levels of viral 2C proteins from panel B. (D) Live-cell imaging showing GFP-LC3 aggregation. RD cells were cotransfected with pEGFP-LC3 and the VR1012 empty vector, Changchun046 2C, Changchun097 2C, or Changchun099 2C. Rapamycin was used as a positive control to induce autophagy. The GFP-LC3 aggregations in the cells were observed by fluorescence microscopy at 48 h posttransfection. Representative images are shown. Magnification, ×400. (E) Bar graph indicating average GFP-LC3 dot formation in RD cells expressing CV-A6 2C. Each bar value indicates the average number of GFP dots in 10 cells per sample. Rapamycin was used as a positive control. (F) Western blotting results showing 2C protein expression in RD cells from panel E. The values in panels A, C, and E are shown as means ± SD.
Apoptosis is commonly observed in enterovirus-induced cell death (31–33). To determine whether CV-A6 2C-mediated cell death was due to 2C-induced apoptosis, RD cells were transfected with vectors expressing exogenous 2C proteins or the control vector, VR1012, or treated with staurosporine (STS) at a final concentration of 4 nM and tested for annexin V positivity. The results showed that STS induced apoptosis at a rate of 6.42%, which was much higher than that detected in cells transfected with VR1012 (Fig. 9B and C). On the other hand, no apoptosis was observed in the cells expressing the lethal-strain-derived Changchun046 2C or Changchun097 2C or the nonlethal-strain-derived Changchun098 2C compared with cells transfected with VR1012 (Fig. 9B and C). These results indicated that the CV-A6 2C protein could not induce apoptotic cell death in RD cells.
A previous study determined that CV-A16 2C induced autophagy in HeLa cells by blocking the fusion of autophagosomes with lysosomes and triggering autophagosome accumulation, shown by the formation of dots of green fluorescent protein (GFP)-tagged microtubule-associated protein 1 light chain 3 (LC3) (34). Thus, the CV-A6 2C protein might also induce RD cell death through autophagy. To validate this hypothesis, GFP-LC3- and CV-A6 2C-expressing vectors were cotransfected into RD cells, and the formation of dots of GFP-LC3 was determined through fluorescence microscopy at 48 h posttransfection. Rapamycin (also known as sirolimus; Selleck Chemicals, Houston, TX, USA) was included as a positive control to trigger autophagy (31). As shown in Fig. 9D and E, the addition of rapamycin, but not its solvent dimethyl sulfoxide (DMSO), enhanced dot formation by GFP-LC3, proving the validity of the assay. Comparing cells transfected with the control vector, RD cells expressing CV-A6 2C showed higher numbers of GFP-LC3 dots (Fig. 9D), indicating that autophagy contributed to the cytotoxicity induced by CV-A6 2C. With similar expression levels (Fig. 9F), the lethal-strain-derived Changchun046 2C and Changchun097 2C induced more GFP-LC3 dot formation than the nonlethal-strain-derived Changchun098 2C (Fig. 9D and E). Taken together, our results indicated that CV-A6 2C triggered cell death through autophagy, contributing to the pathogenesis of CV-A6 strains.
DISCUSSION
Recombination is a common feature of enteroviruses, especially those associated with HFMD. Recently, many studies have reported that multiple HEV-A group viruses, especially non-EV-A71 and non-CV-A16 infections, could coinfect an individual, cocirculate in the same geographical area, and give the viruses the opportunity to undergo recombination (16, 20, 29). Our previous studies have also revealed that circulating CV-A16 and EV-A71 were recombinants based on their prototype viruses (7–9). In the present study, we isolated and fully sequenced four CV-A6 strains that were circulating in Changchun City, northeast China. Although the phylogeny suggested that these four modern CV-A6 strains were closely related to the prototype Gdula, they were in fact recombinants between Gdula (mainly in the P1 region) and CV-A4 (mainly in the P2/P3 regions). A similar recombination pattern was also found in CV-A6 strains isolated in Shanghai in 2013, which shared higher similarity with a recent CV-A4 strain than with the recent CV-A6 strain in the 2C and 3′-UTR regions, and the recombinant CV-A6 strain led to a more generalized rash than that of the nonrecombinant CV-A6 strain (16, 35). These findings suggested that cocirculation of CV-A4 and CV-A6 strains in the same geographic region was a relatively common phenomenon in China (20, 36), and it would increase the occurrence of recombination between the two serotypes. However, CV-A6 strains isolated from Wenzhou (15) exhibited three different recombination breakpoint patterns, with those of most strains located at the 3′ ends of the 5′ UTR and 2A regions, whereas some strains showed a close relationship with CV-A2 and CV-A8 strains and others were located in the P3 region. Gaunt et al. (17) analyzed CV-A6 strains isolated from Edinburgh, Scotland, in 2008 and 2014, and eight recombinant forms (RF-A-H) were circulating worldwide over the past 10 years, with the recombination breakpoints located in the 2A-2C and VP3 regions and between the 5′-UTR and VP1 regions. More recently, it has been reported that recent CV-A6 recombination groups (RF-E, -F, -H, -J, and -K) shared a common ancestor (RF-A), and the recombination breakpoints were located between the 2A-2C and 5′-UTR regions (37). Recombination events can play a significant role in the evolution of enterovirus genomes, and breakpoints detected in these studies (2A-2C regions) are well-known recombination hot spots (38). These results, together with our findings, indicated that nonstructural regions might potentially contribute to the clinical phenotypes and outcomes of CV-A6 infection.
Recombination events can provide advantages to a virus, such as host immune evasion or enhanced infection. Another feature of enteroviruses is that the transmission of the prototype viruses is difficult to detect. In fact, we are unaware of any recent detections of the CV-A6 prototype Gdula or the CV-A16 prototype G-10; only a few cases of the EV-A71 prototype BrCr have been reported (39). The most plausible explanation for losing track of prototypic enteroviruses is that recombination is very beneficial for viral infection and replication, enabling viruses to outcompete their prototypes, whose infectivity is inferior.
The P1 region in modern enteroviruses is conserved with respect to its homologs in the prototype strains. In other words, no intertype recombination could be detected in the P1 region coding for all the structural proteins (VP1, VP2, VP3, and VP4). Our bootscanning results also indicated that the recombination events between Gdula and CV-A4 occurred in the 5′-UTR and 2A regions, but not within P1. Our recent study on EV-D68 indicated that amino acid substitution on VP1 alone affected viral infectivity and contributed to the EV-D68 North American outbreak in 2014 (40). Events such as recombination may introduce dramatic changes to the enteroviral structural proteins, which may lead to failure in virus assembly or impotence in host infection.
The four CV-A6-Changchun strains exhibited distinct lethal potencies in mice. The differences were not due to strain-specific preferences for infecting different tissues or the potency of viral replication in vivo. Phylogenetic analyses of the P2 region separated CV-A6 strains into lethal and nonlethal groups. Furthermore, we identified three unique amino acids in the 2C region that were identical within each group. The 2C protein is the most conserved protein of Picornaviridae viruses. It has been reported that the N-terminal poliovirus 2C protein possesses ATPase and weak GTPase activities (41, 42). The 2C protein of EV-A71 plays an important role in viral replication (42–44), inhibiting activation of the nuclear factor kappa light-chain enhancer of activated B cells (NF-κB) pathway (45). In the present study, we also observed that 2C from CV-A6 alone caused cell death through autophagy instead of apoptosis, and we linked this capability to 3 residues (Fig. 8 and 9). In addition, data from several studies, including the present one, indicated a close relationship between variations of the viral 2C protein and the lethality of CV-A6, at least in a mouse model (29, 30). The 2C protein may benefit CV-A6 by causing cell death and promoting the release of the virion progeny, which would shorten the infection cycle of the virus (Fig. 5). Our observation of 2C-induced autophagy and its association with viral lethality confirms 2C as a potential target for anti-CV-A6 drug development.
In conclusion, we obtained four representative CV-A6-Changchun strains from 39 CV-A6-positive samples, and primary phylogenetic analyses indicated that multiple strains from Shanghai contributed most to the CV-A6 circulating in Changchun. The results of animal testing indicated that the Changchun046, Changchun097, and Changchun099 strains were lethal, whereas the Changchun098 strain was nonlethal. The similarity and bootscanning tests suggested that all four CV-A6-Changchun strains were recombinants of Gdula and prototype CV-A4, or an unknown parental virus similar to CV-A4. These tests failed to reveal a potential reason for distinct lethality. The lethalities of these strains determined in vivo correlated with their potencies in viral replication and release in vitro. In addition, amino acid variations between the lethal and nonlethal groups were detected at several positions; three substitutions in the 2C region were identical within each group and were associated with the capability of inducing autophagy, as well as the pathogenicity of the protein. The present report enriches the epidemiological information on CV-A6 in northeast China, confirming the Gdula/CV-A4 recombination pattern of modern CV-A6 and revealing differing lethalities among CV-A6 strains, to which the viral 2C protein may contribute. The viral 2C protein may be a good target for designing antiviral drugs against CV-A6 infection.
MATERIALS AND METHODS
Ethics statement and sample selection.
The Ethics Committee at the First Hospital of Jilin University approved this study, and the procedures were carried out in accordance with approved guidelines. Informed consent was obtained from the subjects' parents or guardians. Out of 101 stool specimens provided by the Jilin Provincial Center for Disease Control and Prevention (CDC) in 2013 and used for this study, 39 were identified as positive for CV-A6 using a fluorescent PCR kit (Guangzhou Huayin Medical Technology Inc., Guangdong, China). Partial VP1 sequences were retrieved and phylogenetically analyzed. Four representative Changchun CV-A6 strains were selected for DNA isolation, complete genome sequencing, and further analyses.
Reverse transcription-PCR (RT-PCR), PCR amplification, and sequencing.
Extraction of total RNA from stool specimens was performed using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The cDNA was generated in a 20-μl reaction volume using 1 pM each cDNA primer (46) (primers AN32, AN33, AN34, and AN35 [Table 1]) and the EasyScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China), according to the provided instructions. PCRs were performed in 100-μl volumes containing 10 μl of cDNA, 2.5 U of PrimeStar HS DNA polymerase (TaKaRa, Dalian, China), 2 μl of specific forward and reverse primers (10 μM each), 8 μl of deoxynucleotide (dNTP) mixture (2.5 mM each), and 10 μl of 5× PrimeStar buffer (plus MgCl2). The PCR parameters for all reactions were as follows. cDNA was denatured at 94°C for 4 min. Amplification was performed for 35 cycles consisting of a denaturing step for 30 s at 94°C, a primer-annealing step for 30 s at 49°C to 60°C, and a two-part elongation step for 1 to 2 min at 72°C. Extension was carried out at 72°C for 10 min. The reactions were analyzed by electrophoresis on 1.0% agarose gels. The primers used for CV-A6 amplification and detection are listed in Table 1. The locations of primers were designated according to the CV-A6 prototype Gdula strain (GenBank accession no. AY421764). All amplicons were sequenced bidirectionally.
TABLE 1.
Reverse transcription, PCR, and sequencing primers used in this study
| Primer | Sequencea (5′ to 3′) | Location | Region | Reference |
|---|---|---|---|---|
| AN32 | GTYTGCCA | 3009–3002b | VP1 | 46 |
| AN33 | GAYTGCCA | 3009–3002b | VP1 | 46 |
| AN34 | CCRTCRTA | 3111–3104b | VP1 | 46 |
| AN35 | RCTYTGCCA | 3009–3002b | VP1 | 46 |
| 224 | GCIATGYTIGGIACICAYRT | 1977–1996b | VP3 | 46 |
| 222 | CICCIGGIGGIAYRWACAT | 2969–2951b | VP1 | 46 |
| AN89 | CCAGCACTGACAGCAGYNGARAYNGG | 2602–2627b | VP1 | 46 |
| AN88 | TACTGGACCACCTGGNGGNAYRWACAT | 2977–2951b | VP1 | 46 |
| FL-F-1C | TTAAAACAGCCTGTGGGTTG | 1–20c | 5′ UTR | 19 |
| panEV-F | CCCCTGAATGCGGCTAATC | 458–476c | 5′ UTR | 19 |
| panEV-R | GCTGCTTATGGTGACAATC | 629–610c | 5′ UTR | 19 |
| 5′UTR-EV-F-FW2 | CCCTCTTAGTGGCTGCAATC | 1314–1333c | VP2 | 19 |
| VP1-222-FW2-R | TGGGGCAATAGTTAGTGTGA | 1678–1659c | VP2 | 19 |
| 2715-R | TTCACCTCCACAACYCCTACYAGC | 2715–2692c | VP1 | 19 |
| 3311-F | ATAACTAACACTGCAACCGACC | 3272–329c | VP1 | 19 |
| 3441-R | GCCCARTCATTATGAGTGGC | 3441–3422c | 2A | 19 |
| 4069-R | GCMTCCATCYTAGGTATCCCTA | 4098–4077c | 2B | 19 |
| 4613-F | GTGGTCACAGTYATGGACGATC | 4613–4634c | 2C | 19 |
| 5477-R | GTYGCCGTRCGGAGRATRGGTT | 5406–5385c | 3B | 19 |
| 6150-F | GCAGGCRTTGTTCTCYAAGTAT | 6127–6148c | 3D | 19 |
| 5332-F | GTTTTCCCAGTCACGACTTYCARGGBGCKTA | 6785–6815c | 3D | 19 |
| 650-R | GTCACTGGATGGCCAATCC | 632–650c | 5′ UTR | This study |
| 5331-F | CTCTTTGCTGGGTTCCAAG | 5331–5349c | 3A | This study |
| 7434-R | GCTATTCTGGTTATAACAAATTTACC | 7409–7434c | 3′ UTR | This study |
I, inosine; K = T + G, M = A + C, R = A + G, W = T + A, Y = C + T, B = C + G + T, V = A + G + C, N = A + T + C + G.
The locations of all primers are relative to the genome of PV1 Mahoney (GenBank accession number J02281).
Numbering is based on coxsackievirus A6 strain Gdula (accession no. AY421764).
Phylogenetic analyses.
To determine the phylogenetic relationship between Changchun CV-A6 strains and strains from other locations, CV-A6 sequences, including the prototype Gdula, were aligned using the MEGA software package (version 5.0.5) (47). These alignments were subsequently adjusted manually. Prototypes from the HEV-A group were included as reference sequences, whereas EV-D68 Fermon and poliovirus Sabin 1 were used as the outgroup (Table 2). NJ trees were constructed using the Kimura 2-parameter model and 1,000 bootstrap replicates. Bootstrap values greater than 70% were considered statistically significant for grouping. Different sequence lengths and reference sequences were used as indicated in the figure legends.
TABLE 2.
Reference strains used in this study
| Strain | Type | Yr | Location | Length (nt) | GenBank accession no. |
|---|---|---|---|---|---|
| Gdula | CV-A6 | 1949 | New York | 7434 | AY421764 |
| Fleetwood | CV-A2 | 1947 | Delaware | 7398 | AY421760 |
| Olson | CV-A3 | 1948 | New York | 7395 | AY421761 |
| High Point | CV-A4 | 1948 | North Carolina | 7435 | AY421762 |
| Swartz | CV-A5 | 1950 | New York | 7400 | AY421763 |
| Parker | CV-A7 | 1949 | New York | 7404 | AY421765 |
| Donovan | CV-A8 | 1949 | New York | 7396 | AY421766 |
| Kowalik | CV-A10 | 1950 | New York | 7409 | AY421767 |
| Texas-12 | CV-A12 | 1948 | Texas | 7407 | AY421768 |
| G-14 | CV-A14 | 1950 | Republic of South Africa | 7415 | AY421769 |
| G-10 | CV-A16 | 1951 | Republic of South Africa | 7408 | U05876 |
| BrCr | EV-A71A | 1970 | California | 7408 | U22521 |
| UH1/PM/1997 | EV-A71B | 1997 | Malaysia/Kuala Lumpur | 7411 | AM396587 |
| S10862-SAR-98 | EV-A71C | 1998 | Malaysia/Kota Samarahan | 7409 | DQ341359 |
| FRA91-10369 | EV-A76 | 1991 | France/Caen | 7438 | AY697458 |
| BAN00-10359 | EV-A89 | 2000 | Bangladesh/Bhola | 7429 | AY697459 |
| BAN00-10399 | EV-A90 | 1999 | Bangladesh/Comilla | 7425 | AY697460 |
| BAN00-10406 | EV-A91 | 2000 | Bangladesh/Barisal | 7427 | AY697461 |
| RJG7 | EV-A92 | NAa | USA | 7379 | EF667344 |
| Fermon | EV-D68 | 1962 | California | 7367 | AY426531 |
| Sabin1 | Poliovirus 1 | 1961 | USA | 7441 | V01150 |
NA, not applicable.
Cells, antibodies, and virus strains.
Human rhabdomyosarcoma RD cells (no. CCL-136) were obtained from the American Type Culture Collection (Manassas, VA, USA). They were grown in MEM supplemented with 10% fetal bovine serum (FBS) and 3% l-glutamine at 37°C under 5% CO2. The representative strains Changchun046, Changchun097, Changchun098, and Changchun099 were harvested when the cytopathogenic effect (CPE) reached 90%. Viral titers were determined in RD cells by the microplate CPE method and calculated using the Reed-Muench method (48).
The following antibodies were used to detect protein expression: anti-tubulin (Abcam, Cambridge, MA, USA) and anti-hemagglutinin (HA) Covance, (Princeton, NJ, USA). Both antibodies were used according to the manufacturers' protocols.
Neonatal mouse infection test.
The Jilin University Office of Laboratory Animal Management approved our animal care and experimentation procedures, and we carried out the experiments in accordance with accepted guidelines. One-day-old specific-pathogen-free (SPF) ICR neonatal mice (purchased from the Experimental Animal Center, College of Basic Medicine, Jilin University, Changchun, Jilin, China) were used to establish the animal model of viral infection. The neonatal mice were randomly divided into five groups, with three litters per group (n = 8 to 10 mice/litter), and inoculated intracerebrally with the above-mentioned four CV-A6-Changchun strains or MEM (10 μl/mouse). For 21 days following inoculation, all the newborn mice were monitored daily for clinical symptoms and survival rates. The symptoms were scored as follows: 0, healthy; 1, lethargy and inactivity; 2, wasting; 3, limb-shaking weakness; 4, hind limb paralysis; 5, moribund or dead. The mice in the control group were healthy throughout the experiments. The LD50 was calculated using the Reed-Muench method (48).
Histopathological analyses.
Three mice with grade 4 or 5 symptoms from each of the experimental groups, Changchun046 (105.0 CCID50/ml), Changchun097 (104.0 CCID50/ml), Changchun098 (105.0 CCID50/ml), and Changchun099 (105.0CCID50/ml) were subjected to histopathological analyses 5 days postinfection. Three healthy mice from the control group were also included as the negative control. After the mice were anesthetized, nine types of tissue or organ (heart, liver, spleen, lung, kidney, intestine, brain, spine muscle, and hind limb muscle) were harvested and immersion fixed with 10% formaldehyde solution for 5 days. The samples were dehydrated through an ethanol gradient, clarified through dimethylbenzene, and embedded in paraffin. For hematoxylin and eosin (H&E) staining, 4-μm sections of the tissue were obtained. Histopathological analyses of the tissues were performed under a light microscope.
Viral loads in tissues of newborn infected mice.
Injected intracerebrally with Changchun046, Changchun097, Changchun098, Changchun099, or control medium, three challenged mice from each of the above-mentioned groups and three negative-control mice were used to detect viral loads. All samples (heart, liver, spleen, lung, kidney, intestine, brain, spine muscle, hind limb muscle, and blood) were harvested on days 2, 4, and 6 postinfection. All the collected tissue samples were weighed individually and stored at −80°C for further analysis. The collected tissue samples were disrupted, homogenized in sterile phosphate-buffered saline (PBS) by the freeze-thaw/grinding method, and subsequently centrifuged. The supernatants and blood were subjected to RNA extraction, and viral loads were determined by qRT-PCR, as described previously (49). Viral loads were expressed as log10 copies per milligram of tissue or log10 copies per milliliter of blood.
qRT-PCR.
For qRT-PCR, CV-A6-specific primers were designed based on the conserved region of VP1 as follows: CV-A6-F, AATGAGGCGAGTGTGGAAC, and CV-A6-R, AGGTTGGACACAAAAGTGAACT. The SYBR green-based qPCR was carried out on an Mx3005P (Agilent Technologies Stratagene, Santa Clara, CA, USA) using the TransStart Top Green qPCR Super Mix (Transgen Biotech). Each 20-μl reaction mixture contained 10 μl of SYBR green Super Mix (2×), 0.5 μl of 10 μM (each) CV-A6-F and CV-A6-R primers, 7 μl of double-distilled H2O, and 2 μl of cDNA template. The cycling conditions were as follows: 50°C for 2 min and 95°C for 10 min, followed by 45 cycles consisting of 95°C for 30 s and 60°C for 1 min. The copy number of the target cDNA in the qRT-PCR was determined by a standard curve of 10-fold serial dilutions of a vector containing the full-length genome of Changchun046 (ranging from 103 to 109 copies). The absolute RNA copy number was calculated by using standard dilution curves of plasmids containing the target sequence. The sensitivity of the assay or limit of detection was determined to be the lowest copy number that was amplified consistently within the linear portion of the standard curve.
Recombination determination.
To determine possible recombination events, full-length CV-A6-Changchun sequences were aligned with sequences of prototype strains from the HEV-A group and subjected to similarity tests. Reference strains with high similarity (i.e., CV-A6 and CV-A4) were kept for the bootscanning analyses, with a window of 500 bp and a step of 20. Both tests were performed using the SimPlot software package (version 3.5.1) (50). EV-D68 Fermon and poliovirus Sabin 1 were used as the outgroup in both tests.
Protein sequence alignment.
The amino acid sequences of the P2 region of the four CV-A6 strains and the other 33 CV-A6 strains were aligned using the DNAMan software package (version 6.0).
Plasmid construction.
The sequence coding for the full-length Changchun046 2C (accession no. KT779410) was constructed with a C-terminal HA tag and inserted between the SalI and BamHI sites of VR1012 (30), resulting in the recombinant vector VR1012-Changchun046-2C-HA. VR1012-Changchun097-2C-HA and VR1012-Changchun098-2C-HA were constructed using a similar strategy based on Changchun097 2C (accession no. KT779411) and Changchun098 2C (accession no. KT779412), respectively, using the XbaI and BamHI sites of VR1012. A start codon and a stop codon were assigned to each 2C coding region during PCR to help initiate and terminate translation. Plasmid pEGFP-LC3 was constructed by introducing the LC3 coding frame into the pEGFP-C1 vector (BD Biosciences Clontech, Mountain View, CA, USA) with the XhoI and BamHI sites. Transfections were performed using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions.
Cell proliferation assay.
Cell proliferation was assessed using the TransDetect cell-counting kit (CCK) (TransGen Biotech) according to the manufacturer's protocol. RD cells were plated in 24-well plates (1 × 105 cells/well). After 24 h, the plated cells were transfected with VR1012-Changchun046-2C-HA, VR1012-Changchun097-2C-HA, VR1012-Changchun098-2C-HA, and empty VR1012 plasmid. 2C protein from Changchun099 was not included because it shared identical primary amino acid sequences with Changchun097. At 48 h posttransfection, the cells were harvested and resuspended in 600 μl MEM. Each sample (100 μl) was reseeded in a 96-well plate, and 10 μl CCK reagent was added to each well. After incubation at 37°C for another 4 h, the number of cells per well was measured via absorbance at 450 nm. All experiments were performed in triplicate.
One-step growth curve of CV-A6 strains in cells.
RD cells were seeded in a 96-well plate at a density of 3 × 104 cells per well 1 day prior to the assay. The cells were infected with equal amounts of lethal Changchun046 and nonlethal Changchun098 viruses (quantified by qRT-PCR). After 6 h of viral adsorption at 37°C, the supernatant was removed, and the cells were thoroughly washed two times with MEM to remove unbound virus. Then, 100 μl of the maintenance MEM containing 1% FBS was added to each well, and the plates were incubated at 37°C. During incubation, the virus supernatant was harvested at 8, 10, 12, 14, 16, and 18 h postadsorption and stored at −80°C. Total viral RNAs were extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions and quantified by qRT-PCR using CV-A6-specific primers.
FITC annexin V apoptosis detection assay.
The possibility of CV-A6 2C-induced apoptosis was tested using a fluorescein isothiocyanate (FITC) annexin V apoptosis detection kit I (BD Biosciences, San Jose, CA, USA) in RD cells following the manufacturer's protocol. RD cells were seeded in a 12-well plate and transfected with VR1012-Changchun046-2C-HA; VR1012-Changchun097-2C-HA; VR1012-Changchun098-2C-HA; and their control vector, VR1012. Alternatively, RD cells treated with STS (final concentration, 4 nM; Sigma-Aldrich, St. Louis, MO, USA) were also included as the positive control. At 48 h posttransfection (24 h post-STS treatment), cells were harvested, washed twice with cold PBS, and resuspended in 200 μl 1× binding buffer. The cell suspension (100 μl) was transferred to a 5-ml culture tube and incubated with 5 μl of FITC-labeled annexin V for 15 min at room temperature (25°C) in the dark. Five hundred microliters of 1× binding buffer was added to each tube, and the cells were analyzed by BD FACSCalibur flow cytometry (BD Biosciences) to detect FITC fluorescence.
Autophagy detection using fluorescence microscopy.
RD cells were seeded on 24-well culture plates and grown to 60 to 70% confluence. The RD cells were cotransfected with CV-A6 2C and pEGFP-LC3 using Lipofectamine2000 (Invitrogen), according to the manufacturer's guidelines. The fluorescence signals were visualized using a fluorescence microscope (IX83; Olympus, Tokyo, Japan) 48 h posttransfection.
Statistical analyses.
The values for the clinical scores and viral loads were analyzed using a nonparametric one-way analysis of variance (ANOVA) test. The survival rates were evaluated using a log rank test. The cell proliferation data were analyzed using a two-tailed, unpaired Student t test. All results were expressed as means ± standard deviation (SD). A P value of <0.05 was considered significant.
Accession number(s).
All the CV-A6-Changchun sequences obtained during the study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) under accession numbers KT779375 to KT779413.
ACKNOWLEDGMENTS
We thank Editage for providing editing services and Jun-Liang Chang and Yan-Feng Peng for technical assistance.
This work was funded by the Ministry of Science and Technology of the People's Republic of China (2012CB911100 and 2013ZX10001-005 to Xiao-Fang Yu), the National Natural Science Foundation of China (81772169 to Xiao-Fang Yu), the Fundamental Research Funds for Jilin University (2014zkf04 to Shao-Hua Wang), the Sixth Fund for Young Scholars of the First Hospital of Jilin University (00204020001 to Shao-Hua Wang), the National Natural Science Foundation of China (31270202 to Wen-Yan Zhang), the National Natural Science Foundation of China (81401654 to Juan Du), the Fundamental Research Funds for the Central Universities (2017TD-08 to Juan Du), the National Natural Science Foundation of China (81601363 to Ke Zhao), the China Postdoctoral Science Foundation (2017T100212 and 2017M610193 to Ke Zhao), and the Norman Bethune Health Science Center of Jilin University (yb201302 to Ke Zhao). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Bendig JW, Fleming DM. 1996. Epidemiological, virological, and clinical features of an epidemic of hand, foot, and mouth disease in England and Wales. Commun Dis Rep CDR Rev 6:R81–R86. [PubMed] [Google Scholar]
- 2.Chan KP, Goh KT, Chong CY, Teo ES, Lau G, Ling AE. 2003. Epidemic hand, foot and mouth disease caused by human enterovirus 71, Singapore. Emerg Infect Dis 9:78–85. doi: 10.3201/eid1301.020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimizu H, Utama A, Onnimala N, Li C, Li-Bi Z, Yu-Jie M, Pongsuwanna Y, Miyamura T. 2004. Molecular epidemiology of enterovirus 71 infection in the Western Pacific region. Pediatr Int 46:231–235. doi: 10.1046/j.1442-200x.2004.01868.x. [DOI] [PubMed] [Google Scholar]
- 4.Ho M, Chen ER, Hsu KH, Twu SJ, Chen KT, Tsai SF, Wang JR, Shih SR. 1999. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N Engl J Med 341:929–935. [DOI] [PubMed] [Google Scholar]
- 5.Brown BA, Oberste MS, Alexander JP Jr, Kennett ML, Pallansch MA. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J Virol 73:9969–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoke-Fun C, AbuBakar S. 2006. Phylogenetic evidence for inter-typic recombination in the emergence of human enterovirus 71 subgenotypes. BMC Microbiol 6:74. doi: 10.1186/1471-2180-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao K, Han X, Wang G, Hu W, Zhang W, Yu XF. 2011. Circulating coxsackievirus A16 identified as recombinant type A human enterovirus, China. Emerg Infect Dis 17:1537–1540. doi: 10.3201/eid1708.101719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Zhu C, Bao W, Zhao K, Niu J, Yu XF, Zhang W. 2012. Characterization of full-length enterovirus 71 strains from severe and mild disease patients in northeastern China. PLoS One 7:e32405. doi: 10.1371/journal.pone.0032405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei W, Guo H, Li J, Ren S, Wei Z, Bao W, Hu X, Zhao K, Zhang W, Zhou Y, Sun F, Markham R, Yu XF. 2014. Circulating HFMD-associated coxsackievirus A16 is genetically and phenotypically distinct from the prototype CV-A16. PLoS One 9:e94746. doi: 10.1371/journal.pone.0094746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howlett SE. 2018. Coxsackievirus B3-induced myocarditis: new insights into a female advantage. Can J Cardiol 2018:S0828-282X(18)30134-X. doi: 10.1016/j.cjca.2018.01.086. [DOI] [PubMed] [Google Scholar]
- 11.Wieczorek M, Krzysztoszek A. 2017. Molecular characterization of enteroviruses isolated from acute flaccid paralysis cases in Poland, 1999-2014. Pol J Microbiol 65:443–450. doi: 10.5604/17331331.1227670. [DOI] [PubMed] [Google Scholar]
- 12.Puenpa J, Chieochansin T, Linsuwanon P, Korkong S, Thongkomplew S, Vichaiwattana P, Theamboonlers A, Poovorawan Y. 2013. Hand, foot, and mouth disease caused by coxsackievirus A6, Thailand, 2012. Emerg Infect Dis 19:641–643. doi: 10.3201/eid1904.121666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montes M, Artieda J, Pineiro LD, Gastesi M, Diez-Nieves I, Cilla G. 2013. Hand, foot, and mouth disease outbreak and coxsackievirus A6, northern Spain, 2011. Emerg Infect Dis 19. doi: 10.3201/eid1904.121589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang F, Yuan J, Wang X, Li J, Du J, Su H, Zhou B, Jin Q. 2014. Severe hand, foot, and mouth disease and coxsackievirus A6-Shenzhen, China. Clin Infect Dis 59:1504–1505. doi: 10.1093/cid/ciu624. [DOI] [PubMed] [Google Scholar]
- 15.Guo WP, Lin XD, Chen YP, Liu Q, Wang W, Wang CQ, Li MH, Sun XY, Shi M, Holmes EC, Zhang YZ. 2015. Fourteen types of co-circulating recombinant enterovirus were associated with hand, foot, and mouth disease in children from Wenzhou, China. J Clin Virol 70:29–38. doi: 10.1016/j.jcv.2015.06.093. [DOI] [PubMed] [Google Scholar]
- 16.Feng X, Guan W, Guo Y, Yu H, Zhang X, Cheng R, Wang Z, Zhang Z, Zhang J, Li H, Zhuang Y, Zhang H, Lu Z, Li M, Yu H, Bao Y, Hu Y, Yao Z. 2015. A novel recombinant lineage's contribution to the outbreak of coxsackievirus A6-associated hand, foot and mouth disease in Shanghai, China, 2012-2013. Sci Rep 5:11700. doi: 10.1038/srep11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaunt E, Harvala H, Osterback R, Sreenu VB, Thomson E, Waris M, Simmonds P. 2015. Genetic characterization of human coxsackievirus A6 variants associated with atypical hand, foot and mouth disease: a potential role of recombination in emergence and pathogenicity. J Gen Virol 96:1067–1079. doi: 10.1099/vir.0.000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Wang X, Zhang Y, Gong L, Mao H, Feng C, Ojcius DM, Yan J. 2012. Rapid and sensitive identification of RNA from the emerging pathogen, coxsackievirus A6. Virol J 9:298. doi: 10.1186/1743-422X-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YJ, Chang SC, Tsao KC, Shih SR, Yang SL, Lin TY, Huang YC. 2012. Comparative genomic analysis of coxsackievirus A6 strains of different clinical disease entities. PLoS One 7:e52432. doi: 10.1371/journal.pone.0052432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He YQ, Chen L, Xu WB, Yang H, Wang HZ, Zong WP, Xian HX, Chen HL, Yao XJ, Hu ZL, Luo M, Zhang HL, Ma HW, Cheng JQ, Feng QJ, Zhao DJ. 2013. Emergence, circulation, and spatiotemporal phylogenetic analysis of coxsackievirus a6- and coxsackievirus a10-associated hand, foot, and mouth disease infections from 2008 to 2012 in Shenzhen, China. J Clin Microbiol 51:3560–3566. doi: 10.1128/JCM.01231-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di B, Zhang Y, Xie H, Li X, Chen C, Ding P, He P, Wang D, Geng J, Luo L, Bai Z, Yang Z, Wang M. 2014. Circulation of coxsackievirus A6 in hand-foot-mouth disease in Guangzhou, 2010–2012. Virol J 11:157. doi: 10.1186/1743-422X-11-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, Zeng H, Zheng H, Yi L, Guo X, Liu L, Sun L, Tan X, Li H, Ke C, Lin J. 2014. Hand, foot and mouth disease in Guangdong, China, in 2013: new trends in the continuing epidemic. Clin Microbiol Infect 20:O442–O445. doi: 10.1111/1469-0691.12468. [DOI] [PubMed] [Google Scholar]
- 23.Hongyan G, Chengjie M, Qiaozhi Y, Wenhao H, Juan L, Lin P, Yanli X, Hongshan W, Xingwang L. 2014. Hand, foot and mouth disease caused by coxsackievirus A6, Beijing, 2013. Pediatr Infect Dis J 33:1302–1303. doi: 10.1097/INF.0000000000000467. [DOI] [PubMed] [Google Scholar]
- 24.Zeng H, Lu J, Zheng H, Yi L, Guo X, Liu L, Rutherford S, Sun L, Tan X, Li H, Ke C, Lin J. 2015. The epidemiological study of coxsackievirus A6 revealing hand, foot and mouth disease epidemic patterns in Guangdong, China. Sci Rep 5:10550. doi: 10.1038/srep10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan X, Li L, Zhang B, Jorba J, Su X, Ji T, Yang D, Lv L, Li J, Xu W. 2015. Molecular epidemiology of coxsackievirus A6 associated with outbreaks of hand, foot, and mouth disease in Tianjin, China, in 2013. Arch Virol 160:1097–1104. doi: 10.1007/s00705-015-2340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu YQ, Xie GC, Li DD, Pang LL, Xie J, Wang P, Chen Y, Yang J, Cheng WX, Zhang Q, Jin Y, Duan ZJ. 2015. Prevalence of coxsackievirus A6 and enterovirus 71 in hand, foot and mouth disease in Nanjing, China in 2013. Pediatr Infect Dis J 34:951–957. doi: 10.1097/INF.0000000000000794. [DOI] [PubMed] [Google Scholar]
- 27.Han JF, Xu S, Zhang Y, Zhu SY, Wu DL, Yang XD, Liu H, Sun BX, Wu XY, Qin CF. 2014. Hand, foot, and mouth disease outbreak caused by coxsackievirus A6, China, 2013. J Infect 69:303–305. doi: 10.1016/j.jinf.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 28.Sickles GM, Mutterer M, Feorino P, Plager H. 1955. Recently classified types of Coxsackie virus, group A; behavior in tissue culture. Proc Soc Exp Biol Med 90:529–531. doi: 10.3181/00379727-90-22088. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Dong Z, Wei Q, Carr MJ, Li J, Ding S, Tong Y, Li D, Shi W. 2017. A neonatal murine model of coxsackievirus A6 infection for evaluation of antiviral and vaccine efficacy. J Virol 91:e02450-. doi: 10.1128/JVI.02450-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, Mao Q, Li S, Gao F, Zhao H, Liu Y, Wan J, Ye X, Xia N, Cheng T, Liang Z. 2016. A neonatal mouse model for the evaluation of antibodies and vaccines against coxsackievirus A6. Antiviral Res 134:50–57. doi: 10.1016/j.antiviral.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Zhang J, Chen Z, Yang L, Xing X, Ma X, Yang Z. 2013. Both PI3K- and mTOR-signaling pathways take part in CVB3-induced apoptosis of Hela cells. DNA Cell Biol 32:359–370. doi: 10.1089/dna.2013.2003. [DOI] [PubMed] [Google Scholar]
- 32.Xi X, Zhang X, Wang B, Wang T, Wang J, Huang H, Wang J, Jin Q, Zhao Z. 2013. The interplays between autophagy and apoptosis induced by enterovirus 71. PLoS One 8:e56966. doi: 10.1371/journal.pone.0056966. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Zhu G, Zheng Y, Zhang L, Shi Y, Li W, Liu Z, Peng B, Yin J, Liu W, He X. 2013. Coxsackievirus A16 infection triggers apoptosis in RD cells by inducing ER stress. Biochem Biophys Res Commun 441:856–861. doi: 10.1016/j.bbrc.2013.10.142. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y, He X, Zhu G, Tu H, Liu Z, Li W, Han S, Yin J, Peng B, Liu W. 2015. Coxsackievirus A16 elicits incomplete autophagy involving the mTOR and ERK pathways. PLoS One 10:e0122109. doi: 10.1371/journal.pone.0122109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng X, Guan W, Guo Y, Yu H, Zhang X, Cheng R, Wang Z, Zhang Z, Zhang J, Li H, Zhuang Y, Zhang H, Lu Z, Li M, Yu H, Bao Y, Hu Y, Yao Z. 2015. Genome sequence of a novel recombinant coxsackievirus A6 strain from Shanghai, China, 2013. Genome Announc 3:e01347-. doi: 10.1128/genomeA.01347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu QB, Zhang XA, Wo Y, Xu HM, Li XJ, Wang XJ, Ding SJ, Chen XD, He C, Liu LJ, Li H, Yang H, Li TY, Liu W, Cao WC. 2012. Circulation of Coxsackievirus A10 and A6 in hand-foot-mouth disease in China, 2009–2011. PLoS One 7:e52073. doi: 10.1371/journal.pone.0052073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puenpa J, Vongpunsawad S, Osterback R, Waris M, Eriksson E, Albert J, Midgley S, Fischer TK, Eis-Hubinger AM, Cabrerizo M, Gaunt E, Simmonds P, Poovorawan Y. 2016. Molecular epidemiology and the evolution of human coxsackievirus A6. J Gen Virol 97:3225–3231. doi: 10.1099/jgv.0.000619. [DOI] [PubMed] [Google Scholar]
- 38.Lukashev AN, Lashkevich VA, Ivanova OE, Koroleva GA, Hinkkanen AE, Ilonen J. 2005. Recombination in circulating human enterovirus B: independent evolution of structural and non-structural genome regions. J Gen Virol 86:3281–3290. doi: 10.1099/vir.0.81264-0. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, Lu S, Xian J, Ye J, Xiao L, Luo J, Zen K, Liu F. 2013. Complete genome sequence of a human enterovirus 71 strain isolated in Wuhan, China, in 2010. Genome Announc 1:e01112-. doi: 10.1128/genomeA.01112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du J, Zheng B, Zheng W, Li P, Kang J, Hou J, Markham R, Zhao K, Yu XF. 2015. Analysis of enterovirus 68 strains from the 2014 North American outbreak reveals a new clade, indicating viral evolution. PLoS One 10:e0144208. doi: 10.1371/journal.pone.0144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirzayan C, Wimmer E. 1992. Genetic analysis of an NTP-binding motif in poliovirus polypeptide 2C. Virology 189:547–555. doi: 10.1016/0042-6822(92)90578-D. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez PL, Carrasco L. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J Biol Chem 270:10105–10112. doi: 10.1074/jbc.270.17.10105. [DOI] [PubMed] [Google Scholar]
- 43.Tang WF, Yang SY, Wu BW, Jheng JR, Chen YL, Shih CH, Lin KH, Lai HC, Tang P, Horng JT. 2007. Reticulon 3 binds the 2C protein of enterovirus 71 and is required for viral replication. J Biol Chem 282:5888–5898. doi: 10.1074/jbc.M611145200. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Wu Z, Jin Q. 2012. COPI is required for enterovirus 71 replication. PLoS One 7:e38035. doi: 10.1371/journal.pone.0038035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Z, Li H, Zhang Z, Meng J, Mao D, Bai B, Lu B, Mao P, Hu Q, Wang H. 2011. Enterovirus 71 2C protein inhibits TNF-alpha-mediated activation of NF-kappaB by suppressing IkappaB kinase beta phosphorylation. J Immunol 187:2202–2212. doi: 10.4049/jimmunol.1100285. [DOI] [PubMed] [Google Scholar]
- 46.Nix WA, Oberste MS, Pallansch MA. 2006. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol 44:2698–2704. doi: 10.1128/JCM.00542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 49.Chang J, Li J, Liu X, Liu G, Yang J, Wei W, Zhang W, Yu XF. 2015. Broad protection with an inactivated vaccine against primary-isolated lethal enterovirus 71 infection in newborn mice. BMC Microbiol 15:139. doi: 10.1186/s12866-015-0474-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]