ABSTRACT
Influenza A(H1) viruses circulating in swine represent an emerging virus threat, as zoonotic infections occur sporadically following exposure to swine. A fatal infection caused by an H1N1 variant (H1N1v) virus was detected in a patient with reported exposure to swine and who presented with pneumonia, respiratory failure, and cardiac arrest. To understand the genetic and phenotypic characteristics of the virus, genome sequence analysis, antigenic characterization, and ferret pathogenesis and transmissibility experiments were performed. Antigenic analysis of the virus isolated from the fatal case, A/Ohio/09/2015, demonstrated significant antigenic drift away from the classical swine H1N1 variant viruses and H1N1 pandemic 2009 viruses. A substitution in the H1 hemagglutinin (G155E) was identified that likely impacted antigenicity, and reverse genetics was employed to understand the molecular mechanism of antibody escape. Reversion of the substitution to 155G, in a reverse genetics A/Ohio/09/2015 virus, showed that this residue was central to the loss of hemagglutination inhibition by ferret antisera raised against a prototypical H1N1 pandemic 2009 virus (A/California/07/2009), as well as gamma lineage classical swine H1N1 viruses, demonstrating the importance of this residue for antibody recognition of this H1 lineage. When analyzed in the ferret model, A/Ohio/09/2015 and another H1N1v virus, A/Iowa/39/2015, as well as A/California/07/2009, replicated efficiently in the respiratory tract of ferrets. The two H1N1v viruses transmitted efficiently among cohoused ferrets, but respiratory droplet transmission studies showed that A/California/07/2009 transmitted through the air more efficiently. Preexisting immunity to A/California/07/2009 did not fully protect ferrets from challenge with A/Ohio/09/2015.
IMPORTANCE Human infections with classical swine influenza A(H1N1) viruses that circulate in pigs continue to occur in the United States following exposure to swine. To understand the genetic and virologic characteristics of a virus (A/Ohio/09/2015) associated with a fatal infection and a virus associated with a nonfatal infection (A/Iowa/39/2015), we performed genome sequence analysis, antigenic testing, and pathogenicity and transmission studies in a ferret model. Reverse genetics was employed to identify a single antigenic site substitution (HA G155E) responsible for antigenic variation of A/Ohio/09/2015 compared to related classical swine influenza A(H1N1) viruses. Ferrets with preexisting immunity to the pandemic A(H1N1) virus were challenged with A/Ohio/09/2015, demonstrating decreased protection. These data illustrate the potential for currently circulating swine influenza viruses to infect and cause illness in humans with preexisting immunity to H1N1 pandemic 2009 viruses and a need for ongoing risk assessment and development of candidate vaccine viruses for improved pandemic preparedness.
KEYWORDS: antigenic variation, reverse genetic analysis, swine influenza virus, transmissibility, viral pathogenesis
INTRODUCTION
The emergence of a transmissible influenza A virus in humans to which there is little or no preexisting immunity is an ongoing threat to public health. The exposure of humans to animals in which antigenically diverse influenza viruses circulate creates an opportunity for such an event to occur. The segmented nature of the influenza virus genome allows for gene segment reassortment events in coinfected hosts, leading to antigenic shift, while accumulation of substitutions in the surface glycoprotein genes leads to antigenic drift (1). These constant evolutionary mechanisms mandate frequent vaccine composition changes for seasonal influenza vaccines and continuous development of prepandemic candidate vaccine viruses for pandemic preparedness (2, 3).
Classical swine H1N1 influenza viruses were first isolated in 1930 (4) and have been shown to be antigenically similar to the reconstructed 1918 H1N1 pandemic virus (5). Due to their susceptibility to both human and avian influenza viruses, swine have long been believed to be a “mixing vessel” for genetic reassortment and are a potential zoonotic source of influenza A viruses with pandemic potential (6, 7). In fact, this dogma was demonstrated in 2009 when a reassortant H1N1 virus with a classical swine H1 gamma lineage HA gene segment, NA and M gene segments derived from Eurasian swine influenza viruses, and RNA-dependent RNA polymerase gene segments derived from human and avian viruses was identified in humans and spread globally, causing the first influenza pandemic of the 21st century (8, 9). The evolution of the H1N1 pandemic 2009 (H1N1pdm09) virus was complex and stepwise. In the late 1990s, novel A(H3N2) viruses with a triple reassortant internal gene (TRIG) constellation, containing genes from classical swine H1N1 viruses (NP, M, and NS), North American avian influenza viruses (PB2 and PA), and human seasonal H3N2 viruses (HA, NA, and PB1), emerged in North American pigs and were transmitted to humans (10). Subsequent reassortment events led to the emergence of H1N1 and H1N2 viruses in pigs containing the TRIG constellation but with HA and NA genes derived from classical swine H1N1 and/or human seasonal influenza viruses that had entered pigs via reverse zoonoses (11). Prior to 2009, sporadic zoonotic infections with influenza A viruses that circulate in swine (now termed variant influenza viruses and given the subtype nomenclature H1N1v, H1N2v, and H3N2v) were identified (12). Analysis of the H1N1v viruses showed that the majority were antigenically distinct from seasonal H1N1 influenza viruses in humans (12). The H1N1pdm09 virus displaced the previously circulating seasonal H1N1 virus, which originally emerged in 1977 and caused the Russian flu pandemic, and continues to circulate in the human population. Therefore, contemporary H1N1pdm09 viruses are included as a component of annual vaccines. As a result, classical swine gamma lineage H1N1 viruses that persist in the North American swine population have been shown to be antigenically related to the circulating strains of the classical swine origin HA gene segment of the H1N1pdm09 viruses and, thus, human vaccine strains (13).
During 2015, 3 human H1N1v influenza virus infections were detected in the United States (14). In Ohio, an H1N1v virus (A/Ohio/09/2015; OH/09) was isolated from a fatal case of a man likely exposed to infected pigs at a livestock facility (14). Although the individual had underlying cardiovascular disease that may have contributed to the outcome, this case marked the first documented fatality following classical swine H1N1 virus infection in the United States since novel influenza A virus infections became notifiable in the United States in 2005. Another H1N1v virus, A/Iowa/39/2015 (IA/39), was isolated from a patient in Iowa with a history of exposure to swine; the patient was hospitalized but fully recovered. We therefore set out to improve our understanding of the genetic and phenotypic characteristics of these emerging H1N1v viruses. Reverse genetics was used to assess amino acid substitutions within the HA associated with the lack of antigenic cross-reactivity, and the ferret model was employed for risk assessment of these viruses. We compared the virulence and transmissibility of OH/09 and IA/39 viruses to a representative H1N1pdm09 virus, A/California/07/2009 (CA/07), that was used in the seasonal vaccine from 2009 to 2017 and evaluated the protection of prior infection by CA/07 virus from H1N1v infection.
RESULTS
Virus isolation and molecular characterization.
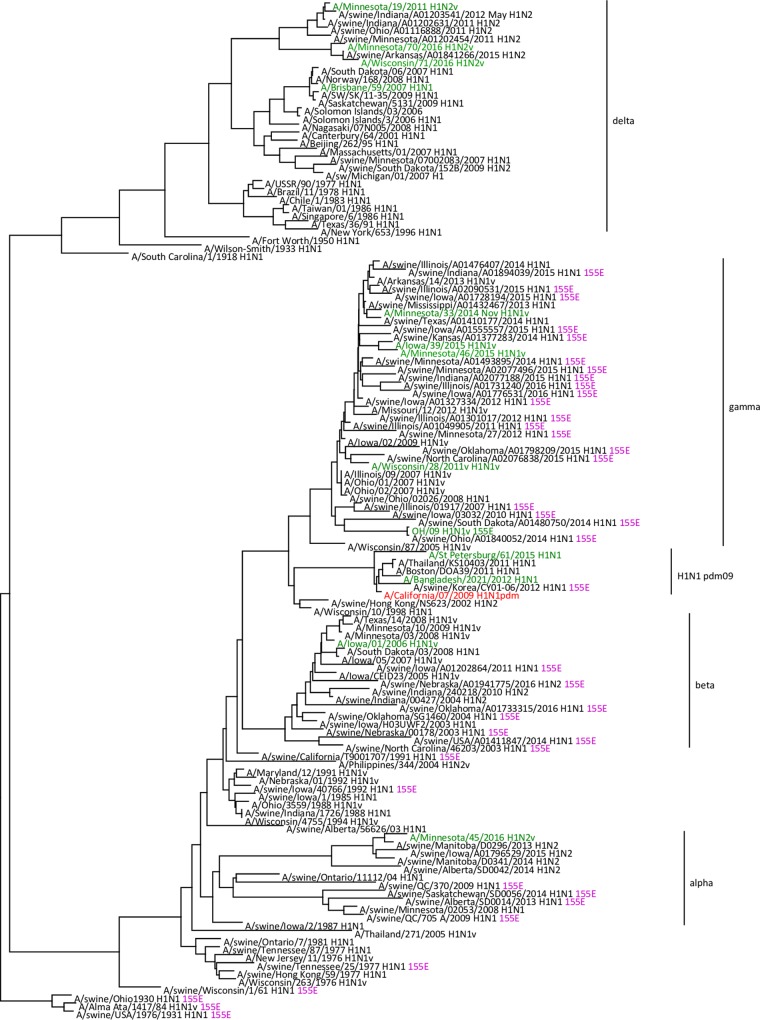
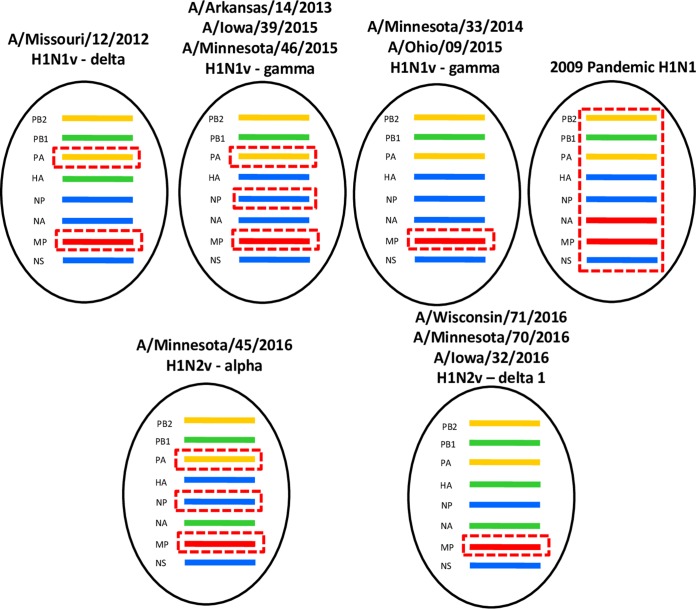
OH/09 virus was isolated from a bronchoalveolar lavage obtained from a patient that presented with influenza-like illness at an intensive care unit. The patient developed acute respiratory disease and died from complications (15). Full genome analysis of the original specimen and virus isolate showed this was an H1N1v virus with an HA gene segment from the classical swine gamma lineage (GISAID accession numbers EPI_ISL_178264 and EPI_ISL_179403). The virus was found to cluster with related 2014 and 2015 swine H1N1 viruses from Ohio and neighboring states (Fig. 1 and Fig. S1A and B in the supplemental material). In a genetic comparison with the nearest H1N1pdm09 vaccine virus, CA/07 X-179A, there were 41 inferred amino acid substitutions in the HA1 portion (globular head domain) of the HA protein. Two of these amino acid substitutions in HA (G155E and D222G; H1 structural numbering) had significant potential to impact antigenicity, receptor binding, and/or transmission. The NA gene segment was determined to belong to the classical swine N1 lineage, while the PB2, PB1, PA, NP, and NS genes belong to the triple reassortant swine lineages (Fig. 2 and Fig. S1C to I). Amino acid sequence analysis of the NA protein did not identify substitutions associated with reduced susceptibility to neuraminidase inhibitors. Like the majority of recent swine and H1v viruses, the M gene segment of OH/09 virus was most closely related to the M gene segments of H1N1pdm09 lineage.
FIG 1.
Evolutionary relationships of the hemagglutinin genes of H1N1v and swine influenza viruses detected in North America. The H1N1pdm09 candidate vaccine virus, A/California/07/2009, is shown in red. Viruses isolated from humans are in green. Viruses possessing HA 155E (shown in purple) are annotated with this residue at the right of each strain name. The scale bar represents nucleotide substitutions per site.
FIG 2.
Genome constellations of H1N1v viruses detected in the United States. Dashed red box indicates genes derived from H1N1pdm09 virus, red indicates genes derived from Eurasian swine lineage, green indicates genes derived from human seasonal lineage, yellow indicates genes derived from avian North American lineage, and blue indicates classical swine H1N1 North American lineage.
IA/39 virus was isolated from a hospitalized patient who had direct contact with swine in the week prior to infection. The patient fully recovered (16). Full genome sequencing of IA/39 (GISAID accession number EPI_ISL_194942) confirmed that this virus was a gamma lineage H1N1v virus containing PA, NP, and M genes derived from H1N1pdm09 virus, PB2 and PA genes derived from triple reassortant swine lineages, and NA and NS derived from the classical swine lineage (Fig. 2 and Fig. S1A to I). The virus gene segments cluster with 2014 and 2015 swine H1N1 viruses from Iowa and neighboring states (Fig. 1). Compared to the sequence of CA/07 virus, there were 35 amino acid substitutions in the HA protein. IA/39 virus shared amino acids at positions 155 (G) and 222 (D) with the CA/07 isolate used in this study. CA/07 virus contained PB1, PB2, and PA genes from triple reassortant swine H1N1 virus lineage, HA, NP, and NS from the classical swine lineage, and the NA and M gene segment from Eurasian swine lineage, a combination of genes that had not been observed in swine and humans prior to 2009 (Fig. 2) (8).
Antigenic characterization and reverse genetics candidate vaccine virus development.
Hemagglutination inhibition (HI) testing with ferret antisera raised against wild-type CA/07 and gamma lineage H1N1v viruses showed significantly reduced titers to OH/09 virus compared to titers with the homologous viruses. HI reactivity of OH/09 virus to adult human pooled sera collected postvaccination with the 2015 to 2016 vaccine was also reduced (Table 1). Based on these data, a candidate vaccine virus for OH/09 was recommended by WHO during the September 2015 Consultation on Influenza Vaccine Composition Meeting (17), and the reverse genetics-derived candidate vaccine virus IDCDC-RG48A (RG48A) was developed. Ferret antisera generated against CA/07 and recent gamma lineage H1N1v viruses also showed significantly reduced titers to RG48A virus compared to titers with the homologous viruses (Table 1). HI reactivity of RG48A virus with the pooled human sera was also significantly reduced. Titers of ferret antisera raised against RG48A virus were reduced to CA/07 and recent gamma lineage H1N1v viruses, indicating two-way antigenic variation between these viruses. HI reactivity of OH/09 virus to postvaccination adult human sera was reduced to RG48A. Reverse genetics virus derived from RG48A virus with only the E155G substitution was also isolated in order to identify the amino acids responsible for this antigenic variation. HI testing with ferret antisera raised against wild-type CA/07 and gamma lineage H1N1v viruses showed no reduction in titers to the reverse genetics-generated OH/09 × PR8 reassortant virus with HA E155G, indicating that this change restored antibody recognition (Table 1). Antisera generated against the OH/09 × PR8 reassortant virus with HA E155G also regained the ability to inhibit hemagglutination of H1N1pdm09 and gamma lineage viruses. Of note, the 155G reverse-engineered OH/09 mutant retained full cross-reactivity with the 155E wild-type OH/09 ferret antisera and vice versa, indicating the single change at position 115 was insufficient to alter antibody recognition. In contrast to OH/09 virus, IA/39 virus cross-reacted with antisera raised to CA/07 and other gamma lineage H1N1 viruses with moderate HI titers, indicating reduced but significantly higher HI titers than those for OH/09. In order to assess which swine influenza viruses carried this mutation, HA sequences of all classical swine H1N1 viruses in the NCBI and GISAID databases (n = 2,775), including those from the alpha, beta, and gamma lineages, were aligned and analyzed. Of these, 11.0% (305/2,775) had the G155E change, indicating its prevalence among swine influenza viruses. These viruses were not localized to any one phylogenetic group but instead were distributed across the alpha (10.1%), beta (16.4%), and gamma (9.5%) lineages of the classical swine H1 viruses (Fig. 1).
TABLE 1.
Hemagglutination inhibition testing of H1N1v, seasonal, and H1N1pdm09 viruses
| Reference virus | Lineage | HI titer for ferret antiserac: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bris/59 | CA/07 | X-179 | BA/2021 | St.P/15 | IA/01 | WI/28 | MO/12 | OH/09 wt | RG48A (155E) | OH/09 (155G) | MN/33 | IA/39 | Human poola | ||
| A/Brisbane/59/2007 | Seasonal H1N1 | 2,560 | 10 | <10 | <10 | 10 | <10 | <10 | <10 | <10 | <10 | 20 | 20 | 10 | 320 |
| A/California/07/2009 | pdm09 | <10b | 5,120 | 1,280 | 320 | 320 | 2,560 | 80 | 40 | 80 | <10 | 2,560 | 1,280 | 160 | 640 |
| A/California/07/09 X-179A | pdm09 | <10 | 640 | 640 | 640 | 320 | 640 | <10 | 20 | 1,280 | 80 | 320 | 640 | 640 | 2,560 |
| A/Bangladesh/2021/2012 | pdm09 (6A) | <10 | 640 | 320 | 1,280 | 160 | 320 | <10 | 20 | 160 | 10 | 640 | 160 | 320 | 640 |
| A/St Petersburg/61/2015 | pdm09 (6B) | <10 | 640 | 320 | 640 | 1,280 | 160 | <10 | <10 | 40 | <10 | 640 | 320 | 80 | 160 |
| A/Iowa/01/2006 | H1N1v (beta) | 10 | 1,280 | 160 | 320 | 10 | 320 | <10 | <10 | 5,120 | 640 | 5,120 | 640 | 640 | 640 |
| A/Wisconsin/28/2011 | H1N1v (gamma) | 10 | 5,120 | 5,120 | 640 | 640 | 5,120 | 640 | 640 | 1,280 | 160 | 5,120 | 5,120 | 5,120 | 1,280 |
| A/Missouri/12/2012 | H1N1v (gamma) | 10 | 5,120 | 5,120 | 5,120 | 1,280 | 5,120 | 5,120 | 5,120 | 5,120 | 1,280 | 5,120 | 5,120 | 5,120 | 5,120 |
| A/Ohio/09/2015 (155E) | H1N1v (gamma) | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | 2,560 | 1,280 | 2,560 | 160 | 320 | 80 |
| A/Ohio/09/2015 RG48A (155E) | H1N1v (gamma) | 10 | 20 | 10 | 80 | 40 | 80 | <10 | 10 | 5,120 | 5,120 | 5,120 | 2,560 | 2,560 | 320 |
| A/Ohio/09/2015 RG (155G) | H1N1v (gamma) | <10 | 5,120 | 2,560 | 320 | 320 | 2,560 | 160 | 160 | 5,120 | 5,120 | 5,120 | 2,560 | 1,280 | 640 |
| A/Minnesota/33/2014 | H1N1v (gamma) | <10 | 5,120 | 2,560 | 640 | 640 | 2,560 | 640 | 320 | 640 | 320 | 5,120 | 5,120 | 1,280 | 640 |
| A/Iowa/39/2015 | H1N1v (gamma) | <10 | 320 | 160 | 320 | 160 | 640 | 10 | 10 | 2,560 | 320 | 5,120 | 5,120 | 5,120 | 1,280 |
Data are from a 2014 to 2015 postvaccine immune serum pool from adult (19 to 49 years of age) vaccinees. A/California/07/2009 was the H1N1 vaccine component.
Titers of <10 indicate the lowest dilution used in the HI assay.
Boldface and underlining indicate the HI titer of the ferret antisera with the homologous reference virus.
Pathogenesis of H1N1v viruses in ferrets.
The ferret model is routinely used in the study of influenza virus pathogenesis and transmission due to several shared similarities between the human and ferret respiratory tracts and a number of shared clinical signs and symptoms of influenza virus infection (18). The results from the ferret model are included in the Influenza Risk Assessment Tool (IRAT), a multiattribute model used to guide the selection of candidate vaccine viruses to aid pandemic preparedness (19). In an effort to better understand the disease caused by emerging H1N1v viruses, 3 groups of 3 ferrets each were intranasally (i.n.) inoculated with 105 PFU of CA/07, OH/09, or IA/39 virus and were euthanized 3 or 4 days later for titration of virus in pulmonary and extrapulmonary tissues. Additional ferrets for each group were inoculated with the same dose and were observed for 14 days for signs of illness, and nasal washes and rectal swabs were collected for measurement of virus shedding. These animals also served as the inoculated donor ferrets in the transmission experiments described later. All animals exhibited mild lethargy and a mean increase in body temperature of ≤1.4°C above the baseline (Table 2). OH/09 virus-infected animals had the greatest weight loss (on average, 13.7% between days 6 and 11 postinoculation [p.i.]) compared with ferrets in the other virus groups, with one ferret requiring euthanasia on day 10 p.i. due to severe weight loss. All animals in the IA/39 and CA/07 virus groups survived the full course of infection. Ferrets inoculated with OH/09 or CA/07 virus, but not those infected by IA/39 virus, showed signs of respiratory infection, such as sneezing and nasal discharge, while only CA/07 virus-infected ferrets exhibited gastrointestinal signs. Each of the viruses was found at high titers in nasal turbinates and trachea of ferrets (Fig. 3). Mean titers of OH/09, IA/39, and CA/07 viruses were 6.1, 5.1, and 4.9 log10 PFU/ml in nasal turbinates and 4.1, 3.9, and 4.9 log10 PFU/g in tracheal tissues, respectively. Virus was detected in the lungs of all ferrets inoculated with IA/39 and CA/07 viruses, with mean titers of 4.3 and 4.4 log10 PFU/g, respectively. OH/09 was detected in the lungs of 2 ferrets, with mean titers of 2.7 log10 PFU/g. Low levels of IA/39 and CA/07 viruses were detected in rectal swabs collected from ferrets 1 to 7 days p.i., while OH/09 virus was below detectable limits (Table 2). Consistently among all of the virus groups, there was no evidence of virus in brain, spleen, kidney, or liver of ferrets (data not shown). These findings indicate that the H1N1v viruses possess the ability to replicate in the respiratory tracts of ferrets while causing mild to moderate morbidity.
TABLE 2.
Comparison of clinical signs observed in ferrets inoculated with H1N1v or H1N1pdm09 virus
| Virus | Inoculation dose (log10 PFU) | % wt lossa | Temp (°C) changeb | Lethargyc | No. of ferrets with clinical sign/total no. of ferrets |
||||
|---|---|---|---|---|---|---|---|---|---|
| Lethalityd | Nasal discharge | Sneezing | Diarrhea | RSe | |||||
| OH/09 | 5.0 | 13.7 (6–11) | 1.4 (1–4) | 1.1 (7/9) | 1/9 | 2/9 | 7/9 | 0/9 | 0/3 |
| IA/39 | 5.0 | 6.0 (6–10) | 0.7 (2–5) | 1.1 (6/6) | 0/6 | 0/6 | 0/6 | 0/6 | 2/6 |
| CA/07 | 5.0 | 10.6 (7–9) | 0.7 (1–5) | 1.1 (3/3) | 0/3 | 1/3 | 1/3 | 3/3 | 3/3 |
Average percent maximum weight loss. Range of days p.i. of observations are indicated in parentheses.
Average maximum temperature change relative to the baseline. Range of days p.i. of observations are indicated in parentheses.
Relative inactivity index is shown (46). Number of ferrets displaying lethargy over total number of ferrets is displayed in parentheses.
Number of animals euthanized during the experiment due to severe weight loss.
Number of ferrets with detectable virus in rectal swabs (RS). Limit of detection is 10 PFU.
FIG 3.
Influenza virus titers in ferret tissues. Three ferrets each were intranasally inoculated with 105 PFU of A/Ohio/09/20015 (A), A/Iowa/39/2015 (B), or A/California/07/2009 (C) virus. Tissues were collected on day 3 p.i. from ferrets inoculated with OH/09 or IA/39 virus and on day 4 p.i. from ferrets inoculated with CA/07 virus. Virus titers in nasal turbinates (NT) are expressed as log10 PFU/ml of tissue homogenate and in trachea (TR) and lungs (LG) as log10 PFU/g of tissue. The limit of detection was 10 PFU. Gray, white, and black bars indicate one of each of the three ferrets tested.
Transmissibility of H1N1v viruses in ferrets.
Whenever a zoonotic virus jumps the species barrier to cause a human infection, it provides an opportunity for the virus to adapt to humans and potentially become capable of sustained transmission. In a previous study, the H1N1pdm09 viruses were shown to transmit efficiently in a direct-contact setting (20). To assess the transmission capabilities of the H1N1v viruses isolated in Ohio and Iowa, experiments using both the direct-contact and respiratory droplet transmission models were performed, each providing the potential for aerosol transmission while the latter prevents transmission by contact. For the direct-contact experiment, groups of three ferrets were i.n. inoculated with 105 PFU of OH/09 or IA/39 virus, and a day later, a naive ferret was placed in the same cage as each inoculated animal. Nasal washes were collected every other day from inoculated and contact animals for virus titration. Virus titers in nasal washes from inoculated ferrets peaked on day 1 or 3 p.i. (≥5.2 log10 PFU/ml) (Table 3) and were cleared by day 7 p.i. (Fig. 4). OH/09 and IA/39 viruses transmitted between all cohoused ferret pairs by day 3 or 5 postcontact (p.c.), respectively, and were cleared by day 9 p.c. Mean peak virus titers in nasal washes of contact ferrets reached ≥5.0 log10 PFU/ml, and seroconversion was also confirmed in all of these animals (Fig. 4 and Table 3). For the respiratory droplet transmission experiment, groups of three ferrets were i.n. inoculated with 105 PFU of OH/09, IA/39, or CA/07 virus. Mean peak titers in nasal washes of inoculated ferrets were again observed on day 1 p.i. at ≥5.1 log10 PFU/ml (Table 3). CA/07 virus transmitted most efficiently, with all three contact ferrets shedding high titers of virus (mean peak of 6.6 log10 PFU/ml) by 3 days p.c. (Fig. 5). IA/39 virus transmitted to 2 of 3 contact animals (mean peak of 5.6 log10 PFU/ml), whereas OH/09 virus transmitted to a single ferret (peak of 5.1 log10 PFU/ml). Overall, the results show that the H1N1v viruses can readily transmit among cohoused ferrets, but they lack the ability to transmit as efficiently as the CA/07 H1N1pdm09 virus by respiratory droplets. Further adaptation of these viruses to humans may result in an H1N1v virus with the ability to transmit as efficiently as the virus that caused the 2009 H1N1 pandemic.
TABLE 3.
Transmission of H1N1v or H1N1pdm09 virus in ferrets
| Virus | Inoculated ferret NW titera | Direct-contact modelb |
Inoculated ferret NW titera | Respiratory droplet modelb |
||
|---|---|---|---|---|---|---|
| Virus detectionc | Seroconversione | Virus detectionc | Seroconversione | |||
| OH/09 | 6.8 ± 1.0 (1) | 3/3 (5.3 ± 0.5; 3, 5) | 3/3 (≥2,560) | 6.8 ± 0.4 (1) | 1/3 (5.1; 5) | 1/3 (2,560) |
| IA/39 | 5.2 ± 0.1 (1, 3) | 3/3 (5.0 ± 0.1; 3, 7) | 3/3 (≥640) | 5.1 ± 0.01 (1) | 2/3 (5.6 ± 0.4; 5, 7) | 2/3 (1,280) |
| CA/07 | NTd | NT | NT | 7.1 ± 0.4 (1) | 3/3 (6.6 ± 0.4; 3) | 3/3 (≥1,280) |
Average maximum nasal wash (NW) titer of inoculated ferrets (log10 PFU/ml) ± SD; days p.i. of peaks are indicated in parentheses.
Number of ferrets in which transmission occurred out of total number of ferrets.
Average maximum nasal wash titer of contact ferrets in which transmission occurred (log10 PFU/ml) ±SD and day p.c. of peaks are indicated in parentheses.
NT, not tested.
Hemagglutination inhibition against homologous virus; titers are shown in parentheses.
FIG 4.
Transmissibility of H1N1v viruses among cohoused ferrets. Three ferrets each were intranasally inoculated with 105 PFU of A/Ohio/09/2015 (A) or A/Iowa/39/2015 (B) virus. The following day, a naive ferret was placed with each inoculated ferret in the same cage. Nasal wash titers from individual inoculated (left side of the graph) and contact (dotted bars; right side of the graph) ferrets were assessed by plaque assay. Limit of detection was 10 PFU. Gray, white, and black bars indicate one of each of the three ferrets tested.
FIG 5.

Respiratory droplet transmissibility of H1N1v and H1N1pdm09 viruses in ferrets. Three ferrets each were intranasally inoculated with 105 PFU of A/Ohio/09/2015 (A), A/Iowa/39/2015 (B), or A/California/07/2009 (C) virus. The following day, a naive ferret was placed in a cage adjacent to each inoculated ferret. Nasal wash titers from individual inoculated (left side of the graph) and contact (dotted bars; right side of the graph) ferrets were collected on the days indicated and assessed by plaque assay. Limit of detection was 10 PFU. Gray, white, and black bars indicate one of each of the three ferrets tested.
Effect of preexposure to CA/07 H1N1pdm09 virus on the outcome of heterologous challenge with OH/09 H1N1v virus.
Nine ferrets exposed to CA/07 virus-infected ferrets in a respiratory droplet transmission setting were used for this study. To confirm that each of the nine ferrets was infected, seroconversion was confirmed by HI assay 3 weeks postexposure to the CA/07 virus-inoculated ferrets (Fig. 6A, left, gray box), and virus titers were quantified in nasal wash samples (Fig. 6B and C, left). The mean peak CA/07 virus titer was observed on day 3 postexposure (≥6.4 log10 PFU/ml), and each of the exposed animals showed HI titers of ≥1,280. Thirty-one days postexposure to CA/07 virus, three ferrets were i.n. inoculated with 105 PFU of CA/07 virus, and the remaining six ferrets were i.n. inoculated with 105 PFU of OH/09 virus. Ferrets rechallenged with the homologous CA/07 virus did not have detectable virus in nasal washes (Fig. 6B, right) through day 4 p.i., and virus was not detected in respiratory tract tissues (nasal turbinates, trachea, and lungs) on day 4 p.i. from these animals (data not shown). Each of the six ferrets challenged with heterologous OH/09 virus had positive nasal wash samples on day 2 p.i. (mean, 2.7 ± 0.4 log10 PFU/ml), and virus was cleared by day 4 p.i. (Fig. 6C, right). Virus was not detected in respiratory tract tissues that were collected on day 4 p.i. from three of these animals (data not shown). Anti-OH/09 HI antibodies (titers from 80 to 640) were detected in sera collected 22 days following secondary inoculation with OH/09 virus (Fig. 6A, right, shaded box). These data indicate that preexposure to CA/07 virus does not fully protect ferrets from infection with the H1N1v OH/09 virus.
FIG 6.
Effect of prior exposure to H1N1pdm09 virus on cross-protection from H1N1v virus challenge in ferrets. (A) Hemagglutination inhibition titers of sera collected from ferrets 22 days after exposure to the A/California/07/2009-inoculated ferrets (primary challenge) or collected from ferrets 22 days after rechallenge with 105 PFU of heterologous virus, A/Ohio/09/2015 (CA/07→OH/09). Black circles show HI titers against A/California/07/2009 virus, and empty circles show titers against A/Ohio/09/2015 virus. Geometric mean hemagglutination inhibition titers (95% confidence interval) are shown. (B and C) Nasal wash titers from ferrets exposed to A/California/07/2009 virus and then rechallenged 31 days later with 105 PFU of A/California/07/2009 virus (n = 3) (B) or A/Ohio/09/2015 (n = 6 on days 2 and 4 p.i.; n = 3 on day 6 p.i.) (C). Viral titers in nasal wash samples collected on the indicated days were assessed by plaque assay. Limit of detection was 10 PFU. Error bars represent ±SD.
DISCUSSION
Since 2005, 433 A(H3N2)v, 20 H1N1v, and 13 H1N2v virus infections have been reported in humans in the United States (14). Patients typically present with mild illness and recover following treatment (14). However, in April 2015 a fatal case of infection with an H1N1v virus, OH/09, was detected in a patient who presented with pneumonia, respiratory failure, and cardiac arrest and who had reported prior exposure to swine. Genomic analysis showed that this virus, like many circulating swine influenza viruses, had acquired the H1N1pdm09 M gene. Full genome sequencing revealed that a second 2015 gamma lineage H1N1v virus, IA/39, contained the PA, NP, and M genes derived from H1N1pdm09 virus. Since the detection of the H1N1pdm09 virus in swine, other reassortant classical swine H1N1 and H1N2 influenza viruses containing various combinations of H1N1pdm09-derived virus genes have also emerged and were detected in humans (21). While the contributions of these genes remain speculative, the increase of reassortant swine viruses containing the M gene segment from H1N1pdm09 virus suggests a gain of fitness in viruses with this gene segment (22, 23). Additionally, the HA genes of viruses that evolved from the classical swine H1 ancestor continue to diverge from one another (24). To better understand how these viruses are evolving, we compared the molecular and antigenic properties of these H1N1v viruses with the prototypical H1N1pdm09 virus, CA/07. We then assessed the ability of these viruses to cause disease and transmit among ferrets to determine their potential risk to humans. Finally, we used a ferret challenge model to measure the immune cross-protection that prior exposure to CA/07 virus would afford ferrets challenged with the antigenically distinct OH/09 virus.
HI testing of ferret antisera raised against CA/07 virus showed significantly reduced titers to the OH/09 egg- and cell-grown virus compared to titers with the homologous virus. The OH/09 virus was also a low reactor relative to other gamma lineage H1N1v viruses based on lack of cross-reactivity of this virus to ferret antisera produced against several previous H1N1v viruses. A similar reduction of the HI titer of postvaccine human sera to OH/09 virus was also detected. Together, these findings indicated that the OH/09 virus was poorly inhibited by antisera to the previous H1N1pdm09 seasonal vaccine strain and all other gamma lineage H1N1 swine influenza viruses used to raise antisera. Genetic analysis of the hemagglutinin gene segment of OH/09 virus identified a G155E substitution within immunodominant antigenic site B, whereas IA/39, like CA/07 virus, had a G at this position. Previous studies have indicated that human H1N1pdm09 viruses with this change show reduced binding in HI assays to convalescent human and ferret antisera raised to the CA/07 vaccine virus (25–27). This mutation has also been associated with increased virulence in mice (28). In addition, this mutation was identified in 11.0% of classical swine H1N1 viruses and was found in each of the three major lineages, suggesting it has evolved separately in multiple genetic groups. Compared to CA/07, both viruses had several amino acid differences, many of which were located in putative antigenic binding sites of the HA1 protein head. Although other changes were observed in HA1, we chose to begin mutation of the reverse genetics-generated OH/09 candidate vaccine virus by introducing the change at position 155, since previous studies of H1N1 swine influenza viruses implicated its importance in antibody recognition and location near the tip of the HA monomer adjacent to a receptor binding site (29). Incorporation of the E155G mutation restored the cross-reactivity of OH/09 E155G with other gamma lineage viruses and CA/07, demonstrating the importance of this mutation in the antigenic relationships of classical swine viruses. The cross-reactivity of antisera raised to both the 155E wild-type OH/09 and the 155G reverse-engineered OH/09 mutant indicated that the single change at position 115 was insufficient to alter antibody recognition between these otherwise identical viruses. Thus, the structural similarity of the viruses likely allowed access of inhibitory antibodies around this site. Because the G155E change is found in all sublineages (alpha, beta, and gamma) of the classical swine influenza viruses, emerging viruses from each of these lineages should be monitored to assess antigenic variability among circulating swine viruses and future H1v viruses detected in humans. Furthermore, the prevalence of this mutation in all H1N1pdm09 viruses detected in humans since the 2009 pandemic was determined to be 0.65%, indicating that this mutation in seasonal viruses is rare but should also be evaluated for changes in antigenicity relative to the H1N1pdm09 component of seasonal vaccines. Identification of this amino acid residue should also be evaluated in both clinical specimens and virus isolates, when possible, to assess if the mutation is selected during passage.
Sequence analysis of both clinical specimens from which OH/09 virus was obtained and the egg-propagated isolate yielded identical protein sequences. Both the clinical specimen and isolate revealed an HA change of D222G in the receptor binding site of OH/09 compared to IA/39 and CA/07 viruses, indicating these amino acid residues did not emerge during egg passage. Substitutions at this position (D222N or G) have been detected occasionally in H1N1 viruses isolated directly from swine, particularly in viruses of the H1N1pdm09 lineage. In humans, the D222G substitution has been detected in viruses from severe and fatal cases of H1N1pdm09 infection, although D222G has also been found in humans with mild disease. This substitution may occur during the course of infection in humans or during serial passage in laboratory host systems (30). In humans, this substitution is detected more often in H1N1pdm09 viruses isolated from the lower respiratory tract, which is a site more likely to be sampled in severe or fatal cases (31). A search of all publicly available H1N1pdm09 viruses since the start of the 2009 pandemic determined 222G was found in 1.31% of viruses. Due to lack of sequence-associated metadata in publicly available databases, it was not possible to reliably determine the number of viruses with this residue that were associated with fatal infections. H1N1pdm09 viruses with this change have not spread to other people but have been shown to increase virulence in a mouse, but not ferret, model of infection (32). Thus, while the D222G substitution can be associated with more severe disease, its overall public health significance remains unclear. Although this study described the first documented fatality following classical swine H1N1 virus infection in the United States since novel influenza A virus infections became notifiable in the United States in 2005, it is recognized that seasonal influenza virus infections result in thousands of deaths in the United States each year. Thus, we cannot rule out that underlying health conditions of the individual contributed to the fatal outcome, as may occur following seasonal influenza virus infection as well. The OH/09 H1N1v virus was sampled from the lower respiratory tract of the infected patient, and while it remains possible that the mutation was selected during replication in the lower respiratory tract, the occurrence of the mutation in classical swine H1N1 viruses detected in contemporary swine viruses is of concern.
Unlike OH/09, IA/39 virus had D at position 222, and this virus was selected for comparative in vivo testing in the ferret model of infection. Ferrets are naturally susceptible to influenza viruses and display symptoms of infection that are comparable to those seen in infected humans, and the respiratory tract of ferrets shares many similarities with the human respiratory tract (18). For these reasons, the ferret has become the model of choice to study influenza viruses. In this study, ferrets were used to assess pathogenicity and transmissibility of H1N1v viruses. Morbidity and mortality in ferrets inoculated with CA/07 or OH/09 virus was comparable to what was observed for other H1N1pdm09 viruses (20), as evidenced by high titers in nasal washes, >10% body weight loss, and the presence of signs of respiratory infection. A milder disease was observed in ferrets inoculated with IA/39 virus; these ferrets had the lowest viral titers in the nasal washes, least amount of weight loss and fever, and no apparent respiratory infection signs. However, IA/39 viruses replicated efficiently in both the upper and lower respiratory tract, similar to what was observed for the H1N1pdm09 virus, CA/07, and other related classical swine H1N1 viruses (20, 33–35). In contrast, OH/09 virus replicated most efficiently in nasal turbinates and least efficiently in the lungs, where the virus was detected in 2 of 3 ferrets. In the ferret model, the tropism of OH/09 virus for the upper (alpha-2,6-linked sialic acid receptors) versus lower (primarily alpha-2,3-linked sialic acid receptors) respiratory tract is in contrast to the hypothesis that the D222G mutation would lead to enhanced specificity for alpha-2,3-linked sialic acid receptors. Although additional studies would be needed to determine genetic correlates of tissue or host tropism, the presence of other HA protein mutations may also play an unknown role in receptor specificity. OH/09 and IA/39 viruses transmitted efficiently between cohoused ferrets, corroborating the observation for other classical swine H1N1 viruses tested in this model (20, 35, 36). In the case of the respiratory droplet experimental design, transmission occurred between 1 of 3 ferret pairs for OH/09 virus and 2 of 3 ferret pairs for IA/39 virus, while CA/07 virus transmitted between all three pairs of ferrets. Overall, these results show that these H1N1v viruses can replicate efficiently in the ferret model, readily transmit in the direct-contact setting, and, although sporadically, are also capable of transmitting by respiratory droplets.
Newly emergent viruses, such as OH/09, that possess features allowing for transmission, and to which there is limited immunity in the human population, are of great public health concern. To assess cross-protection following challenge with antigenically distinct OH/09 virus, ferrets previously exposed to CA/07 (a representative of currently circulating H1 viruses) were used. Animals possessing high HI antibody titers against CA/07 virus were fully protected from secondary infection with the homologous virus. CA/07 virus was not recovered from nasal washes collected on days 2 and 4 p.i. or from respiratory tissues collected on day 4 p.i. In addition, none of the inoculated ferrets displayed signs of morbidity. This result is in agreement with a previous report showing that preinfection with CA/07 virus resulted in sterilizing immunity to homologous challenge and prevented virus transmission to cohoused animals (37). In contrast, ferrets preexposed to CA/07 virus and that possessed no cross-reactive antibodies to OH/09 virus were not fully protected from infection following secondary challenge with this heterologous virus. OH/09 virus was detected in nasal wash samples (6 of 6 ferrets) collected on day 2 p.i. The duration and disease severity of the secondary infection was significantly reduced compared to that seen following the challenge of naive ferrets with OH/09 virus. Cross-protective immunity induced by an antigenically distant H1N1 virus challenge was previously investigated using the ferret and guinea pig models (37, 38). Secondary inoculation of either ferrets or guinea pigs previously infected with an antigenically distinct H1N1 virus resulted in reduced viral titers in nasal washes and enhanced virus clearance. Despite the milder outcome of infection, transmission was still observed. These and the current findings underscore the potential of antigenically distinct viruses to spread in populations with limited preexisting immunity. The protection observed after secondary challenge could be due, in part, to the relatively short interval between infections (31 days), resulting in an elevated nonspecific innate response to primary infection, and/or cross-protective adaptive immunity, such as T cells recognizing shared epitopes. Future studies are warranted to investigate the cross-protective immune responses following heterologous infections. However, in the ferret model, these studies are limited due to the lack of readily available species-specific reagents.
Continuous evolution of swine H1 influenza viruses has resulted in a tremendous diversity of emerging viruses (39) which occasionally cross the species barrier and infect humans following exposure to infected pigs (14). Viruses that are antigenically different from currently and previously circulating human influenza viruses and that are capable of transmission in populations with limited immunity are of particular public health concern. The observations of this study highlight the importance of continued surveillance and monitoring of emerging influenza viruses with the potential for zoonotic transmission events so that we are better prepared for the next pandemic.
MATERIALS AND METHODS
Viruses.
A bronchoalveolar lavage sample obtained from a 27-year-old man with signs of influenza-like illness and respiratory distress was used to inoculate embryonated chicken eggs. Following a 48-h incubation, the allantoic fluid was harvested and hemagglutination (HA) titers were determined using 0.5% turkey red blood cells, confirming virus isolation (A/Ohio/09/2015). While hospitalized, the patient developed primary viral pneumonia, experienced cardiac arrest, and died following respiratory failure. A second case of H1N1v virus, detected in Iowa during 2015, was isolated from a clinical specimen (A/Iowa/39/2015). Virus stocks of OH/09, IA/39, and CA/07 viruses were propagated in the allantoic cavity of 10-day-old embryonated hens' eggs at 35°C for 48 h. Allantoic fluid was pooled from multiple eggs, clarified by centrifugation, and frozen in aliquots at −80°C. Virus titers of each stock were determined by standard plaque assay in MDCK cells (40). The stock viruses were sequenced, and real-time reverse transcription-PCR (RT-PCR) exclusivity tests were performed to rule out the presence of other subtypes of influenza virus. Reverse genetics-generated PR8 reassortants with HA and NA genes of OH/09 (wild type) and with a substitution of HA E155G were constructed as previously described (41).
Sequencing and phylogenetic and molecular analysis.
Each virus stock was sequenced using Illumina MiSeq technology, and genomes were assembled as previously described (42). Codon-complete viral gene segments were aligned with sequences downloaded from the GISAID database following BLAST analysis to identify closely related strains. Phylogenetic trees of aligned gene segments were constructed using the Rainbow tool with neighbor-joining analysis and the Jukes-Cantor substitution model, as previously described (43). Each analysis was run with 1,000 bootstrap iterations for statistical support. Nucleotide sequence alignments used for trees were also translated to amino acid protein sequences to compare each virus to CA/07. Amino acid differences among the viruses were plotted on the HA phylogenetic tree at each branch node.
Antigenic analysis.
Postinfection ferret immune sera were raised to OH/09, OH/09 E155G, IA/39, and CA/07 viruses, as well as select H1N1pdm09 viruses and recent H1N1v and H1N2v viruses. Antisera were harvested from ferrets 14 days postinfection following an intranasal infectious dose of 106 50% egg infectious doses. The panel of antisera was used to test two-way HI titers of each virus to assess cross-reactivity between various genetic lineages of H1N1 and H1N2 viruses. Each serum was treated at a 1:4 dilution with receptor-destroying enzyme (RDE; Denke Sanka) for 18 h at 37°C, followed by adsorption with packed turkey red blood cells. All antigens were back-titered to 4 HAU/25 μl prior to 30 min of incubation at room temperature with 2-fold serial dilutions of antiserum. Turkey red blood cells (0.5%) were used for hemagglutination.
Ferret experiments.
Male Fitch ferrets that were 5 to 7 months old and serologically negative for currently circulating influenza viruses (Triple F Farms, Sayre, PA) were used for this study. All ferrets were housed in Duo-Flo Bioclean mobile units (Lab Products Incorporated, Seaford, DE) during experimentation. Three groups of ferrets were inoculated intranasally (i.n.) with 1 ml of solution containing 105 PFU of CA/07 (n = 3), OH/09 (n = 9), or IA/39 (n = 6) virus diluted in phosphate-buffered saline (PBS). The following day, a serologically naive ferret was placed either in the same cage as each inoculated ferret, to assess virus transmission between ferrets in direct contact (three ferret pairs), or in a cage adjacent to an inoculated ferret to assess respiratory droplet transmission (three ferret pairs) (44). The respiratory droplet transmission model is a more stringent test of virus transmissibility because virus can only be transmitted through exchange of air between inoculated and naive ferrets, while the direct-contact model provides the opportunity for transmission by direct or indirect contact or by inhalation. Seasonal influenza viruses typically transmit very efficiently using either of the transmission models (20). All ferrets were observed for 2 weeks for clinical signs of infection; nasal washes and rectal swabs were collected every 2 days from at least three of the inoculated animals for virus titer determination. Three additional ferrets each were inoculated with the same dose of virus and were euthanized on day 3 or 4 postinoculation (p.i.) for the assessment of virus replication in pulmonary and extrapulmonary tissues, as previously described (45). Convalescent-phase serum was collected from all surviving ferrets 3 weeks p.i. or postcontact (p.c.), and seroconversion was determined by HI assay using homologous virus. To determine the level of protection against OH/09 H1N1v virus infection in ferrets with preexisting immunity to H1N1pdm09 virus, nine serologically negative ferrets were exposed to ferrets inoculated with CA/07 virus in a respiratory droplet transmission setting as described above. Virus titers in nasal washes of the exposed animals were quantified on days 1, 3, 5, and 7 postexposure by standard plaque assay, and seroconversion was confirmed by HI assay 3 weeks postexposure. Thirty-one days postexposure to CA/07 virus, three ferrets were i.n. inoculated with 105 PFU of CA/07 virus, and the remaining six ferrets were i.n. inoculated with 105 PFU of OH/09 virus. All ferrets were observed for clinical signs of infection, and nasal washes were collected on days 2 and 4 p.i. Three of the ferrets inoculated with OH/09 virus and the three ferrets inoculated with CA/07 virus were euthanized on day 4 p.i. for virus titer determination in nasal turbinates, trachea, and lungs. The remaining three ferrets inoculated with OH/09 virus were used to collect nasal wash samples on day 6 p.i. and sera on day 22 p.i. Anti-OH/09 and anti-CA/09 virus antibody were measured in the sera by HI assay and expressed as the reciprocal of the highest dilution of serum that inhibited hemagglutination. Mean nasal wash sample titers (with standard deviations [SD]) and geometric mean (GM) HI titers with 95% confidence intervals (CI) were calculated and graphed using GraphPad Prism 6 software.
The animal work conducted in this study was reviewed and approved by the Centers for Disease Control and Prevention Institutional Animal Care and Use Committee (CDC IACUC; protocol numbers 2621DAVFERC and 2849MAIFERC). CDC IACUC approval was not required for virus propagation in embryonated hens' eggs, because all eggs were destroyed prior to hatching. All animal use adhered to guidance from the Office of Laboratory Animal Welfare (OLAW), National Institutes of Health, responsible for implementation of the PHS Policy Animal Welfare Act (7 U.S.C. sections 2131 to 2159) and the Public Health Service Policy on Humane Care and Use of Laboratory Animals (http://grants.nih.gov/grants/olaw/faqs.htm#App_4).
Supplementary Material
ACKNOWLEDGMENTS
We thank the United States Department of Agriculture, Agricultural Research Service and National Veterinary Services Laboratories, for valuable discussions on influenza viruses circulating in swine and the Comparative Medicine Branch for excellent care of the animals used in this study.
This work was funded in part by the U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, and the Oak Ridge Institute of Science and Education.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention and the Agency for Toxic Substances and Disease Registry.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00095-18.
REFERENCES
- 1.Yoon SW, Webby RJ, Webster RG. 2014. Evolution and ecology of influenza A viruses. Curr Top Microbiol Immunol 385:359–375. [DOI] [PubMed] [Google Scholar]
- 2.Czako R, Subbarao K. 2015. Refining the approach to vaccines against influenza A viruses with pandemic potential. Future Virol 10:1033–1047. doi: 10.2217/fvl.15.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrat F, Flahault A. 2007. Influenza vaccine: the challenge of antigenic drift. Vaccine 25:6852–6862. doi: 10.1016/j.vaccine.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 4.Shope RE. 1931. Swine influenza. III. Filtration experiments and etiology. J Exp Med 54:373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tumpey TM, Garcia-Sastre A, Taubenberger JK, Palese P, Swayne DE, Basler CF. 2004. Pathogenicity and immunogenicity of influenza viruses with genes from the 1918 pandemic virus. Proc Natl Acad Sci U S A 101:3166–3171. doi: 10.1073/pnas.0308391100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma W, Lager KM, Vincent AL, Janke BH, Gramer MR, Richt JA. 2009. The role of swine in the generation of novel influenza viruses. Zoonoses Public Health 56:326–337. doi: 10.1111/j.1863-2378.2008.01217.x. [DOI] [PubMed] [Google Scholar]
- 7.Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge KF, Kawaoka Y, Webster RG. 1994. Potential for transmission of avian influenza viruses to pigs. J Gen Virol 75(Part 9):2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 8.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 10.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K, Krauss S, Webster RG. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 73:8851–8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vincent AL, Ma W, Lager KM, Gramer MR, Richt JA, Janke BH. 2009. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes 39:176–185. doi: 10.1007/s11262-009-0386-6. [DOI] [PubMed] [Google Scholar]
- 12.Shu B, Garten R, Emery S, Balish A, Cooper L, Sessions W, Deyde V, Smith C, Berman L, Klimov A, Lindstrom S, Xu X. 2012. Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990-2010. Virology 422:151–160. doi: 10.1016/j.virol.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 13.Lewis NS, Russell CA, Langat P, Anderson TK, Berger K, Bielejec F, Burke DF, Dudas G, Fonville JM, Fouchier RA, Kellam P, Koel BF, Lemey P, Nguyen T, Nuansrichy B, Peiris JM, Saito T, Simon G, Skepner E, Takemae N, ESNIP3 Consortium, Webby RJ, Van Reeth K, Brookes SM, Larsen L, Watson SJ, Brown IH, Vincent AL. 2016. The global antigenic diversity of swine influenza A viruses. Elife 5:e12217. doi: 10.7554/eLife.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention. 2017. Information on swine influenza/variant influenza virus. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/flu/swineflu/index.htm Accessed on 14 December 2017. [Google Scholar]
- 15.Appiah GD, Blanton L, D'Mello T, Kniss K, Smith S, Mustaquim D, Steffens C, Dhara R, Cohen J, Chaves SS, Bresee J, Wallis T, Xu X, Abd Elal AI, Gubareva L, Wentworth DE, Katz J, Jernigan D, Brammer L, Centers for Disease Control and Prevention. 2015. Influenza activity–United States, 2014-15 season and composition of the 2015-16 influenza vaccine. MMWR Morb Mortal Wkly Rep 64:583–590. [PMC free article] [PubMed] [Google Scholar]
- 16.Blanton L, Kniss K, Smith S, Mustaquim D, Steffens C, Flannery B, Fry AM, Bresee J, Wallis T, Garten R, Xu X, Elal AI, Gubareva L, Wentworth DE, Burns E, Katz J, Jernigan D, Brammer L. 2015. Update: influenza activity–United States and worldwide, May 24-September 5, 2015. MMWR Morb Mortal Wkly Rep 64:1011–1016. doi: 10.15585/mmwr.mm6436a4. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization. 2015. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 18.Belser JA, Katz JM, Tumpey TM. 2011. The ferret as a model organism to study influenza A virus infection. Dis Model Mech 4:575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox NJ, Trock SC, Burke SA. 2014. Pandemic preparedness and the Influenza Risk Assessment Tool (IRAT). Curr Top Microbiol Immunol 385:119–136. [DOI] [PubMed] [Google Scholar]
- 20.Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong W, Wang F, Dong B, Ou C, Meng D, Liu J, Fan ZC. 2015. Novel reassortant influenza viruses between pandemic (H1N1) 2009 and other influenza viruses pose a risk to public health. Microb Pathog 89:62–72. doi: 10.1016/j.micpath.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Chou YY, Albrecht RA, Pica N, Lowen AC, Richt JA, Garcia-Sastre A, Palese P, Hai R. 2011. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J Virol 85:11235–11241. doi: 10.1128/JVI.05794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma J, Shen H, Liu Q, Bawa B, Qi W, Duff M, Lang Y, Lee J, Yu H, Bai J, Tong G, Hesse RA, Richt JA, Ma W. 2015. Pathogenicity and transmissibility of novel reassortant H3N2 influenza viruses with 2009 pandemic H1N1 genes in pigs. J Virol 89:2831–2841. doi: 10.1128/JVI.03355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, Swenson SL, Simon G, Saito T, Berhane Y, Ciacci-Zanella J, Pereda A, Davis CT, Donis RO, Webby RJ, Vincent AL. 2016. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza A viruses. mSphere 1:e00275-16. doi: 10.1128/mSphere.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmeisser F, Friedman R, Besho J, Lugovtsev V, Soto J, Wang W, Weiss C, Williams O, Xie H, Ye Z, Weir JP. 2013. Neutralizing and protective epitopes of the 2009 pandemic influenza H1N1 hemagglutinin. Influenza Other Respir Viruses 7:480–490. doi: 10.1111/irv.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen HK, Nguyen PT, Nguyen TC, Hoang PV, Le TT, Vuong CD, Nguyen AP, Tran LT, Nguyen BG, Le MQ. 2015. Virological characterization of influenza H1N1pdm09 in Vietnam, 2010-2013. Influenza Other Respir Viruses 9:216–224. doi: 10.1111/irv.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klimov AI, Garten R, Russell C, Barr IG, Besselaar TG, Daniels R, Engelhardt OG, Grohmann G, Itamura S, Kelso A, McCauley J, Odagiri T, Smith D, Tashiro M, Xu X, Webby R, Wang D, Ye Z, Yuelong S, Zhang W, Cox N, Writing Committee of the World Health Organization Consultation on Southern Hemisphere Influenza Vaccine Composition for 2012. 2012. WHO recommendations for the viruses to be used in the 2012 southern hemisphere influenza vaccine: epidemiology, antigenic and genetic characteristics of influenza A(H1N1)pdm09, A(H3N2) and B influenza viruses collected from February to September 2011. Vaccine 30:6461–6471. doi: 10.1016/j.vaccine.2012.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ilyushina NA, Khalenkov AM, Seiler JP, Forrest HL, Bovin NV, Marjuki H, Barman S, Webster RG, Webby RJ. 2010. Adaptation of pandemic H1N1 influenza viruses in mice. J Virol 84:8607–8616. doi: 10.1128/JVI.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Both GW, Shi CH, Kilbourne ED. 1983. Hemagglutinin of swine influenza virus: a single amino acid change pleiotropically affects viral antigenicity and replication. Proc Natl Acad Sci U S A 80:6996–7000. doi: 10.1073/pnas.80.22.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melidou A, Gioula G, Exindari M, Chatzidimitriou D, Malisiovas N. 2015. Genetic analysis of post-pandemic 2010-2011 influenza A(H1N1)pdm09 hemagglutinin virus variants that caused mild, severe, and fatal infections in northern Greece. J Med Virol 87:57–67. doi: 10.1002/jmv.23990. [DOI] [PubMed] [Google Scholar]
- 31.Ruggiero T, De Rosa F, Cerutti F, Pagani N, Allice T, Stella ML, Milia MG, Calcagno A, Burdino E, Gregori G, Urbino R, Di Perri G, Ranieri MV, Ghisetti V. 2013. A(H1N1)pdm09 hemagglutinin D222G and D222N variants are frequently harbored by patients requiring extracorporeal membrane oxygenation and advanced respiratory assistance for severe A(H1N1)pdm09 infection. Influenza Other Respir Viruses 7:1416–1426. doi: 10.1111/irv.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abed Y, Pizzorno A, Hamelin ME, Leung A, Joubert P, Couture C, Kobasa D, Boivin G. 2011. The 2009 pandemic H1N1 D222G hemagglutinin mutation alters receptor specificity and increases virulence in mice but not in ferrets. J Infect Dis 204:1008–1016. doi: 10.1093/infdis/jir483. [DOI] [PubMed] [Google Scholar]
- 33.Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T, Sakabe S, Imai M, Hatta Y, Watanabe S, Li C, Yamada S, Fujii K, Murakami S, Imai H, Kakugawa S, Ito M, Takano R, Iwatsuki-Horimoto K, Shimojima M, Horimoto T, Goto H, Takahashi K, Makino A, Ishigaki H, Nakayama M, Okamatsu M, Takahashi K, Warshauer D, Shult PA, Saito R, Suzuki H, Furuta Y, Yamashita M, Mitamura K, Nakano K, Nakamura M, Brockman-Schneider R, Mitamura H, Yamazaki M, Sugaya N, Suresh M, Ozawa M, Neumann G, Gern J, Kida H, Ogasawara K, Kawaoka Y. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munster VJ, de Wit E, van den Brand JM, Herfst S, Schrauwen EJ, Bestebroer TM, van de Vijver D, Boucher CA, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. 2009. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 325:481–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barman S, Krylov PS, Fabrizio TP, Franks J, Turner JC, Seiler P, Wang D, Rehg JE, Erickson GA, Gramer M, Webster RG, Webby RJ. 2012. Pathogenicity and transmissibility of North American triple reassortant swine influenza A viruses in ferrets. PLoS Pathog 8:e1002791. doi: 10.1371/journal.ppat.1002791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belser JA, Wadford DA, Pappas C, Gustin KM, Maines TR, Pearce MB, Zeng H, Swayne DE, Pantin-Jackwood M, Katz JM, Tumpey TM. 2010. Pathogenesis of pandemic influenza A (H1N1) and triple-reassortant swine influenza A (H1) viruses in mice. J Virol 84:4194–4203. doi: 10.1128/JVI.02742-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laurie KL, Carolan LA, Middleton D, Lowther S, Kelso A, Barr IG. 2010. Multiple infections with seasonal influenza A virus induce cross-protective immunity against A(H1N1) pandemic influenza virus in a ferret model. J Infect Dis 202:1011–1020. doi: 10.1086/656188. [DOI] [PubMed] [Google Scholar]
- 38.Steel J, Staeheli P, Mubareka S, Garcia-Sastre A, Palese P, Lowen AC. 2010. Transmission of pandemic H1N1 influenza virus and impact of prior exposure to seasonal strains or interferon treatment. J Virol 84:21–26. doi: 10.1128/JVI.01732-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao S, Anderson TK, Walia RR, Dorman KS, Janas-Martindale A, Vincent AL. 2017. The genomic evolution of H1 influenza A viruses from swine detected in the United States between 2009 and 2016. J Gen Virol 98:2001–2010. doi: 10.1099/jgv.0.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng H, Goldsmith C, Thawatsupha P, Chittaganpitch M, Waicharoen S, Zaki S, Tumpey TM, Katz JM. 2007. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type i interferon response in polarized human bronchial epithelial cells. J Virol 81:12439–12449. doi: 10.1128/JVI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ridenour C, Johnson A, Winne E, Hossain J, Mateu-Petit G, Balish A, Santana W, Kim T, Davis C, Cox NJ, Barr JR, Donis RO, Villanueva J, Williams TL, Chen LM. 2015. Development of influenza A(H7N9) candidate vaccine viruses with improved hemagglutinin antigen yield in eggs. Influenza Other Respir Viruses 9:263–270. doi: 10.1111/irv.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shepard SS, Meno S, Bahl J, Wilson MM, Barnes J, Neuhaus E. 2016. Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genomics 17:708. doi: 10.1186/s12864-016-3030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rith S, Davis CT, Duong V, Sar B, Horm SV, Chin S, Ly S, Laurent D, Richner B, Oboho I, Jang Y, Davis W, Thor S, Balish A, Iuliano AD, Sorn S, Holl D, Sok T, Seng H, Tarantola A, Tsuyuoka R, Parry A, Chea N, Allal L, Kitsutani P, Warren D, Prouty M, Horwood P, Widdowson MA, Lindstrom S, Villanueva J, Donis R, Cox N, Buchy P. 2014. Identification of molecular markers associated with alteration of receptor-binding specificity in a novel genotype of highly pathogenic avian influenza A(H5N1) viruses detected in Cambodia in 2013. J Virol 88:13897–13909. doi: 10.1128/JVI.01887-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, Ortin J, Falcon A, Nguyen TH, Mai LE, Sedyaningsih QER, Harun S, Tumpey TM, Donis RO, Cox NJ, Subbarao K, Katz JM. 2006. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A 103:12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, Chittaganpitch M, Waicharoen S, Nguyen DT, Nguyen T, Nguyen HH, Kim JH, Hoang LT, Kang C, Phuong LS, Lim W, Zaki S, Donis RO, Cox NJ, Katz JM, Tumpey TM. 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol 79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reuman PD, Keely S, Schiff GM. 1989. Assessment of signs of influenza illness in the ferret model. J Virol Methods 24:27–34. doi: 10.1016/0166-0934(89)90004-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.