Abstract
Lengthening the 5′ noncoding sequence on SP6-derived transcripts can increase their translational efficiency by an order of magnitude under some conditions of translation in reticulocyte lysates. This effect was observed upon reiterating three different synthetic oligonucleotides, the sequences of which were designed simply to preclude secondary structure. It seems unlikely that such arbitrarily designed sequences are recognized by sequence-specific translational enhancer proteins. Rather, long 5′ leader sequences appear to accumulate extra 40S ribosomal subunits, which may account for their translational advantage. The buildup of 40S subunits on long, unstructured leader sequences is predicted by the scanning model for initiation. Leader sequences such as these may be ideal for in vitro expression vectors.
The consequences of unusually long 5′ noncoding sequences are hard to assess from mere inspection of natural mRNAs. While some eukaryotic mRNAs with average-to-long leader sequences are known to be translated efficiently (Berkner and Sharp, 1985; Carrington and Freed, 1990; Gallie et al., 1987; Logan and Shenk, 1984; McGarry and Lindquist, 1985), there is no simple correlation between translational efficiency and leader length among natural mRNAs. For example, the adenovirus mRNA that encodes polypeptide IX, with a 5′ noncoding sequence of only 24 nucleotides, appears more efficient than mRNAs bearing the 200-nucleotide tripartite leader (Lawrence and Jackson, 1982). Translation is sometimes improved upon truncating leader sequences (Chinkers et al., 1989; Horiuchi et al., 1990; Ito et al., 1990; Leung et al., 1989; Murray et al., 1988; Rao et al., 1988; Teruya et al., 1990; Timmons and Witte, 1989; Waterhouse et al., 1990), but that is not really a test of length, because in most cases the originally-long leader sequences were GC-rich—hence highly structured—and often burdened with upstream AUG codons. In short, when one natural mRNA is compared with another, or when a full-length leader is compared with a truncated version, the problem is that there are many variables in addition to length. Here I have used synthetic mRNAs to show that translation in vitro improves dramatically as the 5′ noncoding sequence is systematically lengthened—by reiterating a block of 20 nucleotides—provided that the extended leader sequence is unstructured and devoid of spurious AUG codons. I also present experiments that bear on the mechanism by which a long leader sequence facilitates translation.
Materials and methods
The initial set of constructs for this study was derived from plasmid SP64(8336)B13-CAT, described previously (Kozak, 1989a). The 5′ noncoding sequence was lengthened in increments of 20 nucleotides by inserting at the HindIII site (Fig. 1) one or more copies of oligonucleotide 8180, which has the sequence AGCTCACGACCCACTACACA. The resulting plasmids are designated (8180)1B13(8336), (8180)2B13(8336), etc. Series (8180)nB13(8334) was constructed and tested in parallel with series (8180)nB13(8336) to see if a moderately stable hairpin (ΔG − 19 kcal/mol, present in oligonucleotide 8336 but not 8334) downstream from the AUG initiator codon would alter the results. In previous studies a downstream hairpin was shown to preclude leaky scanning when the AUG initiator codon was in a weak context (Kozak, 1990a), or when the leader sequence was unusually short (Kozak, 1991). In the experiments described here, however, the AUG codon was in a favorable context (Kozak, 1986, 1987a, 1987b), and the downstream secondary structure contributed by 8336 had no effect on fidelity except, again, when the leader sequence was very short (compare lanes 1 and 6 in Fig. 1A). Using the same protocol as for oligonucleotide 8180, two other series of plasmids with systematic extensions of the 5′ noncoding sequence were obtained by inserting oligonucleotide 8570 (AGCTACCAGATACAACAACA) or oligonucleotide 2991 (AGCTT[ATCA]13ATC hereafter abbreviated [ATCA]13).
Figure 1.
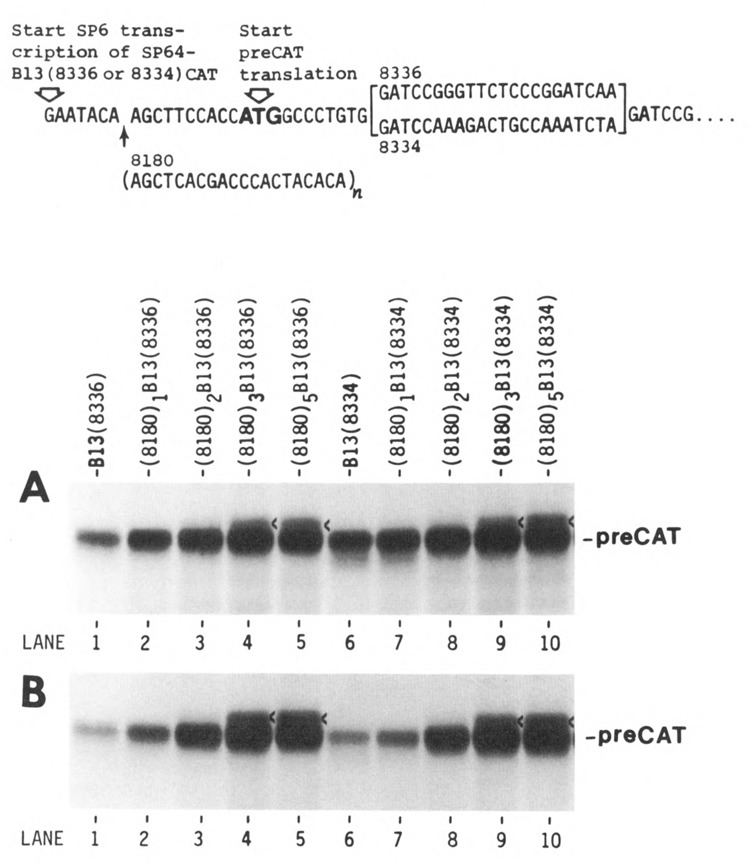
Evaluation of translational efficiency as a function of leader length. Upper. A transcript from the parental plasmid is depicted, with T’s in place of U’s. The preCAT start site is followed by either a structured (8336) or unstructured (8334) adaptor; the sequence downstream from the adaptor was published previously (Kozak, 1989a). The 5′ noncoding sequences on transcripts from the parental plasmids SP64-B13(8336)CAT and SP64-B13(8334)CAT are 17 nucleotides long. Multiples of oligonucleotide 8180, shown below the parental sequence, were inserted at the HindIII site (AAGCTT) to extend the 5′ noncoding sequence in increments of 20 nucleotides. Lower. Autoradiograms are shown of [35S]methionine-labeled proteins synthesized in a reticulocyte translation system under standard salt conditions (A) or with high KCl (B); proteins were fractionated by polyacrylamide gel electrophoresis (Kozak, 1989a). The mRNAs used for translation in panels A and B are indicated above each lane in A. A faint, slower-migrating band in lanes 4, 5, 9, and 10, marked with a caret, is attributable to an upstream ACG codon (see text and Figure 3). The band that runs faster than preCAT in lane 6 reflects a slight degree of leaky scanning (i.e., initiation from the second AUG codon) when the leader sequence is short (Kozak, 1991). Leaky scanning is suppressed by introducing secondary structure downstream (lane 1) or by lengthening the leader sequence (lanes 7–10).
Transcripts were synthesized using SP6 RNA polymerase as described (Kozak, 1989a), except that, after a 15-minute incubation with m7GpppG to allow the initiation of capped transcripts, the GTP concentration was raised to 0.5 mM and incubation continued for another 60 minutes. The mRNAs were purified as described (Kozak, 1989a) and used at 0.5 μg per 30 μl reaction to program in vitro translation. [35S]methionine-labeled “preCAT” protein (i.e., chloramphenicol acetyl transferase with a 22 amino acid N-terminal extension) was synthesized using a rabbit reticulocyte lysate from Bethesda Research Labs under standard conditions (2.2 mM magnesium acetate, 90 mM potassium acetate, and 45 mM KCl; Kozak, 1989a) or with the KCl concentration increased to 78 mM (hereafter designated “high KCl”). The latter manipulation was carried out because differences in translational efficiency are sometimes more obvious when potassium is elevated (Brendler et al., 1981). Use of the stated reaction conditions with a reticulocyte lysate from BRL precludes anomalies that may occur when some other commercial translation systems are used under the rather arbitrary reaction conditions recommended by some suppliers (Kozak, 1990b). A wheat germ translation system, prepared in-house, was used under conditions described previously (Kozak, 1989a). A long leader sequence augmented the synthesis of preCAT protein less dramatically in the high-capacity wheat germ system than in the lower-capacity reticulocyte system, perhaps because an extended leader sequence is most helpful when component(s) of the translational machinery are limiting.
For the analysis of initiation complexes, polypeptide elongation was inhibited by including 0.2 mM sparsomycin, which was a gift from the Division of Cancer Treatment of the National Cancer Institute. The binding of 3H-labeled mRNA to wheat germ or reticulocyte ribosomes was monitored by sedimentation at 4°C through glycerol gradients (10% to 30%) at 39,000 rpm for 2.2 or 3 hours in the Beckman SW41 rotor. To increase the sensitivity of ribosome protection assays, I used transcripts labeled with [α-32P]CTP instead of with 3H-UTP. The sparsomycin-blocked initiation complexes were digested at 4°C for 5 minutes with pancreatic RNase (10 μg/ml, Boehringer Mannheim) and then mixed with an equal volume of 0.4% glutaraldehyde before layering the gradients.
Results and discussion
A long, synthetic 5′ leader sequence improves translational efficiency in vitro
The simplest way to systematically increase leader length without continuously introducing novel sequences is to reiterate a small block of nucleotides. Thus, I introduced 1, 2, 3, or 5 copies of oligonucleotide 8180 near the 5′ end of the transcript shown at the top of Figure 1. The key feature of oligonucleotide 8180 is the scarcity of G residues, which precludes the formation of secondary structure. When transcripts from this series were tested in the reticulocyte system under standard conditions (Fig. 1A), there was a modest 3-fold improvement in protein yield as the leader was lengthened from 17 to 77 nt (lane 1 vs. lane 4); this is displayed quantitatively in Figure 2. Raising the KCl concentration selectively dampened the translation of mRNAs with short leader sequences, resulting in an impressive 10- to 15-fold difference in efficiency between B13 and (8180)3B13 (Fig. 1B, lane 1 vs. lane 4, and Fig. 2). Further lengthening of the 5′ leader to 117 nt caused only a slight improvement in translation relative to the 77 nt leader sequence (Fig. 1, lanes 4 and 5). The enhancing effect of a long 5′ noncoding sequence, shown here in vitro, is consistent with prior studies of the effects of leader length in vivo (Kozak, 1988).
Figure 2.
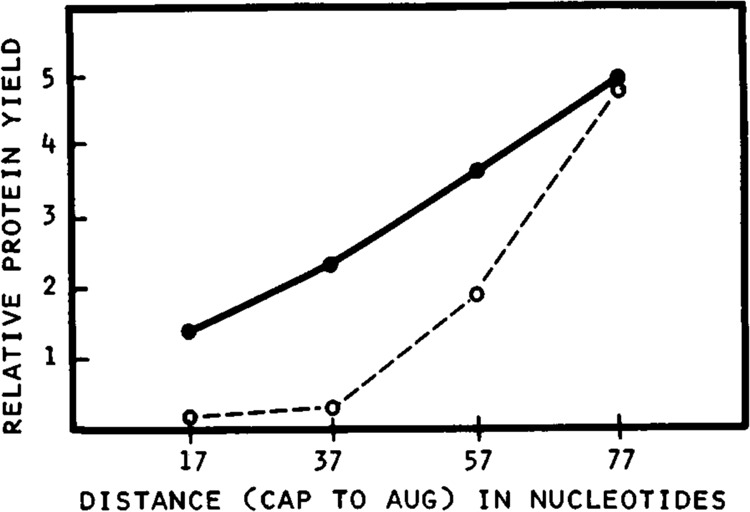
Quantitation of the relationship between leader length and translational efficiency in a reticulocyte cell-free system. The autoradiogram used for quantitation was from an experiment similar to that in Figure 1; the constructs were from series SP64(8180)nB13(8336)CAT. Relative yields of preCAT protein are expressed in optical density units. Symbols: •—• translation at 90 mM potassium acetate and 45 mM KCl (standard conditions); ○- - -○ translation at 90 mM potassium acetate and 78 mM KCl (high KCl).
The only anomaly in Figure 1 is that (8180)3B13 and (8180)5B13 produced a faint extra band just above the preCAT position in lanes 4, 5, 9, and 10. I suspected that the spurious translation product might result from initiation at an ACG triplet in oligonucleotide 8180. To test that hypothesis, I changed the second A in oligonucleotide 8180 to T. As shown in Figure 3, reiteration of the new oligonucleotide 8180T enhanced the yield of preCAT protein considerably (lane 5 vs. lane 1) with no trace of extraneous polypeptide bands.
Figure 3.
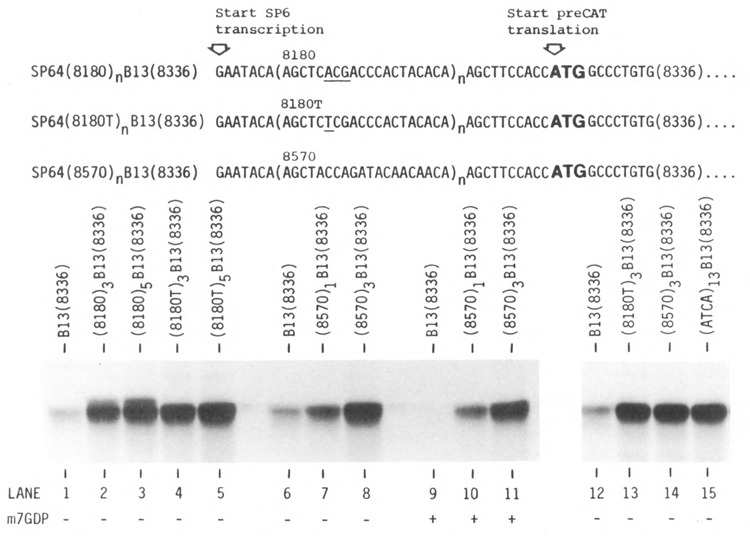
Comparison of translation with three different synthetic leader sequences. The reticulocyte translation system was used under high KCl conditions. The cap analogue m7GDP (0.8 mM) was included in the reactions represented in lanes 9–11. The 5′ sequence of the control transcript B13(8336) is given at the top of Figure 1. The upstream ACG codon in oligonucleotide 8180, which is responsible for the spurious upper band in lanes 2 and 3, is underlined in line 1. Synthesis of the extraneous upper band was abolished when the ACG codon was changed to TCG (oligonucleotide 8180T, line 2, lanes 4 and 5). Each of the transcripts tested in lanes 13–15 contained an insert of 60 nt, for a total leader length of 77 nt. The full 5′ leader sequence of the transcript designated ATCA (lane 15) is given in the text. Lanes 1–11 and lanes 12–15 represent two separate experiments.
Length alone matters
Another oligonucleotide, designated 8570, also supported efficient and exclusive synthesis of preCAT protein (Fig. 3, lane 8). With oligonucleotide 8570, as with oligonucleotide 8180 described above, translational efficiency was proportional to leader length; i.e., a single insert, which extended the leader from 17 to 37 nt, stimulated translation less than a triple insert, which lengthened the leader to 77 nt (Fig. 3, lanes 6–8). The fact that reiteration of two different oligonucleotides stimulated translation to similar extents suggests that the increase in efficiency is due simply to leader length, irrespective of the particular sequence involved. The alternative possibility is that some motif shared by oligonucleotides 8180T and 8570, such as CTAC or TACA, might specifically stimulate translation. To rule out the latter possibility, I designed one more oligonucleotide that lacked the CTAC and TACA motifs. Insertion of the new 60 nt sequence, AGCTT(ATCA)13ATC, conferred the same efficient translation as oligonucleotides (8180T)3 and (8570)3 (Fig. 3, lanes 13–15). The only constraints I observed in designing these three oligonucleotides were that I avoided G residues, in order to preclude the formation of secondary structure, and to some extent I avoided T residues, which are underrepresented in the 5′ noncoding sequences of vertebrate mRNAs (Kozak, 1987b). It seems likely that any moderately long leader sequence which meets those requirements would support efficient translation.
The aforementioned mRNAs with long leader sequences were less sensitive to inhibition by cap analogues than an mRNA with a short (17 nt) leader (Fig. 3, lanes 6–8 vs. 9–11). It may be that interaction with cap binding protein(s) is needed to stabilize mRNA/ribosome complexes only when constraints near the 5′ end of the transcript prevent the 40S subunit from advancing as soon as it touches. Thus, because the long, unstructured 5′ sequences on my transcripts provide easy entry for 40S ribosomal subunits (see below), translation should be and is relatively independent of the m7G cap.
To ensure that the source of RNA polymerase used for in vitro transcription does not influence the outcome of the experiments, I have constructed series 8180T with both a T7 and an SP6 promoter. The resulting transcripts performed identically in translation assays.
Comparison of translational efficiency between synthetic and natural leader sequences
Expression systems that use bacteriophage T7 RNA polymerase to transcribe plasmids that carry portions of the encephalomyocarditis virus (EMCV) leader sequence have been shown to express proteins efficiently in vivo and in vitro (Elroy-Stein et al., 1989). I used one such vector (pT7EMCAT, kindly provided by Drs. O. Elroy-Stein and B. Moss) to compare the translational efficiency of my synthetic leader sequences with that of a natural mRNA. (To assert that the synthetic leaders mediate efficient translation would be meaningless unless “efficient” is defined relative to something.) Figure 4 shows mRNA concentration curves for two constructs from series 8180 (described above) and, by way of comparison, a transcript that carries about 580 nucleotides of the EMCV 5′ noncoding sequence. The CAT protein encoded by T7EMCAT, lacking the N-terminal extension of the preCAT polypeptide, runs appropriately faster in the polyacrylamide gel in Figure 4. Quantitative comparison of protein yields reveals that T7EMCAT mRNA at the lowest concentration is somewhat more efficient than (8180)2B13; but, at higher concentrations of mRNA, (8180)2B13 is the more efficient. Since (8180)3 and (8180)5 transcripts are even more active than (8180)2 (Fig. 1), the synthetic leaders are indeed commendable for in vitro expression studies.
Figure 4.
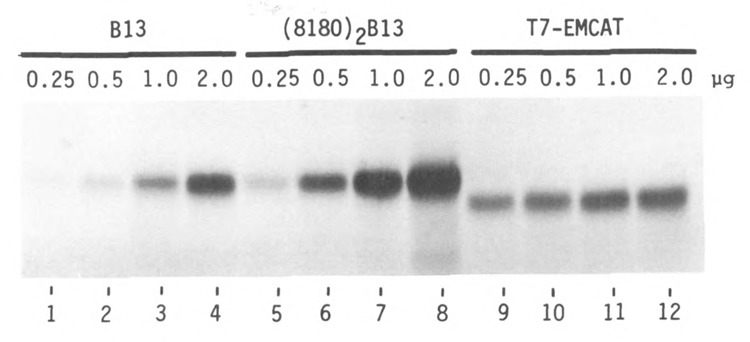
Comparison of an efficient synthetic leader sequence with an efficient natural leader sequence. Transcripts were derived from SP64-B13(8336)CAT (lanes 1–4), SP64(8180)2B13 (lanes 5–8) and pT7EMCAT (lanes 9–12; Elroy-Stein et al., 1989). Each mRNA was tested in the reticulocyte translation system under high KCl conditions; mRNA concentrations (μg per 30 μl reaction) are shown above each lane. Transcripts from the SP64-B13 series also supported translation efficiently in the wheat germ system (not shown), while T7EMCAT transcripts were nonfunctional in that system.
The mechanism by which oligonucleotide 8180 and other synthetic leader sequences augment translation is addressed below. The mechanism by which the EMCV leader sequence functions has just begun to be explored (Jang et al., 1989). The extraordinary amount of mRNA produced in cells engineered to express T7 or T3 RNA polymerase (Fuerst et al., 1989; Zhou et al., 1990) raises the caveat that a transcript such as T7EMCAT need not be translated especially well in vivo to account for the impressive yield of protein.
Effects of 5′leader length on formation of initiation complexes
Leader length had notable effects on ribosome binding when transcripts were incubated, in the presence of 0.2 mM sparsomycin, with reticulocyte (Fig. 5, A–B) or wheat germ ribosomes (Fig. 5, C–D). Although qualitatively similar complexes were obtained in both systems, I prefer the wheat germ system for analyzing initiation complexes because of its higher capacity for binding mRNA (e.g., compare Figure 5A with Figure 5C). The glycerol gradient profiles show that transcripts with a short (17 nt) leader sequence bound a single 80S ribosome (Fig. 5, A and C), while the complexes formed with long (77 nt) leader transcripts showed a heavier-than-80S shoulder (Fig. 5, B and D). The simplest explanation for the latter complexes is that a long leader provides room for extra 40S ribosomal subunits to accumulate upstream from the AUG codon. That interpretation is supported by two further observations: the size of the rapidly sedimenting complexes increased in proportion to the length of the 5′ noncoding sequence (Fig. 5C, D and E); and RNase digestion of rapidly sedimenting complexes like those in Figure 5E indeed released 40S ribosomal subunits bearing fragments of 32P-labeled mRNA (Fig. 5G). As a control, complexes formed with 32P-labeled short leader transcripts were also digested with RNase, and they did not yield a 40S ribosome-protected peak (Fig. 5H). A reasonable hypothesis is that the “preloading” of 40S ribosomal subunits on long leader sequences gives those mRNAs an advantage, especially under conditions of competition; and that may account for their remarkably efficient translation the experiments presented in Figures 1–4.
Figure 5.
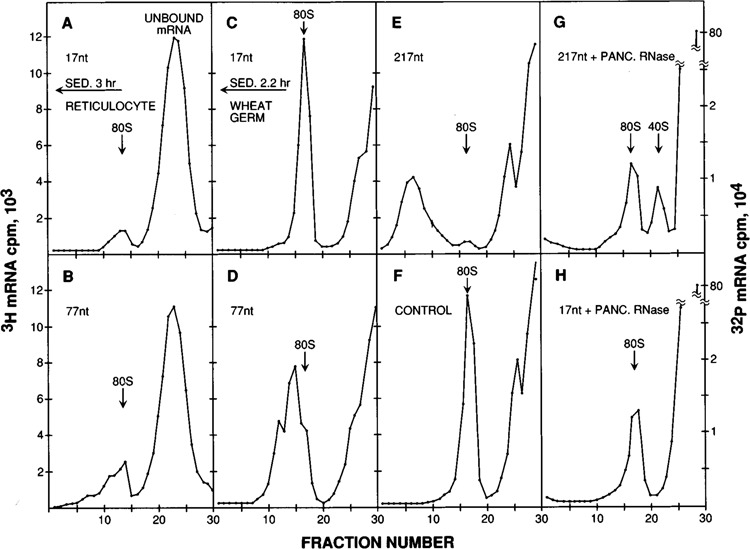
Analysis of initiation complexes by glycerol gradient centrifugation. Radiolabeled mRNAs were incubated for 8 minutes in a sparsomycin-blocked reticulocyte lysate under high KCl conditions (A–B) or in the standard wheat germ system (C–H). The mRNAs used here, from series SP64(8570)nB13(8336)CAT, had 5′ noncoding sequences of 17 (A, C, H), 77 (B, D) or 217 nt (E, G). Sedimentation (right to left) was for 3 (A, B) or 2.2 hours (C–H). The reticulocyte gradients were centrifuged slightly longer than the wheat germ gradients, because in the reticulocyte system the peaks are generally broader and thus harder to resolve. The 60 nt insert ([8570]3) that was present at the 5′ end of the mRNA used in D was relocated to a PvuII site within the CAT coding sequence in the control transcript used in F; the 5′ noncoding sequence on the control transcript was 17 nt long. For the ribosome protection experiments shown in G and H, initiation complexes formed with 32P-labeled mRNA were digested with pancreatic RNase before centrifugation. The 40S-protected peak in G is smaller than might be expected from the size of the starting complexes in E because, once the mRNA has been cleaved by RNase, there is nothing to keep the scanning 40S ribosomes from falling off. The absence of a 40S-protected peak in H validates the interpretation that the 40S peak in G derives from the binding of 40S subunits upstream from the AUG codon on long-leader mRNAs.
Although the translation of long leader transcripts was not very cap-dependent (Fig. 3, lanes 10 and 11 vs. lanes 7 and 8), the binding of ribosomes to those sequences was still end-dependent. This was shown by a control experiment in which three copies of oligonucleotide 8570 were inserted within the CAT coding sequence. As shown in Figure 5F, a transcript thus modified was unable to bind the extra 40S ribosomes that accumulated when (8570)3 was located at the 5′ end (Fig. 5D). (Despite the presence of 17 AUG codons downstream from the PvuII site where oligonucleotide 8570 was inserted, the control transcript described in Figure 5F produced no low molecular weight translation products [data not shown].) Thus, an unstructured sequence 60 nt in length does not mediate direct internal binding of ribosomes. This means that, despite the lack of dependence on the m7G cap, the extra 40S subunits that accumulated on long leader mRNAs must all have entered from the 5′ end. The queuing of 40S ribosomes on long 5′ leader sequences thus constitutes additional evidence in support of the scanning model for initiation (Kozak, 1989b). That translational efficiency does not decline as the 5′ noncoding sequence is lengthened implies that scanning is not rate-limiting, as long as the leader sequence is unstructured.
Queuing of 40S ribosomes might also explain the unexpected initiation from an upstream ACG codon in oligonucleotide 8180 (Fig. 1, lanes 4, 5, 9, 10): if recognition of nonstandard initiator codons is favored when the migration of 40S subunits is slowed (Kozak, 1990a), and if assembly of an 80S ribosome at the AUG codon is slower than the binding of 40S subunits upstream, the 40S subunits forced to wait on these long leader sequences might be expected to utilize weak upstream initiation sites that would otherwise be ignored.
Extrapolations and conclusions
A handful of viral and cellular mRNAs seem to have very efficient 5′ noncoding sequences, as demonstrated by leader shuffling experiments (Berkner and Sharp, 1985; Gallie et al., 1987; Logan and Shenk, 1984; McGarry and Lindquist, 1985). Attempts to pinpoint a motif in any of those sequences that underlies their efficient translation have been notably unsuccessful (McGarry and Lindquist, 1985; Dolph et al., 1990, Sleat et al., 1988; Schoffl et al., 1989). Thus, virtually every portion of those leader sequences has been mutated or deleted or replaced without measurably impairing translation. The apparent absence of a discrete effector motif in viral and cellular 5′ leaders encourages the hypothesis that, apart from the m7G cap and the sequence immediately flanking the AUG codon, a moderately long, unstructured leader irrespective of its particular sequence is necessary and sufficient for efficient initiation. The ability to concoct three unrelated synthetic leader sequences that mediate efficient translation, as described herein, strongly supports that hypothesis. It seems unlikely that such arbitrarily designed sequences are recognized by hypothetical sequence-specific “enhancer proteins.” Rather it seems likely that long leader sequences facilitate initiation by allowing preloading of 40S ribosomal subunits, as discussed in connection with Figure 5.
Although some evidence for the ability of long leader sequences to augment translation in cultured cells has been presented (Kozak, 1988), additional in vivo experiments are needed to test the validity of extrapolating the findings reported here. Other investigators have occasionally reported that lengthening the 5′ noncoding sequence improves translational efficiency in their systems (Levitan et al., 1988), but the facilitating effect of a long leader sequence was missed in most earlier studies (Cigan et al., 1988; Johansen et al., 1984; Rhee and Carmichael, 1989; Sleat et al., 1987), perhaps because secondary structure was inadvertently introduced as the leader was extended, or because the changes in length were outside of the critical range, or because the advantage of a long leader sequence may be slight in the absence of competition. When such complications are precluded, the long and the short of it is that leader length matters.
The magnitude of the stimulation observed in leader-shuffling experiments obviously depends on which mRNA is taken as the baseline. Because my control construct B13(8336) supports translation fairly efficiently to begin with, the fold-stimulation upon switching to a better (longer) leader sequence was less dramatic than that reported by other investigators whose baseline was a virtually untranslatable transcript (Fig. 1 in Falcone and Andrews, 1991). The failure of Falcone and Andrews to include a point of reference (such as the EMCV leader sequence used herein) and their failure to state how much mRNA was used per reaction make it difficult to say just how efficient their best “natural” leader sequence is; and their nonsystematic approach to mutagenesis reveals nothing about why it is efficient. Although Falcone and Andrews did not consider the simple explanation for translational efficiency developed herein, they offered no alternative proposals, and their data are easily accommodated by the rule that a leader sequence need only be moderately long and unstructured to promote efficient initiation of translation.
Previous experiments established that efficient translation of eukaryotic mRNAs requires a methylated 5′ cap (Shatkin, 1976), an unstructured leader sequence (Kozak, 1989c), and an appropriate context around the AUG codon (Kozak, 1986, 1987a, 1987b). Leader length may now be added to the catalogue of structural features in eukaryotic mRNAs that contribute to the fidelity and/or efficiency of initiation. The scanning model for initiation (Kozak, 1989b) rationalizes all of these requirements.
Acknowledgments
I am grateful to Drs. Orna Elroy-Stein and Bernard Moss for a generous gift of plasmid TM1 (pT7EMCAT). Funds were provided by NIH grant GM-33915.
The costs of publishing this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
References
- Berkner K. L. and Sharp P. A. (1985), Nucl Acids Res 13, 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendler T., Godefroy-Colburn T., Carlill R. D., and Thach R. E. (1981), J Biol Chem 256, 11747–11754. [PubMed] [Google Scholar]
- Carrington J. C. and Freed D. D. (1990), J Virol 64, 1590–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinkers M., Garbers D. L., Chang M-S., Lowe D. G., Chin H., Goeddel D. V., and Schulz S. (1989), Nature 338, 78–83. [DOI] [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., and Donahue T. F. (1988), Mol Cell Biol 8, 2964–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolph P. J., Huang J., and Schneider R. J. (1990), J Virol 64, 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elloy-Stein O., Fuerst T. R., and Moss B. (1989), Proc Natl Acad Sci USA 86, 6126–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone D. and Andrews D. W. (1991), Mol Cell Biol 11, 2656–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T. R. and Moss B. (1989), J Mol Biol 206, 333–348. [DOI] [PubMed] [Google Scholar]
- Gallie D. R., Sleat D. E., Watts J. W., Turner P. C., and Wilson T. M. A. (1987), Nucl Acids Res 15, 8693–8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Macon K. J., Kidd V. J., and Volanakis J. E. (1990), J Biol Chem 265, 6521–6524. [PubMed] [Google Scholar]
- Ito K., Kashiwagi K., Watanabe S., Kameji T., Hayashi S., and Igarashi K. (1990), J Biol Chem 265, 13036–13041. [PubMed] [Google Scholar]
- Jang S. K., Davies M. V., Kaufman R. J., and Wimmer E. (1989), J Virol 63, 1651–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen H., Schumperli D., and Rosenberg M. (1984), Proc Natl Acad Sci USA 81, 7698–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1986), Cell 44, 283–292. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1987a), J Mol Biol 196, 947–950. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1987b), Nucl Acids Res 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1988), Mol Cell Biol 8, 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1989a), Mol Cell Biol 9, 5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1989b), J Cell Biol 108, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1989c), Mol Cell Biol 9, 5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1990a), Proc Natl Acad Sci USA 87, 8301–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1990b), Nucl Acids Res 18, 2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1991), Gene Expr 1, 111–115. [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. B. and Jackson K. J. (1982), J Mol Biol 162, 317–334. [DOI] [PubMed] [Google Scholar]
- Leung S., Whitelaw E., and Proudfoot N. J. (1989), Nucl Acids Res 17, 8283–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan E. S., Schofield P. R., Burt D. R., Rhee L. M., Wisden W., Kohler M., Fujita N., Rodriguez H. F., Stephenson A., Darlison M. G., Barnard E. A., and Seeburg P. H. (1988), Nature 335, 76–79. [DOI] [PubMed] [Google Scholar]
- Logan J. and Shenk T. (1984), Proc Natl Acad Sci USA 81, 3655–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry T. J. and Lindquist S. (1985), Cell 42, 903–911. [DOI] [PubMed] [Google Scholar]
- Murray M. B., Zilz N. D., McCreary N. L., MacDonald M. J., and Towle H. C. (1988), J Biol Chem 263, 12770–12777. [PubMed] [Google Scholar]
- Rao C. D., Pech M., Robbins K. C., and Aaronson S. A. (1988), Mol Cell Biol 8, 284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee E. and Carmichael G. G. (1989), J Virol 63, 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoffl F., Rieping M., Baumann G., Bevan M., and Angermuller S. (1989), Mol Gen Genet 217, 246–253. [DOI] [PubMed] [Google Scholar]
- Shatkin A. J. (1976), Cell 9, 645–653. [DOI] [PubMed] [Google Scholar]
- Sleat D. E., Gallie D. R., Jefferson R. A., Bevan M. W., Turner P. C., and Wilson T. M. A. (1987), Gene 60, 217–225. [DOI] [PubMed] [Google Scholar]
- Sleat D. E., Hull R., Turner P. C., and Wilson T. M. A. (1988), Eur J Biochem 175, 75–86. [DOI] [PubMed] [Google Scholar]
- Teruya J. H., Kutsunai S. Y., Spear D. H., Edwards P. A., and Clarke C. F. (1990), Mol Cell Biol 10, 2315–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons M. S. and Witte O. N. (1989), Oncogene 4, 559–567. [PubMed] [Google Scholar]
- Waterhouse P., Khokha R., and Denhardt D. T. (1990), J Biol Chem 265, 5585–5589. [PubMed] [Google Scholar]
- Zhou Y., Giordano T. J., Durbin R. K., and McAllister W. T. (1990), Mol Cell Biol 10, 4529–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]


