ABSTRACT
Natural killer (NK) cells play an important role in the host response against viral infections and cancer development. They are able to kill virus-infected and tumor cells, and they produce different important cytokines that stimulate the antiviral and antitumor adaptive immune response, particularly interferon gamma. NK cells are of particular importance in herpesvirus infections, which is illustrated by systemic and life-threatening herpesvirus disease symptoms in patients with deficiencies in NK cell activity and by the myriad of reports describing herpesvirus NK cell evasion strategies. The latter is particularly obvious for cytomegaloviruses, but increasing evidence indicates that most, if not all, members of the herpesvirus family suppress NK cell activity to some extent. This review discusses the different NK cell evasion strategies described for herpesviruses and how this knowledge may translate to clinical applications.
KEYWORDS: EBV, KSHV, NK cell, cytomegalovirus, evasion, herpesvirus, herpes simplex virus, natural killer cells, pseudorabies virus
NK CELLS
In the growing research field of innate lymphoid cells (ILCs), natural killer (NK) cells, belonging to the group I ILCs, are the best characterized population (1). They may represent up to 15% of peripheral blood lymphocytes, are present in various tissues and lymphoid organs, and can be efficiently recruited to sites of viral infection (1–3). NK cells display a variety of surface receptors capable of triggering or inhibiting their function (i.e., cytotoxicity and cytokine release). Most of the activating receptors recognize ligands which can be induced by cell stress or activation and can therefore be expressed or upregulated in virally infected or tumor cells. These receptors include NKG2D, the natural cytotoxicity receptors (NCRs) (e.g., NKp30, NKp44, and NKp46), and DNAX accessory molecule 1 (DNAM-1). On the other hand, inhibitory receptors typically recognize major histocompatibility complex (MHC) class I molecules, which are expressed by almost all “normal” cells. In humans, the human leukocyte antigen class I (HLA-I)-specific inhibitory receptors are represented by the CD94/NKG2A heterodimeric receptor and a number of killer immunoglobulin-like receptors (KIRs). NK cells also express the activating counterparts of HLA-I-specific receptors (i.e., activating KIRs and CD94/NKG2C). Table 1 gives an overview of some of the better-characterized NK cell receptors and of their known ligands. KIRs, NKG2A, and NKG2C recognize unique patterns of HLA-I alleles and are clonally distributed within the individual cell population, thus contributing to the “diversity” of the NK cell repertoire. How this repertoire is generated and regulated during the individual life span has not been completely understood. Nevertheless, there is evidence suggesting that viral infections, particularly some herpesvirus infections, can take part in the process (4). In humans, human cytomegalovirus (HCMV) infection can induce NKG2C+ NK cell expansion (5), while in mice, murine cytomegalovirus (MCMV) infections have been shown to promote the long-lasting expansion of virus-responsive NK cells expressing the MHC-specific Ly49H receptor (6). These observations, together with the recent findings on the epigenetics of NK cell diversification (7), suggest that NK cells may also display some traits common to “adaptive” or “memory-like” cells, at least for specific NK cell subsets, and indicate a certain level of functional plasticity within the NK cell population. This aspect of NK cell biology has also been suggested by previous studies showing that NK cells could be “educated” to increase their cytotoxic potential during their terminal differentiation stages or to adapt to environmental changes (8).
TABLE 1.
Overview of a selection of activating and inhibitory NK cell receptors in humans and/or mice and their corresponding ligandsa
| Receptor type | Human |
Mouse |
||
|---|---|---|---|---|
| Receptor | Ligand(s) | Receptor | Ligand(s) | |
| Activating | DNAM-1 (CD226) | PVR (Necl5, CD155), nectin 2 (CD112) | DNAM-1 (CD226) | PVR (Necl5, CD155), nectin 2 (CD112) |
| CRTAM (CD355) | Necl-2 | CRTAM (CD355) | Necl-2 | |
| NKG2D | MICA/B, ULBP1 to -6 | NKG2D | Rae-1, MULT-1, H60 | |
| NKp46 (CD335) | Viral HA, and HN, CFP (properdin) | NKp46 | Viral HA, CFP (properdin) | |
| NKp44 (CD336) | Viral HA and HN, PCNA,b MLL5, PDGF-DD | |||
| NKp30 (CD337) | B7-H6, BAT3, HCMV pp65c | |||
| CD16 | Fc portions of IgG | CD16 | Fc portions of IgG | |
| 2B4 (CD244) | CD48 | 2B4 (CD244) | CD48 | |
| Activating KIR | HLA-A11,-Bw4, -C(Asn77-Lys80) | |||
| NKp80 | AICL1 | |||
| NKp65 | KACL | |||
| CD94-NKG2C/E/H | HLA-E (for CD94-NKG2C) | KLRD1-KLRC2/3 (CD94-NKG2C/E) | Qa-1(b) | |
| PILR beta | CD99 | |||
| Tactile (CD96) | CD155 (Necl5, CD155) | Tactile (CD96) | PVR (Necl5, CD155), nectin 1 (CD111) | |
| Inhibitory | Inhibitory KIR | MHC class I polymorphisms | Inhibitory Ly49 | MHC class I |
| NKR-P1A | LLT1 | |||
| NKR-P1B | Clr-b | |||
| CD94-NKG2A/B | HLA-E | KLRD1-KLRC1 (CD94-NKG2A) | Qa-1(b) | |
| TIGIT | PVR (Necl5, CD155), nectin 2 (CD112) | TIGIT | PVR (Necl5, CD155), nectin 2 (CD112) | |
| LIR-1 (ILT-2/CD85j/LILRB1) | HLA (α3), HCMV UL18 | |||
| IRp60 (CD300a) | PS, PE | CD300a | PS, PE | |
| CEACAM1 (CD66) | CEACAM1 (CD66), TIM-3 (HAVCR2) | CEACAM1 | CEACAM1, TIM-3 (HAVCR2) | |
Abbreviations: DNAM-1, DNAX accessory molecule 1; PVR, poliovirus receptor; Necl, nectin-like molecules; CRTAM, class I-restricted T cell-associated molecule; MHC, major histocompatibility complex; MIC, MHC class I chain-related protein; ULBP, UL16 binding proteins; Rae-1, retinoic acid early inducible 1; MULT-1, mouse UL-16-binding protein-like transcript 1; H60, minor histocompatibility protein 60; HCMV, human cytomegalovirus; BAT3, human leukocyte antigen (HLA)-B-associated transcript 3; AICL, activation-induced C-type lectin; KACL, keratinocyte-associated C-type lectin; PILR, paired immunoglobulin-like type 2 receptor; KIR, killer-cell immunoglobulin-like receptor; CRTAM, class I-restricted T cell-associated molecule; HA, hemagglutinin; HN, hemagglutinin-neuraminidase; IgG, immunoglobulin G; LLT1, lectin-like transcript 1; TIGIT,T cell immunoglobulin and immunoreceptor tyrosine-based activation motif (ITIM) domain; PS, phosphatidylserine; PE, phosphatidylethanolamine; CEACAM, carcinoembryonic antigen-related cell adhesion molecules; CFP, complement factor P; PDGF, platelet-derived growth factor; IRp60, inhibitory receptor protein 60; TIM-3, T cell immunoglobulin domain and mucin domain 3; PCNA, proliferating cell nuclear antigen; MLL5, mixed lineage leukemia 5; LIR-1, leukocyte immunoglobulin-like receptor 1; HAVCR2, hepatitis A virus cellular receptor 2; LILRB1, leukocyte immunoglobulin-like receptor, subfamily B member 1; ILT-2, immunoglobulin-like transcript 2.
Triggers inhibitory signaling via NKp44.
Interferes with the activating signaling of NKp30.
Irrespective of functional plasticity, NK cell-mediated recognition and killing of a virus-infected cell is determined by the type and quantity of NK cell receptors engaged by their respective ligands during NK/target cell interaction. The resulting balance in activating/inhibitory signals will dictate whether the NK cell will activate its cytolytic program, resulting in the degranulation of secretory lysosomes (which contain perforins and granzymes) and ultimately leading to the damage and apoptotic death of the target cell. In certain body compartments, including the liver, NK cells can also express FAS-L and tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) and induce apoptosis by triggering death-inducing signaling receptors expressed by target cells. In addition to their cytolytic function, activated NK cells are also an important source of cytokines, particularly interferon gamma (IFN-γ), thereby steering the adaptive immune response toward intracellular infectious agents (e.g., toward a Th1 profile).
Many viruses, including herpesviruses, cause a reduced cell surface expression of MHC class I (i.e., ligands for inhibitory NK cell receptors) in an attempt to lower recognition and elimination of infected cells by CD8+ cytotoxic T lymphocytes (9). In addition, virus infection can trigger cellular stress responses (e.g., the DNA damage response), resulting in the upregulation of stress-related molecules, including different ligands for the activating NK cell receptors NKG2D and DNAM-1. Such virus-induced changes in the NK cell receptor ligand expression render infected cells susceptible targets for NK cells. As a consequence, many viruses have evolved different mechanisms to counteract the NK cell-mediated control of infection.
IMPORTANCE OF NK CELLS IN HERPESVIRUS CONTROL
Although several substantial subfamily-specific differences in genetic content and pathogenesis for the three different Herpesviridae subfamilies (Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae) have been described, a common feature of all herpesviruses is their ability to develop a complex and delicate balance with the immune system of their host, allowing these viruses to establish lifelong infections that are typically not life-threatening in immunocompetent hosts. Not surprisingly, disturbing this subtle balance may have profound consequences. This is particularly the case in patients who suffer from deficiencies in the innate immune response, particularly the type I interferon system and NK cells (10, 11).
Members of all three herpesvirus subfamilies may in fact represent some of the most important viral targets of NK cell activity (11). As a result, patients with different types of NK cell deficiencies have been diagnosed with aggravated, life-threatening, and sometimes fatal infections with herpesviruses of all three subfamilies, including the alphaherpesviruses herpes simplex virus 1 (HSV-1), HSV-2, and varicella-zoster virus (VZV), the betaherpesvirus HCMV, and the gammaherpesvirus Epstein-Barr virus (EBV) (3, 11–13). The pivotal role of NK cells in the herpesvirus-host balance is also exemplified by the numerous evasion mechanisms that herpesviruses have evolved to slow down and suppress the activity of and elimination by NK cells.
HERPESVIRUS NK CELL EVASION STRATEGIES
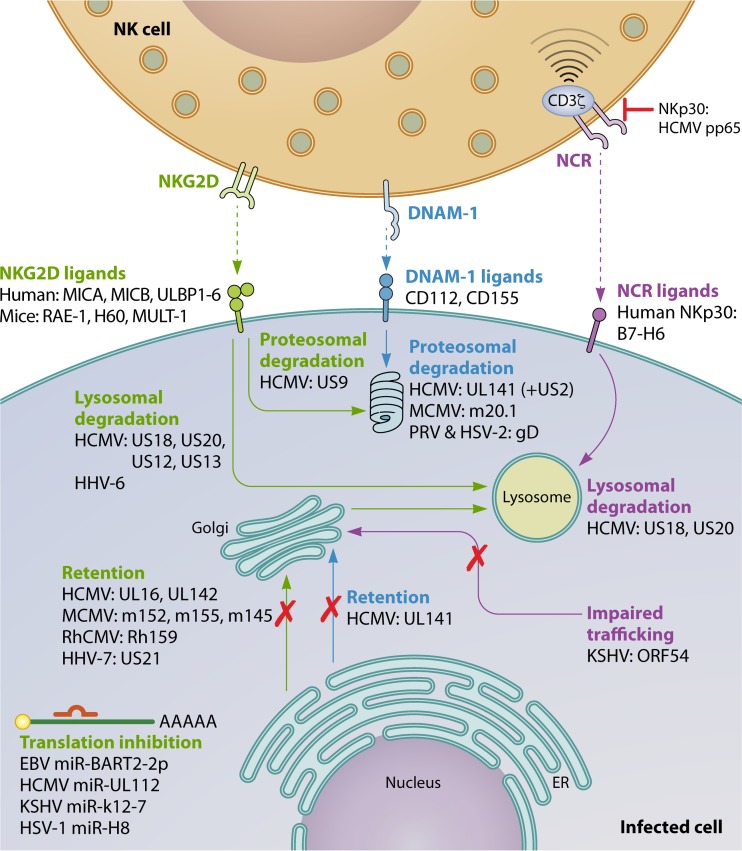
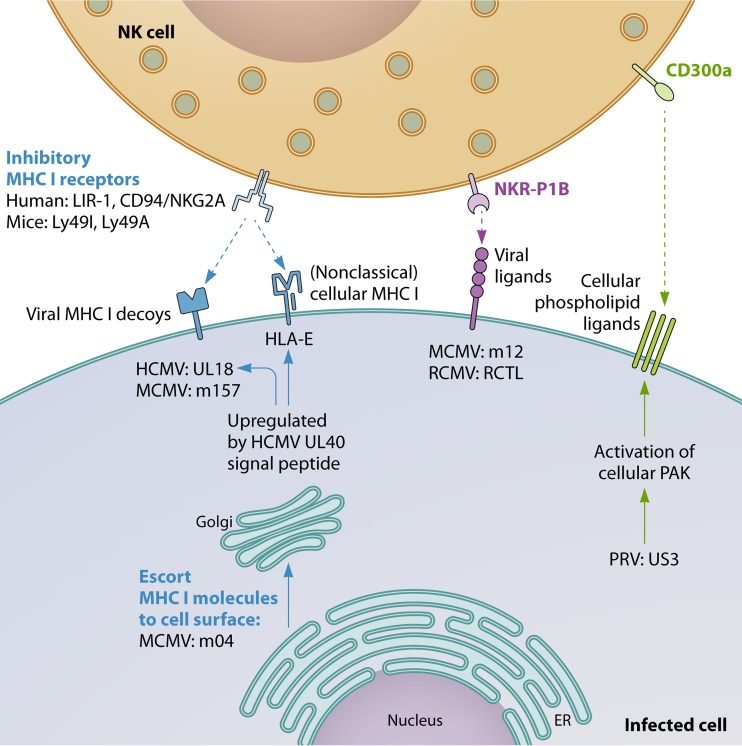
A plethora of different herpesvirus NK cell evasion strategies have been described. Most of these strategies consist of mechanisms to suppress signaling by activating NK cell receptors or mechanisms to trigger inhibitory NK cell signaling. Figures 1 and 2 give an overview of some of the most important herpesvirus NK evasion strategies which involve interference with activating NK signaling (Fig. 1) or increased/retained inhibitory NK cell signaling (Fig. 2). These and other strategies are explained in more detail below.
FIG 1.
Herpesvirus interference with activating NK cell receptors. These and other mechanisms are explained in more detail in the text.
FIG 2.
Herpesvirus modulation of inhibitory NK cell signaling. These and other mechanisms are explained in more detail in the text.
Suppression of NK cell-activating signals. (i) NKG2D.
NKG2D (CD226) is an important immune player to detect danger and is a potent activating receptor. Upregulation of NKG2D ligands may be triggered by the DNA damage response and occurs in transformed cells and virus-infected cells. Engagement of NKG2D on NK cells, but also on cytotoxic T cells, can trigger cytolytic activity (14–16).
The overwhelming amount of data on NKG2D evasion by herpesviruses (and other viruses) indicates that this activating NK cell receptor exerts a particularly strong pressure on these pathogens. Indeed, herpesviruses of all three subfamilies have been reported to encode strategies to suppress NKG2D-mediated recognition of infected cells. As with most NK cell evasion strategies, this is best documented for cytomegaloviruses. Cytomegaloviruses have very large genomes: HCMV has the largest genome of any known human virus, at 236 kbp in size. Consequently, cytomegalovirus genomes encode many proteins that are not essential for replication in vitro but that play an important role in host invasion and persistence, e.g., via interference with the host immune response.
In mice, three ligands for NKG2D have been identified: retinoic acid early inducible 1 (RAE-1), minor histocompatibility protein 60 (H60), and mouse UL-16-binding protein-like transcript 1 (MULT-1). MCMV infection leads to an increased transcription of NKG2D ligands (17, 18), which would be expected to result in potent NKG2D-mediated NK cell reactivity. However, this is strongly counteracted by several MCMV evasion mechanisms. Most of these evasion strategies consist of viral protein-mediated retention of newly synthesized NKG2D ligands in the endoplasmic reticulum (ER)/Golgi apparatus, although triggering of endocytosis of NKG2D ligands has also been described (e.g., RAE-1ε by m138 [see below]).
MCMV glycoprotein gp40 (encoded by m152), which was reported before to downregulate MHC class I and therefore could lead to increased NK cell-mediated attack (19), was found in fact to suppress NK cell activation via interference with NKG2D binding (20). Initially, gp40 was thought to downregulate the NKG2D ligand H60, but subsequent research showed that gp40 prevents cell surface expression of another murine NKG2D ligand, RAE-1 (20, 21). Later, gp40 was found to interact in a pincer-like manner with two sites on the α1 and α2 helices of RAE-1, much like the physiological interaction of NKG2D with RAE-1 (22).
Although m152/gp40 does not affect H60, MCMV also interferes with cell surface expression of this NKG2D ligand, via its m155 glycoprotein. The genes that code for m155 and m152/gp40 both belong to the m145 gene family, which encodes MHC class I like-glycoproteins with limited sequence similarity (±20% amino acid identity) (23–25).
In addition to interfering with cell surface expression of RAE-1 and H60, MCMV also suppresses surface expression of MULT-1, via its m145-encoded glycoprotein, again belonging to the same family (17).
Another MCMV protein, which does not belong to the m145 family, also interferes with the cell surface expression of the murine NKG2D ligands. Indeed, MCMV m138, a viral Fc receptor that binds and thereby inactivates the Fc domain of immunoglobulin G (see below), has been reported to downregulate MULT-1, H60, and a specific variant of RAE-1, RAE-1ε (26, 27).
All these evasion strategies are important for MCMV pathogenesis, as mutants with mutations in any of these NKG2D-interfering viral genes show reduced virulence that can be restored upon NK cell depletion and/or NKG2D blocking (17, 20, 23, 24).
It is also worth noting that MCMV-mediated evasion of NKG2D may depend not only on viral proteins but also on viral microRNA (miRNA), although there is currently no direct evidence supporting this possibility. However, replication of a mutant MCMV lacking two viral miRNAs, miR-M23-2 and miR-m21-1, was selectively reduced in salivary glands, an important organ for viral persistence and transmission, which could be restored by combined depletion of NK cells and CD4+ T cells (28). Hence, although the underlying mechanism for this observation is unclear at this stage, it is possible that like HCMV (see below), MCMV also encodes miRNAs that interfere with NK cell activity and/or NKG2D ligand expression.
In humans, NKG2D ligands include MHC class I chain-related protein A (MICA), MICB, and UL16-binding protein 1 (ULBP1) to ULBP6. Like for MCMV, HCMV infection leads to increased expression of the different stress-induced NKG2D ligands, but this upregulation is effectively counteracted by several viral NK cell evasion mechanisms, for example, via the viral UL16 glycoprotein (29). In fact, ULBPs were discovered by their ability to bind UL16 and were named accordingly as UL16-binding proteins (ULBPs). Expression of ULBPs on the cell surface triggers NK cell cytotoxicity, and UL16 causes intracellular retention of several ULBPs (ULBP1, -2, and -6), thereby diminishing NK cell cytotoxicity (30–34). In addition, UL16 also causes intracellular retention of another important NKG2D ligand, MICB (35). The extracellular domain of UL16 is involved in binding to these various NKG2D ligands, whereas the transmembrane and cytoplasmic domain are involved in retention at the ER and cis-Golgi apparatus (36). UL16 binds MICB but not the closely related MICA, which is due to differences in the extracellular α2 domains of these proteins (37).
Thus, UL16 targets many NKG2D ligands but does not affect them all. However, likely testifying to the evolutionary pressure put by NKG2D on HCMV infection, an increasing number of HCMV NKG2D evasion proteins with a complementary and overlapping palette of NKG2D ligand specificity have been identified. The UL142 glycoprotein of HCMV downregulates both MICA and ULBP3, also via retention at the cis-Golgi apparatus (38–40). By screening a part of the HCMV genome, Fielding et al. identified two additional NKG2D evasins, US18 and US20, which act by targeting MICA for lysosomal degradation (41). Recently, in a follow-up study, these authors identified the US12 gene family of HCMV, consisting of 10 sequentially arranged genes (US12 to -21), as a new major hub of immune regulation and NK cell evasion. They demonstrated that the gene products of several members of the US12 gene family selectively target plasma membrane proteins, including NK cell ligands, adhesion molecules, and cytokine receptors (42). Particularly, they showed that US13 and, in line with their earlier findings, US20 significantly contributed to HCMV-mediated MICA and MICB downregulation and that US12, US13, and US20 contributed to ULBP2 downregulation. They also found that different US12 gene family members appear to cooperate to reach maximal downregulation of NKG2D ligands and other targets (42). In line with their earlier findings on US18 and US20, downregulation of many of the target proteins of the US12 gene family products, including the NKG2D ligands MICB and to a lesser extent ULBP2, could be rescued to some extent by inhibition of the lysosomal degradation pathway (42).
Of the different NKG2D ligands, MICA is the most polymorphic, with >80 known alleles (43). The truncated allele MICA*008 shows the highest prevalence in the human population. Interestingly, MICA*008 is not targeted by some of the HCMV MICA-targeting evasins, such as UL142 (44), because it lacks a cytoplasmic domain due to a frameshift mutation in the transmembrane domain and is glycosylphosphatidylinositol (GPI) anchored to the cell surface instead (45). Since UL142 does not downregulate MICA*008, this NKG2D ligand was long thought to represent an “escape variant” resistant to HCMV evasion strategies (46). However, more recently, Seidel et al. reported that the US9 protein of HCMV specifically downregulates MICA*008 and targets it for proteasomal degradation in overexpression assays, although this effect was not evident in HCMV-infected cells (47). The authors hypothesized that additional, still-unknown viral factors cause intracellular retention of MICA*008 in HCMV-infected cells and that US9 specifically interferes with GPI anchoring of MICA*008, diverting it to the cytosol and proteasome (47).
In addition to the diverse protein-mediated strategies to downregulate NKG2D ligands explained above, HCMV also uses an miRNA strategy to suppress this pathway of NK cell cytotoxicity. Application of an algorithm to predict miRNA targets to HCMV led to the identification of HCMV miR-UL112 as a suppressor of MICB expression (48). Expression of MICB, and of MICA, in host cells is tightly regulated by several cellular microRNAs. Interestingly, HCMV miR-UL122 was found to act synergistically with the cellular microRNA miR-376a to efficiently repress MICB expression (49).
Although experiments in mice using MCMV have convincingly shown that NKG2D ligand downregulation is of vital importance for the virus to be able to replicate and spread in the face of a potent NK cell-driven innate immune response, such experiments are obviously lacking in humans. However, experiments in rhesus macaques using rhesus cytomegalovirus (RhCMV), which is more closely related to HCMV than to MCMV, are in line with the experiments in mice. In RhCMV, the Rh159 causes intracellular retention of MICB (50). Rh159 is an ortholog of HCMV UL148, although the latter does not affect NKG2D ligand expression. An RhCMV mutant strain that lacks Rh159 was unable to establish infection in naive animals. Infection could be restored either by depleting CD8+ cells, which included NK cells, or by introducing the HCMV NKG2D-evasive UL16 gene in the Rh159-deleted RhCMV genome (50), supporting the idea that targeting of NKG2D ligands is of paramount importance to allow cytomegaloviruses to cause successful infection.
Although there is evidence, unsurprisingly, that other herpesviruses also interfere with the NKG2D pathway to various extents during infection, the underlying mechanisms and biological consequences are often less well understood.
Indeed, other betaherpesviruses, such as human herpesvirus 6 (HHV-6) and HHV-7, also downregulate NKG2D ligands (51, 52). The HHV-7 U21 protein binds ULBP1 and reroutes it to lysosomes and also suppresses cell surface expression of MICA and MICB, resulting in reduced NK cell-mediated lysis of U21-expressing cells (51). HHV-6 infection leads to cell surface downregulation of ULBP1, ULBP3, and MICB, thereby reducing NKG2D-induced degranulation of NK cells. Although it is as yet unclear how HHV-6 affects NKG2D ligand expression, the process does not involve intracellular retention but appears to depend on proteasomal degradation (52).
The gammaherpesvirus Kaposi's sarcoma-associated herpesvirus (KSHV) downregulates MICA and MICB via the viral E3 ubiquitin ligase K5 protein, resulting in reduced NK cell-mediated lysis of K5-expressing cells. MICA downregulation involves K5-mediated ubiquitination and internalization of cell surface MICA, but not MICA degradation. Like for HCMV UL142, the truncated MICA allele MICA*008 is not affected by KSHV K5 (53). Interestingly, K5 also downregulates expression of activation-induced C-type lectin (AICL), a ligand of NKp80, another important activating receptor of NK cells (53).
In lymphoblastoid cell lines latently infected with EBV, expression of the latent membrane protein LMP2A leads to downregulation of MICA and ULBP4. Although this suppresses reactivity of EBV-specific CD8+ T cell clones, the potential functional consequences regarding NK cell reactivity have not yet been addressed (54). Also like HCMV, the human gammaherpesviruses KSHV and EBV each encode an miRNA that targets MICB, miR-K12-7 and miR-BART2-5p, respectively, resulting in reduced NK cell-mediated killing (55). The authors hypothesized that MICA might have escaped targeting by herpesviral miRNA attack by mutating and/or shortening its 3′ untranslated region (UTR) (55). Although HCMV, KSHV, and EBV each encode an miRNA that targets MICB, fascinatingly, these viral miRNAs do not share sequence homology, which may point to convergent evolution.
Alphaherpesviruses also affect expression of NKG2D ligands. Indeed, HSV-1 and VZV cause downregulation of particular NKG2D ligands, and we have found recently that infection of cells with the porcine alphaherpesvirus pseudorabies virus (PRV) results in reduced binding of recombinant NKG2D (56, 57; unpublished data). HSV-1 infection of cells leads to reduced cell surface levels of MICA, ULBP1, ULBP2, and ULBP3 (56, 57). Although flow cytometric assays in HSV-1-infected HeLa cells indicated that MICA cell surface downregulation was not caused by total protein reduction, Western blot assays on infected ARPE-19 and human foreskin fibroblast (HFF) cells indicated the opposite, showing reduced protein levels for MICA, ULBP2, and ULBP3 but not ULBP1 (56, 57). Although the underlying reason for these apparent contradictory results is unclear, the authors pointed to differences in experimental setup and methodology to analyze protein expression as possible causes (57). Very little is known about the mechanism of HSV-1-mediated suppression of NKG2D ligand expression, although a late viral gene product(s) appears to be involved in MICA cell surface downregulation (56). VZV infection also modulates NKG2D ligand expression on the cell surface, causing upregulation of MICA expression and reduced expression of ULBP2 and ULBP3 (57). Although alphaherpesviruses have not (yet) been reported to encode an miRNA that specifically targets MICB, like their beta- and gammaherpesvirus counterparts, HSV-1 miRNA miR-H8 has been reported to target the GPI anchoring pathway, thereby reducing expression levels of ULBP2 and ULBP3 but not MICA/B and ULBP1 (58).
(ii) DNAM-1.
Although NKG2D appears to be the activating NK cell receptor that virtually all herpesviruses interfere with, many herpesviruses have also been reported to interfere with several other activating receptors. DNAM-1 evasion via downregulation of its prime ligands CD155 (poliovirus receptor [PVR]) and CD112 (nectin 2) has been described for members of the beta- and alphaherpesviruses. Early reports already showed that fibroblasts infected with attenuated laboratory strains of HCMV such as AD169 and Towne showed a higher sensitivity to NK cell-mediated lysis than cells infected with clinical isolates (59). Tomasec et al. used this information as a starting point to identify the UL141 gene product of HCMV as a protein that protects infected cells from efficient NK cell-mediated lysis (60). UL141 is located at the right end of the unique long (UL) regions in a region (UL/b′) that is deleted in the AD169 and Towne genomes. This deletion also affects other NK cell modulatory genes of HCMV, including UL142 and UL135. HCMV strains encoding UL141 were found to efficiently downregulate cell surface expression of the DNAM-1 ligand CD155, whereas cells infected with strains lacking UL141 upregulated CD155 compared to mock-infected cells (60). In addition, transfection of the UL141 glycoprotein was sufficient to downregulate CD155, by retaining CD155 as an immature protein in the ER (60). Recently, it was found that like HCMV, MCMV also downregulates surface expression of CD155. This downregulation depends on the MCMV m20.1 protein, which, like HCMV UL141, also retains CD155 in the ER and promotes its degradation (61).
Later studies showed that UL141 also suppresses cell surface expression of the other major DNAM-1 ligand, CD112, and targets it for proteasomal degradation (62). However, efficient degradation of CD112 by UL141 requires the assistance of other viral gene products, including US2, which recruits the cellular E3 ligase TRC8 to UL141 and other cellular targets (62, 63).
In several alphaherpesviruses of different species, the gD envelope glycoprotein plays an essential role in host cell entry via its interaction with one of different entry receptors. For some alphaherpesviruses, the DNAM-1 ligands CD112 and CD155 serve as gD receptors (64, 65). For PRV and HSV-2, it was shown that expression of gD in infected or transfected cells resulted in downregulation and degradation of CD112, thereby suppressing NK cell-mediated lysis of these cells (66). Interestingly, like in cells infected with HCMV strains that lack UL141 (60), infection of cells with PRV/HSV-2 strains that lack gD resulted in increased cell surface levels of DNAM-1 ligands (66), suggesting that increased cell surface expression of DNAM-1 ligands may be part of a general host cell response to (herpes)virus infection, possibly involving the cellular DNA damage response.
It is important to note that CD155 and CD112 serve as ligands not only for the activating NK cell receptor DNAM-1 but also for the inhibitory NK cell receptor TIGIT (67). This may have important consequences regarding viral evasion strategies, which will be discussed in the concluding remarks.
(iii) NCRs.
Different mechanisms of evasion from the activity of natural cytotoxicity receptors (NCRs) have been reported for herpesviruses. For example, HCMV targets NKp30 both by suppressing the expression of cellular NKp30 ligands and by interfering with NKp30-mediated signaling. On the one hand, HCMV US18 and US20, which also target MICA (see above), were found to suppress cell surface expression of the NKp30 ligand B7-H6 (42, 68). As in the case of MICA, B7-H6 downregulation involves lysosomal degradation and results in impaired NK cell activation (42). On the other hand, the pp65 major tegument protein of HCMV interacts with NKp30 and induces its dissociation from the CD3zeta signaling chain, thereby abrogating the signaling capacity of NKp30 and reducing NK cell-mediated killing of different targets (69).
When cells latently infected with the gammaherpesvirus KSHV were induced to enter the lytic replication program, expression of the viral open reading frame 54 (ORF54)-encoded protein resulted in downregulation of an unknown NKp44 ligand. The protein encoded by ORF54 is a dUTPase, but its enzymatic activity does not contribute to NKp44 ligand modulation, which, rather, depends on the perturbation of membrane protein trafficking (70).
(iv) CD16 (FcγIII receptor).
NK cells may express the Fcγ receptor CD16, which, upon binding of IgG-opsonized target cells, triggers a strong activating signal inducing the so-called antibody-dependent cell-mediated cytotoxicity (ADCC). Several herpesviruses have been reported to encode viral Fc receptors that may bind and thereby shield the Fc domains of antibodies. These include gE-gI of HSV, VZV, and PRV, gp34, gp95, gpRL13, and gp68 of HCMV, and m138 of MCMV (71–79). At least for HCMV, expression of gp34 and gp68 has been shown to suppress NK cell activation (79). It is currently unclear whether this also holds true for other herpesvirus Fc receptors.
(v) 2B4.
2B4 (or CD244) is a member of the signaling lymphocyte activation molecule (SLAM) family of receptors. In human NK cells, 2B4 generally functions as an activating receptor (with the exception of a specific decidual NK cell population showing inhibitory 2B4). Expression of the adaptor protein SLAM-associated protein (SAP) is required to generate the activating signal. In patients suffering from a rare type of primary immunodeficiency termed X-linked lymphoproliferative disease (XLP), expression or function of SAP is defective and results in inhibitory signaling of 2B4 and of an additional SLAM family member, NK, T, and B cell antigen (NTBA). Remarkably, XLP patients are characterized by an inability to control Epstein-Barr virus infection, which, in these patients, can cause fulminant infectious mononucleosis or promote abnormal inflammatory responses frequently resulting in hemophagocytic lymphohistiocytosis (HLH) (80).
Although most SLAM family members engage in homotypic interactions, 2B4 interacts with CD48 (81). In MCMV-infected cells, the viral m154 protein leads to proteolytic and lysosomal degradation of CD48 and interferes with NK cell cytotoxicity. However, transfection of m154 alone did not affect surface levels of CD48, suggesting that m154 is aided by other viral proteins to downregulate this 2B4 ligand (82). An MCMV mutant lacking m154 was attenuated in vivo, and virulence could be restored upon NK cell depletion (82).
Retaining and increasing NK cell-inhibitory signals.
Herpesviruses have evolved different strategies to reduce the efficiency of elimination of infected cells by cytotoxic T lymphocytes (CTLs), mainly by interfering with MHC I-mediated antigen presentation on infected cells (9). As a consequence, cell surface levels of MHC-I molecules are typically reduced on herpesvirus-infected cells. Since cell surface MHC-I molecules represent the most important “self” antigen recognized by inhibitory NK cell receptors, CTL evasion by herpesviruses comes at the cost of increased susceptibility of infected cells to NK cell-mediated attack. It may therefore not be surprising that herpesviruses, and again in particular betaherpesviruses, often counterbalance this increased susceptibility to “missing self”-mediated NK cell activation by additional sets of evasion strategies. These include the expression of viral MHC-I decoys and the selective retention or upregulation of particular cellular MHC-I variants (particularly HLA-E in humans) that are ineffective in triggering substantial CTL responses but provide an inhibitory signal to NK cells. In addition, some herpesviruses also trigger non-MHC-I-dependent inhibitory NK cell receptors.
(i) Viral MHC-I decoys.
Herpesvirus MHC-I decoys have thus far been described only in cytomegaloviruses. In 1997, Reyburn et al. described one of the first viral strategies to evade NK cell-mediated elimination of infected cells (83). They reported that cell surface expression of the UL18 protein of HCMV, an MHC-I homolog, protected cells against NK cell-mediated attack. N-linked glycosylation of UL18 was involved in this protective effect, and since antibodies against CD94 interfered with protection, the authors assumed that an inhibitory NKG2/CD94 complex on NK cells was involved (83). However, shortly afterwards, Cosman et al. showed that UL18 in fact bound to a novel member of the immunoglobulin superfamily glycoproteins, leukocyte immunoglobulin-like receptor 1 (LIR-1), which was found to represent another MHC-I-binding inhibitory receptor (84). Ultrastructural studies revealed that the affinity of UL18 for LIR-1 was up to >1,000-fold higher than that of MHC-I (85). In line with this, expression of UL18 suppresses the cytotoxicity of LIR-1+ NK cells (86). Quite surprisingly, UL18 increases activation of LIR-1− NK cells via a poorly understood mechanism that is LIR-1 independent (86). More recently, it was found that differences in the UL18 amino acid sequence between HCMV strains affect the ability of LIR-1+ NK cells to control virus spread in vitro (87). Since LIR-1 is widely expressed on different cell populations (e.g., B cells, T cells, macrophages, and dendritic cells [DCs]), UL18 may positively or negatively affect the activity of many other immune cell populations (88).
Like host MHC-I molecules, UL18 associates with β2-microglobulin and, intriguingly, also with endogenous peptides that resemble those presented by MHC-I, which is unique among viral MHC-I homologs (89, 90). The crystal structure of UL18 indicated that the extensive N-glycosylation of UL18 may prevent interactions with other MHC-I binding receptors and also may shield the presented peptide, suggesting a way that a viral protein evolved from its host ancestor to eliminate unwanted interactions while optimizing the interaction with the desired (inhibitory) receptor (85).
The activity of UL18 is regulated by other viral proteins. Indeed, the UL40 signal peptide of HCMV, which upregulates cell surface expression of HLA-E (see the next paragraph), also upregulates UL18 expression (91). In addition, the HCMV US6 protein, which is an inhibitor of the transporter associated with antigen presentation (TAP) and thereby interferes with MHC class I antigen presentation, also influences and is influenced by UL18. On the one hand, UL18 interferes with US6-mediated TAP inhibition, thereby restoring TAP-mediated peptide translocation into the ER. On the other hand, UL18 works together with US6 to interfere with the association between MHC-I and TAP in the ER, thereby suppressing MHC-I peptide loading and cell surface expression, irrespective of TAP inhibition (92).
Like HCMV, MCMV encodes several proteins that share homology with MHC-I. Several of these, such as m145, m152, and m155, do not appear to serve as ligands for inhibitory NK cell receptors but may still affect NK cell activity by downregulating ligands for activating NK cell receptors, including stress-induced NKG2D ligands (see above). In a back-to-back Nature paper with the paper describing the HCMV UL18 MHC-I homolog as an NK cell-evasive viral protein (83), Farrell and coworkers described that the viral m144 MHC-I homolog of MCMV interferes with NK cell-mediated clearance of the virus in vivo and suggested that this may be due to m144 binding to an inhibitory NK cell receptor (93). However, since antibodies against the MHC-I-binding murine inhibitory NK cell receptor Ly49A, -C, -G, or -I did not affect the inhibitory effect of m144 on NK cell activity, it is still unclear exactly how m144 exerts its inhibitory function (94). A flexible region in the α2 domain of m144 has been hypothesized to be involved in targeting still-unknown host cell receptors (95).
An MHC-I-like MCMV protein that has been confirmed to interact with MHC-I-binding NK cell receptors, but with various consequences, is m157. In 2002, Arase and colleagues reported that, depending on the mouse strain, m157 interacts with the inhibitory NK cell receptor Ly49I or the related activating receptor Ly49H, correlating with mouse strain susceptibility or resistance to MCMV infection (96). Indeed, Ly49H-expressing C57BL/6 mice are resistant to MCMV infection and lose this resistance upon depletion of NK cells (97). However, it is important to point out that MCMV isolates from wild mice show very substantial variation in their m157 sequence, and many of these strains are able to replicate in C57BL/6 mice, indicating that the concept of resistance is context dependent (98). In any case, BALB/c and 129/J mice that lack expression of Ly49H are susceptible to MCMV infection. BALB/c mice do not appear to express an m157 receptor at all, whereas in 129/J mice, m157 interacts with the inhibitory NK cell receptor Ly49I, which is expressed on a subset of NK cells (96). The interaction between m157 and either Ly49H or Ly49I occurs at the stalk regions of these proteins (99). One possible explanation for the ability of m157 to bind both inhibitory Ly49I and activating Ly49H is that, via its m157 protein, MCMV may have originally used Ly49I to escape NK cell-mediated elimination, while the host has responded back through the development of a very similar activating receptor, Ly49H, that recognizes the same viral ligand (96, 100).
(ii) Cell surface expression of MHC-I variants.
Cell surface expression of the nonclassical MHC-I molecule HLA-E requires loading by a nonamer peptide derived from the signal sequences of most classical MHC-I molecules. At the cell surface, HLA-E can interact with CD94/NKG2A on NK cells, thereby delivering an inhibitory signal. A database search for the conserved HLA-E-binding peptide revealed a perfect match with a 9-amino-acid sequence in the UL40 protein of HCMV, which was located in the signal sequence of this viral transmembrane glycoprotein (101, 102). Consequently, UL40 expression upregulates cell surface expression of HLA-E, particularly in the presence of IFN-γ (103), and thereby confers protection of cells against lysis by CD94/NKG2A+ NK cells (101, 102). Although the activating CD94/NKG2C receptor may also bind the UL40/HLA-E complex, which would result in NK cell activation, the affinity of CD94/NKG2C for this complex is 6-fold lower than that of CD94/NKG2A (18, 104, 105). As indicated above, the consensus HLA-E-binding epitope in UL40 increases cell surface expression not only of HLA-E but also of the viral NK cell-evading MHC-I decoy protein UL18 (91).
Loading of HLA-E with its endogenous MHC-I-derived peptide depends on transport of the peptide from the cytosol into the ER via the TAP transporter (106, 107). As indicated above, in one of its several strategies to avoid efficient elimination of infected cells by cytotoxic T lymphocytes (CTLs), HCMV infection blocks TAP-mediated transport via its viral US6 protein (108), which would be expected to also affect ER transport of the UL40 peptide. However, the HLA-E-binding peptide in UL40 reaches the ER via a pathway that bypasses TAP, allowing simultaneous inhibition of TAP transport and HLA-E upregulation in HCMV-infected cells. This TAP-independent pathway of UL40 peptide loading on HLA-E is still poorly understood and involves the N-terminal 14 amino acids in the signal peptide of UL40 (91).
As another strategy to suppress CTL activity against infected cells, HCMV encodes two viral proteins, US2 and US11, that direct several MHC-I allelic variants from the ER to the cytoplasm, thereby initiating their degradation (109, 110). Interestingly, HLA-E molecules are resistant to the MHC-I-degrading activity of HCMV US2 and US11 (111, 112).
The m04 (gp34) protein of MCMV interacts with MHC-I in the ER and escorts MHC-I molecules to the cell surface. As such, m04 in fact functions as an antagonist of m152, which retains MHC-I in the ER (113). By enabling MHC-I molecules to reach the cell surface, expression of m04 in MCMV-infected cells allows the engagement of the inhibitory NK cell receptor Ly49A. As a consequence, MCMV mutants that lack m04 show increased susceptibility to NK cell-mediated control in vivo (114). Although it was initially thought that m04 binding to MHC-I might compromise peptide loading, correct MHC-I conformation, and/or T cell receptor engagement, m04 was later shown to indeed serve as a bona fide positive regulator of antigen presentation (113). This suggests that different viruses, possibly influenced by their particular host species, show differences in their cost/benefit balance of suppression of the MHC-I antigen presentation pathway to suppress the cytotoxic T cell response versus allowing sufficient MHC-I to reach the cell surface to reduce NK cell cytotoxicity.
(iii) Triggering MHC-I-independent inhibitory NK cell receptors.
MHC-I molecules represent the most abundant and important ligands for inhibitory NK cell receptors. However, MHC-I-independent inhibitory NK cell receptors have been described and may also be modulated by herpesviruses.
In mouse, NKR-P1 NK cell receptors, which contain both inhibitory and activating members, have been reported to interact with Clr self proteins. Particularly, the broadly expressed self Clr-b is recognized by the inhibitory NKR-P1B (115). MCMV infection leads to loss of Clr-b gene expression, in part by the activity of the viral m122 protein, which would be expected to yield an NK cell-activating signal (116, 117). Recently, however, it was shown that cell surface-expressed m12 directly engages NKR-P1B, thereby suppressing NK cell effector activity in vitro and in vivo (118). Likewise, rat cytomegalovirus (RCMV) infection also leads to rapid suppression of Clr-b expression, but the virus also encodes a viral NKR-P1B decoy ligand, RCMV C-type lectin-like protein (RCTL), that protects cells from NK cell-mediated killing (119).
For the porcine alphaherpesvirus PRV, the conserved US3 protein kinase has been reported to trigger cell surface exposure of phospholipid ligands (phosphatidylserine and/or phosphatidylethanolamine) of the inhibitory receptor CD300a, thereby increasing CD300a binding to the cell surface and reducing the susceptibility of infected cells to NK cell-mediated lysis in vitro (120). The US3-mediated increase in CD300a ligand expression depends on the earlier-described ability of this viral protein kinase to trigger activation of cellular group I p21-activated kinases (PAKs), downstream effectors of Cdc42/Rac1 Rho GTPase signaling (121).
The murine gammaherpesvirus MHV-68 upregulates the inhibitory receptor carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) on alveolar epithelial cells, and there are indications that this upregulation, via homophilic trans interactions with CEACAM1 on NK cells, may result in suppressed NK cell activity (122).
Other herpesvirus NK cell evasion strategies.
Although modulation of the activating/inhibitory NK cell balance represents by far the majority of strategies employed by herpesviruses to interfere with NK cell activity, herpesviruses may also target other aspects of NK cell biology.
For example, viral homologs of cytokines may interfere with NK cell migration/activity. Indeed, the KSHV ORF K4-encoded chemokine vMIP-II (vCCL2) inhibits the migration of NK cells by blocking the binding of host chemokines Fractalkine and RANTES to their respective receptors, and viral interleukin-10 (vIL-10) encoded by EBV BCRF1 impairs NK cell-mediated killing of infected B cells (123, 124).
HCMV may also interfere with efficient formation of an immunological synapse between NK cells and infected host cells, which is required for efficient target cell lysis, by pUL135-mediated remodeling of the actin cytoskeleton in the infected cell (125).
Also, cytomegaloviruses suppress expression of TRAIL death receptors (DRs) in infected cells, which are required for TRAIL-induced apoptosis by NK cells. In particular, UL141 of HCMV, which also suppresses cell surface expression of DNAM-1 ligands (see above), binds human TRAIL DRs and retains them in the ER (126). Likewise, MCMV m166 protein, which is unrelated to HCMV UL141, suppresses expression of TRAIL DRs in infected cells, thus contributing to NK cell evasion in vivo (127). The underlying mechanism is not fully understood, although it likely does not involve (direct) interaction of m166 with TRAIL DRs (127).
CONCLUDING REMARKS
Seeing the impressive array of methods that herpesviruses use to suppress recognition and elimination of infected cells by NK cells, it is clear that these innate lymphoid cells pose a very significant threat to herpesviruses. With regard to the number of viral NK cell evasive strategies, cytomegaloviruses overshadow all the other herpesviruses. This may point to a particularly prominent role for NK cells in cytomegalovirus biology, may reflect the larger coding capacity of these viruses, and/or may simply indicate that NK cell evasion strategies have been underresearched for the other herpesviruses. It is worth noting that, irrespective of the molecular methods used, most if not all herpesviruses appear to suppress stress-induced ligands of the activating NK cell receptor NKG2D.
Among the many activating and inhibitory receptors on the surface of NK cells, some are so-called “paired receptors.” Such paired receptors share a highly similar extracellular domain, often resulting in overlapping ligand specificity, but display different cytoplasmic domains with opposing functions in immune regulation, activating versus inhibiting. Although the ligand specificities of the paired receptors typically overlap, they may display different ligand affinities, where the inhibitory members often show a higher ligand affinity. Examples of these include CD94/NKG2A (C-type lectin), KIR2DL1 (KIR family), and CD300a (immunoglobulin superfamily), which display higher binding affinity to their respective ligands HLA-E, HLA-CLys80, and phosphatidylethanolamine (PE) compared to their activating counterparts CD94/NKG2C, KIR2DS1, and CD300c (100, 128). These paired receptors may have consequences for viral NK cell evasion. For example, as indicated above, the US3 protein kinase of PRV triggers increased expression of phospholipid ligands for the inhibitory NK cell receptor CD300a (120). Since the corresponding activating paired NK cell receptor CD300c also binds these phospholipids, this may interfere with efficient viral NK cell evasion. In this context, it will be interesting to dissect exactly which phospholipid ligands for CD300a/c are upregulated in response to US3, since CD300c displays lower affinity for PE than CD300a, but the affinities of both receptors for phosphatidylserine are comparable (128). Regarding the paired inhibitory CD94/NKG2A and activating CD94/NKG2C receptors, both may engage the HCMV UL40 peptide/HLA-E complex, leading to NK cell inhibition or activation, respectively. Although this could suggest that CD94/NKG2C may be part of an evolutionary response to viral immune evasion, this is difficult to reconcile with the 6-fold-lower affinity of the activating CD94/NKG2C receptor toward UL40 peptide/HLA-E compared to the affinity of CD94/NKG2A (18, 104, 105).
Another example of activating/inhibitory NK cell receptors with overlapping ligand specificity are the stimulatory NK cell receptors DNAM-1 and Tactile (CD96) on the one hand and the inhibitory NK cell receptor TIGIT on the other hand, which all are widely distributed on NK cells. CD155 (PVR) is a ligand for all three of these receptors (100). A hypothetical model has been put forward on how the combination of these three different receptors protects healthy cells from NK cell-mediated killing while contributing to the NK cell-mediated killing of tumors in virus-infected cells. In normal, healthy cells, CD155 is expressed at low levels on the surface, probably sufficient to allow the high-affinity inhibitory TIGIT receptor to engage and elicit an inhibitory signal to NK cells. As indicated above, stress responses, including transformation or (herpes)virus infection, may trigger increased expression of CD155, allowing engagement of the lower-affinity activating DNAM-1 and Tactile receptors, contributing to NK cell-mediated killing of the target cells. Efficient but incomplete suppression of cell surface levels of CD155, e.g., via expression of HCMV UL141, may then revert the situation to apparently normal, allowing the virus to avoid recognition by DNAM-1 and Tactile but possibly still permitting engagement of TIGIT (67, 100, 129).
A class of NK cell receptors that has currently been understudied with regard to their interaction with herpesviruses are KIRs. KIRs are a very diverse family of MHC-I-binding receptors that are expressed on subsets of NK and T cells and that can be activating or inhibitory. KIRs are composed of two (2D) or three (3D) extracellular immunoglobulin domains. Their cytoplasmic domain has a varying length, and individual KIRs are therefore designated long (L) or short (S) tailed. With some exceptions (e.g., KIR2DL4), KIR members with a long cytoplasmic domain are inhibitory, since they contain immunoreceptor tyrosine-based inhibitory motif (ITIM) domains, whereas short-tailed KIRs are typically activating via their interaction with activating adaptor proteins (e.g., DAP12). Despite the fact that little is known about herpesvirus evasion of KIRs, there is substantial evidence that these receptors are important in herpesvirus infections, particularly for HCMV. For example, there is evidence that the KIR repertoire in the host adapts to CMV infection. Indeed, the spectrum of KIR expression on NK cells of CMV-seronegative donors was found to be random, whereas seropositive donors showed a preference in KIR2DS2, KIR2DS4, and KIR3DS1 expression (130). Also, there is evidence that particular KIR genes protect against CMV reactivation during late-term pregnancies, which may have important implications for congenital CMV infections (131). KIRs also play an important role in CMV infections in the context of transplantations. In solid organ transplant recipients, CMV is the most common clinically relevant donor-derived infection and remains a major source of morbidity and mortality. A retrospective, single-center cohort study on different types of solid organ transplant patients found that a combination of the activating KIR2DS2 and weak inhibitory KIR2DL3, which share the ligand HLA-C1, in the recipient was associated with a decreased risk of CMV viremia when both donor and recipient were homozygous for HLA-C1 (132). Hence, a better understanding of the NK receptors important for the control of CMV and identification of additional NK evasion strategies may improve the outcome after organ transplantation.
Further unraveling the fascinatingly complex interplay of herpesviruses with NK cells may have important consequences for several scientific fields. Indeed, proof-of-principle studies have already shown potential application of modulation of NK cell activation by herpesviruses in vaccine development, construction of therapeutic viral vectors, and even xenotransplantation. With regard to vaccine development, there are indications that cytomegalovirus strains that are genetically engineered to trigger increased NK cell responses, e.g., via expression of NKG2D ligands, may be promising. Indeed, an MCMV strain expressing the mouse NKG2D ligand RAE-1γ was substantially attenuated while eliciting strong and long-lasting protective immunity, and an HCMV strain expressing the human NKG2D ligand ULBP2 showed increased susceptibility to NK cell cytotoxicity and triggered an effective HCMV-specific T cell response in a humanized mouse model (133). Importantly, although NK cells belong to the innate branch of the immune response, there is increasing evidence that NK cells may display some form of memory toward antigens/pathogens to which the host was previously exposed. This appears to be particularly the case for cytomegalovirus infections (134). In both mice and humans, there is increasing evidence that a cytomegalovirus infection triggers a memory NK cell response and a long-lasting imprinting of the NK cell receptor repertoire (134). A better understanding of NK cell memory and of whether similar responses may or may not be triggered by other (herpes)viruses will be crucial to understand the potential of this fascinating new aspect of NK cell biology for vaccine development.
Talimogene laherparepvec (T-VEC), an attenuated HSV-based oncolytic vector, is the first viral vector to obtain approval for therapeutic use in particular cases of melanoma (135). One important aspect regarding the efficacy of these oncolytic vectors is that they need to display an optimized interaction with the immune system, often aimed at avoiding premature immune-mediated clearance of the vector while retaining tumor-specific immune responses. In this context, the suppressed NK cell-mediated clearance of a genetically engineered vesicular stomatitis virus-based oncolytic vector that expresses the HCMV DNAM-1-evasive gene UL141 was associated with increased oncolytic potency in a rat model of hepatocellular carcinoma (136). This approach may also hold promise toward the optimization of particular HSV-based oncolytic vectors, since the HSV-based vector rQNestin34.5 is prematurely cleared by recipient NK cells in mouse models of glioblastoma (137).
Insights into herpesvirus evasion of NK cells may also be of significance in unrelated research fields, such as xenotransplantation. Swine has been considered an ideal source of donor organs for xenotransplantation in humans (138). However, vigorous immune responses to xenografts, including NK cell-mediated responses, severely limit clinical applications. Expression of the NK cell-evasive UL18 gene of HCMV in swine endothelial cells was found to suppress human NK cell-mediated reactivity toward these porcine cells (139).
In conclusion, the extraordinary diversity of NK cell evasion strategies displayed by herpesviruses illustrates the substantial evolutionary pressure that this branch of the immune system has put on these pathogens. Since the first description, 2 decades ago, of (herpes)viral strategies to suppress NK cell reactivity (83, 93), a myriad of different evasion mechanisms has been unraveled, and several additional strategies likely still await discovery. A fascinating but challenging next step will be to translate this increasing wealth of knowledge into prophylactic and therapeutic applications.
ACKNOWLEDGMENTS
This research was supported by grants from the Special Research Fund of Ghent University (G.O.A. grant 01G01317), F.W.O.-Vlaanderen (grant G.0176.15N), and the Belgian Science Policy (BELSPO) via the BELVIR consortium (IAP, phase VII). Steffi De Pelsmaeker was supported by a Ph.D. grant from the agency Flanders Innovation & Entrepreneurship (VLAIO).
We thank César de Medina Pérez for help in the design of the figures.
REFERENCES
- 1.Montaldo E, Vacca P, Vitale C, Moretta F, Locatelli F, Mingari MC, Moretta L. 2016. Human innate lymphoid cells. Immunol Lett 179:2–8. doi: 10.1016/j.imlet.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. 2017. The broad spectrum of human natural killer cell diversity. Immunity 47:820–833. doi: 10.1016/j.immuni.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biron CA, Byron KS, Sullivan JL. 1989. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med 320:1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 4.Strauss-Albee DM, Fukuyama J, Liang EC, Yao Y, Jarrell JA, Drake AL, Kinuthia J, Montgomery RR, John-Stewart G, Holmes S, Blish CA. 2015. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci Transl Med 7:297ra115. doi: 10.1126/scitranslmed.aac5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gumá M, Angulo A, Vilches C, Gómez-Lozano N, Malats N, López-Botet M. 2004. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 6.O'Sullivan TE, Sun JC, Lanier LL. 2015. Natural killer cell memory. Immunity 43:634–645. doi: 10.1016/j.immuni.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, Han H, Chiang SCC, Foley B, Mattsson K, Larsson S, Schaffer M, Malmberg K, Ljunggren H, Miller JS, Bryceson YT. 2015. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42:443–456. doi: 10.1016/j.immuni.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun JC. 2010. Re-educating natural killer cells. J Exp Med 207:2049–2052. doi: 10.1084/jem.20101748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuren ABC, Costa AI, Wiertz EJHJ. 2016. Recent advances in viral evasion of the MHC class I processing pathway. Curr Opin Immunol 40:43–50. doi: 10.1016/j.coi.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Jouanguy E, Zhang S-Y, Chapgier A, Sancho-Shimizu V, Puel A, Picard C, Boisson-Dupuis S, Abel L, Casanova J-L. 2007. Human primary immunodeficiencies of type I interferons. Biochimie 89:878–883. doi: 10.1016/j.biochi.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 11.Orange JS. 2013. Natural killer cell deficiency. J Allergy Clin Immunol 132:515–525. doi: 10.1016/j.jaci.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Etzioni A, Eidenschenk C, Katz R, Beck R, Casanova JL, Pollack S. 2005. Fatal varicella associated with selective natural killer cell deficiency. J Pediatr 146:423–425. doi: 10.1016/j.jpeds.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 13.Almerigogna F, Fassio F, Giudizi MG, Biagiotti R, Manuelli C, Chiappini E, Galli L, Romagnani S, De Martino M. 2011. Natural killer cell deficiencies in a consecutive series of children with herpetic encephalitis. Int J Immunopathol Pharmacol 24:231–238. doi: 10.1177/039463201102400128. [DOI] [PubMed] [Google Scholar]
- 14.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. 1999. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 15.Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. 2000. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol 1:119–126. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- 16.Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. 2001. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol 2:255–260. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- 17.Krmpotic A, Hasan M, Loewendorf A, Saulig T, Halenius A, Lenac T, Polic B, Bubic I, Kriegeskorte A, Pernjak-Pugel E, Messerle M, Hengel H, Busch DH, Koszinowski UH, Jonjic S. 2005. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J Exp Med 201:211–220. doi: 10.1084/jem.20041617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun JC, Lanier LL. 2009. The natural selection of herpesviruses and virus-specific NK cell receptors. Viruses 1:362–382. doi: 10.3390/v1030362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziegler H, Thäle R, Lucin P, Muranyi W, Flohr T, Hengel H, Farrell H, Rawlinson W, Koszinowski UH. 1997. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity 6:57–66. doi: 10.1016/S1074-7613(00)80242-3. [DOI] [PubMed] [Google Scholar]
- 20.Krmpotić A, Busch DH, Bubić I, Gebhardt F, Hengel H, Hasan M, Scalzo AA, Koszinowski UH, Jonjić S. 2002. MCMV glycoprotein gp40 confers virus resistance to CD8+ T cells and NK cells in vivo. Nat Immunol 3:529–535. doi: 10.1038/ni799. [DOI] [PubMed] [Google Scholar]
- 21.Lodoen M, Ogasawara K, Hamerman JA, Arase H, Houchins JP, Mocarski ES, Lanier LL. 2003. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J Exp Med 197:1245–1253. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R, Natarajan K, Revilleza MJR, Boyd LF, Zhi L, Zhao H, Robinson H, Margulies DH. 2012. Structural basis of mouse cytomegalovirus m152/gp40 interaction with RAE1γ reveals a paradigm for MHC/MHC interaction in immune evasion. Proc Natl Acad Sci U S A 109:E3578–E3587. doi: 10.1073/pnas.1214088109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodoen MB, Abenes G, Umamoto S, Houchins JP, Liu F, Lanier LL. 2004. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. J Exp Med 200:1075–1081. doi: 10.1084/jem.20040583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hasan M, Krmpotic A, Ruzsics Z, Bubic I, Lenac T, Halenius A, Loewendorf A, Messerle M, Hengel H, Jonjic S, Koszinowski UH. 2005. Selective down-regulation of the NKG2D ligand H60 by mouse cytomegalovirus m155 glycoprotein. J Virol 79:2920–2930. doi: 10.1128/JVI.79.5.2920-2930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawlinson WD, Farrell HE, Barrell BG. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol 70:8833–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenac T, Budt M, Arapovic J, Hasan M, Zimmermann A, Simic H, Krmpotic A, Messerle M, Ruzsics Z, Koszinowski UH, Hengel H, Jonjic S. 2006. The herpesviral Fc receptor fcr-1 down-regulates the NKG2D ligands MULT-1 and H60. J Exp Med 203:1843–1850. doi: 10.1084/jem.20060514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arapović J, Lenac Roviš T, Reddy AB, Krmpotić A, Jonjić S. 2009. Promiscuity of MCMV immunoevasin of NKG2D: m138/fcr-1 down-modulates RAE-1ε in addition to MULT-1 and H60. Mol Immunol 47:114–122. doi: 10.1016/j.molimm.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Dölken L, Krmpotic A, Kothe S, Tuddenham L, Tanguy M, Marcinowski L, Ruzsics Z, Elefant N, Altuvia Y, Margalit H, Koszinowski UH, Jonjic S, Pfeffer S. 2010. Cytomegalovirus microRNAs facilitate persistent virus infection in salivary glands. PLoS Pathog 6:e1001150. doi: 10.1371/journal.ppat.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rolle A, Mousavi-Jazi M, Eriksson M, Odeberg J, Soderberg-Naucler C, Cosman D, Karre K, Cerboni C. 2003. Effects of human cytomegalovirus infection on ligands for the activating NKG2D receptor of NK cells: up-regulation of UL16-binding protein (ULBP)1 and ULBP2 is counteracted by the viral UL16 protein. J Immunol 171:902–908. doi: 10.4049/jimmunol.171.2.902. [DOI] [PubMed] [Google Scholar]
- 30.Cosman D, Müllberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, Kubin M, Chalupny NJ. 2001. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity 14:123–133. doi: 10.1016/S1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 31.Kubin M, Cassiano L, Chalupny J, Chin W, Cosman D, Fanslow W, Müllberg J, Rousseau AM, Ulrich D, Armitage R. 2001. ULBP1, 2, 3: novel MHC class I-related molecules that bind to human cytomegalovirus glycoprotein UL16, activate NK cells. Eur J Immunol 31:1428–1437. doi:. [DOI] [PubMed] [Google Scholar]
- 32.Welte SA, Sinzger C, Lutz SZ, Singh-Jasuja H, Sampaio KL, Eknigk U, Rammensee HG, Steinle A. 2003. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur J Immunol 33:194–203. doi: 10.1002/immu.200390022. [DOI] [PubMed] [Google Scholar]
- 33.Valés-Gómez M, Browne H, Reyburn HT. 2003. Expression of the UL16 glycoprotein of human cytomegalovirus protects the virus-infected cell from attack by natural killer cells. BMC Immunol 4:4. doi: 10.1186/1471-2172-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eagle RA, Traherne JA, Hair JR, Jafferji I, Trowsdale J. 2009. ULBP6/RAET1L is an additional human NKG2D ligand. Eur J Immunol 39:3207–3216. doi: 10.1002/eji.200939502. [DOI] [PubMed] [Google Scholar]
- 35.Wu J, Chalupny NJ, Manley TJ, Riddell SR, Cosman D, Spies T. 2003. Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glycoprotein. J Immunol 170:4196–4200. doi: 10.4049/jimmunol.170.8.4196. [DOI] [PubMed] [Google Scholar]
- 36.Dunn C, Chalupny NJ, Sutherland CL, Dosch S, Sivakumar PV, Johnson DC, Cosman D. 2003. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J Exp Med 197:1427–1439. doi: 10.1084/jem.20022059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spreu J, Stehle T, Steinle A. 2006. Human cytomegalovirus-encoded UL16 discriminates MIC molecules by their alpha2 domains. J Immunol 177:3143–3149. doi: 10.4049/jimmunol.177.5.3143. [DOI] [PubMed] [Google Scholar]
- 38.Chalupny NJ, Rein-Weston A, Dosch S, Cosman D. 2006. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem Biophys Res Commun 346:175–181. doi: 10.1016/j.bbrc.2006.05.092. [DOI] [PubMed] [Google Scholar]
- 39.Ashiru O, Bennett NJ, Boyle LH, Thomas M, Trowsdale J, Wills MR. 2009. NKG2D ligand MICA is retained in the cis-Golgi apparatus by human cytomegalovirus protein UL142. J Virol 83:12345–12354. doi: 10.1128/JVI.01175-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennett NJ, Ashiru O, Morgan FJE, Pang Y, Okecha G, Eagle RA, Trowsdale J, Sissons JGP, Wills MR. 2010. Intracellular sequestration of the NKG2D ligand ULBP3 by human cytomegalovirus. J Immunol 185:1093–1102. doi: 10.4049/jimmunol.1000789. [DOI] [PubMed] [Google Scholar]
- 41.Fielding CA, Aicheler R, Stanton RJ, Wang ECY, Han S, Seirafian S, Davies J, McSharry BP, Weekes MP, Antrobus PR, Prod'homme V, Blanchet FP, Sugrue D, Cuff S, Roberts D, Davison AJ, Lehner PJ, Wilkinson GWG, Tomasec P. 2014. Two novel human cytomegalovirus NK cell evasion functions target MICA for lysosomal degradation. PLoS Pathog 10:e1004058. doi: 10.1371/journal.ppat.1004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fielding CA, Weekes MP, Nobre LV, Ruckova E, Wilkie GS, Paulo JA, Chang C, Suarez NM, Davies JA, Antrobus R, Stanton RJ, Aicheler RJ, Nichols H, Vojtesek B, Trowsdale J, Davison AJ, Gygi SP, Tomasec P, Lehner PJ, Wilkinson GWG. 2017. Control of immune ligands by members of a cytomegalovirus gene expansion suppresses natural killer cell activation. Elife 6:e22206. doi: 10.7554/eLife.22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernández-Messina L, Reyburn HT, Valés-Gómez M. 2012. Human NKG2D-ligands: cell biology strategies to ensure immune recognition. Front Immunol 3:299. doi: 10.3389/fimmu.2012.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Lazaro AM, Lavingia B, Stastny P. 2001. Typing for all known MICA alleles by group-specific PRC and SSOP. Hum Immunol 62:620–631. doi: 10.1016/S0198-8859(01)00241-5. [DOI] [PubMed] [Google Scholar]
- 45.Ashiru O, López-Cobo S, Fernández-Messina L, Pontes-Quero S, Pandolfi R, Reyburn HT, Valés-Gómez M. 2013. A GPI anchor explains the unique biological features of the common NKG2D-ligand allele MICA*008. Biochem J 454:295–302. doi: 10.1042/BJ20130194. [DOI] [PubMed] [Google Scholar]
- 46.Zou Y, Bresnahan W, Taylor RT, Stastny P. 2005. Effect of human cytomegalovirus on expression of MHC class I-related chains A. J Immunol 174:3098–3104. doi: 10.4049/jimmunol.174.5.3098. [DOI] [PubMed] [Google Scholar]
- 47.Seidel E, Thuy V, Le K, Bar-on Y, Tsukerman P, Enk J, Yamin R, Stein N. 2015. Dynamic co-evolution of host and pathogen: HCMV downregulates the prevalent allele MICA*008 to escape elimination by NK cells. Cell Rep doi: 10.1016/j.celrep.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O. 2007. Host immune system gene targeting by a viral miRNA. Science 317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nachmani D, Lankry D, Wolf DG, Mandelboim O. 2010. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat Immunol 11:806–813. doi: 10.1038/ni.1916. [DOI] [PubMed] [Google Scholar]
- 50.Sturgill ER, Malouli D, Hansen SG, Burwitz BJ, Seo S, Schneider CL, Womack JL, Verweij MC, Ventura AB, Bhusari A, Jeffries KM, Legasse AW, Axthelm MK, Hudson AW, Sacha JB, Picker LJ, Früh K. 2016. Natural killer cell evasion is essential for infection by rhesus cytomegalovirus. PLoS Pathog 12:e1005868. doi: 10.1371/journal.ppat.1005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider CL, Hudson AW. 2011. The human herpesvirus-7 (HHV-7) U21 immunoevasin subverts NK-mediated cytoxicity through modulation of MICA and MICB. PLoS Pathog 7:e1002362. doi: 10.1371/journal.ppat.1002362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmiedel D, Tai J, Levi-Schaffer F, Dovrat S, Mandelboim O. 2016. Human Herpesvirus 6 downregulates the expression of activating ligands during lytic infection to escape elimination by natural killer cells. J Virol 90:9608–9617. doi: 10.1128/JVI.01164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas M, Boname JM, Field S, Nejentsev S, Salio M, Cerundolo V, Wills M, Lehner PJ. 2008. Down-regulation of NKG2D and NKp80 ligands by Kaposi's sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc Natl Acad Sci U S A 105:1656–1661. doi: 10.1073/pnas.0707883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rancan C, Schirrmann L, Hüls C, Zeidler R, Moosmann A. 2015. Latent membrane protein LMP2A impairs recognition of EBV-infected cells by CD8+ T cells. PLoS Pathog 11:e1004906. doi: 10.1371/journal.ppat.1004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5:376–385. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Schepis D, D'Amato M, Studahl M, Bergström T, Kärre K, Berg L. 2009. Herpes simplex virus infection downmodulates NKG2D ligand expression. Scand J Immunol 69:429–436. doi: 10.1111/j.1365-3083.2009.02241.x. [DOI] [PubMed] [Google Scholar]
- 57.Campbell TM, McSharry BP, Steain M, Slobedman B, Abendroth A. 2015. Varicella-zoster virus and herpes simplex virus 1 differentially modulate NKG2D ligand expression during productive infection. J Virol 89:7932–7943. doi: 10.1128/JVI.00292-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Enk J, Levi A, Weisblum Y, Yamin R, Charpak-Amikam Y, Wolf DG, Mandelboim O. 2016. HSV1 microRNA modulation of GPI anchoring and downstream immune evasion. Cell Rep 17:949–956. doi: 10.1016/j.celrep.2016.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cerboni C, Mousavi-Jazi M, Linde A, Söderström K, Brytting M, Wahren B, Karre K, Carbone E. 2000. Human cytomegalovirus strain-dependent changes in NK cell recognition of infected fibroblasts. J Immunol 164:4775–4782. doi: 10.4049/jimmunol.164.9.4775. [DOI] [PubMed] [Google Scholar]
- 60.Tomasec P, Wang ECY, Davison AJ, Vojtesek B, Armstrong M, Griffin C, McSharry BP, Morris RJ, Llewellyn-Lacey S, Rickards C, Nomoto A, Sinzger C, Wilkinson GWG. 2005. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat Immunol 6:181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lenac Rovis T, Kucan Brlic P, Kaynan N, Juranic Lisnic V, Brizic I, Jordan S, Tomic A, Kvestak D, Babic M, Tsukerman P, Colonna M, Koszinowski U, Messerle M, Mandelboim O, Krmpotic A, Jonjic S. 2016. Inflammatory monocytes and NK cells play a crucial role in DNAM-1-dependent control of cytomegalovirus infection. J Exp Med 213:1835–1850. doi: 10.1084/jem.20151899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prod'homme V, Sugrue DM, Stanton RJ, Nomoto A, Davies J, Rickards CR, Cochrane D, Moore M, Wilkinson GWG, Tomasec P. 2010. Human cytomegalovirus UL141 promotes efficient downregulation of the natural killer cell activating ligand CD112. J Gen Virol 91:2034–2039. doi: 10.1099/vir.0.021931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsu JL, van den Boomen DJH, Tomasec P, Weekes MP, Antrobus R, Stanton RJ, Ruckova E, Sugrue D, Wilkie GS, Davison AJ, Wilkinson GWG, Lehner PJ. 2015. Plasma membrane profiling defines an expanded class of cell surface proteins selectively targeted for degradation by HCMV US2 in cooperation with UL141. PLoS Pathog 11:e1004811. doi: 10.1371/journal.ppat.1004811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spear PG. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol 6:401–410. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 65.Campadelli-Fiume G, Menotti L. 2007. Entry of alphaherpesviruses into the cell, p 93–111. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 66.Grauwet K, Cantoni C, Parodi M, De Maria A, Devriendt B, Pende D, Moretta L, Vitale M, Favoreel HW. 2014. Modulation of CD112 by the alphaherpesvirus gD protein suppresses DNAM-1-dependent NK cell-mediated lysis of infected cells. Proc Natl Acad Sci U S A 111:16118–16123. doi: 10.1073/pnas.1409485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stanietsky N, Simic H, Arapovic J, Toporik A, Levy O, Novik A, Levine Z, Beiman M, Dassa L, Achdout H, Stern-Ginossar N, Tsukerman P, Jonjic S, Mandelboim O. 2009. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci U S A 106:17858–17863. doi: 10.1073/pnas.0903474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Charpak-Amikam Y, Kubsch T, Seidel E, Oiknine-Djian E, Cavaletto N, Yamin R, Schmiedel D, Wolf D, Gribaudo G, Messerle M, Cicin-Sain L, Mandelboim O. 2017. Human cytomegalovirus escapes immune recognition by NK cells through the downregulation of B7-H6 by the viral genes US18 and US20. Sci Rep 7:8661. doi: 10.1038/s41598-017-08866-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arnon TI, Achdout H, Levi O, Markel G, Saleh N, Katz G, Gazit R, Gonen-Gross T, Hanna J, Nahari E, Porgador A, Honigman A, Plachter B, Mevorach D, Wolf DG, Mandelboim O. 2005. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol 6:515–523. [DOI] [PubMed] [Google Scholar]
- 70.Madrid AS, Ganem D. 2012. Kaposi's sarcoma-associated herpesvirus ORF54/dUTPase downregulates a ligand for the NK activating receptor NKp44. J Virol 86:8693–8704. doi: 10.1128/JVI.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson DC, Frame MC, Ligas MW, Cross a, Stow MND. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol 62:1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Litwin V, Jackson W, Grose C. 1992. Receptor properties of two varicella-zoster virus glycoproteins, gpI and gpIV, homologous to herpes simplex virus gE and gI. J Virol 66:3643–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Favoreel HW, Nauwynck HJ, Van Oostveldt P, Mettenleiter TC, Pensaert MB. 1997. Antibody-induced and cytoskeleton-mediated redistribution and shedding of viral glycoproteins, expressed on pseudorabies virus-infected cells. J Virol 71:8254–8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Atalay R, Zimmermann A, Wagner M, Borst E, Benz C, Messerle M, Hengel H. 2002. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcgamma receptor homologs. J Virol 76:8596–8608. doi: 10.1128/JVI.76.17.8596-8608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thäle R, Lucin P, Schneider K, Eggers M, Koszinowski UH. 1994. Identification and expression of a murine cytomegalovirus early gene coding for an Fc receptor. J Virol 68:7757–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sprague ER, Reinhard H, Cheung EJ, Farley AH, Trujillo RD, Hengel H, Bjorkman PJ. 2008. The human cytomegalovirus Fc receptor gp68 binds the Fc CH2-CH3 interface of immunoglobulin G. J Virol 82:3490–3499. doi: 10.1128/JVI.01476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cortese M, Calo S, D'Aurizio R, Lilja A, Pacchiani N, Merola M. 2012. Recombinant human cytomegalovirus (HCMV) RL13 binds human immunoglobulin G Fc. PLoS One 7:e50166. doi: 10.1371/journal.pone.0050166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Corrales-Aguilar E, Hoffmann K, Hengel H. 2014. CMV-encoded Fcγ receptors: modulators at the interface of innate and adaptive immunity. Semin Immunopathol 36:627–640. doi: 10.1007/s00281-014-0448-2. [DOI] [PubMed] [Google Scholar]
- 79.Corrales-Aguilar E, Trilling M, Hunold K, Fiedler M, Le VTK, Reinhard H, Ehrhardt K, Mercé-Maldonado E, Aliyev E, Zimmermann A, Johnson DC, Hengel H. 2014. Human cytomegalovirus Fcγ binding proteins gp34 and gp68 antagonize Fcγ receptors I, II and III. PLoS Pathog 10:e1004131. doi: 10.1371/journal.ppat.1004131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parolini S, Bottino C, Falco M, Augugliaro R, Giliani S, Franceschini R, Ochs HD, Wolf H, Bonnefoy JY, Biassoni R, Moretta L, Notarangelo LD, Moretta A. 2000. X-linked lymphoproliferative disease. 2B4 molecules displaying inhibitory rather than activating function are responsible for the inability of natural killer cells to kill Epstein-Barr virus-infected cells. J Exp Med 192:337–346. doi: 10.1084/jem.192.3.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Waggoner SN, Kumar V. 2012. Evolving role of 2B4/CD244 in T and NK cell responses during virus infection. Front Immunol 3:377. doi: 10.3389/fimmu.2012.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zarama A, Pérez-Carmona N, Farré D, Tomic A, Borst EM, Messerle M, Jonjic S, Engel P, Angulo A. 2014. Cytomegalovirus m154 hinders CD48 cell-surface expression and promotes viral escape from host natural killer cell control. PLoS Pathog 10:e1004000. doi: 10.1371/journal.ppat.1004000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reyburn HT, Mandelboim O, Valés-Gómez M, Davis DM, Pazmany L, Strominger JL. 1997. The class I MHC homologue of human cytomegalovirus inhibits attack by natural killer cells. Nature 386:514–517. doi: 10.1038/386514a0. [DOI] [PubMed] [Google Scholar]
- 84.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu ML. 1997. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity 7:273–282. doi: 10.1016/S1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 85.Yang Z, Bjorkman PJ. 2008. Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor. Proc Natl Acad Sci U S A 105:10095–10100. doi: 10.1073/pnas.0804551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Prod'homme V, Griffin C, Aicheler RJ, Wang ECY, McSharry BP, Rickards CR, Stanton RJ, Borysiewicz LK, López-Botet M, Wilkinson GWG, Tomasec P. 2007. The human cytomegalovirus MHC class I homolog UL18 inhibits LIR-1+ but activates LIR-1− NK cells. J Immunol 178:4473–4481. doi: 10.4049/jimmunol.178.7.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen K, Stanton R, Banat J, Wills M. 2016. Leukocyte immunoglobulin-like receptor 1-expressing human natural killer cell subsets differentially recognize isolates of human cytomegalovirus through the viral major histocompatibility complex class I homolog UL18. J Virol 90:3123–3137. doi: 10.1128/JVI.02614-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wagner CS, Ljunggren H-G, Achour A. 2008. Immune modulation by the human cytomegalovirus-encoded molecule UL18, a mystery yet to be solved. J Immunol 180:19–24. doi: 10.4049/jimmunol.180.1.19. [DOI] [PubMed] [Google Scholar]
- 89.Browne H, Smith G, Beck S, Minson T. 1990. A complex between the MHC class I homologue encoded by human cytomegalovirus and beta 2 microglobulin. Nature 347:770–772. doi: 10.1038/347770a0. [DOI] [PubMed] [Google Scholar]
- 90.Fahnestock ML, Johnson JL, Renny Feldman RM, Neveu JM, Lane WS, Bjorkman JP. 1995. The MHC class I homolog encoded by human cytomegalovirus binds endogenous peptides. Immunity 3:583–590. doi: 10.1016/1074-7613(95)90129-9. [DOI] [PubMed] [Google Scholar]
- 91.Prod'homme V, Tomasec P, Cunningham C, Lemberg MK, Stanton RJ, McSharry BP, Wang ECY, Cuff S, Martoglio B, Davison AJ, Braud VM, Wilkinson GWG. 2012. Human cytomegalovirus UL40 signal peptide regulates cell surface expression of the NK cell ligands HLA-E and gpUL18. J Immunol 188:2794–2804. doi: 10.4049/jimmunol.1102068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim Y, Park B, Cho S, Shin J, Cho K, Jun Y, Ahn K. 2008. Human cytomegalovirus UL18 utilizes US6 for evading the NK and T-cell responses. PLoS Pathog 4:e1000123. doi: 10.1371/journal.ppat.1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Farrell HE, Vally H, Lynch DM, Fleming P, Shellam GR, Scalzo AA, Davis-Poynter NJ. 1997. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature 386:510–514. doi: 10.1038/386510a0. [DOI] [PubMed] [Google Scholar]
- 94.Kubota A, Kubota S, Farrell HE, Davis-Poynter N, Takei F. 1999. Inhibition of NK cells by murine CMV-encoded class I MHC homologue m144. Cell Immunol 191:145–151. doi: 10.1006/cimm.1998.1424. [DOI] [PubMed] [Google Scholar]
- 95.Natarajan K, Hicks A, Mans J, Robinson H, Guan R, Mariuzza RA, Margulies DH. 2006. Crystal structure of the murine cytomegalovirus MHC-I homolog m144. J Mol Biol 358:157–171. doi: 10.1016/j.jmb.2006.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 97.Scalzo AA, Fitzgerald NA, Wallace CR, Gibbons AE, Smart YC, Burton RC, Shellam GR. 1992. The effect of the Cmv-1 resistance gene, which is linked to the natural killer cell gene complex, is mediated by natural killer cells. J Immunol 149:581–589. [PubMed] [Google Scholar]
- 98.Voigt V, Forbes CA, Tonkin JN, Degli-Esposti MA, Smith HRC, Yokoyama WM, Scalzo AA. 2003. Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc Natl Acad Sci U S A 100:13483–13488. doi: 10.1073/pnas.2233572100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Berry R, Ng N, Saunders PM, Vivian JP, Lin J, Deuss FA, Corbett AJ, Forbes CA, Widjaja JM, Sullivan LC, McAlister AD, Perugini MA, Call MJ, Scalzo AA, Degli-Esposti MA, Coudert JD, Beddoe T, Brooks AG, Rossjohn J. 2013. Targeting of a natural killer cell receptor family by a viral immunoevasin. Nat Immunol 14:699–705. doi: 10.1038/ni.2605. [DOI] [PubMed] [Google Scholar]
- 100.Stanietsky N, Mandelboim O. 2010. Paired NK cell receptors controlling NK cytotoxicity. FEBS Lett 584:4895–4900. doi: 10.1016/j.febslet.2010.08.047. [DOI] [PubMed] [Google Scholar]
- 101.Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S, Cerundolo V, Borysiewicz LK, McMichael AJ, Wilkinson GW. 2000. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287:1031. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 102.Ulbrecht M, Martinozzi S, Grzeschik M, Hengel H, Ellwart JW, Pla M, Weiss EH. 2000. The human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J Immunol 164:5019–5022. doi: 10.4049/jimmunol.164.10.5019. [DOI] [PubMed] [Google Scholar]
- 103.Cerboni C, Mousavi-Jazi M, Wakiguchi H, Carbone E, Kärre K, Söderström K. 2001. Synergistic effect of IFN-gamma and human cytomegalovirus protein UL40 in the HLA-E-dependent protection from NK cell-mediated cytotoxicity. Eur J Immunol 31:2926–2935. doi:. [DOI] [PubMed] [Google Scholar]
- 104.Llano M, Lee N, Navarro F, García P, Albar JP, Geraghty DE, López-Botet M. 1998. HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: preferential response to an HLA-G-derived nonamer. Eur J Immunol 28:2854–2863. doi:. [DOI] [PubMed] [Google Scholar]
- 105.Kaiser BK, Pizarro JC, Kerns J, Strong RK. 2008. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc Natl Acad Sci U S A 105:6696–6701. doi: 10.1073/pnas.0802736105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Braud VM, Allan DSJ, Wilson D, McMichael AJ. 1998. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol 8:1–10. doi: 10.1016/S0960-9822(98)70014-4. [DOI] [PubMed] [Google Scholar]
- 107.Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. 1998. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol 160:4951–4960. [PubMed] [Google Scholar]
- 108.Ahn K, Gruhler A, Galocha B, Jones TR, Wiertz EJHJ, Ploegh HL, Peterson PA, Yang Y, Früh K. 1997. The ER-luminal domain of the HCMV glypcoprotein US6 inhibits peptide translocation by TAP. Immunity 6:613–621. doi: 10.1016/S1074-7613(00)80349-0. [DOI] [PubMed] [Google Scholar]
- 109.Wiertz EJHJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769–779. doi: 10.1016/S0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 110.Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 111.Schust DJ, Tortorella D, Seebach J, Phan C, Ploegh HL. 1998. Trophoblast class I major histocompatibility complex (MHC) products are resistant to rapid degradation imposed by the human cytomegalovirus (HCMV) gene products US2 and US11. J Exp Med 188:497–503. doi: 10.1084/jem.188.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Llano M, Gumá M, Ortega M, Angulo A, López-Botet M. 2003. Differential effects of US2, US6, and US11 human cytomegalovirus proteins of HLA class Ia and HLA-E expression: impact on target susceptibility to NK cell subsets. Eur J Immunol 33:2744–2754. doi: 10.1002/eji.200324182. [DOI] [PubMed] [Google Scholar]
- 113.Holtappels R, Gillert-Marien D, Thomas D, Podlech J, Deegen P, Herter S, Oehrlein-Karpi SA, Strand D, Wagner M, Reddehase MJ. 2006. Cytomegalovirus encodes a positive regulator of antigen presentation. J Virol 80:7613–7624. doi: 10.1128/JVI.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Babić M, Pyzik M, Zafirova B, Mitrović M, Butorac V, Lanier LL, Krmpotić A, Vidal SM, Jonjić S. 2010. Cytomegalovirus immunoevasin reveals the physiological role of “missing self” recognition in natural killer cell dependent virus control in vivo. J Exp Med 207:2663–2673. doi: 10.1084/jem.20100921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carlyle JR, Jamieson AM, Gasser S, Clingan CS, Arase H, Raulet DH. 2004. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci U S A 101:3527–3532. doi: 10.1073/pnas.0308304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Aguilar OA, Mesci A, Ma J, Chen P, Kirkham CL, Hundrieser J, Voigt S, Allan DSJ, Carlyle JR. 2015. Modulation of Clr ligand expression and NKR-P1 receptor function during murine cytomegalovirus infection. J Innate Immun 7:584–600. doi: 10.1159/000382032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kirkham CL, Aguilar OA, Yu T, Tanaka M, Mesci A, Chu KL, Fine JH, Mossman KL, Bremner R, Allan DSJ, Carlyle JR. 2017. Interferon-dependent induction of Clr-b during mouse cytomegalovirus infection protects bystander cells from natural killer cells via NKR-P1B-mediated inhibition. J Innate Immun 9:343–358. doi: 10.1159/000454926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aguilar OA, Berry R, Rahim MMA, Reichel JJ, Popović B, Tanaka M, Fu Z, Balaji GR, Lau TNH, Tu MM, Kirkham CL, Mahmoud AB, Mesci A, Krmpotić A, Allan DSJ, Makrigiannis AP, Jonjić S, Rossjohn J, Carlyle JR. 2017. A viral immunoevasin controls innate immunity by targeting the prototypical natural killer cell receptor family. Cell 169:58–71. doi: 10.1016/j.cell.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 119.Voigt S, Mesci A, Ettinger J, Fine JH, Chen P, Chou W, Carlyle JR. 2007. Cytomegalovirus evasion of innate immunity by subversion of the NKR-P1B:Clr-b missing-self axis. Immunity 26:617–627. doi: 10.1016/j.immuni.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 120.Grauwet K, Vitale M, De Pelsmaeker S, Jacob T, Laval K, Moretta L, Parodi M, Parolini S, Cantoni C, Favoreel HW. 2016. Pseudorabies virus US3 protein kinase protects infected cells from NK cell-mediated lysis via increased binding of the inhibitory NK cell receptor CD300a. J Virol 90:1522–1533. doi: 10.1128/JVI.02902-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Van den Broeke C, Radu M, Deruelle M, Nauwynck H, Hofmann C, Jaffer ZM, Chernoff J, Favoreel HW. 2009. Alphaherpesvirus US3-mediated reorganization of the actin cytoskeleton is mediated by group A p21-activated kinases. Proc Natl Acad Sci U S A 106:8707–8712. doi: 10.1073/pnas.0900436106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Adler H, Steer B, Juskewitz E, Kammerer R. 2014. Murine gammaherpesvirus 68 (MHV-68) escapes from NK-cell-mediated immune surveillance by a CEACAM1-mediated immune evasion mechanism. Eur J Immunol 44:2521–2522. doi: 10.1002/eji.201444593. [DOI] [PubMed] [Google Scholar]
- 123.Yamin R, Kaynan NS, Glasner A, Vitenshtein A, Tsukerman P, Bauman Y, Ophir Y, Elias S, Bar-On Y, Gur C, Mandelboim O. 2013. The viral KSHV chemokine vMIP-II inhibits the migration of naive and activated human NK cells by antagonizing two distinct chemokine receptors. PLoS Pathog 9:e1003568. doi: 10.1371/journal.ppat.1003568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jochum S, Moosmann A, Lang S, Hammerschmidt W, Zeidler R. 2012. The EBV immunoevasins vIL-10 and BNLF2a protect newly infected B cells from immune recognition and elimination. PLoS Pathog 8:e1002704. doi: 10.1371/journal.ppat.1002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stanton RJ, Prod'Homme V, Purbhoo MA, Moore M, Aicheler RJ, Heinzmann M, Bailer SM, Haas J, Antrobus R, Weekes MP, Lehner PJ, Vojtesek B, Miners KL, Man S, Wilkie GS, Davison AJ, Wang ECY, Tomasec P, Wilkinson GWG. 2014. HCMV pUL135 remodels the actin cytoskeleton to impair immune recognition of infected cells. Cell Host Microbe 16:201–214. doi: 10.1016/j.chom.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Smith W, Tomasec P, Aicheler R, Loewendorf A, Nemcovicova I, Wang ECY, Stanton RJ, MacAuley M, Norris P, Willen L, Ruckova E, Nomoto A, Schneider P, Hahn G, Zajonc DM, Ware CF, Wilkinson GWG, Benedict CA. 2013. Human cytomegalovirus glycoprotein UL141 targets the TRAIL death receptors to thwart host innate antiviral defenses. Cell Host Microbe 13:324–335. doi: 10.1016/j.chom.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Verma S, Loewendorf A, Wang Q, McDonald B, Redwood A, Benedict CA. 2014. Inhibition of the TRAIL death receptor by CMV reveals its importance in NK cell-mediated antiviral defense. PLoS Pathog 10:e1004268. doi: 10.1371/journal.ppat.1004268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dimitrova M, Zenarruzabeitia O, Borrego F, Simhadri VR. 2016. CD300c is uniquely expressed on CD56bright natural killer cells and differs from CD300a upon ligand recognition. Sci Rep 6:23942. doi: 10.1038/srep23942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, Tom I, Ivelja S, Refino CJ, Clark H, Eaton D, Grogan JL. 2009. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol 10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 130.Béziat V, Liu LL, Malmberg J-A, Ivarsson MA, Sohlberg E, Björklund AT, Retière C, Sverremark-Ekström E, Traherne J, Ljungman P, Schaffer M, Price DA, Trowsdale J, Michaëlsson J, Ljunggren H, Malmberg K. 2013. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121:2678–2688. doi: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Alvarado-Hernández DL, Benítez-Sánchez A, Rodríguez-Cuevas JS, Rosales-Saavedra T, Guerra-Palomares SE, Comas-García A, Noyola DE, García-Sepúlveda CA. 2016. Killer-cell immunoglobulin-like receptors and cytomegalovirus reactivation during late pregnancy. Int J Immunogenet 43:189–199. doi: 10.1111/iji.12271. [DOI] [PubMed] [Google Scholar]
- 132.van Duin D, Avery RK, Hemachandra S, Yen-Lieberman B, Zhang A, Jain A, Butler RS, Barnard J, Schold JD, Fung J, Askar M. 2014. KIR and HLA interactions are associated with control of primary CMV infection in solid organ transplant recipients. Am J Transplant 14:156–162. doi: 10.1111/ajt.12532. [DOI] [PubMed] [Google Scholar]
- 133.Slavuljica I, Busche A, Babić M, Mitrović M, Gašparović I, Cekinović D, Car EM, Pugel EP, Ciković A, Lisnić VJ, Britt WJ, Koszinowski U, Messerle M, Krmpotić A, Jonjić S. 2010. Recombinant mouse cytomegalovirus expressing a ligand for the NKG2D receptor is attenuated and has improved vaccine properties. J Clin Invest 120:4532–4545. doi: 10.1172/JCI43961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Peng H, Tian Z. 2017. Natural killer cell memory: progress and implications. Front Immunol 8:1143. doi: 10.3389/fimmu.2017.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bommareddy PK, Patel A, Hossain S, Kaufman HL. 2017. Talimogene laherparepvec (T-VEC) and other oncolytic viruses for the treatment of melanoma. Am J Clin Dermatol 18:1–15. doi: 10.1007/s40257-016-0238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Altomonte J, Wu L, Meseck M, Chen L, Ebert O, Garcia-Sastre A, Fallon J, Mandeli J, Woo SLC. 2009. Enhanced oncolytic potency of vesicular stomatitis virus through vector-mediated inhibition of NK and NKT cells. Cancer Gene Ther 16:266–278. doi: 10.1038/cgt.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Alvarez-Breckenridge CA, Yu J, Price R, Wojton J, Pradarelli J, Mao H, Wei M, Wang Y, He S, Hardcastle J, Fernandez SA, Kaur B, Lawler SE, Vivier E, Mandelboim O, Moretta A, Caligiuri MA, Chiocca EA. 2012. NK cells impede glioblastoma virotherapy through NKp30 and NKp46 natural cytotoxicity receptors. Nat Med 18:1827–1834. doi: 10.1038/nm.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yung GLP, Rieben R, Bühler L, Schuurman HJ, Seebach JD. 2017. Xenotransplantation: where do we stand in 2016? Swiss Med Wkly 147:w14403. [DOI] [PubMed] [Google Scholar]
- 139.Kim JS, Choi SE, Yun IH, Kim JY, Ahn C, Kim SJ, Ha J, Hwang ES, Cha CY, Miyagawa S, Park CG. 2004. Human cytomegalovirus UL18 alleviated human NK-mediated swine endothelial cell lysis. Biochem Biophys Res Commun 315:144–150. doi: 10.1016/j.bbrc.2004.01.027. [DOI] [PubMed] [Google Scholar]