Abstract
Development of noninvasive, reliable biomarkers for lung cancer diagnosis has many clinical benefits knowing that most of lung cancer patients are diagnosed at the late stage. For this purpose, we conducted proteomic analyses of 231 human urine samples in healthy individuals (n = 33), benign pulmonary diseases (n = 40), lung cancer (n = 33), bladder cancer (n = 17), cervical cancer (n = 25), colorectal cancer (n = 22), esophageal cancer (n = 14), and gastric cancer (n = 47) patients collected from multiple medical centers. By random forest modeling, we nominated a list of urine proteins that could separate lung cancers from other cases. With a feature selection algorithm, we selected a panel of five urinary biomarkers (FTL: Ferritin light chain; MAPK1IP1L: Mitogen-Activated Protein Kinase 1 Interacting Protein 1 Like; FGB: Fibrinogen Beta Chain; RAB33B: RAB33B, Member RAS Oncogene Family; RAB15: RAB15, Member RAS Oncogene Family) and established a combinatorial model that can correctly classify the majority of lung cancer cases both in the training set (n = 46) and the test sets (n = 14–47 per set) with an AUC ranging from 0.8747 to 0.9853. A combination of five urinary biomarkers not only discriminates lung cancer patients from control groups but also differentiates lung cancer from other common tumors. The biomarker panel and the predictive model, when validated by more samples in a multi-center setting, may be used as an auxiliary diagnostic tool along with imaging technology for lung cancer detection.
Keywords: Lung cancer, Machine learning, Urinary biomarkers
Highlights
-
•
A case-control study of biomarker discovery for lung cancer diagnosis was conducted.
-
•
Human urine profiles in control cases and cancers were characterized.
-
•
A list of candidate biomarkers was nominated and evaluated.
-
•
A panel of urinary biomarkers was established and tumor-specificity was evaluated.
Cancer diagnosis with a noninvasive method at the early stage of the disease is highly desirable. Here, we analyzed hundreds of human urine samples from healthy individuals, patients with benign pulmonary diseases, and 6 types of cancers by proteomics and developed a panel of five urinary proteins that can separate the lung cancer from benign pulmonary diseases as well as the other 5 cancers (bladder, cervical, colorectal, esophageal and gastric) with a good sensitivity and disease specificity. Further validation experiments with expanded sample numbers are required to investigate whether this method can be applied in a clinical setting for the diagnosis of lung cancer.
1. Introduction
Lung cancer is the second most common cancer among males and females worldwide and the most common cancer in China (Torre et al., 2016b, Torre et al., 2016a). It is the leading cause of cancer death in both men and women in the United States (Torre et al., 2016a). In 2012, there were approximately 1.8 million new cases and 1.6 million cancer deaths documented, which highlight a global public health concern (Stewart et al., 2014). Non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) are the two main histologic subtypes of lung cancer with the NSCLC as the most common subtype, accounting for about 83% of all lung cancers (Miller et al., 2016).
Computed tomography (CT) screening is the main test for lung cancer screening but is associated with a high false positive rate (Aberle et al., 2013). Disease stage significantly affects cancer treatment and survivorship. The 5-year survival rate is 55% for patients diagnosed at the early stage and 4% at the advanced stage (Miller et al., 2016). Unfortunately, majority of cases are diagnosed at the advanced stage due to the lack of symptoms and reliable biomarkers at the early stage (Miller et al., 2016).
Searching noninvasive biomarkers for clinical diagnosis is a continuous effort but success has been limited (Zhang and Chan, 2005). Current clinically used tumor markers for lung cancer screening including AFP (alpha fetoprotein), CA 19-9 (carbohydrate antigen 19-9), CA 125 (carcinoma antigen 125), CA 15-3 (carcinoma antigen 15-3), and CEA (carcino-embryonic antigen) lack sensitivity and specificity (Li et al., 2012, Harmsma et al., 2013). Some earlier proteomic studies towards lung cancer diagnosis based on urine or serum specimens have identified a few putative biomarkers, but the specificity against other tumors is poor or has not been investigated (Zhang et al., 2015, Nolen et al., 2015, Patz et al., 2007, Yildiz et al., 2007). In this study, we employed proteomics technology implemented with machine learning statistics to search for sensitive, lung cancer-specific diagnostic biomarkers from patient urines as a commonly used, noninvasive matrix as an alternative to blood.
2. Materials and Methods
2.1. Patient Specimens
At the biomarker discovery stage, a total of 46 urine specimens in the training set from healthy controls (CTL, n = 23) and lung cancer patients (LC, n = 23) were collected at Tianjin Baodi Hospital, Tianjin, China. Healthy controls were age- (>50 year) and gender-matched (frequency matching with random sampling) to lung cancer cases (Fig. 1a). Urine samples were collected from Non-small cell lung cancer (NSCLC) patients at the time they were diagnosed with lung cancer and had no anticancer treatment. Urine samples were collected from healthy donors who had no known lung diseases and had negative clinical tumor markers (AFP: alpha fetoprotein, CA 19-9: carbohydrate antigen 19-9, CA 125: carcinoma antigen 125, CA 15-3: carcinoma antigen 15-3, and CEA: carcino-embryonic antigen). A blood test monitored the levels of urea nitrogen, creatinine, and uric acid to exclude any cases that may have renal dysfunction. For validation purposes, an independent case-control test set (10 CTL, 10 LC; Fig. 1a, test set 1) with same criteria was obtained from the same Hospital. In addition to healthy donors, urines from benign pulmonary conditions (pneumonia, n = 23; COPD: Chronic Obstructive Pulmonary Disease, n = 17) were also sampled in the same Hospital. Clinical details of healthy controls, benign lung diseases, and lung cancer patients were summarized in Table 1. To validate if the biomarkers found in this study is lung cancer-specific, additional urine samples were collected from patients of bladder cancer (BC, n = 17), cervical cancer (CCA, n = 25), colorectal cancer (CRC, n = 22), esophageal cancer (EC, n = 14), or gastric cancer (GC, n = 47) in three hospitals (GC and CRC: Affiliated Hospital of Academy of Military Medical Sciences, Beijing, China; CCA: No.1 Affiliated Hospital of Medical School, Xi'an Jiaotong University, Xi'an, China; BC and EC: Tianjin Baodi Hospital, Tianjin, China). All participants have provided signed informed consent and samples were collected with ethics approval from institutional review board of hospitals participating in this study. Our research strictly followed the standards indicated by the Declaration of Helsinki.
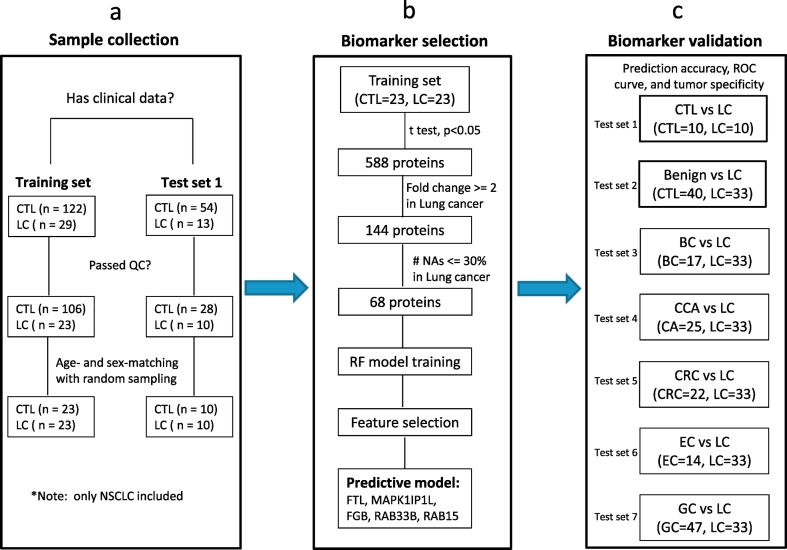
Fig. 1.
Flow diagram of lung cancer biomarker study.
(a) A total of 218 urine specimens were randomly collected from healthy donors or NSCLC patients. After QC filtering and age/sex-matching, a pair of 23 or 10 case-control urine samples was selected in the training set or test set (test set 1), one for biomarker discovery and the other one for biomarker validation, respectively. (b) Student's t-test revealed a total of 588 proteins with a p value <.05 in the training set, 144 were up-regulated with at least 2 folds in the cancer group. Finally, 68 proteins were retained by restricting the number of missing values in <30% of lung cancer cases. A random forest model was developed upon the training set with 68 proteins. By running feature selection algorithm, five biomarkers were selected and incorporated into a predictive model. (c) The biomarker panel and the predictive model were evaluated on 7 independent test sets to determine how well the model can predict lung cancer from healthy individuals and benign lung diseases (test set 1–2) or from other cancers (test set 3–7). Abbreviations: CTL, healthy controls; LC, lung cancer; BC, bladder cancer; CCA, cervical cancer; CRC, colorectal cancer; EC, esophageal cancer; GC, gastric cancer; NSCLC, non-small-cell lung cancer; QC, quality control.
Table 1.
Clinical profiles and demographics of healthy controls and lung cancer patients.
| Demographics | Training set |
Test set |
Benign lung diseases |
|||
|---|---|---|---|---|---|---|
| CTL (n = 23) | LC (n = 23) | CTL (n = 10) | LC (n = 10) | COPD (n = 17) | Pneumonia (n = 23) | |
| Age, years | 55.61 ± 8.02 | 65.65 ± 11.2 | 55.8 ± 3.49 | 65.7 ± 8.96 | 73.88 ± 10.07 | 60.39 ± 22 |
| Sex | ||||||
| Male | 16 | 16 | 7 | 7 | 13 | 16 |
| Female | 7 | 7 | 3 | 3 | 4 | 7 |
| Clinical stage | ||||||
| 1 | 1 | 2 | ||||
| 2 | 4 | 1 | ||||
| 3 | 10 | 3 | ||||
| 4 | 8 | 4 | ||||
| Subtype | ||||||
| ADC | 10 | 2 | ||||
| SCC | 13 | 8 | ||||
ADC, adenocarcinoma; SCC, squamous cell carcinoma; CTL, healthy controls; LC, lung cancer; COPD, Chronic Obstructive Pulmonary Disease.
2.2. Urinary Proteome Measurements by LC-MS/MS
Nano LC-MS/MS (liquid chromatography tandem mass spectrometry) analysis of human urine samples was conducted as previously (Supplementary Fig. S1a) (Leng et al., 2017). Briefly, about 10 ml of midstream urine was centrifuged at 200,000g for 70 min. After ultracentrifugation, pellet was reduced with DTT to remove the uromodulin (the most abundant urinary protein) (Pisitkun et al., 2006, Raimondo et al., 2013). After heating at 65 °C for 30 min, pellet was washed with wash buffer (10 mM TEA, 100 mM NaCl, pH 7.4) twice and ultra-centrifuged for 30 min. The pellet was dissolved in SDS buffer (1% SDS, 50 mM Tris, pH 8.5) and resolved on an SDS-PAGE gel. Gel was cut into six pieces and then subjected to in-gel trypsin digestion.
Six gel fractions were combined into 2–3 injections. Tryptic peptides were resolved on a home-made, capillary column packed with C18 particles and analyzed by Thermo Fisher Orbitrap mass spectrometers coupled with online Easy-nLC 1000 nano-HPLC system (Thermo Fisher Scientific). LC-MS/MS data were processed in Proteome Discoverer 1.4 software (Thermo Fisher Scientific) and searched against Human Refseq protein database (Released on 2013/07/04) on Mascot search engine (Version 2.3, Matrix Science Inc) with appropriate mass tolerances (precursor ions: 20 ppm; fragment ions: 0.02 or 0.5 Da). Variable modifications including cysteine carbamidomethylation, methionine oxidation, and protein N-terminal acetylation were incorporated in the search. A maximum of one miscleavage of trypsin was allowed. All peptides below 1% false discovery rate were retained and grouped into gene products. Each protein to be reported requires a minimum of one unique and strict peptide (i.e. sequence-specific peptide with a mascot ion score higher than 20 at the gene level). All keratins were removed from the list. Protein abundance was measured as iBAQ (intensity-based absolute quantification) - a label-free quantification algorithm (Schwanhausser et al., 2011). For batch-to-batch comparison, iBAQ was converted into iFOT (intensity-based Fraction of Total) representing a normalized intensity of a protein identified in an LC-MS/MS run (Liu et al., 2013). For visualization purpose, the number of iFOT is multiplied by 105. Tryptic digests of 293T cell as QC (quality control) samples were routinely assessed by LC-MS/MS to guarantee the instrument reproducibility. Sample metadata were summarized in Supplementary Table S1. All raw files and search results have been deposited in ProteomeXchange via iProX (ww.iprox.org) with the identification no.: PXD008846 or IPX0001153000.
2.3. Statistical Analysis and Biomarker Selection
Missing value imputation was performed on all data sets after normalization by using k-nearest neighbors (KNN) algorithm (R package: impute, Version 1.47.0) (Troyanskaya et al., 2001). Proteins that have <10% of missing numbers in each class were imputed and substituted with the mean of its five closest neighbors. All other missing values were set to be 0.0099. This has resulted in a list of 7408 protein IDs (Supplementary Table S2). Gene ontology analysis was performed by WebGestalt – a functional enrichment analysis web tool (Wang et al., 2017). Ward's hierarchical clustering analysis was implemented in the R statistical software with the “hclust” function using average linkage as distance metric.
Statistical analysis was performed on the training set to identify potential urine biomarkers for lung cancer. Student's t-test resulted in 588 differentially expressed proteins between healthy controls and lung cancer patients at p < 0.05 (Fig. 1b). For practical purposes, the candidates were further narrowed down to 68 proteins that were up-regulated by >2-fold in the lung cancer group and were detected in >70% of times in the lung cancer patients. Random forest, an ensemble, supervised machine learning algorithm, implemented with feature selection method was used to select variables (proteins) and build a classifier (a predictive model based on a panel of proteins) upon the training set (Genuer et al., 2010).
To evaluate the prediction accuracy, the predictive model with a panel of proteins was tested on an independent validation set comprised of 10 healthy controls and 10 lung cancer patients with matching age and sex. To investigate whether the selected proteins can separate lung cancer cases from benign lung diseases or other types of tumors, the model was further tested on other validation sets which contained lung cancer cases, benign pulmonary conditions (pneumonia and COPD), and one of other five cancers (Fig. 1c).
3. Results
3.1. Urine Proteomes in Controls and Six Cancers
In total, we assayed 383 urine specimens and selected 231 urine subjects (in the training set and 7 test sets, Fig. 1) that passed QC and after age/sex-matching (Supplementary Table S2). We have achieved high batch-to-batch instrument reproducibility of iFOT measured with high Pearson correlation coefficients (0.88 on average) between QC samples. A total of 7408 proteins (Supplementary Table S2) were identified and quantified (mean iFOT: 12.42; standard deviation: 17.09). On average, we were able to identify and quantify 1248.69 ± 314.79 of proteins in one urine sample. Gene ontology analysis revealed that membrane and vesicle are the two main components of urine proteins (Supplementary Fig. S1b). We observed some well-known exosomal markers such as PDCD6IP (also known as Alix), HSPA8 (also known as HSC70), and TSG101 as well as tetraspanin proteins and RAB proteins (Supplementary Table S2 and Fig. S1c) (Yoshioka et al., 2013, Bobrie et al., 2012, Greening et al., 2015). The relatively high abundance of these proteins indicates exosomal proteins are one of the main categories in human urine sediments. Unsupervised hierarchical clustering analysis of urine profiles has separated some groups well, suggesting that urine profiling may have disease-specific features (e.g. LC and benign cases, Supplemental Fig. S2). Student's t-test on the training set identified 588 significantly altered proteins with p values <0.05 at 93.8% of statistical power (Fig. 1b). Of these proteins, 144 were up-regulated by >2 folds in lung cancer patients. Considering the practical purpose of clinical biomarkers, we further removed low abundant proteins from the list and chose 68 candidates that were relatively abundant and were detectable in >70% of lung cancer urine specimens.
3.2. Candidate Biomarker Selection, Model Development, and Biomarker Panel Selection
With the 68 proteins selected above, we next ran a random forest model (Genuer et al., 2010, Breiman, 2001) to determine if the urine profiling had cancer-specific features for lung cancer diagnosis (Fig. 1b). Due to the relatively small sample sizes in two data sets, 2/3 of individuals in the training set were selected to grow decision trees by bootstrapping (random sampling with replacement) while the remaining samples were left out as out of bag (oob) samples for cross-validation purpose to estimate the classification error and measure the variable importance (Supplementary Fig. S3). As evaluated from 1000 forests, random forest model with these proteins was able to predict lung cancer cases correctly at ~95% of the time both on the oob samples (1/3 of training samples) and the test set (CTL = 10, LC = 10) (Fig. 2a). The top 5 most important variables are: FTL (Ferritin light chain), MAPK1IP1L (Mitogen-Activated Protein Kinase 1 Interacting Protein 1 Like), FGB (Fibrinogen Beta Chain), GNS [glucosamine (N-acetyl)-6-sulfatase] and RAB33B, (Member RAS Oncogene Family) (Fig. 2b). The top 15 proteins ranked by variable importance were able to correctly separate lung cancer patients from healthy individuals well in either the training set (n = 46) or the test set (n = 20) with the area under the ROC curve (AUC) of >0.75 when they were combined (Supplementary Fig. S4). Among them, FTL, FGB and C9 (Complement C9) have been reported in serum or urine lung cancer biomarker studies previously (Li et al., 2012, Kim et al., 2016, Ahn and Cho, 2013, Nolen et al., 2015, Patz et al., 2007).
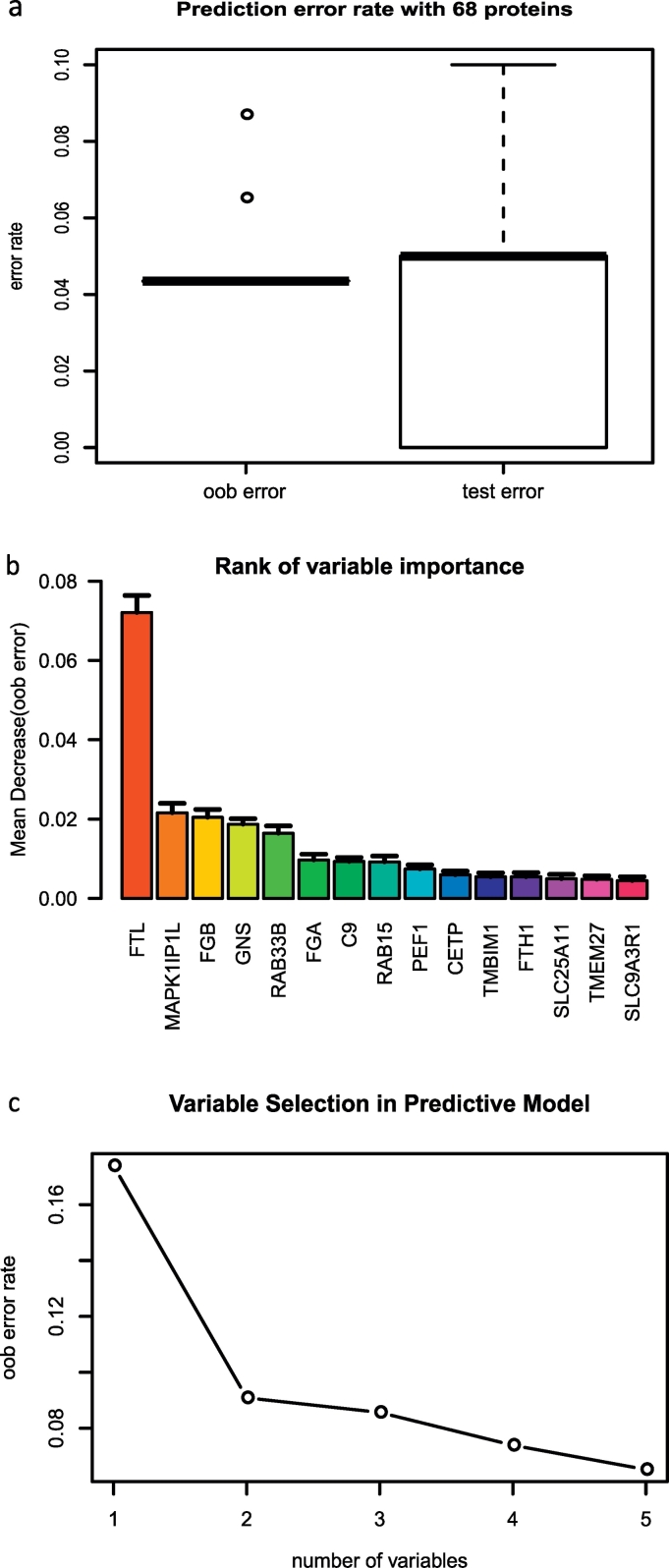
Fig. 2.
Prediction performance, variable importance, and variable selection during model building.
(a) Evaluation of prediction errors by cross-validation. (b) The top 15 most important variables ranked by mean decrease in accuracy estimated from 1′000 forests on oob (out of bag) samples. (c) Selection of 5 variables in the predictive model based on the classification error rate on oob samples.
To build a simple random forest model with a manageable size of variables, feature selection algorithm was implemented to remove the redundancy as variables may be highly correlated (Genuer et al., 2010). The algorithm selected 5 proteins (FTL, MAPK1IP1L, FGB, RAB33B, and RAB15) in the predictive model with ~6.5% of mean classification error rate on the oob samples (i.e. cross-validation error) (Fig. 2c). With these 5 proteins, the predictive model could correctly separate most of lung cancer cases from the controls in the training set as sufficient as the random forest model using all 68 proteins (Supplementary Fig. S5).
3.3. Evaluation of Biomarker Panel in Healthy Individuals and Benign Pulmonary Conditions
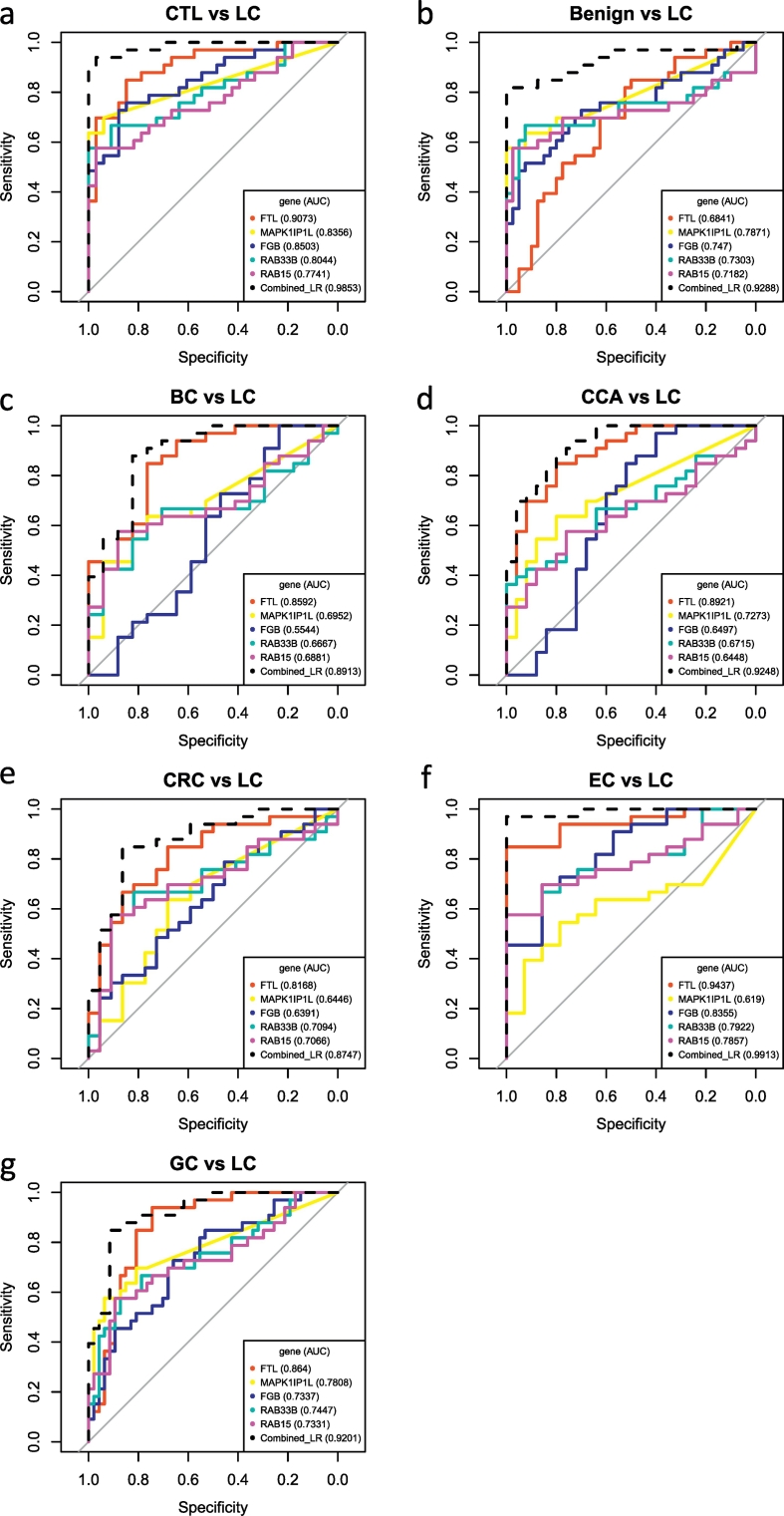
The five proteins selected in the predictive model were then assessed on an independent, blinded data set (test set 1, Fig. 1) comprised of 10 healthy controls and 10 lung cancer cases. The predictive model was able to correctly classify 9 LC urine samples and 9 healthy controls with 10% of prediction error on either CTL or LC samples (Table 2). Compared with the five clinically used tumor markers (AFP, CA 19-9, CA 125, CA 15-3, and CEA), we found that 8 out of 33 lung cancer patients had normal blood levels of all these proteins, indicating a high false negative rate by using these markers. The protein FTL in the model had the best discriminating power with an AUC of 0.9073 while the combined model (logistic regression model with 5 proteins) had reached an AUC of 0.9853 (Fig. 3a). Other four proteins were also able to separate two groups well with AUCs: 0.8356 (MAPK1IP1L), 0.8503 (FGB), 0.8044 (RAB33B), and 0.7741 (RAB15), respectively. Since these five proteins are over-expressed and are relatively abundant in the lung cancer group (Fig. 4), they are most likely to be detectable if biochemical assays such as ELISA (enzyme-linked immunosorbent assay) are adopted.
Table 2.
Random forest model in predicting lung cancer against controls and other cancers.
| CTL vs LC | Benign vs LC | BC vs LC | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Predicted | Predicted | Predicted | |||||||
| Group | CTL | LC | Group | Benign | LC | Group | BC | LC | |
| Actual | CTL | 9 | 1 | Benign | 28 | 12 | BC | 10 | 7 |
| LC | 1 | 9 | LC | 1 | 32 | LC | 2 | 31 | |
| Error | 0.1 | Error | 0.178 | Error | 0.18 | ||||
| Sensitivity | 90% | Sensitivity | 96.97% | Sensitivity | 93.94% | ||||
| Specificity | 90% | Specificity | 70% | Specificity | 58.82% | ||||
| CCA vs LC | CRC vs LC | EC vs LC | |||||||
| Predicted | Predicted | Predicted | |||||||
| Group | CCA | LC | Group | CRC | LC | Group | EC | LC | |
| Actual | CCA | 18 | 7 | CRC | 12 | 10 | EC | 12 | 2 |
| LC | 1 | 32 | LC | 1 | 32 | LC | 1 | 32 | |
| Error | 0.138 | Error | 0.2 | Error | 0.064 | ||||
| Sensitivity | 96.97% | Sensitivity | 96.97% | Sensitivity | 96.97% | ||||
| Specificity | 72% | Specificity | 54.55% | Specificity | 85.71% | ||||
| GC vs LC | |||||||||
| Predicted | |||||||||
| Group | GC | LC | |||||||
| Actual | GC | 38 | 9 | ||||||
| LC | 1 | 32 | |||||||
| Error | 0.125 | ||||||||
| Sensitivity | 96.97% | ||||||||
| Specificity | 80.85% | ||||||||
Sensitivity = number of true positives / (number of true positives + number of false negatives); Specificity = number of true negatives / (number of true negatives + number of false positives); CTL, healthy controls; LC, lung cancer; BC, bladder cancer; CCA, cervical cancer; CRC, colorectal cancer; EC, esophageal cancer; GC, gastric cancer. All test sets except for the test set 1 (CTL vs LC) compares all lung cancer patients to benign diseases or other cancers.
Fig. 3.
AUCs of five individual markers and the combinatorial logistic model in classifying lung cancer against (a) CTL, (b) Benign lung diseases, (c) BC, (d) CCA, (e) CRC, (f) EC, and (g) GC.
The logistic model (dashed line) combines all five markers. Abbreviations: CTL, healthy controls; LC, lung cancer; BC, bladder cancer; CCA, cervical cancer; CRC, colorectal cancer; EC, esophageal cancer; GC, gastric cancer. Note: lung cancer cases in the training set and test set were combined.
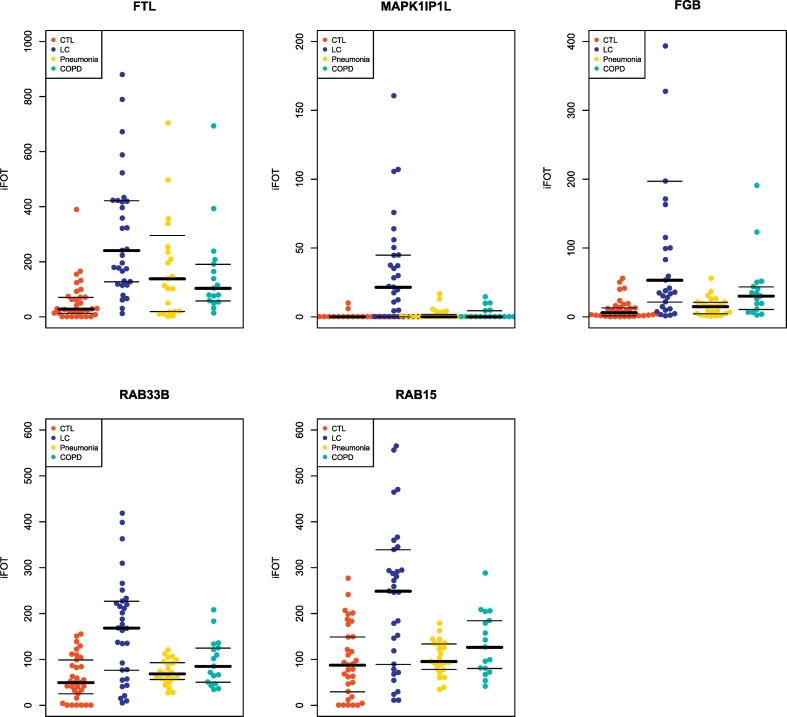
Fig. 4.
Relative abundance of five proteins in healthy controls, benign pulmonary conditions, and lung cancer patients.
iFOT of five urinary proteins in the CTL (n = 33), pneumonia (n = 23), COPD (n = 17), and LC (n = 33) groups. Abbreviations: CTL, healthy controls; COPD, Chronic Obstructive Pulmonary Disease; LC, lung cancer.
To investigate if these proteins can separate lung cancers from benign lung diseases, we assayed 40 urine samples from patients who were diagnosed with either pneumonia (n = 23) or COPD (n = 17). The biomarker panel recognized 32 lung cancer cases with high sensitivity but with medium specificity (Table 2, Fig. 3b). Further inspection of their abundances indicated that some proteins were mildly elevated in pneumonia or COPD, suggesting that their correlation to inflammation may account for reduced specificity (Fig. 4).
3.4. The Biomarker Panel in Differentiating Lung Cancer from Other Cancers
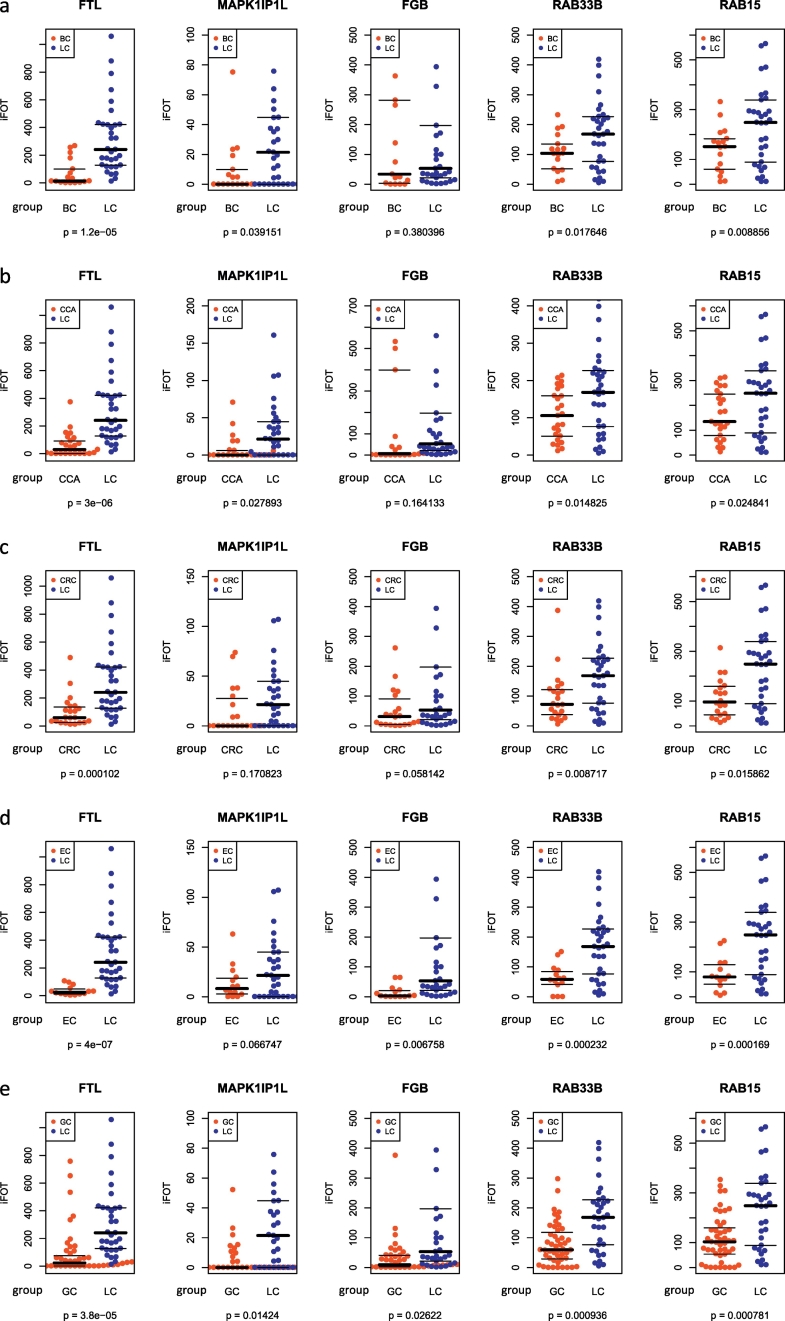
Only very few studies have evaluated the specificity of the biomarkers in classifying lung cancer against other diseases. One study conducted by Nolen et al. found that the disease selectivity of their biomarkers was moderate or poor in discriminating lung cancer against breast cancer and prostate cancer (Nolen et al., 2015). To assess the cancer specificity, we tested our model and five proteins in predicting lung cancer against other five common cancers in the remaining data sets (test set 3–7, Fig. 1C). The predictive model classified lung cancer cases with 18%, 13.8%, 20%, 6.4% and 12.5% classification errors in predicting LCs against BCs, CCAs, CRCs, ECs, and GCs, respectively (Table 2). The model was able to discriminate the LCs from other cancers with great sensitivity in all test sets (>93%) and high specificity in three test sets (CCA vs LC: 72%; EC vs LC: 85.71%; GC vs LC: 80.85%). A single marker, FTL, could distinguish LCs from other cancers with AUCs >0.81 in 5 test sets (Fig. 3c-g). The logistic model in which five protein markers were combined achieved a highest value of AUC in these data sets (Fig. 3c-g).
We further performed case-by-case comparison to examine whether the levels of these proteins were differentially expressed in six cancer groups. As shown in Fig. 5, FTL is the most significant marker, which differentiated LCs from all other cancers with the smallest p values (Student's t-test) comparing with other four markers; RAB33B and RAB15 were also significantly over-expressed in the LC group across all test sets (p < 0.05), while MAPK1IP1L and FGB exhibited significant difference in some test sets. These results indicate that the biomarker panel is tumor-specific when it predicts lung cancer patients against healthy controls with good sensitivity and specificity. It is worth mentioning that the disease specificity may not be evaluated precisely since comparisons were made between all LCs (n = 33) and one of other cancers thus were not completely in an independent manner although the RF model during training had no prior knowledge of disease specificity towards other cancers since no other cancer cases were included in the training set.
Fig. 5.
Relative abundances of five proteins in lung cancer and other cancers.
Comparison of the levels of the individual markers between (A) BC and LC, (B) CCA and LC, (C) CRC and LC, (D) EC and LC, and (E) GC and LC. Statistical analysis was performed by Student's t-test. Abbreviations: LC, lung cancer; BC, bladder cancer; CCA, cervical cancer; CRC, colorectal cancer; EC, esophageal cancer; GC, gastric cancer.
3.5. Other Urinary Proteins Highly Correlated with Panel Biomarkers
The predictive model has eliminated the redundancy in the step of feature selection in order to keep the model simple and efficient for the prediction purpose. For this reason, we did correlation analysis to recover other variables that were highly associated with the proteins in the panel (Supplementary Fig. S6). Five proteins were found to be closely related to some of these markers with a minimum of a Pearson correlation coefficient, r ≥ 0.7. RAB14 (Member RAS Oncogene Family), together with RAB15 and RAB33B are Rab GTPases, a family of small GTPases which mainly functions in controlling intracellular membrane trafficking (Hutagalung and Novick, 2011, Zhen and Stenmark, 2015, Stenmark, 2009). The high correlation of FGG (Fibrinogen Gamma Chain), and FGA (Fibrinogen Alpha Chain) with FGB was expected as they are components of fibrinogen, a glycoprotein that is essential for blood clot formation (Mosesson, 2005). FTH1 (Ferritin Heavy Chain 1) and FTL are the subunits of the ferritin protein (Wang et al., 2010). Ferritin as a biomarker for lung cancer diagnosis has been investigated in an earlier study (Li et al., 2012). ATP6V1E1 (ATPase H+ Transporting V1 Subunit E1) is highly correlated with MAPK1IP1L. Proteins as functional molecules in the cell are usually interconnected; in this respect, Rab GTPases, fibrinogen, and ferritin are the three major upregulated protein families found in this study. However, the functional connection of these protein families to lung cancer remains unknown.
4. Discussion
Lung cancer as a devastating disease continues to be a main health challenge worldwide. Although much effort towards cancer diagnostics and treatment has been made, lung cancer mortality has not been significantly improved over the past several decades (Torre et al., 2016b). Patients who are diagnosed at the late stage of the disease often face very limited treatment options and poor prognosis (Scheff and Schneider, 2013). Imaging technology, CT for instance, has demonstrated high sensitivity for lung cancer screening but also suffers from low specificity (Aberle et al., 2013). Furthermore, due to the high cost and demand for technical expertise, CT for lung cancer screening is only limited to those who live in developed countries, and covers only a small population who are high risk individuals such as smokers (Bach et al., 2007, Aberle et al., 2013). Therefore, searching and developing low-cost, reliable biomarkers for lung cancer screening in a large population is highly desirable.
Blood and urine are two frequently researched biomatrices for discovery of biomarkers of human diseases as both can be sampled frequently and non-invasively. Urine as body fluid, however, has several advantages over blood: 1) it can be easily obtained in large volumes; and 2) urinary proteome is less complex and has a relatively lower dynamic range, thus those low abundant but functionally important proteins such as exosomal proteins can be reliably measured by LC-MS/MS (Jakobsen et al., 2015, Hoorn et al., 2005, Barratt and Topham, 2007). For those reasons, we have assayed >300 human urine samples and identified over 7000 proteins in healthy donors, benign pulmonary conditions, and six common cancers. We have validated that urine profiling has diagnostic features for lung cancer screening and nominated a list of candidate markers for future validation, providing a rich resource for urinary biomarker studies.
As single biomarker may hardly achieve satisfactory discriminating power due to the tumor heterogeneity, seeking multiple biomarkers and developing a combinatorial model for cancer detection is hence a desirable strategy as demonstrated by some earlier studies (Patz et al., 2007, Radon et al., 2015). By virtue of the advanced analytical instruments and statistical algorithms, we have revealed a list of candidate urinary markers and selected five of them to build a predictive model for lung cancer diagnosis. With this model, we are able to identify majority of lung cancer patients from control cases with great sensitivity and specificity. The random forest model has achieved low classification errors both on oob samples and independent samples. The individual markers can separate different cases with good AUCs and the combinatorial panel has resulted in a higher AUC value than any single markers in different data sets. More importantly, these markers when combined present a tumor-specific profile in discriminating lung cancer against other cancers although the individual proteins may have a limited discriminating power. It is worth noting that two proteins (RAB 15 and RAB33B) on the panel have an exosomal origin (Yoshioka et al., 2013, Bobrie et al., 2012, Greening et al., 2015). The elevation of the RAB proteins in lung cancer may be associated with tumorigenesis and thus may account for the tumor specificity (Tzeng and Wang, 2016, Zhen and Stenmark, 2015). In summary, the panel marker we found in this study could benefit a large population and be applied to clinical diagnostics of NSCLC for general purpose in the future after a validation trial with expanded sample numbers in a multi-center setting.
It is worth noting that several other proteins that are not included in our panel should also be placed on the candidate list, some of which has already been investigated including C9 and Ferritin (Li et al., 2012, Kim et al., 2016, Ahn and Cho, 2013, Nolen et al., 2015). Among all these candidates, RAB14, RAB15 and RAB33B belong to the family of small GTPases; FGG, FGA, and FGB are subunits of fibrinogen; while FTH1 and FTL come from ferritin protein.
While the biomarker panel and the predictive model is powerful in discriminating lung cancer against control cases and other cancers, the current research should be further expanded onto a larger population with more clinical profiles including age, smoking status, subtypes, disease stage, and race that were not or not fully explored in this study. On the other hand, decision made based upon these proteins should be treated with caution when applied to clinical screening, since some of the proteins may be originated from inflammation as demonstrated by their limited specificity in discriminating NSCLC patients from benign controls and several other cancers. The potential clinical usefulness of the biomarker panel should be combined with routine image screening tests to rule out the false positives. Meanwhile, developing a simple model with clear cut-off values on these proteins is also highly desirable since the random forest model is a tree-based ensemble method. The cut-off values of these proteins often vary in trees and forests. We are well aware of the limited number of lung cancer cases in the test set at this moment and the lack of an independent validation set, which is necessary for future study and requires a significantly increased number of sample sizes and thus much more efforts beyond this study that is at a relatively early stage. Further development and validation by independent, routine techniques that are more operationally feasible also seems indispensable for clinic uses in future.
The following are the supplementary data related to this article.
Characterization of the human urinary proteome. (a) Urine proteomics workflow, (b) Cellular component categories of urinary proteome, and (c) Relative abundance of some well-studied exosomal markers.
Ward's hierarchical clustering analysis of (a) all proteins, (b) 280 most variable proteins, and (c) 588 differentially expressed proteins. Abbreviations: CTL, healthy controls; LC, lung cancer; BC, bladder cancer; CCA, cervical cancer; CRC, colorectal cancer; EC, esophageal cancer; GC, gastric cancer. Note: most variable proteins are those who have a CV (coefficient of variation) >200% and express in at least 50% of all cases.
Flow chart of model development and variable selection. Random forest model implemented with feature selection method was used to select variables and build a predictive model upon the training set. About 2/3 of cases in the training set were randomly sampled to grow decision trees by bootstrapping and the remaining samples (~1/3) were left out as oob (out of bag) samples for evaluation of model performance. Parameters used in the model building: p, number of variables in the training set = 68; mtry, number of variables randomly selected at every split = square root of p; ntree, number of trees to grow in one forest = 1001; nforest, total number of forests to grow = 1′000.
AUCs of top 15 most important urinary proteins in classifying lung cancer patients and healthy individuals. AUCs of top 15 candidates in separating the healthy controls and lung cancers in the training set, test set, or when they were combined. Abbreviation: AUC, area under the ROC curve.
Multidimensional scaling plot of two groups with (a) all variables or (b) five variables selected in the predictive model. Abbreviations: CTL, healthy controls; LC, lung cancer.
Proteins highly correlated to five markers in the predictive model. Five proteins (RAB14, FGG, FGA, FTH1 and ATP6V1E1) that have a Pearson correlation coefficient, r ≥ 0.7 to one of five markers are shown.
Metadata of urine samples used in this study.
Sheet 1: Relative abundances (iFOT) of 7408 proteins in all groups; Sheet 2: iFOT of 588 differentially expressed proteins in all groups; Sheet 3: iFOT of 68 proteins in all groups. Abbreviations: CTL, healthy controls; LC, lung cancer; BC, bladder cancer; CCA, cervical cancer; CRC, colorectal cancer; EC, esophageal cancer; GC, gastric cancer; COPD: Chronic Obstructive Pulmonary Disease.
Acknowledgments
Acknowledgments
We are grateful to all participating patients in donating urine specimen and the hospitals in collecting samples and patients' health information.
Funding Sources
This project was supported by an internal research grant from the Joint Center for Translational Medicine between Beijing Proteome Research Center and Tianjin Baodi Hospital, and an International Collaboration Grant 2014DFB30010 from the Ministry of Science and Technology of China. The funders had no roles in study design, data collection, data analysis and interpretation, or writing the manuscript.
Disclosure
The authors declare no conflicts of interests.
Authors' Contributions
Conception and design: C Zhang, Y Wang, G Wang, B Zhen, J Qin.
Acquisition of data: C Zhang, W Leng, C Sun, T Lu, Z Chen, X Men.
Data analysis and interpretation: C Zhang.
Manuscript writing: C Zhang, W Leng, Y Wang, J Qin.
Final approval of manuscript: All authors.
Contributor Information
Guangshun Wang, Email: wgsTMUBH@163.com.
Bei Zhen, Email: zp1963@sina.com.
Jun Qin, Email: jqin1965@126.com.
References
- Aberle D.R., Abtin F., Brown K. Computed tomography screening for lung cancer: has it finally arrived? Implications of the national lung screening trial. J. Clin. Oncol. 2013;31:1002–1008. doi: 10.1200/JCO.2012.43.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J.-M., Cho J.-Y. Current serum lung cancer biomarkers. J. Mol. Biomark. Diagn. 2013;S4:001. [Google Scholar]
- Bach P.B., Jett J.R., Pastorino U., Tockman M.S., Swensen S.J., Begg C.B. Computed tomography screening and lung cancer outcomes. JAMA. 2007;297:953–961. doi: 10.1001/jama.297.9.953. [DOI] [PubMed] [Google Scholar]
- Barratt J., Topham P. Urine proteomics: the present and future of measuring urinary protein components in disease. CMAJ. 2007;177:361–368. doi: 10.1503/cmaj.061590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrie A., Colombo M., Krumeich S., Raposo G., Thery C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles. 2012:1. doi: 10.3402/jev.v1i0.18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Genuer R., Poggi J.M., Tuleau-Malot C. Variable selection using random forests. Pattern Recogn. Lett. 2010;31:2225–2236. [Google Scholar]
- Greening D.W., Xu R., Ji H., Tauro B.J., Simpson R.J. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- Harmsma M., Schutte B., Ramaekers F.C. Serum markers in small cell lung cancer: opportunities for improvement. Biochim. Biophys. Acta. 2013;1836:255–272. doi: 10.1016/j.bbcan.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Hoorn E.J., Pisitkun T., Zietse R., Gross P., Frokiaer J., Wang N.S., Gonzales P.A., Star R.A., Knepper M.A. Prospects for urinary proteomics: exosomes as a source of urinary biomarkers. Nephrol. (Carlton) 2005;10:283–290. doi: 10.1111/j.1440-1797.2005.00387.x. [DOI] [PubMed] [Google Scholar]
- Hutagalung A.H., Novick P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen K.R., Paulsen B.S., Baek R., Varming K., Sorensen B.S., Jorgensen M.M. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J. Extracell Vesicles. 2015;4:26659. doi: 10.3402/jev.v4.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.I., Ahn J.M., Sung H.J., Na S.S., Hwang J., Kim Y., Cho J.Y. Meta-markers for the differential diagnosis of lung cancer and lung disease. J. Proteome. 2016;148:36–43. doi: 10.1016/j.jprot.2016.04.052. [DOI] [PubMed] [Google Scholar]
- Leng W., Ni X., Sun C., Lu T., Malovannaya A., Jung S.Y., Huang Y., Qiu Y., Sun G., Holt M.V., Ding C., Sun W., Men X., Shi T., Zhu W., Wang Y., He F., Zhen B., Wang G., Qin J. Proof-of-concept workflow for establishing reference intervals of human urine proteome for monitoring physiological and pathological changes. EBioMedicine. 2017;18:300–310. doi: 10.1016/j.ebiom.2017.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Asmitananda T., Gao L., Gai D., Song Z., Zhang Y., Ren H., Yang T., Chen T., Chen M. Biomarkers in the lung cancer diagnosis: a clinical perspective. Neoplasma. 2012;59:500–507. doi: 10.4149/neo_2012_064. [DOI] [PubMed] [Google Scholar]
- Liu Q., Ding C., Liu W., Song L., Liu M., Qi L., Fu T., Malovannaya A., Wang Y., Qin J., Zhen B. In-depth proteomic characterization of endogenous nuclear receptors in mouse liver. Mol. Cell. Proteomics. 2013;12:473–484. doi: 10.1074/mcp.M112.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.D., Siegel R.L., Lin C.C., Mariotto A.B., Kramer J.L., Rowland J.H., Stein K.D., Alteri R., Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- Mosesson M.W. Fibrinogen and fibrin structure and functions. J. Thromb. Haemost. 2005;3:1894–1904. doi: 10.1111/j.1538-7836.2005.01365.x. [DOI] [PubMed] [Google Scholar]
- Nolen B.M., Lomakin A., Marrangoni A., Velikokhatnaya L., Prosser D., Lokshin A.E. Urinary protein biomarkers in the early detection of lung cancer. Cancer Prev. Res. (Phila.) 2015;8:111–119. doi: 10.1158/1940-6207.CAPR-14-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patz E.F., Campa M.J., Gottlin E.B., Kusmartseva I., Guan X.R., Herndon J.E., II Panel of serum biomarkers for the diagnosis of lung cancer. J. Clin. Oncol. 2007;25:5578–5583. doi: 10.1200/JCO.2007.13.5392. [DOI] [PubMed] [Google Scholar]
- Pisitkun T., Johnstone R., Knepper M.A. Discovery of urinary biomarkers. Mol. Cell. Proteomics. 2006;5:1760–1771. doi: 10.1074/mcp.R600004-MCP200. [DOI] [PubMed] [Google Scholar]
- Radon T.P., Massat N.J., Jones R., Alrawashdeh W., Dumartin L., Ennis D., Duffy S.W., Kocher H.M., Pereira S.P., Guarner Posthumous L., Murta-Nascimento C., Real F.X., Malats N., Neoptolemos J., Costello E., Greenhalf W., Lemoine N.R., Crnogorac-jurcevic T. Identification of a three-biomarker panel in urine for early detection of pancreatic adenocarcinoma. Clin. Cancer Res. 2015;21:3512–3521. doi: 10.1158/1078-0432.CCR-14-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondo F., Morosi L., Corbetta S., Chinello C., Brambilla P., Della Mina P., Villa A., Albo G., Battaglia C., Bosari S., Magni F., Pitto M. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol. BioSyst. 2013;9:1220–1233. doi: 10.1039/c3mb25582d. [DOI] [PubMed] [Google Scholar]
- Scheff R.J., Schneider B.J. Non-small-cell lung cancer: treatment of late stage disease: chemotherapeutics and new frontiers. Semin. Intervent Radiol. 2013;30:191–198. doi: 10.1055/s-0033-1342961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stewart B.W.W., Chris, International agency for research on cancer & world health organization . International Agency for Research on Cancer WHO Press; Lyon, France, Geneva, Switzerland: 2014. World Cancer Report; p. 2014. [Google Scholar]
- Torre L.A., Siegel R.L., Jemal A. Lung cancer statistics. Adv. Exp. Med. Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- Torre L.A., Siegel R.L., Ward E.M., Jemal A. Global Cancer incidence and mortality rates and trends—an update. Cancer Epidemiol. Biomark. Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- Troyanskaya O., Cantor M., Sherlock G., Brown P., Hastie T., Tibshirani R., Botstein D., Altman R.B. Missing value estimation methods for DNA microarrays. Bioinforma. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Tzeng H.T., Wang Y.C. Rab-mediated vesicle trafficking in cancer. J. Biomed. Sci. 2016;23:70. doi: 10.1186/s12929-016-0287-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Knovich M.A., Coffman L.G., Torti F.M., Torti S.V. Serum ferritin: past, present and future. Biochim. Biophys. Acta. 2010;1800:760–769. doi: 10.1016/j.bbagen.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Vasaikar S., Shi Z., Greer M., Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz P.B., Shyr Y., Rahman J.S., Wardwell N.R., Zimmerman L.J., Shakhtour B., Gray W.H., Chen S., Li M., Roder H., Liebler D.C., Bigbee W.L., Siegfried J.M., Weissfeld J.L., Gonzalez A.L., Ninan M., Johnson D.H., Carbone D.P., Caprioli R.M., Massion P.P. Diagnostic accuracy of MALDI mass spectrometric analysis of unfractionated serum in lung cancer. J. Thorac. Oncol. 2007;2:893–901. doi: 10.1097/JTO.0b013e31814b8be7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka Y., Konishi Y., Kosaka N., Katsuda T., Kato T., Ochiya T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Chan D.W. Cancer proteomics: in pursuit of “true” biomarker discovery. Cancer Epidemiol. Biomark. Prev. 2005;14:2283–2286. doi: 10.1158/1055-9965.EPI-05-0774. [DOI] [PubMed] [Google Scholar]
- Zhang H., Cao J., Li L., Liu Y., Zhao H., Li N., Li B., Zhang A., Huang H., Chen S., Dong M., Yu L., Zhang J., Chen L. Identification of urine protein biomarkers with the potential for early detection of lung cancer. Sci. Rep. 2015;5:11805. doi: 10.1038/srep11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y., Stenmark H. Cellular functions of Rab GTPases at a glance. J. Cell Sci. 2015;128:3171–3176. doi: 10.1242/jcs.166074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of the human urinary proteome. (a) Urine proteomics workflow, (b) Cellular component categories of urinary proteome, and (c) Relative abundance of some well-studied exosomal markers.
Ward's hierarchical clustering analysis of (a) all proteins, (b) 280 most variable proteins, and (c) 588 differentially expressed proteins. Abbreviations: CTL, healthy controls; LC, lung cancer; BC, bladder cancer; CCA, cervical cancer; CRC, colorectal cancer; EC, esophageal cancer; GC, gastric cancer. Note: most variable proteins are those who have a CV (coefficient of variation) >200% and express in at least 50% of all cases.
Flow chart of model development and variable selection. Random forest model implemented with feature selection method was used to select variables and build a predictive model upon the training set. About 2/3 of cases in the training set were randomly sampled to grow decision trees by bootstrapping and the remaining samples (~1/3) were left out as oob (out of bag) samples for evaluation of model performance. Parameters used in the model building: p, number of variables in the training set = 68; mtry, number of variables randomly selected at every split = square root of p; ntree, number of trees to grow in one forest = 1001; nforest, total number of forests to grow = 1′000.
AUCs of top 15 most important urinary proteins in classifying lung cancer patients and healthy individuals. AUCs of top 15 candidates in separating the healthy controls and lung cancers in the training set, test set, or when they were combined. Abbreviation: AUC, area under the ROC curve.
Multidimensional scaling plot of two groups with (a) all variables or (b) five variables selected in the predictive model. Abbreviations: CTL, healthy controls; LC, lung cancer.
Proteins highly correlated to five markers in the predictive model. Five proteins (RAB14, FGG, FGA, FTH1 and ATP6V1E1) that have a Pearson correlation coefficient, r ≥ 0.7 to one of five markers are shown.
Metadata of urine samples used in this study.
Sheet 1: Relative abundances (iFOT) of 7408 proteins in all groups; Sheet 2: iFOT of 588 differentially expressed proteins in all groups; Sheet 3: iFOT of 68 proteins in all groups. Abbreviations: CTL, healthy controls; LC, lung cancer; BC, bladder cancer; CCA, cervical cancer; CRC, colorectal cancer; EC, esophageal cancer; GC, gastric cancer; COPD: Chronic Obstructive Pulmonary Disease.