Abstract
We have identified a common nonpathological polymorphism of the human tyrosinase gene. In Caucasians codon 402 can be either CGA (arginine) [p = .85] or CAA (glutamine) [p = .15]. This polymorphism also occurs in American Blacks, but the codon 402CAA (Gin) allele was not detected in Oriental populations. The substitution of glutamine for arginine at codon 402 results in moderate thermoinstability of the corresponding tyrosinase polypeptide. Tyrosinase enzymatic activity expressed in HeLa cells transfected with a codon 402Gln tyrosinase cDNA is reduced by approximately 75 percent when cells are cultured at 37°C as compared to 31°C, whereas enzymatic activity of codon 402Arg tyrosinase is not temperature-sensitive. However, the genotype at codon 402 of tryosinase is not correlated with the apparent pigmentation phenotype in normal Caucasians.
The coloration of hair, skin, and eyes in mammals results principally from the synthesis of melanin pigments in melanocytes of the hair follicles, epidermal-dermal junction of the skin, retinal pigment epithelium, and the iris. The first two steps of melanin biosynthesis, the hydroxylation of L-tyrosine to 3,4-dihydroxyphenylalanine (dopa) and the subsequent dehydrogenation of dopa to dopaquinone, are both catalyzed by a single enzyme, tyrosinase (monophenol monooxygenase; monophenol, L-dopa: oxygen oxidoreductase; EC. 1.14.18.1; Lerner et al., 1949). Tyrosinase is a copper-containing glycoprotein of approximately 60 kDa molecular weight (Nishioka, 1978; reviewed in Hearing and Jimenez, 1989). Recently, cDNAs for mouse (Yamamoto et al., 1987; Müller et al., 1988) and human (Kwon et al., 1987; Shibahara et al., 1988; Bouchard et al., 1989) tyrosinases have been cloned and the 529-amino acid sequence of the tyrosinase polypeptide deduced.
Tyrosinase activity is deficient in patients with classic type I oculocutaneous albinism (reviewed in King and Summers, 1988), and we and others have identified a number of different tyrosinase gene mutations in patients with this disorder (Tomita et al., 1989; Giebel et al., 1990, 1991b; Kikuchi et al., 1990; Spritz et al., 1990, 1991; Takeda et al., 1990). Here we describe a common nonpathologic polymorphism of human tyrosinase that is associated with moderate thermoinstability of the corresponding tyrosinase polypeptide but has no apparent effect on the resulting pigmentation phenotype.
Materials and methods
Polymerase chain reaction (PCR) and DNA sequencing
Genomic DNA was isolated from peripheral blood leukocytes of unrelated normal individuals and patients with type I OCA. A 269-bp DNA fragment containing exon 4 of the tyrosinase gene plus portions of the adjacent intervening sequences (bases 2565–2834 in the sequence of Giebel et al., 1991a) was amplified from 0.1 μg DNA by 40 cycles of PCR using oligonucleotide primers 5′-TAATATATGCCTTATTTTAC-3′ and 5′-TAAAGTTTTGTGTTATCTCA 3′ (Giebel et al., 1991a) and Taq DNA polymerase (Perkin-Elmer Cetus, Norwalk, CT) exactly as described by Saiki et al. (1988). Each cycle consisted of 30 seconds at 94°C, 1 minute at 50°C, and 2 minutes at 72°C. The amplified PCR products were purified by electrophoresis in 4% polyacrylamide gels and cloned in bacteriophage vectors M13mp18 or mp19. The nucleotide sequences of at least six independent clones were determined for each individual.
Allele-specific oligonucleotide hybridizations
Exon 4 fragments were PCR-amplified from 0.1 μg genomic DNA from unrelated normal individuals and patients with type I OCA as described above. One-fifth (20 μl) of each reaction was applied to MAGNA nylon membranes (Micron Separations) with a Bio-Dot SF microfiltration apparatus (Bio-Rad). Allele-specific oligonucleotide hybridizations of replicate filters were carried out as described (Kogan and Gitschier, 1990) using 5′ end-radiolabeled 19-mer oligonucleotide probes corresponding to the codon 402CGA (Arg) (5′-GCAGTGGCTCCGAAGGCAC-3′) and codon 402CAA (Gln) (5′-GCAGTGGCTCCAAAGGCAC-3′) alleles.
Construction of codon 402Gln tyrosinase expression plasmid
The codon 402CAA (Gln) polymorphic substitution was introduced into the human tyrosinase cDNA expression plasmid pcTYR, which contains CGA (Arg) at codon 402 (Bouchard et al., 1989), by replacing the Pvu II/Bgl II fragment of pcTYR with the corresponding fragment of cDNA clone pMEL34, which contains CAA (Gln) at this site (Kwon et al., 1987). The nucleotide sequence of the polymorphic expression plasmid pcTYR-402Gln was verified by double-stranded DNA sequencing (Zhang et al., 1988).
Transient transfection of HeLa cells and assay of tyrosinase
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% bovine calf serum (HyClone Laboratories) supplemented with 2 mM L-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. The human tryosinase cDNA expression plasmid pcTYR (Bouchard et al., 1989) or pcTYR-402Gln plus the chloramphenicol acetyltransferase expression plasmid pSV2CAT (Gorman et al., 1982) were transiently transfected into triplicate plates of cultured HeLa cells by the Ca-phosphate precipitation procedure (Graham and van der Eb, 1973). Cells were harvested 64 hours after transfection; monolayers were washed three times with cold PBS, and the cells were scraped and pelleted. The cells were resuspended in cold 20 mM Na-phosphate buffer pH 6.8 containing 0.5% Triton X-100 and were sonicated for 2 minutes on ice. Sonicated cell extracts were centrifuged at 10,000 × g for 15 minutes at 4°C and the supernatants were dialysed twice against 5 mM Na-phosphate buffer pH 6.8.
Fluorometric assay of tyrosinase was as described (Husain et al., 1982) except that 4 mM ascorbic acid was included in the reaction mixture as reductant for tyrosine hydroxylation (Tripathi et al., 1988) and to reduce any dopaquinone formed by tyrosinase back to dopa; measurement of dopa thus provided a true estimate of the tyrosine hydroxylase activity of tyrosinase. The 55 μl reactions contained 0.1 mM L-tyrosine, 5 μM L-dopa, 4 mM ascorbic acid, 40 mM Na-phosphate buffer pH 6.8, and 15 μl cell extract. Incubation time was fixed at 4 hours, and production of dopa was measured by specific fluorescence at 360 nm excitation and 490 nm emission wavelengths. One unit of tyrosinase activity was defined as 1 pmol of dopa formed per minute. CAT activity was measured exactly according to Gorman et al. (1982). The protein concentrations of the cell extracts were determined by the microassay method of Peterson (1977). Tyrosinase specific activities were corrected for slight plate-to-plate variation in transfection efficiency by dividing by the CAT activity (% 14C-chloramphenicol converted to the three forms of acetyl-chloramphenicol; Gorman et al., 1982) in each extract.
Tyrosinase enzymatic activities were determined at both 31°C and 37°C. Activity of the codon 402Gln tyrosinase, produced in HeLa cells transfected with pcTyr-402Gln and cultured at 31°C, was not decreased by assay at 37°C (data not shown). Because the optimal temperature of the tyrosinase assay is 37°C (Lerner et al., 1949 and Lerner, 1953), only data for tyrosinase assays at that temperature are reported.
Results
Observation of a novel polymorphism at codon 402 of tyrosinase
Comparison of various published human tyrosinase cDNA (Kwon et al., 1987; Shibahara et al., 1988; Bouchard et al., 1989) and genomic (Giebel et al., 1991a) DNA sequences revealed two discrepancies. We previously reported that one of these differences, TCT (Ser) versus TAT (Tyr) at codon 192, is a common polymorphism among Caucasian individuals (Giebel and Spritz, 1990). The second difference is at codon 402, CGA (Arg) versus CAA (Gln). Several human tyrosinase cDNA (Shibahara et al., 1988; Bouchard et al., 1989) and genomic (Giebel et al., 1991a) sequences specify CGA at this site, whereas the sequence of another human tyrosinase cDNA, pMEL34 (Kwon et al., 1987) specifies CAA. In an initial attempt to resolve this apparent discrepancy we repeated the DNA sequence analysis of pMEL34. This analysis confirmed that codon 402 is indeed CAA in pMEL34 (data not shown).
In addition, during the course of DNA sequence analyses of tyrosinase genes from a number of normal individuals and patients with type I (tyrosinase-deficient) OCA, we observed both CGA (Arg) and CAA (Gln) at codon 402 (Fig. 1). Both forms of codon 402 occurred in association with both normal and various different albino mutant alleles. Therefore, this difference seemed likely to be a nonpathologic polymorphism of the human tyrosinase gene.
Figure 1.
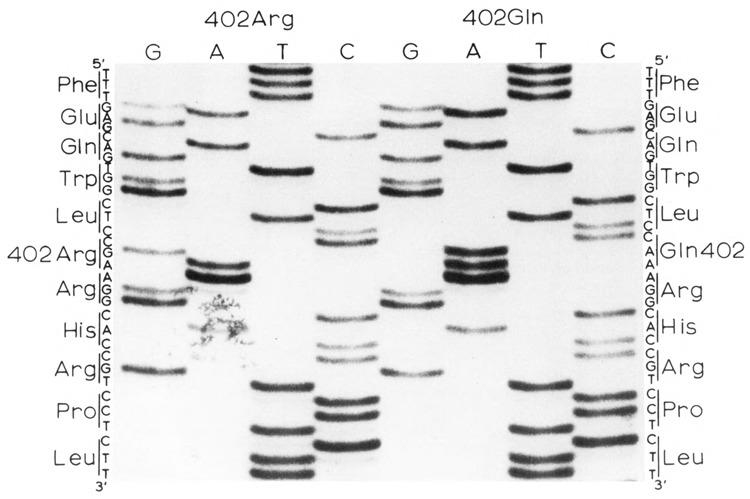
DNA sequences in the region of the codon 402 polymorphism. The sequence of the coding strand is shown.
Frequency of the codon 402 polymorphism in different ethnic groups
To determine the frequency of alleles at the codon 402 CGA/CAA polymorphism, we screened DNAs of 14 unrelated individuals of northern European Caucasian ethnic origin and individuals of several different non-Caucasian ethnic groups by allele-specific oligonucleotide hybridization. DNA fragments containing exon 4 of the tyrosinase gene were amplified by PCR of genomic DNA and hybridized to oligonucleotide probes specific for either CGA or CAA at codon 402 (Fig. 2). Twenty-three alleles (82%) were CGA (Arg) at codon 402, and five (18%) were CAA (Gln). As shown in Table 1, when combined with DNA sequence data on an additional 31 alleles from unrelated Caucasians, the overall frequency of the codon 402 CGA (Arg) allele among Caucasians was .85 and that of the codon 402 CAA (Gln) allele was .15. Thus, the codon 402 CGA (Arg) versus CAA (Gln) difference is a relatively common polymorphism among Caucasians of northern European descent. We have previously described another common tyrosinase gene polymorphism among Caucasians at codon 192 (Giebel and Spritz, 1990). Among the normal Caucasians studied here, we observed all four of the possible codon 192/codon 402 haplotypes in the approximate proportions expected, based on the individual frequencies of the two polymorphisms. Together, these data suggest that both the codon 192 and codon 402 polymorphisms are ancient in the Caucasian population.
Figure 2.
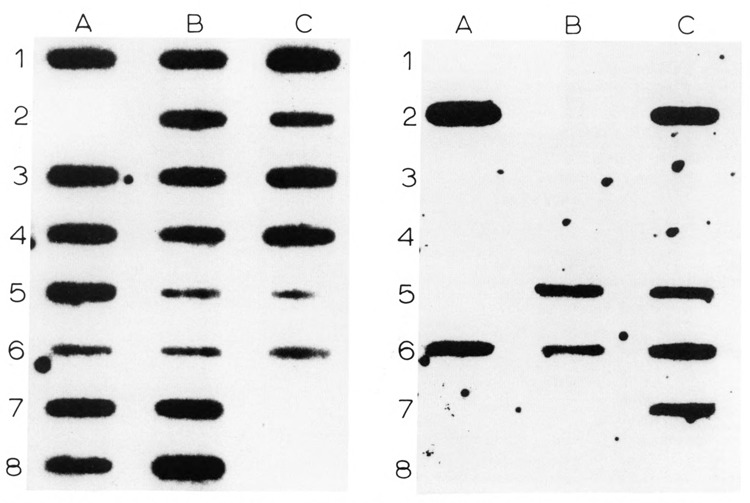
Allele-specific oligonucleotide hybridization of the codon 402 polymorphism. Left: Codon 402CGA (Arg) probe. Right: Codon 402CAA (Gln) probe. Normal Caucasians: A1–7, B1–2, B5–7, C2–3, C5–6; normal Chinese: A8, B3, B8; normal Japanese: C4; Indian: B4, Cl; normal human tyrosinase cDNA pMel34: C7. No sample was applied to C8. Other individuals tested are not shown.
Table 1.
Frequency of codon 402 polymorphism in different ethnic groups.
| Ethnic group | No. alleles | CGA (Arg) (%) | CAA (Gln) (%) |
|---|---|---|---|
| Caucasians | 59 | 50(84.7) | 9(15.3) |
| Non-Caucasians | |||
| Chinese | 66 | 66(100) | 0(0) |
| Japanese | 2 | 2(100) | 0(0) |
| Asian Indian | 4 | 4(100) | 0(0) |
| Afghan | 1 | 1 | 0 |
| African Pygmy | 4 | 4(100) | 0(0) |
| Australian Aborigine | 4 | 4(100) | 0(0) |
| American Blacks | 52 | 48(92) | 4(8) |
The codon 402 CAA polymorphism is also not uncommon among American Blacks. Among the 25 American Blacks studied, 22 were homozygous for CGA (Arg), and four were heterozygous for CAA (Gln) at codon 402. Thus, the frequency of the codon 402CGA (Arg) allele among American Blacks was .92, and that of the codon 402CAA (Gln) allele was .08. Although the frequency of the codon 402CAA (Gln) allele among American Blacks was lower than among Caucasians, this difference is not significant (p = .194; Fisher’s exact test). We also studied two Australian Aborigine and two African Pygmy individuals; all four were homozygous for CGA (Arg) at codon 402. Although the number of individuals studied here is too small to be certain, it may be that the codon 402CAA (Gln) allele is rare or absent from indigenous Black populations, but was introduced into the gene pool of American Blacks by admixture with “Caucasian genes” during historic times (Karaboh et al., 1990).
In contrast, the codon 402 CAA (Gln) allele appears to be essentially absent from at least some Asian populations. We studied 33 Chinese, one Japanese, two Asian Indian, and one Afghan individual; all were homozygous for CGA (Arg) at codon 402. Codon 402 was thus monomorphic among the Oriental individuals that we studied, a highly significant difference from the Caucasian population (p = .0005). It is perhaps noteworthy that the codon 192 polymorphism also appears to be uncommon among Orientals (Giebel and Spritz, 1990).
Properties of codon 402Arg and codon 402Gln human tyrosinases
To determine whether the polymorphism of arginine versus glutamine at codon 402 has any effect on the catalytic properties or stability of the corresponding tyrosinase polypeptides, we expressed the respective forms of human tyrosinase in cultured HeLa cells. Human tyrosinase cDNA expression plasmid pcTYR (Bouchard et al., 1989) encoded arginine, and plasmid pcTYR-402Gln encoded glutamine at codon 402; both plasmids encode tyrosine at polymorphic codon 192. HeLa cells were transiently cotransfected with plasmids pcTYR or pcTYR-402Gln along with pSV2CAT and cultured for 64 hours at either 31°C or 37°C. Cell extracts were prepared; and tyrosinase (tyrosine hydroxylase) activities, CAT activities, and protein concentrations were determined. As shown in Table 2, under standard assay conditions of 0.1 mM L-tyrosine, 5 μM dopa, pH 6.8 (Husain et al., 1982) there was no difference between the activities of codon 402Arg 31°C or 37°C and codon 402Gln tyrosinase produced at 31°C. In contrast, the activity of codon 402Gln tyrosinase produced at 37°C was much lower—only 27% of that produced at 31°C. These data demonstrate that the codon 402Gln substitution results in moderate thermoinstability of the corresponding tyrosinase polypeptide.
Table 2.
Activity of codon 402Arg versus codon 402Gln tyrosinases in transfected HeLa cells. HeLa cells were transfected with an expression vector containing either codon 402Arg (pcTyr) or codon 402Gln (pcTyr-402Gln) tyrosinase cDNA plus psV2CAT and cultured at either 31°C or 37°C. Cells were harvested 64 hours following transfection, and cell extracts were assayed for tyrosinase (tyrosine hydroxylase) and CAT activities at 37°C. The activity of codon 402Arg tyrosinase produced at each temperature was defined as 100 percent.
| Tyrosinase specific activity* | ||
|---|---|---|
| Temperature of transfection and culture | ||
| 31°C | 37°C | |
| 402Arg | 5.7 ± 0.6 (100%) | 5.6 ± 0.3 (100%) |
| 402Gln | 6.0 ± 1.0 (105%) | 1.5 ± 0.3 (26.8%) |
Tyrosine specific activities are expressed as pMol dopa formed per minute per mg of protein. The values shown were corrected for slight plate-to-plate variation in transfection efficiency by dividing by the CAT activity in each plate, which was not affected by the temperature of transfection and culture. Each experiment was performed in triplicate.
We next characterized the effect of the codon 402Gln substitution on the kinetic properties of tyrosinase. We studied the characteristic catalytic lag of tyrosinase activity (Pomerantz, 1966) by assaying tyrosinase (tyrosine hydroxylase) activities in the extracts of transfected HeLa cells using a fixed concentration (0.1 mM) of L-tyrosine at varying time intervals at pH 6.8 in the absence of added dopa at 37°C. As shown in Figure 3, in extracts of HeLa cells transfected and cultured at 31°C, a lag of 30–60 minutes in enzyme activity was observed for both the codon 402Arg and codon 402Gln tyrosinases. In contrast, in extracts of HeLa cells transfected and cultured at 37°C, the lag of codon 402Arg tyrosinase activity was unchanged, whereas the lag of codon 402Gln tyrosinase activity was increased to 4 hours, most likely resulting principally from the approximately 75 percent reduction in the amount of active enzyme at the higher temperature.
Figure 3.
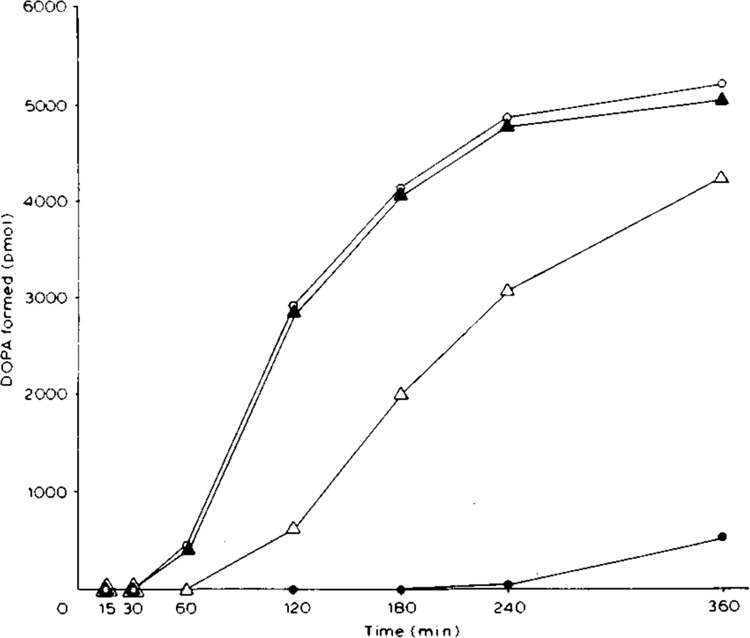
Time course of tyrosinase (tyrosine hydroxylase) activity in extracts of transfected HeLa cells. The 55 μl reaction mixtures contained 0.1 mM L-tyrosine, 4 mM ascorbic acid, 40 mM Na-phosphate pH 6.8, 15 μl cell extract. ○, cells transfected with pcTYR (402Arg) at 37°C; •. pcTYR-402Gln at 37°C; ▴, pcTYR (402Arg) at 31 °C; ▵, pcTYR-402Gln at 31°C. Protein concentrations in the four cell extracts were 7.9, 7.1, 7.7, and 6.1 mg/ml, respectively.
We analyzed inhibition of the different forms of tyrosinase by high concentrations of L-tyrosine (Pomerantz, 1966) by assaying tyrosinase activities in the HeLa cell extracts at a fixed time interval (6 hours) over varying tyrosine concentrations in the absence of exogenous dopa. As shown in Figure 4, concentrations of tyrosine greater than 0.4 mM inhibited tyrosinase activity in all of the transfected HeLa cell extracts. The properties of the codon 402 Arg and codon 402Gln tyrosinases produced at 31°C were essentially identical. However, activity of the codon 402Gln tyrosinase produced at 37°C was severely inhibited by concentrations of tyrosine greater than 0.1 mM. Km values for L-tyrosine were determined by double-reciprocal plot (Lineweaver and Burk, 1934). The Km for L-tyrosine of codon 402 Arg tyrosinase produced at both 31°C and at 37°C was 0.09 mM, the same as that of codon 402Gln tyrosinase produced at 31°C. In contrast, the Km of codon 402Gln tyrosinase produced at 37°C was much higher, 0.25 mM. The Vmax of codon 402Arg tyrosinase produced at both 31°C (17.6 pMol/minute) and 37°C (17.8 pMol/minute) was similar to that of codon .402Gln tyrosinase produced at 31°C (11.2 pMol/minute). In contrast, the Vmax of codon 402Gln tyrosinase produced at 37°C was much lower (0.76 pMol/minute). Together, these data indicate that codon 402Gln tyrosinase is thermosensitive, most likely undergoing significant structural unfolding at 37°C, with a consequent substantial decrease of affinity for L-tyrosine.
Figure 4.
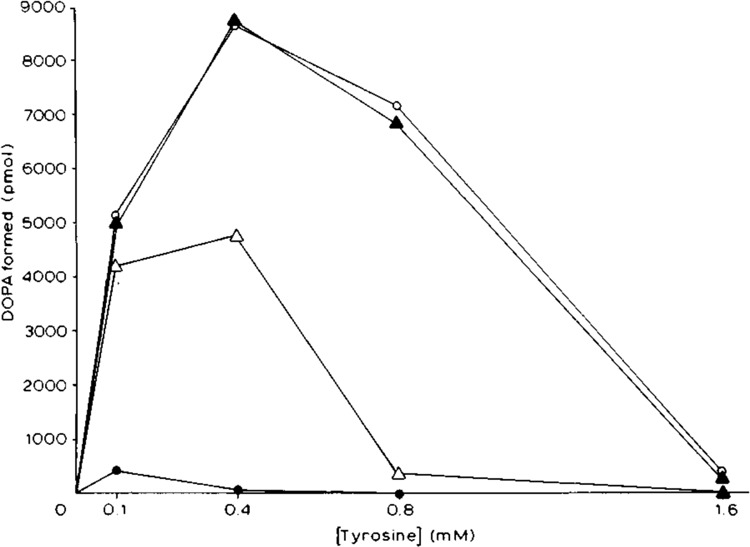
Effect of L-tyrosine concentration on tyrosinase (tyrosine hydroxylase) activity in transfected HeLa cells. Assay conditions were as described in the legend to Figure 3, except that the time of incubation was fixed at 6 hours, and the concentration of L-tyrosine was varied. For symbols, see legend to Figure 3. To determine the Km and Vmax, tyrosinase activities were assayed at 0.05, 0.1, 0.2, 0.3, and 0.4 mM L-tyrOsine (○; ▴; and ▵) or 0.05, 0.1, and 0.15 mM L-tyrosine (•) concentrations at 37°C (data not shown).
Discussion
Tyrosinase is the copper-containing enzyme that catalyzes the first two steps in the melanin biosynthetic pathway (Lerner et al., 1949). Cloning of human tyrosinase cDNAs (Kwon et al., 1987; Shibahara et al., 1988; Bouchard et al., 1989) permitted deduction of the 529-amino acid sequence of the tyrosinase polypeptide. We have previously identified a nonpathologic polymorphism of human tyrosinase—at amino acid 192—for two uncharged polar residues, serine versus tyrosine (Giebel and Spritz, 1990). Here we describe a second nonpathologic human tyrosinase polymorphism: codon 402 can encode either arginine (CGA) or glutamine (CAA), respectively basic and neutral residues. Among Caucasians the frequency of the codon 402CGA (Arg) allele is approximately .85, and that of the codon 402CAA (Gln) allele is .15. Codon 402 encodes arginine in the tyrosinases of Streptomyces glaucescens (Huber et al., 1985), mouse (Yamamoto et al., 1987; Müller et al., 1988), and most humans, as well as the homologous portion of the human tyrosinase-related brown locus polypeptide (Shibahara et al., 1986). Therefore, it seems likely that CGA represents the ancestral sequence at this site; the change of codon 402 from CGA to CAA may have resulted from deamination of 5-methylcytidine at a CpG dinucleotide on the noncoding strand.
The substitution of glutamine for arginine at codon 402 results in the loss of one positive charge; the predicted isoelectric pH of the tyrosinase polypeptide would therefore be shifted from 5.92 to 5.82. Enzyme activity and kinetic data indicate that the principal effect of this substitution is a decrease in thermal stability. Catalytic activity of the codon 402Gln and codon 402Arg tyrosinases are almost identical at 31°C, but at 37°C activity of codon 402Gln tyrosinase is reduced by approximately 75 percent because of protein unfolding. Thus, enzymatic activity of codon 402Gln tyrosinase might be significantly reduced at body temperature in vivo. However, even individuals homozygous for the codon 402CAA (Gln) allele would probably have approximately 25 percent of the tyrosinase activity of individuals homozygous for the codon 402CGA (Arg) allele, far in excess of the very low or absent activity found in tyrosinase-deficient albinos (King and Summers, (1988). We observed no apparent correlation between the codon 402 tyrosinase genotype and the apparent pigmentation phenotype among normal Caucasian individuals, even among persons homozygous for the codon 402 CAA (Gln) allele. Therefore, the in vivo pigmentation phenotype may not be sensitive to a 75 percent decrease in tyrosinase activity. Alternatively, the activity of codon 402Gln tyrosinase in vivo might be increased by various positive regulators of tyrosinase biosynthesis or enzymatic activity such as cAMP (Johnson and Pastan, 1972), dopa (Pomerantz, 1966; Husain et al., 1982), and citrate (Devi et al., 1989).
Codons 363 to 420 of the tyrosinase polypeptide exhibit considerable amino acid sequence similarity to hemocyanin, another copper-containing protein, and this region has been very highly conserved among tyrosinases of all species studied. Therefore, this region of tyrosinase has been suggested as a possible binding site for copper (Lerch, 1988). The codon 402 polymorphism occurs near the center of this region. Moreover, six (28.6%) of the 21 known tyrosinase missense substitutions in patients with type I (tyrosinase-deficient) oculocutaneous albinism also occur within this region. Of particular interest, two of the substitutions in this region result in severely thermosensitive forms of tyrosinase: codon 420 His→Arg in the Himalayan (c h/c h ) mouse (Kwon et al., 1989) and codon 422 Arg→Gln in some albino humans (Giebel et al., 1991b). Thus, substitutions in this region may be particularly likely to result in decreased structural stability of the corresponding tyrosinase poly-peptides, especially those substitutions that occur on the genetic background of the codon 402CAA (Gln) allele.
Acknowledgments
The authors thank B. Kwon for the human tyrosinase cDNA pMEL34, and B. Bouchard and A. Houghton for tyrosinase cDNA expression plasmid pcTYR.
This work was supported by Clinical Research Grant 6-408 from the March of Dimes Birth Defects Foundation and Grant AR-39892 from the National Institutes of Health. This is paper number 3177 from the Laboratory of Genetics, University of Wisconsin-Madison.
The costs of publishing this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
References
- Bouchard B., Fuller B. B., Vijayasaradhi S., and Houghton A. N. (1989), J Exp Med 169, 2029–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi C. C., Tripathi R. K., and Ramaiah A. (1989), Pigm Cell Res 2, 117–122. [DOI] [PubMed] [Google Scholar]
- Giebel L. B. and Spritz R. A. (1990), Nucl Acids Res 18, 3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebel L. B., Strunk K. M., King R. A., Hanifin J. M., and Spritz R. A. (1990), Proc Natl Acad Sci USA 87, 3225–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebel L. B., Strunk K. M., and Spritz R. A. (1991a), Genomics 9, 435–445. [DOI] [PubMed] [Google Scholar]
- Giebel L. B., Tripathi R. K., King R. A., and Spritz R. A. (1991b), J Clin Invest 87, 1119–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., and Howard B. H. (1982), Mol Cell Biol 2, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. and van der Eb A. (1973), Virology 52, 456–457. [DOI] [PubMed] [Google Scholar]
- Hearing V. J. and Jimenez M. (1989), Pigm Cell Res 2, 75–85. [DOI] [PubMed] [Google Scholar]
- Huber M., Hintermann G., and Lerch K. (1985), Biochemistry 24, 6038–6044. [DOI] [PubMed] [Google Scholar]
- Husian I., Vijayan E., Ramaiah A., Pasricha J. S., and Madan N. C. (1982), J Invest Dermatol 78, 243–252. [DOI] [PubMed] [Google Scholar]
- Johnson G. S. and Pastan I. (1972), Nature New Biol 237, 267–268. [DOI] [PubMed] [Google Scholar]
- Kamboh M. I., Chakraborty R., and Ferrell R. E. (1990), Am J Hum Genet 47 [Suppl], A138. [Google Scholar]
- Kikuchi H., Hara S., Ishiguro S., Tamai M., and Watanabe M. (1990), Hum Genet 85, 123–124. [DOI] [PubMed] [Google Scholar]
- King R. A. and Summers C. G. (1988), Dermatol Clin 6, 217–228. [PubMed] [Google Scholar]
- Kogan S. C. and Gitschier J. (1990), in PCR Protocols (Innis M. A., Gelfand D. H., Sninsky J. J., and White T. J., eds.), Academic Press, Inc., San Diego, pp. 288–299. [Google Scholar]
- Kwon B. S., Haq A. K., Pomerantz S. H., and Halaban R. (1987), Proc Natl Acad Sci USA 84, 7473–7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon B. S., Halaban R., and Chintamaneni C. (1989), Biochem Biophys Res Comm 161, 252–260. [DOI] [PubMed] [Google Scholar]
- Lerch K. (1988), Adv Pigment Cell Res, Prog Clin Biol Res 256, 85–98. [PubMed] [Google Scholar]
- Lerner A. B., Fitzpatrick T. B., Calkins E., and Summerson W. H. (1949), J Biol Chem 178, 185–195.18112101 [Google Scholar]
- Lerner A. B. (1953), in Advances in Enzymol, vol. 14 (Nord F. F., ed.), Interscience Publishers, Inc., New York, pp. 73–128. [Google Scholar]
- Lineweaver H. and Burk D. (1934), J Amer Chem Soc 56, 658–666. [Google Scholar]
- Müller G., Ruppert S., Schmid F., and Schütz G. (1988), EMBO J 7, 2723–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K. (1978), Eur J Biochem 85, 137–146. [DOI] [PubMed] [Google Scholar]
- Peterson G. L. (1977), Anal Biochem 83, 346–356. [DOI] [PubMed] [Google Scholar]
- Pomerantz S. H. (1966), J Biol Chem 241, 161–168. [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., and Erlich H. A. (1988), Science 239, 487–491. [DOI] [PubMed] [Google Scholar]
- Shibahara S., Tomita Y., Sakakura T., Nager C., Chaudhri B., and Müller R. (1986), Nucl Acids Res 14, 2413–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara S., Tomita Y., Tagami H., Müller R. M., and Cohen T. (1988), Tohoku J Exp Med 156, 403–414. [DOI] [PubMed] [Google Scholar]
- Spritz R. A., Strunk K. M., Giebel L. B., and King R. A. (1990), New Engl J Med 322, 1724–1728. [DOI] [PubMed] [Google Scholar]
- Spritz R. A., Strunk K. M., Hsieh C-L., Sekhon G. S., and Francke U. (1991), Am J Hum Genet 48, 318–324. [PMC free article] [PubMed] [Google Scholar]
- Takeda A., Tomita Y., Matsunaga J., Tagami H., and Shibahara S. (1990), J Biol Chem 265, 17792–17797. [PubMed] [Google Scholar]
- Tomita Y., Takeda A., Okinaga S., Tagami H., and Shibahara S. (1989), Biochem Biophys Res Comm 164, 990–996. [DOI] [PubMed] [Google Scholar]
- Tripathi R. K., Chaya Devi C., and Ramaiah A. (1988), Biochem J 252, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Takeuchi S., Kudo T., Makino K., Nakata A., Shinoda T., and Takeuchi T. (1987), Jpn J Genet 62, 271–274. [Google Scholar]
- Zhang H., Scholl R., Browse J., and Somerville C. (1988), Nucl Acids Res 16, 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]


