Abstract
The functional consequences of unusually short 5′ noncoding sequences on eukaryotic mRNAs are explored here by using an in vitro transcription and translation system. As the distance of the first AUG codon from the m7G cap was decreased from 32 to 3 nucleotides, the yield of protein initiated from the first AUG codon progressively decreased, with a corresponding increase in initiation from the second AUG codon. The leakiness attributable to a too-short leader sequence was offset, however, by introducing secondary structure downstream from the first AUG codon.
Vertebrate mRNAs have 5′ noncoding sequences that range in length from three to several hundred nucleotides, with an average length of about 90 nucleotides (Kozak, 1987a). From inspection of natural mRNAs, the consequences of an extremely short 5′ leader sequence are not immediately obvious. There are a few cellular mRNAs in which ribosomes initiate at two AUG codons, the first of which lies very close to the cap (Kelley et al., 1982; Peterson and Piatigorsky, 1986; Rose et al., 1977; Strubin et al., 1986); but the first AUG codon in those mRNAs also happens to be in an unfavorable context for initiation (Kozak, 1986), which makes it hard to identify the cause of the leaky scanning. A similar problem complicates attempts to explain why Phaseolus vulgaris lectin protein (Hoffman, 1984) and sea raven antifreeze protein (Hayes et al., 1989) are initiated predominantly from the third AUG codon: the first and second AUG codons occur in suboptimal contexts and are close to the cap. A considerable number of viral and cellular mRNAs have 5′ noncoding sequences of only 7 to 13 nucleotides (Cate et al., 1986; Chung et al., 1986; Dasgupta et al., 1975; Fluhr et al., 1986; Jameson et al., 1984; Kozak, 1977; Peleman et al., 1989; Reynolds et al., 1989; Rose, 1978; Shinshi et al., 1990; Virgin et al., 1985). The absence of any recognized translational defect in those mRNAs would seem at first sight to contradict the hypothesis that a short leader sequence is deleterious. Experiments described here reveal, however, that progressive shortening of the 5′ noncoding sequence on synthetic transcripts does promote leaky scanning. Other experiments suggest an explanation for why initiation is not leaky with the aforementioned natural mRNAs; namely, that downstream secondary structure seems to counter the deleterious effect of a short leader sequence, much as a properly positioned downstream stem-loop structure suppresses the leakiness that results from an unfavorable primary sequence context around the AUG codon (Kozak, 1990). The explanation in both situations might be that downstream secondary structure slows movement of the 40S ribosomal subunit away from the 5′ end of the mRNA, thereby providing more time for the first AUG codon to be recognized.
Materials and methods
Plasmid constructions
The constructs used here were derived from plasmids SP64(8334)B13-CAT and SP64(8336)B13-CAT, described previously (Kozak, 1989). Transcription of the parental plasmids with SP6 RNA polymerase produces mRNAs that have 17 nucleotides between the m7G cap and the first (pre-CAT) ATG codon. To obtain transcripts with shorter leader sequences, I introduced at the HindIII site of SP64(8334 or 8336)B13-CAT the oligonucleotide
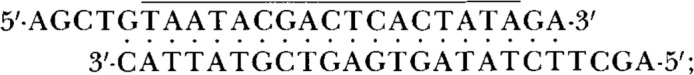
the overlined portion of this oligonucleotide constitutes a promoter for bacteriophage T7 RNA polymerase (Rosenberg et al., 1987). The sequence (including the start site for translation of preCAT protein) between the Hindlll and BamHI sites of SP64/T7(8334 or 8336)B13-CAT was next replaced by one of three synthetic oligo-nucleotides of the form
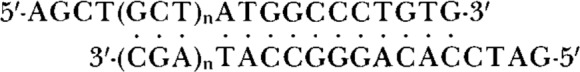
where n = 0, 1 or 2. Transcription of the resulting plasmids with T7 RNA polymerase produces leader sequences of 6, 9, and 12 nucleotides, as illustrated for T6, T9, and T12 in Figure 1. To obtain a construct that would generate a leader sequence of only 3 nucleotides, I cut SP64/T7(8334 or 8336)B13-CAT with Hindlll, digested with mung bean nuclease (New England BioLabs) to remove the single-stranded extension, cut the DNA again with BamHI to delete the fragment that carries the preCAT start site, and inserted in its place the synthetic oligonucleotide
Figure 1.

T7-generated transcripts with short leader sequences. The four plasmids in this series are designated T3, T6, T9, and T12, where the numbers indicate the lengths of the leader sequences when the plasmids are transcribed by T7 RNA polymerase. The oligonucleotide insert that carries the bacteriophage T7 promoter is shown in lower case letters in the top line. Below, the beginning of the transcribed sequence is shown, with T’s in place of U’s, for all four constructs. Initiation at the SP6 promoter, which was retained upstream from the T7 promoter, produces mRNAs with leader sequences 29 nucleotides longer than those on the corresponding T7 transcripts; the SP6-generated mRNAs from this series were used only where explicitly stated in the text. In addition to the authentic AUG initiator codon for CAT protein, each transcript has an upstream AUG triplet that enables synthesis of an N-terminally extended “preCAT” protein. To adjust the reading frame between the two AUG codons, a 22-nucleotide adaptor (upper line, bracketed) was inserted. Adaptor 8336 introduces a stem-loop structure (AG -19 kcal/mol) downstream from the preCAT start site; the alternative adaptor 8334 has no deliberate secondary structure.
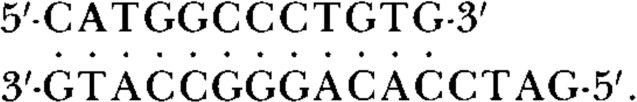
The resulting construct is designated T3. The procedures used to transform E. coli RR1, purify plasmids, and verify their DNA sequences have been described (Kozak, 1989).
In vitro transcription and translation
CsCl-purified plasmids linearized with Aval were used as templates for in vitro transcription reactions. Capped transcripts were synthesized using SP6 or T7 RNA polymerase (Bethesda Research Labs) at 37°C as described (Kozak, 1989) except that, after a 15-minute incubation with m7GpppG to allow initiation of capped transcripts, the GTP concentration was raised to 0.5 mM and incubation continued for another 60 minutes. All transcripts were labeled with [3H]UTP to facilitate measurement of the amount of mRNA used in the translation reactions.
Synthesis of pS]methionine-labeled preCAT and CAT proteins in wheat germ extracts at 23°C was done as described (Kozak, 1989). The mRNA concentration was 0.5 gg per 25 gl reaction. Translation of the transcripts described herein was strongly dependent on the m7G cap. Equal aliquots of each translation reaction were analyzed by polyacrylamide gel electrophoresis (Kozak, 1989). Autoradiograms that had been exposed for 12 hours were quantified by densitometry.
Results and discussion
Figure 1 depicts a set of mRNAs designed to evaluate the fidelity of initiation when the 5′ noncoding sequence is very short. These transcripts contain two AUG codons in-frame with the chloramphenicol acetyltransferase (CAT) coding sequence. The yield of an elongated “pre-CAT” protein versus the authentic CAT protein provides an easy measure of initiation from the first versus the second AUG codon. Inasmuch as the context around the first AUG codon (G in position −3 and G in position +4) is favorable for initiation (Kozak, 1986), preCAT should be the exclusive translation product- or nearly so-unless the proximity of the first AUG codon to the 5′ end of the mRNA impairs its recognition.
Figure 2 shows the results of translating these mRNAs in the wheat germ cell-free system. T3(8334) mRNA, which has only 3 nucleotides between the m7G cap and the first AUG codon, directed synthesis of preCAT and CAT proteins in a ratio of 1.5 : 2 (Fig. 2, lane 1), which indicates considerable leakiness despite the favorable context around the preCAT start site. The gradual reduction of CAT translation (lower band in Fig. 2, lanes 1–4) indicates that the tendency of ribosomes to bypass the first AUG codon diminished as the 5′ noncoding sequence was increased from 3 to 6, 9, or 12 nucleotides. Indeed, when the leader was lengthened to 32 nucleotides by using the upstream SP6 promoter (Fig. 1, top line), preCAT protein initiated from the first AUG codon was the sole translation product (Fig. 2, lane 5). The quantitative presentation of these results in Figure 3 confirms a progressive decrease in initiation of preCAT from the first AUG codon, and a corresponding increase in initiation of CAT from the second AUG codon, as the first AUG is moved closer to the m7G cap.
Figure 2.
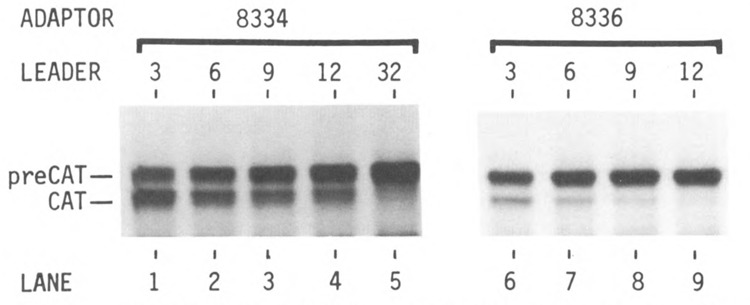
Demonstration that short leader sequences encourage leaky scanning in the wheat germ translation system. [:«S]methionine-labeled proteins were fractionated by polyacrylamide gel electrophoresis as described previously (Kozak, 1989). The yield of CAT protein (lower band in the autoradiogram) is a measure of the extent to which ribosomes bypass the first AUG codon and initiate instead at the second AUG. With T7-derived mRNAs from plasmids T3(8334), T6(8334), T9(8334), and T12(8334), initiation from the second AUG codon declined as the 5′ non-coding sequence was lengthened from 3 to 12 nucleotides (lanes 1–4). In lanes 6–9, the presence of the structure-prone oligonucleotide 8336 downstream from the preCAT start site suppressed the tendency of ribosomes to bypass the first AUG codon.
Figure 3.
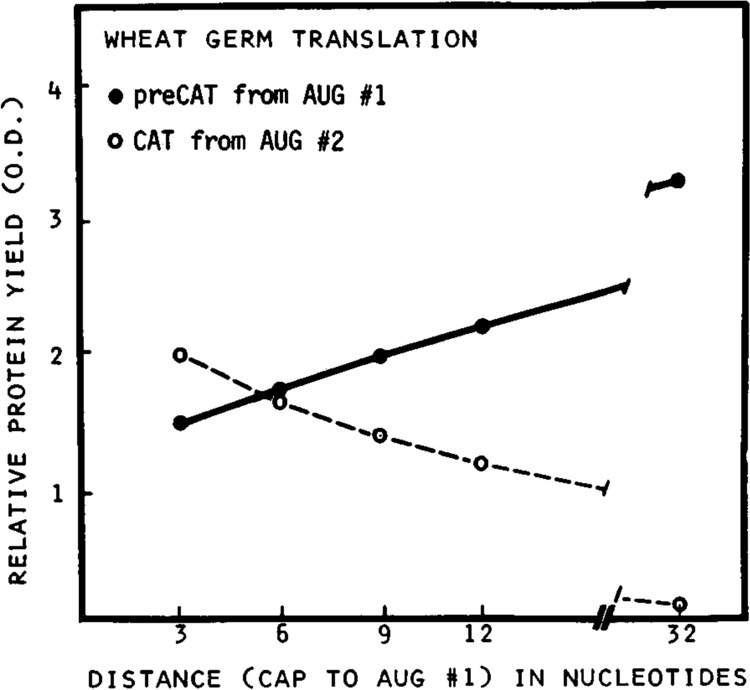
Initiation of translation from the first and second AUG codons as a function of leader length. T7-derived transcripts from plasmids T3(8334), T6(8334), T9(8334), and T12(8334) were translated in the wheat germ system. T3(8334) was also transcribed with SP6 polymerase to obtain the indicated 32-nucleotide leader sequence. [35S]methionine-labeled preCAT (•) and CAT (○) proteins were fractionated by polyacrylamide gel electrophoresis as in Figure 2; the resulting autoradiogram was quantified by densitometry. Protein yields in optical density units are plotted as a function of distance from the m7G cap to the preCAT start site (AUG #1).
There is a way to suppress leaky scanning while retaining a short leader sequence: namely, by introducing a structure-prone oligonucleotide downstream from the first AUG codon (Fig. 2, lanes 6–9). This echoes the ability of downstream secondary structure to suppress the leakiness that otherwise results when the primary sequence around the first AUG codon is unfavorable for initiation (Kozak, 1990). Results similar to those shown for the wheat germ system in Figure 2 were also obtained with the reticulocyte translation system (data not shown).
These experiments expand the catalogue of structural features in eukaryotic mRNAs that affect the fidelity and efficiency of initiation. In higher eukaryotes, selection of the correct start site has been shown previously to depend on (1) the primary sequence around the AUG codon (Kozak, 1986); (2) the position of the AUG codon (i.e., whether or not it is first); and (3) the presence of downstream secondary structure, which may facilitate recognition of the preceding AUG codon by slowing scanning (Kozak, 1990). Here I show that a too-short leader sequence adversely affects the fidelity of initiation. Thus, despite the favorable primary sequence around the first (preCAT) AUG codon in plasmids T3, T6, T9, and T12, ribosomes tend increasingly to bypass the preCAT start site as the first AUG codon is moved closer to the cap (Figs. 2 and 3). This tendency, which I have called leaky scanning, is completely suppressed when the leader sequence is 32 nucleotides long or when a modest amount of secondary structure is introduced downstream from the first AUG codon. The occurrence of leaky scanning when the 5′ noncoding sequence is short clearly reflects more than absence of the preferred primary sequence context. Although the preferred context for initiation has been shown to extend as far as 9 nucleotides upstream from the AUG codon (Kozak, 1987a), the nucleotides in positions −4 to −9 contribute significantly only when the critical purine in position −3 is absent (Kozak, 1986, 1987b). Inasmuch as T3, T6, T9, and T12 all have G in position −3, as well as the preferred G in position +4, the local context is strong enough that presence or absence of the preferred sequence in positions −4 to −9 should not matter. Thus, irrespective of what the sequence is, it seems the 5′ leader has to be a certain length for the first AUG codon to be selected faithfully. That conclusion is in keeping with some previous studies documenting impaired translation of yeast phosphoglycerate kinase (van den Heuvel et al., 1989) and SV40 late proteins (Dabrowski and Alwine, 1988; Grass and Manley, 1987; Sedman et al., 1990) when the 5′ noncoding sequence is short. Further lengthening of the 5′ leader sequence beyond what is required for the fidelity of initiation can dramatically increase the efficiency of translation, as reported elsewhere (Kozak, 1991).
Acknowledgment
Research in my laboratory is supported by NIH grant GM33915.
The costs of publishing this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
References
- Cate R. L., Mattaliano R. J., Hession C., Tizard R., Farber N. M., Cheung A., Ninfa E. G., Frey A. Z., Gash D. J., Chow E. P., Fisher R. A., Bertonis J. M., Torres G., Wallner B. P., Ramachandran K. L., Ragin R. C., Manganaro T. F., MacLaughlin D. T., and Donahoe P. K. (1986), Cell 45, 685–698. [DOI] [PubMed] [Google Scholar]
- Chung B.-C., Matteson K. J., and Miller W. L. (1986), Proc Natl Acad Sci USA 83, 4243–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowski C. and Alwine J. C. (1988), J Virol 62, 3182–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta R., Shih D. S., Saris C., and Kaesberg P. (1975), Nature 256, 624–628. [DOI] [PubMed] [Google Scholar]
- Fluhr R., Moses P., Morelli G., Coruzzi G., and Chua N-H. (1986), EMBO J 5, 2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass D. S. and Manley J. L. (1987), J Virol 61, 2331–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes P. H., Scott G. K., Ng N., Hew C., and Davies P. L. (1989), J Biol Chem 264, 18761–18767. [PubMed] [Google Scholar]
- Hoffman L. M. (1984), J Mol Appl Genet 2, 447–453. [PubMed] [Google Scholar]
- Jameson L., Chin W. W., Hollenberg A. N., Chang A. S., and Habener J. F. (1984), J Biol Chem 259, 15474–15480. [PubMed] [Google Scholar]
- Kelley D. E., Coleclough C., and Perry R. P. (1982), Cell 29, 681–689. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1977), Nature 269, 390–394. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1986), Cell 44, 283–292. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1987a), Nucl Acids Res 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1987b), J Mol Biol 196, 947–950. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1989), Mol Cell Biol 9, 5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1990), Proc Natl Acad Sci USA 87, 8301–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1991), Gene Expr 1, 117–125. [PMC free article] [PubMed] [Google Scholar]
- Peleman J., Saito K., Cottyn B., Engler G., Seurinck J., Van Montagu M., and Inze D. (1989), Gene 84, 359–369. [DOI] [PubMed] [Google Scholar]
- Peterson C. A. and Piatigorsky J. (1986), Gene 45, 139–147. [DOI] [PubMed] [Google Scholar]
- Reynolds D. S., Gurley D. S., Stevens R. L., Sugarbaker D. J., Austen K. F., and Serafin W. E. (1989), Proc Natl Acad Sci USA 86, 9480–9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. K. (1978), Cell 14, 345–353. [DOI] [PubMed] [Google Scholar]
- Rose S. M., Kuehl W. M., and Smith G. P. (1977), Cell 12, 453–462. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D., Lin S., Dunn J. J., and Studier F. W. (1987), Gene 56, 125–135. [DOI] [PubMed] [Google Scholar]
- Sedman S. A., Gelembiuk G. W., and Mertz J. E. (1990), J Virol 64, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinshi H., Neuhaus J-M., Ryals J., and Meins F. Jr. (1990), Plant Mol Biol 14, 357–368. [DOI] [PubMed] [Google Scholar]
- Strubin M., Long E. O., and Mach B. (1986), Cell 47, 19–625. [DOI] [PubMed] [Google Scholar]
- van den Heuvel J. J., Bergkamp R., Planta R. J., and Raué H. A. (1989), Gene 79, 83–95. [DOI] [PubMed] [Google Scholar]
- Virgin J. B., Silver B. J., Thomason A. R., and Nilson J. H. (1985), J Biol Chem 260, 7072–7077. [PubMed] [Google Scholar]


