ABSTRACT
Mammarenaviruses cause chronic infections in their natural rodent hosts. Infected rodents shed infectious virus into excreta. Humans are infected through mucosal exposure to aerosols or direct contact of abraded skin with fomites, resulting in a wide range of manifestations from asymptomatic or mild febrile illness to severe life-threatening hemorrhagic fever. The mammarenavirus matrix Z protein has been shown to be a main driving force of virus budding and to act as a negative regulator of viral RNA synthesis. To gain a better understanding of how the Z protein exerts its several different functions, we investigated the interaction between Z and viral polymerase L protein using the prototypic mammarenavirus, lymphocytic choriomeningitis virus (LCMV). We found that in the presence of an active viral ribonucleoprotein (vRNP), the Z protein translocated from nonionic detergent-resistant, membrane-rich structures to a subcellular compartment with a different membrane composition susceptible to disruption by nonionic detergents. Alanine (A) substitution of a highly conserved leucine (L) at position 72 in LCMV Z protein abrogated Z-L interaction. The L72A mutation did not affect the stability or budding activity of Z when expressed alone, but in the presence of an active vRNP, mutation L72A promoted rapid degradation of Z via a proteasome- and lysosome-independent pathway. Accordingly, L72A mutation in the Z protein resulted in nonviable LCMV. Our findings have uncovered novel aspects of the dynamics of the Z protein for which a highly conserved L residue was strictly required.
IMPORTANCE Several mammarenaviruses, chiefly Lassa virus (LASV), cause hemorrhagic fever disease in humans and pose important public health concerns in their regions of endemicity. Moreover, mounting evidence indicates that the worldwide-distributed, prototypic mammarenavirus, lymphocytic choriomeningitis virus (LCMV), is a neglected human pathogen of clinical significance. The mammarenavirus matrix Z protein plays critical roles in different steps of the viral life cycle by interacting with viral and host cellular components. Here we report that alanine substitution of a highly conserved leucine residue, located at position 72 in LCMV Z protein, abrogated Z-L interaction. The L72A mutation did not affect Z budding activity but promoted its rapid degradation in the presence of an active viral ribonucleoprotein (vRNP). Our findings have uncovered novel aspects of the dynamics of the Z protein for which a highly conserved L residue was strictly required.
KEYWORDS: arenavirus, matrix protein, RNA-dependent RNA polymerase, protein stability
INTRODUCTION
Mammarenaviruses cause chronic infections in their natural rodent hosts worldwide (1), and human infections occur through mucosal exposure to aerosols or by direct contact of abraded skin with fomites. Several mammarenaviruses cause viral hemorrhagic fevers (VHFs) in humans and pose important public health problems in their areas of endemicity (2–6). Mammarenaviruses are classified into two main groups, Old World (OW) and New World (NW) mammarenaviruses (1). The OW Lassa virus (LASV), the causative agent of Lassa fever (LF), is the mammarenavirus with the highest impact on human health. LASV infects several hundred thousand individuals annually in Western Africa, resulting in a high number of LF cases associated with high morbidity and lethality (7). Moreover, mounting evidence indicates that the worldwide-distributed OW mammarenavirus lymphocytic choriomeningitis virus (LCMV) is a neglected human pathogen of clinical significance, especially in congenital infections (8–12). In addition, LCMV poses a particular threat to immunocompromised individuals, as has been illustrated by fatal cases of LCMV infection associated with organ transplants (13, 14). Concerns about human-pathogenic mammarenaviruses are exacerbated because of the lack of FDA-licensed vaccines. The live attenuated vaccine strain Candid#1 of the NW mammarenavirus Junin virus (JUNV), the causative agent of Argentine HF (AHF), has been shown to be effective to protect against AHF, but Candid#1 is licensed only in Argentina and does not confer protection against LF. Likewise, current antimammarenaviral therapy is limited to an off-label use of the nucleoside analogue ribavirin that is only partially effective and can cause significant side effects (15–17). However, the small-molecule compounds favipiravir and ST-193 have shown promising results in animal models of LF (18, 19), but their efficacy in severe human LF cases remains to be determined (20). Therefore, it is important to develop novel antimammarenavirus strategies. This task would be facilitated by a detailed understanding of the molecular and cellular biology of mammarenaviruses.
Mammarenaviruses are enveloped viruses with a bisegmented negative-strand RNA genome (1). Each genome segment, L and S, directs the synthesis of two proteins from open readings frames located in opposite direction and separated by a noncoding intergenic region (IGR) (1). The S RNA segment encodes the viral nucleoprotein (NP) and glycoprotein precursor (GPC). GPC is cotranslationally cleaved by signal peptidase to generate a stable signal peptide (SSP) and posttranslationally processed by site 1 protease (S1P) to generate the mature virion surface glycoproteins GP1 and GP2, which together with SSP form the GP complex that mediates receptor recognition and cell entry. The L RNA segment encodes the viral RNA-dependent RNA polymerase (L) and the matrix Z protein (21, 22), a key structural viral protein that is involved in several different steps of the virus life cycle. The Z protein interacts with the GP complex (23) and plays a critical role in assembly of infectious viral particles. Z is also a main driving force of mammarenavirus budding, a process facilitated by the interaction of L (late) domains within the C-terminal region of Z and components of endosomal sorting complex transport (ESCRT) pathway of the cell (21, 24, 25). Both Z interaction with the GP complex and Z-mediated budding require myristolyation of glycine residue at position 2 to promote Z interaction with membranes (26). Additionally, the Z protein exhibits a dose-dependent inhibitory effect on viral RNA synthesis, both RNA replication and gene transcription, mediated by the virus ribonucleoprotein (vRNP) complex composed of the viral genomic RNA species and viral transacting factors NP and L (27, 28). The Z protein of the NW mammarenaviruses Tacaribe virus (TACV) (29) and Machupo virus (MACV) (30) have been shown to directly interact with the L protein, which was proposed to be the mechanism by which Z inhibited viral RNA synthesis. However, interaction between the OW Z and L proteins and its contribution to virus multiplication have not been examined in detail. Here we demonstrate that Z proteins of OW LCMV and LASV directly interact with the corresponding viral L protein. We identified a highly conserved leucine residue at position 72 in LCMV Z as playing a critical role in Z-L interaction. Z protein with L72A substitution failed to interact with L but exhibited expression levels and budding activity similar to those of wild-type Z protein. Unexpectedly, the L72A mutation promoted rapid degradation of Z in the presence of active vRNP via a process that used a proteasome- and lysosome-independent pathway. Consistent with these results, mutation L72A in the Z protein resulted in nonviable LCMV. Our findings have uncovered novel features of the dynamics of the Z protein, for which a highly conserved L residue is strictly required.
RESULTS
OW mammarenavirus Z proteins interact with homologous L proteins.
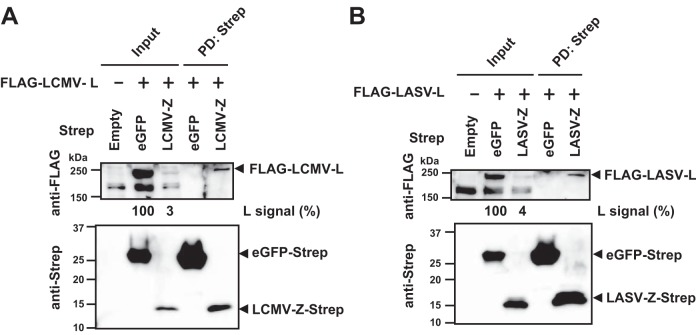
We asked whether similar to the findings with NW mammarenaviruses (29, 30), we could detect an interaction between Z and L proteins of the OW mammarenavirus LCMV. For this, we cotransfected 293T cells with Strep-tagged Z protein- and FLAG-tagged L protein-expressing plasmids and examined Z-L interaction by pulldown (PD) assay. As a negative control, we used Strep-tagged enhanced green fluorescent protein (eGFP). The L protein was readily detected by Western blotting in the complex PD by the Z protein, but not eGFP (Fig. 1A). We obtained similar results for the OW LASV (Fig. 1B). We observed that L protein expression was reduced when coexpressed with the Z protein but not with eGFP. The mechanisms by which Z protein mediates reduced expression levels of L protein remain to be determined.
FIG 1.
Interaction between L and Z proteins of OW mammarenaviruses LCMV and LASV. 293T cells were transfected with a plasmid expressing Strep-tagged eGFP or the Z protein together with a plasmid expressing FLAG-tagged L protein. Plus and minus signs indicate plasmid presence and absence in the transfection mix. Plasmids for LCMV and LASV were used for panels A and B, respectively. An empty vector, pCAGGS, was used to identify nonspecific protein bands in Western blots. At 72 h posttransfection, cells were lysed and PD was performed using Strep-Tactin. Protein levels in clarified whole-cell lysate (Input) and eluate (PD: Strep) were examined by Western blotting. Numbers at the bottoms of anti-FLAG Western blots correspond to relative signal intensity of FLAG-tagged LCMV- or LASV-L protein in whole-cell lysate (Input) [L signal (%)]. The signal intensity of the L protein coexpressed with eGFP-Strep was set to 100%.
A highly conserved leucine residue at position 72 in LCMV Z critically contributes to Z-L interaction.
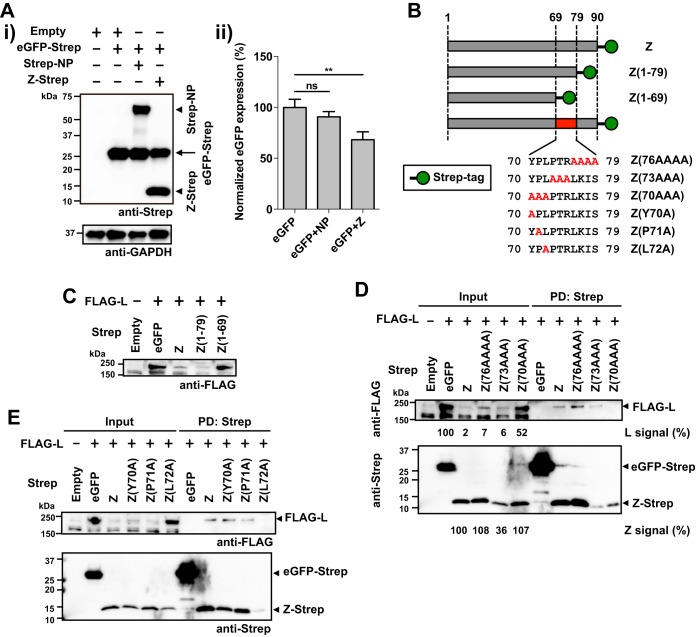
Interaction between the Z protein and eIF4E has been linked to the Z protein's ability to negatively regulate translation efficiency of both viral and cellular mRNAs (31, 32). Consistent with this, we observed that coexpression of Z with eGFP resulted in modestly, but significantly, decreased expression levels of eGFP (Fig. 2A), whereas we showed that coexpression of Z with the L protein resulted in large reductions in expression levels of L protein (Fig. 1A and B). These findings indicate that besides its potential role in modulating translation efficiency of the L mRNA, the Z protein affected expression levels of the L protein by other mechanism that likely required Z-L protein-protein interaction (PPI). To examine this issue, we conducted experiments to define the region and residues in the Z protein that mediated Z-L PPI. We coexpressed FLAG-tagged L protein with a collection of Strep-tagged Z proteins with C-terminal truncations (Fig. 2B and C). Consistent with our previous results, L protein expression was reduced when coexpressed with the Z protein, but not with eGFP (Fig. 2C). Similarly, C-terminal deletion of 11 amino acids (aa) within the Z protein (aa 80 to 90) did not affect Z's ability to cause reduced expression levels of L protein when coexpressed in transfected cells (Fig. 2C). However, a Z mutant protein with further 10-aa C-terminal deletion (aa 70 to 79) did not affect expression levels of L protein in cotransfected cells (Fig. 2C). To further investigate the contribution of aa 70 to 79 of the Z protein to Z-L interaction, we generated a series of plasmids expressing Strep-tagged mutant Z proteins containing three or four alanine substitutions covering aa 70 to 79 (Fig. 2B) and used them to examine their interaction with the L protein by PD assay. The L protein was pulled down with wild-type (WT) Z protein and Z mutants Z(76AAAA) and Z(73AAA) (Fig. 2D). In contrast, the L protein was barely detectable in the PD with Z(70AAA) or with eGFP (Fig. 2D), indicating that aa 70 to 72 are critical for Z-L interaction. Accordingly, expression levels of L protein were largely reduced when coexpressed with WT Z protein and Z muant Z(76AAAA) or Z(73AAA), whereas coexpression with Z(70AAA) resulted in only a modest decrease in L level (Fig. 2D). To assess the contribution of aa 70 to 72 to Z-L interaction, we introduced individual alanine substitution at each of those positions (Fig. 2B). Alanine substitution at position 70 or 71 did not affect Z-L interaction, whereas levels of L protein were drastically reduced in the complex PD by Z(L72A) (Fig. 2E). PD samples contained lower levels of Z(L72A) than the other Z mutants tested. This might indicate that Z(L72A) is intrinsically less stable and some Z(L72A) degradation occurred during the incubation with the Strep-Tactin resin.
FIG 2.
The leucine (L) residue at position 72 of LCMV Z is critical for Z-L interaction. (A) Reduction of eGFP expression by coexpression with the Z protein. 293T cells were transfected with 0.5 μg of pC-eGPF-Strep together with 0.5 μg of pC-Empty (Empty), pC-Strep-NP (Strep-NP), or pC-Z-Strep (Z-Strep). At 48 h posttransfection, cell lysates were prepared and protein expression was examined by Western blotting. Representative Western blot data from three independent experiments are shown (i). Data represent means ± SD of results from three independent experiments (ii). Mean signal intensity of eGPF-Strep expressed alone was set to 100%. ** P value of < 0.01. ns, not significant. (B) Schematic diagram of amino acid composition of the C termini of the WT and mutant Z proteins. (C to E) Mapping of amino acid residues required for Z-L interaction. 293T cells were cotransfected with FLAG-L and Strep-tagged WT or the indicated mutant Z, and at 48 hm posttransfection cell lysates were prepared and protein expression in whole-cell lysates and Strep tag-mediated PD was examined as described for Fig. 1. Numbers at the bottoms of the anti-FLAG and anti-Strep Western blots correspond to relative signal intensity of L [L signal (%)] or Z protein [Z signal (%)], respectively, in whole-cell lysate (Input) (D). Signal intensity of FLAG-tagged L protein coexpressed with eGFP-Strep or that of Strep-tagged WT Z protein was set to 100%.
Functional characterization of the LCMV Z L72A mutant.
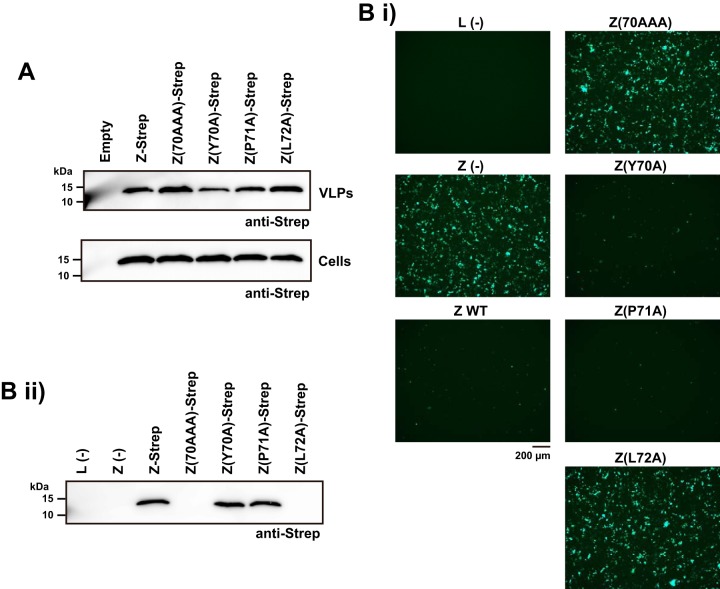
Mammarenavirus Z proteins contain canonical L domains within their C termini that are essential for normal virus budding activity (33, 34). Consistent with L72 not being part of the PPPY L domain motif present within the C terminus of LCMV Z (21), we predicted that the L72A substitution should not affect Z protein budding activity. To address this issue, we examined the ability of the mutant Z proteins to produce virion-like particles (VLPs) by budding assay. For this, we transfected 293T cells with expressing plasmids for the WT or each of the mutant Z proteins, and 48 h later, we collected VLPs from tissue culture supernatant (TCS) by ultracentrifugation and prepared total cell lysates. We determined VLP-associated and total intracellular Z protein levels by Western blotting. Both the Z(70AAA) and Z(L72A) mutants produced levels of VLPs similar to that of the WT Z protein (Fig. 3A). This result also indicated that L72A substitution did not cause a major disruption of the overall Z structure.
FIG 3.
Effect of L72A substitution on Z's functions. (A) Effect of L72 substitutions on Z-mediated budding. 293T cells were transfected with WT or the indicated mutant Z protein-expressing plasmids. At 48 h posttransfection, VLPs present in tissue culture supernatants were collected by ultracentrifugation and total cell lysates prepared. Levels of Z protein in VLPs and cell lysates were determined by Western blotting using an antibody to Strep. (B) Effect of L72 substitution on Z's ability to inhibit MG activity. 293T cells were transfected with pT7-MG/eGFP and pC-T7pol, pC-NP, and pC-L, together with or without [Z(−)], a plasmid expressing the WT or the indicated mutant Z proteins. For a negative control, pC-L was omitted from the transfection mix [L(−)]. At 72 h posttransfection, eGFP expression was examined by epifluorescence (i); whole-cell lysates were prepared and Z protein expression levels in clarified cell lysates were examined by Western blotting using an antibody to Strep (ii).
To investigate the impact of L72A substitution on the Z's ability to inhibit viral RNA synthesis, we tested whether mutant Z proteins could interfere with the activity of an LCMV minigenome (MG). For this, we transfected 293T cells with plasmids expressing the different components of an LCMV MG, which included a T7 polymerase-expressing plasmid, a plasmid expressing under the control of the T7 promoter an LCMV MG RNA coding for eGPF, and plasmids expressing the minimal, NP and L, viral trans-acting factors required for RNA synthesis directed by the vRNP, with or without a plasmid expressing WT or mutant Z protein. Expression of eGFP served as a surrogate for viral genome replication and gene transcription. Consistent with previous findings (27), MG-directed expression of the reporter eGFP was inhibited by the WT Z protein (Fig. 3Bi). In contrast, Z mutants containing L72A or 70AAA substitutions did not cause a reduction on MG activity, whereas Y70A and P71A substitutions did not affect Z's ability to inhibit vRNP activity in the MG cell-based assay (Fig. 3Bi). To confirm that similar levels of the mutant Z proteins were expressed, we prepared total cell lysates and determined Z protein expression levels by Western blotting using an antibody to Strep-tag (Fig. 3Bii). Both the Z(70AAA) and Z(L72A) mutants were expressed to levels similar to that of the WT Z when expressed individually (Fig. 3A). In contrast, and unexpectedly, levels of the Z(70AAA) and Z(L72A) mutant proteins were barely detectable when coexpressed with the MG components (Fig. 3Bii). This finding prevented us from concluding that L72, or Z-L interaction, is required for the Z protein's ability to inhibit viral RNA synthesis.
Role of L72 in LCMV Z protein stability.
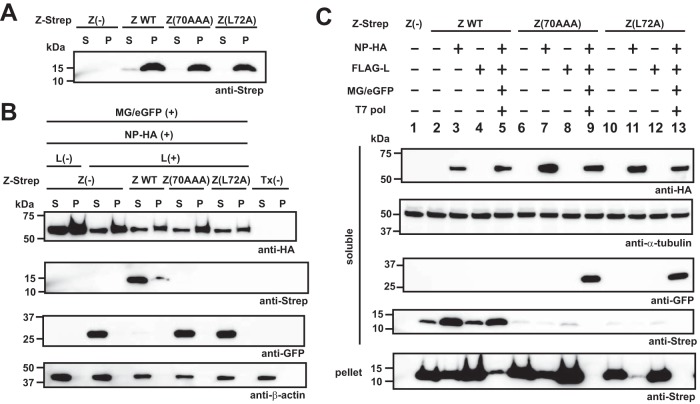
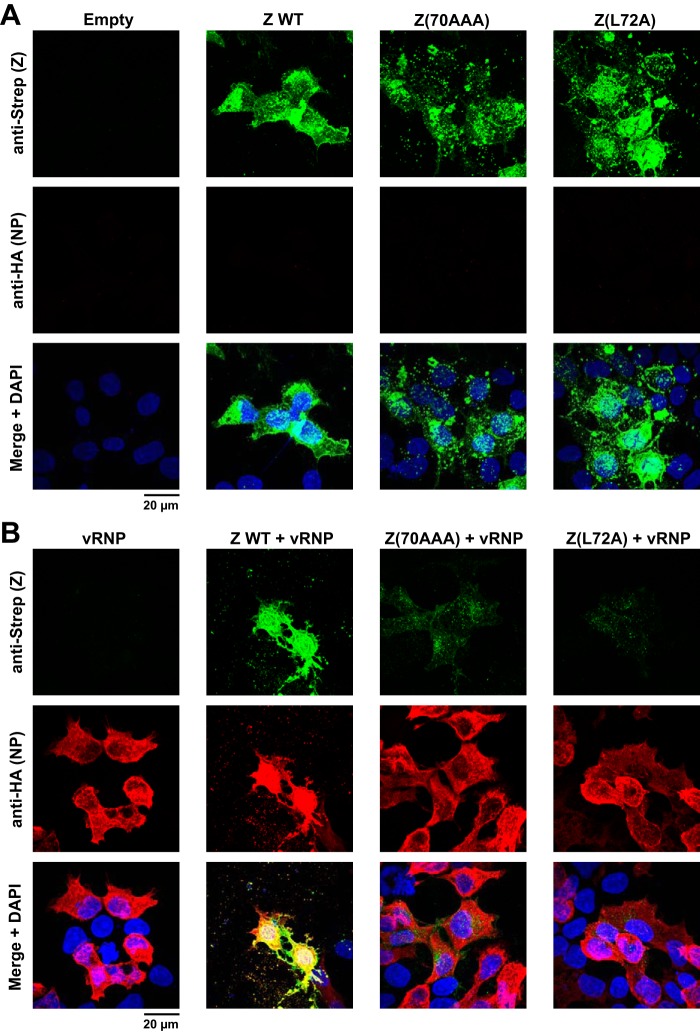
A possible explanation for the loss of mutant Z proteins in the presence of active vRNP is that Z protein associated with vRNP traffics to a membrane-rich, nonionic-detergent-resistant fraction. To address this issue, we examined solubility of the WT and mutant Z proteins under different lysis conditions and in the presence or absence of an active vRNP. We transfected 293T cells with plasmids expressing the WT or each of the mutant Z proteins, and at 72 h posttransfection, cells were lysed with PD lysis buffer (+) and soluble (S) and insoluble (pellet [P]) fractions were separated by centrifugation. The pellet was lysed with 2× SDS-loading buffer containing 4% SDS. Equivalent amounts of the fractions were subjected to SDS-PAGE, and levels of the WT or mutant Z proteins in the S and P fractions were determined by Western blotting (Fig. 4A). When individually expressed, the WT and mutant Z proteins were highly enriched in the P fraction. Next, we asked whether the mutant Z proteins could be detected in the insoluble fraction in the presence of the active ribonucleoprotein complex, which could account for the loss of the mutant Z proteins in Fig. 3Bii. In contrast to the solubility of the WT Z protein expressed alone, most of the WT Z protein was present in the soluble fraction in the presence of vRNP (Fig. 4B). On the other hand, neither Z(70AAA) nor Z(L72A) was detected in either the soluble or insoluble fractions. Intriguingly, the NP was almost evenly distributed in soluble and insoluble fractions, and this distribution was not altered by the formation of vRNP complex (Fig. 4B). These data indicate that the mutant Z protein containing L72A substitution was quickly degraded in the presence of vRNP complex. To further investigate the requirements for the rapid degradation of the mutant Z proteins, we compared Z levels in the soluble and insoluble fractions when the Z proteins were expressed only with the NP or the L protein or with all MG components. Most of the WT Z protein was present in P fraction when expressed alone, but levels of WT Z protein in the S fraction increased very significantly in cells expressing an active vRNP (Fig. 4C, lanes 2 and 5). The subcellular distribution of WT Z protein was not significantly altered when the protein was coexpressed with the L protein (Fig. 4C, lane 4). In contrast, coexpression of WT Z protein with NP resulted in increased levels of WT Z protein in the S fraction and concomitantly decreased Z protein levels in the P fraction (Fig. 4C, lane 3). In the absence of other viral proteins, the mutant Z(70AAA) was expressed to levels similar to that of WT Z protein and present mostly in the P fraction, whereas expression of Z(70AAA) was barely detectable in the presence of active vRNP (Fig. 4C, lanes 6 and 9). As with the WT Z protein, coexpression of Z(70AAA) with L did not change the expression levels or subcellular distribution of Z(70AAA) (Fig. 4C, lane 8), whereas coexpression of Z(70AAA) with NP resulted in reduced expression levels of Z(70AAA) in the P fraction without a corresponding increase in Z(70AAA) expression levels in the S fraction (Fig. 4C, lane 7). Mutant Z(L72A) exhibited behavior similar to that of Z(70AAA), but overall expression levels of Z(L72A) were slightly lower than for Z(70AAA) (Fig. 4C, lanes 10 to 13). We also examined transfected cells by immunofluorescence staining to assess at the single-cell level the expression of WT and mutant Z proteins in the absence or presence of an active vRNP. Consistent with our Western blot results, WT and mutant Z(70AAA) and Z(L72A) Z proteins exhibited similar expression patterns when individually expressed (Fig. 5A). In contrast, when expressed in the presence of an active vRNP, levels of Z(70AAA) and Z(L72A), but not of WT, Z proteins were drastically reduced (Fig. 5B). Taken together, our results indicated that the pattern of subcellular distribution of Z protein is influenced by the presence of an active vRNP and that a mutant Z protein that is unable to interact with the L protein is targeted for rapid degradation.
FIG 4.
Effect of L72A mutation on Z protein stability in the presence of an active vRNP. (A) Solubility of WT and mutant Z proteins. 293T cells seeded in a 12-well plate were transfected with plasmids expressing Strep-tagged WT or the indicated mutant Z proteins. At 72 h posttransfection, the transfected cells were lysed with PD lysis buffer (+) and soluble (S) and insoluble fractions separated by centrifugation at 21,130 × g at 4°C for 10 min. Pellet containing the insoluble fraction (P) was lysed with 2× SDS loading buffer. Z protein levels in the S and P fractions were analyzed by Western blotting. (B and C) Expression of mutant Z proteins in the presence of an active vRNP. 293T cells seeded in a 12-well plate were transfected with pT7-MG/eGFP (MG/eGFP) and pC-T7pol (T7 pol), pC-NP-HA (NP-HA), and either pC-L (L) or pC-FALG-L (FLAG-L) together with or without a plasmid expressing WT or the indicated mutant Z proteins or remained untransfected [Tx(−)]. Plus and minus signs indicate plasmid presence and absence in the transfection mix. At 72 h posttransfection, protein levels in the S and P fractions were analyzed by Western blotting.
FIG 5.
Subcellular distribution of the mutant Z proteins in the presence or absence of an active vRNP. 293T cells seeded on coverslips in a 24-well plate (1 × 105 cells/well) and cultured overnight were transfected with 25 ng of plasmid expressing WT or the indicated mutant Z proteins together with (B) or without (A) plasmids required for intracellular reconstruction of an active vRNP (vRNP) (pT7-MG/CAT, pC-T7pol, pC-L, and pC-NP). Forty-eight hours later, transfected cells were fixed and Strep-tagged Z proteins and HA-tagged NP expression detected by IF using a mouse monoclonal antibody to Strep and a rabbit polyclonal antibody to HA, respectively. Nuclei were detected by DAPI staining. Stained cells were analyzed with a confocal microscope.
Effects of proteasome and lysosome activity inhibitors on Z(70AAA) and Z(L72A) protein stability in the presence of an active vRNP.
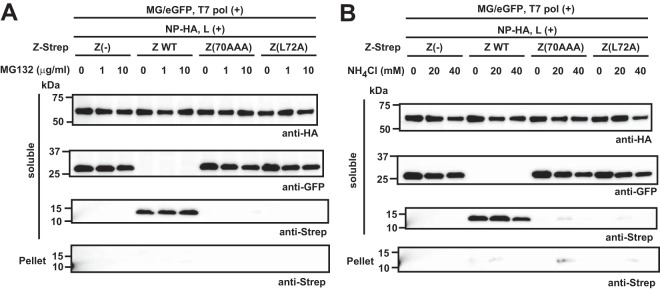
We hypothesized that the presence of an active vRNP activated a pathway that targeted Z(70AAA) and Z(L72A) for rapid degradation. To assess our hypothesis, we investigated if either of the two major protein degradation pathways, proteasome and lysosome, was involved in this process. For this, we examined the effects of the proteasome inhibitor MG132 and lysosome inhibitor ammonium chloride (NH4Cl) on levels of expression of the mutant Z proteins in the presence of an active vRNP. We transfected 293T cells with plasmids expressing the WT and mutant Z proteins together with plasmids required for the formation of an active vRNP, and at 5 or 19 h posttransfection, we treated cells with NH4Cl or MG132, respectively. At 48 h posttransfection, we determined Z protein expression levels in the S and P fractions. Neither NH4Cl nor MG132 treatment prevented the rapid degradation of Z(70AAA) and Z(L72A) in the presence of an active vRNP (Fig. 6).
FIG 6.
Neither the proteasome nor the lysosome pathway is involved in degradation of the Z mutants in the presence of an active vRNP. Effect of the proteasome inhibitor MG132 (A) or the lysosome inhibitor ammonium chloride (NH4Cl) (B) on Z's stability in the presence of an active vRNP. 293T cells were transfected with plasmids required for formation of active vRNP (pT7-MG/eGFP [MG/eGFP], pC-T7pol [T7 pol], pC-L [L], and pC-NP-HA [NP-HA]) together with or without a plasmid expressing the WT or the indicated mutant Z proteins. MG132 (A) and NH4Cl (B) were added to culture media at 19 h or 5 h, respectively, posttransfection. At 48 h posttransfection, cells were harvested and Z protein expression levels in the S and P fractions were examined by Western blotting.
Role of the highly conserved L72 in LCMV Z on production of infectious viral progeny.
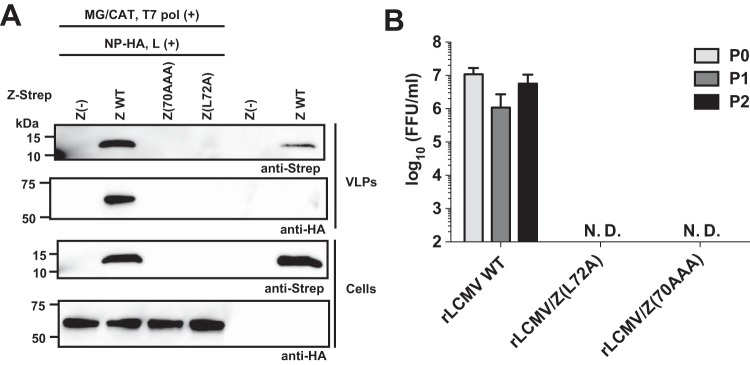
An alternative explanation for the rapid disappearance of Z(70AAA) and Z(L72A) proteins when expressed in the context of an active vRNP might be that an interaction between these mutant Z proteins and vRNP promoted rapid formation and cell egress of VLP, resulting in barely detectable intracellular levels of the mutant Z proteins. To investigate this possibility, we examined Z levels in TCS when expressed in the absence or presence of an active vRNP. As we expected, the WT Z protein efficiently produced VLPs in the presence and absence of MG components (Fig. 7A). In contrast, when expressed in the presence of an active vRNP, neither Z(70AAA) nor Z(L72A) was detected in TCS or whole-cell lysates, indicating that the loss of the mutant Z proteins in the presence of active ribonucleoprotein was not due to accelerated VLP egress (Fig. 7A). Consistent with this finding, using our well-established LCMV reverse genetics system (35–37) we were unable to rescue a viable rLCMV carrying Z(70AAA) or Z(L72A) Z mutants, whereas WT rLCMV was efficiently rescued (Fig. 7B), indicating that L72 was essential for viability of LCMV.
FIG 7.
L72 in LCMV Z protein is critical for production of infectious LCMV. (A) The presence of an active vRNP enhances production of VLPs. 293T cells in a 12-well plate were transfected with 100 ng of a plasmid expressing WT or the indicated mutant Z proteins together with (+) or without (−) plasmids required for the intracellular formation of an active vRNP (pT7-MG/CAT [MG/CAT], pC-T7pol [T7 pol], pC-L [L], and pC-NP-HA [NP-HA]). At 48 h posttransfection, VLPs present in tissue culture supernatants were collected by ultracentrifugation and total cell lysates prepared. Levels of Z and NP in VLPs and cell lysates were determined by Western blotting using antibodies to Strep and HA, respectively. (B) L72A substitution in the Z protein prevented rescue of viable LCMV. BHK-21 cells were transfected with plasmids directing RNA Pol1-mediated intracellular synthesis of S and L RNA genome species containing the WT or the indicated Z mutations, together with plasmids for expression of the trans-acting viral factors NP and L. At 6 days posttransfection, TCS was collected (P0) and used to infect fresh monolayers of BHK-21 cells. At 72 h p.i., TCS were collected (P1) and used to infect fresh monolayers of BHK-21 cells. At 72 h after infection with P1, TCS were collected (P2). Virus titers in TCS from P0, P1, and P2 were determined by IFFA. N.D., not detected.
DISCUSSION
In the present study, we have shown that, as is the case with the NW virus proteins, the OW virus Z proteins interact with their corresponding L proteins, and a highly conserved L residue among all known mammarenavirus Z proteins (L72 in LCMV Z) was strictly required for Z-L interaction. The presence of isoleucine (I) at this position in the Z protein of clade C NW mammarenaviruses Latino virus (LATV) and Oliveros virus (OLVV) represent exceptions among known mammarenaviruses (38), suggesting that the I residue can functionally substitute for the highly conserved L residue. However, similar to L72A substitution, L72I substitution in LCMV Z protein resulted in its rapid degradation in the presence of an active vRNP (data not shown), raising the possibility that the assignment of an I residue to this position in LATV and OLVV Z proteins may represent sequencing errors. Our results have also shown that the subcellular distribution of Z is strongly influenced in the presence of an active vRNP. Our results support the view that the interaction of Z with an active vRNP results in translocation of Z associated with nonionic-detergent-resistant, membrane-rich structures to a subcellular compartment with a different membrane composition susceptible to disruption by nonionic detergents. NP was present both in soluble and nonsoluble fractions, and coexpression of NP with Z resulted in decreased and increased levels of the Z protein in the nonsoluble and soluble fractions, respectively, suggesting that Z may be able to shuttle between these subcellular compartments but in the presence of active ribonucleoprotein, the direction of Z protein trafficking is favored into the soluble compartment.
L protein levels were significantly reduced in the PD complex with Z(L72A) compared to the PD with Z WT. However, compared to Z WT, levels of Z(L72A) protein were also reduced in the PD complex, which prevented us from unequivocally asserting the requirement of L72 for Z-L interaction. However, L(70AAA) interaction with L was significantly less efficient than that of L(73AAA), which was also present at low level in the corresponding PD sample. Moreover, Z(Y70A) and Z(P71A) mutants efficiently interacted with the L protein. These results would suggest that L72 plays a critical role in Z-L interaction.
Although the L residue at position 72 in LCMV Z is highly conserved among mammarenaviruses, its role in Z's function remains to be determined, and this could be complicated by the rapid degradation of Z proteins with mutations affecting this highly conserved L residue when expressed in the presence of an active vRNP. Thus, L79A mutation in TACV Z protein, the counterpart to L72A in LCMV Z protein, disrupted Z-NP interaction and incorporation of the GP complex into VLPs without affecting the budding activity of the Z protein (39). However, the effect of L79A mutation on TACV Z protein stability was not examined, and it remains to be determined whether the conserved L residue in the C-terminal region of the Z protein is universally required among mammarenaviruses for Z stability in the presence of an active vRNP.
Results from treatment with either MG132 or NH4Cl indicated that neither the ubiquitin-proteasome nor the lysosome proteolysis major pathways that mediate protein degradation contributed to the rapid degradation of the Z(70AAA) and Z(L72A) mutants in the presence of an active vRNP. Proteasome- and lysosome-independent protein degradation has been reported so far only for a limited number of proteins, including human thyroid peroxidase (40) and mutant forms of BiP (41), but the involved proteases remain largely uncharacterized.
The crystal structure of LASV Z protein revealed that Z can be present as both monomeric (42, 43) and dodecamer (44) forms. The Z monomer assembles into a dodecamer structure consisting of six intermediate-state dimers in a concentration- and time-dependent manner, which is associated with dynamic conformational change in the C-terminal tail (44). Intriguingly, L71 in LASV Z (counterpart of L72 of LCMV Z) is significantly altered by this conformational change. However, the relevance of oligomeric forms of the Z protein in mammarenavirus multiplication remains to be determined.
Our results show that solubility of the Z protein, and thereby likely its subcellular location and interactome, is strongly affected in the presence of an active vRNP. Therefore, findings obtained from studies involving Z expression by itself may not accurately capture some Z interactions that take place during mammarenavirus natural infection. Recently, an interactome analysis for JUNV Z protein has been reported (45). A limited degree of overlap was observed between host proteins identified in virions released from infected cells and those in VLPs from cells, in which the Z protein was expressed alone, suggesting that the host proteins found in VLPs may include proteins that are not involved in facilitating production of infectious viral progeny.
NW arenavirus Z-L interaction has been proposed to be required for the NW arenavirus Z protein to inhibit viral RNA synthesis (29, 30), and a TACV Z mutant with a C-terminal deletion region that included L79, the counterpart of L72 in LCMV Z protein, was unable to bind to TACV L protein and no longer inhibited the TACV MG activity (29). However, this study did not examine the effect of L79A mutation on TACV Z protein stability in the context of the MG assay. Therefore, it is plausible that as with the LCMV Z mutants 70AAA and L72A we describe here, Z's inability to bind L resulted also in rapid degradation of the TACV Z mutant protein when in the presence of an active vRNP.
The NW arenavirus Machupo virus (MACV), but not LASV, Z protein interacted with MACV L polymerase, and accordingly, LASV Z protein did not inhibit MACV MG activity (30), suggesting that as with Z-NP interactions (46), Z-L interactions are also subjected to viral species restrictions. Studies aimed at a detailed biochemical and functional characterization of Z-L and Z-NP interaction among different mammarenavirus species will contribute to a better understanding of the network of mammarenavirus protein interactions that modulate the activity of the vRNP. This knowledge can uncover novel viral protein interaction interfaces amenable for antiviral drug targeting.
MATERIALS AND METHODS
Cells.
Baby hamster kidney BHK-21 (American Type Culture Collection [ATCC] CCL-10), grivet Vero E6 (ATCC CRL-1586), and human 293T (also known as HEK 293T) (ATCC CRL-3216) cells were grown in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Waltham, MA) containing 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 mg/ml of streptomycin, and 100 U/ml of penicillin at 37°C and 5% CO2.
Plasmids.
Plasmids expressing untagged LCMV NP (pC-NP), C-terminally hemagglutinin (HA)-tagged LCMV NP (pC-NP-HA), N-terminally Strep-tagged LCMV NP (pC-Strep-NP), untagged LCMV L (pC-L), and N-terminally FLAG-tagged LCMV L (pC-FLAG-L) and LASV L (pC-FLAG-LASV-L) proteins have been described previously (27, 47–49). Plasmid expressing a C-terminally Strep-tagged enhanced green fluorescent protein (eGFP) was generated by PCR-based mutagenesis inserting a linker and the Strep-tag sequence immediately downstream of eGPF open reading frame (ORF) of pCAGGS-eGFP. Plasmids expressing C-terminally Strep-tagged Z protein of LCMV (pC-LCMV-Z-Strep) and LASV (pC-LASV-Z-Strep) were generated using procedures similar to those used for pC-LASV-Z-FLAG (50). Plasmids expressing mutant Z proteins were generated by PCR-based mutagenesis introducing corresponding mutations into pC-LCMV-Z-Strep. A plasmid that directs intracellular synthesis, under the control of an T7 promoter, of an LCMV minigenome (MG) RNA derived from the S segment (pT7-MG/eGFP) was generated by substituting the eGFP ORF for a chloramphenicol acetyltransferase (CAT) ORF in pT7-MG/CAT (previously described as pMG#7Δ2G [51]). Plasmids for rescue of rLCMV were generated based on pPol1S Cl-13 and pPol1L Cl-13 plasmids, which direct RNA polymerase I (Pol1)-mediated intracellular synthesis of S and L, respectively, RNA genome species of Cl-13 strain of LCMV (35, 36). pPol1L/Z(70AAA) and pPol1L/Z(L72A) were generated by PCR-based mutagenesis introducing triple mutations (Y70A, P71A, and L72A) and single mutation (L72A), respectively, into the Z gene of pPol1L Cl-13.
Pulldown of Strep-tagged proteins.
293T cells seeded in 100-mm culture dishes at 4.5 × 106/dish and cultured overnight were transfected with 2 μg of plasmid expressing C-terminally Strep-tagged eGFP or WT or mutant Z proteins together with 8 μg of plasmid expressing N-terminally FLAG-tagged L protein using 2 μl of 1 mg/ml polyethyleneimine Max (PEI Max) (Polysciences, Warrington, PA)/μg of DNA. At 5 h posttransfection, transfection medium was replaced with fresh medium and incubated at 37°C and 5% CO2. At 72 h posttransfection, cells were washed two times with ice-cold phosphate-buffered saline (PBS), and cells were lysed with 1 ml of PD lysis buffer (+) (250 mM NaCl, 50 mM Tris-HCl [pH 7.5], 0.5% Triton X-100, 10% glycerol, 1 mM MgCl2, 1 μM CaCl2, 1 μM ZnCl2) supplemented with halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA) and 5 μg/ml of DNase I (Worthington Biochemical Corporation, Lakewood, NJ). Lysates were clarified by centrifugation at 21,130 × g at 4°C for 10 min to remove cell debris. Clarified cell lysates were incubated with Strep-Tactin Sepharose resin (Qiagen, Germantown, MD) at 4°C. After 2 h of incubation, the resin was washed three times with PD lysis buffer (+) and once with PD lysis buffer without Triton X-100 [PD lysis buffer (−)]. After centrifugation at 9,391 × g and 4°C for 30 s, the last wash buffer was removed, and protein complexes associated with the resin were eluted with 50 μl of PD lysis buffer (−) containing 2.5 mM desthiobiotin.
Western blotting.
Clarified lysates were mixed at a 1:1 ratio with 2× SDS-loading buffer (100 mM Tris [pH 6.8], 20% 2-mercaptoethanol, 4% SDS, 0.2% bromophenol blue, 20% glycerol) and boiled for 10 min. Proteins samples were fractionated by SDS-PAGE using 4 to 20% gradient polyacrylamide gels (Mini-PROTEAN TGX gels; Bio-Rad, Hercules, CA), and proteins were transferred by electroblotting onto polyvinylidene difluoride membranes (Immobilon transfer membranes; Millipore, Billerica, MA). To detect each protein, membranes were reacted with mouse monoclonal antibodies (MAbs) to Strep (Qiagen, Germantown, MD), eGFP (TaKaRa Bio USA, Mountain View, CA), and β-actin (Santa Cruz Biotechnology, Dallas, TX) or rabbit polyclonal antibodies to glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Millipore, Billerica, MA), HA (Santa Cruz Biotechnology, Dallas, TX), or α-tubulin (Cell Signaling Technologies, Danvers, MA), respectively, followed by incubation with appropriate horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G (IgG) antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). SuperSignal West Pico or Femto chemiluminescent substrate (Thermo Fisher Scientific) was used to elicit chemiluminescent signals that were visualized using an ImageQuant LAS 4000 imager (GE Healthcare Bio-Sciences, Pittsburgh, PA).
Budding assay.
Budding assay was performed as described previously (50). 293T cells seeded (3.5 × 105/well) in a 12-well plate were transfected with 0.5 μg of empty pCAGGS vector or plasmid expressing Strep-tagged WT or mutant Z protein using Lipofectamine 2000. At 5 h posttransfection, media were replaced with fresh media and incubated at 37°C and 5% CO2. At 48 h posttransfection, virion-like particle (VLP)-containing TCS and cells were collected. Total cell lysate was prepared by lysing the cells with lysis buffer (1% NP-40, 50 mM Tris-HCl [pH 8.0], 62.5 mM NaCl, 0.4% sodium deoxycholate). After clarification of TCS from cell debris by centrifugation at 400 × g and 4°C for 10 min, VLPs were collected by ultracentrifugation at 100,000 × g and 4°C for 30 min through a 20% sucrose cushion. VLPs were resuspended in PBS. Z expression in total cell lysate and TCS (containing VLPs) was analyzed by Western blotting.
LCMV MG assay.
The LCMV minigenome (MG) assay was done as described previously (21). Briefly, 293T cells seeded (4 × 105/well) in a 12-well plate and cultured overnight were transfected with 0.5 μg of pT7-MG/eGFP, 0.5 μg of a plasmid expressing T7 polymerase (pC-T7), 0.3 μg of either pC-NP or pC-NP-HA, and 0.3 μg of either pC-L or pC-FLAG-L together with or without 0.5 μg of a plasmid expressing the WT or mutant Z protein using 2.5 μl of Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA)/μg of DNA. At 5 h posttransfection, media were replaced with fresh media and incubated at 37°C and 5% CO2. At 72 h posttransfection, eGFP expression was examined by a fluorescence microscope, Axiovert S100 (Carl Zeiss, Jena, Germany). Expression levels of each protein were determined by Western blotting.
Preparation of soluble and insoluble fractions to PD lysis buffer (+).
Cells in a 12-well plate were washed with PBS and lysed with 300 μl of PD lysis buffer (+). To separate soluble fraction from insoluble fraction, lysate was centrifuged at 21,130 × g at 4°C for 10 min. The supernatant (soluble fraction) was collected and mixed (1:1 ratio) with 2× SDS loading buffer. Pellet (insoluble fraction) was solubilized in 600 μl of 2× SDS loading buffer by passing through a 25-gauge needle. Samples from S and P fractions corresponding to equivalent numbers of cells were examined by Western blotting to assess protein distribution between S and P fractions.
Immunofluorescence assay.
293T cells seeded on coverslips in a 24-well plate (1 × 105/well) and cultured overnight were transfected with 25 ng of plasmid expressing WT or mutant Z proteins together with (Fig. 5B) or without (Fig. 5A) plasmids required for formation of active MG ribonucleoprotein (125 ng of pT7-MG/CAT, 125 ng of pC-T7pol, 75 ng of pC-L, and 75 ng of pC-NP). At 48 h posttransfection, cells were fixed with 4% paraformaldehyde (PFA) in PBS. After cell permeabilization and blocking by treatment with 1% normal goat serum in dilution buffer (DB: 0.3% Triton X-100 in PBS containing 3% bovine serum albumin [BSA]), cells were incubated with primary mouse anti-Strep (Qiagen) and rabbit anti-HA (Santa Cruz Biotechnology, Dallas, TX) antibodies, followed by secondary anti-mouse and anti-rabbit IgG antibody conjugated with Alexa Fluor 488 and Alexa Fluor 568, respectively. To visualize nuclei, 4′,6-diamidino-2-phenylindole (DAPI) Fluoromount-G (Southern Biotech, Birmingham, AL) was used to mount coverslips on a slide glass. Stained cells were examined by confocal microscopy (LSM 710; Zeiss) and data analyzed by ZEN software (Zeiss).
Rescue of rLCMV.
The different recombinant LCMVs (rLCMVs) used in this study were rescued as described previously (35–37), with minor modifications. BHK-21 cells seeded in a 6-well plate at 7.5 × 105/well were cultured overnight and transfected with plasmids pPol1S Cl-13 (0.8 μg) and pPol1L Cl-13 (1.4 μg), together with plasmids pC-NP (0.8 μg) and pC-L (1.0 μg), using 2.5 μl of Lipofectamine 2000 (Invitrogen)/μg of DNA. For rescue of rLCMVs expressing either L(70AAA) [rLCMV/Z(70AAA)] or Z(L72A) [rLCMV/Z(L72A)], pPol1L/Z(70AAA) or pPol1L/Z(L72A), respectively, was used instead of pPol1L Cl-13. After 5 h of transfection, the transfection mix was removed and fresh medium added. At 3 days posttransfection, tissue culture supernatant (TCS) was removed, 3 ml of fresh medium was added, and cells were cultured for another 3 days. The TCS was collected at 6 days posttransfection and clarified by centrifugation at 400 × g at 4°C to remove cell debris. This clarified TCS was designated P0 and used to infect a fresh monolayer of BHK-21 cells in a 6-well plate, and the cells were cultured for another 3 days. At 72 h after infection with P0 TCS, TCS was collected and clarified by centrifugation at 400 × g at 4°C for 10 min to remove cell debris (P1). These steps were repeated to obtain P2 TCS. Virus titers in TCS from P0, P1, and P2 were determined by immunofocus forming assay (IFFA).
Virus titration.
LCMV titers were determined by IFFA as described previously (52). Briefly, 10-fold serial virus dilutions were used to infect Vero E6 cell monolayers in a 96-well plate, and at 20 h postinfection (p.i.), cells were fixed with 4% PFA in PBS. After cell permeabilization by treatment with DB, cells were stained with a rat MAb to NP (VL-4; Bio X Cell, West Lebanon, NH) conjugated with Alexa Fluor 488 (VL-4-AF488; protein labeling kit; Life Technologies, Carlsbad, CA). The limit of detection is 100 focus-forming units (FFU) per ml.
Statistics.
Data were analyzed for P values by a two-tailed unpaired t test using GraphPad Prism software.
ACKNOWLEDGMENTS
We thank B. Cubitt for her help with the quantitative analysis of the Western blot results.
This research was supported by NIH/NIAID grants AI047140 and AI077719 to J.C.D.L.T. and by funds from the Japan Society for the Promotion of Science, Daiichi Sankyo Foundation of Life Science, and KANAE Foundation for the Promotion of Medical Science to M.I.
Footnotes
This is manuscript 29631 from The Scripps Research Institute.
REFERENCES
- 1.Buchmeier MJ, Peters CJ, de la Torre JC. 2007. Arenaviridae: the viruses and their replication, p 1791–1851. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Enria DA, Briggiler AM, Sanchez Z. 2008. Treatment of Argentine hemorrhagic fever. Antiviral Res 78:132–139. doi: 10.1016/j.antiviral.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geisbert TW, Jahrling PB. 2004. Exotic emerging viral diseases: progress and challenges. Nat Med 10:S110–S121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- 4.Khan SH, Goba A, Chu M, Roth C, Healing T, Marx A, Fair J, Guttieri MC, Ferro P, Imes T, Monagin C, Garry RF, Bausch DG, Mano River Union Lassa Fever Network. 2008. New opportunities for field research on the pathogenesis and treatment of Lassa fever. Antiviral Res 78:103–115. doi: 10.1016/j.antiviral.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 5.McCormick JB, Fisher-Hock SP. 2002. Lassa fever. Curr Top Microbiol Immunol 262:75–110. [DOI] [PubMed] [Google Scholar]
- 6.Peters CJ. 2002. Human infection with arenaviruses in the Americas. Curr Top Microbiol Immunol 262:65–74. [DOI] [PubMed] [Google Scholar]
- 7.Richmond JK, Baglole DJ. 2003. Lassa fever: epidemiology, clinical features, and social consequences. BMJ 327:1271–1275. doi: 10.1136/bmj.327.7426.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton LL, Mets MB. 1999. Lymphocytic choriomeningitis virus: pediatric pathogen and fetal teratogen. Pediatr Infect Dis J 18:540–541. doi: 10.1097/00006454-199906000-00013. [DOI] [PubMed] [Google Scholar]
- 9.Barton LL, Mets MB. 2001. Congenital lymphocytic choriomeningitis virus infection: decade of rediscovery. Clin Infect Dis 33:370–374. doi: 10.1086/321897. [DOI] [PubMed] [Google Scholar]
- 10.Barton LL, Mets MB, Beauchamp CL. 2002. Lymphocytic choriomeningitis virus: emerging fetal teratogen. Am J Obstet Gynecol 187:1715–1716. doi: 10.1067/mob.2002.126297. [DOI] [PubMed] [Google Scholar]
- 11.Jahrling PB, Peters CJ. 1992. Lymphocytic choriomeningitis virus. A neglected pathogen of man. Arch Pathol Lab Med 116:486–488. [PubMed] [Google Scholar]
- 12.Mets MB, Barton LL, Khan AS, Ksiazek TG. 2000. Lymphocytic choriomeningitis virus: an underdiagnosed cause of congenital chorioretinitis. Am J Ophthalmol 130:209–215. doi: 10.1016/S0002-9394(00)00570-5. [DOI] [PubMed] [Google Scholar]
- 13.Fischer SA, Graham MB, Kuehnert MJ, Kotton CN, Srinivasan A, Marty FM, Comer JA, Guarner J, Paddock CD, DeMeo DL, Shieh W-J, Erickson BR, Bandy U, DeMaria A Jr, Davis JP, Delmonico FL, Pavlin B, Likos A, Vincent MJ, Sealy TK, Goldsmith CS, Jernigan DB, Rollin PE, Packard MM, Patel M, Rowland C, Helfand RF, Nichol ST, Fishman JA, Ksiazek T, Zaki SR, LCMV in Transplant Recipients Investigation Team. 2006. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N Engl J Med 354:2235–2249. doi: 10.1056/NEJMoa053240. [DOI] [PubMed] [Google Scholar]
- 14.Peters CJ. 2006. Lymphocytic choriomeningitis virus—an old enemy up to new tricks. N Engl J Med 354:2208–2211. doi: 10.1056/NEJMp068021. [DOI] [PubMed] [Google Scholar]
- 15.Damonte EB, Coto CE. 2002. Treatment of arenavirus infections: from basic studies to the challenge of antiviral therapy. Adv Virus Res 58:125–155. doi: 10.1016/S0065-3527(02)58004-0. [DOI] [PubMed] [Google Scholar]
- 16.Moreno H, Gallego I, Sevilla N, de la Torre JC, Domingo E, Martin V. 2011. Ribavirin can be mutagenic for arenaviruses. J Virol 85:7246–7255. doi: 10.1128/JVI.00614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker WB. 2005. Metabolism and antiviral activity of ribavirin. Virus Res 107:165–171. doi: 10.1016/j.virusres.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Cashman KA, Smith MA, Twenhafel NA, Larson RA, Jones KF, Allen RD III, Dai D, Chinsangaram J, Bolken TC, Hruby DE, Amberg SM, Hensley LE, Guttieri MC. 2011. Evaluation of Lassa antiviral compound ST-193 in a guinea pig model. Antiviral Res 90:70–79. doi: 10.1016/j.antiviral.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Safronetz D, Rosenke K, Westover JB, Martellaro C, Okumura A, Furuta Y, Geisbert J, Saturday G, Komeno T, Geisbert TW, Feldmann H, Gowen BB. 2015. The broad-spectrum antiviral favipiravir protects guinea pigs from lethal Lassa virus infection post-disease onset. Sci Rep 5:14775. doi: 10.1038/srep14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raabe VN, Kann G, Ribner BS, Morales A, Varkey JB, Mehta AK, Lyon GM, Vanairsdale S, Faber K, Becker S, Eickmann M, Strecker T, Brown S, Patel K, De Leuw P, Schuettfort G, Stephan C, Rabenau H, Klena JD, Rollin PE, McElroy A, Stroher U, Nichol S, Kraft CS, Wolf T, Emory Serious Communicable Diseases Unit. 2017. Favipiravir and ribavirin treatment of epidemiologically linked cases of Lassa fever. Clin Infect Dis 65:855–859. doi: 10.1093/cid/cix406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez M, Craven RC, de la Torre JC. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci U S A 100:12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strecker T, Eichler R, Meulen J, Weissenhorn W, Klenk HD, Garten W, Lenz O. 2003. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles. J Virol 77:10700–10705. doi: 10.1128/JVI.77.19.10700-10705.2003. (Erratum, 77:12927, doi:10.1128/JVI.77.23.12927.2003.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capul AA, Perez M, Burke E, Kunz S, Buchmeier MJ, de la Torre JC. 2007. Arenavirus Z-glycoprotein association requires Z myristoylation but not functional RING or late domains. J Virol 81:9451–9460. doi: 10.1128/JVI.00499-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urata S, Noda T, Kawaoka Y, Yokosawa H, Yasuda J. 2006. Cellular factors required for Lassa virus budding. J Virol 80:4191–4195. doi: 10.1128/JVI.80.8.4191-4195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urata S, Yasuda J, de la Torre JC. 2009. The Z protein of the New World arenavirus Tacaribe virus has bona fide budding activity that does not depend on known late domain motifs. J Virol 83:12651–12655. doi: 10.1128/JVI.01012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez M, Greenwald DL, de la Torre JC. 2004. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J Virol 78:11443–11448. doi: 10.1128/JVI.78.20.11443-11448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee KJ, Novella IS, Teng MN, Oldstone MB, de La Torre JC. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol 74:3470–3477. doi: 10.1128/JVI.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López N, Jacamo R, Franze-Fernandez MT. 2001. Transcription and RNA replication of Tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J Virol 75:12241–12251. doi: 10.1128/JVI.75.24.12241-12251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jácamo R, Lopez N, Wilda M, Franze-Fernandez MT. 2003. Tacaribe virus Z protein interacts with the L polymerase protein to inhibit viral RNA synthesis. J Virol 77:10383–10393. doi: 10.1128/JVI.77.19.10383-10393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kranzusch PJ, Whelan SP. 2011. Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex. Proc Natl Acad Sci U S A 108:19743–19748. doi: 10.1073/pnas.1112742108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell Dwyer EJ, Lai H, MacDonald RC, Salvato MS, Borden KL. 2000. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J Virol 74:3293–3300. doi: 10.1128/JVI.74.7.3293-3300.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kentsis A, Dwyer EC, Perez JM, Sharma M, Chen A, Pan ZQ, Borden KL. 2001. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J Mol Biol 312:609–623. doi: 10.1006/jmbi.2001.5003. [DOI] [PubMed] [Google Scholar]
- 33.Urata S, de la Torre JC. 2011. Arenavirus budding. Adv Virol 2011:180326. doi: 10.1155/2011/180326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urata S, Yasuda J. 2012. Molecular mechanism of arenavirus assembly and budding. Viruses 4:2049–2079. doi: 10.3390/v4102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emonet SF, Garidou L, McGavern DB, de la Torre JC. 2009. Generation of recombinant lymphocytic choriomeningitis viruses with trisegmented genomes stably expressing two additional genes of interest. Proc Natl Acad Sci U S A 106:3473–3478. doi: 10.1073/pnas.0900088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flatz L, Bergthaler A, de la Torre JC, Pinschewer DD. 2006. Recovery of an arenavirus entirely from RNA polymerase I/II-driven cDNA. Proc Natl Acad Sci U S A 103:4663–4668. doi: 10.1073/pnas.0600652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sánchez AB, de la Torre JC. 2006. Rescue of the prototypic Arenavirus LCMV entirely from plasmid. Virology 350:370–380. doi: 10.1016/j.virol.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Capul AA, de la Torre JC, Buchmeier MJ. 2011. Conserved residues in Lassa fever virus Z protein modulate viral infectivity at the level of the ribonucleoprotein. J Virol 85:3172–3178. doi: 10.1128/JVI.02081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casabona JC, Levingston Macleod JM, Loureiro ME, Gomez GA, Lopez N. 2009. The RING domain and the L79 residue of Z protein are involved in both the rescue of nucleocapsids and the incorporation of glycoproteins into infectious chimeric arenavirus-like particles. J Virol 83:7029–7039. doi: 10.1128/JVI.00329-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fayadat L, Siffroi-Fernandez S, Lanet J, Franc JL. 2000. Degradation of human thyroperoxidase in the endoplasmic reticulum involves two different pathways depending on the folding state of the protein. J Biol Chem 275:15948–15954. doi: 10.1074/jbc.M905763199. [DOI] [PubMed] [Google Scholar]
- 41.Donoso G, Herzog V, Schmitz A. 2005. Misfolded BiP is degraded by a proteasome-independent endoplasmic-reticulum-associated degradation pathway. Biochem J 387:897–903. doi: 10.1042/BJ20041312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volpon L, Osborne MJ, Borden KL. 2008. NMR assignment of the arenaviral protein Z from Lassa fever virus. Biomol NMR Assign 2:81–84. doi: 10.1007/s12104-008-9090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volpon L, Osborne MJ, Capul AA, de la Torre JC, Borden KL. 2010. Structural characterization of the Z RING-eIF4E complex reveals a distinct mode of control for eIF4E. Proc Natl Acad Sci U S A 107:5441–5446. doi: 10.1073/pnas.0909877107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hastie KM, Zandonatti M, Liu T, Li S, Woods VL Jr, Saphire EO. 2016. Crystal structure of the oligomeric form of Lassa virus matrix protein Z. J Virol 90:4556–4562. doi: 10.1128/JVI.02896-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziegler CM, Eisenhauer P, Kelly JA, Dang LN, Beganovic V, Bruce EA, King BR, Shirley DJ, Weir ME, Ballif BA, Botten J. 2017. A proteomic survey of Junin virus interactions with human proteins reveals host factors required for arenavirus replication. J Virol doi: 10.1128/JVI.01565-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ortiz-Riaño E, Cheng BY, de la Torre JC, Martinez-Sobrido L. 2011. The C-terminal region of lymphocytic choriomeningitis virus nucleoprotein contains distinct and segregable functional domains involved in NP-Z interaction and counteraction of the type I interferon response. J Virol 85:13038–13048. doi: 10.1128/JVI.05834-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwasaki M, Ngo N, Cubitt B, de la Torre JC. 2015. Efficient interaction between arenavirus nucleoprotein (NP) and RNA-dependent RNA polymerase (L) is mediated by the virus nucleocapsid (NP-RNA) template. J Virol 89:5734–5738. doi: 10.1128/JVI.00103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee KJ, Perez M, Pinschewer DD, de la Torre JC. 2002. Identification of the lymphocytic choriomeningitis virus (LCMV) proteins required to rescue LCMV RNA analogs into LCMV-like particles. J Virol 76:6393–6397. doi: 10.1128/JVI.76.12.6393-6397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodrigo WW, de la Torre JC, Martinez-Sobrido L. 2011. Use of single-cycle infectious lymphocytic choriomeningitis virus to study hemorrhagic fever arenaviruses. J Virol 85:1684–1695. doi: 10.1128/JVI.02229-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urata S, Ngo N, de la Torre JC. 2012. The PI3K/Akt pathway contributes to arenavirus budding. J Virol 86:4578–4585. doi: 10.1128/JVI.06604-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perez M, de la Torre JC. 2003. Characterization of the genomic promoter of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 77:1184–1194. doi: 10.1128/JVI.77.2.1184-1194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Battegay M. 1993. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24 well plates. ALTEX 10:6–14. [PubMed] [Google Scholar]