ABSTRACT
Human papillomavirus 16 (HPV16) is the leading cause of cervical cancer. For initial infection, HPV16 utilizes a novel endocytic pathway for host cell entry. Unique among viruses, uptake occurs asynchronously over a protracted period of time, with half-times between 9 and 12 h. To trigger endocytic uptake, the virus particles need to undergo a series of structural modifications after initial binding to heparan sulfate proteoglycans (HSPGs). These changes involve proteolytic cleavage of the major capsid protein L1 by kallikrein-8 (KLK8), exposure of the N terminus of the minor capsid protein L2 by cyclophilins, and cleavage of this N terminus by furin. Overall, the structural changes are thought to facilitate the engagement of an elusive secondary receptor for internalization. Here, we addressed whether structural changes are the rate-limiting steps during infectious internalization of HPV16 by using structurally primed HPV16 particles. Our findings indicate that the structural modifications mediated by cyclophilins and furin, which lead to exposure and cleavage, respectively, of the L2 N terminus contribute to the slow and asynchronous internalization kinetics, whereas conformational changes elicited by HSPG binding and KLK8 cleavage did not. However, these structural modifications accounted for only 30 to 50% of the delay in internalization. Therefore, we propose that limited internalization receptor availability for engagement of HPV16 causes slow and asynchronous internalization in addition to rate-limiting structural changes in the viral capsid.
IMPORTANCE HPVs are the main cause of anogenital cancers. Their unique biology is linked to the differentiation program of skin or mucosa. Here, we analyzed another unique aspect of HPV infections using the prototype HPV16. After initial cell binding, HPVs display an unusually protracted residence time on the plasma membrane prior to asynchronous uptake. As viruses typically do not expose themselves to host immune sensing, we analyzed the underlying reasons for this unusual behavior. This study provides evidence that both extracellular structural modifications and possibly a limited availability of the internalization receptor contribute to the slow internalization process of the virus. These findings indicate that perhaps a unique niche for initial infection that could allow for rapid infection exists. In addition, our results may help to develop novel, preventive antiviral measures.
KEYWORDS: virus entry, papillomavirus, structural modification, furin, kinetic
INTRODUCTION
Papillomaviruses (PV) are a large family of small, nonenveloped DNA viruses comprising more than 190 types (1), which cause species- and tissue-specific infections (2). Most importantly, they are human pathogens that infect skin and mucosa. Although most infections are asymptomatic or cause benign warts, they also lead to a type-dependent development of invasive cancers upon long-term persistence (2). In particular, high-risk human papillomavirus 16 (HPV16) alone causes over 60% of all cervical cancers (2).
The molecular mechanism facilitating the transmission of HPV is understudied. However, it is clear that it involves the nonenveloped capsids, which are built by L1 and L2, the major and minor capsid proteins, respectively. L1 self-assembles into an icosahedral (T=7) capsid, which consists of 72 pentamers, also called capsomers (3, 4). Hydrophobic interactions stabilize the capsomers, which are interlinked by a C-terminal arm invading the neighboring pentamer to form the capsid (5, 6). After initial assembly, the capsid is stabilized by interpentameric disulfide bonds, which are formed during particle maturation (7, 8). On average, 12 to 72 copies of L2 are incorporated into the capsid, where they locate mostly within the luminal cavities of the capsomers (9). The minor capsid protein L2 aids in genome encapsidation, which in turn is chromatinized by cellular histones (10). The presence of the viral DNA (vDNA) further compacts and stabilizes the capsid (11, 12).
Capsids are generated during the unique life cycle of HPVs, which is tightly linked to the differentiation program of epithelial tissues. HPVs infect basal stem cells or transiently amplifying cells of wounded epithelia (13). In basal cells, the viral genome is maintained at a low copy number (14). Only upon differentiation into suprabasal cells, viral genome amplification and virus assembly occurs, whereupon infectious particles are shed from the cornified layers (2, 15). This renders a quantitative purification of uniform HPVs from tissues extremely challenging. Therefore, most research on the structure and entry of HPVs relies on so-called pseudoviruses (PsVs). These are L1/L2 particles that harbor a chromatinized reporter plasmid instead of the viral genome (7, 10). PsVs are immunologically indistinct from authentic viruses (2), which indicates that PsVs constitute an excellent and relevant tool to study host cell entry of HPVs.
During the multistep procedure of entry into host cells, the protective capsids undergo sequential structural changes to eventually release the viral genomes into the host nuclei for viral gene expression and replication, a process termed uncoating (16). Like for other viruses, uncoating of HPV virions occurs through interactions with cellular factors and within different chemical milieus of discrete cellular compartments. For HPVs, uncoating already starts with binding. HPV16 binds primarily to heparan sulfate proteoglycans (HSPGs) on the host cell surface or in the extracellular matrix (ECM) (17–23). In the ECM, it can also bind laminin 332 as a transient coreceptor (24). Importantly, the interaction of HPV16 with glycosaminoglycan (GAG) chains of HSPGs triggers an initial conformational change in the L1 capsid (22). This is followed by a cascade of structural alterations that eventually lead to a receptor switch from HSPGs to an elusive secondary receptor (25). First, the HSPG-induced conformational change in the capsid facilitates a subsequent cleavage of L1 by the trypsin-like serine protease kallikrein-8 (KLK8), presumably allowing access of further enzymes (26). Next, cyclophilins externalize the N terminus of L2 from the capsid lumen (27). Initial work suggested that externalization of the L2 N terminus renders it accessible for the cellular protein convertase furin, which removes 12 amino acids proximal to the N terminus and thereby exposes a conserved, cross-neutralizing epitope of L2 termed RG-1 (28, 29). However, the importance of cyclophilin activity at the plasma membrane for HPV16 infection has recently been questioned (30), as efficient furin cleavage of L2 was observed even upon inhibition of cyclophilins. This leaves the interdependence of cyclophilin action and furin cleavage in question. Nevertheless, these changes are thought to represent the terminal modification of HPV capsids and to result in primed virus particles that are ready to interact with the elusive secondary receptor (25, 31, 32).
Secondary receptor engagement likely triggers the uptake of HPV16 by a clathrin-, caveolin-, lipid raft-, and dynamin-independent but actin-dependent endocytic pathway (33, 34). The virus is routed through the endolysosomal system, where L1 capsomers and a subviral complex of L2 and vDNA are almost completely separated by cyclophilins to enable trafficking of L2/vDNA to the Golgi complex (33, 35–37). After penetration, the subviral complex gains access to the nuclear space only upon nuclear envelope breakdown (NEBD) during mitosis (38, 39).
Live-cell imaging of HPV16 uptake revealed that, while single endocytic events are quick and occur within minutes after commitment to it, they occur over a protracted period of time in a seemingly stochastic manner (33). In different studies, the half-time of uptake has been estimated at 4 to 12 h following binding to cells (19, 22, 25, 33, 40–42). Interestingly, our previous work indicates that infectious internalization exhibits a lag time of about 2 to 4 h (33). The underlying reason for asynchronous internalization or the lag time is still unknown. Nevertheless, it is reasonable to assume that extracellular structural changes or accessibility of the elusive secondary receptor causes asynchronous internalization.
To date, it is unclear whether the enzymatic reactions responsible for the structural alterations occur stochastically at different capsomers, whether subsequent changes require a threshold number of modified capsomers, or whether individual capsomers are fully structurally modified in a sequential manner. In addition to the kinetics of biochemical reactions, the availability of the cellular interaction partners for the structural changes may limit processing kinetically.
As suggested above, the availability of the secondary receptor for virus endocytosis may play a role. However, addressing receptor availability is challenging, mostly because its identity still remains elusive (32). Based on the requirement for HPV16 entry, various factors, including alpha 6-integrin, epidermal growth factor receptors, tetraspanin CD151, and annexin A2 heterotetramer, have been put forward as potential candidates (33, 43–49). However, biochemical data demonstrating direct interactions of receptor candidates and virus particles are mostly lacking. Moreover, it is possible that several receptor candidates are needed to form a complex or internalization platform for HPV16 endocytosis (32). This complex, in turn, may exist in a preassembled form or may assemble only upon induction.
Since the secondary receptor (or receptor complex) that triggers virus endocytosis remains poorly defined, our study focused on whether the extracellular structural modifications of HPV16 virions are important for asynchronous uptake. For this, we used structurally primed HPV16 particles that reflect different stages of conformational changes, including (i) furin-precleaved HPV16 (FPC-HPV16) particles (50, 51), (ii) HPV16 particles that expose the L2 N terminus independently of cyclophilins (L2-GP-N HPV16) (27), (iii) KLK8-precleaved HPV16 (26), and (iv) combinations thereof.
Our results indicated that externalization of the L2 N terminus by cyclophilin B contributed only to a minor extent to asynchronous uptake. However, furin cleavage was clearly rate limiting. Since any further structural changes on the cell surface did not contribute to asynchronous internalization, it is likely that the availability of the secondary receptor(s) contributes to a large extent to the asynchronous internalization of HPV16.
RESULTS
FPC-HPV16 shares important structural characteristics with HPV16.
To test whether conformational changes within the HPV16 capsid would be rate limiting for infectious internalization, we used structurally primed virions that mimic individual steps of the conformational and structural alterations during entry. If any of those changes were rate limiting, primed virions should enter more synchronously and quickly.
First, FPC-HPV16 virions were employed, which are thought to represent terminally modified virions and are supposed to interact directly with the secondary receptor (complex) (50, 51). Previously, FPC-HPV16 has been produced by exogenously adding recombinant furin during the maturation of PsVs (50) or by using furin-overexpressing producer cells (51). Here, the two methods were used in parallel to result in indistinguishable virion preparations and infectivities (data not shown). In the following, only the results from FPC-HPV16 virions produced in furin-overexpressing cells are given.
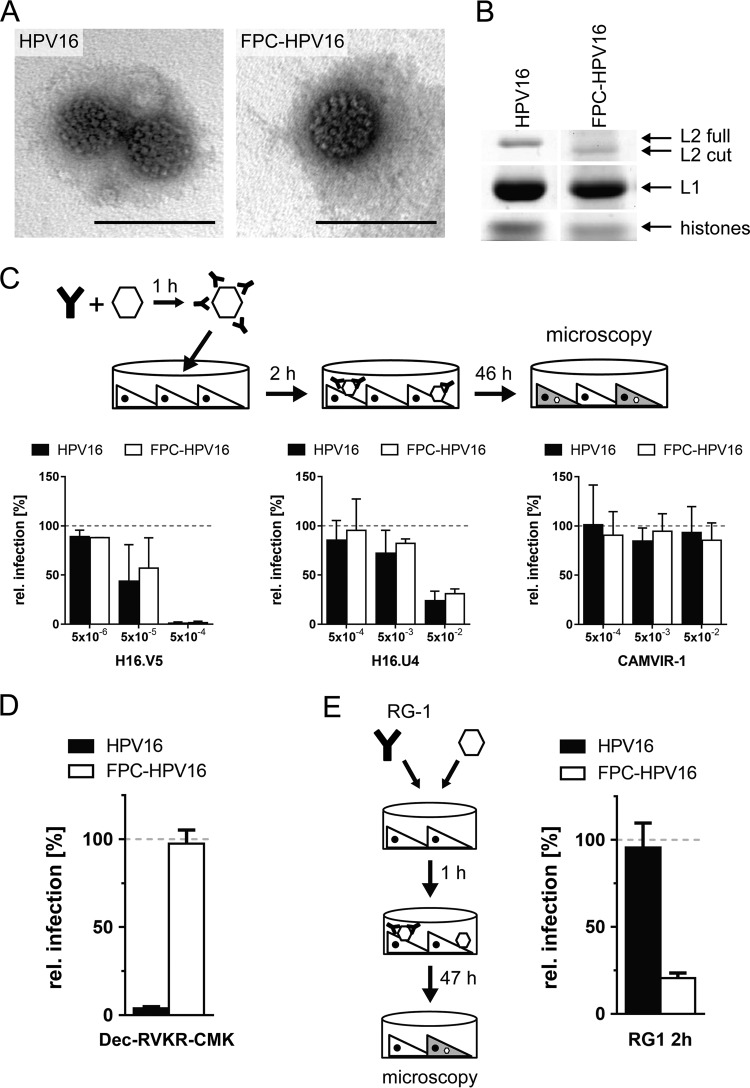
Initially, the FPC-HPV16 particles were thoroughly characterized in comparison to HPV16 particles. By electron microscopy after negative staining, HPV16 and FPC-HPV16 were morphologically indistinct, suggesting correct particle assembly (Fig. 1A). To assess potentially more subtle assembly defects as well as the effectiveness of furin cleavage of L2, the composition of virions was analyzed after SDS-PAGE and Coomassie blue staining by densitometry. Quantification of L2 (∼75 kDa), L1 (∼50 kDa), and cellular histones (10 to 25 kDa) suggested that similar amounts of L2 and histones in relation to L1 were incorporated into HPV16 and FPC-HPV16 virions as expected (Fig. 1B) (51). Furin precleavage appeared to be quantitative in FPC-HPV16 particles, as any uncleaved L2 signal was undetectable (Fig. 1B; note the slightly higher mobility of L2 for FPC-HPV16 than for HPV16).
FIG 1.
FPC-HPV16 assemble correctly. (A) HPV16 and FPC-HPV16 particles were analyzed by electron microscopy after negative staining. Depicted are representative images of virions. Bars, 100 nm. (B) HPV16 and FPC-HPV16 particle composition by SDS-PAGE and Coomassie blue staining. Note the shift in the size of the furin-cleaved L2 protein (L2 cut). (C) Surface epitopes of HPV16-GFP (black) and FPC-HPV16-GFP (white) particles were targeted with neutralizing antibody H16.V5 (left) or H16.U4 (middle) or nonneutralizing control antibody CAMVIR-1 (right) at indicated dilutions. Presented are infection values (relative proportion of GFP-expressing cells) in HeLa cells relative to untreated controls in percentages ± standard deviations (SD). (D) Dependence on furin of HPV16 (black) or FPC-HPV16 (white) infection was tested in the presence of 10 μM furin inhibitor Dec-RVKR-CMK or, as a control, dimethyl sulfoxide (DMSO). Depicted are infection values relative to DMSO-treated controls in percentages ± SD. (E) To analyze structural processing, HPV16 or FPC-HPV16 and the L2-neutralizing RG-1 antibody were added to HeLa cells. Depicted are infection values relative to mouse serum-treated controls in percentages ± SD.
To test for the conformational integrity of L1 capsomers in FPC-HPV16, neutralizing antibodies against defined epitopes (H16.V5 or H16.U4 [52, 53]) or, as a control, the nonneutralizing antibody CAMVIR-1 was employed in inhibition experiments. H16.V5 binds an epitope at the top of the pentamer (5, 52–54), and H16.U4 binds to a C-terminal conformational epitope (5, 55), whereas CAMVIR-1 binds to a linear epitope (amino acids 206 to 210 of HPV16 L1 [56, 57]). After antibody binding to virions, the viruses were added to HeLa cells, and their residual infectivity was determined. The presence of H16.V5 or H16.U4 reduced infection in a dose-dependent manner for FPC-HPV16 and HPV16 to a similar extent, whereas CAMVIR-1 did not affect infection as expected (Fig. 1C) (52, 53, 56, 57). This meant that both V5 and U4 epitopes were exposed in FPC-HPV16 particles, similar to what was seen in HPV16 particles.
Next, we tested whether furin precleavage was functional. As HPV16 requires furin-cleavage and FPC-HPV16 should not (28), infectivity assays were carried out in the presence of the furin inhibitor Dec-RVKR-CMK. The infectivity of FPC-HPV16 remained unaffected in the presence of Dec-RVKR-CMK, whereas HPV16 infectivity was abolished as expected (Fig. 1D) (28). As a second measure, the exposure of the so-called RG-1 epitope was tested. The cross-neutralizing RG-1 epitope of L2 is exposed only upon furin cleavage (29). Using antibody neutralization directed against this RG-1 epitope prior to infection (58), FPC-HPV16 infection was efficiently neutralized with only a residual infectivity remaining, whereas HPV16 infection was unperturbed (Fig. 1E). This indicated that in FPC-HPV16 particles the RG-1 epitope was exposed, whereas in HPV16 it was not.
Overall, our results indicated that FPC-HPV16 particles assembled correctly, exposed crucial epitopes on the capsid surface, and were efficiently and functionally precleaved by furin.
FPC-HPV16 enters host cells by the same pathway as HPV16.
A prerequisite for studying the internalization kinetics of FPC-HPV16 is that it would enter host cells by the same mechanism as HPV16. To this end, we analyzed for the first time the entry mechanism of FPC-HPV16 in comparison to HPV16 by testing key cellular factors involved in this process. For this, we made use of small-molecule inhibitors directed against cellular functions required for HPV16 entry. In particular, we tested factors important for clathrin-, caveolin-, flotillin-, cholesterol-, and dynamin-independent but actin-dependent endocytosis, endosomal acidification, and nuclear envelope breakdown for nuclear import of HPV16 (33, 39).
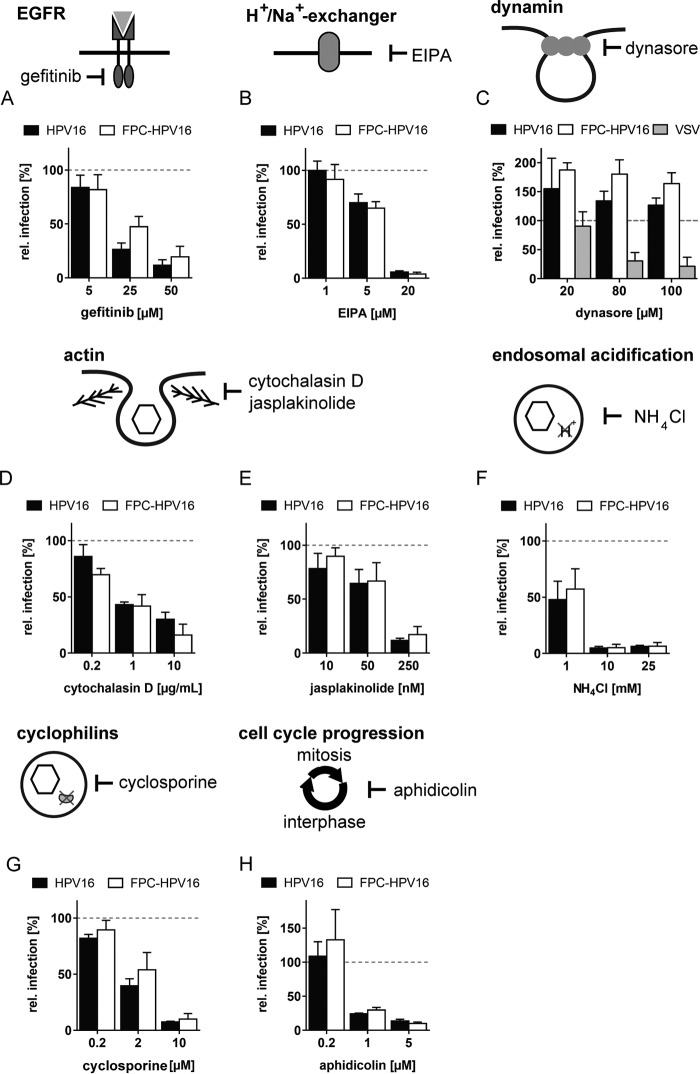
Endocytosis of HPV16 requires an activation of epidermal growth factor receptor (EGFR) signaling and the activity of Na+/H+ exchangers (33, 45). EGFR signaling was blocked using gefitinib (Iressa), which targets the tyrosine kinase domain of the EGFR family (59). HPV16 and FPC-HPV16 exhibited a similar, dose-dependent reduction of infection to 12% ± 5% and 14% ± 10%, respectively (Fig. 2A). The same was true when the function of the Na+/H+ exchanger was blocked with the amiloride derivative EIPA [5-(N-ethyl-N-isopropyl)amiloride], reducing infection to 6% ± 1% and 4% ± 2% for HPV16 and FPC-HPV16, respectively (Fig. 2B).
FIG 2.
FPC-HPV16 and HPV16 enter host cells by the same entry pathway. HeLa cells were infected with HPV16 (black), FPC-HPV16 (white), or VSV (gray) in the presence of the indicated concentrations of gefitinib (A), EIPA (B), dynasore (C), cytochalasin D (D), jasplakinolide (E), NH4Cl (F), cyclosporine (G), or aphidicolin (H). Depicted are infection values relative to solvent-treated controls in percentages ± SD.
Many endocytic pathways depend on dynamin-2 to mediate scission of forming vesicles (60, 61). However, HPV16 endocytosis occurs independently of dynamin-2 (33). To test whether FPC-HPV16 endocytosis would additionally require dynamin, cells were infected in the presence of dynasore, a selective dynamin inhibitor (62). As a positive control, the functionality of the drug was assessed using vesicular stomatitis virus (VSV) infection, which occurs by clathrin-mediated endocytosis requiring dynamin (63). As expected, a dose-dependent reduction of VSV infection was observed in the presence of dynamin (Fig. 2C). In contrast, both HPV16 and FPC-HPV16 infections were increased by the presence of dynasore as previously reported (Fig. 2C) (33, 34). Increase of HPV16 infection has been attributed to an upregulation of dynamin-independent pathways (33, 34). In conclusion, FPC-HPV16, like HPV16, was internalized by dynamin-2-independent endocytosis.
Actin dynamics mediate vesicle scission during HPV16 endocytosis (33). The actin depolymerizing agent cytochalasin D reduced the relative infectivity of HPV16 and FPC-HPV16 dose dependently to 30% ± 6% and 21% ± 10%, respectively (Fig. 2D). Similarly, jasplakinolide, an inhibitor of actin depolymerization, resulted in a reduction of infection to 12% ± 2% and 19% ± 9% for HPV16 and FPC-HPV16, respectively (Fig. 2E). Thus, actin dynamics were crucial for endocytosis of both HPV16 and FPC-HPV16. Based on the inhibitory profile, infectious uptake of FPC-HPV16 occurred via an endocytic machinery similar to that of HPV16.
After internalization, HPV16 traffics through the endolysosomal system and depends on endosomal acidification presumably for uncoating (33, 35). To analyze whether FPC-HPV16 also requires acid activation in the endosomal pathway, we neutralized the endosomal pH with NH4Cl, followed by infection with HPV16 and FPC-HPV16. Infection was completely inhibited for both HPV16 and FPC-HPV16 in the presence of 10 mM NH4Cl (Fig. 2F). In addition to endosomal acidification, HPV uncoating depends on the activity of cyclophilins in endosomes. Bienkowska-Haba and colleagues showed that cyclophilins separate L1 capsomers from a subviral complex consisting of L2 and the chromatinized genome in early endosomes (35). Incidentally, cyclophilin activity is additionally required for the exposure of the L2 N terminus for subsequent cleavage by furin at the plasma membrane (27). Cyclosporine, an inhibitor of cyclophilins, was used to analyze the requirement for cyclophilins during FPC-HPV16 infection. HPV16 and FPC-HPV16 infections were inhibited similarly in the presence of cyclosporine (Fig. 2G). Thus, FPC-HPV16 likely required cyclophilins for uncoating such as HPV16.
In addition to the trafficking events directing HPV16 to the perinuclear space, we tested whether the import of FPC-HPV16 into the nucleus would occur in a similar fashion. Nuclear import of HPV16 genomes depends on nuclear envelope breakdown (NEBD) (38, 39). Aphidicolin, an inhibitor of progression through the synthesis phase (S phase), prevents HPV16 vDNA delivery to the nucleus (39). When cell cycle progression was blocked by aphidicolin, a dose-dependent reduction of infection to 14% ± 3% and 10% ± 2% was observed for HPV16 and FPC-HPV16, respectively (Fig. 2H). This suggested that NEBD was also essential for nuclear import of FPC-HPV16.
Taken together, our data indicated that endocytosis and entry of FPC-HPV16 occurred by a mechanism similar to that of HPV16, allowing kinetic studies of FPC-HPV16 particles.
Furin processing is rate limiting for virus uptake.
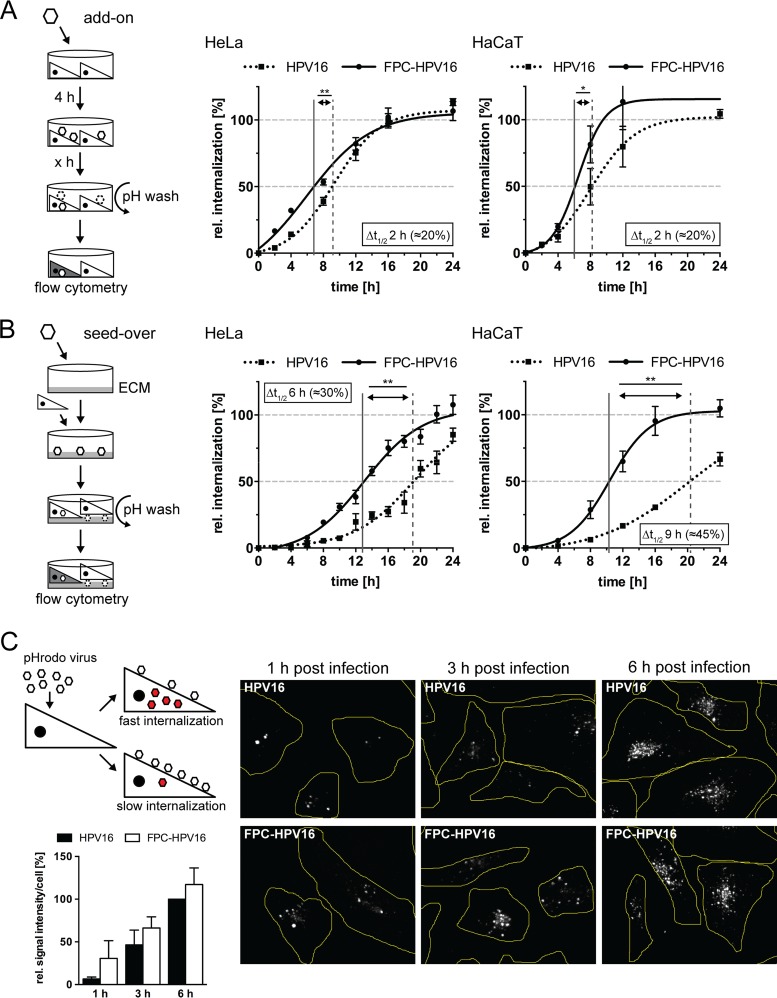
To determine whether structural preprocessing would lead to a faster and more synchronous internalization of viruses, we compared the infectious internalization kinetics of HPV16 and preprocessed HPV16. For this, we used a previously established assay to exclusively measure the internalization of infectious virions (33). Briefly, virions were bound to cells in the cold. After binding, the cells were shifted to 37°C to allow endocytosis. At various time points following temperature shift, extracellular virus particles were rendered noninfectious by a high-pH wash. The infectivity of virions already internalized at the time point of the pH wash was then scored by measuring the relative percentage of green fluorescent protein (GFP)-positive cells at 48 h postinfection (p.i.). By using low multiplicities of infection (MOIs), i.e., less than one infectious unit per cell, the entry dynamics of the average virion entering a cell is assessed rather than that of the fastest of several infectious virions. Consistent with our previous work (22, 33), HPV16 infectious internalization followed a sigmoidal time curve with a lag time of about 2 h in HeLa and HaCaT cells (Fig. 3A, dotted line), and with a half-time of about 9 h for HPV16 in HeLa and HaCaT cells (Fig, 3A, dotted line), FPC-HPV16 exhibited a different internalization curve with no or reduced lag phase in HeLa and HaCaT cells, respectively, and a linear increase within the first 10 h of infectious internalization (Fig. 3A, solid lines). The infectious internalization for FPC-HPV16 in HeLa and HaCaT cells had a half-time of about 6 to 7 h and thus was 20 to 25% faster than HPV16 (Fig. 3A, solid lines). This suggested that N-terminal cleavage of L2 by furin restricts uptake and contributes to an initial lag phase, which thereby represents a rate-limiting step for HPV16 internalization. Notably, virus uptake still occurred asynchronously, indicating that additional factors for slow and asynchronous virus uptake existed.
FIG 3.
Infectious internalization of FPC-HPV16 is faster and more synchronous than that of HPV16. (A) In an add-on experiment, HPV16 (dotted) or FPC-HPV16 (solid) was used to infect HeLa (left) or HaCaT (right) cells. At indicated times postinfection, extracellular virus was inactivated by a high-pH wash (pH 10.5). Depicted are the half-times of infectious internalization and the difference in the half-time between the two viruses in hours and percentages. (B) A seed-over experiment was performed as described for panel A. Infectious internalization values were normalized to the 48-h samples and are depicted in percentages ± standard errors of the means (SEM) for all experiments. Curves were fitted with the nonlinear regression function of GraphPad Prism v6. Statistical significance was tested by two-tailed unpaired Student's t test in GraphPad Prism v6; P values are indicated by asterisks: *, P < 0.05; **, P < 0.01. (C) pHrodo-HPV16 and pHrodo-FPC-HPV16 particles were added to cells, and the pHrodo signal was imaged at different time points p.i. and analyzed as relative intensity per cell.
Alternatively to binding to HSPGs, HPV16 can interact with laminin 332 in the extracellular matrix (24). We had previously observed that infectious internalization of ECM-bound HPV16 is significantly slower than that of cell surface-bound virus with a half-time of about 18 h (22, 33). Presumably, ECM binding restricts the diffusion of particles and sampling of the cell surface (32, 64). To corroborate our findings on the internalization kinetics, we compared our findings for uptake of cell surface-bound virus in the “add-on” assay with those for uptake of ECM-bound virions in what we called the “seed-over” assay. For this, FPC-HPV16 and HPV16 were bound to HaCaT-derived ECM. Subsequently, HeLa or HaCaT cells were seeded on top of the ECM with bound virus. As before, a high-pH wash was performed at different times after seeding, followed by infectivity analysis of the internalized virus. Again, HPV16 infectious internalization followed a sigmoidal time curve notably with an increased lag time of about 4 to 6 h (Fig. 3B, dotted line). The half-times of infectious internalization were about 19 h and 20 h for HPV16 in HeLa and HaCaT, respectively, as expected (Fig. 3B, dotted line) (22). FPC-HPV16 also showed a sigmoidal internalization curve, however, with a reduced lag phase of about 4 h. The linear increase of infectious internalization started as early as 8 h p.i., i.e., roughly 4 h earlier than for HPV16 (Fig. 3B, solid versus dotted line). This resulted in half-times of infectious internalization of 11 to 13 h, which were 30 to 45% faster than for HPV16, similar to the results in the add-on experiments (Fig. 3B, solid line). However, FPC-HPV16 still exhibited consistently asynchronous internalization kinetics, suggesting the existence of additional determinants thereof.
The infectious internalization assay indicated an overall faster uptake of FPC-HPV16. However, as this assay relies on successful delivery of the pseudogenomes to the nucleus from already internalized virions, we cannot formally rule out any effect of later entry steps. Therefore, our findings were corroborated using another internalization assay. In this assay, virus particles that were covalently labeled with a pH-sensitive fluorophore (pHrodo) were employed. pHrodo emits fluorescence exclusively in acidic environments, such as endosomes (65). pHrodo-HPV16 and pHrodo-FPC-HPV16 particles were added to cells, and the pHrodo signal was imaged at different time points p.i. and analyzed as relative intensity per cell. At 1 and 3 h after addition to cells, a stronger fluorescence signal was observed for FPC-HPV16 than for HPV16 (Fig. 3C). The increase of fluorescence intensities leveled off at 6 h p.i. for both viruses (not shown), probably because the degradation of L1/fluorophore in lysosomes counteracted a further increase from additional incoming virions (33). A clear increase of FPC-HV16-derived fluorescence was visible in all individual experiments, despite the variability in the degree of increase.
As such, these results were consistent with faster internalization like in the infectious internalization assay. Notably, the kinetics of the pHrodo assay were faster than for infectious internalization. This likely reflects the higher particle numbers that had to be used to score the pHrodo signals. The higher particle number necessitated measuring not only the average infectious particle but also additional noninfectious particles. Moreover, it is likely that the fastest virions gave rise to a fluorescence signal, thus explaining the faster kinetics.
Overall, the results indicated that furin cleavage was a rate-limiting step for HPV16 infectious internalization, and precleavage partially reduced or even eliminated the lag phase prior to internalization. However, further determinants clearly contributed to the protracted, asynchronous mode of virus internalization. Possibly, these determinants include further rate-limiting structural modifications and/or a limited availability of the secondary receptor (complex). Interestingly, the prolonged lag time of ECM- versus cell-bound virions may suggest mechanistic differences of HPV16 entry depending on their initial binding.
KLK8 cleavage is not rate limiting for HPV16 internalization.
To analyze further structural modifications that may affect the kinetics of internalization in addition to and independently of furin cleavage, we focused on those generated by KLK8 and CyPB, which are crucial for L2 exposure and cleavage. HSPG-induced conformational changes were omitted from further analysis, as our previous work already showed that they are not rate limiting for infectious internalization (22). Since the proteolytic processing of L1 by KLK8 follows the HSPG-induced changes, it was analyzed next.
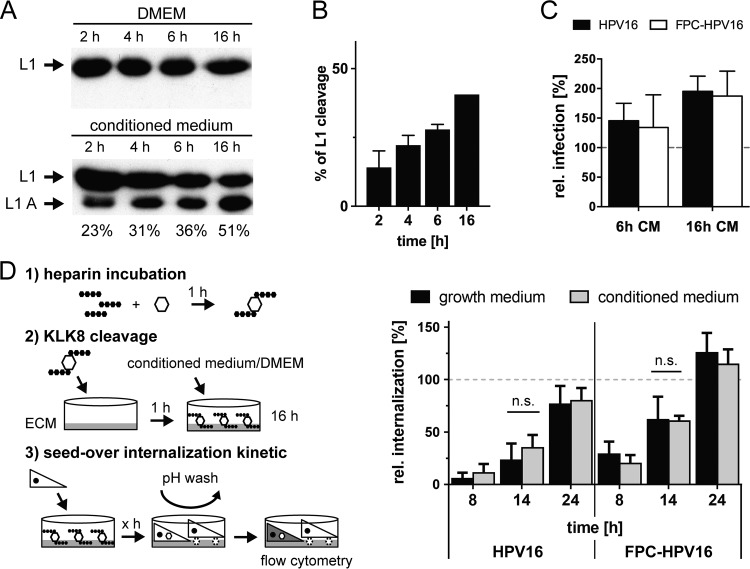
KLK8 cleaves HPV16 L1 extracellularly after a crucial interaction of virions with HS and promotes efficient exposure of the L2 RG-1 epitope (26). In order to preprocess virions, the particles were thus pretreated with heparin, bound to ECM, and incubated with conditioned medium, a supernatant of cells that contains secreted KLK8 according to published procedures (26). Treatment with cell culture medium alone serves as a negative control, as it fails to mediate L1 cleavage (Fig. 4A) (26). To contribute to asynchronous internalization, KLK8 cleavage would have to occur asynchronously as well. Therefore, the kinetics of KLK8 cleavage were analyzed after different times of virus incubation with conditioned medium. In Western blotting, cleaved L1 (L1 A) showed a faster mobility of about 5 kDa compared to full-length L1 (Fig. 4, L1 A) (26). After 2 h, 30% of total L1 was converted into L1 A, which reached a maximum of 50% at 16 h (Fig. 4B). These results indicated that KLK8 cleavage occurred in part protractedly. Since only about 30 to 50% of the L1 molecules could be cleaved in vitro, the cleavage site may be obscured for the remaining L1 molecules in virus particles. At this point, we analyzed the infectivity resulting from the KLK8-precleaved ECM-bound virus particles. Interestingly, infectivity increased proportionally to the preincubation time with conditioned medium and resulted in a relative increase of infection to about 180% at maximal KLK8 cleavage (Fig. 4C). This effect was observed for HPV particles as well as for FPC-HPV particles. Overall this indicated that KLK8 cleavage may be inefficient during infection of tissue culture cells.
FIG 4.
KLK8 cleavage is not rate limiting for HPV16 internalization. (A) Kinetics of L1 cleavage by KLK8 was analyzed by incubation of HPV16 PsVs with 10 mg/ml heparin, binding to ECM, and incubation with conditioned medium or, as a control, DMEM. Samples were analyzed at the indicated times following conditioned medium incubation by SDS-PAGE and Western blotting using L1-specific CAMVIR-1 antibody. L1 cleavage bands (L1 A) were quantified by densitometry using Fiji. (B) Depicted is the increase of the cleaved L1 band (L1 A) normalized to the average maximal cleavage reached after 16 h of incubation with the batches of conditioned medium used in the experiments in percentages ± SD (n = 2). (C) Infection levels of HPV16 or FPC-HPV16 after 6 or 16 h of preincubation with conditioned medium (CM) relative to levels after preincubation with DMEM in percentages ± SD. (D) Schematic depiction of the experimental setup for KLK8 pretreatment. Virus particles are pretreated with heparin for structural activation (1) and then bound to HaCaT ECM and treated with conditioned medium or DMEM for 16 h (2). Seed-over infectious internalization (3) was performed as described before. Infection of HPV16 (left) or FPC-HPV16 (right) preincubated with growth medium (black) or conditioned medium (gray) for 16 h as described for panel A in a seed-over setup (compare with Fig. 3B). Depicted are infectious internalization values normalized to the 48-h samples in percentages ± SD.
To analyze if particles with maximal KLK8 cleavage increased the kinetics of uptake, KLK8-cleaved HPV16 and FPC-HPV16 were analyzed in a seed-over experiment. However, KLK8-precleaved viruses did not enter more synchronously or quickly (Fig. 4D). Hence, we conclude that KLK8 processing was not rate limiting for internalization.
Conformational changes by cyclophilins modestly accelerate virus uptake.
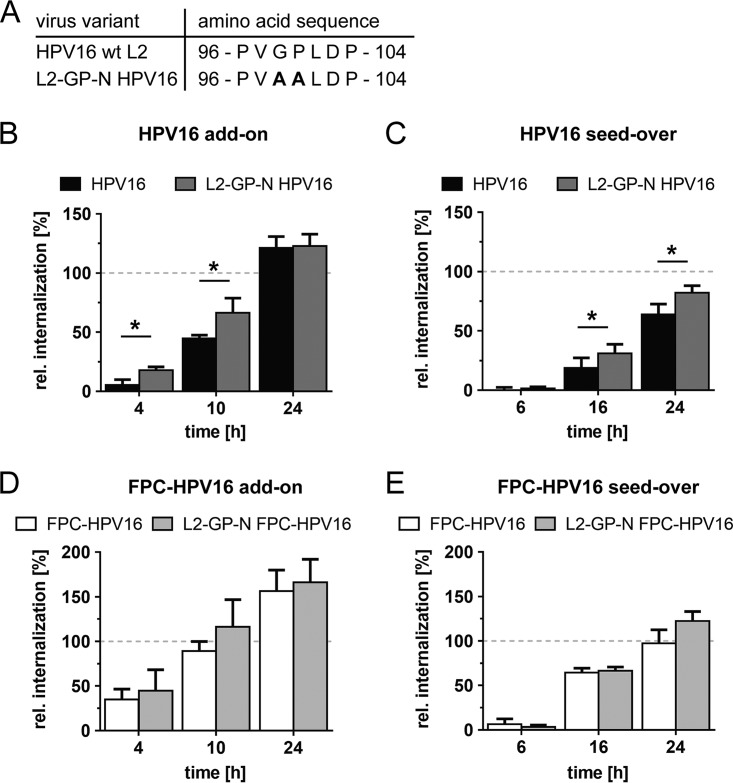
The exposure of the cross-neutralizing L2 epitope RG-1 was originally shown to be dependent on the peptidyl-prolyl isomerase function of cyclophilins through externalization of the L2 N terminus (27, 35). Mutations in L2 that result in a higher flexibility of the molecule (Fig. 5A) render RG-1 exposure of the resulting virus particles (L2-GP-N HPV16) cyclophilin independent (27). Despite recent evidence that furin cleavage itself can occur without the help of cyclophilins (30), we assessed whether higher flexibility of L2 would influence the rate of internalization. Using L2-GP-N HPV16 particles, we analyzed whether they internalized at different rates for selected time points at which FPC-HPV16 internalization kinetics differed most from HPV16. The uptake kinetics of L2-GP-N HPV16 were slightly but significantly faster than those of HPV16 (Fig. 5B). At 4 h and 10 h p.i., the numbers of internalized L2-GP-N HPV16 were increased about 3.5-fold and 1.5-fold, respectively. Similar effects were observed in seed-over experiments, resulting in 1.7-fold and 1.3-fold increases of L2-GP-N HPV16 internalization at 16 h and at 24 h p.i., respectively (Fig. 5C). Thus, exposure of the L2 N terminus may be partially rate limiting, with an estimated difference in half-time of infectious internalization of about 10%.
FIG 5.
CyPB activity modestly accelerates HPV16 internalization. (A) Amino acid sequences of the putative L2 CyPB binding motif (HPV16 versus HPV16 L2-GP-N). (B) The infectious internalization kinetics of HPV16 (black) and HPV16 L2-GP-N (dark gray) were compared in add-on experiments (as described for Fig. 3A). (C) Infectious internalization of HPV16 (black) and HPV16 L2-GP-N (dark gray) was analyzed in seed-over experiments (as described for Fig. 3B). (D) The infectious internalization kinetics of FPC-HPV16 (white) and FPC-HPV16 L2-GP-N (light gray) were compared in add-on experiments. (E) Infectious internalization of FPC-HPV16 HPV16 (white) and HPV16 L2-GP-N (light gray) were analyzed in seed-over experiments. Infectious internalization values are shown relative to 48-h infection samples in percentages ± SD for all experiments.
Next, we asked whether the contributions of cyclophilins to asynchronous internalization would be additive to those of L2 cleavage. Interestingly, when comparing L2-GP-N FPC-HPV16 to FPC-HPV16, the internalization rates were indistinct (Fig. 5D and E). This meant that once furin cleavage had occurred, cyclophilins were no longer rate limiting for uptake. This is consistent with the notion that furin-precleaved particles represent terminally modified virions.
Asynchronous uptake is independent of the cell cycle.
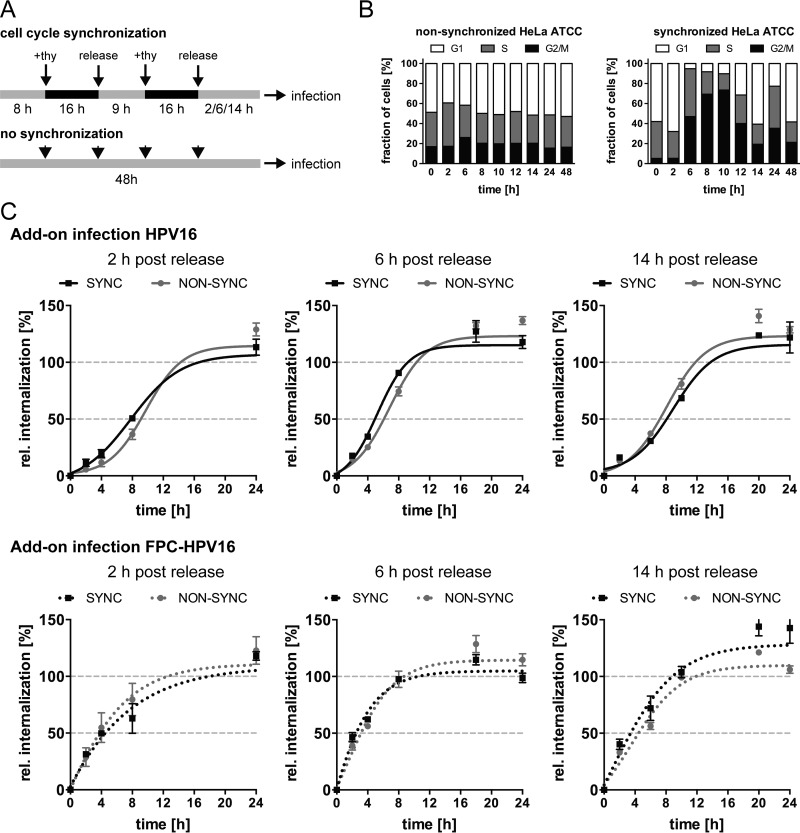
Despite the contribution of structural alterations of the viral capsid to asynchronous internalization, our data indicated that additional factors must play a role. Previous work from the Banks laboratory (66) showed that cell cycle synchronization accelerates initial infection. To assess whether cell cycle synchronization would impact HPV16 uptake, the kinetics of infectious internalization were analyzed in synchronized cells. Following the published protocol, cells were synchronized by a double thymidine arrest in S phase, whereas unsynchronized cells were used as controls (Fig. 6A). As expected, about 8 to 10 h postrelease from S phase, a wave of mitoses was observed (Fig. 6B). Cells were infected 2 h, 6 h, and 14 h postrelease with HPV16 and FPC-HPV16 under add-on conditions to cover time points further, close to and past the ensuing mitotic wave after S-phase release. The rates of HPV16 and FPC-HPV16 internalization remained unaffected in synchronized compared to unsynchronized cells under all tested conditions, suggesting that cell cycle synchronization failed to facilitate a more synchronous HPV16 endocytosis (Fig. 6C). Interestingly, 6 h postrelease from thymidine blocking, a similarly shortened half-time of infectious internalization was observed for both HPV16 and FPC-HPV16 and in both synchronized and nonsynchronized cells. This meant that this effect was not due to furin cleavage or cell cycle synchronization. However, it may indicate that cells were primed for uptake by the medium change that occurred at the time of release from the S-phase block.
FIG 6.
Infectious internalization kinetics of HPV16 are unaffected by cell cycle synchronization. (A) Schematic depiction of cell cycle synchronization by double thymidine block. (B) Synchronized and nonsynchronized cells were fixed with ethyl alcohol (EtOH) and analyzed for their cell cycles states at different times postrelease. Cell cycle phases were designated according to cellular DNA content by PI staining and flow-cytometric analysis. G1, a single set of chromosomes (i.e., DNA content = 1); G2/M, duplicated chromosomes (DNA content = 2); S, replicating chromosomes between the two states. Shown are the values from two independent experiments. (C) Infectious internalization of HPV16 (upper panels) and FPC-HPV16 (lower panels) with synchronized (black dotted line) and nonsynchronized (gray dotted line) cells was analyzed in add-on experiments. Cells were infected at 2 h, 6 h, or 14 h postrelease from thymidine block. Curves were fitted with the nonlinear regression function of GraphPad Prism v6. Relative infectious internalization values were normalized to 48-h infection samples and are depicted as percentages ± SEM.
FPC-HPV infection is independent of structural activation by HSPGs.
Since cell cycle synchronization did not shed more light on a potential cellular regulator of virus uptake, we turned back to the question of secondary-receptor engagement and its potential role in restricting virus uptake. Binding of HPV16 to HSPGs leads to a first conformational change (22, 26, 67), which in itself is insufficient for infectious internalization (25). The subsequent structural modifications of the capsid are thought to enable the engagement of an internalization receptor (complex) independent of HSPGs (32). According to this model, the affinity of the virus to HSPGs is reduced by the structural alterations to allow transfer to the elusive secondary receptor (25). FPC-HPV16 as terminally restructured particles should therefore be able to infect cells independently of HSPGs.
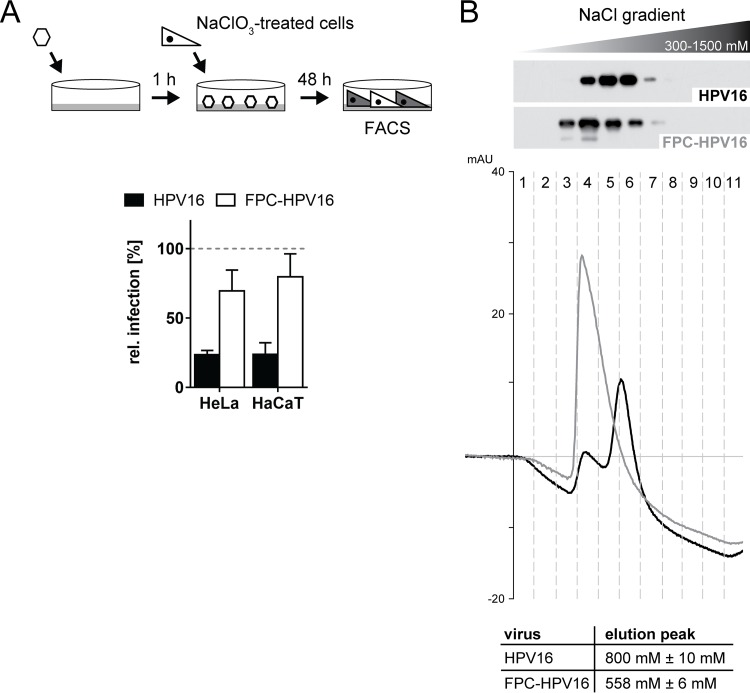
To test this, FPC-HPV16 was used to infect cells devoid of functional HSPGs. For this, the virus was bound to laminin 332 as a transient receptor in the ECM. HeLa and HaCaT cells were treated with NaClO3 to result in strongly reduced levels of GAG sulfation (68), so that they became nonpermissive for virus infection depending on sulfated HSPGs (22). These cells were subsequently seeded on top of the virus. As expected, HPV16 control infection was strongly reduced upon NaClO3 treatment (Fig. 7A, black bars) (22). In contrast, FPC-HPV caused infection of NaClO3-treated cells reaching about 75 to 80% of the infectivity in untreated HeLa or HaCaT cells (Fig. 7A, white bars). This strongly indicated that FPC-HPV16 infection was independent of HSPGs in line with being terminally restructured.
FIG 7.
FPC-HPV infection occurs independently of HSPGs, and particles show reduced binding affinity to heparin. (A) HPV16 (black) or FPC-HPV16 (white) infection of NaClO3-treated HeLa or HaCaT cells in the seed-over setup (as described for Fig. 3B). Infectious values are shown in percentages ± SD relative to untreated cell samples for all experiments. (B) Heparin binding affinity of HPV16 (black) and FPC-HPV16 (gray) was analyzed using HiTrap heparin HP columns (GE) and subsequent elution by increasing salt concentrations (300 mM to 1,500 mM). Depicted is a representative Western blot for HPV16 L1 of the eluted fractions 1 to 11 of HPV16 (upper panel) or FPC-HPV16 (lower panel) during salt gradient elution. Representative chromatograms of the HPLC runs were aligned to the corresponding elution fractions (HPV16, black line; FPC-HPV16, gray line). Salt concentrations corresponding to the main elution peak from at least 3 independent experiments are summarized in the table.
FPC-HPV16 exhibits reduced binding affinity to highly sulfated heparan sulfates.
As infection with FPC-HPV16 was independent of HSPG interaction, we addressed whether preprocessed particles indeed exhibited a decreased affinity to HSPGs as previously proposed (25, 31, 32). For this, heparin, representing the highly sulfated domains of HS, was used as a proxy (22). The affinities of FPC-HPV16 and HPV16 to heparin were analyzed by binding virus particles to a HiTrap heparin HP column and by subsequently eluting the viruses with a linear NaCl gradient. Eluted fractions were collected and tested for L1 content by Western blotting. HPV16 was eluted in fractions 4 to 7 after applying the gradient (Fig. 7B, upper panel), whereas FPC-HPV16 particles were already observed in fraction 3 (Fig. 7B, lower panel). The elution of FPC-HPV16 with lower salt concentrations indicated that the affinity of FPC-HPV16 to heparin was lower. The more-detailed chromatograms confirmed this result. The main elution peak for HPV16 was detected at 800 ± 10 mM NaCl, whereas the main peak for FPC-HPV16 particles was already detected at 558 ± 6 mM NaCl. These data provided the first biochemical evidence that HPV16 particles exhibit a reduced affinity to highly sulfated HS once they are processed by furin.
The accessibility of the secondary receptor for HPV16 interaction is limited.
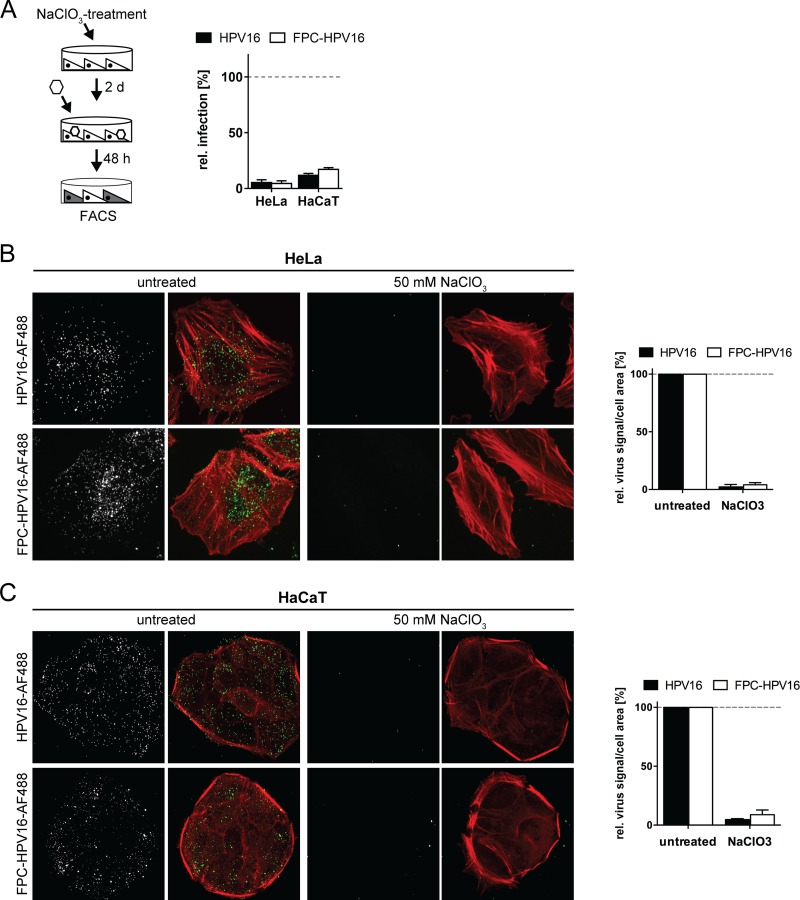
After verification of the proposed functional properties of FPC-HPV16, the next test assessed whether FPC-HPV16 particles would bind to cells exclusively via the secondary receptor. Again, NaClO3-treated cells were employed as a model for dysfunctional HSPGs. In add-on experiments, HPV16 infection was abrogated in NaClO3-treated HeLa and HaCaT cells, most likely because binding to cells was impaired (Fig. 8A) (22). FPC-HPV16 was also unable to infect NaClO3-treated HeLa or HaCaT cells (Fig. 8A). While FPC-HPV16 infection occurred in the seed-over setup through binding to laminin 332 (Fig. 7A), these results indicated that FPC-HPV16 was unable to directly bind to NaClO3-treated cells.
FIG 8.
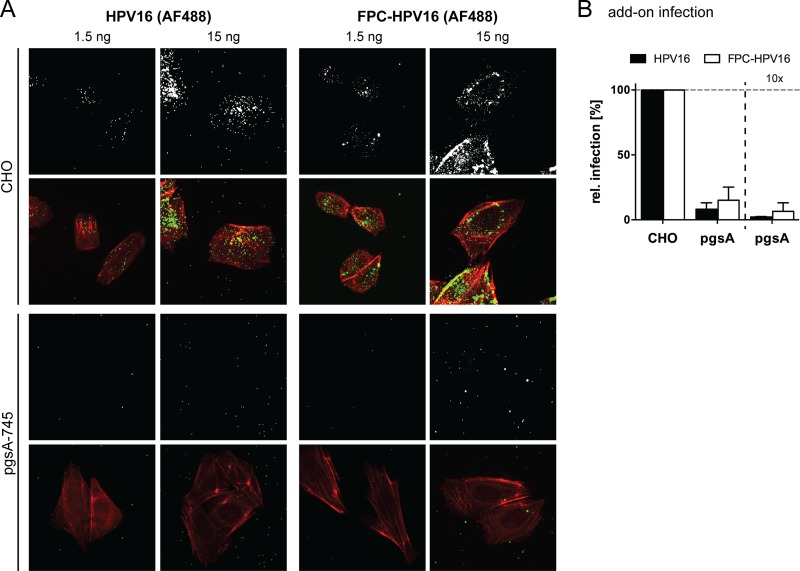
The accessibility of the secondary receptor for virus interaction is limited. (A) HeLa or HaCaT cells were treated with 50 mM NaClO3 for 2 days and subsequently infected with HPV16 (black) or FPC-HPV16 (white) in add-on experiments (as described for Fig. 3A). Relative infection values were normalized to 48-h infection samples of untreated cells in percentages ± SD. FACS, fluorescence-activated cell sorting. (B) AF488-labeled HPV16 (gray/green, upper panels) or FPC-HPV16 (gray/green, lower panels) particles were bound to untreated or NaClO3-treated HeLa cells. Cells were fixed and stained for F-actin with Atto647N-phalloidin (red). Depicted are representative maximum intensity projections of confocal image stacks from at least three independent experiments. Virus signals were quantified relative to cell area for more than 150 cells per condition. (C) As described for panel B, using HaCaT cells.
This was surprising, since previous work suggested that FPC-HPV16 can bind directly to HSPG-deficient cells (50). To verify that in fact virus binding to cells was inefficient, we tested virus binding to NaClO3-treated cells using virions covalently labeled with fluorophores (64). Fluorescence microscopy showed that HPV16 and FPC-HPV16 readily bound to HeLa and HaCaT cells (Fig. 8B and C). However, binding was almost completely lost in NaClO3-treated cells for both HPV16 and FPC-HPV16 (Fig. 8B and C). The same was true when binding and infection was analyzed in pgsA-745 cells (Fig. 9), which lack GAGs due to a mutation in the xylosyltransferase gene (69). Although these findings supported primarily FPC-HPV16's ineffectiveness to bind to cells in the absence of highly sulfated proteoglycans, it is important to note that the quantifications showed a tendency of FPC-HPV16 to interact slightly better with HS-deficient cells than did HPV16.
FIG 9.
(A) Indicated amounts of AF488-labeled HPV16 (gray/green, left panels) or FPC-HPV16 (gray/green, right panels) particles were bound to CHO wild-type (upper panels) or pgsA-745 (lower panels) cells for 1 h. Cells were fixed, stained for F-actin (red), and imaged as described for Fig. 8B. Shown are maximum intensity projections of representative cells from 1 of at least 3 independent experiments. (B) Add-on infection of pgsA cells with different amounts of HPV16 (black) and FPC-HPV16 (white) compared to parental CHO cells. Shown are average relative infection values in percentages ± SD from 3 independent experiments.
Most importantly, however, the overall results suggest that while infection by FPC-HPV16 occurs efficiently in the absence of HS, these particles bound only inefficiently to cells. This may indicate that the secondary receptor (complex) is, at least for the most part, unavailable for incoming FPC-HPV16 particles.
DISCUSSION
In this work, the underlying cause for slow and asynchronous uptake of HPV16 was studied. Using structurally primed virions, furin cleavage was identified as an important rate-limiting factor for HPV16 internalization. In addition, cyclophilin activity, likely during L2 N-terminal exposure, was modestly rate limiting. However, structural priming accounted for only up to one-half of the slow uptake. Thus, our work not only linked structural priming to slow and asynchronous internalization of HPV16 but also highlighted the existence of further rate-limiting steps. With these findings, we propose a model in which initial structural modifications in combination with secondary-receptor availability are impeding rapid virus uptake. This may point to a unique niche of HPV16 infection, e.g., a wounding scenario during initial infection, which may provide sufficient enzyme activity for the structural alterations and/or the availability of the secondary receptor (complex).
With internalization half-times ranging between 4 and 15 h, the slow and asynchronous HPV uptake into cells is well established (19, 22, 25, 33, 41, 42, 70). However, the wide range of time spans for internalization that have been reported is surely noteworthy. Most likely, they are the result of different experimental setups. For example, HPV internalization kinetics were characterized by indirectly assessing postbinding antibody neutralization (19, 42, 70), by immunofluorescence analysis of remaining cell surface virus (41), or by accumulation of intracellular virus signal (25). Particularly, analysis by immunofluorescence does not differentiate between infectious and noninfectious particles, thus very likely overestimating the relevant degree of uptake. The infectious internalization assay of this study analyzes exclusively the uptake of virus particles that give rise to infection featuring lower multiplicities of infection (MOI) and longer half-times (22, 33). This notion is supported by our own experiments using the pHrodo assay (Fig. 3C). Therefore, the timing of internalization may be more accurately described by the infectious internalization assay. One drawback of this assay is that it cannot fully exclude kinetic effects on later stages of entry, since it relies on the delivery of the vDNA to the nucleus. Membrane penetration and nuclear import are the main additional reasons for asynchronous entry of papillomaviruses, and both are coordinated by the onset of mitosis (39, 71, 72). Since mitosis occurred in all samples asynchronously and cell cycle synchronization did not affect infectious internalization, any major impact on these late stages appears unlikely but remains a formal possibility. However, both the pHrodo assay and the infectious internalization assay determined that infectious uptake occurred generally faster for FPC-HPV16 particles.
In the infectious internalization assay, HPV16 internalization exhibited pronounced lag phases of 4 h and 8 h in add-on and seed-over experiments, respectively. The significant delay in the seed-over experiments may simply be due to removal of cell surface receptors by trypsinization, necessitating reexpression to facilitate virus uptake. Alternatively, the different mobility of cell surface-bound versus ECM-bound virions may affect the kinetics. Cell surface-bound virions would more easily be able to sample the plasma membrane for the presence of a secondary receptor (complex) during lateral diffusion than the ECM-bound virions. Thus, they would stochastically interact more frequently with required cellular receptors. Also, if the virus were partially covered by or embedded in the ECM, ECM remodeling might be required to facilitate transfer to the plasma membrane. In support of this notion, previous work suggests that matrix metalloproteinases (MMPs), which are typically responsible for ECM remodeling, are required for HPV16 infection (26, 45). As initial ECM rather than cell binding appears to be the major route of infection in vivo (21, 73), it will be interesting to address in more detail in the future the question of ECM remodeling and whether wound healing contributes to it.
Independent of the infection scenario, the internalization kinetics was unequivocally reduced in furin-precleaved virus. This not only indicated that furin cleavage is a rate-limiting step during HPV16 entry but also hinted that the amount of active furin in the cellular supernatants may be limiting. This is supported by findings of Bronnimann and colleagues (30), who reported that HPV16 cleavage by furin and the infectivity of the particles are more efficient if furin is enriched in the cultivation medium. It has been reported that membrane-anchored furin can be shed from the cell surface by unknown proteases while keeping its activity (74). To date it remains unknown whether the membrane-bound or the soluble form of furin acts on HPV16 particles or whether a specific activation of furin or of (re-)localization to the plasma membrane would be needed. Furin has more than 490 predicted targets (75); among them are proteases involved in extracellular remodeling, like MMPs, ADAMs (A disintegrin and metalloproteinases), and ADAMTSs (A disintegrin and metalloproteinases with thrombosporin motifs), and in wound healing (76). It is therefore tempting to speculate that HPV16 encounters active furin specifically during wound healing of epidermal tissue, which is thought to provide the virus with access to the initial target cells/ECM.
Despite the notion that too little furin is present in tissue culture cells to facilitate rapid or efficient processing, it is important to note that evidence for the stoichiometry and the extent of structural modifications during entry is still lacking. Do modifications occur strictly sequentially? How quickly can structural modifications be achieved? Do all pentamers of a virion or only a subset have to be modified? Initial evidence exists at least for the latter: recent structural data suggest that conformational changes occur upon HS binding primarily on the pentavalently coordinated vertex pentamers (67).
As an alternative to the model where structural changes occur postbinding, it has been hypothesized that HPV16 particles may be preprocessed during terminal differentiation of infected epithelia and are released as terminally restructured particles during desquamation, potentially leading to quicker entry (32, 77, 78). Nevertheless, this alternative model raises at least two major questions. First, our data showed that preprocessed viruses (e.g., FPC-HPV16) bind less efficiently to heparan sulfate moieties than unprocessed virions. Why would a virus have evolved to use a less efficient receptor-binding particle at a critical step of infection? Or is it that preprocessed particles bind more efficiently to the extracellular matrix? At least in the mouse vaginal challenge model, the latter does not occur (21). However, whether the basement membrane of the mouse vagina provides a niche fully equivalent to the human counterpart still needs to be verified. Second, the preexposure of the cross-neutralizing RG-1 epitope of L2 that occurs during structural processing seems counterintuitive. It is generally thought that exposure of the RG-1 epitope occurs shortly before internalization to hide the epitope within the particle (58), which in turn may be part of the immune evasion mechanism of HPVs during host cell entry (79, 80). So how would HPVs efficiently evade the immune system, if the virus particles—even if preprocessed—do not enter quickly but remain accessible for immune detection, as our data indicate?
Despite the identification of furin cleavage as a major rate-limiting step for HPV16 internalization, the question remains as to which other processes may contribute to the slow, asynchronous uptake. Neither HSPG binding nor KLK8 cleavage played a role (Fig. 4) (22). Increased flexibility of the L2 N terminus (L2-GP-N mutant) (27) was rate limiting only prior to particle processing by furin (Fig. 5). The latter observation suggests a sequential processing of HPV16, with furin cleavage as the final processing step, consistent with the favorite model (31, 32). Bronnimann and colleagues (30) recently suggested that cyclophilin and furin may not act interdependently, as L2 processing by furin was observable upon cyclophilin inhibition. Exposure of the cross-neutralizing RG-1 epitope, however, required furin cleavage as well as cyclophilin activity. To date, it remains uncertain to which degree furin cleavage and L2 isomerization would be interdependent or would have to occur sequentially. Here, we did not assess whether furin cleavage occurs independently from L2 isomerization or not. Nevertheless, our data indicate that L2 isomerization does not contribute to protracted internalization per se but only prior to furin cleavage.
Although speculative, further rate-limiting structure modifications of the virus particle could be envisioned and would provide a rationale for the remaining asynchronism of HPV16 uptake. In addition, there are further determinants that may affect the kinetics of HPV16 uptake. For example, the role of the L2 N-terminal peptide released by furin is unclear. An essential role during host cell entry is unlikely, since FPC-HPV16 particles lacking this peptide are fully capable of entering cells. However, an accessory role such as timely activation of HPV16 endocytosis may be possible. Alternatively, the peptide may simply have to be removed to activate L2 for entry. The generation or removal of the peptide may thus be important for efficiency or kinetics or plays a role during in vivo infection. Hence, potential functions of the L2 N-terminal peptide should be addressed in future studies.
Also important for rate-limiting HPV16 uptake would be the presence and accessibility of the secondary receptor (complex). We found that FPC-HPV16 particles exhibited infrequent binding to cells in the absence of sulfated HSPG upon NaClO3 treatment or in HSPG-deficient pgsA-745 cells, which indicated that the secondary receptor is not readily available or is bound only with low affinity. Our data seemingly contradict a previous report, in which binding of FPC-HPV16 particles to NaClO3-treated or pgsA cells was observed (50). These differences may result from experimental setups. In the earlier study by Day and colleagues (50), FPC-HPV16 with only partially cleaved L2 was used, whereas in our study virtually all L2 molecules were processed. Speculatively, the degree of furin cleavage may be crucial for efficient engagement of the secondary receptor. Perhaps the conformational changes elicited during entry are necessitated on a specific subset of capsomers, whereas modification of all capsomers is not favorable. Similar to what is observed for KLK8 cleavage (26), furin cleavage may thus occur only partially during infection (as has been observed in reference 30).
Another difference was that previous work used 10-fold more virus but failed to quantify the extent of binding. However, even higher particle numbers failed to significantly increase the number of bound particles on the cell surface in comparison to HPV16 in immunofluorescence assays (Fig. 9 and data not shown) or Western blotting (data not shown).
In conclusion, the secondary receptor may not be bound efficiently by fully processed FPC-HPV16 or may in fact not be accessible for binding.
The availability or accessibility of the elusive secondary receptor is an intriguing question and may also be crucial for its identification. So far, integrin α6 (ITGα6), tetraspanin CD151, epidermal growth factor receptor (EGFR), and annexin A2 heterotetramer (A2t) were identified as crucial plasma membrane molecules for endocytosis (33, 43–49, 81, 82), but whether they act as or regulate the secondary receptor is unknown. In a synthetic model, the presence of a receptor complex including all of these components has been proposed (32). A number of scenarios may serve as rationales for rate limitation of uptake by these secondary receptor candidates: (i) absence from the plasma membrane, which requires induction of expression; (ii) need for a triggered lateral formation of a complex in the membrane; (iii) dissociation of a receptor from complexes upon stimulation, and so on. Induction of any of these processes may occur by signaling through the EGFR, which is observed quickly with or after HPV16 binding (33, 45). Moreover, the requirement of tetraspanins for HPV16 infection is intriguing (47). Tetraspanins are prime candidates for the formation of nano- and microdomains, the tetraspanin-enriched microdomains (TEMs), and are able to interact with integrins (reviewed in references 83 and 84). TEMs have also been linked to host cell entry of other viruses, e.g., hepatitis C virus (HCV) and human cytomegalovirus (HCMV) (85, 86). The interaction of the internalization receptor candidate tetraspanin CD151 with ITGα3 and ITGα6 and with other tetraspanins has been linked to HPV16 internalization (47). Therefore, an efficient assembly of a TEM-dependent HPV16 entry platform requires a prolonged time period. Alternatively, receptor candidates may be trapped in preexisting functional cellular complexes unavailable to engage HPV16. TEMs are known to compartmentalize the plasma membrane by clustering proteins in specific membrane domains, thereby modulating their availability for other processes or interactions (87). As the TEM composition is dynamic (88, 89), release of HPV16 receptor candidates from TEM may be required for uptake. However, the latter possibility seems less likely, as HPV16 has been shown to cointernalize with CD151 (47).
Examples for such preexisting structures are hemidesmosomes, which establish adhesive connections of the basal cells with the ECM and basement membrane (reviewed in reference 90). Hemidesmosomes are complexes that incorporate, e.g., ITGα6/β4 and CD151. Disassembly of hemidesmosomes is required during migration and wound healing processes (91). It is possible that HPV16 interaction with ITGα6 and CD151 may require induction of hemidesmosome disassembly.
To date it is still unknown whether furin secretion and/or release is increased during wound healing, but it is tempting to speculate that endocytosis of HPV16 in its biological niche occurs during wound healing. This specific environment would be enriched with a plethora of secreted proteases to mediate extracellular matrix remodeling during wound healing processes, which could at the same time allow for a more efficient processing of HPV16 particles and the presentation of the secondary receptor (complex).
While our work shows that furin cleavage of HPV16 L2 and potentially the availability of the internalization receptor were limiting the rate of virus uptake, the major question remains whether a biological niche that would promote rapid uptake exists. Future work will therefore have to explore this in more-complex infection systems like organotypic raft culture, in vivo models, and dedicated epithelial wounding scenarios.
MATERIALS AND METHODS
Cell lines, antibodies, and reagents.
HeLa cells were from ATCC. HaCaT cells originated from N. Fusenig (DKFZ, Heidelberg, Germany [92]); these and 293TT cells were a kind gift from J. T. Schiller (NIH, NCI, Bethesda, MD, USA [10]). Parental CHO and pgsA cells originated from J. D. Esko (UCSD, San Diego, CA, USA [69]). 293TTF cells for FPC-HPV16 production and RG-1 antibody were a kind gift from R. Roden (Johns Hopkins University, Baltimore, MD, USA [51]). CAMVIR-1 antibody was from Santa Cruz Biotechnology. Horseradish peroxidase (HRP)-anti-mouse secondary antibody was from GE Healthcare. H16.V5 and H16.U4 were a kind gift from N. Christensen (PSU, Hershey, PA, USA [52, 53]). Furin inhibitor Dec-RVKR-CMK was from Bachem. Gefitinib was from Tocris bioscience. EIPA, cytochalasin D, aphidicolin, NH4Cl, NaClO3, heparin, and thymidine were from Sigma-Aldrich. Cyclosporine and jasplakinolide were from Calbiochem. Dynasore was from Merck. Furin convertase was from Alexis Biochemicals. Alexa Fluor 488 succinimidylester was from Thermo Scientific. Atto647N-phalloidin was from Sigma-Aldrich. RedDot2 was from VWR.
Viruses.
HPV16 PsVs containing a GFP reporter plasmid (HPV16-GFP) were produced using 293TT cells as previously described (7, 93), whereas FPC-HPV16 PsVs were produced using furin-overexpressing cells (293TTF) (51) or as an alternative, in 293TT cells, followed by addition of exogenous furin prior to virus maturation (50). In brief, 293TTF or 293TT cells were transfected with p16SheLL and pCIneo. After 48 h, cells were harvested and lysed. For optimal furin activity, cell lysates were incubated for 24 h and supplemented with 10 mM HEPES (pH 7.6) and 2 mM CaCl2 with or without addition of 20 U exogenous furin, respectively (94). For optimal maturation, lysates were incubated for a further 24 h with 25 mM ammonium sulfate (pH 9.0) (8). PsVs were purified on a 25% to 39% linear OptiPrep gradient (Sigma-Aldrich). For microscopy, HPV16 or FPC-HPV16 were covalently labeled with Alexa Fluor dyes as previously described (64). For production of L2-GP-N HPV16 PsVs (27), the L2 sequence in p16SheLL was mutated by Quikchange II site-directed mutagenesis (Agilent Technologies) using primers 16L2_99A100A_fw (5′-TGGACCCCGTGGCCGCCAGCGACCCCAG-3′) and 16L2_99A100A_rev (5′-CTGGGGTCGCTGGCGGCCACGGGGTCCA-3′). HPV16L2-GP-N PsV were produced as above. Recombinant vesicular stomatitis virus (VSV strain Indiana) expressing GFP (VSV-GFP) was produced as previously described (63).
Analysis of particle composition.
About 1 μg of HPV16 PsV was separated by SDS-PAGE and stained with Coomassie brilliant blue. Linear regression fitting of signal intensity values from densitometry analysis by Fiji was carried out using protein standards (ImageJ distribution [95]).
Infection assays.
For add-on experiments, 5 × 104 cells were seeded 16 h prior to infection with 1 to 5 ng PsVs to result in 20% infected (GFP-expressing) cells. Cells were harvested and analyzed for infection (GFP expression) at 48 h p.i. by flow cytometry. For seed-over experiments, HaCaT-derived extracellular matrix (ECM) was produced as previously described (22). Subsequently, 5 to 15 ng HPV16 PsVs was bound to ECM for 1 h at 37°C in growth medium before 5 × 104 HeLa cells were seeded onto the ECM-bound virus particles. Infectivity was analyzed as described above. For VSV infections, 5 × 104 HeLa cells were infected 24 h with VSV-GFP at a multiplicity of infection of 5 PFU/cell. Cells were infected for 1 h at 37°C followed by a medium change. Infection (GFP expression) was analyzed 6 h p.i. Raw infection values were normalized to untreated control infections to result in relative infection values.
Cell perturbations with small-molecule inhibitors.
Cells were pretreated with inhibitors at the indicated concentrations for 30 min or for aphidicolin for 16 h prior to infection. Cells were infected as described above. To reduce cytotoxicity, inhibitors, except aphidicolin, were replaced by growth medium with 10 mM NH4Cl (10 mM HEPES, pH 7.6) at 12 h p.i. as described previously (33, 39). To decrease glycosaminoglycan sulfation, HeLa or HaCaT cells were pretreated with 50 mM NaClO3 in growth medium for 2 days prior to experimentation.
Antibody neutralization assays.
About 16 h prior to experimentation, 5,000 HeLa cells per well were seeded in optical bottom 96-well plates. HPV16 and FPC-HPV16 were incubated with the L1 antibodies H16.V5, H16.U4, and CAMVIR-1 (dilution range, 1:500 to 1:500,000) for 1 h at room temperature in Dulbecco's modified Eagle medium (DMEM) prior to their addition to cells. Infectivity was scored at 48 h p.i. For this, cell nuclei were stained with RedDot2 before plates were analyzed by automated microscopy (Zeiss Axio Observer Z1, equipped with a Yokogawa CSU22 spinning disc module and a CoolSnap HQ camera; Visitron Systems GmbH) followed by automated computational image analysis as described previously (33). Raw infection values were normalized to untreated control infections to result in relative infection values. For RG-1 neutralization, RG-1 antibody (1:2,500) and PsVs were added to 5,000 HeLa cells per well of a 96-well plate. After 2 h, the inoculum was replaced by growth medium and cells were processed for infection analysis by automated microscopy at 48 h p.i. as described above.
Infectious internalization kinetics.
For add-on internalization experiments, 5 × 104 cells were seeded as above. PsVs (5 to 10 ng) were added to cells on ice and incubated at 4°C for 4 h to allow binding. The medium was removed, and plates were supplemented with warm growth medium and transferred to 37°C. The samples were washed for 90 s with 0.1 M CAPS (N-cyclohexyl-3-aminopropanesulfonic acid) (pH 10.5) buffer at the indicated times postwarming, which renders extracellular virus particles noninfectious (33). Cells were washed twice with phosphate-buffered saline (PBS) before fresh growth medium was added. Cells were fixed with 4% paraformaldehyde (PFA) at 48 h p.i. and analyzed for infection by flow cytometry. For seed-over internalization experiments, we followed the seed-over infection protocol. At the indicated times following cell seeding, cells were treated with CAPS buffer as above and processed for infection analysis at 48 h p.i.
pHrodo internalization assay.
To analyze internalization of virus particles that were labeled with a pH-sensitive dye (pHrodo), 7.5 × 104 cells were seeded on coverslips 24 h prior to experimentation. Similar particle numbers were added to 350 μl growth medium and bound for 1 h at 37°C. Medium was changed, and at least 10 fields of view per sample were imaged as confocal slices with a 40× objective on a spinning disk microscope (as above) at indicated time points. For analysis, average intensity projections were assembled using Fiji (ImageJ distribution [95]). The total virus signal per projections was analyzed using CellProfiler v2.2.0 (96, 97). Intensity values per time point were summarized using Pivot tables and normalized to the number of cells, determined by manual counting from brightfield images.
KLK8 cleavage kinetics.
To analyze the kinetics of KLK8 cleavage, HPV16 samples were preincubated with 10 mg/ml heparin for 2 h at room temperature, bound to ECM, and incubated with conditioned medium or growth medium for the indicated times as described previously (26). Conditioned medium was obtained by harvesting supernatants from HaCaT cells grown for 72 h and stored at 4°C. Lysates were prepared in 5× SDS-loading buffer and analyzed by SDS-PAGE and Western blotting using CAMVIR-1. In addition, samples preincubated for 6 h or 16 h with conditioned medium were subjected to seed-over infections using HeLa cells.
Cell cycle synchronization.
Prior to infections, the cell cycle of 7.5 × 105 HeLa cells was synchronized by a double-thymidine block. In brief, 8 h postseeding, cells were treated with 2 mM thymidine for 16 h, after which thymidine was washed out. After 9 h, cells were blocked again with 2 mM thymidine for a further 16 h. After the thymidine washout, cells were infected with HPV16 or FPC-HPV16 in an add-on experiment to analyze infectious internalization kinetics upon cell cycle synchronization. To assign certain cell cycle states to the cell populations, propidium iodide (PI) staining was performed for several time points postrelease to determine the cellular DNA content of the cells by flow cytometry. Cell cycle phase distributions were determined by Gaussian fitting of the cell populations with FlowJo v9.9.6. The sum of all cells at each time point was used for normalization.
Virus binding.
To assess virus binding, 1 to 2.5 ng AF488-labeled HPV16 or FPC-HPV16 was added to 7.5 × 104 to 10 × 104 cells for 1 h. After fixation in 4% PFA, F-actin was stained with Atto647N-phalloidin and analyzed by microscopy as described above. For quantification of virus binding, z-stacks were acquired using a magnification of ×40. For qualitative analysis, cells were imaged at a magnification of ×63. Maximum intensity projections were analyzed using CellProfiler v2.2.0 (96, 97). In brief, cell areas and virus particles were detected by image segmentation, and average virus intensities were normalized against cell area and depicted relative to virus intensities per cell area in untreated samples. More than 150 cells were analyzed per condition and independent experiment.
Heparin affinity assay.
After equilibration with 10 column volumes of 300 mM NaCl–10 mM phosphate buffer (pH 7.4), 50 ng HPV16 or FPC-HPV16 PsVs was bound to HiTrap heparin HP (1-ml) columns (GE) and washed with 10 column volumes. For elution, a linear NaCl gradient (300 to 1,500 mM) was applied. Five hundred-microliter fractions were collected and frozen in liquid nitrogen. Proteins were precipitated with 0.02% sodium deoxycholate and 10% trichloroacetic acid (TCA) overnight at 4°C. Protein pellets were resuspended in SDS loading buffer. Samples were analyzed by Western blotting, and L1 was detected with CAMVIR-1.
Electron microscopy.
About 1 × 106 to 5 × 106 PsVs in PBS–0.8 M NaCl were absorbed for 1 min on Formvar-coated, carbon-sputtered grids. Particles were contrasted for 7 min with 1% phosphotungstic acid. Samples were analyzed directly after drying. The sample was analyzed at 80 kV on a FEI-Tecnai 12 electron microscope (FEI, Eindhoven, Netherlands). Images of selected areas were documented with an Olympus Veleta 4k charge-coupled-device (CCD) camera.
ACKNOWLEDGMENTS
We thank all members of the Schelhaas laboratory for helpful comments on the manuscript. We also thank R. Roden (Johns Hopkins University, Baltimore, MD, USA) and N. Christensen (PSU College of Medicine, Hershey, PA, USA) for providing reagents and M. Termathe and S. Leidel (MPI, Münster, Germany) for technical support of the HPLC experiments.
This work was supported by funding to M.S. by the German Research Foundation (DFG EXC 1003 [partly]), within the Infect-ERA initiative by funding from the Federal Ministry for Education and Research (BMBF, 031L0095A), and by the International Graduate School 1409 (GRK1409).
REFERENCES
- 1.Bzhalava D, Eklund C, Dillner J. 2015. International standardization and classification of human papillomavirus types. Virology 476:341–344. doi: 10.1016/j.virol.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 2.Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. 2012. The biology and life-cycle of human papillomaviruses. Vaccine 30(Suppl 5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 3.Klug A, Finch JT. 1965. Structure of viruses of the papilloma-polyoma type. I. Human wart virus. J Mol Biol 11:403–423. [DOI] [PubMed] [Google Scholar]
- 4.Baker TS, Newcomb WW, Olson NH, Cowsert LM, Olson C, Brown JC. 1991. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J 60:1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Modis Y, Trus BL, Harrison SC. 2002. Atomic model of the papillomavirus capsid. EMBO J 21:4754–4762. doi: 10.1093/emboj/cdf494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolf M, Garcea RL, Grigorieff N, Harrison SC. 2010. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A 107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck CB, Thompson CD, Pang YY, Lowy DR, Schiller JT. 2005. Maturation of papillomavirus capsids. J Virol 79:2839–2846. doi: 10.1128/JVI.79.5.2839-2846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardone G, Moyer AL, Cheng N, Thompson CD, Dvoretzky I, Lowy DR, Schiller JT, Steven AC, Buck CB, Trus BL. 2014. Maturation of the human papillomavirus 16 capsid. mBio 5(4):e01104-. doi: 10.1128/mBio.01104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buck CB, Cheng N, Thompson CD, Lowy DR, Steven AC, Schiller JT, Trus BL. 2008. Arrangement of L2 within the papillomavirus capsid. J Virol 82:5190–5197. doi: 10.1128/JVI.02726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2004. Efficient intracellular assembly of papillomaviral vectors. J Virol 78:751–757. doi: 10.1128/JVI.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fligge C, Schafer F, Selinka HC, Sapp C, Sapp M. 2001. DNA-induced structural changes in the papillomavirus capsid. J Virol 75:7727–7731. doi: 10.1128/JVI.75.16.7727-7731.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schafer F, Florin L, Sapp M. 2002. DNA binding of L1 is required for human papillomavirus morphogenesis in vivo. Virology 295:172–181. doi: 10.1006/viro.2002.1361. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt A, Rochat A, Zeltner R, Borenstein L, Barrandon Y, Wettstein FO, Iftner T. 1996. The primary target cells of the high-risk cottontail rabbit papillomavirus colocalize with hair follicle stem cells. J Virol 70:1912–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBride AA. 2008. Replication and partitioning of papillomavirus genomes. Adv Virus Res 72:155–205. doi: 10.1016/S0065-3527(08)00404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Longworth MS, Laimins LA. 2004. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev 68:362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerqueira C, Schelhaas M. 2012. Principles of polyoma- and papillomavirus uncoating. Med Microbiol Immunol 201:427–436. doi: 10.1007/s00430-012-0262-1. [DOI] [PubMed] [Google Scholar]
- 17.Joyce JG, Tung JS, Przysiecki CT, Cook JC, Lehman ED, Sands JA, Jansen KU, Keller PM. 1999. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J Biol Chem 274:5810–5822. doi: 10.1074/jbc.274.9.5810. [DOI] [PubMed] [Google Scholar]
- 18.Combita AL, Touze A, Bousarghin L, Sizaret PY, Munoz N, Coursaget P. 2001. Gene transfer using human papillomavirus pseudovirions varies according to virus genotype and requires cell surface heparan sulfate. FEMS Microbiol Lett 204:183–188. doi: 10.1111/j.1574-6968.2001.tb10883.x. [DOI] [PubMed] [Google Scholar]
- 19.Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J Virol 75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson KM, Kines RC, Roberts JN, Lowy DR, Schiller JT, Day PM. 2009. Role of heparan sulfate in attachment to and infection of the murine female genital tract by human papillomavirus. J Virol 83:2067–2074. doi: 10.1128/JVI.02190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kines RC, Thompson CD, Lowy DR, Schiller JT, Day PM. 2009. The initial steps leading to papillomavirus infection occur on the basement membrane prior to cell surface binding. Proc Natl Acad Sci U S A 106:20458–20463. doi: 10.1073/pnas.0908502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerqueira C, Liu Y, Kuhling L, Chai W, Hafezi W, van Kuppevelt TH, Kuhn JE, Feizi T, Schelhaas M. 2013. Heparin increases the infectivity of human papillomavirus type 16 independent of cell surface proteoglycans and induces L1 epitope exposure. Cell Microbiol 15:1818–1836. doi: 10.1111/cmi.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards KF, Bienkowska-Haba M, Dasgupta J, Chen XS, Sapp M. 2013. Multiple heparan sulfate binding site engagements are required for the infectious entry of human papillomavirus type 16. J Virol 87:11426–11437. doi: 10.1128/JVI.01721-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Culp TD, Budgeon LR, Marinkovich MP, Meneguzzi G, Christensen ND. 2006. Keratinocyte-secreted laminin 5 can function as a transient receptor for human papillomaviruses by binding virions and transferring them to adjacent cells. J Virol 80:8940–8950. doi: 10.1128/JVI.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selinka HC, Florin L, Patel HD, Freitag K, Schmidtke M, Makarov VA, Sapp M. 2007. Inhibition of transfer to secondary receptors by heparan sulfate-binding drug or antibody induces noninfectious uptake of human papillomavirus. J Virol 81:10970–10980. doi: 10.1128/JVI.00998-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerqueira C, Samperio Ventayol P, Vogeley C, Schelhaas M. 2015. Kallikrein-8 proteolytically processes human papillomaviruses in the extracellular space to facilitate entry into host cells. J Virol 89:7038–7052. doi: 10.1128/JVI.00234-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bienkowska-Haba M, Patel HD, Sapp M. 2009. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog 5:e1000524. doi: 10.1371/journal.ppat.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards RM, Lowy DR, Schiller JT, Day PM. 2006. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc Natl Acad Sci U S A 103:1522–1527. doi: 10.1073/pnas.0508815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Day PM, Gambhira R, Roden RB, Lowy DR, Schiller JT. 2008. Mechanisms of human papillomavirus type 16 neutralization by l2 cross-neutralizing and l1 type-specific antibodies. J Virol 82:4638–4646. doi: 10.1128/JVI.00143-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bronnimann MP, Calton CM, Chiquette SF, Li S, Lu M, Chapman JA, Bratton KN, Schlegel AM, Campos SK. 2016. Furin cleavage of L2 during papillomavirus infection: minimal dependence on cyclophilins. J Virol 90:6224–6234. doi: 10.1128/JVI.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Day PM, Schelhaas M. 2014. Concepts of papillomavirus entry into host cells. Curr Opin Virol 4C:24–31. doi: 10.1016/j.coviro.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raff AB, Woodham AW, Raff LM, Skeate JG, Yan L, Da Silva DM, Schelhaas M, Kast WM. 2013. The evolving field of human papillomavirus receptor research: a review of binding and entry. J Virol 87:6062–6072. doi: 10.1128/JVI.00330-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schelhaas M, Shah B, Holzer M, Blattmann P, Kuhling L, Day PM, Schiller JT, Helenius A. 2012. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog 8:e1002657. doi: 10.1371/journal.ppat.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spoden G, Kuhling L, Cordes N, Frenzel B, Sapp M, Boller K, Florin L, Schelhaas M. 2013. Human papillomavirus types 16, 18, and 31 share similar endocytic requirements for entry. J Virol 87:7765–7773. doi: 10.1128/JVI.00370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bienkowska-Haba M, Williams C, Kim SM, Garcea RL, Sapp M. 2012. Cyclophilins facilitate dissociation of the HPV16 capsid protein L1 from the L2/DNA complex following virus entry. J Virol 86:9875–9887. doi: 10.1128/JVI.00980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Day PM, Thompson CD, Schowalter RM, Lowy DR, Schiller JT. 2013. Identification of a role for the trans-Golgi network in human papillomavirus 16 pseudovirus infection. J Virol 87:3862–3870. doi: 10.1128/JVI.03222-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie MA, Poffenberger AC, Nelson CD, Atwood WJ, DiMaio D. 2013. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci U S A 110:7452–7457. doi: 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. 2009. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog 5:e1000318. doi: 10.1371/journal.ppat.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aydin I, Weber S, Snijder B, Samperio Ventayol P, Kuhbacher A, Becker M, Day PM, Schiller JT, Kann M, Pelkmans L, Helenius A, Schelhaas M. 2014. Large scale RNAi reveals the requirement of nuclear envelope breakdown for nuclear import of human papillomaviruses. PLoS Pathog 10:e1004162. doi: 10.1371/journal.ppat.1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Day PM, Lowy DR, Schiller JT. 2003. Papillomaviruses infect cells via a clathrin-dependent pathway. Virology 307:1–11. doi: 10.1016/S0042-6822(02)00143-5. [DOI] [PubMed] [Google Scholar]
- 41.Selinka HC, Giroglou T, Nowak T, Christensen ND, Sapp M. 2003. Further evidence that papillomavirus capsids exist in two distinct conformations. J Virol 77:12961–12967. doi: 10.1128/JVI.77.24.12961-12967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith JL, Campos SK, Ozbun MA. 2007. Human papillomavirus type 31 uses a caveolin 1- and dynamin 2-mediated entry pathway for infection of human keratinocytes. J Virol 81:9922–9931. doi: 10.1128/JVI.00988-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evander M, Frazer IH, Payne E, Qi YM, Hengst K, McMillan NA. 1997. Identification of the alpha6 integrin as a candidate receptor for papillomaviruses. J Virol 71:2449–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoon CS, Kim KD, Park SN, Cheong SW. 2001. alpha(6) Integrin is the main receptor of human papillomavirus type 16 VLP. Biochem Biophys Res Commun 283:668–673. doi: 10.1006/bbrc.2001.4838. [DOI] [PubMed] [Google Scholar]
- 45.Surviladze Z, Dziduszko A, Ozbun MA. 2012. Essential roles for soluble virion-associated heparan sulfonated proteoglycans and growth factors in human papillomavirus infections. PLoS Pathog 8:e1002519. doi: 10.1371/journal.ppat.1002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spoden G, Freitag K, Husmann M, Boller K, Sapp M, Lambert C, Florin L. 2008. Clathrin- and caveolin-independent entry of human papillomavirus type 16–involvement of tetraspanin-enriched microdomains (TEMs). PLoS One 3:e3313. doi: 10.1371/journal.pone.0003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheffer KD, Gawlitza A, Spoden GA, Zhang XA, Lambert C, Berditchevski F, Florin L. 2013. Tetraspanin CD151 mediates papillomavirus type 16 endocytosis. J Virol 87:3435–3446. doi: 10.1128/JVI.02906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dziduszko A, Ozbun MA. 2013. Annexin A2 and S100A10 regulate human papillomavirus type 16 entry and intracellular trafficking in human keratinocytes. J Virol 87:7502–7515. doi: 10.1128/JVI.00519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodham AW, Da Silva DM, Skeate JG, Raff AB, Ambroso MR, Brand HE, Isas JM, Langen R, Kast WM. 2012. The S100A10 subunit of the annexin A2 heterotetramer facilitates L2-mediated human papillomavirus infection. PLoS One 7:e43519. doi: 10.1371/journal.pone.0043519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Day PM, Lowy DR, Schiller JT. 2008. Heparan sulfate-independent cell binding and infection with furin-precleaved papillomavirus capsids. J Virol 82:12565–12568. doi: 10.1128/JVI.01631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang JW, Jagu S, Kwak K, Wang C, Peng S, Kirnbauer R, Roden RB. 2014. Preparation and properties of a papillomavirus infectious intermediate and its utility for neutralization studies. Virology 449:304–316. doi: 10.1016/j.virol.2013.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. 1996. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology 223:174–184. doi: 10.1006/viro.1996.0466. [DOI] [PubMed] [Google Scholar]
- 53.Christensen ND, Cladel NM, Reed CA, Budgeon LR, Embers ME, Skulsky DM, McClements WL, Ludmerer SW, Jansen KU. 2001. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 291:324–334. doi: 10.1006/viro.2001.1220. [DOI] [PubMed] [Google Scholar]
- 54.Day PM, Baker CC, Lowy DR, Schiller JT. 2004. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc Natl Acad Sci U S A 101:14252–14257. doi: 10.1073/pnas.0404229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carter JJ, Wipf GC, Benki SF, Christensen ND, Galloway DA. 2003. Identification of a human papillomavirus type 16-specific epitope on the C-terminal arm of the major capsid protein L1. J Virol 77:11625–11632. doi: 10.1128/JVI.77.21.11625-11632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McLean CS, Churcher MJ, Meinke J, Smith GL, Higgins G, Stanley M, Minson AC. 1990. Production and characterisation of a monoclonal antibody to human papillomavirus type 16 using recombinant vaccinia virus. J Clin Pathol 43:488–492. doi: 10.1136/jcp.43.6.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roden RB, Armstrong A, Haderer P, Christensen ND, Hubbert NL, Lowy DR, Schiller JT, Kirnbauer R. 1997. Characterization of a human papillomavirus type 16 variant-dependent neutralizing epitope. J Virol 71:6247–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gambhira R, Karanam B, Jagu S, Roberts JN, Buck CB, Bossis I, Alphs H, Culp T, Christensen ND, Roden RB. 2007. A protective and broadly cross-neutralizing epitope of human papillomavirus L2. J Virol 81:13927–13931. doi: 10.1128/JVI.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barker AJ, Gibson KH, Grundy W, Godfrey AA, Barlow JJ, Healy MP, Woodburn JR, Ashton SE, Curry BJ, Scarlett L, Henthorn L, Richards L. 2001. Studies leading to the identification of ZD1839 (iressa™): an orally active, selective epidermal growth factor receptor tyrosine kinase inhibitor targeted to the treatment of cancer. Bioorg Med Chem Lett 11:1911–1914. doi: 10.1016/S0960-894X(01)00344-4. [DOI] [PubMed] [Google Scholar]
- 60.Doherty GJ, McMahon HT. 2009. Mechanisms of endocytosis. Annu Rev Biochem 78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 61.Mercer J, Schelhaas M, Helenius A. 2010. Virus entry by endocytosis. Annu Rev Biochem 79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 62.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell 10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 63.Johannsdottir HK, Mancini R, Kartenbeck J, Amato L, Helenius A. 2009. Host cell factors and functions involved in vesicular stomatitis virus entry. J Virol 83:440–453. doi: 10.1128/JVI.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schelhaas M, Ewers H, Rajamaki ML, Day PM, Schiller JT, Helenius A. 2008. Human papillomavirus type 16 entry: retrograde cell surface transport along actin-rich protrusions. PLoS Pathog 4:e1000148. doi: 10.1371/journal.ppat.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samperio Ventayol P, Schelhaas M. 2015. Fluorescently labeled human papillomavirus pseudovirions for use in virus entry experiments. Curr Protoc Microbiol 37:14B.4.1–22. doi: 10.1002/9780471729259.mc14b04s37. [DOI] [PubMed] [Google Scholar]
- 66.Broniarczyk J, Massimi P, Bergant M, Banks L. 2015. Human papillomavirus infectious entry and trafficking is a rapid process. J Virol 89:8727–8732. doi: 10.1128/JVI.00722-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S. 2016. Cryoelectron microscopy maps of human papillomavirus 16 reveal L2 densities and heparin binding site. Structure 25:253–263. doi: 10.1016/j.str.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 68.Humphries DE, Silbert JE. 1988. Chlorate: a reversible inhibitor of proteoglycan sulfation. Biochem Biophys Res Commun 154:365–371. doi: 10.1016/0006-291X(88)90694-8. [DOI] [PubMed] [Google Scholar]
- 69.Esko JD, Stewart TE, Taylor WH. 1985. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc Natl Acad Sci U S A 82:3197–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buck CB, Thompson CD, Roberts JN, Muller M, Lowy DR, Schiller JT. 2006. Carrageenan is a potent inhibitor of papillomavirus infection. PLoS Pathog 2:e69. doi: 10.1371/journal.ppat.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aydin I, Villalonga-Planells R, Greune L, Bronnimann MP, Calton CM, Becker M, Lai KY, Campos SK, Schmidt MA, Schelhaas M. 2017. A central region in the minor capsid protein of papillomaviruses facilitates viral genome tethering and membrane penetration for mitotic nuclear entry. PLoS Pathog 13:e1006308. doi: 10.1371/journal.ppat.1006308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calton CM, Bronnimann MP, Manson AR, Li S, Chapman JA, Suarez-Berumen M, Williamson TR, Molugu SK, Bernal RA, Campos SK. 2017. Translocation of the papillomavirus L2/vDNA complex across the limiting membrane requires the onset of mitosis. PLoS Pathog 13:e1006200. doi: 10.1371/journal.ppat.1006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roberts JN, Buck CB, Thompson CD, Kines R, Bernardo M, Choyke PL, Lowy DR, Schiller JT. 2007. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nat Med 13:857–861. doi: 10.1038/nm1598. [DOI] [PubMed] [Google Scholar]
- 74.Denault J-B, Bissonnette L, Longpré J-M, Charest G, Lavigne P, Leduc R. 2002. Ectodomain shedding of furin: kinetics and role of the cysteine-rich region. FEBS Lett 527:309–314. doi: 10.1016/S0014-5793(02)03249-0. [DOI] [PubMed] [Google Scholar]
- 75.Shiryaev SA, Chernov AV, Golubkov VS, Thomsen ER, Chudin E, Chee MS, Kozlov IA, Strongin AY, Cieplak P. 2013. High-resolution analysis and functional mapping of cleavage sites and substrate proteins of furin in the human proteome. PLoS One 8:e54290. doi: 10.1371/journal.pone.0054290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang P, Tortorella M, England K, Malfait AM, Thomas G, Arner EC, Pei D. 2004. Proprotein convertase furin interacts with and cleaves pro-ADAMTS4 (Aggrecanase-1) in the trans-Golgi network. J Biol Chem 279:15434–15440. doi: 10.1074/jbc.M312797200. [DOI] [PubMed] [Google Scholar]
- 77.Conway MJ, Cruz L, Alam S, Christensen ND, Meyers C. 2011. Differentiation-dependent interpentameric disulfide bond stabilizes native human papillomavirus type 16. PLoS One 6:e22427. doi: 10.1371/journal.pone.0022427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cruz L, Biryukov J, Conway MJ, Meyers C. 2015. Cleavage of the HPV16 minor capsid protein L2 during virion morphogenesis ablates the requirement for cellular furin during de novo infection. Viruses 7:5813–5830. doi: 10.3390/v7112910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stanley M. 2008. Immunobiology of HPV and HPV vaccines. Gynecol Oncol 109:S15–S21. doi: 10.1016/j.ygyno.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 80.Day PM, Schiller JT. 2009. The role of furin in papillomavirus infection. Future Microbiol 4:1255–1262. doi: 10.2217/fmb.09.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McMillan NA, Payne E, Frazer IH, Evander M. 1999. Expression of the alpha6 integrin confers papillomavirus binding upon receptor-negative B-cells. Virology 261:271–279. doi: 10.1006/viro.1999.9825. [DOI] [PubMed] [Google Scholar]
- 82.Woodham AW, Taylor JR, Jimenez AI, Skeate JG, Schmidt T, Brand HE, Da Silva DM, Kast WM. 2015. Small molecule inhibitors of the annexin A2 heterotetramer prevent human papillomavirus type 16 infection. J Antimicrob Chemother 70:1686–1690. doi: 10.1093/jac/dkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Berditchevski F. 2001. Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci 114:4143–4151. [DOI] [PubMed] [Google Scholar]
- 84.Hemler ME. 2005. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 85.Hochdorfer D, Florin L, Sinzger C, Lieber D. 2016. Tetraspanin CD151 promotes initial events in human cytomegalovirus infection. J Virol 90:6430–6442. doi: 10.1128/JVI.00145-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 87.Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, Sanchez-Madrid F. 2009. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol 19:434–446. doi: 10.1016/j.tcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 88.Charrin S, Manie S, Thiele C, Billard M, Gerlier D, Boucheix C, Rubinstein E. 2003. A physical and functional link between cholesterol and tetraspanins. Eur J Immunol 33:2479–2489. doi: 10.1002/eji.200323884. [DOI] [PubMed] [Google Scholar]
- 89.Espenel C, Margeat E, Dosset P, Arduise C, Le Grimellec C, Royer CA, Boucheix C, Rubinstein E, Milhiet PE. 2008. Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J Cell Biol 182:765–776. doi: 10.1083/jcb.200803010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Walko G, Castañón MJ, Wiche G. 2015. Molecular architecture and function of the hemidesmosome. Cell Tissue Res 360:363–378. doi: 10.1007/s00441-014-2061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hopkinson SB, Hamill KJ, Wu Y, Eisenberg JL, Hiroyasu S, Jones JC. 2014. Focal contact and hemidesmosomal proteins in keratinocyte migration and wound repair. Adv Wound Care (New Rochelle) 3:247–263. doi: 10.1089/wound.2013.0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol 106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2005. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol Med 119:445–462. [DOI] [PubMed] [Google Scholar]
- 94.Molloy SS, Bresnahan PA, Leppla SH, Klimpel KR, Thomas G. 1992. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J Biol Chem 267:16396–16402. [PubMed] [Google Scholar]
- 95.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. 2011. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]