Abstract
Long non-coding RNAs (lncRNAs) belong to a recently discovered class of molecules proposed to regulate various cellular processes. Here, we systematically analyzed their expression in human subcutaneous white adipose tissue (WAT) and found that a limited set was differentially expressed in obesity and/or the insulin resistant state. Two lncRNAs herein termed adipocyte-specific metabolic related lncRNAs, ASMER-1 and ASMER-2 were enriched in adipocytes and regulated by both obesity and insulin resistance. Knockdown of either ASMER-1 or ASMER-2 by antisense oligonucleotides in in vitro differentiated human adipocytes revealed that both genes regulated adipogenesis, lipid mobilization and adiponectin secretion. The observed effects could be attributed to crosstalk between ASMERs and genes within the master regulatory pathways for adipocyte function including PPARG and INSR. Altogether, our data demonstrate that lncRNAs are modulators of the metabolic and secretory functions in human fat cells and provide an emerging link between WAT and common metabolic conditions.
Keywords: White adipose tissue, Adipocytes, Long non-coding RNAs, Metabolic traits, Lipolysis, Adiponectin
Highlights
-
•
Systematic analysis of lncRNA expression in human subcutaneous white adipose tissue.
-
•
Identification of adipocyte-enriched lncRNAs correlating with metabolic disorders.
-
•
Loss-of-function studies revealed the roles of two lncRNAs in regulating adipogenesis and function of fat cells.
Long non-coding RNAs (LncRNAs) have emerged as important factors that impact gene expression. Recent studies have suggested significant regulatory roles of lncRNAs in fat cells. However, their clinical relevance and their mechanism of action in human fat cells are still not well defined. We employed a translational research approach to identify a group of fat cell-specific lncRNAs linked to obesity and/or insulin resistance and were able to demonstrate their complex effects on fat cell function. One identified lncRNA may constitute a therapeutic target for future metabolic disorders directed against fat cell lipolysis.
1. Introduction
Long non-coding RNAs (lncRNAs) are defined as RNA transcripts longer than 200 nucleotides that do not code a polypeptide. Some of these molecules regulate various biological processes including gene imprinting, chromatin alteration and the allosteric modification of enzyme activity (Ponting et al., 2009; Quinn and Chang, 2016; Rinn and Chang, 2012). While some lncRNAs have primarily been implicated in cancer and neurodegenerative disorders (Batista and Chang, 2013; Esteller, 2011), recent studies, mostly in rodent models, also suggest the potential involvement of lncRNAs in metabolic diseases (reviewed in (Losko et al., 2016)). Thus, alterations in the nutritional status of mice (i.e. fasting, refeeding and overfeeding), result in altered expression of several lncRNAs in tissues relevant for metabolism, i.e. liver, white adipose tissue (WAT) and skeletal muscle (Yang et al., 2016), but only effects in liver were investigated in detail. LncRNAs may also control the function of pancreatic β cells of mice and humans (Akerman et al., 2017; Losko et al., 2016). However, much less is known about the potential roles of lncRNAs in modulating the function of WAT where studies have primarily focused on their effects on in vitro differentiation of adipose precursor cells (i.e. adipogenesis) (Wei et al., 2016). During human adipocyte differentiation the expression of lncRNAs co-cluster with the expression of coding genes and regulatory factors such as microRNAs and enhancers (Ehrlund et al., 2017), indicating that lncRNAs may have potentially critical functional roles in adipocytes. Furthermore, a recent genome-wide association study on cardiometabolic traits demonstrated that several WAT-expressed lncRNAs were associated with single nucleotide polymorphisms linked to cardiometabolic disease (Ballantyne et al., 2016). These studies suggest that WAT-expressed lncRNAs may affect other processes in fat cells besides adipogenesis.
WAT plays a pivotal role in metabolic disease not only based on its energy storage/release capacity but also because of its endocrine function, as it secretes numerous proteins termed adipokines (Kershaw and Flier, 2004; Sethi and Vidal-Puig, 2007). The metabolic and endocrine functions of WAT may impact the regulation of several other organs such as brain, liver and skeletal muscle (Kershaw and Flier, 2004; Sethi and Vidal-Puig, 2007). Alterations in WAT function are well-documented in obesity and insulin resistance (Guilherme et al., 2008). Moreover, WAT from different body regions is linked explicitly to metabolic disease, with visceral WAT being more pernicious than the subcutaneous depot (Wajchenberg, 2000). Nevertheless, regional differences within subcutaneous WAT could also be of importance as several investigators have shown that the abdominal region is positively, and the gluteofemoral region is negatively associated with cardiometabolic disease (Snijder et al., 2003; Yusuf et al., 2005). While the role of lncRNAs in explaining these differences are not known, it was recently reported that the lncRNA HOTAIR, with potent positive effects on adipogenesis, was expressed in gluteal but not in abdominal subcutaneous WAT (Divoux et al., 2014).
In order to assess the potential roles of lncRNAs in human WAT, we presently investigated their expression in human WAT and association with obesity and insulin resistance (Fig. 1). By combining RNA sequencing and gene microarray analyses of WAT from two clinical cohorts of obese and non-obese individuals with significant between subject variations in insulin sensitivity, we identified a set of potentially relevant lncRNAs. To identify lncRNAs that were potentially important for fat cell function we compared the expression of each gene in isolated fat cells and the stromal vascular fraction (SVF) from the same WAT biopsy using Real-time PCR. Key findings were then confirmed by probing previously generated 5′ Cap Analysis of Gene Expression (CAGE) data from human adipocyte precursor cells during differentiation from our previous study (Ehrlund et al., 2017). Finally, the functional role of the adipocyte-specific lncRNAs displaying the most prominent clinical associations was investigated in differentiated fat cells following knockdown with antisense oligonucleotides where we focused on lipid mobilization (lipolysis) and endocrine function (adiponectin release).
Fig. 1.
Flowchart represents the strategy used for gene expression analysis. These initial data used in the analysis were obtained from two well-characterized clinical cohorts; Affymetrix microarray data from visceral and subcutaneous adipose tissue of 80 obese individuals with or without insulin resistance and adipose RNA-sequencing data from 15 lean and 13 obese subjects. Identified candidate long non-coding RNAs (lncRNAs) for further studies. This involved global expression during human in vitro adipogenesis determined by single molecule RNA-seq using 5′-Cap analysis gene expression (CAGE) and expression analysis in different cellular fractions of adipose tissue using quantitative Real-time PCR. hADSCs = human adipose tissue-derived stem cells; WAT = white adipose tissue; SVF = stromal vascular fraction of adipose tissue.
2. Materials and Methods
2.1. Cohorts
The study included two cohorts consisting of 108 women who were recruited from the general adult population in the Stockholm (Sweden) area (Supplementary Table 1). Cohort 1 comprised 15 lean and 13 obese women who were matched for age. Cohort 2 consisted 80 obese age-matched women with or without insulin resistance and has been described in detail before (Arner et al., 2016). Obesity was defined as body mass index (BMI) > 30 kg/m2. The study was approved by the regional ethics board, and written informed consent was obtained from each subject. Subjects in cohort 1 were investigated in the morning after an overnight fast when abdominal subcutaneous WAT was obtained by fine-needle aspiration (Kolaczynski et al., 1994). One part of WAT was frozen and stored at −70 °C for subsequent RNA extraction. The remaining tissue was subjected to collagenase treatment and isolated fat cells were used to measure lipolysis and lipogenesis as described in detail (Lofgren et al., 2005). The basal rates as well as rate stimulated by isoprenaline, a synthetic catecholamine (lipolysis), and insulin (lipogenesis) were measured. In cohort 2, abdominal subcutaneous and visceral (from greater omentum) WAT was obtained at the beginning of bariatric surgery. A venous blood sample was collected alongside for clinical chemistry measures as described (Arner et al., 2016). For comparisons between different fractions of the WAT, Samples of abdominal subcutaneous adipose tissue (about 100–200 g) were obtained from cosmetic liposuction of 11 healthy female subjects. The mean and (range) of age and BMI were 38 (20–50) years and 26 (22–29) kg/m2, respectively.
2.2. Mapping of Affymetrix Probesets to the Annotation of FANTOM CAGE Associated Transcriptome (FANTOM-CAT)
To compare our previously published microarray data (GSE101492) (Arner et al., 2016) with the RNA-seq data (generated from this study) on the same set of gene models, we remapped the Affymetrix HTA 2.0 probe sets onto the FANTOM-CAT gene models (robust level, n = 53,220 genes) (Hon et al., 2017). Briefly, coordinates of the transcript exons of each FANTOM-CAT gene on hg19 were obtained from http://fantom.gsc.riken.jp/cat/. Coordinates of all probes of Affymetrix GeneChip™ Human Transcriptome Array (HTA) 2.0 (mapped to hg19) were obtained from manufacture's website (http://www.affymetrix.com/support/technical/byproduct.affx?product=human_transcriptome). The coordinates of FANTOM-CAT exons and Affymetrix HTA 2.0 probes were intersected using bedtools (Quinlan and Hall, 2010). A FANTOM-CAT gene is defined as successfully mapped to an Affymetrix probeset when ≥50% of the probes within the probe set intersect with the exons of the transcripts of the corresponding FANTOM-CAT gene. Of the 65,967 probe sets, 38,213 and 6133 of them can be successfully mapped to single and multiple FANTOM-CAT genes, respectively. On the other side of the 53,220 FANTOM-CAT genes, 16,620 and 9263 of them can be successfully mapped to single and multiple Affymetrix probesets, respectively. Total number of the probes, number of mapped probes and number of associated FANTOM-CAT IDs can be found in Supplementary Table 2.
2.3. RNA Sequencing Analysis (RNA-Sequencing)
Total RNA was extracted from samples of cohort 1 using the RNeasy Lipid Tissue Mini Kit (74,804, Qiagen, Hilden, Germany) as per manufacturer's instructions. RNA concentration and purity were measured using a using Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Lafayette, USA). The quality of the extracted RNA samples was investigated using the Agilent Bioanalyzer (Agilent, Santa Clara, CA) and all RNA samples submitted for sequencing had an RNA Integrity Number (RIN) above 8. RNA libraries for sequencing were prepared using TruSeq RNA kits (Illumina, CA, USA) according to the manufacturer's instructions with the following changes. The protocols were automated using an MBS 1200 pipetting station (Nordiag AB, Sweden). All purification steps and gel-cuts were replaced by the magnetic bead clean-up methods as previously described (Borgstrom et al., 2011). The samples were sequenced on an Illumina HiSeq 2000 as paired-end reads to 100 bp.
To obtain the abundance estimation for each gene, the transcript abundance quantification was first conducted using the ultrafast quasi-alignment tool Kallisto (Bray et al., 2016), which pseudo-aligned the sequencing reads from each sample to the assembly based on the FANTOM-CAT. Then the gene-level estimates, representing the overall transcriptional output of each gene, were obtained by summing the corresponding transcript-level estimates using a Bioconductor package, Tximport (Soneson et al., 2015). Differential gene expression analysis was conducted at the gene level using EdgeR, after applying the filter for at least half of the samples above the detection level (cpm > 1), selecting for genes with a false discovery rate (FDR) of <0.05 (Robinson et al., 2010). Identified genes were interrogated for their functional classes and importance in biology using the pathway analysis with the Ingenuity Pathway Analysis tool (IPA, Ingenuity Systems, Inc., Redwood City, CA, USA). Canonical pathways with a p-value (corrected using the Benjamini-Hochberg method) <0.01 (expected FDR < 1%) were significantly enriched for differentially expressed genes.
2.4. Weighted Gene Co-Expression Network Analysis
Weighted gene co-expression network analysis (WGCNA) was performed on the 17,000 filtered genes from the RNA-seq data of obese and lean subjects using a R package (Langfelder and Horvath, 2008). The automatic one-step network construction and module detection method with default settings were used, which include an unsigned type of topological overlap matrix (TOM), a power β of 6, a minimal module size of 30, and a branch merge cut height of 0.25. All modules were represented by a colour. The module eigengene was used to represent each module, which was calculated by the first principal component. Using the module eigengene, the Module-Trait relationships were estimated by calculating the Pearson's correlations between the module eigengene and the clinical traits included in the analysis. Those Module-Trait relationships were used to select potential biologically interesting modules for downstream analysis.
2.5. Transcriptome Analysis with Affymetrix Microarray
Gene expression profiling in cohort 2 was performed using GeneChip® Human Transcriptome Array (HTA)-2.0 and has been published previously (GSE101492) (Arner et al., 2016). For the current study, the raw data were analyzed with packages available from Bioconductor (http://www.bioconductor.org). Normalization and calculation of gene expression were performed with the robust multichip average expression measure using the oligo package (Carvalho and Irizarry, 2010). Before further analysis, collapseRows R function (Miller et al., 2011) was used to convert and collapse the transcript abundance quantification detected by Affymetrix probesets to mapped FANTOM-CAT IDs. Differential gene expression analysis was conducted at the gene level using Limma, selecting for genes with a FDR < 0.05 (Smyth, 2004).
2.6. Cell Culture
Human adipose-derived stromal cells (hADSCs) were isolated from subcutaneous abdominal WAT from a male donor (16 years old, BMI 24 kg/m2). hADSCs were cultured and differentiated into adipocytes using described protocols (Gao et al., 2017).
2.7. LncRNA Knockdown
In vitro differentiated hADSCs (day 0 or day 8 post-induction) were transfected with LNA™ GapmeR antisense oligonucleotides (Exiqon, Vedbaek, Denmark) using a Neon™ transfection system (MPK5000, Invitrogen, Göteborg, Sweden) as per manufacturer's instructions. Target sequences for each antisense oligonucleotides are listed below; Anti-ASMER1_1: AGAGTTGCAGTCCACA. Anti-ASMER1_2: TACGGGCTAAAAGCTA. Anti-ASMER2_1: TTACACGAAGCCTTTG. Anti-ASMER2_2: TTTGATCCACTTTGCC. Negative control A was used as Anti-Control. Transfection took place using 2 pulses of 20 ms 1300 V pulses using the Neon™ 100 μl kit (MPK10096, Invitrogen) after which cells were plated at a seeding density of 1 × 105 cells/well for 24-well plates (mRNA and lipolysis analysis) and 5 × 104 cells/well for 48-well plates (adiponectin release analysis).
2.8. RNA Expression Analysis
Total RNA was extracted from samples as described above. cDNA synthesis followed using the iScript™ cDNA Synthesis Kit (1708891, Bio-Rad, Sundbyberg, Sweden) as per manufacturer's instructions before SYBR green qRT-PCR analysis on an iCycler IQ (Bio-Rad). More information about all probes used can be obtained on request. Relative gene expression was calculated using the comparative ∆Ct method with the selected internal controls. 18S was the internal control for analysis of cell type specific expression and expression during adipogensis. B2M was used for other presented experiments.
2.9. Rate of Lipolysis
Lipolysis measurements were made using the Free Glycerol Determination Kit (FG0100, Sigma-Aldrich, Stockholm, SWE) as per manufacturer's instructions. Briefly, media was collected from mature adipocytes on day 13 post-differentiation. Addition of the Amplex® Ultra Red (A36006, Invitrogen) was first made to the Free Glycerol reagent before incubation with adipocyte medium for 5 min at 37 °C in a 110 μl reaction. Glycerol release into the media was quantified on an Infinite M200 microplate reader (Tecan, Männedorf, CHE) at a wavelength of 590 nm. A glycerol standard curve run alongside each assay was used to determine final glycerol concentration. Glycerol is an endpoint of hydrolysis (lipolysis) of the adipocyte triglycerides.
2.10. Adiponectin Release
Adiponectin release into the cultured cell media was determined using an ELISA (10-1193-01, Mercodia, Uppsala, Sweden) measuring all forms of the protein as per manufacturer's instructions. Briefly, media was collected from mature adipocytes on day 13 post-differentiation. 25 μl of media was added to 100 μl of assay buffer, added to the α-human adiponectin coated plate and shaken for 1 h at room temperature. After appropriate wash steps, 100 μl of the HRP-conjugated α-human adiponectin antibody was added and stirred for 1 h at room temperature. Amplification of the adiponectin signal was achieved through the addition of 200 μl of Substrate TMB for 15 min before the reaction was stopped and optical density read at 450 nm on an Infinite M200 microplate reader. An adiponectin standard curve run alongside each assay was used to determine final adiponectin concentration.
2.11. Insulin-Stimulated Lipogenesis
The cells in 48 well-plate were first washed one time with DMEM without glucose (Biochrom, Berlin, Germany) and incubated in insulin-free DMEM (Biochrom) supplemented with 1 μmol glucose for 3 h. Following the starvation, the cells were incubated for 2 h in the presence or absence of 10–7 mol/l insulin and d-[3-3H]glucose (37 MBq/ml; Perkin Elmer-Cetus, Norwalk, CT) diluted 1:1000. Subsequently, the cells were washed three times with cold PBS and lysed in 0.1% SDS/H2O. Lysate (10 μl) was saved for determination of protein concentration. The rest of the lysate was transferred to cuvettes containing scintillation fluid [toluene with 5 g/l 2.5-diphenyloxazol and 0.3 g/l 1.4-bis(4-methyl-5-phenyl-2-oxazolyl)-benzene; all from Sigma-Aldrich, St. Louis, MO], and counts per minute were recorded after overnight phase separation. At micro-molar glucose concentrations, glucose transport but not further metabolism of glucose to lipids is the rate-limiting step for lipogenesis in fat cells when using this method (Arner and Engfeldt, 1987).
2.12. Statistical Analyses
Unless otherwise stated, comparisons were performed using Student's paired t-test or ANOVA. Associations were evaluated using Pearson correlations. Receiver operator curve (ROC) analyses were used to assess the power of each lncRNA to discriminate between the control state and metabolic disease states. A p < 0.05 was considered statistically significant. Error bars in figures are S.E.M. Analyses were performed using GraphPad Prism version 7.01 for Windows (GraphPad Software, La Jolla California USA).
3. Results
3.1. Identification of Adipocyte Enriched lncRNAs that are Associated with Obesity
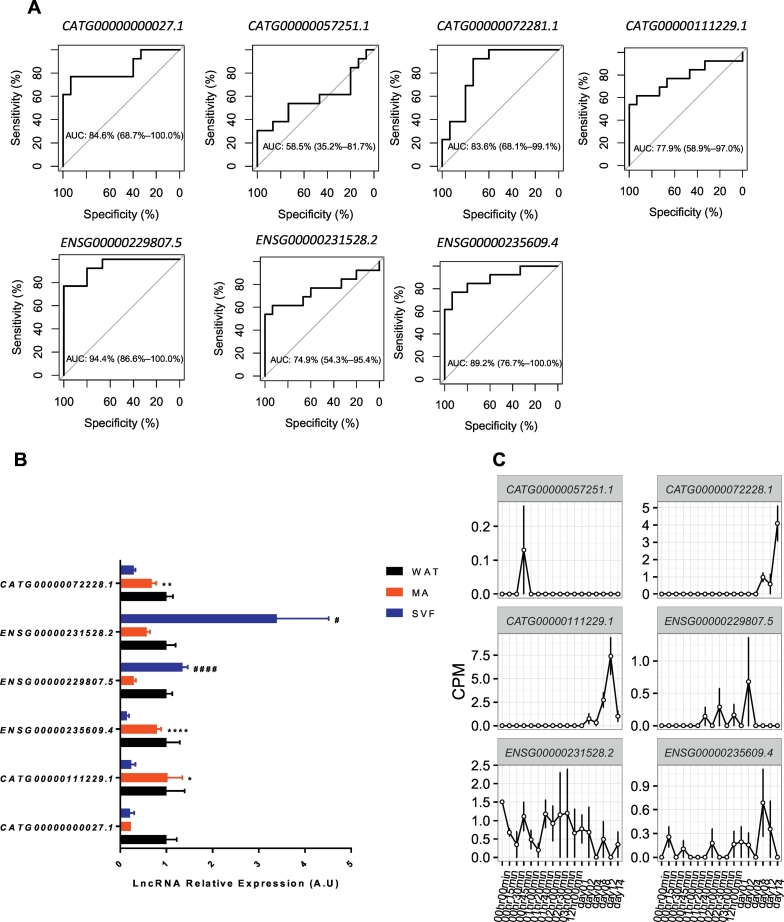
We investigated the transcriptomic expression profiles in WAT from cohort 1 using RNA-seq, based on the Robust gene models annotation in the FANTOM-CAT for coding (i.e. mRNA) and non-coding (i.e. lncRNAs) genes (Hon et al., 2017). First, we filtered for genes that were expressed >1 count per million (cpm) in at least half of the samples, resulting in 3277 lncRNAs and 13876 mRNAs for further analysis. We then identified 965 differentially expressed genes in WAT between obese and non-obese subjects (FDR < 5%) (Supplementary Table 3). The majority of these (841 out of 965) were annotated to protein-encoding mRNAs (Supplementary Fig. 1A). Gene set enrichment analysis, using an FDR of <1%, showed an overrepresentation for pathways known to be perturbed in obesity, including peroxisome proliferation activated receptor-gamma (PPARG), nuclear factor kappa B-signalling and phosphatidylinositol-3 kinase/AKT signalling. The complete list of pathways is presented in Supplementary Fig. 1B. The RNA-seq data identified 86 differentially expressed lncRNAs, However, as this data was produced with a non-strand specific protocol, we decided to only focus on the 34 intergenic lncRNAs (Supplementary Fig. 1C) to avoid the possible bias caused by the expression signal from the antisense strand. However, by manually examining the gene model of the 34 identified intergenic lncRNAs, we noticed the existence of overlapping genes at the 5′ and/or 3′ end for some of these identified intergenic lncRNAs. Thus, we further manually curated the gene models at their genomic loci to exclude the lncRNA loci with transcript models overlapped at both 5′ and/or 3′ end according to the FANTOM-CAT annotation (Hon et al., 2017). All the removed intergenic lncRNAs are marked with an asterisk in Supplementary Table 3. The genome landscape for one of the removed lncRNAs is presented in Supplementary Fig. 1D. After this, seven lncRNAs remained for further analysis. Except for CATG00000057251.1, the expression levels of each of these lncRNAs could effectively distinguish between obese and non-obese subjects, as demonstrated by the receiver operator curve (ROC) (Fig. 2A). WAT is a heterogeneous organ composed of many different cell types, herein, adipocytes were the focus. We, therefore, compared the expression of these lncRNAs by Real-time PCR in isolated mature adipocytes and SVF from the same subjects. CATG00000057251.1 could not be detected despite the use of several different primers. Among the remaining six lncRNAs, CATG00000111229.1, CATG00000072281.1 and ENSG00000235609.4 were significantly enriched in fat cells (p ≤ 0.05, Student's t-test) (Fig. 2B). To determine their possible regulation during adipogenesis, we examined the expression of the seven obesity-related lncRNAs in previously generated CAGE data from human subcutaneous adipose-derived stem cells (hADSC) (Ehrlund et al., 2017). These cells display a complex transcriptional regulatory network during differentiation where the expression patterns of various transcriptional regulators can be subdivided into early, intermediate, late, transiently or constitutively expressed (Ehrlund et al., 2017). The temporal expression analysis during the 14 day-long in vitro differentiation of hADSC period showed that one was transient, one was reduced while four increased during adipogenesis (including CATG00000111229.1 and ENSG00000235609.4); one gene (CATG00000000027.1) was not detected in the dataset (Fig. 2C). This pattern is suggesting a role in terminal adipocyte differentiation and function.
Fig. 2.
Identification of differentially expressed adipocyte specific lncRNAs in obese individuals versus lean controls. A. ROC curve of 7 intergenic lncRNAs which were significantly altered in white adipose tissue (WAT) of obese individuals comparing to lean controls. B. Analysis of cell type specific expression in WAT for the 6 identified lncRNAs in obese WAT using Real-time PCR. MA = mature Adipocyte, SVF = stromal vascular fraction. n = 11 per group, significant enrichment in the MA fraction compared with the SVF is indicated by * and significant enrichment in the SVF compared to the MA fraction by #, *p < 0.05, **p < 0.01, ****p < 0.0001, ####p < 0.0001. (for lncRNA CATG00000057251.1, the expression could not be detected). C. Analysis of expression during human adipose tissue-derived stem cells (hADSCs) in vitro differentiation to adipocytes for the 6 lncRNAs regulated in obese WAT by CAGE, The expression of CATG00000000027.1 was not detected by CAGE.
3.2. Identification of Adipocyte Enriched lncRNAs Associated with Insulin Resistance
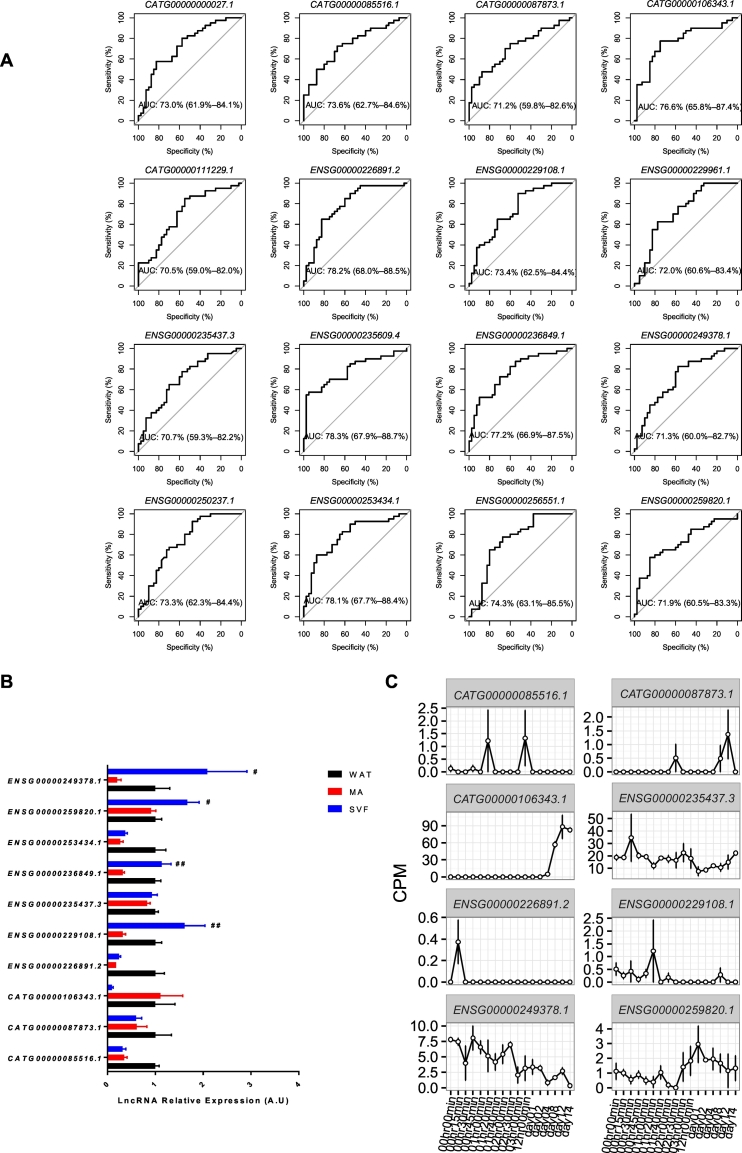
The findings above suggest potential functional roles of lncRNAs in obesity and related metabolic complications. To further investigate this hypothesis, we reanalysed our previously published microarray data to compare lncRNAs expression in subcutaneous and visceral WAT from 40 insulin resistant and 40 insulin sensitive obese women (cohort 2) (Arner et al., 2016). The level of insulin sensitivity was assessed in vivo using the well-established homeostasis model evaluating insulin resistance, termed HOMA (Matthews et al., 1985). Our published work on WAT gene expression profiles was exclusively focused on coding mRNAs and reported that the differences in the mRNA expression between insulin resistant and insulin sensitive subject were more marked in subcutaneous WAT than in visceral WAT. Usually, visceral WAT is considered more pernicious than subcutaneous in the context of obesity complications (Guilherme et al., 2008; Kershaw and Flier, 2004; Sethi and Vidal-Puig, 2007; Wajchenberg, 2000). The region-specific differences in the gene transcriptome were mirrored in the current analysis where we did not find any lncRNA in visceral WAT that was differentially expressed between insulin sensitive and resistant obese (data not shown). The reason why the expression of lncRNAs (as for mRNAs) is not significantly altered by insulin resistance in visceral WAT is currently unknown. A possible explanation is that only obese subjects were investigated in cohort 2 and obesity per se has a significant effect on visceral WAT gene transcription, beyond insulin resistance. Evidence for this assumption has recently been presented (Ryden et al., 2016). The observation that insulin resistance did not impact visceral WAT lncRNA expression prompted us to revisit our microarray dataset on the lncRNAs in subcutaneous WAT. To enable the comparisons between this published microarray dataset, the RNA-seq and CAGE datasets mentioned above, we mapped the microarray probes (Affymetrix HTA 2.0) onto the FANTOM-CAT gene models (Details in Materials and Methods). In total, 44,346 probe sets could be mapped to 29,239 FANTOM-CAT genes (Supplementary Table 2). A comparison of the expression profiles between subcutaneous WAT from insulin-resistant and insulin-sensitive individuals identified 744 differentially expressed FANTOM-CAT genes in WAT (corresponding to 863 probe sets, adjusted p ≤ 0.05) (Supplementary Table 4). Similar to our findings in obesity, the majority of the differentially expressed genes (642 of 744) were annotated to protein-encoding mRNA as illustrated in Supplementary Fig. 1C. Forty-four lncRNAs were identified as significantly altered in WAT of obese insulin-resistant women (Supplementary Fig. 1C). By manual curation as described above, 16 lncRNAs remained for further analysis (Fig. 3A). Based on the ROC curve in Fig. 3A, we demonstrated that all these lncRNAs explicitly associated with insulin resistance. Interestingly, CATG00000111229.1, CATG00000000027.1 and ENSG00000235609.4 previously identified in obese individuals of cohort 1, were also altered in insulin resistance. Their expression among different adipose fractions and their transcriptional dynamics during differentiation were already presented in Fig. 2B and Fig. 2C. The expression of the remaining 13 lncRNAs in different adipose fractions was examined by Real-time PCR (Fig. 3B). Three of them could not be analyzed due to failure to design specific primers. None of the other 10 lncRNAs displayed enrichment in the mature adipocyte. When the expression pattern during differentiation was examined (Fig. 3C) three exhibited late expression during adipogenesis, three genes had a transient gene expression and one gene decreased after adipogenic induction. The remaining two lncRNAs were not detected in hADSCs although they were expressed in WAT. Using the same arguments as above for obesity, the three more late expressed lncRNAs might play a role in fine-tuning adipocyte differentiation and function.
Fig. 3.
Identification of differentially expressed adipocyte specific lncRNAs in insulin resistance individuals versus insulin sensitive individuals. A. ROC curve of 16 intergenic lncRNAs that were significantly regulated in WAT of obese insulin resistant individuals. B. Analysis of cell type specific expression in WAT for the 10 differentially regulated lncRNAs genes regulated in obese WAT using Real-time PCR. n = 11 per group, # SVF versus WAT, significant enrichment in the SVF compared to the MA fraction is indicated by #, #p < 0.05, ##p < 0.01. See legend to Fig. 2 for an explanation of symbols. For lncRNA ENSG00000229961.1, ENSG00000250237.1 and ENSG00000256551.1, we could not detect the expression. C. Analysis of expression during human adipose tissue-derived stem cells (hADSCs) over in vitro differentiation to adipocytes for the 8 lncRNAs regulated in WAT of obese insulin resistance individuals by CAGE. The CAGE data for CATG00000111229.1 and ENSG00000235609.4 are in Fig. 2C. The expression of the other 6 lncRNAs was not detected by CAGE.
3.3. Comparison of lncRNAs in WAT Associated with Obesity and Insulin Resistance
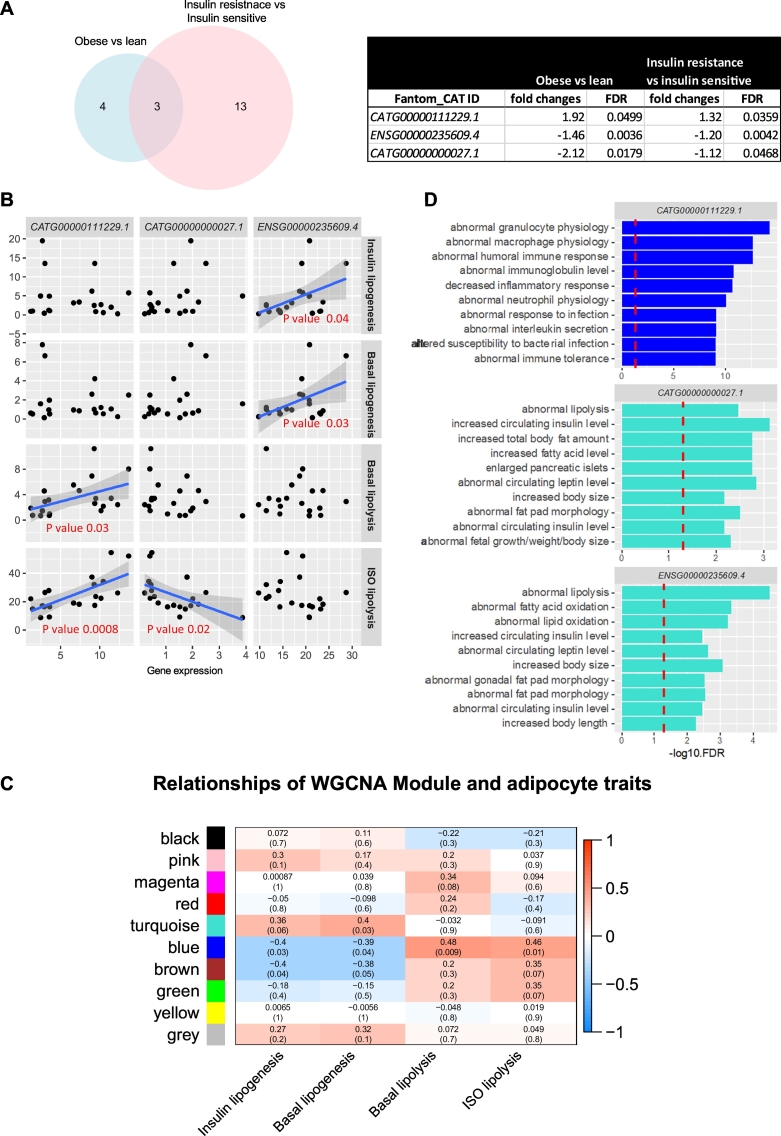
When overlapping the data in Fig. 2, Fig. 3, three lncRNAs were common among the significantly regulated genes (Fig. 4A). The altered expression of the three ncRNAs was validated and confirmed in cohort 2 samples by Real-time PCR (Supplementary Fig. 2). We further investigated the correlation between the expression of these lncRNAs and different adipocyte phenotypes using cohort 1 (Fig. 4B). We combined the obese and non-obese subjects in these analyses in order to obtain sufficient statistical power. CATG00000111229.1 was significantly and positively correlated with basal and stimulated hydrolysis of triglycerides (lipolysis), measured as glycerol release, whereas ENSG00000235609.4 was positively associated with basal and stimulated lipogenesis (conversion of glucose to lipids). Herein, lipogenesis was measured under conditions when glucose uptake is the rate-limiting step for the further metabolism of glucose to lipid (Arner and Engfeldt, 1987). CATG00000000027.1 was weakly and negatively correlated with stimulated lipolysis. These data suggest diverse roles of the three identified lncRNAs in the regulation of the fat cell lipid/glucose metabolism. To shed light on the involved regulatory pathways for these three lncRNAs, weighted gene co-expression network analysis (WGCNA) was performed on the 17,000 filtered genes from RNA-seq data of cohort 1 (see details in Material and Methods). This analysis identified 10 modules of co-expressed genes, one module (blue) was highly correlated with lipolysis and one (turquoise) was associated with lipogenesis (Fig. 4C). Interestingly, CATG00000111229.1 was a member of the blue module while the other two lncRNAs belonged to the turquoise module. We next performed pathway enrichment analysis as guilt-by-association to predict the non-coding RNA function. All genes in one module constituted a cluster of genes that share a similar expression. Therefore, genes in the blue module that were strongly (p < 0.005) and positively correlated with expression of CATG00000111229.1 (Supplementary Table 5) were selected. A similar approach was used for the other two lncRNAs in the turquoise module (Fig. 4D, genes positively correlated with expression of ENSG00000235609.4 presented in Supplementary Table 6) was used. Based on the correlation with different genes the function of a specific lncRNA can be predicted. This approach revealed that CATG00000111229.1 was involved in inflammation pathways and the other two lncRNAs in regulating lipid metabolism (Fig. 4D).
Fig. 4.
Identification of adipocyte specific lncRNAs associated with insulin resistance. A. Venn diagram (left panel) of the overlap of affected lncRNAs in obese versus lean and obese insulin resistant versus insulin sensitive subjects; the detailed information for the 3 overlapped lncRNAs are presented in the table (right panel). B. Dot plot demonstrating the correlation between the expression of 3 lncRNAs and 4 selected clinical parameters insulin stimulated lipogenesis, basal lipogenesis, basal lipolysis and lipolysis stimulated by a catecholamine (Isoprenaline) in the lean vs obese cohort, respectively. Regression lines for each sample group were added. C. Heatmap of correlations with p-values in parentheses between modules and clinical parameters. Degrees of associations are indicted by colours, positive correlation, red; negative correlation, blue; no correlation, white. D. Bar plots showing the pathways characterized in mouse phenotypes for highly positive correlated genes with our selected lncRNAs within each module (indicated by bar colour).
3.4. Effect of Targeted Knockdown of lncRNAs ASMER-1 and ASMER-2 on Mature Adipocyte Function
To gain insights into the functional role of identified lncRNAs, we focused on ENSG00000235609.4 and CATG00000111229.1, which from here on in are referred as adipocyte specific metabolic related lncRNA 1 (ASMER-1) and ASMER-2. This is because they were significantly enriched in the mature fat cells and regulated by both clinical conditions examined. CATG00000000027.1 was not detected in our hADSCs cells used for functional analysis and was therefore excluded. ASMER-1 and ASMER-2 displayed distinct evolutionary constraints and cellular localization. ASMER-1 is conserved at the exon level whereas ASMER-2 is not conserved at any level, illustrated with PhyloP scores track in the genomic landscape of ASMER-1 and ASMER-2 loci (Fig. 5A). RNA from nuclear and cytoplasmic fractions of in vitro differentiated hADSCs at day 13 was used to determine the subcellular localization of ASMER-1 and ASMER-2. Real-time PCR analysis revealed a roughly equal distribution of ASMER-1 in both subcellular compartments of fully adipocyte differentiated hADSCs (Fig. 5B), while high cytoplasmic enrichment was found for ASMER-2 (Fig. 5B).
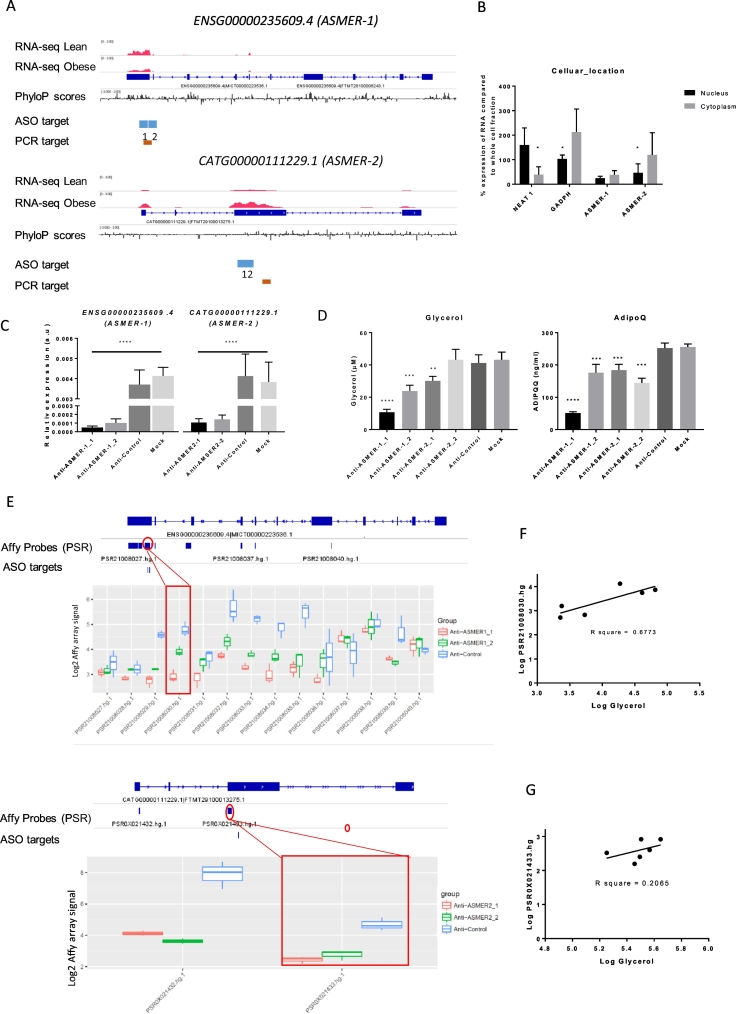
Fig. 5.
Functional characterization of the lncRNAs ASMER-1 and ASMER-2 in the adipocyte. A. Schematic of the genomic landscape of ENSG00000235609.4 (ASMER-1) and CATG00000111229.1 (ASMER-2) loci respectively. Images from the Integrative Genomics Viewer (IGV) depict RNA-seq signal as the density of mapped RNA-seq reads. Tracks 1 and 2 show in red the example of the non–strand-specific RNA-seq signal of poly(A)+ RNA from adipose tissues of one obese and lean subject. Tracks 3 shows the FANTOM-CAT annotation of the transcript structure. Track 4 depicts the conservation scores by phyloP (phylogenetic p-values) for multiple alignments of 99 vertebrate genomes to the human genome, downloaded from UCSC browser. Track 5 indicates the targeted positions of the antisense oligos. Track 6 shows the position of the targeted amplicon for PCR primers to evaluate the gene expression by Real-time PCR. B. RNA was extracted from the nuclei or cytoplasm of in in vitro differentiated hADSCs at day 13. The RNA was used for the realtime-PCR analysis of ASMER-1, ASMER-2, NEAT1 (nuclear retained), and GAPDH mRNAs (cytoplasm retained). C. Relative expression of ASMER-1 and ASMER-2 at day13 in in vitro differentiated hADSCs treated with two antisense oligonucleotides for each of ASMER-1 and ASMER-2 at day 8, respectively, n = 4, ****p < 0.0001. Scrambled oligos were used as a control (Anti-Control). Mock samples were treated with all reagents except antisense oligonucleotides. D. glycerol release and Adiponectin release were measured in the medium of in vitro differentiated hADSCs transfected with either control (Anti-Control), Mock, ASMER-1 (Anti-ASMER-1_1or_2) or ASMER-2 antisense oligo (Anti-ASMER-2_1 or _2) n = 4, ****p < 0.0001, ***p < 0.001, **p < 0.01. E. Knockdown efficiency was verified by examining the microarray detected gene expression at the level of probe selection regions (PSRs). Track 1 shows the FANTOM-CAT annotation of the transcript structure. Track 2 depicts the genomic positions of PSRs for ASMER-1 (14 regions) and ASMER-2 (2 regions). Track 3 indicates the targeted positions of the antisense oligos. Track 4 plots the log microarray signal of each PSR associated with ASMER-1 and ASMER-2, respectively, after silencing of ASMER-1 (two antisense oligonucleotides) or ASMER-2 (two antisense oligonucleotides) together with Control. F–G. Linear regression analysis between glycerol release and array signal of one representative PSR for ASMER-1 (F) and ASMER-2 (G).
To further characterize the functional role of ASMER-1 and -2 in mature adipocytes, loss-of-function studies were performed using two antisense oligos for each gene in in vitro differentiated hADSCs. Cells were transfected at day 8 post-induction and samples were collected at day 13. A schematic presentation of the approximate position of the designed antisense oligo for silencing and the Real-time PCR amplicon for measuring the gene expression are shown in Fig. 5A. The expression of both ASMER-1 and -2 was markedly attenuated following treatment with either of the selected antisense oligonucleotides (Fig. 5C). With regard to glycerol release (lipolysis measure), knockdown of ASMER-1 resulted in a significant decrease with both oligonucleotides (although one more pronounced than the other) but with only one of the ASMER-2 oligonucleotides (p ≤ 0.01, Student's t-test, Fig. 5D). Knockdown of both ASMERs resulted in a significant decrease in adiponectin release (p ≤ 0.001, Student's t-test, Fig. 5D) but again one of the ASMER-1 oligonucleotides resulted in a more pronounced effect. We also investigated the impact of lncRNA knockdown on lipogenesis. However, inhibition of ASMER-1 or ASMER-2 did not influence insulin induced lipogenesis (n = 6), Supplementary Fig. 3.
In order to gain further insight into the mechanism by which ASMER-1 and -2 affect fat cell function, we performed gene microarray analysis of the knockdown experiments (3 samples for each condition). To be able to evaluate the knockdown efficiency at the whole transcript level, normalized microarray signals at the probe selection region (PSR) were extracted and plotted along the FANTOM-CAT transcriptome model (Fig. 5E). These data indicated a substantial knockdown effect for both ASMERs with the two different antisense oligos, respectively. However, one antisense oligo for ASMER-1 displayed a stronger silencing effect (Fig. 5E, upper panel). Interestingly, the knocking down efficiency of the different antisense oligos correlated well with glycerol release (Fig. 5F for ASMER-1 and Fig. 5G for ASMER-2). Knockdown of ASMER-1 did not affect the gene expression of ASMER-2 and the other way around (data not shown).
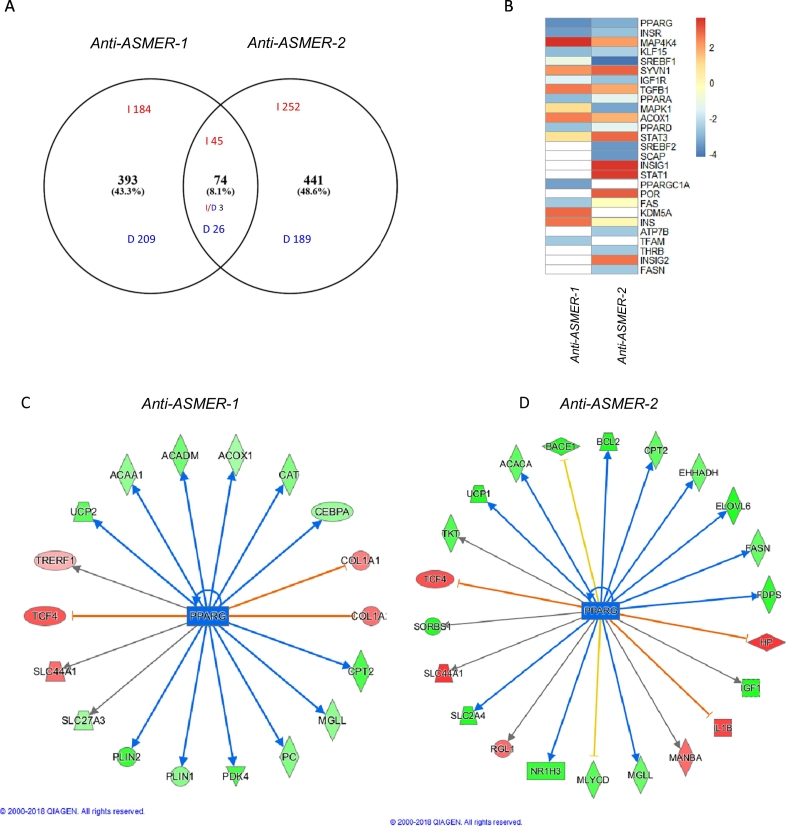
To be able to identify the response genes and avoid effects of differences in knockdown efficiency, samples from two antisense oligos were combined as one group and compared with control samples. The response genes were selected using a cut off 0.3 for log-fold change and a nominal p value < 0.01 (FDR < 33%). A similar number of genes (~500) was altered following silencing of each ASMERs (Supplementary Table 7–8) where around 10% of the genes were shared (Fig. 6A). Interestingly, the pathway analysis using IPA revealed that silencing of either ASMERs impacted on several signalling pathways of importance for adipocyte function, such as PPARG, INSR and MAP4K4 (Fig. 6B). However, while these pathways were shared, it was apparently not the same set of genes that were regulated by ASMER-1 or -2 silencing. An example is given for PPAR gamma signalling (Fig. 6C and D). Several genes involved in lipolysis as well as fatty acid and energy metabolism were identified for either ASMER-1 or -2, but only a few were common. ASMER-1 knock-down increased the expression of PLIN1 and MGLL encoding a lipid droplet protein and a lipase, respectively, while ASMER-2 knockdown only affected MGLL expression. This might explain why knocking down the former gene had a more consistent effect on glycerol release than knocking down ASMER-2 (Fig. 5D).
Fig. 6.
Global transcriptome analysis after targeted knockdown for ASMER-1 and ASMER-2 in in vitro differentiated mature adipocyte. A. Venn-diagram of the overlapping genes affected by transfecting antisense oligonucleotides against ASMER-1 or ASMER-2 in in vitro differentiated hADSCs. B. Identification of upstream mediators that underlie the altered transcriptome after silencing of ASMER-1 or ASMER-2 through pathway enrichment analysis using Ingenuity Pathway Analysis (IPA). C and D. Significantly changed PPARG downstream genes identified by IPA after silencing of ASMER-1 (C) and ASMER-2 (D). Red and green highlight the up-regulated and down-regulated proteins, respectively, and the colour depth is correlated to the fold change. Orange and blue lines with arrows indicate indirect activation and inhibition, respectively. Yellow and grey dashed lines with arrows depict inconsistent effects and no prediction, respectively.
3.5. Effect of Targeted Knockdown of lncRNAs ASMER-1 and ASMER-2 on Adipogenesis
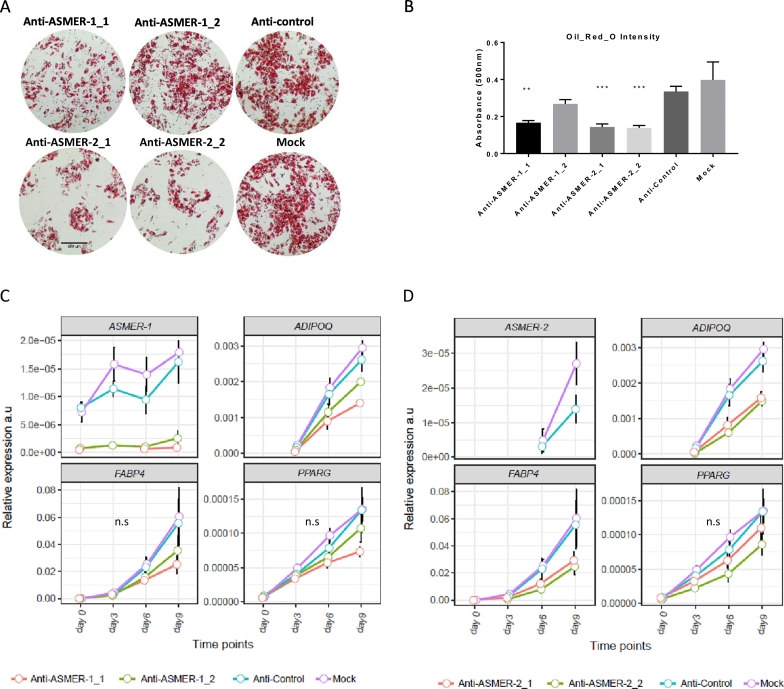
To investigate the functional role of ASMER-1 and -2 in the process of adipogenesis, the antisense oligos mentioned above were used to silence either ASMERs at day 0 (prior to adipogenic induction). Following induction, expression of several critical adipogenic markers was measured at day 9 and triglyceride accumulation was quantified by Oil-Red-O staining at day 13 (Fig. 7A-B). Silencing attenuated triglyceride accumulation at day 13 with much stronger effects for ASMER-2. A marked reduction in the expression of adipogenic genes was observed for both ASMERs (Fig. 7C for ASMER-1, Fig. 7D for ASMER-2), again with ASMER-2 antisense oligos resulting in a stronger repressive effect (Fig. 7D) compared with ASMER-1 (Fig. 7C).
Fig. 7.
Functional characterization of the lncRNAs ASMER-1 and ASMER-2 in the adipogenesis. A. Micrographs of Oil-Red-O staining of the hADSC cell after differentiation at day 13 with silencing of ASMER-1 (two antisense oligonucleotides), ASMER-2 (two antisense oligonucleotides), Control and Mock at day 0 of differentiation. Scale bar is 300 μm. B. Lipid content was measured by quantification of Oil-Red-O staining (absorbance at 495 nm), n = 4, ***p < 0.001, **p < 0.01. C. hADSCs were induced to differentiate at day 0 after silencing of ASMER-1 (two antisense oligonucleotides). Gene expression of ASMER-1 as well as established genes regulated during adipogenesis (ADIPOQ, FABP4 and PPARG) were determined by Real-time PCR at different time points. n.s = not significant. D. After silencing of ASMER-2 (two antisense oligonucleotides) at day 0. Gene expression of ASMER-2 as well as established genes regulated during adipogenesis (ADIPOQ, FABP4, and PPARG) were determined by Real-time PCR at different time points. Silencing the expression of AMSER-2 resulted in significantly lower expression levels of 3 measured genes. n.s = not significant.
4. Discussion
The primary goal of this project was to identify adipocyte-specific lncRNAs linked to obesity and insulin resistance and study their role in controlling two primary functions of human adipocytes, lipolysis and adipokine release. We started with a data mining process which identified a number of lncRNAs in human adipose tissue that correlated with obesity and/or insulin resistance. This identified two lncRNAs that were enriched in fat cells and associated with both examined clinical conditions. We then investigated their impact on mature cell function and adipogenesis by loss-of-function experiments using antisense oligonucleotides. Finally, downstream signalling of the two mentioned lncRNAs was studied with global transcriptome analysis.
It might be surprising that a rather low number of lncRNAs were specifically associated with obesity and/or insulin resistance. This is, on the other hand, in line with the findings in other tissues and clinical conditions as reviewed (Batista and Chang, 2013; Esteller, 2011; Losko et al., 2016). Furthermore, a study in a rodent model of perturbed metabolism also found a small number of differentially expressed lncRNAs in the liver (Li et al., 2015). It is, therefore, possible that a limited set of WAT lncRNAs are clinically relevant in the context of obesity and/or insulin resistance. Admittedly, we cannot exclude an insufficient sensitivity to detect lowly expressed lncRNAs with current transcriptome technologies. Another caveat is that we only examined intergenic lncRNAs. The latter genes were chosen because our data mining tools (gene expression arrays and RNA-seq) only allowed identification of these lncRNA genes. The strict filtering process employed to select these intergenic lncRNAs could also have missed some relevant hits. Nevertheless, the 20 lncRNAs shown herein to be specifically related to these disorders by ROC curve analysis might serve as potentially significant pathophysiological metabolic markers. Interestingly, none of the previously studied lncRNA involved in adipogenesis was identified as being of clinical relevance for obesity and insulin resistance.
Based on the guilt-by-association with the coding gene expression, we hypothesized that some adipocyte-expressed lncRNAs linked to obesity and insulin resistance could also associate with essential processes in fat cells functions including lipolysis (Arner and Langin, 2014) and lipogenesis (Guilherme et al., 2008). Indeed, the adipose expression of the selected lncRNAs correlated with both measures in human fat cells. However, the pattern of correlation was not uniform (Fig. 4B) and the association with gene regulatory pathways also differed (Fig. 4C, D). On the other hand, this is not surprising bearing in mind the deviations in gene expression pattern induced by lncRNA silencing discussed below and the different expression pattern for each of the lncRNAs observed during adipogenesis (Figs. 2C and 3C).
Association studies can only suggest causality. We therefore also examined the effect of gene depletion in differentiated human fat cells. We focused on ASMER-1 and ASMER-2, as they were significantly enriched in the mature fat cells and, secondly, regulated by both clinical conditions examined. The third potentially important lncRNA CATG00000000027.1 was not detected in our hADSCs and could therefore not be evaluated functionally. As a metabolic functional marker, we used glycerol release which constitutes the end product of triglyceride breakdown and is not re-utilized by fat cells. Fat cell lipolysis is altered in obesity and insulin resistance (Arner and Langin, 2014). As an endocrine marker, we chose adiponectin release, a hormone strongly associated with insulin resistance (Caselli, 2014).
A problem with gene silencing with antisense oligonucleotides is off-target effects. We, therefore, used two different antisense oligos for each lncRNA, each targeting different DNA sequence in the lncRNA genes (Fig. 5A, E). Interestingly, very similar results were obtained following silencing of either gene, namely inhibition of both lipolysis and adiponectin release. For all oligos, the effect was statistically significant for adiponectin, but only 3 oligos were significantly effective for lipolysis. This is likely to be secondary to knockdown efficiency as there was a strong correlation between this measure and the anti-lipolytic effect for both lncRNAs (Fig. 5F, G). Although the phenotypic impact of ASMER-1 and -2 depletion were similar, it is likely that they regulate lipolysis and endocrine function through different mechanisms as evidenced by their effects on gene expression. The difference between ASMER-1 and -2 is further emphasized by their subcellular localization (Fig. 5B) and effects on adipogenesis where ASMER-2 appears to be more critical than ASMER-1 for adipocyte differentiation.
There are some caveats with this study. We only investigated women so there might be undetected gender differences. The latter might be important for different WAT depots given that visceral adipose is more abundant among men. We cannot exclude effects of obesity on the distribution of different cell types within WAT and that lncRNAs display a distinct gene expression pattern in non-adipocytes. This is, however, not relevant for ASMER-1 and -2, which were most abundantly expressed in fat cells, the target cell type in our study. Our fat cell line was derived from a male donor. While in vitro cultured hADSC may display gender-based differences, we have compared such cells obtained from male and female donors. This uncharted study showed that they have similar functional phenotypes (basal or stimulated lipolysis and lipogenesis as well as the release of adipokines). Because the precise mechanism of action of lncRNAs is not yet well defined, further investigations are needed at the molecular level. It is likely that the regulation is complex. A large number of genes are regulated by lncRNA depletion and may constitute direct or indirect direct targets for ASMER-1 and -2. The complete gene lists are therefore publically available (Supplementary Table 7–8). Finally, we did not observe any effect of ASMERs silencing on adipocyte lipogenesis in spite of the clinical correlation between gene expression and lipogenesis rates. The reason for this discrepancy is not known for the moment. However, it is possible that ASMERs influence lipogenesis indirectly in vivo.
Can fat cell lncRNAs constitute targets for treatment of obesity and/or insulin resistance (Boon et al., 2016)? ASMER-2 could be a candidate for lipolysis. Its expression was upregulated in obesity and insulin resistance. These conditions associated with increased basal lipolysis (Arner and Langin, 2014). Knock-down at the fat cell level decreased basal lipolysis. Thus, anti ASMER-2 drugs might lower lipolysis and improve metabolic complications to obesity. This speculation needs, of course, to be supported by future investigations.
In conclusion, this study shows that a number of hitherto unknown lncRNAs in subcutaneous WAT are actively and specifically related to obesity and/or insulin resistance and correlate with the metabolic functions of fat cells. Two lncRNAs, ASMER-1 and -2 may be particularly important as they are fat cell-enriched and have similar effects on lipolysis, endocrine function and adipocyte differentiation. Thus, lncRNAs may constitute specific markers and targets for future therapeutic strategies in metabolic disorders.
Funding Sources
This study was supported by grants from EU/EFPIA Innovative Medicines Initiative Joint Undertaking (EMIF grant n°115372), the Novo Nordisk Foundation including the Tripartite Immuno-metabolism Consortium (TrIC) (Grant number: NNF15CC0018486) and the MSAM consortium (Grant number: NNF15SA0018346), the Swedish Diabetes Association (Grant number: DIA2013-003), CIMED, the Swedish Research Council (Grant number: K2012-55X-01034-46-5), the Stockholm County Council (Grant number: 20160040), EFSD and the Diabetes Program at Karolinska Institutet. AK is a fellow of Novo Nordisk (Copenhagen, Denmark) Postdoctoral Fellowship Program at Karolinska Institutet. HG supported as a Naomi Berrie fellow (Columbia University, NY, USA) in Diabetes research.
Conflicts of Interest
The authors declare no conflict of interest associated with this publication.
Author Contributions
P.A. and H.G. designed the study. P.A., H.G. and A.K. performed data analysis, the literature search and data interpretation as well as designed the figures. P.A., H.G. and A.K. wrote the first version of the manuscript. I.D. P.A. and M.R. generated and analyzed the clinical data. H.G. and A.K. performed the human adipocyte experiments and related data analysis. H.J. and C-C. H. performed bioinformatic analyses. P.A., M.R. and E.D. provided financial support for this study. P.A. and H.G. are guarantors of the data. All authors read, gave input, and approved the final version of the manuscript.
Acknowledgements
The authors would like to acknowledge support from Science for Life Laboratory, the National Genomics Infrastructure, NGI, and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.03.010.
Contributor Information
Hui Gao, Email: hui.gao@ki.se.
Peter Arner, Email: peter.arner@ki.se.
Appendix A. Supplementary Data
Supplementary table 1-8.
Supplementary figures
References
- Akerman I., Tu Z., Beucher A., Rolando D.M., Sauty-Colace C., Benazra M., Nakic N., Yang J., Wang H., Pasquali L. Human pancreatic beta cell lncRNAs control cell-specific regulatory networks. Cell Metab. 2017;25:400–411. doi: 10.1016/j.cmet.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner P., Engfeldt P. Fasting-mediated alteration studies in insulin action on lipolysis and lipogenesis in obese women. Am. J. Phys. 1987;253:E193–201. doi: 10.1152/ajpendo.1987.253.2.E193. [DOI] [PubMed] [Google Scholar]
- Arner P., Langin D. Lipolysis in lipid turnover, cancer cachexia, and obesity-induced insulin resistance. Trends Endocrinol. Metab. 2014;25:255–262. doi: 10.1016/j.tem.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Arner P., Sahlqvist A.S., Sinha I., Xu H., Yao X., Waterworth D., Rajpal D., Loomis A.K., Freudenberg J.M., Johnson T. The epigenetic signature of systemic insulin resistance in obese women. Diabetologia. 2016;59:2393–2405. doi: 10.1007/s00125-016-4074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantyne R.L., Zhang X., Nunez S., Xue C., Zhao W., Reed E., Salaheen D., Foulkes A.S., Li M., Reilly M.P. Genome-wide interrogation reveals hundreds of long intergenic noncoding RNAs that associate with cardiometabolic traits. Hum. Mol. Genet. 2016;25:3125–3141. doi: 10.1093/hmg/ddw154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon R.A., Jae N., Holdt L., Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J. Am. Coll. Cardiol. 2016;67:1214–1226. doi: 10.1016/j.jacc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- Borgstrom E., Lundin S., Lundeberg J. Large scale library generation for high throughput sequencing. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Carvalho B.S., Irizarry R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli C. Role of adiponectin system in insulin resistance. Mol. Genet. Metab. 2014;113:155–160. doi: 10.1016/j.ymgme.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Divoux A., Karastergiou K., Xie H., Guo W., Perera R.J., Fried S.K., Smith S.R. Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity (Silver Spring) 2014;22:1781–1785. doi: 10.1002/oby.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlund A., Mejhert N., Bjork C., Andersson R., Kulyte A., Astrom G., Itoh M., Kawaji H., Lassmann T., Daub C.O. Transcriptional dynamics during human adipogenesis and its link to adipose morphology and distribution. Diabetes. 2017;66:218–230. doi: 10.2337/db16-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Gao H., Volat F., Sandhow L., Galitzky J., Nguyen T., Esteve D., Astrom G., Mejhert N., Ledoux S., Thalamas C. CD36 is a marker of human adipocyte progenitors with pronounced adipogenic and triglyceride accumulation potential. Stem Cells. 2017;35:1799–1814. doi: 10.1002/stem.2635. [DOI] [PubMed] [Google Scholar]
- Guilherme A., Virbasius J.V., Puri V., Czech M.P. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw E.E., Flier J.S. Adipose tissue as an endocrine organ. J. Clin. Endocrinol. Metab. 2004;89:2548–2556. doi: 10.1210/jc.2004-0395. [DOI] [PubMed] [Google Scholar]
- Kolaczynski J.W., Morales L.M., Moore J.H., Jr., Considine R.V., Pietrzkowski Z., Noto P.F., Colberg J., Caro J.F. A new technique for biopsy of human abdominal fat under local anaesthesia with lidocaine. Int. J. Obes. Relat. Metab. Disord. 1994;18:161–166. [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Ruan X., Yang L., Kiesewetter K., Zhao Y., Luo H., Chen Y., Gucek M., Zhu J., Cao H. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015;21:455–467. doi: 10.1016/j.cmet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofgren P., Hoffstedt J., Naslund E., Wiren M., Arner P. Prospective and controlled studies of the actions of insulin and catecholamine in fat cells of obese women following weight reduction. Diabetologia. 2005;48:2334–2342. doi: 10.1007/s00125-005-1961-6. [DOI] [PubMed] [Google Scholar]
- Losko M., Kotlinowski J., Jura J. Long noncoding RNAs in metabolic syndrome related disorders. Mediat. Inflamm. 2016;2016 doi: 10.1155/2016/5365209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews D.R., Hosker J.P., Rudenski A.S., Naylor B.A., Treacher D.F., Turner R.C. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Miller J.A., Cai C., Langfelder P., Geschwind D.H., Kurian S.M., Salomon D.R., Horvath S. Strategies for aggregating gene expression data: the collapseRows R function. BMC Bioinformatics. 2011;12:322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryden M., Hrydziuszko O., Mileti E., Raman A., Bornholdt J., Boyd M., Toft E., Qvist V., Naslund E., Thorell A. The adipose transcriptional response to insulin is determined by obesity, not insulin sensitivity. Cell Rep. 2016;16:2317–2326. doi: 10.1016/j.celrep.2016.07.070. [DOI] [PubMed] [Google Scholar]
- Sethi J.K., Vidal-Puig A.J. Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J. Lipid Res. 2007;48:1253–1262. doi: 10.1194/jlr.R700005-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004:3. doi: 10.2202/1544-6115.1027. (Article3) [DOI] [PubMed] [Google Scholar]
- Snijder M.B., Dekker J.M., Visser M., Bouter L.M., Stehouwer C.D., Kostense P.J., Yudkin J.S., Heine R.J., Nijpels G., Seidell J.C. Associations of hip and thigh circumferences independent of waist circumference with the incidence of type 2 diabetes: the Hoorn Study. Am. J. Clin. Nutr. 2003;77:1192–1197. doi: 10.1093/ajcn/77.5.1192. [DOI] [PubMed] [Google Scholar]
- Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajchenberg B.L. Subcutaneous and visceral adipose tissue: their relation to the metabolic syndrome. Endocr. Rev. 2000;21:697–738. doi: 10.1210/edrv.21.6.0415. [DOI] [PubMed] [Google Scholar]
- Wei S., Du M., Jiang Z., Hausman G.J., Zhang L., Dodson M.V. Long noncoding RNAs in regulating adipogenesis: new RNAs shed lights on obesity. Cell. Mol. Life Sci. 2016;73:2079–2087. doi: 10.1007/s00018-016-2169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Li P., Yang W., Ruan X., Kiesewetter K., Zhu J., Cao H. Integrative transcriptome analyses of metabolic responses in mice define pivotal LncRNA metabolic regulators. Cell Metab. 2016;24:627–639. doi: 10.1016/j.cmet.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf S., Hawken S., Ounpuu S., Bautista L., Franzosi M.G., Commerford P., Lang C.C., Rumboldt Z., Onen C.L., Lisheng L. Obesity and the risk of myocardial infarction in 27,000 participants from 52 countries: a case-control study. Lancet. 2005;366:1640–1649. doi: 10.1016/S0140-6736(05)67663-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1-8.
Supplementary figures