ABSTRACT
Chronic infection by hepatitis B virus (HBV) is the major contributor to liver disease worldwide. Though HBV replicates via a nuclear episomal DNA (covalently closed circular DNA [cccDNA]), integration of HBV DNA into the host cell genome is regularly observed in the liver in infected patients. While reported as a prooncogenic alteration, the mechanism(s) and timing of HBV DNA integration are not well understood, chiefly due to the lack of in vitro infection models that have detectable integration events. In this study, we have established an in vitro system in which integration can be reliably detected following HBV infection. We measured HBV DNA integration using inverse nested PCR in primary human hepatocytes, HepaRG-NTCP, HepG2-NTCP, and Huh7-NTCP cells after HBV infection. Integration was detected in all cell types at a rate of >1 per 10,000 cells, with the most consistent detection in Huh7-NTCP cells. The integration rate remained stable between 3 and 9 days postinfection. HBV DNA integration was efficiently blocked by treatment with a 200 nM concentration of the HBV entry inhibitor Myrcludex B, but not with 10 μM tenofovir, 100 U of interferon alpha, or a 1 μM concentration of the capsid assembly inhibitor GLS4. This suggests that integration of HBV DNA occurs immediately after infection of hepatocytes and is likely independent of de novo HBV genome replication in this model. Site analysis revealed that HBV DNA integrations were distributed over the entire human genome. Further, integrated HBV DNA sequences were consistent with double-stranded linear HBV DNA being the major precursor. Thus, we have established an in vitro system to interrogate the mechanisms of HBV DNA integration.
IMPORTANCE Hepatitis B virus (HBV) is a common blood-borne pathogen and, following a chronic infection, can cause liver cancer and liver cirrhosis. Integration of HBV DNA into the host genome occurs in all known members of the Hepadnaviridae family, despite this form not being necessary for viral replication. HBV DNA integration has been reported to drive liver cancer formation and persistence of virus infection. However, when and the mechanism(s) by which HBV DNA integration occurs are not clear. In this study, we have developed and characterized an in vitro system to reliably detect HBV DNA integrations that result from a true HBV infection event and that closely resemble those found in patient tissues. Using this model, we showed that integration occurs when the infection is first established. Importantly, we provide here a system to analyze molecular factors involved in HBV integration, which can be used to develop strategies to halt its formation.
KEYWORDS: HBV DNA integration, Myrcludex B, hepatocellular carcinoma, nonhomologous end joining, microhomology-mediated end joining, HBV double-stranded linear DNA, inverse nested PCR
INTRODUCTION
Chronic infection with hepatitis B virus (HBV) is one of the most widespread causes of liver cirrhosis and primary liver cancer (hepatocellular carcinoma [HCC]). While highly effective vaccines can prevent new infection, there is currently no cure for the ∼240 million patients worldwide with chronic hepatitis B infection, the main contributor toward viral hepatitis-associated morbidity and mortality (1). HBV-associated disease progression is largely driven by persistence of HBV infection and the subsequent chronic antiviral inflammatory response. Another reported driver of HCC is the integration of HBV DNA into the host cell genome, which has been suggested to induce chromosomal instability (2, 3), cis activation of cellular genes (2, 4–6), insertional mutagenesis into tumor suppressors (5–7), and persistent expression of mutant HBV proteins that drive cellular stress (8, 9).
HBV DNA integration is thought to occur as a by-product of HBV viral replication, as it is not required to support production of new virions. The viral replication cycle starts when HBV enters hepatocytes using sodium taurocholate cotransporting polypeptide (NTCP) as a receptor (10, 11). Following entry, the HBV nucleocapsid containing the relaxed circular DNA (rcDNA) or, more rarely, the double-stranded linear DNA (dslDNA) genome is released into the cytoplasm and transported to the nucleus (12). Intranuclear HBV DNA is converted by host DNA repair proteins into covalently closed circular DNA (cccDNA), the stable episomal transcriptional template for HBV mRNAs (13). Viral pregenomic RNA (pgRNA) is also transcribed from cccDNA and is encapsidated into viral capsids with the HBV polymerase (12). Reverse transcription of the pgRNA occurs within the nucleocapsid, resulting in rcDNA or occasionally dslDNA forms. These nucleocapsids are either (i) enveloped and secreted as virions, the default pathway required for viral spread, or (ii) transported to the nucleus to add to the intranuclear cccDNA pool (12).
An additional possible fate for intranuclear dslDNA HBV genomes is integration into the host cell genome at the site of double-stranded DNA breaks by nonhomologous end joining (NHEJ) (14). While HBV dslDNA is 18 nucleotides (nt) longer than genome length, the HBV core antigen (HBcAg) promoter is separated from its open reading frame (ORF), leading to a replication-incompetent form of the virus. Importantly, however, integrated HBV DNA can act as a template for the expression of HBV surface antigen (15), which has been described to be a factor in HBV-specific immune tolerance and subsequent infection persistence. Integration is observed at a frequency of 1 in 102 to 104 cells in the woodchuck and duck models of HBV infection (16–18) and in chronically infected HBV patients (19–21). Though integration into the host cell genome has been observed in infection with all known members of the hepadnavirus family (16, 18, 19, 22, 23), many of the molecular mechanisms, functions, and cellular consequences of this phenomenon are presently unknown (24). One major limiting factor is the lack of a cellular model for human HBV DNA infection where integration can be consistently detected and subsequently analyzed.
In this study, we found that HBV DNA integration following in vitro infection occurs in all HBV-susceptible cell types, including primary human hepatocytes (PHH). From the sequence of integrations, we show that the DNA sequence of integration junctions produced in vitro closely resemble those found in nontumor patient tissues with respect to both HBV and cellular sequences, suggesting that the in vitro systems reflect the same biological pathways as those used in a true HBV infection. We used these results to develop an optimal system (based on hepatoma cell lines) in which hundreds of integration events can be generated and detected relatively easily, thereby allowing the interrogation of the molecular mechanisms of HBV DNA integration.
RESULTS
HBV DNA integration was detected following NTCP-dependent HBV infection in all cell types tested.
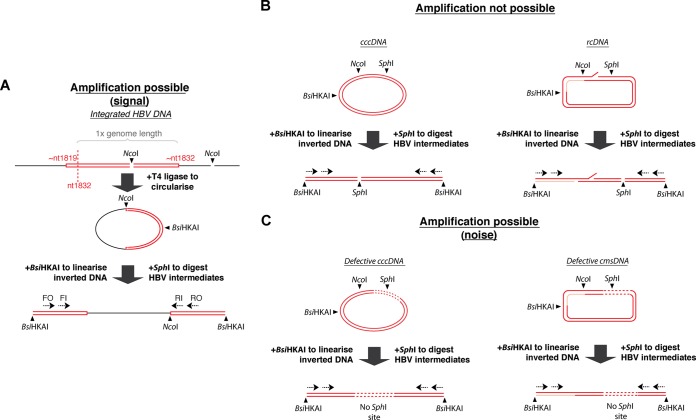
We first tested multiple HBV-susceptible cell types, including PHH and HepaRG-NTCP, HepG2-NTCP, and Huh7-NTCP cells. We detected integration of HBV dslDNA by inverse nested PCR (invPCR), a previously described PCR-based method that has the sensitivity to detect single copies of virus-cell junctions (25). An outline of the technique is shown in Fig. 1A. The greater-than-genome-length dslDNA forms a virus-cell junction, the right arm of which is excised using NcoI (one site in the HBV genome and one site in the downstream human sequence). This excised fragment is then ligated with T4 ligase, linearized with BsiHKAI, and amplified using HBV-specific primers.
FIG 1.
Detection of HBV DNA integration by invPCR. HBV DNA integration was detected using the invPCR assay (A). The right virus-cell junction formed by integration of HBV dslDNA (red rectangle, with approximate ends based on the nucleotide numbering of HBV DNA sequence described under GenBank accession no. U95551.1) into the host cell genome (black) is excised with NcoI digestion. This fragment is circularized with the addition of T4 ligase and then digested again with BsiHKAI to linearize the inverted product. Nested primers specific (dashed arrows) are then used to PCR amplify the unknown cellular region contained between the two HBV sequences. To limit amplification of nonintegrated HBV DNA forms that compete for PCR amplification of virus-cell junctions, SphI is added to the digestion reactions (B). For full-length HBV DNA intermediates (cccDNA and rcDNA), the SphI site rendered the inverted HBV DNA unamplifable due to a double-stranded DNA break. Further for rcDNA, the gap (red) and nick regions decrease amplification efficiency. There are also a minority of amplifiable nonintegrated HBV DNA forms, including cccDNA and closed minus-strand DNA (cmsDNA), that are defective (contain large deletions, shown as dashed lines) and consequently lack the SphI site (C). These competed with the amplification of our intended signal (integrated HBV DNA) and were depleted by treatment with reverse transcriptase inhibitors and induction of cell mitosis (Fig. 2A).
The amplification of many HBV DNA forms, such as HBV cccDNA and rcDNA, was inhibited by digestion with SphI (Fig. 1B), which cuts downstream of the expected right-hand junction of the HBV DNA sequence. Further, rcDNA forms are much less efficiently amplified due to incomplete ligation at the nick/gap region. Single-stranded minus-strand HBV DNA (data not shown) acts similarly to rcDNA and is likewise poorly amplified. However, invPCR is able to detect a minority of the closed minus-strand rcDNA (26) and cccDNA HBV DNA forms that are defective and contain deletions of the SphI site (Fig. 1C). We depleted these forms by treatment with reverse transcriptase inhibitors and, where possible, induction of a round of mitosis by splitting the HBV-infected cells (which has been shown previously to deplete episomal cccDNA [27]) via transfer from a 12-well to a 6-well plate (Fig. 2A).
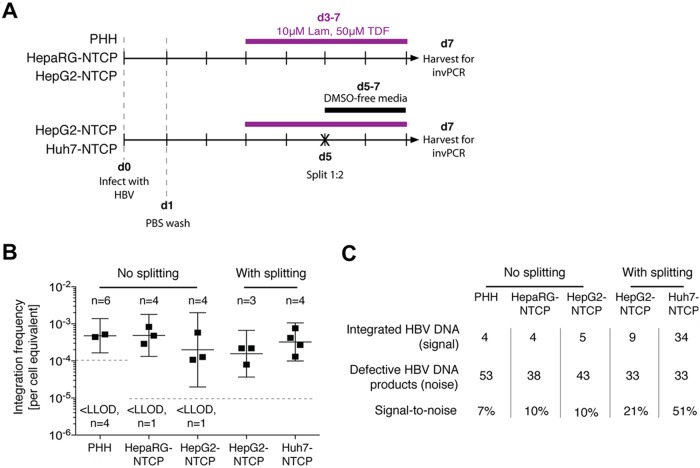
FIG 2.
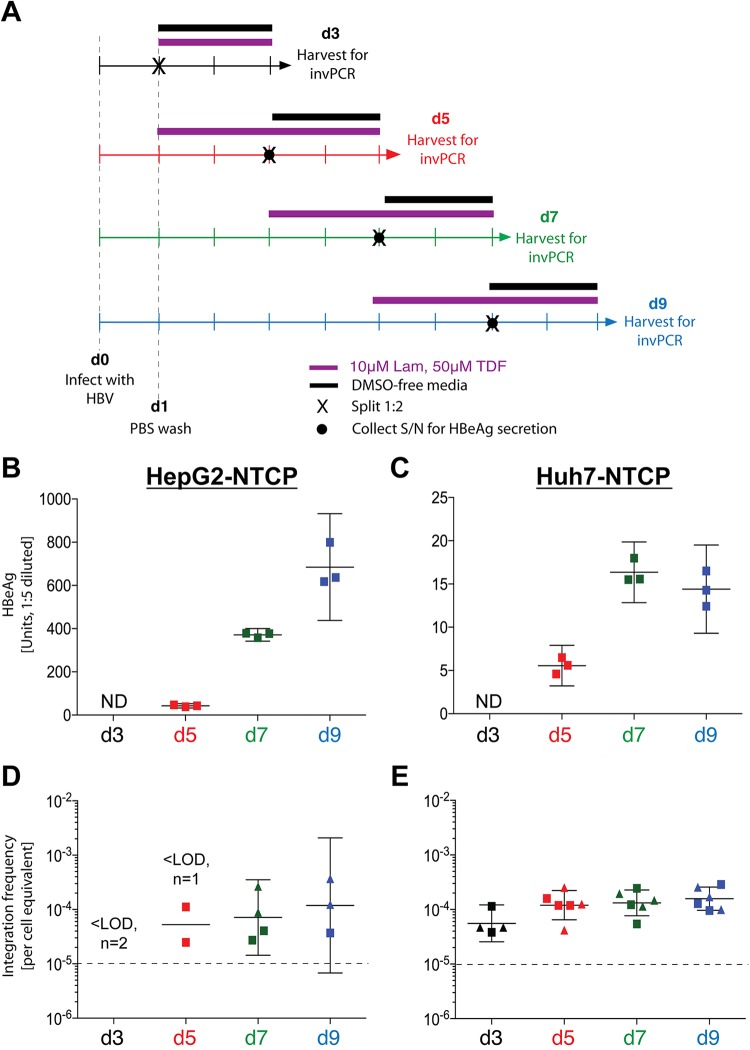
HBV DNA integration occurs in all HBV-susceptible cell types. In the initial experimental setup (A), cells were infected at day 0 with an HBV inoculum of 1,000 VGE/cell, which was washed off 1 day after. All cell types were maintained in culture medium containing 10 μM lamivudine and 50 μM tenofovir (Lam and TDF, respectively) from 3 dpi (d3) until harvest at 7 dpi (purple bar). For hepatoma cell lines, HBV-infected cells were split by transferring them from a 12-well to a 6-well plate (black cross) and maintained in a DMSO-free medium to stimulate a round of mitosis. At 7 dpi, cells were collected for DNA extraction and subsequent invPCR analysis to determine the frequency of HBV DNA integration (B; the mean ± 95% confidence interval [CI] are shown for each cell type). Number of independent infections (n) and number of samples below lower limit of detection (LLOD; 10−5 for PHH and 1.6 × 10−4 for all other cell types) are displayed. The total number of products amplified from virus-cell junctions and from defective HBV DNA forms (C) and the ratio of these two product types (signal-to-noise) are shown for each cell type (detailed information for each independent infection is shown in Table 1).
In this experimental setup, HBV DNA integration was detected in all cell types (Fig. 2B), with putative integration frequencies ranging from ∼10−3 to ∼10−4 integrations per cell equivalent. No significant differences in integration rate were observed between the cell types. Further, the induction of cell mitosis did not significantly alter the observed integration rate. Together, these results suggest that HBV DNA integrations occurs in all cell types that support HBV infection and, at least in hepatoma cells, are stable through cell mitosis (unlike episomal forms).
Importantly, no HBV DNA integration was detected (lower limit of detection = 1 integration per 105 cell equivalents) when invPCR was performed on DNA extracted from (i) mock-infected HepG2-NTCP cells, (ii) HBV-inoculated HepG2 cells not expressing the HBV receptor NTCP, (iii) HBV-infected HepG2-NTCP cells that were not inverted (by excluding NcoI in the inversion protocol), or (iv) 108 viral genome equivalents (VGE) of HBV inoculum. These technical negative controls show that the detected integration events resulted from true HBV infection and were not PCR-generated artifacts.
We assessed the ratio of virus-cell junctions (generated by HBV DNA integration) to virus-virus products (generated by defective forms of HBV DNA) that were amplified by invPCR (Fig. 2C; detailed information is provided in Table 1). In PHH and HepaRG-NTCP and nonsplit cells, products formed by defective HBV DNA titrated to similar concentrations as products formed by HBV integration. This limited efficient detection of virus-cell junctions; 10 to 20 PCR products required sequencing to find a single virus-cell junction. On the other hand, the splitting procedure in the hepatoma cell lines depleted amplifiable defective HBV DNA while maintaining the number of virus-cell junctions (after normalizing to DNA input into the invPCR assay), thereby increasing the signal-to-noise ratio in the detection of integrated HBV DNA (Fig. 2C).
TABLE 1.
Detection of virus-cell junctions generated by HBV DNA integration in human liver cell types that support HBV
| Condition | Cell type | n | Index for secreted HBeAg (dilution)a | No. of virus-cell junctions | No. of virus-virus products | Ratio (%) | Integration rateb (per 104 cell equivalents) |
|---|---|---|---|---|---|---|---|
| Without splitting | Primary human hepatocytesc | 6 | 112 (1:5) | 3 | 9 | 25 | 5.2 |
| 53 (1:5) | 1 | 4 | 20 | 4.4 | |||
| 26 (1:5) | 0 | 22 | 0 | <1.0 | |||
| 224 (1:5) | 0 | 5 | 0 | <1.0 | |||
| 137 (1:5) | 0 | 8 | 0 | <1.0 | |||
| 155 (1:5) | 0 | 5 | 0 | <1.0 | |||
| HepaRG-NTCP | 4 | 16 (1:5) | 1 | 7 | 12.5 | 0.83 | |
| 24 (1:5) | 1 | 4 | 20 | 0.29 | |||
| 250 (1:2) | 2 | 11 | 15.4 | 4.8 | |||
| 240 (1:2) | 0 | 16 | 0 | <0.16 | |||
| HepG2-NTCP | 4 | 281 (1:5) | 1 | 14 | 7.1 | 1.07 | |
| 288 (1:5) | 0 | 9 | 0 | <0.16 | |||
| 299 (1:5) | 2 | 6 | 33.3 | 5.8 | |||
| 316 (1:5) | 2 | 14 | 14.3 | 1.2 | |||
| With splitting | HepG2-NTCP | 3 | 410 (1:5) | 3 | 8 | 27.3 | 0.80 |
| 345 (1:5) | 3 | 11 | 21.4 | 2.2 | |||
| 365 (1:5) | 3 | 14 | 20 | 2.2 | |||
| Huh7-NTCP | 4 | 75 (1:2) | 13 | 12 | 52 | 7.6 | |
| 89 (1:2) | 7 | 3 | 70 | 2.7 | |||
| 55 (1:2) | 9 | 7 | 56 | 1.3 | |||
| 54 (1:2) | 5 | 11 | 31 | 4.2 |
Supernatant was diluted at either 1:2 or 1:5 prior to HBeAg detection by immunoassay.
Putative integration rate was determined by dividing the number of integration junctions at a particular dilution of inverted DNA by the cell equivalents in that dilution.
Represents 2 donors, with 3 independent infections for each donor.
While it is unclear whether cell mitosis produces two cells completely free of cccDNA, induction of mitosis is one of the only known ways to reduce cccDNA in HBV-infected cells (27). After induction of mitosis in HepG2-NTCP cells, we found an ∼1-log reduction (but not complete eradication) of defective HBV DNA products amplified by the invPCR. Whether the incomplete depletion of these defective HBV DNA products is due to their redistribution to the cytoplasm is not clear. Despite this, as consistent detection of HBV DNA integration would enable accurate characterization of any changes in integration rate (e.g., due to cellular factors, viral factors, and drug treatments), we used Huh7-NTCP and HepG2-NTCP cells in all of the remaining experiments.
True HBV integration rate is >1 integration per 1,000 cells.
The observed integration rate of 1 per ∼104 cells is a lower bound, which could be suppressed by other factors, including the number of cells that take up HBV virions, the efficiency of DNA fragment inversion during the invPCR technique, and the lack of restriction enzyme sites required for invPCR detection of integrated HBV DNA. We aimed to quantify the impact of some of these factors to estimate the true rate of HBV DNA integration more accurately.
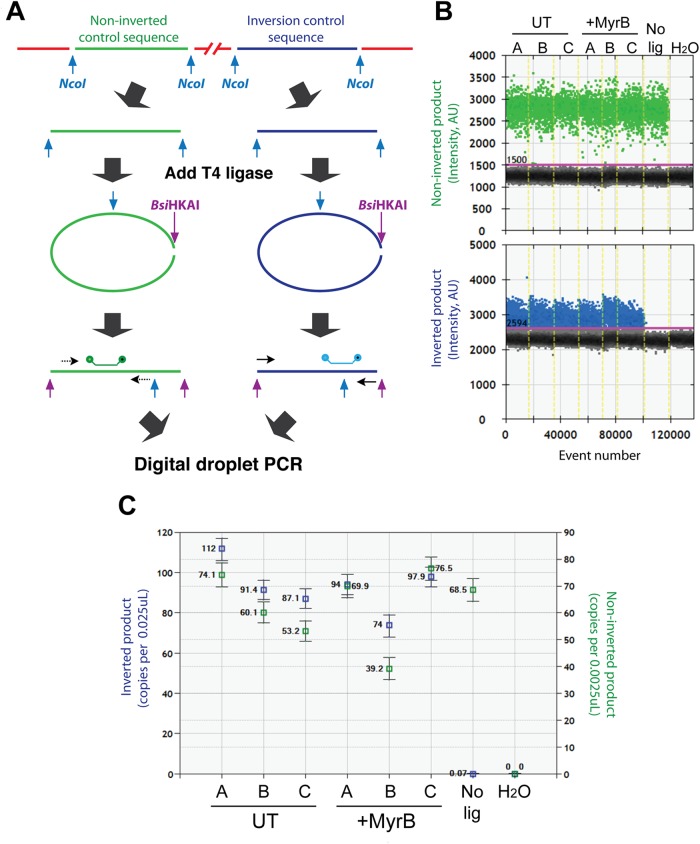
First, we confirmed using a digital droplet PCR (ddPCR) assay that DNA recovery during the invPCR procedure and inversion efficiency was high and was unlikely to contribute greatly to a lower observed integration rate. We found no significant difference between the number of cell equivalents in inverted DNA extracts from untreated and Myrcludex B (MyrB)-treated groups (Fig. 3), showing that lower DNA input was not the cause of the decrease in integration rate observed in MyrB-treated cells. Inversion efficiency (defined by the ratio between the number of noninverted and inverted products) was close to 100% in both untreated and MyrB-treated samples, while control samples in which ligase was not added in the inversion reaction had a ratio close to the limit of detection (∼0.1%). This suggests that differences in inversion efficiency were not a significant factor in detection of integrated HBV DNA.
FIG 3.
Determination of inversion efficiency by digital droplet PCR. (A) During the inversion procedure, cellular regions also undergo inversion. Primers and probes specific for regions in the beta-globin and procollagen 1 alpha genes were, respectively, designed such that they would amplify no matter the inversion state (green) or only when the 1-kb fragment was inverted (blue). (B) Duplex ddPCR was carried out on untreated (UT) and Myrcludex B-treated (+MyrB) samples (n = 3) depicted in Fig. 8, including a no-ligation negative control (No lig) and a water negative control (H2O). (C) After a threshold was defined according to water controls, absolute numbers of each noninverted (green) and inverted product (blue) were calculated. For each data point, absolute copy numbers ± 95% Poisson error are shown.
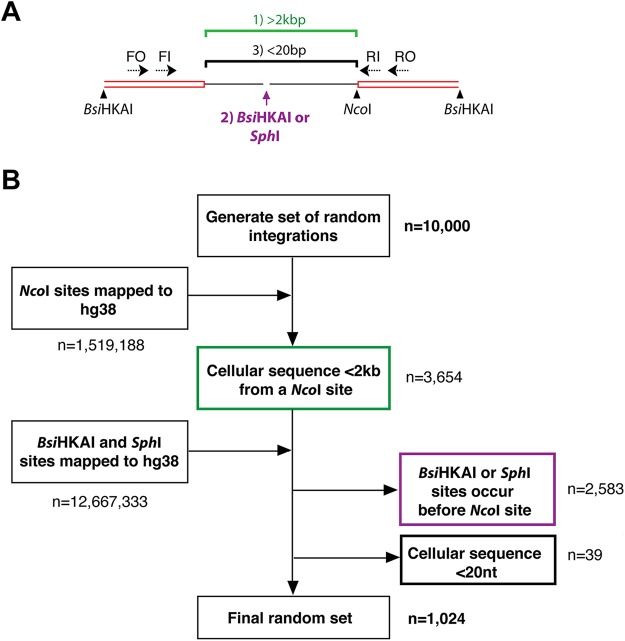
Next, to find the percentage of integrations that theoretically would be detectable by invPCR, we developed an in silico simulation of random HBV DNA integration. HBV DNA integrations were randomly distributed throughout the cellular genome in silico. We then applied three detection criteria to these integrated genomes (Fig. 4A). (i) The NcoI restriction enzyme site occurs <2 kb downstream of the integration junction. We have found that invPCR products of >2 kb were rarely detected by invPCR (in this and previous studies), possibly due to a combination of poor circularization efficiency and less efficient PCR amplification of long DNA fragments (data not shown). (ii) The restriction sites for BsiHKAI and SphI did not occur between the integration junction and the NcoI site. If the cellular region between the integration and downstream NcoI site contains sites recognized by BsiHKAI and SphI, then the inverted fragment is cleaved during the linearization reaction and rendered unamplifable by the HBV-specific primers. (iii) The NcoI restriction enzyme site occurs >20 nt downstream of the integration junction. With fragments smaller than 20 nt, it is difficult to unambiguously map the sequence to specific sites in the human genome and to determine if the fragment is human or HBV derived.
FIG 4.
In silico modeling to estimate the fraction of integration junctions detected by invPCR. There are 3 main sequence conditions in the cellular DNA that could prevent the detection of the integration junction by invPCR (A): (i) the cellular region being >2 kbp (green), (ii) the occurrence of BsiHKAI or SphI sites within the cellular region (purple), or (iii) the cellular sequence being <20 bp (blue). These three conditions were used to filter out integrations randomly generated in silico, occurring randomly throughout the human genome (B).
If any single one of these criteria was not met by a randomly generated integration, then it was classed as undetectable. Following the application of these criteria to the 10,000 randomly generated integrations, only 1,024 integrations were classed as detectable, suggesting that invPCR could identify at most ∼10% of all integrations (Fig. 4B). Taken together, the results show that the lower bound of HBV DNA integration in all cell lines is likely on the order of 1 integration per 1,000 cells, given the observed integration frequency by invPCR (∼10−4) and theoretical detection frequency of integrations shown by our in silico model (10−1).
HBV DNA integration in Huh7-NTCP cells occurs within 3 days of HBV infection.
We then established a timeline of when integration occurs by harvesting cells at 3, 5, 7, and 9 days postinfection (dpi) (Fig. 5A). HBV e antigen (HBeAg) secretion showed that cells were efficiently infected, with HepG2-NTCP cells secreting HBeAg at a level almost an order of magnitude higher than Huh7-NTCP cells (Fig. 5B and C). This difference is a function of cell type and their expression of HBeAg and not an indicator of number of infected cells, as shown previously (10). After invPCR analysis, we found that the integration rates in HepG2-NTCP and Huh7-NTCP cells did not appear to increase past 5 and 3 dpi, respectively (Fig. 5D and E), both plateauing at ∼1 integration per 104 cells. HepG2-NTCP cells showed high standard deviations within samples and varied widely between replicates (mainly due to few integrations being detected within each sample). We therefore continued further analyses using Huh7-NTCP cells, which showed consistently detectable HBV DNA integrations.
FIG 5.
Kinetics of HBV antigen expression and HBV DNA integration after HBV infection of HepG2-NTCP and Huh7-NTCP cells. Four infection time courses were carried out on HepG2-NTCP and Huh7-NTCP cells in triplicate (A). Cells were infected at day 0 with an HBV inoculum of 1,000 VGE/cell and washed off 1 day after. Cells were harvested at 3 (black arrow), 5 (red arrow), 7 (green arrow), and 9 (blue arrow) dpi. As per previous experiments, cells were maintained in medium supplemented with 10 μM lamivudine and 50 μM tenofovir (purple bar), transferred to 6-well plates (black cross), and maintained in DMSO-free medium (black bar) prior to harvest. Immediately prior to transfer to a 6-well plate (except for the d3 time course, due to signal interference from input inoculum), cell supernatant (S/N) was collected (black circle) and analyzed by immunoassay for secreted HBeAg (B and C for HepG2-NTCP and Huh7-NTCPs, respectively; mean ± 95% CI for 3 replicates shown for each time course). At the final time point, total DNA was extracted from cells and analyzed by invPCR for HBV DNA integration (D and E for HepG2-NTCP and Huh7-NTCP cells, respectively; geometric mean ± 95% CI). For invPCR, each data point represents a separate 1:3 dilution of inverted DNA extracted (only positive dilutions are shown) from 2 separate infections (indicated by either a square or triangle). ND, not determined; LOD, lower limit of detection (10−5 cell equivalents; dashed line).
Integration can be prevented by MyrB but not by drugs acting downstream of cccDNA formation.
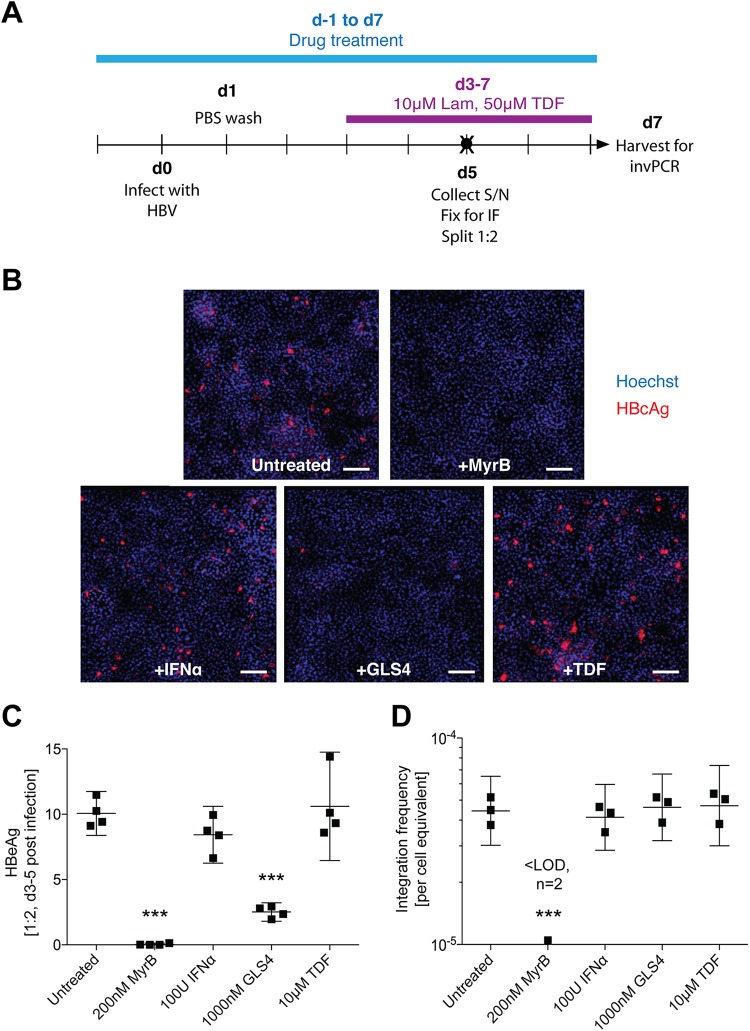
We determined at which step during the viral replication cycle integration occurs and tested the effect of anti-HBV compounds on HBV integration. As shown above (Fig. 5), integration occurs early during HBV infection. While we had included reverse transcriptase inhibitors in the experiments described above to inhibit amplifiable nonintegrated forms of HBV DNA, the cells were treated after infection past the period that integration would have likely taken place.
Given this, at 1 day prior to HBV infection (Fig. 6A), we applied various drugs that inhibit HBV replication at different points: HBV entry inhibition using MyrB at a dose shown to almost completely prevent virus infection via the blockade of the HBV-binding site of NTCP (10), stimulation of antiviral innate immune response using interferon alpha, HBV capsid assembly inhibition using GLS4, and reverse transcription inhibition using tenofovir. The dose of GLS4 used in this experiment (1 μM) has been shown to completely inhibit capsid assembly as one of its multiple modes of action (28), so this experiment is designed to show the contribution of intracellular cycling of de novo nucleocapsids to HBV DNA integration in this model. Further, in these experiments we infected cells with a smaller inoculum (500 VGE/cell) in the anticipation that some treatments could induce increased HBV DNA integration.
FIG 6.
Effect of anti-HBV compounds on HBV DNA integration frequency. Huh7-NTCP cells were infected as per the d7 time course in Fig. 5A (A) after 1 day of preincubation with various anti-HBV drugs (blue bar), including MyrB, interferon alpha (IFN-α), GLS4, and tenofovir. At 5 dpi, one set of replicates was analyzed by immunofluorescence (B). HBcAg staining is shown in red, and Hoechst nuclear staining is shown in blue. The scale bar represents 250 μm. Cell supernatant was also collected at 5 dpi (black circle in panel A) and analyzed by immunoassay for secreted HBeAg (C; mean ± 95% CI; ***, P < 0.001, compared to untreated control by one-way analysis of variance [ANOVA]; n = 4). At 7 dpi, total DNA was extracted from cells and analyzed by invPCR for HBV DNA integration (D; geometric mean ± 95% CI; ***, P < 0.001 by one-way ANOVA; n = 3).
Immunofluorescence (IF) showed that ∼5% of cells in the untreated group were positive for HBV core antigen (HBcAg) expression at 5 dpi (Fig. 6B). Consistent with previous studies, MyrB prevented HBV entry and subsequent antigen expression in almost all cells. Further, GLS4 abrogated most of the HBcAg signal, showing efficient inhibition of intracellular HBV capsids (29). Treatment with interferon alpha or tenofovir did not affect HBcAg expression, as expected from prior studies (30, 31). In general, levels of HBeAg secretion reflected the number of HBcAg-positive cells, except that GLS4-treated cells still showed some HBeAg secretion (at ∼25% of the untreated group) despite very few HBcAg-positive cells, likely due to the disruption (and the subsequent degradation) of cytoplasmic HBV capsids (Fig. 6C). invPCR results were consistent with the lower infection rate, with untreated cells showing an integration rate of ∼5 per 105 cells, half that of previous experiments which used an inoculum of 1,000 VGE/cell (Fig. 6D). Importantly, we found that only MyrB inhibited HBV DNA integration; no other treatments had significantly different integration rates from untreated controls, suggesting that the observed integration in this model occurs early in the replication cycle of HBV, likely using the input virus as a substrate.
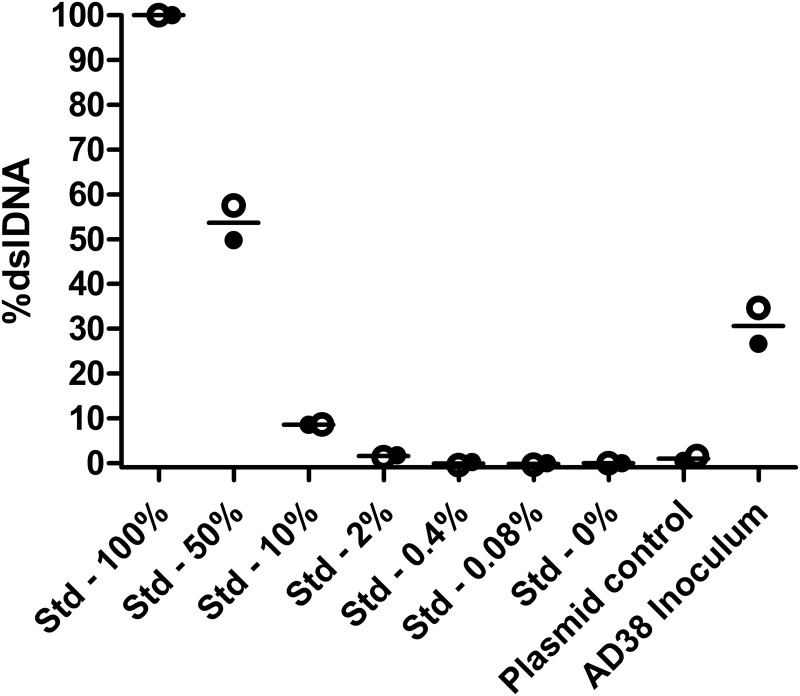
Following this conclusion, we used a previously developed clamp-based quantitative PCR (qPCR) assay to quantify the ratio of dslDNA (32) within our HBV inoculum and found that ∼30% of virions contained dslDNA (Fig. 7), consistent with previously reported Southern blot studies of HepAD38-derived HBV DNA (33). This ratio is within the range found in patient sera (previously shown to range from 3 to 36% [32]), suggesting that the observed integration is not due to an artificially high level of dslDNA being used for inoculation.
FIG 7.
Determination of the ratio of double-stranded linear to total HBV DNA in viral inoculum. A standard curve (Std samples) was generated for a proportional mixture of PCR amplicons that mimicked dslDNA or rcDNA forms, which were used to set 100% and 0%, respectively. Two independent repetitions were carried out (filled and hollow points represent independent experiments; lines represent means between the two experiments) and showed that the HepAD38-derived inoculum had a mean dslDNA percentage of 30%. A Midiprep-purified plasmid containing a 1.3-mer of the HBV genome (64) (which should generate the long/rcDNA form) was used as a negative control. The lower limit of quantification, as shown by the plasmid control, was ∼1%.
Integrated HBV DNA events in vitro closely resemble those found in patient tissues.
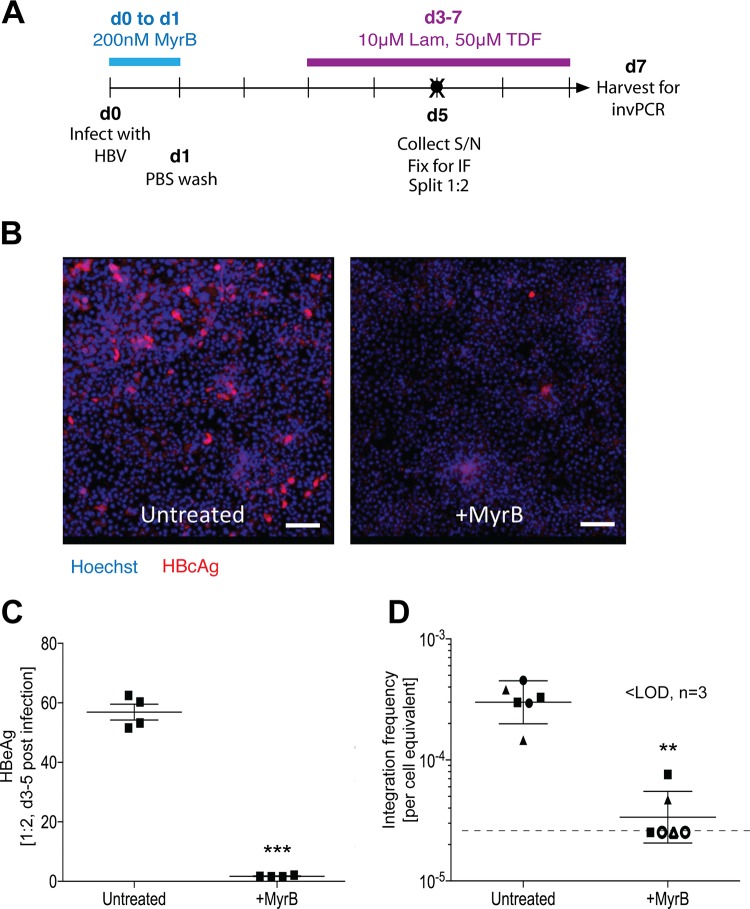
Finally, we analyzed the sequences of the virus-cell junctions detected in our in vitro model and compared them to those previously detected in liver tissues of patients with chronic HBV infection (19–21) to determine whether our system recapitulated in vivo molecular pathways. We also used integrations generated by the in silico model described above to account for detection biases based on the invPCR technique (e.g., the distribution of restriction enzyme sites over the host genome).
We generated large numbers of virus-cell junctions in the Huh7-NTCP cell line using optimized time postinfection (Fig. 8; identical to d7 in Fig. 5A) and a single optimal dilution of inverted DNA for PCR to produce ∼1 PCR product per well. We treated cells with MyrB as a negative control and (as expected) observed significantly lower numbers of HBV-expressing cells, as shown by both immunofluorescence detecting HBcAg (Fig. 8B) and HBeAg secretion in the supernatant (Fig. 8C), confirming that HBV infection occurred through NTCP-dependent entry pathways. A total of 197 virus-cell junctions were sequenced, from which we calculated a mean integration rate of 3.17 × 10−4 (standard deviation, ±1.03 × 10−4) per cell equivalent, or 1 integration event per ∼3 × 103 cell equivalents. MyrB reduced integration down to the lower limit of detection with an average rate of 1 integration per ∼4 × 104 cell equivalents.
FIG 8.
Generation of HBV integration junctions for sequence analysis. Huh7-NTCP cells were infected as per the d7 time course in Fig. 5A (A), with a negative control that was treated with MyrB during the inoculation period (blue bar). At 5dpi, one set of replicates was analyzed by immunofluorescence (B). HBcAg staining is shown in red, and Hoechst nuclear staining is shown in blue. The scale bar represents 250 μm. Cell supernatant was also collected at 5 dpi (black circle in panel A) and analyzed by immunoassay for secreted HBeAg (C; mean ± 95% CI; ***, P < 0.001 by paired t test; n = 4). At 7 dpi, total DNA was extracted from cells and analyzed by invPCR for HBV DNA integration (D; geometric mean ± 95% CI; **, P < 0.01 by paired t test; n = 4). For invPCR, each data point represents a separate inversion from 3 separate infections (indicated by either a square, triangle, or circle). Samples below the lower limit of detection (dashed line) of 2.5 × 10−5 cell equivalents are shown as hollow data points.
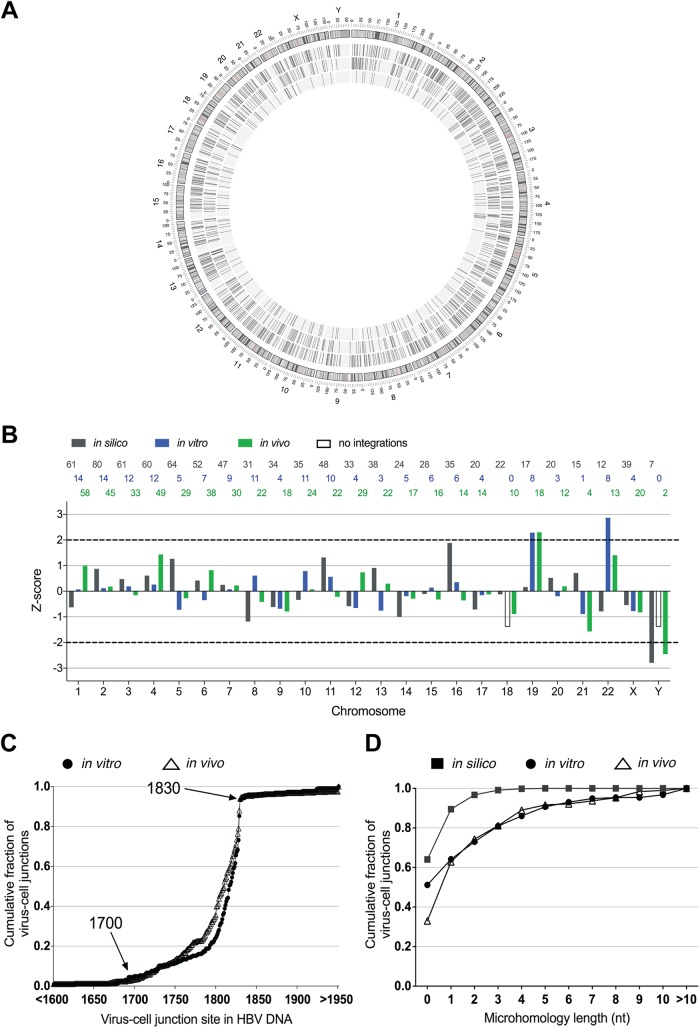
We mapped the distribution of HBV DNA integrations (in silico data set, n = 883; in vitro data set, n = 161 integrations; in vivo data set, n = 559 integrations) with respect to the cellular chromosomes; integration frequencies are shown in Table 2. As previously reported, HBV DNA integration was detected across the whole genome, with no obvious genomic hot spots for integration (Fig. 9A). After normalization of the number of integrations in each data set, length of each chromosome, we confirmed by Z-score analysis that there were generally no preferred chromosomal preferences for HBV DNA integration (Fig. 9B), though slight but significant enrichment (∼2-fold) was seen in chromosomes 19 (in vitro and in vivo data sets) and 22 (in vitro data set).
TABLE 2.
Distribution of HBV DNA integrations across the cellular genome
| Chromosome |
In silico |
In vitro |
In vivo |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No.a | % integrations per % of genome | Z-score | No. | % integrations per % of genome | Z-score | No. | % integrations per % of genome | Z-score | |
| 1 | 61 | 0.86 | −0.63 | 14 | 1.08 | 0.07 | 58 | 1.29 | 1.01 |
| 2 | 80 | 1.16 | 0.88 | 14 | 1.11 | 0.12 | 45 | 1.03 | 0.18 |
| 3 | 61 | 1.08 | 0.48 | 12 | 1.16 | 0.19 | 33 | 0.92 | −0.16 |
| 4 | 60 | 1.10 | 0.61 | 12 | 1.21 | 0.26 | 49 | 1.42 | 1.44 |
| 5 | 64 | 1.23 | 1.27 | 5 | 0.53 | −0.72 | 29 | 0.88 | −0.27 |
| 6 | 52 | 1.06 | 0.42 | 7 | 0.79 | −0.35 | 38 | 1.23 | 0.82 |
| 7 | 47 | 1.03 | 0.25 | 9 | 1.08 | 0.08 | 30 | 1.04 | 0.23 |
| 8 | 31 | 0.75 | −1.18 | 11 | 1.45 | 0.61 | 22 | 0.84 | −0.42 |
| 9 | 34 | 0.86 | −0.62 | 4 | 0.55 | −0.68 | 18 | 0.72 | −0.79 |
| 10 | 35 | 0.91 | −0.34 | 11 | 1.58 | 0.79 | 24 | 0.99 | 0.07 |
| 11 | 48 | 1.24 | 1.32 | 10 | 1.42 | 0.56 | 22 | 0.90 | −0.22 |
| 12 | 33 | 0.87 | −0.58 | 4 | 0.58 | −0.65 | 29 | 1.20 | 0.74 |
| 13 | 38 | 1.16 | 0.91 | 3 | 0.50 | −0.76 | 22 | 1.06 | 0.3 |
| 14 | 24 | 0.78 | −1 | 5 | 0.90 | −0.19 | 17 | 0.88 | −0.29 |
| 15 | 28 | 0.96 | −0.11 | 6 | 1.13 | 0.14 | 16 | 0.87 | −0.32 |
| 16 | 35 | 1.36 | 1.88 | 6 | 1.27 | 0.35 | 14 | 0.86 | −0.36 |
| 17 | 20 | 0.84 | −0.71 | 4 | 0.92 | −0.15 | 14 | 0.93 | −0.13 |
| 18 | 22 | 0.96 | −0.12 | 0 | 0.00 | −1.48 | 10 | 0.69 | −0.89 |
| 19 | 17 | 1.01 | 0.16 | 8 | 2.62 | 2.29 | 18 | 1.70 | 2.3 |
| 20 | 20 | 1.09 | 0.52 | 3 | 0.89 | −0.2 | 12 | 1.03 | 0.19 |
| 21 | 15 | 1.12 | 0.71 | 1 | 0.41 | −0.89 | 4 | 0.47 | −1.57 |
| 22 | 12 | 0.83 | −0.79 | 8 | 3.02 | 2.86 | 13 | 1.41 | 1.41 |
| X | 39 | 0.87 | −0.54 | 4 | 0.49 | −0.77 | 20 | 0.71 | −0.82 |
| Y | 7 | 0.43 | −2.79 | 0 | 0.00 | −1.48 | 2 | 0.19 | −2.45 |
Number of integrations.
FIG 9.
Sequence analysis of in vitro-generated virus-cell junction HBV DNA integration events in in silico, in vitro, and in vivo data sets were represented graphically in a circular form using Circos (A). The outer rim of the Circos plot represents the ideograms of each chromosome. The inner rings show in silico (outermost), in vivo (middle), and in vitro (innermost) HBV integrations. HBV DNA integrations from in silico (gray), in vitro (green), and in vivo (blue) data sets were mapped to each cellular chromosome, normalized to the chromosome length and number of integrations per data set (B). These values were converted into a Z-score, with 0 representing the mean of normalized integration frequency for all chromosomes and 1 representing 1 SD. The number of integrations found in each data set within chromosome is shown on the top of the graph in its respective color. The cumulative frequency of the integration breakpoints for either in vitro (filled circles) or in vivo (hollow triangles) data sets was plotted with respect to the HBV nucleotide position as described under GenBank accession no. U95551.1. (C). As expected from previous studies, enrichment of the integration events was observed at nucleotides 1700 to 1830 (aligning to the 3′ end of the HBx ORF and 5′ end of the precore/core ORF). Homologous sequence lengths between the human and HBV genome at the integration sites were plotted as cumulative fractions for the in silico (filled squares; n = 833), in vitro (filled circles; n = 191), and in vivo (hollow triangles; n = 224) data sets (D). To determine the expected homologous sequence distribution, viral integration breakpoints were randomly produced in the in silico model based on the frequency distribution of HBV junctions observed in the in vitro and in vivo data sets.
Next, we mapped the sites of integration with respect to the HBV genome (Fig. 9C). We found that the distributions of HBV breakpoints were similar between in vitro and in vivo groups and the majority of HBV DNA integrations end prior to position 1832 (the expected end of the dslDNA form) with 5′ truncations of 0 to ∼100 nt. It is likely that the small offset between the curves of the in vitro and in vivo groups represents the slight differences in sequences of the infecting HBV strain (genotype D in the in vitro group and a variety of genotypes in the in vivo group as described in Materials and Methods). Further, we randomly assigned each in silico-generated integration an HBV breakpoint based on this distribution to act as a null hypothesis to determine whether significant sequence homology was observed in the virus-cell junctions.
We found that the majority of integration junctions were likely generated by NHEJ repair mechanisms, as the integrated HBV sequence shared limited homology to the cellular sequence for in vitro and in vivo data sets (Fig. 9D). On the other hand, given data from our in silico data set (Fig. 9D), we would expect ∼90% of virus-cell junctions to show <2 nt of homology between the HBV and cellular region by chance. However, we observed that the in vivo and in vitro data sets (while similar to each other) contained only ∼60% of virus-cell junctions that show <2 nt of homology. Therefore, the difference of these figures led us to the conclusion that ∼30% of virus-cell junctions are formed through a biological process requiring 2 nt or more of homology, indicative of microhomology-mediated end joining (MMEJ; also known as alternative NHEJ). This suggests that in both our in vitro model and patient tissues, a mixture of DNA repair mechanisms may be involved in HBV DNA integration.
In summary, our sequence analysis data show that the HBV DNA integration in our in vitro model is likely to be generated by the same biological pathways as those in HBV-infected patients.
DISCUSSION
This study represents the most in-depth characterization of in vitro-generated HBV DNA integrations to date. With these data, we have shown that HBV integration occurs within days of infection in different human liver cell types that support HBV infection (including PHH), consistent with observations in HBV patients (21, 34–36) and animal models (18, 27, 37). Further, we have established a suitable cell culture model that supports consistently detectable HBV DNA integration at a rate of ∼1 integration per 10,000 cells, similar to the rate reported in animal model studies (18, 27, 37). In this system, HBV DNA integration occurs early in the viral replication cycle, as it can be directly blocked by the HBV entry inhibitor MyrB, but not by 10 μM tenofovir, 100 U of interferon alpha, or a 1 μM concentration of the capsid assembly inhibitor GLS4. Finally, we showed that the sequences of integrated HBV DNA generated by our system closely resemble those found in patient tissues.
Based on these findings, several conclusions can be drawn regarding HBV DNA integration and HBV replication. First, one of the main results is that integration can be inhibited by MyrB, providing strong evidence that integration requires entry through an NTCP-dependent pathway. The outcome of this is that any nonvirion contaminants in the inoculum (e.g., secreted naked nucleocapsids, exosomes, etc.) are excluded from the cause of the detected integration. While previous work in the duck HBV model suggested that dslDNA-containing virions are the main substrates for HBV DNA integration (18, 23), the contribution of various enveloped forms of HBV (such as RNA-, splice variant DNA-, single-stranded DNA [ssDNA]-, and rcDNA-containing virions) toward human HBV DNA integration requires further study. Several reports have indicated that RNA can act as a template for double-stranded DNA break repair in eukaryotic cells (38, 39). Viral misappropriation of this pathway may lead to HBV sequences being inserted into the cellular genome. Further, as the viral polymerase (which has reverse transcriptase functions) is delivered along with the virion, viral RNA could conceivably be converted into cDNA within the cell, providing a template for HBV DNA integration. As HBV DNA integration occurs somewhat rarely, HBV RNA or RNA-DNA hybrid structures could be used to repair cellular DNA breaks, leading to integration of a HBV DNA sequence, even if these processes do not occur efficiently. Indeed, this in vitro HBV infection system now allows further molecular examination of the true substrates of HBV DNA integration.
Second, sequence analysis of both in vivo and in vitro virus-cell junctions suggests that the integration occurs through both NHEJ and MMEJ. We found that the majority of virus-cell DNA junctions (approximately two-thirds) display little shared homology (0 to 1 nt) between the HBV and cellular sequences. Thus, extended tracts of homology (e.g., previously reported virus-cell junctions with longer sequence homology, up to 11 nt [40]) are not indicative of the majority of virus-cell junctions. In the remaining third of virus-cell DNA junctions, we observed enriched homology (2 to 6 nt) at the site of integration, consistent with the MMEJ pathway (41). As NHEJ and MMEJ occur throughout the cell cycle (42), our sequence analysis agrees with our data showing that the cellular mitotic state does not affect the integration rate: the integration rates in terminally differentiated hepatocyte-like cells and PHH and less differentiated hepatoma cell lines undergoing mitosis are approximately equal and do not change when mitosis is induced.
MMEJ has been suggested by some groups as a pathway for HBV integration (3, 40, 43, 44) and is associated with chromosomal translocation (41) and other cancer-associated changes (45, 46). Thus, it is plausible that HBV DNA integrations occur preferentially in hepatocytes with precancerous alterations in DNA repair, explaining why the majority of HCCs contain integrated HBV DNA (5, 19, 47, 48), compared to 1 in 103 to 104 cells in the general hepatocyte population of an infected liver (18, 37). This is consistent with the “mutator phenotype” hypothesis of carcinogenesis, wherein an increased mutation rate (e.g., through more error-prone DNA repair machinery) is an inevitable selective advantage in the preneoplastic microenvironment (49, 50). Alternatively, while we show that cell turnover itself does not increase HBV DNA integration (Fig. 2B), it may be possible that the mitosis of HCC or HCC-initiating cells that is associated with clonal expansion results in daughter cells that are more susceptible to de novo HBV infection and thereby additional chances for integration to occur. The lack of physiologically relevant models for HCC development in chronic human HBV infection currently limits the study of these hypotheses.
Further, previous studies have reported the observation of repeated virus-cell junctions in separate infections (40, 43), suggesting that HBV preferentially integrates into particular cellular DNA sequences (e.g., long interspersed transposable elements). While some integrations were indeed observed in or near repetitive DNA elements, we found no convincing evidence of sequence preference in our analysis of 297 in vitro-generated virus-cell junctions above chance compared to our in silico model. Only 10 repeated virus-cell junctions were observed (likely due to the mitosis of a hepatocyte containing the virus-cell junction), with only one being repeated in a separate DNA extraction (data not shown). Therefore, our data strengthen the hypothesis that HBV DNA integration occurs at the site of randomly dispersed double-stranded DNA breaks. Slightly nonuniform HBV DNA integration over the cellular genome was observed, however, with ∼2-fold enrichment of HBV DNA integration into chromosomes 19 and 22 in both the in vitro and in vivo data sets. Future studies are needed to determine whether this enrichment may be due to increased DNA damage in these chromosomes, a hypothesis that may be tested in further studies using our model.
Finally, HBV DNA integration preferentially occurs at early stages of the viral replication cycle, given the timing of integration (<3 dpi) and the plateauing integration rate by 5 dpi (despite constant HBV antigen expression). Importantly, our HBV inhibitor experiments show that blocking viral replication at stages past virus entry do not affect integration rates, suggesting that the majority of HBV DNA integration in this short-term infection model likely uses the incoming viral DNA as a major substrate. This hypothesis can be further examined in our model by using replication-deficient HBV virions (e.g., polymerase-defective or HBcAg-knockout HBV mutants) as an inoculum. Whether nuclear import of viral capsids occurs over a long-term (decades-long) infection within a particular hepatocyte in a chronic HBV patient is not known and would be technically impossible to show in this model.
Moreover, the extent to which the Huh7-NTCP cells reflect nuclear import of nucleocapsids within a true infection is unclear, and this may constitute a shortcoming of our system. The concept of nuclear import of de novo nucleocapsids has been established in the duck model of HBV DNA, in which extensive intracellular amplification of viral cccDNA occurs (51). We are not aware of any data showing (or ruling out) the amplification of cccDNA in Huh7-NTCP cells. However, there is evidence by others showing that intracellular amplification of cccDNA does not occur in more physiologically accurate systems, such as PHH and HepaRG cells. Recent studies in the humanized mouse model (27, 52) show that increases in cccDNA in the liver occur mainly through an extracellular pathway (i.e., new infection of hepatocytes) and can be blocked using entry inhibitors. Similar findings have been described for HepaRG cells (53). Therefore, the role that intracellular cccDNA amplification plays in HBV DNA integration and whether the Huh7-NTCP system differs significantly from PHH and HepaRG cells in this respect are still open questions to be answered by the field at large.
Due to the extensive amplification of HBV DNA intermediates by our invPCR assay, we were forced (for practical reasons) to use hepatoma cells rather than nontransformed hepatocyte-like cells in the majority of our experiments. While we posit that defective closed minus-strand HBV DNA and cccDNA may contribute to the amplified invPCR products containing only HBV sequences (Fig. 1C), the true source of these forms is not known. If indeed our hypothesis is correct, then the higher intracellular replication observed in HepG2-NTCP and HepaRG-NTCP cells and PHH than in Huh7-NTCP cells (10, 11) may account for why the last provides an optimal model for detection of integrated HBV DNA. An alternate possibility is that conversion of rcDNA from input virions to nuclear closed minus-strand rcDNA is less efficient in Huh7-NTCP cells. Though important to improve the system in future, the identity of the cell-specific factors of these intermediate forms lies outside the scope of this study.
Other limitations of this model include the inability to control for the number of cells that have taken up HBV. Previous reports have shown that following HBV inoculation, Huh7-NTCP cells exhibit a lower proportion of HBcAg-positive cells than HepG2-NTCP cells inoculated with the same virus dose (10). However, the true number of cells that have internalized the virus is likely higher than the observed proportion of HBcAg-positive cells, as the detection by immunofluorescence (IF) is dependent on expression level, secretion level, and background associated with each cell type. For example, while HBV-infected Huh7-NTCP cells secrete higher levels of HBsAg (as measured by HBsAg enzyme-linked immunosorbent assay [ELISA] of the cell supernatant compared to HepG2-NTCP cells), IF staining for HBsAg is poor (with many HBcAg-positive cells appearing HBsAg negative). On the other hand, HepG2-NTCP cells show the opposite phenotype, with low HBsAg secretion and high IF signals for both HBcAg and HBsAg. Further, the susceptibility of Huh7-NTCP to hepatitis delta virus (HDV; uses the same receptor as HBV) is equal to or greater than that of HepG2-NTCP cells (10). This would suggest that virus internalizations via HBsAg-NTCP interactions are similar between these two cell lines, despite different numbers of HBcAg-positive cells after HBV inoculation. Finally, should the HBV virion be internalized and its genome integrate into the cellular DNA, the integrated HBV DNA is not likely to express HBcAg or HBeAg due to the separation of the HBc/e ORF from its native promoter in the dslDNA form, making HBcAg immunofluorescence a poor measure. In summary, the numbers of cells that have internalized virions during the inoculation process between different cell types cannot be directly compared using immunofluorescence.
Indeed, the rate of HBV internalization is incredibly difficult to measure and measures for this (e.g., visualization of infectious HBV particles) are major technical limitations in the HBV field. Quantitative PCR measurements for HBV DNA of inoculated cells are complicated by the fact that the input virus is detected despite multiple washes with phosphate-buffered saline (PBS) and medium exchanges. We and others have found that total DNA extracted from Myrcludex B-treated cells generally contains the same amount of total HBV DNA as untreated cells prior to high HBV replication (occurring around 3 to 5 days postinfection). Therefore, measuring the extent of HBV virion uptake remains an ongoing technical issue.
Another technical shortcoming is the inability to determine the full integrated HBV DNA sequence with these assays. The nature of the invPCR technique does not allow simultaneous identification of both left and right virus-cell DNA junctions of the integrated HBV DNA. Profiling these in separate inversion reactions has a low probability of detecting the same integration event, as very few copies (1 or 2) of each integration event exist in the DNA extract of the infected cells due to limited clonal expansion in our infection model. As with the right end, the left end of the HBV integration is likely to not exactly match the dslDNA ends but instead to include terminal truncations of approximately hundreds of base pairs, as has been shown previously in woodchuck and chimpanzee models of HBV infection (16, 22).
While whole-genome sequencing has been used in other studies to detect integrated HBV DNA, these have all been carried out for chronically infected HBV patients with HCC where hepatocytes containing HBV DNA integrations have undergone extensive clonal expansion (clones of >10,000 cells), thereby considerably increasing the likelihood of detection. In our system, whole-genome deep sequencing would not have the sensitivity required for the detection of the virus-cell junctions with low copy numbers generated by our experimental layout. Even given a relatively deep coverage of 100×, we would expect a 1-in-10 chance to detect a single-copy integration in our experimental setup with an integration rate of 10−3 integrations per cell. Therefore, invPCR (though imperfect) gives a greater specificity and sensitivity to detect HBV DNA integration events at the low copy numbers generated by this infection system.
Apart from the elucidation of the specific viral substrates and the impact of HBV proteins (as described above), this in vitro system can also be used as a test bed for potential inhibitors of HBV integration and to determine specific cellular factors involved in HBV DNA integration. We hypothesize that factors associated with DNA repair (e.g., NHEJ and MMEJ machinery) and the induction of double-stranded breaks (e.g., DNA guardian proteins) may play a role in the rate of HBV integration. This model would also be useful in discovering the mechanisms of specific anti-HBV drugs (e.g., distinguishing between factors directly involved in the conversion of rcDNA to cccDNA and those that inhibit entry steps upstream of cccDNA formation, as the latter factors would inhibit both cccDNA copy number and HBV DNA integration rates).
Our results show that HBV DNA integration occurs within days of the initial infection at a true rate of ∼1 in 103 cells, a rate consistent with animal models of acute HBV infection (18, 37). This presents a large burden of integrated HBV DNA in the liver, as shown by the estimations below.
Assuming that the integration rate is ≈1 in 103 cells (estimate 1), there are ≈2 × 1011 cells in a liver (estimate 2), 2% of the genome is coding (estimate 3), and the human genome has ∼2 × 104 genes (estimate 4). Given estimates 1 and 2, the number of integrations per liver is ≈2 ×108 (estimate 5). Given estimates 3 and 5, the number of integrations in coding regions per liver is ≈1 × 106 (estimate 6). Given estimates 4 and 6, the average number of integrations per gene per liver is ≈5 × 102 (estimate 7). In other words, for any given gene, there are ∼500 hepatocytes in the liver containing an HBV DNA integration.
Therefore, every patient exposed to HBV, including those not currently considered for treatment (e.g., patients in the immune-tolerance phase and acute HBV patients), could have an increased risk for HCC. Indeed, virological clearance of HBV does not completely negate HCC risk (54), though the direct role of integrated HBV DNA is still not clear. Large epidemiological studies are still needed to explore this hypothesis, but its confirmation would strengthen the argument for earlier treatment of chronic HBV infection to limit HCC risk (21).
In conclusion, we have established a novel in vitro system of HBV DNA integration resulting from NTCP-dependent HBV infection, which has given insight into aspects of HBV replication (e.g., dynamics of HBV capsid nuclear import) and pathogenesis (e.g., the time points of potential HBV-associated HCC risk). HBV DNA integration has so far been an underresearched field, but our system would allow the identification and in-depth analysis of cellular and viral factors that may alter pathways involved in integration. From this, we hope to provide greater insight into why HBV DNA integrates, particularly its function in virus persistence, replication, and pathogenesis.
MATERIALS AND METHODS
Cell culture and HBV infection.
PHH, differentiated HepaRG-hNTCP cells (10), and human hepatoma cell lines (HepG2-NTCP and Huh7-NTCP cells [10]) were used for in vitro infection. PHH were isolated from liver resections of patients undergoing partial hepatectomy as previously described (55) and plated onto coverslips in a 24-well format. All tissue donors gave written informed consent for the experimental use of liver specimens, and the protocol was approved by the ethics review committee of Hannover Medical School (252-2008). PHH and HepaRG-NTCP cells were cultivated in William's E medium supplemented with 1.5% dimethyl sulfoxide (DMSO), as previously described (56, 57). Hepatoma cell lines were maintained in DMSO-free Dulbecco's modified Eagle's medium, as previously described (10). All cell lines were cultured in a 12-well format for HBV infection.
For HBV infection, supernatant from HepAD38 (58) was used as an inoculum, after purification as previously described (59). Briefly, inocula were purified by heparin affinity chromatography, concentrated ∼100-fold in 10% fetal calf serum, and stored at −80°C until use. Heparin purification has been shown previously to effectively remove naked nuclecapsids from the final eluted sample, leaving only HBV virions and some contaminating subviral particles (60). HBV inoculum concentration, expressed as VGE per milliliter, was quantified using the COBAS AmpliPrep/COBAS TaqMan HBV test (Roche) by the Heidelberg University Clinic Virology Diagnostics Department. Cells were infected with purified HBV at 1,000 (or 500, for drug treatment experiments) VGE/cell in 500 μl (or 250 μl for PHH) of culture medium supplemented with 4% polyethylene glycol 8000 (Sigma-Aldrich, St. Louis, MO) and 1.5% or 2.5% DMSO (for nonhepatoma or hepatoma cells, respectively). Cells were washed twice with 1× phosphate-buffered saline (PBS) at 24 h postinfection. Culture medium was replaced every 2 days following HBV infection until harvest.
To deplete HBV cccDNA forms in preparation for invPCR, cells were treated with 50 μM tenofovir disoproxil (CDS021622; Sigma-Aldrich) and 10 μM lamivudine (L1295; Sigma-Aldrich) from 4 days prior to harvest. For further HBV DNA depletion in HBV-infected hepatoma cell lines, cells were trypsinized 2 days prior to harvest and transferred from a 12-well to 6-well plate in DMSO-free Dulbecco's modified Eagle's medium to induce a round of mitosis, which has been reported to induce loss of HBV cccDNA (27). To measure HBV replication, secreted HBeAg in culture supernatants (prediluted either 1:2 or 1:5 in 1× PBS) was detected via immunoassay (ADIVA Centaur HBeAg; Siemens, Munich, Germany) by the Heidelberg University Clinic Analytical Centre.
For drug treatments, Huh7-NTCP cells were treated with 200 nM Myrcludex B (MyrB; synthesized by solid-phase synthesis in-house, as previously described [61]), 100 U/ml of interferon alpha (PeproTech GmbH, Hamburg, Germany), a 1 μM concentration of the HBV capsid assembly inhibitor GLS4 (Novira, Doylestown, PA), or 10 μM tenofovir 1 day prior to HBV infection with an inoculum of ∼500 VGE/cell. Drugs were included in the cell culture medium until cell harvest.
Detection of HBcAg-positive cells by immunofluorescence.
Cells were washed with 1 ml of 1× PBS and fixed with 300 μl of 4% paraformaldehyde for 20 min at room temperature. After permeabilization of cells with 0.25% Triton X-100 in 1× PBS, HBcAg was detected using a 1:3,000 dilution of polyclonal rabbit anti-HBcAg antibody (B0586; Dako, Denmark) (57). Cells were incubated with primary antibody overnight at 4°C, washed thrice with 1 ml of 1× PBS, and incubated in the dark for 1 h at room temperature with an AF545-conjugated goat anti-rabbit secondary antibody (A-11010; Invitrogen/Thermo Fisher Scientific, Carlsbad, CA) at a dilution of 1:500 and 2 μg/ml of Hoechst 33342 (H1399; Invitrogen). Fluorescence microscopy images were taken at a magnification of ×40. For each sample, an area the size of 5 by 5 fields of view at this magnification were acquired and merged by NIS Elements Advanced software (Nikon, Minato, Tokyo, Japan).
Inverse nested PCR.
At harvest for DNA extraction, cells were trypsinized, resuspended in 1 ml of culture medium, pelleted, and resuspended in 400 μl of digestion buffer containing 100 mM NaCl, 0.5% SDS, 50 mM Tris HCl (pH 7.5), 10 mM EDTA, and 2 mg/ml of proteinase K. Total cellular DNA was extracted with phenol-chloroform and analyzed for integrated HBV DNA using invPCR (Fig. 1A), as previously described (25). Sanger sequencing was performed by GATC Biotech (Germany). The integration rate was determined by dividing the number of virus-cell junctions detected by the number of cell equivalents within a particular dilution of DNA template. An annotated table (Table S1) of all detected virus-cell junctions is provided in the supplemental material.
Digital droplet PCR to determine cell number and inversion efficiency.
To find the inversion efficiency, we designed specific digital droplet PCR assays that could detect inversion of DNA sequences in single-copy cellular genes (Fig. 3A). Half a microliter of the 40 μl of inverted DNA was used in a 20-μl duplex ddPCR assay volume, composed of 1× ddPCR Supermix for Probes (1863010; Bio-Rad, Hercules, CA) and 0.15 nM each primer and probe. The forward and reverse primer and probe sequences for the uninverted control fragment of beta globin gene were 5′-GAAGAGCCAAGGACAGGTAC-3′, 5′-TGAGGTTGCTAGTGAACACAG-3′, and 5′-6-carboxy-2,4,4,5,7,7-hexachlorofluorescein (HEX)-AGGGTTGGCCAATCTACTCC-3′-BHQ1, respectively. The forward and reverse primer and probe sequences for the inverted control fragment of the procollagen 1 alpha gene were 5′-CTGGTGAACGTGGTGCAG-3′, 5′-CTGGGAGGTAGGGGTAGGAA-3′, and 5′-6-carboxyfluorescein (FAM)-TCTATCCCATGGAGATAGGG-3′BHQ1, respectively. The ddPCR assay was carried out essentially according to the manufacturer's protocol (with some modifications in PCR conditions) on a QX200 digital droplet PCR system (Bio-Rad). We modified the conditions for PCR amplification within the droplets to match our primers and probes to the following steps: an initial 10-min denaturation, enzyme activation, and droplet stabilization step at 95°C, followed by 40 cycles of a 10-s denaturation step at 95°C, a 15-s annealing step at 54°C, and a 20-s elongation step at 68°C, finished with a 10-min enzyme deactivation step at 95°C. Products were then stored at 4°C until droplet reading and quantification using both HEX and FAM channels.
In silico simulation of HBV DNA integration.
HBV integrations were generated in silico by first selecting a set of 10,000 random genomic positions using BEDTools (v2.26.0) (62) (with random seed 71346). Restriction enzyme sites for NcoI, BsiHKAI, and SphI were mapped by aligning the restriction sites to the human reference genome, GRCh38.p7 (hg38), using Bowtie (v1.1.1) (63). Overlap of restriction sites with the 2-kb windows downstream of selected genomic positions was determined using the BEDtools intersect tool. Three selection criteria were used to filter out those positions that would not be detected by invPCR: (i) presence of an NcoI site <2 kb downstream of the junction, (ii) absence of BsiHKAI and SphI sites between the junction and the NcoI site, and (iii) the sequence between junction and NcoI site being >20 nt.
PNA clamp-mediated qPCR to quantify HBV dslDNA/rcDNA ratio.
Peptide nucleic acid (PNA)-mediated clamp qPCR was carried out as previously described (32) on 106 copies of PCR amplicons corresponding to the long and short amplicons (corresponding to the final rcDNA and dslDNA forms generated by the assay, respectively) in various ratios as a standard curve; DNA was extracted from ∼2 μl of HepAD38-derived inocula using the innuPREP virus DNA kit (Analytik Jena AG, Jena, Germany) or a Midiprep-purified plasmid containing a 1.3-mer of the HBV genome (64) (corresponding to the long/rcDNA form) as a negative control. The ratio of dslDNA was calculated as the difference in threshold cycles (2ΔCT) between PNA-containing and PNA-deficient reactions, normalizing using pure long- and short-form reactions to 0% and 100%, respectively. PNA-mediated clamp qPCR was performed in duplicate on independent DNA extracts of HepAD38-derived inoculum.
Data sets and bioinformatics analysis.
We analyzed 4 data sets of virus-cell junctions detected using invPCR: 3 from previously published studies from primary liver tissues (n = 45) (19–21) and 1 from the in vitro HBV infection model described here (Huh7-NTCP only). The in vivo data set had a male-to-female ratio of 14:27. Of the 45 patients investigated, 5 were infected with HBV genotype A, 9 with HBV genotype B, 10 with genotype C, 12 with genotype D, and 4 with genotype E. FASTA sequences were aligned against human (GRCh38.p7) and HBV genomes (GenBank accession no. U95551.1, genotype D) using a standalone version of BLAST ncbi-blast-2.6.0+ (65) with a custom-created database. Sequences were excluded from downstream analysis (i) if sequences fully aligned either only to the human genome or only to the HBV genome, (ii) if the alignment was <20 nt, or (iii) if there was <80% identity between query and reference genome sequences. Further, Repeat Masker (66) was used to annotate repeat elements and mark sequences that fully fall into the repetitive regions. The functional annotation of the integrated breakpoints was performed by ANNOVAR (67). Detected integration events were plotted against a circular representation of the human genome (Circos plot) using Circos (68).
Accession number(s).
The complete in vitro nucleotide sequences are available in GenBank under accession numbers MH057851 to MH058006.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yi Ni and Florian A. Lempp for reagents (cell lines and HBV inoculum) and William S. Mason for sequence data used in this study. We acknowledge Katrin Schoeneweis, Anja Rippert, Franziska Schlund, and Sarah Engelhardt for technical assistance. We are grateful to William S. Mason, Florian Lempp, and Benno Zehnder for their critical readings of the manuscript and fruitful discussions, Miriam Kleinig for proofreading, and Ralf Bartenschlager for continuous support.
S.U. is a coapplicant and coinventor on patents protecting Myrcludex B as an HBV/HDV entry inhibitor. The rest of the authors have no conflicts of interest.
This work received funding from the German Centre for Infection Research (DZIF), TTU Hepatitis Projects 5.806, 5.807, and 5.704, and the Deutsche Forschungsgemeinschaft (DFG) TRR179 (TP 15). M.A.B. was supported by funding from University of Sydney Australian Postgraduate Award, Sydney Catalyst and Cancer Institute NSW.
T.T. conceived the project, designed and carried out the experiments, analyzed the data, generated figures, and wrote the manuscript; M.A.B. conducted the sequence analysis, generated figures, and assisted in writing the manuscript; F.W.R.V. supplied the primary human hepatocytes and contributed to execution of the experiments; N.A.S. provided funding for M.A.B., contributed to data analysis and interpretation of the results, and assisted in writing the manuscript; S.U. provided funding for T.T. and the project, contributed intellectual input for project and experimental design, and assisted in writing the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02007-17.
REFERENCES
- 1.Stanaway JD, Flaxman AD, Naghavi M, Fitzmaurice C, Vos T, Abubakar I, Abu-Raddad LJ, Assadi R, Bhala N, Cowie B, Forouzanfour MH, Groeger J, Mohd Hanafiah K, Jacobsen KH, James SL, MacLachlan J, Malekzadeh R, Martin NK, Mokdad AA, Mokdad AH, Murray CJ, Plass D, Rana S, Rein DB, Richardus JH, Sanabria J, Saylan M, Shahraz S, So S, Vlassov VV, Weiderpass E, Wiersma ST, Younis M, Yu C, El Sayed Zaki M, Cooke GS. 2016. The global burden of viral hepatitis from 1990 to 2013: findings from the Global Burden of Disease Study 2013. Lancet 388:1081–1088. doi: 10.1016/S0140-6736(16)30579-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding D, Lou X, Hua D, Yu W, Li L, Wang J, Gao F, Zhao N, Ren G, Li L, Lin B. 2012. Recurrent targeted genes of hepatitis B virus in the liver cancer genomes identified by a next-generation sequencing-based approach. PLoS Genet 8:e1003065. doi: 10.1371/journal.pgen.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan H, Yang Y, Zhang L, Tang G, Wang Y, Xue G, Zhou W, Sun S. 2015. Characterization of the genotype and integration patterns of hepatitis B virus in early- and late-onset hepatocellular carcinoma. Hepatology 61:1821–1831. doi: 10.1002/hep.27722. [DOI] [PubMed] [Google Scholar]
- 4.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H, Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. 2012. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 5.Paterlini-Brechot P, Saigo K, Murakami Y, Chami M, Gozuacik D, Mugnier C, Lagorce D, Brechot C. 2003. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 22:3911–3916. doi: 10.1038/sj.onc.1206492. [DOI] [PubMed] [Google Scholar]
- 6.Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, Mulawadi FH, Wong KF, Liu AM, Poon RT, Fan ST, Chan KL, Gong Z, Hu Y, Lin Z, Wang G, Zhang Q, Barber TD, Chou WC, Aggarwal A, Hao K, Zhou W, Zhang C, Hardwick J, Buser C, Xu J, Kan Z, Dai H, Mao M, Reinhard C, Wang J, Luk JM. 2012. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 44:765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 7.Brechot C. 2004. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology 127:S56–S61. doi: 10.1053/j.gastro.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 8.Wang HC, Huang W, Lai MD, Su IJ. 2006. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci 97:683–688. doi: 10.1111/j.1349-7006.2006.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang HC, Wu HC, Chen CF, Fausto N, Lei HY, Su IJ. 2003. Different types of ground glass hepatocytes in chronic hepatitis B virus infection contain specific pre-S mutants that may induce endoplasmic reticulum stress. Am J Pathol 163:2441–2449. doi: 10.1016/S0002-9440(10)63599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, Stindt J, Koniger C, Nassal M, Kubitz R, Sultmann H, Urban S. 2014. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 11.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1:e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blondot ML, Bruss V, Kann M. 2016. Intracellular transport and egress of hepatitis B virus. J Hepatol 64:S49–S59. doi: 10.1016/j.jhep.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Nassal M. 2015. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 14.Bill CA, Summers J. 2004. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc Natl Acad Sci U S A 101:11135–11140. doi: 10.1073/pnas.0403925101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wooddell CI, Yuen MF, Chan HL, Gish RG, Locarnini SA, Chavez D, Ferrari C, Given BD, Hamilton J, Kanner SB, Lai CL, Lau JYN, Schluep T, Xu Z, Lanford RE, Lewis DL. 2017. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med 9:eaan0241. doi: 10.1126/scitranslmed.aan0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mason WS, Jilbert AR, Summers J. 2005. Clonal expansion of hepatocytes during chronic woodchuck hepatitis virus infection. Proc Natl Acad Sci U S A 102:1139–1144. doi: 10.1073/pnas.0409332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Summers J, Mason WS. 2004. Residual integrated viral DNA after hepadnavirus clearance by nucleoside analog therapy. Proc Natl Acad Sci U S A 101:638–640. doi: 10.1073/pnas.0307422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang W, Summers J. 1999. Integration of hepadnavirus DNA in infected liver: evidence for a linear precursor. J Virol 73:9710–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason WS, Liu C, Aldrich CE, Litwin S, Yeh MM. 2010. Clonal expansion of normal-appearing human hepatocytes during chronic hepatitis B virus infection. J Virol 84:8308–8315. doi: 10.1128/JVI.00833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu T, Mason WS, Clouston AD, Shackel NA, McCaughan GW, Yeh MM, Schiff ER, Ruszkiewicz AR, Chen JW, Harley HA, Stroeher UH, Jilbert AR. 2015. Clonal expansion of hepatocytes with a selective advantage occurs during all stages of chronic hepatitis B virus infection. J Viral Hepat 22:737–753. doi: 10.1111/jvh.12380. [DOI] [PubMed] [Google Scholar]
- 21.Mason WS, Gill US, Litwin S, Zhou Y, Peri S, Pop O, Hong ML, Naik S, Quaglia A, Bertoletti A, Kennedy PT. 2016. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology 151:986–998.e4. doi: 10.1053/j.gastro.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mason WS, Low HC, Xu C, Aldrich CE, Scougall CA, Grosse A, Clouston A, Chavez D, Litwin S, Peri S, Jilbert AR, Lanford RE. 2009. Detection of clonally expanded hepatocytes in chimpanzees with chronic hepatitis B virus infection. J Virol 83:8396–8408. doi: 10.1128/JVI.00700-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong SS, Jensen AD, Chang CJ, Rogler CE. 1999. Double-stranded linear duck hepatitis B virus (DHBV) stably integrates at a higher frequency than wild-type DHBV in LMH chicken hepatoma cells. J Virol 73:1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu T, Budzinska MA, Shackel NA, Urban S. 2017. HBV DNA integration: molecular mechanisms and clinical implications. Viruses 9(4):E75. doi: 10.3390/v9040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu T, Jilbert AR. 2017. Detection of hepatocyte clones containing integrated hepatitis B virus DNA using inverse nested PCR. Methods Mol Biol 1540:97–118. doi: 10.1007/978-1-4939-6700-1_9. [DOI] [PubMed] [Google Scholar]
- 26.Luo J, Cui X, Gao L, Hu J. 21 June 2017. Identification of intermediate in hepatitis B virus CCC DNA formation and sensitive and selective CCC DNA detection. J Virol doi: 10.1128/JVI.00539-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allweiss L, Volz T, Giersch K, Kah J, Raffa G, Petersen J, Lohse AW, Beninati C, Pollicino T, Urban S, Lutgehetmann M, Dandri M. 20 April 2017. Proliferation of primary human hepatocytes and prevention of hepatitis B virus reinfection efficiently deplete nuclear cccDNA in vivo. Gut doi: 10.1136/gutjnl-2016-312162. [DOI] [PubMed] [Google Scholar]
- 28.Berke JM, Dehertogh P, Vergauwen K, Van Damme E, Mostmans W, Vandyck K, Pauwels F. 5 June 2017. Capsid assembly modulators have a dual mechanism of action in primary human hepatocytes infected with hepatitis B virus. Antimicrob Agents Chemother doi: 10.1128/AAC.00560-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu G, Liu B, Zhang Y, Li J, Arzumanyan A, Clayton MM, Schinazi RF, Wang Z, Goldmann S, Ren Q, Zhang F, Feitelson MA. 2013. Preclinical characterization of GLS4, an inhibitor of hepatitis B virus core particle assembly. Antimicrob Agents Chemother 57:5344–5354. doi: 10.1128/AAC.01091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isorce N, Testoni B, Locatelli M, Fresquet J, Rivoire M, Luangsay S, Zoulim F, Durantel D. 2016. Antiviral activity of various interferons and pro-inflammatory cytokines in non-transformed cultured hepatocytes infected with hepatitis B virus. Antiviral Res 130:36–45. doi: 10.1016/j.antiviral.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 31.Alfaiate D, Lucifora J, Abeywickrama-Samarakoon N, Michelet M, Testoni B, Cortay JC, Sureau C, Zoulim F, Deny P, Durantel D. 2016. HDV RNA replication is associated with HBV repression and interferon-stimulated genes induction in super-infected hepatocytes. Antiviral Res 136:19–31. doi: 10.1016/j.antiviral.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Zhao XL, Yang JR, Lin SZ, Ma H, Guo F, Yang RF, Zhang HH, Han JC, Wei L, Pan XB. 2016. Serum viral duplex-linear DNA proportion increases with the progression of liver disease in patients infected with HBV. Gut 65:502–511. doi: 10.1136/gutjnl-2014-308989. [DOI] [PubMed] [Google Scholar]
- 33.Gao W, Hu J. 2007. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J Virol 81:6164–6174. doi: 10.1128/JVI.02721-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scotto J, Hadchouel M, Hery C, Alvarez F, Yvart J, Tiollais P, Bernard O, Brechot C. 1983. Hepatitis B virus DNA in children's liver diseases: detection by blot hybridisation in liver and serum. Gut 24:618–624. doi: 10.1136/gut.24.7.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yaginuma K, Kobayashi H, Kobayashi M, Morishima T, Matsuyama K, Koike K. 1987. Multiple integration site of hepatitis B virus DNA in hepatocellular carcinoma and chronic active hepatitis tissues from children. J Virol 61:1808–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimbi GC, Kramvis A, Kew MC. 2005. Integration of hepatitis B virus DNA into chromosomal DNA during acute hepatitis B. World J Gastroenterol 11:6416–6421. doi: 10.3748/wjg.v11.i41.6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Summers J, Jilbert AR, Yang W, Aldrich CE, Saputelli J, Litwin S, Toll E, Mason WS. 2003. Hepatocyte turnover during resolution of a transient hepadnaviral infection. Proc Natl Acad Sci U S A 100:11652–11659. doi: 10.1073/pnas.1635109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakraborty A, Tapryal N, Venkova T, Horikoshi N, Pandita RK, Sarker AH, Sarkar PS, Pandita TK, Hazra TK. 2016. Classical non-homologous end-joining pathway utilizes nascent RNA for error-free double-strand break repair of transcribed genes. Nat Commun 7:13049. doi: 10.1038/ncomms13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, Storici F. 2014. Transcript-RNA-templated DNA recombination and repair. Nature 515:436–439. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chauhan R, Churchill ND, Mulrooney-Cousins PM, Michalak TI. 2017. Initial sites of hepadnavirus integration into host genome in human hepatocytes and in the woodchuck model of hepatitis B-associated hepatocellular carcinoma. Oncogenesis 6:e317. doi: 10.1038/oncsis.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, Jasin M, Brunet E. 2014. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell 55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao Z, Bozzella M, Seluanov A, Gorbunova V. 2008. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lau CC, Sun T, Ching AK, He M, Li JW, Wong AM, Co NN, Chan AW, Li PS, Lung RW, Tong JH, Lai PB, Chan HL, To KF, Chan TF, Wong N. 2014. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell 25:335–349. doi: 10.1016/j.ccr.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 44.Zhao LH, Liu X, Yan HX, Li WY, Zeng X, Yang Y, Zhao J, Liu SP, Zhuang XH, Lin C, Qin CJ, Zhao Y, Pan ZY, Huang G, Liu H, Zhang J, Wang RY, Yang Y, Wen W, Lv GS, Zhang HL, Wu H, Huang S, Wang MD, Tang L, Cao HZ, Wang L, Lee TP, Jiang H, Tan YX, Yuan SX, Hou GJ, Tao QF, Xu QG, Zhang XQ, Wu MC, Xu X, Wang J, Yang HM, Zhou WP, Wang HY. 2016. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun 7:12992. doi: 10.1038/ncomms12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sfeir A, Symington LS. 2015. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem Sci 40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McVey M, Lee SE. 2008. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet 24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buendia MA. 1992. Hepatitis B viruses and hepatocellular carcinoma. Adv Cancer Res 59:167–226. doi: 10.1016/S0065-230X(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 48.Robinson WS. 1994. Molecular events in the pathogenesis of hepadnavirus-associated hepatocellular carcinoma. Annu Rev Med 45:297–323. doi: 10.1146/annurev.med.45.1.297. [DOI] [PubMed] [Google Scholar]
- 49.Loeb LA. 2001. A mutator phenotype in cancer. Cancer Res 61:3230–3239. [PubMed] [Google Scholar]
- 50.Beckman RA. 2010. Efficiency of carcinogenesis: is the mutator phenotype inevitable? Semin Cancer Biol 20:340–352. doi: 10.1016/j.semcancer.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 51.Wu TT, Coates L, Aldrich CE, Summers J, Mason WS. 1990. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology 175:255–261. doi: 10.1016/0042-6822(90)90206-7. [DOI] [PubMed] [Google Scholar]
- 52.Volz T, Allweiss L, Ben MM, Warlich M, Lohse AW, Pollok JM, Alexandrov A, Urban S, Petersen J, Lutgehetmann M, Dandri M. 2013. The entry inhibitor Myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with hepatitis B virus. J Hepatol 58:861–867. doi: 10.1016/j.jhep.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Hantz O, Parent R, Durantel D, Gripon P, Guguen-Guillouzo C, Zoulim F. 2009. Persistence of the hepatitis B virus covalently closed circular DNA in HepaRG human hepatocyte-like cells. J Gen Virol 90:127–135. doi: 10.1099/vir.0.004861-0. [DOI] [PubMed] [Google Scholar]
- 54.Simonetti J, Bulkow L, McMahon BJ, Homan C, Snowball M, Negus S, Williams J, Livingston SE. 2010. Clearance of hepatitis B surface antigen and risk of hepatocellular carcinoma in a cohort chronically infected with hepatitis B virus. Hepatology 51:1531–1537. doi: 10.1002/hep.23464. [DOI] [PubMed] [Google Scholar]
- 55.Kleine M, Riemer M, Krech T, DeTemple D, Jager MD, Lehner F, Manns MP, Klempnauer J, Borlak J, Bektas H, Vondran FW. 2014. Explanted diseased livers—a possible source of metabolic competent primary human hepatocytes. PLoS One 9:e101386. doi: 10.1371/journal.pone.0101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. 2002. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A 99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schulze A, Mills K, Weiss TS, Urban S. 2012. Hepatocyte polarization is essential for the productive entry of the hepatitis B virus. Hepatology 55:373–383. doi: 10.1002/hep.24707. [DOI] [PubMed] [Google Scholar]
- 58.Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, King RW. 1997. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother 41:1715–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lempp FA, Mutz P, Lipps C, Wirth D, Bartenschlager R, Urban S. 2016. Evidence that hepatitis B virus replication in mouse cells is limited by the lack of a host cell dependency factor. J Hepatol 64:556–564. doi: 10.1016/j.jhep.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 60.Seitz S, Iancu C, Volz T, Mier W, Dandri M, Urban S, Bartenschlager R. 2016. A slow maturation process renders hepatitis B virus infectious. Cell Host Microbe 20:25–35. doi: 10.1016/j.chom.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 61.Schulze A, Schieck A, Ni Y, Mier W, Urban S. 2010. Fine mapping of pre-S sequence requirements for hepatitis B virus large envelope protein-mediated receptor interaction. J Virol 84:1989–2000. doi: 10.1128/JVI.01902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delaney WE IV, Isom HC. 1998. Hepatitis B virus replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology 28:1134–1146. doi: 10.1002/hep.510280432. [DOI] [PubMed] [Google Scholar]
- 65.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Res 41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smit A, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0. Institute for Systems Biology, Seattle, WA: http://www.repeatmasker.org. [Google Scholar]
- 67.Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.