ABSTRACT
H7N9 virus has caused five infection waves since it emerged in 2013. The highest number of human cases was seen in wave 5; however, the underlying reasons have not been thoroughly elucidated. In this study, the geographical distribution, phylogeny, and genetic evolution of 240 H7N9 viruses in wave 5, including 35 new isolates from patients and poultry in nine provinces, were comprehensively analyzed together with strains from first four waves. Geographical distribution analysis indicated that the newly emerging highly pathogenic (HP) and low-pathogenicity (LP) H7N9 viruses were cocirculating, causing human and poultry infections across China. Genetic analysis indicated that dynamic reassortment of the internal genes among LP-H7N9/H9N2/H6Ny and HP-H7N9, as well as of the surface genes, between the Yangtze and Pearl River Delta lineages resulted in at least 36 genotypes, with three major genotypes (G1 [A/chicken/Jiangsu/SC537/2013-like], G3 [A/Chicken/Zhongshan/ZS/2017-like], and G11 [A/Anhui/40094/2015-like]). The HP-H7N9 genotype likely evolved from G1 LP-H7N9 by the insertion of a KRTA motif at the cleavage site (CS) and then evolved into 15 genotypes with four different CS motifs, including PKGKRTAR/G, PKGKRIAR/G, PKRKRAAR/G, and PKRKRTAR/G. Approximately 46% (28/61) of HP strains belonged to G3. Importantly, neuraminidase (NA) inhibitor (NAI) resistance (R292K in NA) and mammalian adaptation (e.g., E627K and A588V in PB2) mutations were found in a few non-human-derived HP-H7N9 strains. In summary, the enhanced prevalence and diverse genetic characteristics that occurred with mammalian-adapted and NAI-resistant mutations may have contributed to increased numbers of human infections in wave 5.
IMPORTANCE The highest numbers of human H7N9 infections were observed during wave 5 from October 2016 to September 2017. Our results showed that HP-H7N9 and LP-H7N9 had spread virtually throughout China and underwent dynamic reassortment with different subtypes (H7N9/H9N2 and H6Ny) and lineages (Yangtze and Pearl River Delta lineages), resulting in totals of 36 and 3 major genotypes, respectively. Notably, the NAI drug-resistant (R292K in NA) and mammalian-adapted (e.g., E627K in PB2) mutations were found in HP-H7N9 not only from human isolates but also from poultry and environmental isolates, indicating increased risks for human infections. The broad dissemination of LP- and HP-H7N9 with high levels of genetic diversity and host adaptation and drug-resistant mutations likely accounted for the sharp increases in the number of human infections during wave 5. Therefore, more strategies are needed against the further spread and damage of H7N9 in the world.
KEYWORDS: H7N9, HPAIV, wave five, avian influenza virus, dynamic reassortment, evolution, genetic diversity, origin
INTRODUCTION
Prior to 2013, the H7 subtype avian influenza viruses (AIVs) were mainly found in birds and were known to cause occasional human infections with mild illness (1). In March 2013, a novel H7N9 AIV emerged and caused severe disease in humans in China (2–7), with a case fatality rate of 39.1% (612 deaths/1,564 cases) as of 30 October 2017 (http://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en/). Previous studies have shown that the novel H7N9 was low-pathogenicity (LP) avian influenza virus (LPAIV) and had an increased ability to bind to human-like sialic acid receptors (8–11). In addition, they also possessed several host adaptation mutations (e.g., T271A, K526R, E627K, or D701N) in the PB2 protein, which were associated with increased virus replication rates and disease severity in mammals and/or humans (12–14). So far, poultry-to-human transmission has been the primary route for human H7N9 infections (15, 16). The majority of human cases had histories of exposure to live-poultry or AIV-contaminated environments, such as live-poultry markets (LPMs) (http://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en/) (5, 15–17). Due to potential for further adaptation to mammals and possible human-to-human transmission, the World Health Organization (WHO) previously classified H7N9 as “an unusually dangerous virus for humans” (http://www.sciencemag.org/news/2013/04/h7n9-unusually-dangerous-virus-international-group-experts-concludes). Several asymptomatic and mild infections with the H7N9 LPAIVs, which may increase mammalian adaptation and transmissibility between humans, have also been reported in humans (18, 19).
There have been five infection waves since H7N9 emerged in March 2013. Notably, the fifth wave started earlier than the previous four, with a sudden and steep increase in the number of human cases (http://www.who.int/influenza/human_animal_interface/avian_influenza/riskassessment_AH7N9_201702/en/). More importantly, a high-pathogenicity (HP) avian influenza virus (HPAIV) H7N9 variant emerged during the fifth wave (wave 5) and has been identified as the causative pathogen in several human infections (20, 21). Furthermore, there have been reports of H7N9 infections in several family clusters, as well as in patients in the same hospital ward (http://www.who.int/influenza/human_animal_interface/avian_influenza/archive/en/). A comprehensive study of the geographical distribution and genetics of H7N9 viruses in wave 5 is needed to understand the underlying causes for the increased numbers of cases.
In this study, we sequenced 35 H7N9 AIVs collected from patients and poultry in LPMs or poultry farms in nine Chinese provinces during wave 5. Together with publicly available data, full-length genome sequences from a total of 240 H7N9 AIVs in wave 5 from 26 provinces and municipalities were systematically analyzed, and the results are presented here.
RESULTS
A sudden increase in the number of human H7N9 cases in wave 5.
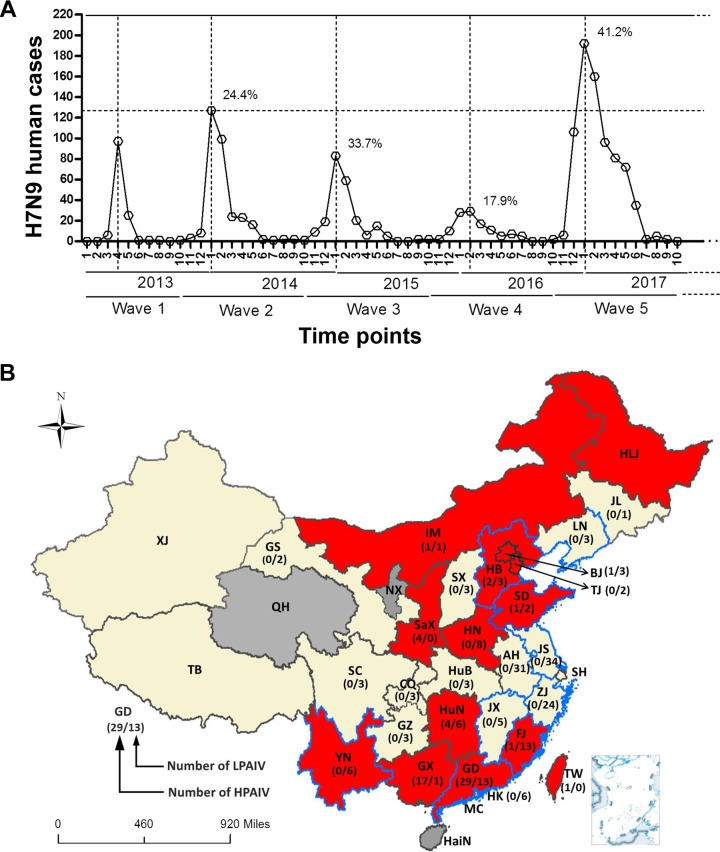
Human infections with H7N9 AIV emerge in waves during the winter months, with peaks around January, and there have been five waves to date (Fig. 1A). The fifth wave began in October 2016, and the number of infections suddenly increased and peaked in January 2017 but remained at high levels through May 2017. Over 760 human cases were reported between October 2016 and September 2017, which is substantially higher than the rates in waves 1 to 4 (Fig. 1A).
FIG 1.
The temporal and spatial distributions of H7N9 human infections during wave 5. (A) The numbers of human H7N9 cases in each wave are listed by month. The fatality rates in January of wave 2 to 5 are shown. (B) Three provinces (QH, NX, and HaiN) without human or poultry H7N9 infections during wave 5 are colored in dark gray. The other provinces reporting human or poultry H7N9 infections are colored in red or faint yellow. Red indicates that HP-H7N9 was found in these provinces. The blue border indicates the new H7N9 viruses isolated from poultry and humans by our group. The numbers of HP- and LP-H7N9 strains analyzed in the present study are listed within the brackets as indicated in the provinces. Abbreviations in the map are as follows: AH, Anhui; BJ, Beijing; CQ, Chongqing; FJ, Fujian; GD, Guangdong; GS, Gansu; GX, Guangxi; GZ, Guizhou; HaiN, Hainan; HB, Hebei; HK, Hong Kong; HLJ, Heilongjiang; HN, Henan; HuB, Hubei; HuN, Hunan; IM, Inner Mongolia; JL, Jilin; JS, Jiangsu; JX, Jiangxi; LN, Liaoning; MC, Macao; NX, Ningxia; QH, Qinghai; SaX, Shaanxi; SC, Sichuan; SD, Shandong; SH, Shanghai; SX, Shanxi; TB, Tibet; TJ, Tianjin; TW, Taiwan; XJ, Xinjiang; YN, Yunnan; ZJ, Zhejiang.
Similarly to previous waves, almost all cases of human H7N9 infections corresponded to a history of exposure to poultry or AIV-contaminated environments during wave 5, with the cases starting at the centers of the Yangtze River Delta (Zhejiang and Jiangsu) and the Pearl River Delta (Guangdong). Over 71% of cases were reported after December 2016 were found in areas around those two centers (including Zhejiang, Jiangsu, Shanghai, Anhui, Guangdong, Hong Kong, Macao, Fujian, Hunan, Jiangxi, and Guangxi). Approximately 140 cases were reported in Guizhou, Hubei, Yunnan, Sichuan, Chongqing, Shandong, Henan, and Taiwan provinces or municipalities, where are geographically distant from the two centers. Over 80 human cases were reported in even more distant regions, such as Liaoning, Jilin, Inner Mongolia, Hebei, Tianjin, Beijing, Shanxi, Shaanxi, Gansu, Xinjiang, and Tibet provinces or municipalities, after February 2017. In contrast to previous waves (22), human infections with H7N9 during wave 5 were widespread throughout virtually all of China, although the reports from Heilongjiang Province merely noted an H7N9 outbreak in poultry. Only three provinces (Ningxia, Qinghai, and Hainan) did not report H7N9 infections during wave 5 (Fig. 1B).
Importantly, HP-H7N9 variants have emerged in poultry or environmental samples from LPMs or have caused outbreaks in poultry farms from several provinces. These include Guangdong, Guangxi, Fujian, Hunan, Hebei, Tianjin, Beijing, Henan, Shandong, Shaanxi, Inner Mongolia, and Heilongjiang, and human infections were reported in Guangdong, Taiwan, Guangxi, Hunan, Shaanxi, Hebei, Henan, Fujian, and Yunnan (Fig. 1B). LP-H7N9 was additionally found in Jiangsu, Zhejiang, Jiangxi, and Liaoning provinces in LPMs or poultry farms. The prevalence of HP-H7N9 and LP-H7N9 in poultry farms and LPMs from the south (Guangdong) to the north (Inner Mongolia) of China closely matched the geographical distribution of reported human cases (Fig. 1; see also Table S1 in the supplemental material).
LP- and HP-H7N9 are cocirculating in China during wave 5.
A total of 35 whole-genome sequences of H7N9 viruses in wave 5 were obtained by our AIV surveillance network (23). These sequences originated from nine provinces, including Guangdong (n = 12, 9 from human cases and 3 from poultry in LPMs), Jiangsu (n = 4, from poultry in LPMs), Jiangxi (n = 4, from poultry in LPMs), Liaoning (n = 2, from poultry in poultry farms), Shandong (n = 3, from poultry in poultry farms), Yunnan (n = 2, from human cases), Zhejiang (n = 3, from poultry in LPMs), Fujian (n = 4, 3 from human cases and 1 from a poultry farm), and Hebei (n = 1, from a poultry farm) (Fig. 1) (Table 1). Genome sequences from 827 additional H7N9 AIVs were also obtained from the Global Initiative on Sharing All Influenza Data (GISAID), including 205 viruses isolated from wave 5 and the remaining from waves 1 to 4. In total, our data set included 240 H7N9 strains from 26 provinces or municipalities in wave 5 (Fig. 1B; see also Table S1), with 200 strains from humans. By inspecting the hemagglutinin (HA) protein alignment, a total of 61 strains (Guangdong, n = 29; Guangxi, n = 17; Hunan, n = 4; Shaanxi, n = 4; Hebei, n = 2; Beijing, n = 1; Fujian, n = 1; Inner Mongolia, n = 1; Shandong, n = 1; Taiwan, n = 1) were found to contain an insertion of multiple basic amino acids at the cleavage site (CS) (Fig. 1B) (Table 1) and thus should be regarded as HP-H7N9 variants. The remaining H7N9 sequences were found to be LP. Despite the emergence and spread of HP-H7N9, LP-H7N9 has still been dominant in China during wave 5. Taking into account all reported regions with HP-H7N9 or outbreaks in poultry with human cases, there were a total of 14 provinces and municipalities (including 4 provinces and municipalities without HP-H7N9 strain information) from the north (Inner Mongolia and Heilongjiang) to the south (Guangdong) of China which has reported HP-H7N9 (Fig. 1), suggesting that LP- and HP-H7N9 were cocirculating and causing human infections across China.
TABLE 1.
Molecular characteristics of the H7N9 viruses isolated from poultry and humansa
| H7N9 virus | HA (H3 numbering) |
NA (N2 numbering) |
M2 |
PB2 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cleavage site | Residue 186 | Residues 224–228 | Residue 292 | Stalk deletion | Residue 31 | Residue 526 | Residue 588 | Residue 591 | Residue 627 | Residue 701 | |
| A/GD/Th008/2017 | PKRKRTARG | V | NGQSG | K | 69–73 | N | K | V | Q | E | D |
| A/SZ/Th001/2016 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | K | D |
| A/SZ/Th002/2016 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | K | D |
| A/SZ/Th003/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| A/SZ/Th004/2017 | PKG----RG | V | NGLSG | K | 69–73 | N | K | V | Q | E | D |
| A/GD/Th005/2017 | PKGKRIARG | V | NGQSG | R | 69–73 | N | R | A | Q | K | D |
| A/SZ/Th006/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | K | D |
| A/SZ/Th007/2017 | PKRKRTARG | V | NGQSG | K | 69–73 | N | K | V | Q | K | D |
| A/SZ/Th008/2017 | PKRKRTARG | V | NGHSG | K | 69–73 | N | R | A | Q | E | N |
| A/FJ/QZ-Th001/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | K | D |
| A/FJ/QZ-Th002/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | K | D |
| A/FJ/QZ-Th005/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | K | D |
| A/YN/YN001/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | K | D |
| A/YN/YN002/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | K | D |
| CK/GD/SZBJ0011-O/2017 | PKRKRTARG | V | NGQSG | K | 69–73 | N | K | V | Q | E | D |
| EN/GD/SZBA-E1/2017 | PKGKRTARG | V | NGQSG | R | 69–73 | N | K | V | Q | K | D |
| CK/JS/KS001-O/2016 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| CK/JS/KS004-O/2016 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| CK/JS/KS008-O/2016 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| DK/JX/ShRSX002-O/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| EN/JX/ShRXZ038/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| CK/JX/ShRGF080-P/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| CK/JX/ShRGF219-O/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| CK/ZJ/HZ142/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| CK/ZJ/HZ293/2016 | PKG----RG | V | NGLSG | R | 69–73 | N | K | I | Q | E | D |
| CK/ZJ/HZ298/2016 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| CK/GD/DGCPLB032/2016 | PKG----RG | V | NGQSG | R | 69–73 | N | K | V | Q | E | D |
| CK/LN/LN-083/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | R | A | Q | E | D |
| CK/SD/QD2-758/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| CK/SD/QD4-739/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| CK/SD/DZ056/2017 | PKRKRTARG | V | NGQSG | R | 69–73 | N | R | A | Q | E | D |
| CK/LN/SY059/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| CK/HB/XT001/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| CK/JS/DT001/2017 | PKG----RG | V | NGLSG | R | 69–73 | N | K | V | Q | E | D |
| CK/FJ/NP001/2017 | PKRKRTARG | V | NGQSG | R | 69–73 | N | R | A | Q | E | D |
A total of 35 H7N9 viruses were isolated and sequenced in the present study. CK, chicken; DK, duck; EN, environment; FJ, Fujian; GD, Guangdong; HB, Hebei; JS, Jiangsu; JX, Jiangxi; LN, Liaoning; SD, Shandong; SZ, Shenzhen; YN, Yunnan; ZJ, Zhenjiang.
Molecular characterizations of H7N9 AIVs in wave 5.
Similarly to viruses in previous waves, H7N9 viruses in wave 5 possessed the amantadine resistance mutation (S31N) in M2. The neuraminidase inhibitor (NAI)-resistant R292K mutation was also found in four new human strains. Interestingly, CK/GD/SZBJ0011-O/2017 isolated from LPM also possessed K292. To our knowledge, this is the first report that the NAI-resistant H7N9 strain was isolated from poultry (Table 1). All of the new human isolates possessed at least one of the four mammalian adaptation mutations, i.e., K526R, A588V, E627K, or D701N, in PB2. Several human strains (A/SZ/Th001/2016, A/SZ/Th002/2016, A/SZ/Th006/2017, A/SZ/Th007/2017, A/YN/YN001/2017, A/YN/YN002/2017, A/FJ/QZ-Th001/2017, A/FJ/QZ-Th002/2017, and A/FJ/QZ-Th005/2017) possessed both the A588V and E627K mutations. A/GD/Th005/2017 had both K526R and E627K mutations, while A/SZ/Th008/2017 had both the K526R and D701N mutations. Importantly, apart from CK/ZJ/HZ293/2016, which possesses I588, all of the avian-origin isolates possessed either of the mammalian adaptive residues V588 and R526 in PB2. Furthermore, the E627K mutation was found in EN/GD/SZBA-E1/2017, an environmental strain which also possessed the A588V mutation (Table 1).
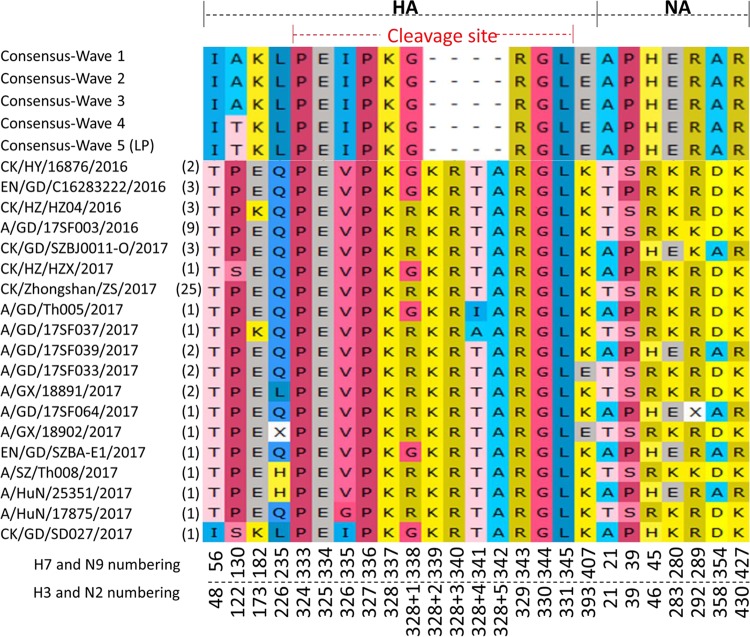
Consistent with data from previous waves, V186 and L226 in HA were found in all of the LP-H7N9 AIVs from human samples during wave 5. In contrast, the majority of HP-H7N9 viruses from humans and poultry possessed Q226 in HA, except for five human and one poultry strains (A/SZ/Th008/2017 and A/HuN/25351/2017 possessed H226, which had not been seen before in H7N9; two A/GX/18891/2017-like human strains and the poultry-origin CK/GD/SD027/2017 strain possessed L226; A/GX/18902/2017 possessed either residue H or residue L at position 226 of HA). There were four motif patterns at the CS in the HA protein of the HP H7N9 viruses, PKRKRTAR/G, PKGKRTAR/G, PKGKRIAR/G, and PKRKRAAR/G (Fig. 2). The majority of the HP strains possessed PKRKRTAR/G. Interestingly, the PKGKRTAR/G motif was just found in poultry-derived viruses and the last two motifs named above were possessed by only a single human strain each, A/GD/Th005/2017 and A/GD/17SF037/2017, respectively (Fig. 2) (Table 1). All of the H7N9 viruses from waves 1 to 5 possessed the same five-amino-acid-deletion (amino acids 69 to 73; N2 numbering) in the NA stalk.
FIG 2.
Distinctive amino acid variations in HA and NA of HP-H7N9 compared to LP-H7N9. The amino acid sequences of HA and NA were aligned between HP-H7N9 and the consensus sequences of LP-H7N9. The specific residues in HA and NA are listed. “X” of A/GX/18902/2017 at site 235 indicates an H or L residue, and “X” of A/GD/17SF064/2017 at site 289 indicates a K or R residue. The numbers in brackets represent the numbers of HP-H7N9 strains. CK, chicken; DK, duck; EN, environment; GD, Guangdong; GX, Guangxi; HuN, Hunan; HY, Heyuan; HZ, Huizhou; SZ, Shenzhen.
Distinctive amino acid substitutions in the HA and NA of HPAIV H7N9.
Apart from the four-amino-acid insertion, several specific amino acid substitutions between the HP- and LP-H7N9 AIVs were also identified (Fig. 2). For example, amino acid residues at positions 56, 130, 182, 235 (H3 numbering as 226), 335, and 407 in the HA protein of the HP strains were different from those of the LP strains (Fig. 2). In addition, amino acids at positions 21, 39, 45, 280, 354, and 427 were also specific to NA of HP-H7N9, although a few HP-H7N9 strains, e.g., CK/GD/SZBJ0011-O/2017-like and A/GD/17SF039/2017-like, possessed the same amino acids as LP-H7N9. Interestingly, A/GD/17SF064/2017 was associated with either a K or R residue mutation caused by a single nucleotide polymorphism (SNP) at site 289 (N2 numbering as 292) of NA (Fig. 2).
Origin and dynamic reassortment of H7N9 AIVs in wave 5.
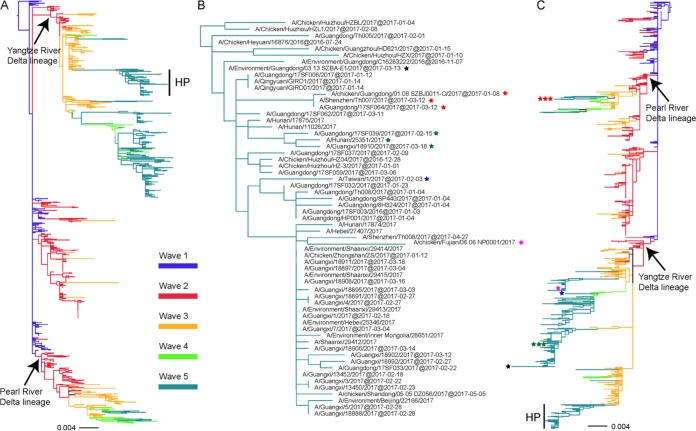
On the basis of the phylogenies, both the HA and NA of all the H7N9 strains identified in wave 5 clustered within the Yangtze River Delta lineage (n = 229) and the Pearl River Delta lineage (n = 7), while four strains possessed a hybrid combination with HA from the Yangtze River Delta lineage and NA from the Pearl River Delta lineage (Fig. 3; see also Fig. S1 in the supplemental material). Although the 61 HP-H7N9 AIVs have four different amino acid motifs at the CS, all of the HP viruses were closely related, forming a single cluster in the Yangtze River Delta lineage in the HA tree (Fig. 3A and B; see also Fig. S1A). It should be noted that the Guangxi strains formed an independent clade (Fig. 3B), suggesting that there might have been more than one virus introduction event from Guangdong to Guangxi. However, the NA genes of the 61 HP strains were not clustered together in the phylogenetic tree (Fig. 3C; see also Fig. S1B). A total of 58 strains fell within the Yangtze River Delta lineage, among which 52 HP strains clustered together and shared the same origin. CK/FJ/NP001/2017, EN/GD/SZBA-E1/2017, A/GD/17SF039/2017, A/GX/18910/2017, A/HuN/25351/2017, and A/Taiwan/1/2017 did not fall within this cluster (Fig. 3C; see also Fig. S1B). Remarkably, the NA gene sequences of three HP strains, A/SZ/Th007/2017, A/GD/17SF064/2017, and CK/GD/SZBJ0011-O/2017, fell within the Pearl River Delta lineage (Fig. 3C; see also Fig. S1B). Therefore, the HA genes of the HP strains shared a single origin in the Yangtze River Delta lineage, whereas the NA gene had multiple different origins.
FIG 3.
Phylogenies of HA and NA genes of H7N9 viruses from different waves. All of the H7N9 viruses from waves 1 to 5, including our 35 new isolates, were used to perform the phylogenetic analysis. Panel A shows the HA gene tree, and the HP-H7N9 viruses in panel A were magnified as shown in panel B. Panel C shows the NA gene tree. The HP strains with different NA lineages are marked as stars with different colors in panels B and C; stars with the same color represent the same strain. H7N9 viruses from waves 1, 2, 3, 4, and 5 are colored blue, red, orange, light green, and lime green, respectively.
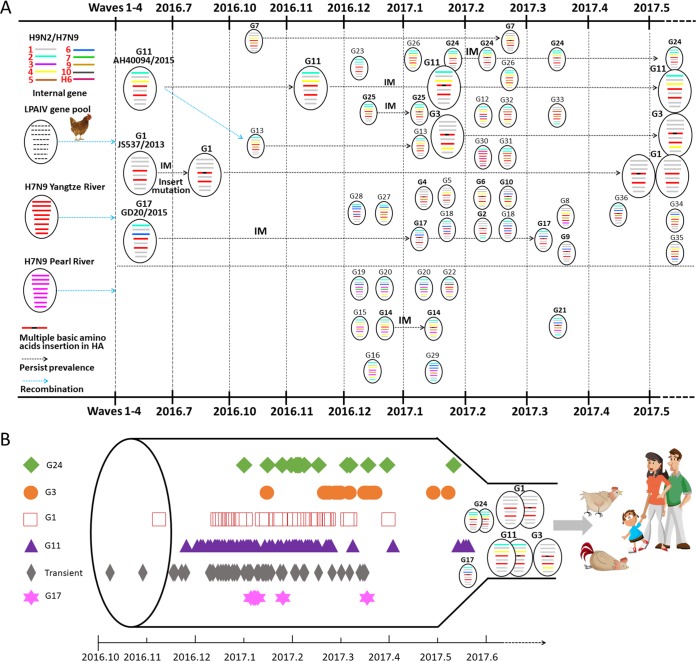
Phylogenetic analyses of the internal genes revealed high diversity compared to the HA and NA genes (Fig. 4; see also Fig. S1C to H) (Table 2; see also Table S1). After designating different H7N9 lineages for each internal gene, we classified the 240 isolates of wave 5 into 36 genotypes, 27 of which had emerged for the first time, suggesting continuing dynamic reassortment of H7N9 AIVs in wave 5 (Fig. 4A) (Table 2; see also Table S1). Nine genotypes, G1, G2, G3, G5, G9, G11, G17, G23, and G25, emerged before wave 5 and were still circulating in the influenza season current at the time of writing. In particular, G1 (n = 38), known as CK/JS/SC537/2013 (JS537) in a previous study, emerged in wave 1 and continued to circulate during wave 5. Similarly, G11 (n = 98), represented by A/AH/40094/2015 (AH40094), emerged in wave 3 and still circulating in wave 5. Additionally, the strains in G3 (n = 28, represented by CK/Zhongshan/ZS/2017) that were detected during wave 5 all belonged to HP-H7N9 and were widely spread in nine provinces or municipalities (Guangdong, Guangxi, Hunan, Fujian, Hebei, Beijing, Shandong, Shaanxi, and Inner Mongolia). These three genotypes caused the majority of human and poultry infections and seemed to be the major genotypes in wave 5 (Fig. 4B). In detail, 229 isolates from wave 5 and the Yangtze River Delta HA/NA lineage belonged to 29 genotypes, seven strains of the Pearl River Delta HA/NA lineage clustered into five genotypes (G15, G16, G19, G20, and G29), and the four hybrid HA/NA strains belonged to two genotypes (G14 and G21) (Table 2; see also Table S1).
FIG 4.
Schematic representation of the evolutionary pathway of the H7N9 viruses during wave 5. (A) The schematic evolutionary pathway of H7N9 in wave 5 on the basis of the timeline of the first and last detection for each genotype during wave 5. Three dominant genotypes (G1, G3, and G11) were expressed as bigger “virions.” The eight gene segments (shown as horizontal bars starting from top to bottom of the “virion”) are PB2, PB1, PA, HA, NP, NA, M, and NS. Different colors of gene segments represent different virus lineages. (B) The dominant genotypes, the genotypes that included more than eight strains, and the transient genotypes are listed along with their isolation times. Symbols represent the corresponding genotypes of the H7N9 viruses.
TABLE 2.
Genotypes of H7N9 viruses in wave 5
| Genotype | HA | NA | PB2 | PB1 | PA | NP | MP | NS | No. of strains |
First detection | Reference straina | Accession no. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LPAIV | HPAIV | ||||||||||||
| G1 | Yangtze | Yangtze | 1 | 1 | 1 | 1 | 1 | 1 | 29 | 9 | 16 April 2013 | CK/JS/SC537/2013 | EPI_ISL_142930 |
| G2 | Yangtze | Yangtze | 1 | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 1 March 2016 | CK/GD/DG16419/2016 | EPI_ISL_249139 |
| G3 | Yangtze | Yangtze | 1 | 1 | 1 | 1 | 4 | 1 | 0 | 28 | 26 February 2015 | A/GD-92/2015 | EPI_ISL_198746 |
| G4 | Yangtze | Yangtze | 1 | 1 | 1 | 9 | 1 | 1 | 0 | 1 | 15 January 2017 | A/CK/GuZ/HD621/2017 | EPI_ISL_248816 |
| G5 | Yangtze | Yangtze | 1 | 1 | 4 | 1 | 1 | 1 | 2 | 0 | 28 October 2014 | EN/JS/98337/2014 | EPI_ISL_192340 |
| G6 | Yangtze | Yangtze | 1 | 1 | 4 | 1 | 4 | 1 | 0 | 1 | 22 February 2017 | A/GD/17SF033/2017 | EPI_ISL_267759 |
| G7 | Yangtze | Yangtze | 1 | 1 | 4 | 9 | 1 | 1 | 1 | 1 | 7 October 2016 | A/ZJ/6/2016 | EPI_ISL_242866 |
| G8 | Yangtze | Yangtze | 1 | 1 | 4 | 9 | 3 | 1 | 1 | 0 | 3 March 2017 | A/GX/6/2017 | EPI_ISL_268525 |
| G9 | Yangtze | Yangtze | 1 | 1 | 6 | 1 | 1 | 1 | 0 | 1 | 31 December 2014 | A/JS/18828/2014 | EPI_ISL_192278 |
| G10 | Yangtze | Yangtze | 1 | 1 | 6 | 7 | 4 | 1 | 0 | 1 | 27 February 2017 | A/GX/18892/2017 | EPI_ISL_268502 |
| G11 | Yangtze | Yangtze | 2 | 1 | 4 | 1 | 1 | 1 | 97 | 1 | 23 May 2015 | A/AH/40094/2015 | EPI_ISL_192471 |
| G12 | Yangtze | Yangtze | 2 | 1 | 4 | 6 | 4 | 1 | 1 | 0 | 16 February 2017 | CK/JX/ShRGF219-O/2017 | EPI_ISL_259763 |
| G13 | Yangtze | Yangtze | 2 | 1 | 4 | 9 | 1 | 1 | 9 | 0 | 26 October 2016 | A/JS/60467/2016 | EPI_ISL_242873 |
| G14 | Yangtze | Pearl | 2 | 1 | 4 | 9 | 4 | 1 | 1 | 1 | 10 December 2016 | CK/GD/DGCPLB032/2016 | EPI_ISL_259747 |
| G15 | Pearl | Pearl | 2 | 1 | 4 | 9 | 4 | 1 | 1 | 0 | 13 December 2016 | A/GD/60061/2016 | EPI_ISL_242888 |
| G16 | Pearl | Pearl | 2 | 1 | 4 | 9 | 4 | 2 | 1 | 0 | 28 December 2016 | A/HK/VB16189623/2016 | EPI_ISL_240520 |
| G17 | Yangtze | Yangtze | 2 | 1 | 6 | 1 | 1 | 1 | 0 | 8 | 15 January 2015 | A/GD-20/2015 | EPI_ISL_198726 |
| G18 | Yangtze | Yangtze | 2 | 1 | 6 | 6 | 1 | 1 | 3 | 0 | 5 January 2017 | A/HK/125/2017 | EPI_ISL_259269 |
| G19 | Pearl | Pearl | 2 | 1 | 6 | 7 | 1 | 1 | 1 | 0 | 9 December 2016 | A/GD/60060/2016 | EPI_ISL_242871 |
| G20 | Pearl | Pearl | 2 | 1 | 6 | 7 | 4 | 1 | 3 | 0 | 28 December 2016 | A/GD/60923/2016 | EPI_ISL_242870 |
| G21 | Yangtze | Pearl | 2 | 1 | 6 | 9 | 1 | 2 | 0 | 2 | 12 March 2017 | A/GD/17SF064/2017 | EPI_ISL_267764 |
| G22 | Yangtze | Yangtze | 2 | 1 | 10 | 9 | 4 | 1 | 1 | 0 | 16 January 2017 | A/SZ/Th003/2017 | EPI_ISL_250313 |
| G23 | Yangtze | Yangtze | 2 | 2 | 1 | 1 | 1 | 1 | 3 | 0 | 13 January 2016 | A/FJ/1/2016 | EPI_ISL_233628 |
| G24 | Yangtze | Yangtze | 2 | 2 | 4 | 1 | 1 | 1 | 12 | 2 | 1 January 2017 | A/FJ/02151/2017 | EPI_ISL_242842 |
| G25 | Yangtze | Yangtze | 2 | 2 | 4 | 9 | 1 | 1 | 1 | 3 | 28 February 2016 | CK/GD/CZ9/2016 | EPI_ISL_249282 |
| G26 | Yangtze | Yangtze | 2 | 2 | 4 | 9 | 4 | 1 | 3 | 0 | 11 January 2017 | A/HuN/02287/2017 | EPI_ISL_242845 |
| G27 | Yangtze | Yangtze | 2 | 6 | 4 | 9 | 4 | 1 | 1 | 0 | 30 December 2016 | A/SZ/Th002/2016 | EPI_ISL_250425 |
| G28 | Yangtze | Yangtze | 2 | 6 | 6 | 6 | 1 | 1 | 1 | 0 | 19 December 2016 | A/HK/VB16184091/2016 | EPI_ISL_239994 |
| G29 | Pearl | Pearl | 2 | 6 | 6 | 9 | 1 | 2 | 1 | 0 | 1 January 2017 | CK/GD/JMS26/2017 | —b |
| G30 | Yangtze | Yangtze | 2 | H6 | H6 | H6 | 1 | H6 | 1 | 0 | 16 February 2017 | DK/JX/ShRSX002-O/2017 | EPI_ISL_259751 |
| G31 | Yangtze | Yangtze | 2 | 2 | 5 | 9 | 1 | 1 | 1 | 0 | 4 February 2017 | A/HuN/25360/2017 | EPI_ISL_285569 |
| G32 | Yangtze | Yangtze | 2 | 1 | 5 | 1 | 4 | 1 | 1 | 0 | 22 February 2017 | A/GZ/18980/2017 | EPI_ISL_285148 |
| G33 | Yangtze | Yangtze | 1 | 1 | 5 | 9 | 1 | 2 | 1 | 0 | 22 March 2017 | A/CQ-Yuzhong/1500/2017 | EPI_ISL_285024 |
| G34 | Yangtze | Yangtze | 1 | 6 | 4 | 9 | 1 | 1 | 1 | 0 | 17 June 2017 | A/SC/29764/2017 | EPI_ISL_285299 |
| G35 | Yangtze | Yangtze | 1 | 1 | 4 | 9 | 6 | 1 | 1 | 0 | 25 June 2017 | A/YN/32293/2017 | EPI_ISL_285307 |
| G36 | Yangtze | Yangtze | 2 | 1 | 6 | 9 | 1 | 1 | 0 | 1 | 11 April 2017 | A/HuN/25351/2017 | EPI_ISL_269517 |
AH, Anhui; CK, chicken; CQ, Chongqing; DK, duck; EN, environment; FJ, Fujian; GD, Guangdong; GuZ, Guangzhou; GX, Guangxi; GZ, Guizhou; HK, Hong Kong; HuN, Hunan; JS, Jiangsu; JX, Jiangxi; SC, Sichuan; SZ, Shenzhen; YN, Yunnan; ZJ, Zhenjiang.
—, not available.
The 61 HP-H7N9 strains with full-length genomes belonged to 15 genotypes (Fig. 4) (Table 2; see also Table S1); among them, G1, G7, G11, G14, G24, and G25 were also seen in LP-H7N9 AIVs and G2, G3, G4, G6, G9, G10, G17, G21, and G36 were found only in the HP strains that emerged in wave 5. Therefore, the internal genes of HP-H7N9 viruses also underwent a dynamic reassortment process, although G1 (n = 9, CK/Heyuan/16876/2016-like) and G3 (n = 28, CK/Zhongshan/ZS/2017-like) accounted for the ∼61% of the HP strains.
DISCUSSION
H7N9 AIVs have caused the largest number of human cases in wave 5. Similarly to previous waves, human infections were first identified in the Yangtze River Delta and Pearl River Delta regions and then were gradually identified in other regions (22). However, in contrast with previous waves, over 100 H7N9 human cases were reported in the regions around the two centers as early as December 2016. Additionally, over 220 human cases were reported during wave 5 in regions far from the two centers, some of which were important poultry breeding regions. Meanwhile, HP- and LP-H7N9 viruses were also identified in poultry from the south (Guangdong) to the north (Inner Mongolia and Heilongjiang) of China (Fig. 1; see also Table S1 in the supplemental material) (http://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza/2017/). These results indicated a much broader distribution of H7N9 viruses in China in wave 5, and the potential infection and transmission of both LP- and HP-H7N9 in poultry farms warrant further investigation.
The positivity rates for H7N9 were 15.7%, 6.5%, and 0.5% in LPMs from the Yangtze River Delta (Jiangsu, Shanghai and Zhejiang), the South Central region (Hubei, Hunan, Anhui and Fujian), and the Pearl River Delta (Guangdong), respectively, between November 2014 and April 2016 (24). In December 2016, the positivity rates for H7N9 were 15.8% and 9.4% in the LPMs of Jiangsu and Guangdong provinces, respectively (http://www.who.int/influenza/human_animal_interface/avian_influenza/riskassessment_AH7N9_201702/en/). These results indicated that the positivity rate of H7N9 in Jiangsu Province was consistent with previous observations from the start of wave 5, whereas a higher infection rate was seen in Guangdong Province than had been seen in previous waves (24) (http://www.who.int/influenza/human_animal_interface/avian_influenza/riskassessment_AH7N9_201702/en/). An increased positivity rate in provinces across China was probably one of the underlying reasons for the increased numbers of human infections in wave 5.
The emergence of HP-H7N9 undoubtedly poses a serious threat to both agriculture and public health. Although there was no evidence that HP-H7N9 was more virulent than LP-H7N9 for humans (20) or more transmissible as a consequence of enhanced infectivity of the respiratory epithelium in nasal passages (25), poultry infected by HP-H7N9 become sick and may shed high concentrations of viruses that would increase the risk of human infections. Human infections caused by HP-H7N9 AIVs have been identified in the Guangdong, Guangxi, Fujian, Hunan, Shaanxi, Hebei, Henan, and Yunnan provinces of mainland China and also in Taiwan (http://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en/) (26), but several additional provinces, such as Heilongjiang, Inner Mongolia, Shandong, Beijing, and Tianjin, have also reported HP-H7N9 outbreaks in poultry or detection in LPMs (Fig. 1; see also Table S1) (http://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza/2017/). The majority of the circulating H7N9 strains in wave 5 were LP, suggesting that LP-H7N9 AIVs are still dominant in China. However, it seems that HP-H7N9 has spread into the north (Inner Mongolia and Heilongjiang) from the south (Guangdong) of China, and continued transmission and cocirculation with LP-H7N9 in China warrant further investigation.
HP-H7N9 AIVs contain the characteristic insertion of multiple basic amino acids at the CS of the HA protein (20). At least four different motifs were identified at the CS of the HA protein, suggesting various levels of potential virulence of the HP-H7N9 strain with respect to poultry and mammals (1, 27, 28). In addition, apart from the insertion of multiple basic amino acids, several amino acid substitutions in HA have also been found only in the novel HP-H7N9 AIVs. Interestingly, almost all of the HP-H7N9 viruses have Q226 (H3 numbering) in HA, indicating preferential binding to avian virus-type receptors (8, 29). Therefore, the HP-H7N9 variant likely first emerged in poultry and has not yet adapted to human as well as LP-H7N9. However, two human-origin HP-H7N9 isolates possessed H226, which had not been found before, and thus the receptor binding properties associated with this mutation need further characterization. Furthermore, two human-derived strains and one poultry-derived HP-H7N9 strain also possessed L226 (Fig. 2), indicating the ability to bind to human-type receptors (30). All these mutations suggested that HP-H7N9 was adapting to humans, and genetic mutations associated with pathogenicity need to be closely monitored.
The functional balance between the HA and NA proteins has been previously described (31–36). Along with the occurrence of specific mutations in the HA protein, there are several residues that have been identified specifically in the NA protein of the HP-H7N9 strains, but the potential biological functions of these mutations require further studies. However, it should be noted that eight HP-H7N9 strains possessed an LP-H7N9-like NA, which is indicative of multiple origins.
The HA and NA genes of LP-H7N9 isolates shared the same origin and were relatively stable (37). Similarly to the HA gene of LP-H7N9 AIVs, the HP strains clustered together and shared a single origin Yangtze or Pearl River Delta lineage. However, the NA gene sequences of HP-H7N9 failed to cluster together. Alternatively, they fell within different parts of the Yangtze River Delta lineage, as well as within the Pearl River Delta lineage, suggesting that the NA gene might have also undergone a dynamic reassortment, which has not been seen in LP-H7N9.
The evolution of internal genes of LP-H7N9 was characteristic of a dynamic reassortment process (37–39). Our phylogenetic analyses showed that the LP-H7N9 AIVs were still undergoing dynamic reassortment in wave 5. The HP strains also underwent dynamic reassortment and recruited internal genes from the LPAIV reservoir, such as LP-H7N9/H9N2. On the basis of the previous classification method (37), the viruses in waves 1 to 5 were systematically analyzed. At least 36 genotypes were assembled via dynamic reassortment and were cocirculating in wave 5 (Fig. 4) (Table 2; see also Table S1). Three major genotypes (G1, G3, and G11) were identified. The G1 (JS537-like) viruses have been prevalent since wave 1 and may be ancestors for the HP variants emerging in wave 5 (Fig. 4), while the derived G3 genotype was dominant for HP strains. G11 (AH40094-like) viruses were also dominant and emerged as early as wave 3; however, only 1 of 98 G11 strains belonged to HP.
The R292K amino acid substitution in NA conferring NAI resistance was mainly found in clinics after treatments with NAIs (e.g., oseltamivir) (20, 40). Fourteen HP-H7N9 isolates in the present study possessed the R292K mutation. Notably, one was isolated from poultry (CK/GD/SZBJ0011-O/2017). Moreover, one NAI-resistant HP-H7N9 strain with an R292K mutation was reported that could transmit among ferrets (41). The E627K mutation in the PB2 protein is associated with increased replication efficiency and pathogenicity of AIV in mammalian hosts (12–14) and was seen in one environmental HP strain (EN/GD/SZBA-E1/2017). Furthermore, mammalian adaptation mutations A588V and K526R in PB2 were also found in the avian-origin isolates. These results suggest that there is increasing NAI resistance and adaptation of HP/LP-H7N9 to mammals.
In summary, we performed a systematic analysis of the genetic evolution of H7N9 AIVs in wave 5 compared to previous waves in China. Our results revealed a much broader distribution of H7N9 with high genetic diversity in China than previously known, which may account for the sharp increase in human infections with this virus. Although LP-H7N9 was still dominant in China, the rapid spread of and variations seen in HP-H7N9 are causes for concern. Intensive surveillance of H7N9 AIV in China is required to monitor the continuing dynamic reassortment of both LP- and HP-H7N9, as well as the spread of HP-H7N9 across China.
MATERIALS AND METHODS
H7N9 infection confirmation and whole-genome sequencing.
Oropharyngeal or sputum samples were collected from suspected human cases at Shenzhen Third People's Hospital and Yunnan Center for Disease Control and Prevention. Environmental samples and oropharyngeal and cloacal swabs from apparently healthy or diseased poultry in LPMs and poultry farms located close to the suspected patients were also collected. Viral RNA was extracted using a Magabio plus virus RNA purification kit (automatic nucleic acid purification system NPA-32+; Bioer, China). Clinical samples were detected by quantitative reverse transcription-PCR (qRT-PCR) using avian influenza H7N9 detection kits (Mabsky Bio-tech Co., Ltd. and Zhengzhou Zhongdao Biotechnology Co., Ltd.), following manufacturer instructions. Whole-viral-genome amplification and Sanger sequencing were performed using specific primers (see Table S2 in the supplemental material).
Geographical distribution analysis.
On the basis of data from the National Health and Family Planning Commission of the People's Republic of China (http://www.nhfpc.gov.cn/jkj/new_index.shtml), FluTrackers (https://flutrackers.com/forum/forum/china-h7n9-outbreak-tracking), and WHO (http://www.who.int/influenza/human_animal_interface/avian_influenza/archive/en/), human H7N9 cases in mainland China were analyzed by month since 2013. In addition to the H7N9-positive regions identified in the present study, all provinces and municipalities reporting human and poultry infections during wave 5 were systematically analyzed.
Genetic analysis.
The gene sequences were edited using DNASTAR (Version 5.01) and BioEdit (Version 7.0.5.2). Protein sequences of 35 new H7N9 viruses were obtained using DNASTAR (Version 5.01) and confirmed by the use of the Annotation Program from GenBank (https://www.ncbi.nlm.nih.gov/genomes/FLU/annotation/). Nucleotide and amino acid sequences for all 240 H7N9 viruses in wave 5, including 205 whole genomes downloaded from GISAID (http://platform.gisaid.org), and for 35 new isolates were aligned using DNAMAN (Version 6.0). Biologically important amino acid residue mutations associated with pathogenicity, replication, transmissibility, host adaptation, and drug resistance (12–14, 42–51) were inspected.
Phylogenetic and genotyping analysis.
Full-length H7N9 genome sequences from previous waves were also downloaded from GenBank and GISAID. For internal genes, the reference strains were retrieved via BLAST comparisons of the H7N9 gene sequences to those obtained from GISAID. For each gene, multiple-sequence alignment was performed using Muscle (52). Phylogenetic analysis was performed using RAxML (53). GTRGAMMA was applied as the nucleotide substitution model, and 1,000 bootstrap replicates were run. For phylogenetic trees of the internal genes, different lineages of H7N9 AIVs were classified if they had a common ancestor from no later than 2013. Generally, on the basis of the classification of different lineages of each internal gene and bootstrap values of >80%, the genotype of each H7N9 AIV from wave 2 to wave 5 was designated and genotypes from wave 5 were analyzed in detail.
Ethics statement.
The study was performed in accordance with guidelines approved by the Ethics Committees from Shenzhen Third People's Hospital (SZTHEC2016001) and Yunnan Center for Disease Control and Prevention Ethics Committee (YNCDC2017001).
Accession number(s).
Full-length genome sequences of the 35 H7N9 viruses from our study have been deposited in the GISAID EpiFlu database with accession numbers EPI_ISL_250311 to -17, EPI_ISL_250424 and -25, EPI_ISL_259747 to -63, EPI_ISL_266936, EPI_ISL_266938, and EPI_ISL_276781 to -87.
Supplementary Material
ACKNOWLEDGMENTS
We thank the data submitters from the GISAID and GenBank Flu databases for the AIV sequences.
This study was conceived and designed by Yuhai Bi, Weifeng Shi, and George F. Gao, who led the research groups on the genetic evolution and pathogenesis of pathogenic microbes. Chuansong Quan, Weifeng Shi, Yang Yang, Yongchun Yang, Xiaoqing Liu, Wen Xu, Hong Li, Juan Li, Qianli Wang, Zhou Tong, Cheng Zhang, Sufang Ma, Zhenghai Ma, Guanghua Fu, Yu Huang, Houhui Song, Liuqing Yang, Zewu Zhang, William J. Liu, Yingxia Liu, Wenjun Liu, and Yuhai Bi collected the samples. Chuansong Quan, Weifeng Shi, and Yang Yang amplified and sequenced the whole genome of the viruses. Yuhai Bi, Weifeng Shi, Chuansong Quan, and Gary Wong analyzed the data and completed the revision.
This work was supported by the National Key Research and Development Project of China (2016YFE0205800), National Science and Technology Major Project (2016ZX10004222 and 2018ZX10713001-010), Ministry of Science and Technology of China (MOST) 973 Project (2015CB910501), intramural special grants for influenza virus research from the Chinese Academy of Sciences (KJZD-EW-L15), Sanming Project of Medicine in Shenzhen (ZDSYS201504301534057), Shenzhen Science and Technology Research and Development Project (JCYJ20160427151920801 and JCYJ20151029151932602), National Natural Science Foundation of China (NSFC, 81470096), Science Foundation of Two Sides of Strait (U1305212), and External Cooperation Program of Chinese Academy of Sciences (153211KYSB20160001). George F. Gao is a leading principal investigator of the NSFC Innovative Research Group (81621091). Weifeng Shi is supported by the Taishan Scholars program of Shandong Province (ts201511056). Yuhai Bi is supported by the Youth Innovation Promotion Association of Chinese Academy of Sciences (CAS) (2017122).
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00301-18.
REFERENCES
- 1.Belser JA, Bridges CB, Katz JM, Tumpey TM. 2009. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg Infect Dis 15:859–865. doi: 10.3201/eid1506.090072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 3.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF. 2013. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 4.Gao HN, Lu HZ, Cao B, Du B, Shang H, Gan JH, Lu SH, Yang YD, Fang Q, Shen YZ, Xi XM, Gu Q, Zhou XM, Qu HP, Yan Z, Li FM, Zhao W, Gao ZC, Wang GF, Ruan LX, Wang WH, Ye J, Cao HF, Li XW, Zhang WH, Fang XC, He J, Liang WF, Xie J, Zeng M, Wu XZ, Li J, Xia Q, Jin ZC, Chen Q, Tang C, Zhang ZY, Hou BM, Feng ZX, Sheng JF, Zhong NS, Li LJ. 2013. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med 368:2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- 5.Cowling BJ, Jin L, Lau EH, Liao Q, Wu P, Jiang H, Tsang TK, Zheng J, Fang VJ, Chang Z, Ni MY, Zhang Q, Ip DK, Yu J, Li Y, Wang L, Tu W, Meng L, Wu JT, Luo H, Li Q, Shu Y, Li Z, Feng Z, Yang W, Wang Y, Leung GM, Yu H. 2013. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet 382:129–137. doi: 10.1016/S0140-6736(13)61171-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Xiao H, Wu Y, Liu D, Qi X, Shi Y, Gao GF. 2014. H7N9: a low pathogenic avian influenza A virus infecting humans. Curr Opin Virol 5:91–97. doi: 10.1016/j.coviro.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao GF, Wu Y. 2013. Haunted with and hunting for viruses. Sci China Life Sci 56:675–677. doi: 10.1007/s11427-013-4525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z, Qin C, Sun H, Liu J, Haywood J, Liu W, Gong W, Wang D, Shu Y, Wang Y, Yan J, Gao GF. 2013. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342:243–247. doi: 10.1126/science.1242917. [DOI] [PubMed] [Google Scholar]
- 9.Xiong X, Martin SR, Haire LF, Wharton SA, Daniels RS, Bennett MS, McCauley JW, Collins PJ, Walker PA, Skehel JJ, Gamblin SJ. 2013. Receptor binding by an H7N9 influenza virus from humans. Nature 499:496–499. doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- 10.Xu R, de Vries RP, Zhu X, Nycholat CM, McBride R, Yu W, Paulson JC, Wilson IA. 2013. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science 342:1230–1235. doi: 10.1126/science.1243761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J. 2013. Structural analysis of the hemagglutinin from the recent 2013 H7N9 influenza virus. J Virol 87:12433–12446. doi: 10.1128/JVI.01854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song W, Wang P, Mok BW, Lau SY, Huang X, Wu WL, Zheng M, Wen X, Yang S, Chen Y, Li L, Yuen KY, Chen H. 2014. The K526R substitution in viral protein PB2 enhances the effects of E627K on influenza virus replication. Nat Commun 5:5509. doi: 10.1038/ncomms6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mok CK, Lee HH, Lestra M, Nicholls JM, Chan MC, Sia SF, Zhu H, Poon LL, Guan Y, Peiris JS. 2014. Amino acid substitutions in polymerase basic protein 2 gene contribute to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J Virol 88:3568–3576. doi: 10.1128/JVI.02740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi Y, Xie Q, Zhang S, Li Y, Xiao H, Jin T, Zheng W, Li J, Jia X, Sun L, Liu J, Qin C, Gao GF, Liu W. 2015. Assessment of the internal genes of influenza A (H7N9) virus contributing to high pathogenicity in mice. J Virol 89:2–13. doi: 10.1128/JVI.02390-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao GF. 2014. Influenza and the live poultry trade. Science 344:235. doi: 10.1126/science.1254664. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Yu X, Pu X, Xie L, Sun Y, Xiao H, Wang F, Din H, Wu Y, Liu D, Zhao G, Liu J, Pan J. 2013. Environmental connections of novel avian-origin H7N9 influenza virus infection and virus adaptation to the human. Sci China Life Sci 56:485–492. doi: 10.1007/s11427-013-4491-3. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Jiang H, Wu P, Uyeki TM, Feng L, Lai S, Wang L, Huo X, Xu K, Chen E, He J, Kang M, Zhang R, Zhang J, Wu J, Hu S, Zhang H, Liu X, Fu W, Ou J, Wu S, Qin Y, Zhang Z, Shi Y, Artois J, Fang VJ, Zhu H, Guan Y, Gilbert M, Horby PW, Leung GM, Gao GF, Cowling BJ, Yu H. 2017. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013–17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis 17:822–832. doi: 10.1016/S1473-3099(17)30323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu H, Wu JT, Cowling BJ, Liao Q, Fang VJ, Zhou S, Wu P, Zhou H, Lau EH, Guo D, Ni MY, Peng Z, Feng L, Jiang H, Luo H, Li Q, Feng Z, Wang Y, Yang W, Leung GM. 2014. Effect of closure of live poultry markets on poultry-to-person transmission of avian influenza A H7N9 virus: an ecological study. Lancet 383:541–548. doi: 10.1016/S0140-6736(13)61904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin YP, Yang ZF, Liang Y, Li ZT, Bond HS, Chua H, Luo YS, Chen Y, Chen TT, Guan WD, Lai JCC, Siu YL, Pan SH, Peiris JSM, Cowling BJ, PunMok CK. 2016. Population seroprevalence of antibody to influenza A(H7N9) virus, Guangzhou, China. BMC Infect Dis 16:632. doi: 10.1186/s12879-016-1983-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang F, Bi Y, Wang J, Wong G, Shi W, Hu F, Yang Y, Yang L, Deng X, Jiang S, He X, Liu Y, Yin C, Zhong N, Gao GF. 2017. Human infections with recently-emerging highly pathogenic H7N9 avian influenza virus in China. J Infect 75:71–75. doi: 10.1016/j.jinf.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Qi W, Jia W, Liu D, Li J, Bi Y, Xie S, Li B, Hu T, Du Y, Xing L, Zhang J, Zhang F, Wei X, Eden JS, Li H, Tian H, Li W, Su G, Lao G, Xu C, Xu B, Liu W, Zhang G, Ren T, Holmes EC, Cui J, Shi W, Gao GF, Liao M. 15 January 2018. Emergence and adaptation of a novel highly pathogenic H7N9 influenza virus in birds and humans from a 2013 human-infecting low-pathogenic ancestor. J Virol doi: 10.1128/JVI.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, Yang L, Zhu W, Zhang Y, Zou S, Bo H, Gao R, Dong J, Huang W, Guo J, Li Z, Zhao X, Li X, Xin L, Zhou J, Chen T, Dong L, Wei H, Liu L, Tang J, Lan Y, Yang J, Shu Y. 2016. Two outbreak sources of influenza A (H7N9) viruses have been established in China. J Virol 90:5561–5573. doi: 10.1128/JVI.03173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bi Y, Shi W, Chen J, Chen Q, Ma Z, Wong G, Tian W, Yin R, Fu G, Yang Y, Liu WJ, Quan C, Wang Q, He S, Li X, Xia Q, Wang L, Pan Z, Li L, Li H, Xu W, Luo Y, Zeng H, Dai L, Xiao H, Sharshov K, Shestopalov A, Shi Y, Yan J, Li X, Liu Y, Lei F, Liu W, Gao GF. 2017. CASCIRE surveillance network and work on avian influenza viruses. Sci China Life Sci 60:1386–1391. doi: 10.1007/s11427-017-9251-2. [DOI] [PubMed] [Google Scholar]
- 24.Bi Y, Chen Q, Wang Q, Chen J, Jin T, Wong G, Quan C, Liu J, Wu J, Yin R, Zhao L, Li M, Ding Z, Zou R, Xu W, Li H, Wang H, Tian K, Fu G, Huang Y, Shestopalov A, Li S, Xu B, Yu H, Luo T, Lu L, Xu X, Luo Y, Liu Y, Shi W, Liu D, Gao GF. 2016. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe 20:810–821. doi: 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 25.Meliopoulos VA, Karlsson EA, Kercher L, Cline T, Freiden P, Duan S, Vogel P, Webby RJ, Guan Y, Peiris M, Thomas PG, Schultz-Cherry S. 2014. Human H7N9 and H5N1 influenza viruses differ in induction of cytokines and tissue tropism. J Virol 88:12982–12991. doi: 10.1128/JVI.01571-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, Zhu W, Li X, Chen M, Wu J, Yu P, Qi S, Huang Y, Shi W, Dong J, Zhao X, Huang W, Li Z, Zeng X, Bo H, Chen T, Chen W, Liu J, Zhang Y, Liang Z, Shi W, Shu Y, Wang D. 1 December 2017. Genesis and spread of newly emerged highly pathogenic H7N9 avian viruses in mainland China. J Virol doi: 10.1128/JVI.01277-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawaoka Y, Webster RG. 1988. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc Natl Acad Sci U S A 85:324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Sun Y, Sun H, Pu J, Bi Y, Shi Y, Lu X, Li J, Zhu Q, Gao GF, Yang H, Liu J. 2012. A single amino acid at the hemagglutinin cleavage site contributes to the pathogenicity and neurovirulence of H5N1 influenza virus in mice. J Virol 86:6924–6931. doi: 10.1128/JVI.07142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu X, Shi Y, Zhang W, Zhang Y, Qi J, Gao GF. 2013. Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut). Protein Cell 4:502–511. doi: 10.1007/s13238-013-3906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi J, Deng G, Kong H, Gu C, Ma S, Yin X, Zeng X, Cui P, Chen Y, Yang H. 2017. H7N9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell Res 12:1409–1421. doi: 10.1038/cr.2017.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, Kawaoka Y. 2000. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J Virol 74:6015–6020. doi: 10.1128/JVI.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu R, Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, Wilson IA. 2012. Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. J Virol 86:9221–9232. doi: 10.1128/JVI.00697-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casalegno JS, Ferraris O, Escuret V, Bouscambert M, Bergeron C, Lines L, Excoffier T, Valette M, Frobert E, Pillet S, Pozzetto B, Lina B, Ottmann M. 2014. Functional balance between the hemagglutinin and neuraminidase of influenza A(H1N1)pdm09 HA D222 variants. PLoS One 9:e104009. doi: 10.1371/journal.pone.0104009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gen F, Yamada S, Kato K, Akashi H, Kawaoka Y, Horimoto T. 2013. Attenuation of an influenza A virus due to alteration of its hemagglutinin-neuraminidase functional balance in mice. Arch Virol 158:1003–1011. doi: 10.1007/s00705-012-1577-3. [DOI] [PubMed] [Google Scholar]
- 35.Yen HL, Liang CH, Wu CY, Forrest HL, Ferguson A, Choy KT, Jones J, Wong DD, Cheung PP, Hsu CH, Li OT, Yuen KM, Chan RW, Poon LL, Chan MC, Nicholls JM, Krauss S, Wong CH, Guan Y, Webster RG, Webby RJ, Peiris M. 2011. Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc Natl Acad Sci U S A 108:14264–14269. doi: 10.1073/pnas.1111000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W, Qi J, Shi Y, Li Q, Gao F, Sun Y, Lu X, Lu Q, Vavricka CJ, Liu D, Yan J, Gao GF. 2010. Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus. Protein Cell 1:459–467. doi: 10.1007/s13238-010-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui LB, Liu D, Shi WF, Pan JC, Qi X, Li XB, Guo XL, Zhou MH, Li W, Li J, Haywood J, Xiao HX, Yu XF, Pu XY, Wu Y, Yu HY, Zhao KC, Zhu YF, Wu B, Jin T, Shi ZY, Tang FY, Zhu FC, Sun QL, Wu LH, Yang RF, Yan JH, Lei FM, Zhu BL, Liu WJ, Ma JC, Wang H, Gao GF. 2014. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun 5:3142. doi: 10.1038/ncomms4142. [DOI] [PubMed] [Google Scholar]
- 38.Lam TT, Zhou B, Wang J, Chai Y, Shen Y, Chen X, Ma C, Hong W, Chen Y, Zhang Y, Duan L, Chen P, Jiang J, Li L, Poon LL, Webby RJ, Smith DK, Leung GM, Peiris JS, Holmes EC, Guan Y, Zhu H. 2015. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature 522:102–105. doi: 10.1038/nature14348. [DOI] [PubMed] [Google Scholar]
- 39.Wang D, Yang L, Gao R, Zhang X, Tan Y, Wu A, Zhu W, Zhou J, Zou S, Li X, Sun Y, Zhang Y, Liu Y, Liu T, Xiong Y, Xu J, Chen L, Weng Y, Qi X, Guo J, Dong J, Huang W, Dong L, Zhao X, Liu L, Lu J, Lan Y, Wei H, Xin L, Chen Y, Xu C, Chen T, Zhu Y, Jiang T, Feng Z, Yang W, Wang Y, Zhu H, Guan Y, Gao GF, Li D, Han J, Wang S, Wu G, Shu Y. 2014. Genetic tuning of the novel avian influenza A(H7N9) virus during interspecies transmission, China, 2013. Euro Surveill 19:20836. doi: 10.2807/1560-7917.ES2014.19.25.20836. [DOI] [PubMed] [Google Scholar]
- 40.Wu Y, Bi Y, Vavricka CJ, Sun X, Zhang Y, Gao F, Zhao M, Xiao H, Qin C, He J, Liu W, Yan J, Qi J, Gao GF. 2013. Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses. Cell Res 23:1347–1355. doi: 10.1038/cr.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imai M, Watanabe T, Kiso M, Nakajima N, Yamayoshi S, Iwatsuki-Horimoto K, Hatta M, Yamada S, Ito M, Sakai-Tagawa Y, Shirakura M, Takashita E, Fujisaki S, McBride R, Thompson AJ, Takahashi K, Maemura T, Mitake H, Chiba S, Zhong G, Fan S, Oishi K, Yasuhara A, Takada K, Nakao T, Fukuyama S, Yamashita M, Lopes TJS, Neumann G, Odagiri T, Watanabe S, Shu Y, Paulson JC, Hasegawa H, Kawaoka Y. 2017. A highly pathogenic avian H7N9 influenza virus isolated from a human is lethal in some ferrets infected via respiratory droplets. Cell Host Microbe 22:615–626 e618. doi: 10.1016/j.chom.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao C, Ma W, Sun N, Huang L, Li Y, Zeng Z, Wen Y, Zhang Z, Li H, Li Q, Yu Y, Zheng Y, Liu S, Hu P, Zhang X, Ning Z, Qi W, Liao M. 2016. PB2-588 V promotes the mammalian adaptation of H10N8, H7N9 and H9N2 avian influenza viruses. Sci Rep 6:19474. doi: 10.1038/srep19474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naffakh N, Tomoiu A, Rameix-Welti MA, van der Werf S. 2008. Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu Rev Microbiol 62:403–424. doi: 10.1146/annurev.micro.62.081307.162746. [DOI] [PubMed] [Google Scholar]
- 44.Chen GW, Chang SC, Mok CK, Lo YL, Kung YN, Huang JH, Shih YH, Wang JY, Chiang C, Chen CJ, Shih SR. 2006. Genomic signatures of human versus avian influenza A viruses. Emerg Infect Dis 12:1353–1360. doi: 10.3201/eid1209.060276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinhauer DA. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 46.Jiao P, Tian G, Li Y, Deng G, Jiang Y, Liu C, Liu W, Bu Z, Kawaoka Y, Chen H. 2008. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol 82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tada T, Suzuki K, Sakurai Y, Kubo M, Okada H, Itoh T, Tsukamoto K. 2011. Emergence of avian influenza viruses with enhanced transcription activity by a single amino acid substitution in the nucleoprotein during replication in chicken brains. J Virol 85:10354–10363. doi: 10.1128/JVI.00605-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song MS, Pascua PN, Lee JH, Baek YH, Lee OJ, Kim CJ, Kim H, Webby RJ, Webster RG, Choi YK. 2009. The polymerase acidic protein gene of influenza a virus contributes to pathogenicity in a mouse model. J Virol 83:12325–12335. doi: 10.1128/JVI.01373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zamarin D, García-Sastre A, Xiao X, Wang R, Palese P. 2005. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PloS Pathog 1:e4. doi: 10.1371/journal.ppat.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ping J, Dankar SK, Forbes NE, Keleta L, Zhou Y, Tyler S, Brown EG. 2010. PB2 and hemagglutinin mutations are major determinants of host range and virulence in mouse-adapted influenza A virus. J Virol 84:10606–10618. doi: 10.1128/JVI.01187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mok CK, Yen HL, Yu MY, Yuen KM, Sia SF, Chan MC, Qin G, Tu WW, Peiris JS. 2011. Amino acid residues 253 and 591 of the PB2 protein of avian influenza virus A H9N2 contribute to mammalian pathogenesis. J Virol 85:9641–9645. doi: 10.1128/JVI.00702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.