Abstract
Apoptosis-inducing factor (AIF) is a mitochondrial oxidoreductase that contributes to cell death programmes and participates in the assembly of the respiratory chain. Importantly, AIF deficiency leads to severe mitochondrial dysfunction, causing muscle atrophy and neurodegeneration in model organisms as well as in humans. The purpose of this review is to describe functions of AIF and AIF-interacting proteins as regulators of cell death and mitochondrial bioenergetics. We describe how AIF deficiency induces pathogenic processes that alter metabolism and ultimately compromise cellular homeostasis. We report the currently known AIFM1 mutations identified in humans and discuss the variability of AIFM1-related disorders in terms of onset, organ involvement and symptoms. Finally, we summarize how the study of AIFM1-linked pathologies may help to further expand our understanding of rare inherited forms of mitochondrial diseases.
Keywords: Apoptosis-inducing factor (AIF), Cell death, Mitochondria, Mitochondrial diseases, Oxidative phosphorylation (OXPHOS)
Highlights
-
•
AIF is a mitochondrial NADH-dependent oxidoreductase.
-
•
Nuclear translocation of AIF occurs during cell death and has been associated with human disorders.
-
•
Under physiological settings, AIF participates to the biogenesis of the respiratory complexes.
-
•
AIFM1 mutations have been identified in patients with impaired mitochondrial bioenergetics.
-
•
Inherited AIFM1 mutations lead to a variety of clinical manifestations, including severe childhood-onset mitochondrial diseases.
1. Introduction
The X-linked AIFM1 gene encodes for a 67-kDa AIF polypeptide that is imported into the mitochondria, where it is processed to an inner-membrane-tethered protein. Proteolytic cleavage between Met53 and Ala54 generates a 62-kDa mature form, which is mainly exposed to the intermembrane space (IMS) (Otera et al., 2005), although a small pool of AIF protein is also loosely associated with the cytosolic side of the mitochondrial outer membrane (MOM) (Yu et al., 2009). AIF comprises a NADH/NADPH-binding domain and two flanking flavin adenine dinucleotide (FAD)-binding motifs. The NADH-binding region possesses a sequence homology to bacterial NADH-dependent ferredoxin reductase and to the phylogenetically related yeast NADH:ubiquinone oxidoreductase Ndi1p (Mate et al., 2002; Elguindy and Nakamaru-Ogiso, 2015). The folded AIF polypeptide undergoes dimerization upon NADH reduction, while the subsequent conformational change stabilizes the reduced form of the molecule, increasing AIF resistance to oxidation (Sevrioukova, 2009; Churbanova and Sevrioukova, 2008; Villanueva et al., 2015). The C-terminal region contributes to the dimeric interface and determines the binding with other molecules, including the DNA. The overall crystal structure indicates that AIF has a glutathione reductase fold and shares a structural homology to the ferredoxin reductase BphA4 (Mate et al., 2002). AIF exhibits NADH-dependent oxidoreductase activity in vitro, however the electron transfer to oxidized substrates occurs with much less efficiency compared to other flavoproteins (Sevrioukova, 2011; Churbanova and Sevrioukova, 2008). While AIF redox capacity determines its mitochondrial housekeeping function, the oxidoreductase activity is instead dispensable during cell demise. In the next sections, we will summarize the contribution of AIF to cell death (Fig. 1A). We will outline a few distinct pathways with unique AIF-dependent biochemical signatures and discuss AIF participation to detrimental cascades occurring in a few disease settings. Then, we will describe the importance of AIF in the maintenance of the respiratory complexes and, therefore, mitochondrial activity (Fig. 1A). Finally, we will provide an overview of the mutations in the AIFM1 gene that have been causally linked to inherited pathological conditions in humans.
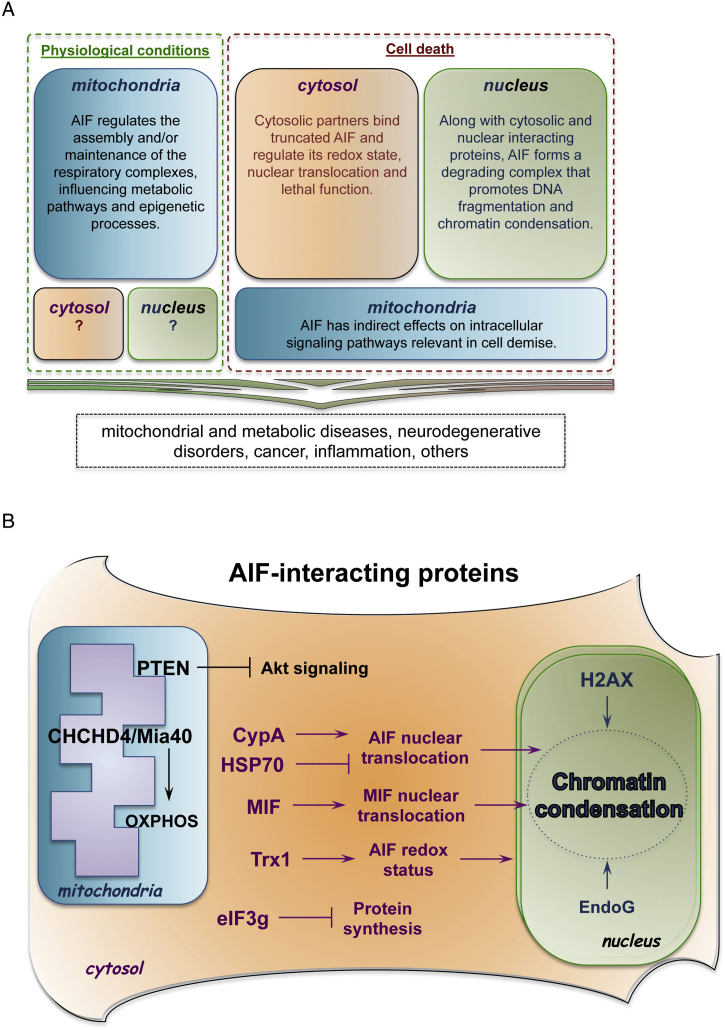
Fig. 1.
Pathophysiological functions of AIF. (A) Under physiological settings, AIF has a vital role in mitochondrial bioenergetics, since it supports the normal oxidative phosphorylation of the cell. Consequently, mitochondrial AIF has an impact on multiple catabolic and anabolic pathways, as well as on epigenetic processes that depend on mitochondrial metabolites. It remains unclear whether soluble AIF molecules are present in other subcellular compartments under physiological conditions. Upon detrimental signals, mitochondrial AIF indirectly modulates intracellular signaling pathways (e.g., PTEN/Akt), while cytosolic AIF binds molecular partners that determine its nuclear translocation. In the nucleus, AIF forms a degrading complex responsible for chromatinolysis and cell death. These molecular processes have been associated to an array of pathological conditions, including inherited diseases. (B) Schematic summaries of AIF-interacting proteins. Mitochondrial intermembrane space: AIF binding to CHCHD4/Mia40 contributes to the oxidative folding of electron transport chain subunits. Moreover, AIF physically interacts with mitochondria-localized PTEN, inhibits PTEN oxidation and indirectly influences Akt signaling pathway. Cytosol: upon release from the mitochondria, AIF interacts with several cytosolic proteins. The binding kinetics between HSP70 and CypA determines AIF nuclear translocation rate. In the cytosol and in the nucleus, TRX1 interaction prevents AIF oxidation in cells exposed to stress and undergoing apoptosis. Another recently identified AIF-binding partner is MIF. In a functional complex with AIF, the endonuclease MIF translocates to the nucleus and mediates chromatinolysis. During apoptosis, AIF interacts with the eukaryotic translational initiation factor EIF3g and, consequently, inhibits protein synthesis. Nucleus: along with CypA, AIF interacts with histone H2AX and forms a degrading complex that regulates DNA cleavage. At least in invertebrates, AIF regulates EndoG activity and promotes chromatin condensation.
2. AIF from Apoptosis to Unique Death Pathways, with an Overview of its Lethal Contribution across Disorders
AIF was originally identified as a soluble 57-kDa fragment that, during apoptosis and upon dissipation of the mitochondrial membrane potential, is released from the mitochondria and translocates to the nucleus in a caspase-independent manner (Susin et al., 1999; Susin et al., 1996). Subsequent studies have shown that, during cell death, the accumulation of AIF in the cytosol requires both permeabilization of the MOM and the concomitant cleavage of the membrane-bound protein between Leu101 and Gly102 (Otera et al., 2005; Susin et al., 1999; Susin et al., 1996). It is still under debate whether a cytosolic protease or a resident mitochondrial processing peptidase is responsible for the release of truncated AIF from the IMS in response to death stimuli. One line of evidence suggests that calcium-dependent cysteine proteases, calpains, can induce AIF release from permeabilized mitochondria (Polster et al., 2005; Cao et al., 2007), which along with other cysteine proteases (i.e., lysosomal cathepsins) cleave AIF, at least in vitro (Yuste et al., 2005). In the cytosol, truncated AIF physically interacts with multiple proteins, including heat shock 70-kDa protein (HSP70) and cyclophilin A (CypA) (Fig. 1B). In an antagonistic and redox-controlled manner, these two proteins regulate the cellular localization of truncated AIF protein. In this regard, HSP70 promotes AIF retention in the cytoplasm (Ravagnan et al., 2001), whereas CypA facilitates the redistribution of AIF monomer to the nucleus (Zhu et al., 2007a; Cande et al., 2004). Based on recent biochemical evidence, it seems that thioredoxin TRX1 reduces AIF in the cytosol and eventually in the nucleus, in part modulating AIF nuclear activity during cell death (Nakao et al., 2015; Shelar et al., 2015). This interaction has several evolutionarily conserved aspects, since knockdown of thioredoxin-2 (Dm Trx-2) contributes to AIF-induced cell death in Drosophila melanogaster (Joza et al., 2008). Although the picture is not complete, it is possible that the thioredoxin-mediated regulation of AIF redox status determines the binding to HSP70 or CypA, in line with previous data reporting that CypA preferentially binds oxidized AIF (Cande et al., 2004). Similarly, it remains unclear whether the two nuclear localization sequences (NLS), within the FAD and C-terminal domains, actively mediate an ATP-dependent nuclear import of AIF/CypA complex. During apoptosis, AIF nuclear translocation occurs downstream of mitochondrial permeabilization and, therefore, in cells undergoing a possible energy crisis. In this and other settings, it is reasonable to assume that AIF translocation does not require active transport, but may be facilitated by the increased permeability of the nuclear pore complex during cell death (Bano et al., 2010; Ferrando-May et al., 2001). In the nucleus, AIF promotes DNA degradation into 20- to 50-kb fragments and, consequently, induces chromatin condensation required for cell disassembly (Susin et al., 1999). Notably, the apoptogenic effect of AIF does not require NADH and is not abrogated by inactivation of the FAD binding site (Miramar et al., 2001). Since AIF does not possess any obvious nuclease activity, it primarily functions to recruit and modulate the activity of a non-specific endonuclease that directly cleaves DNA. In the nematode Caenorhabditis elegans, the endonuclease G (EndoG) ortholog CPS-6 is released from the mitochondria and binds to the mitochondrial worm ortholog of AIF (WAH-1), mediating DNA degradation in the nucleus during programmed cell death (Wang et al., 2002; Parrish et al., 2001). This process has a few evolutionarily conserved aspects (Li et al., 2001), however both AIF and EndoG can stimulate chromatin fragmentation in mammalian cells independently of one another (Artus et al., 2010; Wang et al., 2016b). Notably, downregulation of wah-1 is epistatic to cps-6 loss-of-function, suggesting that CPS-6 and WAH-1 work in the same cell death pathway downstream of caspase CED-3. Mechanistically, it seems that reduced WAH-1 protein enhances CPS-6 dimerization in response to oxidative stress and cell death stimuli, hence controlling its nuclease activity and pro-apoptotic function (Lin et al., 2016). It is worth noting that recent in vitro as well as in vivo studies have challenged the contribution of EndoG in AIF-dependent DNA degradation. This is especially relevant in certain types of caspase-independent cell death, where AIF nuclear translocation is the commitment point in the cascade of events that lead to demise of injured cells. This form of cell demise (and recently designated parthanatos; reviewed in (Fatokun et al., 2014)) occurs as a consequence of the poly(ADP-ribose) polymerase (PARP) overactivation. PARPs are mainly, but not exclusively, in the nucleus and are multidomain enzymes with a wide spectrum of regulatory functions (reviewed in (Bai, 2015)). In response to DNA damage and formation of single-strand breaks, PARPs cleave NAD+ and modified acceptor proteins or synthesized branched poly(ADP-ribose) (PAR) polymers. These PAR polymers can work as signaling species (Andrabi et al., 2006) as well as inhibitors of enzymes involved in critical metabolic processes, such as glycolysis (Andrabi et al., 2014; Alano et al., 2010). As a downstream effect, PAR polymers may influence AIF processing and release through their direct binding at the C-terminal of the membrane-bound AIF (Wang et al., 2011). Alternatively, PARP-1 activity may exhaust NAD+ pool, which would indirectly dissipate the mitochondrial membrane potential and facilitate AIF release (Alano et al., 2010; Andrabi et al., 2014). In cells treated with DNA-alkylating agents or exposed to excitotoxic conditions, sustained PARP-1 activity produces excessive PAR polymers that trigger AIF release from the mitochondria through a yet unidentified mechanism (Yu et al., 2002). Recently, it was shown that the macrophage migration inhibitory factor (MIF), a previously unrecognized nuclease, binds cytosolic AIF and translocates to the nucleus, where it cleaves genomic DNA and causes cell death (Wang et al., 2016b). These intriguing findings give a different perspective on the processes downstream mitochondrial AIF release, despite that many of the underlying mechanisms remain elusive. In this regard, it would be interesting to determine whether cytosolic MIF contributes to DNA fragmentation in other types of cell injuries and if it is mutually exclusive with EndoG in these death paradigms. Moreover, it is tempting to speculate that, apart from shuttling from the cytosol to the nucleus, AIF may regulate MIF targeting and binding to distinct chromatin domains, for example DNA double-strand break enriched in phosphorylated H2AX (Artus et al., 2010). Thus, AIF contributes to cell demise in association with endonucleases that directly degrade chromosomal DNA.
Given its crucial role as a death effector in various cell demise paradigms, it is not surprising that the pro-death function of AIF contributes to a large spectrum of pathological conditions. In brain ischemia, neuronal damage following haemorrhagic or ischemic stroke occurs as a result of hypoxia, glucose deprivation, formation of free radicals and glutamate-mediated N-methyl-d-aspartate (NMDA) receptor overactivation, with the latter that ultimately causes massive intracellular Ca2+ overload and, as a consequence, triggers several neurotoxic programs (Bano and Ankarcrona, 2018; Moskowitz et al., 2010; Curcio et al., 2016; Lai et al., 2014; Berliocchi et al., 2005). It is likely that nuclear translocation of AIF can occur through alternative routes, with a population of injured neurons that initiate a classic apoptotic pathway culminating with caspase activation, while other cells die following DNA damage and consequent PARP-dependent parthanatos cascade. Despite the complexity of this scenario, ample evidence suggests that mitochondrial AIF release represents the conclusive step toward neuronal loss. In this regard, downregulation of AIF or its binding partners (i.e., CypA, MIF) involved in chromatinolysis significantly reduces the brain damage following ischemic insults (Zhu et al., 2007a; Zhu et al., 2007b; Culmsee et al., 2005; Wang et al., 2016b; Wang et al., 2011). Apart from this evident neuron-specific role, one emerging aspect is the contribution of AIF to hematopoietic development and immune response, primarily through its role in mitochondrial bioenergetics (Burguillos et al., 2011; Milasta et al., 2016; Cabon et al., 2018). In this context, it is tempting to speculate that AIF dysfunction may influence neurodegenerative processes in a non-cell-autonomous manner, by altering for example glia metabolism and inflammation. Finally, a large body of data suggests that AIF is prognostic of disease progression in certain tumors. Indeed, high AIF expression proved to be predictive of a longer overall survival in patients with B-cell lymphoma (Troutaud et al., 2010), but was not found to be prognostic in colorectal cancer (Perraud et al., 2011) or pediatric glioma (Smith et al., 2011). Similarly, AIF overexpression was observed in patients with chronic myeloid leukemia and associated with worse prognosis and diminished responsiveness to tyrosine kinase inhibitors (Quintas-Cardama et al., 2012). Certainly, more research efforts are warranted for a comprehensive understanding of the role of AIF in disease onset, progression and therapy response in humans.
3. AIF Contribution to Mitochondrial Respiration: Assembly Factor or Import Regulator?
Over the last two decades, the biological implications of AIF in modulating chromatin condensation and cell death have attracted the interest of many scientists in the field of apoptosis (Sevrioukova, 2011; Hangen et al., 2010; Fatokun et al., 2014). However, AIF's role as an evolutionarily conserved death executioner was not sufficient to explain some of the phenotypes observed in Aifm1 deficient mice. While a pioneering study in embryonic stem cells (ES) supported that genetic inactivation of Aifm1 alters apoptosis during differentiation and tissue morphogenesis (Joza et al., 2001), a different perspective on AIF function came from the characterization of the Harlequin mutant mouse model and the fact that some of the phenotypes could not be simply reconciled as a consequence of aberrant cell death programs. The Harlequin mutation derives from a proviral insertion in the X-linked Aifm1 locus, causing 80% reduction in Aifm1 mRNA expression (Klein et al., 2002). As a consequence of AIF deficiency, Harlequin mutant neurons display higher oxidative stress and increased accumulation of bromodeoxyuridine (BrdU) associated with aberrant expression of cell cycle regulators and progressive cerebellar degeneration. These breakthrough observations raised the possibility that AIF has additional functions beyond its established contribution to cell death processes. In support of this hypothesis, a series of studies in cell culture models pinpointed to a housekeeping role in mitochondrial respiration that is of key importance for cell survival (Vahsen et al., 2004). The authors observed that gene silencing of AIF by small interfering RNA (siRNA) in HeLa cells as well as genetic deletion of Aifm1 in mouse ES cells led to diminished oxygen consumption rate (OCR) and enhanced lactate production. Further investigation in cells and tissues lacking AIF unveiled that the primary mitochondrial defect is the loss of respiratory complex subunits, with complex I carrying the most severe damage (Vahsen et al., 2004). This line of evidence was subsequently confirmed in aif1 mutant (Δaif1) Saccharomyces cerevisiae, which displays altered growth in nonfermentable carbon sources (i.e., glycerol, lactate) compared to wild type strains. Equally important, Dm AIF knockout in Drosophila melanogaster larvae show impaired oxidative phosphorylation (OXPHOS) due to loss of complex I and complex IV activities, resulting in larval growth arrest and lethality (Joza et al., 2008). Similarly, wah-1 downregulation leads to decreased mitochondrial respiration, affects development and significantly reduces survival and fitness of the nematode C. elegans (Troulinaki et al., 2018). To understand these dual functions of AIF, it is important to disentangle its pro-apoptotic function from its homeostatic contribution in the mitochondria. This was achieved in AIF deficient cortical neurons, where aberrant mitochondrial morphology and cristae organization was rescued by overexpression of mutated AIF proteins that are anchored to the IMS and cannot translocate to the nucleus (Cheung et al., 2006). Together, these lines of evidence suggest that AIF is an evolutionarily conserved regulator of mitochondrial bioenergetics (Fig. 1A). As one possible explanation for this archaic metabolic function, it has been proposed that AIF might transfer equivalents to the electron transport chain and, as a consequence, would maintain the NAD+/NADH pool and/or the NADH-dependent proton pumping across the mitochondrial inner membrane (MIM) (Elguindy and Nakamaru-Ogiso, 2015). This is supported by the fact that AIF shares homology with the yeast Ndi1, a NADH-oxidoreductase that sustains mitochondrial membrane potential and can bypass complex I lesions (De Corby et al., 2007; Jafari et al., 2016). As an alternative mechanism, AIF may regulate mitochondrial function by participating in the assembly and/or stabilization of the respiratory complexes. Knowing that the absence of AIF causes only modest expression changes of nuclear-encoded mitochondrial OXPHOS genes (Vahsen et al., 2004), the post-transcriptional regulation of OXPHOS subunits seems a more likely scenario. Consistent with this view, AIF regulates the biogenesis of the electron transport chain through its physical interaction with coiled-coil-helix-coiled-coil-helix domain containing 4 (CHCHD4) (Meyer et al., 2015; Hangen et al., 2015). The IMS oxidoreductase CHCHD4 is the mammalian homolog of yeast Mia40, a crucial component of the mitochondrial import machinery that catalyzes the oxidation of small precursor proteins through the transient formation of disulphide bridges and the transfer of reducing equivalents to oxygen as a final acceptor (Banci et al., 2009; Terziyska et al., 2005; Chacinska et al., 2004). In a functional complex with the sulfhydryl oxidase Erv1, IMS soluble as well as membrane anchored Mia40 works as a docking receptor for incoming reduced intermediates that are retained in the IMS upon oxidation and proper conformational folding (Sideris and Tokatlidis, 2010). The discovery that AIF binds CHCHD4 has shed new light on an unrecognized function of AIF as a molecular partner of the mitochondrial import machinery. The physical interaction between the two proteins occurs at the N-terminal domain of CHCHD4 and is enhanced by the proper folding of AIF as well as the presence of NADH or NADPH rather than oxidized cofactors (Hangen et al., 2015). While AIF deficiency enhances the IMS degradation of CHCHD4, downregulation of CHCHD4 does not affect AIF expression. Importantly, overexpression of CHCHD4 rescues the loss of OXPHOS subunits in various cultured cells lacking AIF (Hangen et al., 2015; Meyer et al., 2015), hence linking AIF deficiency with the loss of oxidative folding in the IMS. This new line of evidence supports that AIF indirectly assists the assembly of the OXPHOS system, since it contributes to the proper folding of OXPHOS subunits and other factors in a CHCHD4-dependent fashion.
These studies provide a series of coherent answers to the role of AIF in mitochondrial bioenergetics. However, it is reasonable to think that loss of CHCHD4 impairs other mitochondrial processes, apart from ATP synthesis. In this regard, it has been recently shown that CHCHD4 regulates the heterodimerization, but not the mitochondrial import, of MICU1 and MICU2, two regulatory components of the mitochondrial calcium uniporter complex (Petrungaro et al., 2015; Perocchi et al., 2010). As a result, impaired association of MICU1/MICU2 heterodimers to the mitochondrial calcium uniporter (MCU) enhances mitochondrial Ca2+ uptake, with potential biological consequences indirectly linked to CHCHD4 function. On the other hand, AIF may influence mitochondrial function through the regulation of other cellular processes, given the broad number of AIF binding partners. For example, AIF interacts with the eukaryotic translation initiation factor 3 subunit g (eIF3g) and, as a result, inhibits protein synthesis during apoptosis (Kim et al., 2006) (Fig. 1B). Assuming that a small pool of AIF protein is available in the cytosol or bound to the OMM, it may be possible that AIF associates with eIF3g and finely tunes protein synthesis during normal cell growth. Similarly, cellular metabolism may be dependent on the redox status of AIF substrates, as in the case of phosphatase and tensin homolog (PTEN), which is maintained in a reduced form by AIF in the mitochondria and regulates Akt signaling pathway upon its redistribution to the cytosol and nucleus (Shen et al., 2015) (Fig. 1B). Given the recent development in the field, we may have merely scratched the surface of the many pleiotropic roles of AIF in cellular homeostasis and survival. In fact, dissecting adaptive or “pre-conditioning” responses secondary to a deficiency in mitochondrial respiration from “truly” anti-apoptotic effects in AIF-deficient mice in vivo (Cheung et al., 2006) remains a challenge. As an example, AIF release from mitochondria was observed in cardiomyocytes after salt-induced heart failure and ischemia/reperfusion injury (Siu et al., 2007; Kim et al., 2003), indicating a pro-death role of AIF in myocytes. However, muscle-specific AIF knockout mice develop skeletal muscle atrophy and cardiomyopathy secondary to complex I deficiency (Joza et al., 2005). Moreover, AIF deficiency results in higher sensitivity to oxidative stress and necrotic-like cell death in cardiomyocytes, as observed in Harlequin mutant mice, which are also more prone to heart damage following ischemia/reperfusion insults (van Empel et al., 2005). In the context of brain ischemia, Harlequin mutant mice show reduced infarct volumes after focal cerebral ischemia or hypoxia/ischemia-induced neuronal injury (Zhu et al., 2007b; Culmsee et al., 2005) but display systemic metabolic alterations such as mild acidosis, likely secondary to compensatory glycolysis activation. Forebrain-specific AIF null mice have been shown to activate several adaptive mechanisms in vivo, including increased glycolysis and mitochondrial biogenesis (Germain et al., 2013). These compensatory mechanisms were associated with AMP-activated protein kinase (AMPK) activation, and were abrogated by deletion of one allele of the upstream kinase LKB1 (Germain et al., 2013). Taken as a whole, these studies support the widespread view of AIF as a death factor with a critical role in cell metabolism and survival (Hangen et al., 2010).
4. AIF Mutations and their Link to Mitochondrial Diseases
Mitochondrial diseases are a heterogeneous group of clinically relevant metabolic disorders that manifest with a wide range of symptoms (Gorman et al., 2016; Koopman et al., 2016; Turnbull and Rustin, 2016). Although dysfunction of one specific organ has been observed in a few cases, mitochondrial diseases typically involve multiple tissues, with organs that are highly dependent on ATP the most severely affected. These syndromes have often childhood onset, although they can also occur with certain prevalence in adolescents as well as in adults (Koopman et al., 2016; Turnbull and Rustin, 2016; Gorman et al., 2016). The molecular etiology of these pathologies is very complex, since both nuclear and mitochondrial DNA genes contribute to the ~1500 proteins necessary for proper mitochondrial function (Calvo et al., 2016). As a consequence, mitochondrial diseases may have autosomal, X-linked and maternal inheritance, with rare sporadic cases that can occur especially in adults. Impaired mitochondrial bioenergetics and aberrant metabolic intermediates are the primary defects observed in patients and are generally considered useful biomarkers. Apart from these evident metabolic signatures, diagnosis relies mostly on next-generation sequencing, which has significantly contributed to the identification of more than 300 mutations linked to mitochondrial diseases with a variable spectrum of subtle and serious biological outcomes (Turnbull and Rustin, 2016; Koopman et al., 2016; DiMauro et al., 2013). Even more puzzling is the loose connection between disease expressions and genetic defects. For example, different pathogenic mutations on a single gene can cause a variety of conditions with a marked array of clinical presentations. On the other hand, loss-of-function mutations in different loci may be responsible for the same clinical manifestation, as in the case of Leigh syndrome, one of the most common forms of mitochondrial disease in children (Lake et al., 2016). Given the current gaps of knowledge, our understanding of the pathophysiology of these diseases is as yet not sufficient to provide relevant therapeutic alternatives (Koopman et al., 2016; Wang et al., 2016a; Viscomi et al., 2015).
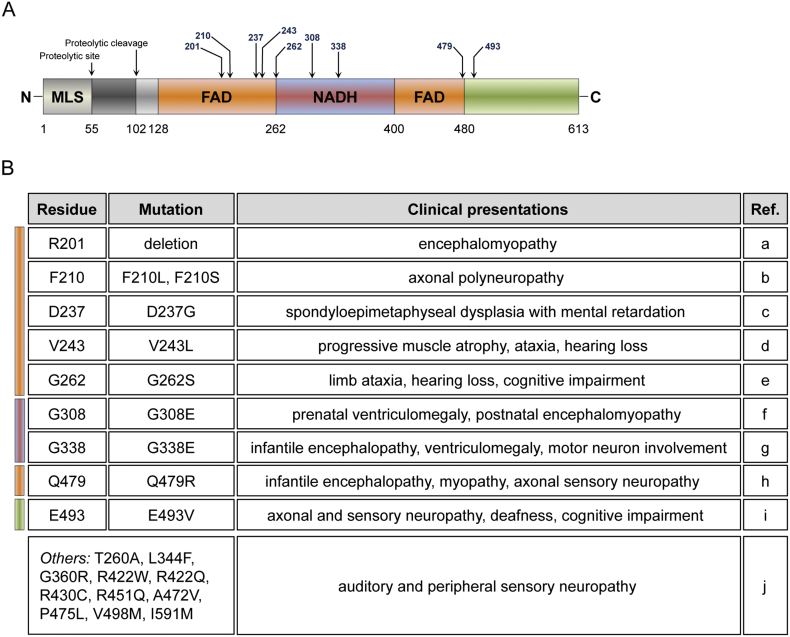
In this challenging context, over the last years an increasing number of clinical reports have described mutations in the AIFM1 gene that are causally linked to mitochondrial diseases. The first pathogenic mutation was identified in male infant patients born from monozygotic twin sisters (Ghezzi et al., 2010). Both patients displayed involuntary movements, abnormal MRI signal intensity in the subcortical basal ganglia, motor peripheral neuropathy and muscular atrophy. Along with increased lactate levels in the plasma, their muscle biopsies showed ragged-red fibers and a loss of cytochrome c oxidase (COX) staining, clearly indicating a substantial impairment of the OXPHOS system. In exon 5 of the X-linked AIFM1 gene, sequence analyses identified a trinucleotide deletion that causes the loss of the arginine 201 (R201 del) within one the FAD-binding domains (Fig. 2A). Based on in vitro data, it seems that the resulting AIF variant has a folding profile comparable to the wild type AIF, however AIF mutant polypeptide shows weaker FAD binding and, as a consequence, aberrant redox properties (Sevrioukova, 2016; Ghezzi et al., 2010). From this breakthrough discovery, later whole exome sequencing led to the identification of additional AIFM1 mutations responsible for pathological conditions (Fig. 2A-2B). In two brothers from healthy parents, a maternally inherited missense mutation in exon 9 of AIFM1 gene resulted in AIF variant with a glutamate replacing a glycine at residue 308 (G308E) (Berger et al., 2011). As the deletion of R201, G308E is a loss-of-function mutation that profoundly perturbs AIF redox activity and affinity for NADH (Sevrioukova, 2016). From the clinical standpoint, AIF (G308E) mutant has detrimental effect in humans. The two brothers exhibited enlarged brain lateral ventricles at prenatal stage and developed muscle weakness and atrophy within the first two months after birth. At the cellular level, immunohistochemistry of muscle biopsy confirmed cytochrome c oxidase deficiency accompanied by reduced complex I activity (Berger et al., 2011). Another missense mutation in the AIFM1 gene has been described in an Italian-American family, where a glutamate-to-valine substitution at residue 493 (E493V) caused hereditary Cowchock syndrome (Rinaldi et al., 2012), a form of Charcot-Marie-Tooth disease with childhood-onset. In a consistent manner, males of the family displayed recessive axonal motor and sensory neuropathy, progressive intellectual decline and muscle wasting, although without ragged-red fibers and substantial histochemical differences in cytochrome c oxidase as well as succinate dehydrogenase activity (Rinaldi et al., 2012). Compared to the other two mutations, AIF (E493V) variant induces only modest structural changes and does not cause major OXPHOS defects. Further studies in vitro and in vivo will help to define the molecular mechanisms underlying the pathophysiology of this AIF mutant. Since the original discovery of AIF (R201 del) variant, several additional reports have described AIFM1 missense nucleotide substitutions associated with a spectrum of clinical presentations, including slowly progressive mitochondrial encephalomyopathy, familial and sporadic auditory neuropathy, early-onset severe infantile motor neuron dysfunction and hypomyelinating leukodystrophy associated with neurodegeneration (Morton et al., 2017; Miyake et al., 2017; Hu et al., 2017; Diodato et al., 2016; Mierzewska et al., 2017; Ardissone et al., 2015; Kettwig et al., 2015; Sancho et al., 2017). Of the currently described mutations, twelve mutated residues (i.e., R201 del, F210 L and F210S, D237G, V243 L, T260A, G262S, R422W and R422Q, R430C, R451Q, A472V, P475L, Q479R) are in the two FAD-binding segments; four mutations (i.e., G308E, G338E, L344F, G360R) are within the NADH-binding domain and three mutations (i.e., E493V, V498 M, I591M) are located in the C-terminal region of the AIF protein (Fig. 2A-2B). While the increasing number of pathogenic AIF variants highlights the importance of AIF in human pathophysiology, it further indicates the high variability in terms of clinical manifestations that mutations in a single gene can induce. Definitely, this has important implications for the development of therapeutic interventions for mitochondrial diseases, especially if they have to cover such a heterogeneous group of pathologies. Of the recently identified AIF variants in patients, only six (i.e., R201 del, V243 L, G262S, G308E, G338E and E493V) have been studied in vitro for their chemical and biophysical properties (Sevrioukova, 2016; Rinaldi et al., 2012; Ghezzi et al., 2010). To date, little is known about the impact of mutant AIF proteins on mitochondrial oxidative phosphorylation, function and cell death pathways. In the future, it will be important to use novel experimental cellular and animal models that may help to unveil important molecular components contributing to the pathogenesis of AIF-related mitochondrial diseases.
Fig. 2.
Disease-causing mutations in the AIF protein. (A) Schematic drawing of human AIF protein and the annotated disease-causing mutations identified in patients. Mutated amino acids are indicated with arrows and numbers. In the scheme, it is also indicated the mitochondrial localization sequence (MLS) and the calpain cleavage site (proteolytic cleavage) of the membrane-bound protein. FAD and NADH represent the FAD-binding domains and NADH-binding motif, respectively. Numbers at the bottom correspond to the first and last amino acid of each domain. (B) Table reports the identified disease-causing mutations and describes the clinical presentations in patients. Colour-code rectangles indicate AIF domains. The table includes also a group of mutations (others) that have been identified in unrelated families of individuals with X-linked progressive auditory and peripheral sensory neuropathy. References (Ref.) are the following: (a) (Ghezzi et al., 2010); (b) (Hu et al., 2017; Sancho et al., 2017); (c) (Mierzewska et al., 2017; Miyake et al., 2017); (d) (Kettwig et al., 2015); (e) (Ardissone et al., 2015); (f) (Berger et al., 2011); (g) (Diodato et al., 2016); (h) (Morton et al., 2017); (i) (Rinaldi et al., 2012); (j) (Zong et al., 2015).
5. Conclusion and Future Questions
It is now emerging that many of the genes and proteins participating in mitochondrial apoptosis and cell death pathways may have acquired cell death-inducing properties as secondary, not primary functions. Cytochrome c is an essential component of the mitochondrial respiratory chain, responsible for shuttling of electrons from complex III to IV. Cytochrome c is also essential for apoptosome formation and caspase activation downstream of MOM permeabilization during apoptosis (Li et al., 1997), a process that evolved later in evolution. Omi/Htra2, EndoG and, as we discussed above, also AIF have key functions in maintaining mitochondrial function as well as cell death execution (Kilbride and Prehn, 2013; Hogg and Prehn, 2013). In the mitochondria, the oxidation of reduced substrates (i.e., NADH and FADH2) and the consequent transfer of equivalents to the electron transport chain create a proton gradient across the MIM that drives ATP production. In addition to their role in ATP production, mitochondria are multifunctional organelles that shape intracellular signaling pathways and produce intermediate metabolites (i.e., fatty acids, amino acids, nucleotides, hormones) relevant for biosynthetic processes (Zong et al., 2016). Considering the importance of mitochondria in cellular metabolism, the clinical variability of mitochondria-associated diseases, as exemplified by the spectrum of AIFM1 mutations, remains a fascinating matter of discussion. Given that these pathologies all exhibit altered OXPHOS as a unique common denominator, one would expect more consistent and overlapping signs, symptoms and age onset in patients. Surprisingly, that is not the case, complicating considerably the management of these conditions. As a possible explanation, cells may sustain diverse adaptations (e.g., metabolic and transcriptional changes) that confer resistance despite the high burden due to mitochondrial impairment. In some cases, these adaptations may induce a long-lasting epigenetic response that activates protective signaling pathways. Up to a certain threshold, mild mitochondrial deficiency triggers a transcriptional stress programme that induces beneficial metabolic changes and sustains lifespan extension in invertebrates as well as in mammals (Lin and Haynes, 2016; Wang and Hekimi, 2015; Troulinaki and Bano, 2012). Differences in the ability to activate protective and adaptive pathways may explain some of the heterogeneity observed. Other important considerations are the functional relationship between mitochondrial dysfunction, environmental and nutritional risk factors and the genomic landscape of each organism. In this regard, it may be that clinical manifestations occur as a result of synthetic lethality, when for example a loss-of-function mutation affecting mitochondria depends on the synergistic epistasis with other deleterious alleles, environmental stress and age to cause decreased fitness and, ultimately, pathology. In our opinion, AIFM1-related diseases fit in this scenario, since AIFM1 mutations are causally linked to a broad range of clinical conditions that cannot be explained simply because of decreased expression of AIF protein. In support of this hypothesis, aberrant biochemical properties of AIF variants do not fully correlate with the corresponding associated clinical outcomes (Sevrioukova, 2016). Harlequin mutant mice also display experimental variability across individual animals in terms of disease symptoms and course (Benit et al., 2008), which has been explained in part by the mixed genetic background as for other AIF deficient models (Pospisilik et al., 2007). We believe that the study of these newly identified disease-causing AIFM1 mutations may help us to gain insights into the physiological and pathological contribution of AIF. Moreover, it may improve our understanding on molecular processes that are common with other chronic and inherited mitochondria-associated diseases.
6. Search Strategy and Selection Criteria
PubMed and Scopus were used as the main search engines. References were searched using the following terms: apoptosis, Apoptosis-inducing factor (AIF), cell death, mitochondria, mitochondrial diseases. Only articles published in English were included in this manuscript.
Acknowledgments
Acknowledgements
We would like to thank Dr. Orla Watters for her constructive inputs. We are grateful to the DZNE and the Helmholtz Zukunftsthema “Aging and Metabolic Programming (AMPro)” who supported this work. The authors received support through the “Bundesministerium für Bildung und Forschung” (BMBF, Germany) and Science Foundation Ireland (14/JP-ND/B3077; 17/COEN/3474) under the aegis of the EU Joint Programme-Neurodegenerative Disease Research (JPND-www.jpnd.eu; Cellular Bioenergetics of Neurodegenerative Diseases, CeBioND) and the Centres of Excellence in Neurodegeneration Research Pathfinder III initiative. DB is members of the DFG Cluster of Excellence ImmunoSensation. JHMP is a Principal Investigator of the Science Foundation Ireland FutureNeuro Research Centre (16/RC/3948).
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- Alano C.C., Garnier P., Ying W., Higashi Y., Kauppinen T.M., Swanson R.A. NAD+ depletion is necessary and sufficient for poly(ADP-ribose) polymerase-1-mediated neuronal death. J. Neurosci. 2010;30:2967–2978. doi: 10.1523/JNEUROSCI.5552-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi S.A., Kim N.S., Yu S.W., Wang H., Koh D.W., Sasaki M., Klaus J.A., Otsuka T., Zhang Z., Koehler R.C., Hurn P.D., Poirier G.G., Dawson V.L., Dawson T.M. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi S.A., Umanah G.K., Chang C., Stevens D.A., Karuppagounder S.S., Gagne J.P., Poirier G.G., Dawson V.L., Dawson T.M. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc. Natl. Acad. Sci. U. S. A. 2014;111:10209–10214. doi: 10.1073/pnas.1405158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardissone A., Piscosquito G., Legati A., Langella T., Lamantea E., Garavaglia B., Salsano E., Farina L., Moroni I., Pareyson D., Ghezzi D. A slowly progressive mitochondrial encephalomyopathy widens the spectrum of AIFM1 disorders. Neurology. 2015;84:2193–2195. doi: 10.1212/WNL.0000000000001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus C., Boujrad H., Bouharrour A., Brunelle M.N., Hoos S., Yuste V.J., Lenormand P., Rousselle J.C., Namane A., England P., Lorenzo H.K., Susin S.A. AIF promotes chromatinolysis and caspase-independent programmed necrosis by interacting with histone H2AX. EMBO J. 2010;29:1585–1599. doi: 10.1038/emboj.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P. Biology of poly(ADP-ribose) polymerases: the factotums of cell maintenance. Mol. Cell. 2015;58:947–958. doi: 10.1016/j.molcel.2015.01.034. [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gallo A., Martinelli M., Sideris D.P., Katrakili N., Tokatlidis K. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 2009;16:198–206. doi: 10.1038/nsmb.1553. [DOI] [PubMed] [Google Scholar]
- Bano D., Ankarcrona M. Beyond the critical point: an overview of excitotoxicity, calcium overload and the downstream consequences. Neurosci. Lett. 2018;10(663):79–85. doi: 10.1016/j.neulet.2017.08.048. [DOI] [PubMed] [Google Scholar]
- Bano D., Dinsdale D., Cabrera-Socorro A., Maida S., Lambacher N., Mccoll B., Ferrando-May E., Hengartner M.O., Nicotera P. Alteration of the nuclear pore complex in Ca(2+)-mediated cell death. Cell Death Differ. 2010;17:119–133. doi: 10.1038/cdd.2009.112. [DOI] [PubMed] [Google Scholar]
- Benit P., Goncalves S., Dassa E.P., Briere J.J., Rustin P. The variability of the harlequin mouse phenotype resembles that of human mitochondrial-complex I-deficiency syndromes. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger I., Ben-Neriah Z., Dor-Wolman T., Shaag A., Saada A., Zenvirt S., Raas-Rothschild A., Nadjari M., Kaestner K.H., Elpeleg O. Early prenatal ventriculomegaly due to an AIFM1 mutation identified by linkage analysis and whole exome sequencing. Mol. Genet. Metab. 2011;104:517–520. doi: 10.1016/j.ymgme.2011.09.020. [DOI] [PubMed] [Google Scholar]
- Berliocchi L., Bano D., Nicotera P. Ca2+ signals and death programmes in neurons. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2005;360:2255–2258. doi: 10.1098/rstb.2005.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burguillos M.A., Hajji N., Englund E., Persson A., Cenci A.M., Machado A., Cano J., Joseph B., Venero J.L. Apoptosis-inducing factor mediates dopaminergic cell death in response to LPS-induced inflammatory stimulus: evidence in Parkinson's disease patients. Neurobiol. Dis. 2011;41:177–188. doi: 10.1016/j.nbd.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Cabon L., Bertaux A., Brunelle-Navas M.N., Nemazanyy I., Scourzic L., Delavallee L., Vela L., Baritaud M., Bouchet S., Lopez C., Quang Van V., Garbin K., Chateau D., Gilard F., Sarfati M., Mercher T., Bernard O.A., Susin S.A. AIF loss deregulates hematopoiesis and reveals different adaptive metabolic responses in bone marrow cells and thymocytes. Cell Death Differ. 2018 doi: 10.1038/s41418-017-0035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S.E., Clauser K.R., Mootha V.K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande C., Vahsen N., Kouranti I., Schmitt E., Daugas E., Spahr C., Luban J., Kroemer R.T., Giordanetto F., Garrido C., Penninger J.M., Kroemer G. AIF and cyclophilin a cooperate in apoptosis-associated chromatinolysis. Oncogene. 2004;23:1514–1521. doi: 10.1038/sj.onc.1207279. [DOI] [PubMed] [Google Scholar]
- Cao G., Xing J., Xiao X., Liou A.K., Gao Y., Yin X.M., Clark R.S., Graham S.H., Chen J. Critical role of calpain I in mitochondrial release of apoptosis-inducing factor in ischemic neuronal injury. J. Neurosci. 2007;27:9278–9293. doi: 10.1523/JNEUROSCI.2826-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuan Szklarz L.K., Schulze-Specking A., Truscott K.N., Guiard B., Meisinger C., Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung E.C., Joza N., Steenaart N.A., Mcclellan K.A., Neuspiel M., Mcnamara S., Maclaurin J.G., Rippstein P., Park D.S., Shore G.C., Mcbride H.M., Penninger J.M., Slack R.S. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. EMBO J. 2006;25:4061–4073. doi: 10.1038/sj.emboj.7601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churbanova I.Y., Sevrioukova I.F. Redox-dependent changes in molecular properties of mitochondrial apoptosis-inducing factor. J. Biol. Chem. 2008;283:5622–5631. doi: 10.1074/jbc.M709147200. [DOI] [PubMed] [Google Scholar]
- Culmsee C., Zhu C., Landshamer S., Becattini B., Wagner E., Pellecchia M., Blomgren K., Plesnila N. Apoptosis-inducing factor triggered by poly(ADP-ribose) polymerase and bid mediates neuronal cell death after oxygen-glucose deprivation and focal cerebral ischemia. J. Neurosci. 2005;25:10262–10272. doi: 10.1523/JNEUROSCI.2818-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M., Salazar I.L., Mele M., Canzoniero L.M., Duarte C.B. Calpains and neuronal damage in the ischemic brain: the swiss knife in synaptic injury. Prog. Neurobiol. 2016;143:1–35. doi: 10.1016/j.pneurobio.2016.06.001. [DOI] [PubMed] [Google Scholar]
- De Corby A., Gaskova D., Sayles L.C., Lemire B.D. Expression of Ndi1p, an alternative NADH:ubiquinone oxidoreductase, increases mitochondrial membrane potential in a C. Elegans model of mitochondrial disease. Biochim. Biophys. Acta. 2007;1767:1157–1163. doi: 10.1016/j.bbabio.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Dimauro S., Schon E.A., Carelli V., Hirano M. The clinical maze of mitochondrial neurology. Nat. Rev. Neurol. 2013;9:429–444. doi: 10.1038/nrneurol.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diodato D., Tasca G., Verrigni D., D'amico A., Rizza T., Tozzi G., Martinelli D., Verardo M., Invernizzi F., Nasca A., Bellacchio E., Ghezzi D., Piemonte F., Dionisi-Vici C., Carrozzo R., Bertini E. A novel AIFM1 mutation expands the phenotype to an infantile motor neuron disease. Eur. J. Hum. Genet. 2016;24:463–466. doi: 10.1038/ejhg.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elguindy M.M., Nakamaru-Ogiso E. Apoptosis-inducing factor (AIF) and its family member protein, AMID, are rotenone-sensitive NADH:ubiquinone oxidoreductases (NDH-2) J. Biol. Chem. 2015;290:20815–20826. doi: 10.1074/jbc.M115.641498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatokun A.A., Dawson V.L., Dawson T.M. Parthanatos: mitochondrial-linked mechanisms and therapeutic opportunities. Br. J. Pharmacol. 2014;171:2000–2016. doi: 10.1111/bph.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando-May E., Cordes V., Biller-Ckovric I., Mirkovic J., Gorlich D., Nicotera P. Caspases mediate nucleoporin cleavage, but not early redistribution of nuclear transport factors and modulation of nuclear permeability in apoptosis. Cell Death Differ. 2001;8:495–505. doi: 10.1038/sj.cdd.4400837. [DOI] [PubMed] [Google Scholar]
- Germain M., Nguyen A.P., Khacho M., Patten D.A., Screaton R.A., Park D.S., Slack R.S. LKB1-regulated adaptive mechanisms are essential for neuronal survival following mitochondrial dysfunction. Hum. Mol. Genet. 2013;22:952–962. doi: 10.1093/hmg/dds500. [DOI] [PubMed] [Google Scholar]
- Ghezzi D., Sevrioukova I., Invernizzi F., Lamperti C., Mora M., D'adamo P., Novara F., Zuffardi O., Uziel G., Zeviani M. Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor. Am. J. Hum. Genet. 2010;86:639–649. doi: 10.1016/j.ajhg.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman G.S., Chinnery P.F., Dimauro S., Hirano M., Koga Y., Mcfarland R., Suomalainen A., Thorburn D.R., Zeviani M., Turnbull D.M. Mitochondrial diseases. Nat. Rev. Dis. Prim. 2016;2:16080. doi: 10.1038/nrdp.2016.80. [DOI] [PubMed] [Google Scholar]
- Hangen E., Blomgren K., Benit P., Kroemer G., Modjtahedi N. Life with or without AIF. Trends Biochem. Sci. 2010;35:278–287. doi: 10.1016/j.tibs.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Hangen E., Feraud O., Lachkar S., Mou H., Doti N., Fimia G.M., Lam N.V., Zhu C., Godin I., Muller K., Chatzi A., Nuebel E., Ciccosanti F., Flamant S., Benit P., Perfettini J.L., Sauvat A., Bennaceur-Griscelli A., Ser-Le Roux K., Gonin P., Tokatlidis K., Rustin P., Piacentini M., Ruvo M., Blomgren K., Kroemer G., Modjtahedi N. Interaction between AIF and CHCHD4 regulates respiratory chain biogenesis. Mol. Cell. 2015;58:1001–1014. doi: 10.1016/j.molcel.2015.04.020. [DOI] [PubMed] [Google Scholar]
- Hogg M.C., Prehn J.H. Endonuclease-G and the pathways to dopaminergic neurodegeneration: a question of location? EMBO J. 2013;32:3014–3016. doi: 10.1038/emboj.2013.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Wang M., Castoro R., Simmons M., Dortch R., Yawn R., Li J. Eur J Neurol. 2017. A novel missense mutation in AIFM1 results in axonal polyneuropathy and misassembly of OXPHOS complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari G., Wasko B.M., Kaeberlein M., Crofts A.R. New functional and biophysical insights into the mitochondrial Rieske iron-sulfur protein from genetic suppressor analysis in C. elegans. Worm. 2016;5 doi: 10.1080/21624054.2016.1174803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joza N., Susin S.A., Daugas E., Stanford W.L., Cho S.K., Li C.Y., Sasaki T., Elia A.J., Cheng H.Y., Ravagnan L., Ferri K.F., Zamzami N., Wakeham A., Hakem R., Yoshida H., Kong Y.Y., Mak T.W., Zuniga-Pflucker J.C., Kroemer G., Penninger J.M. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410:549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- Joza N., Oudit G.Y., Brown D., Benit P., Kassiri Z., Vahsen N., Benoit L., Patel M.M., Nowikovsky K., Vassault A., Backx P.H., Wada T., Kroemer G., Rustin P., Penninger J.M. Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Mol. Cell. Biol. 2005;25:10261–10272. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joza N., Galindo K., Pospisilik J.A., Benit P., Rangachari M., Kanitz E.E., Nakashima Y., Neely G.G., Rustin P., Abrams J.M., Kroemer G., Penninger J.M. The molecular archaeology of a mitochondrial death effector: AIF in Drosophila. Cell Death Differ. 2008;15:1009–1018. doi: 10.1038/cdd.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettwig M., Schubach M., Zimmermann F.A., Klinge L., Mayr J.A., Biskup S., Sperl W., Gartner J., Huppke P. From ventriculomegaly to severe muscular atrophy: expansion of the clinical spectrum related to mutations in AIFM1. Mitochondrion. 2015;21C:12–18. doi: 10.1016/j.mito.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Kilbride S.M., Prehn J.H. Central roles of apoptotic proteins in mitochondrial function. Oncogene. 2013;32:2703–2711. doi: 10.1038/onc.2012.348. [DOI] [PubMed] [Google Scholar]
- Kim G.T., Chun Y.S., Park J.W., Kim M.S. Role of apoptosis-inducing factor in myocardial cell death by ischemia-reperfusion. Biochem. Biophys. Res. Commun. 2003;309:619–624. doi: 10.1016/j.bbrc.2003.08.045. [DOI] [PubMed] [Google Scholar]
- Kim J.T., Kim K.D., Song E.Y., Lee H.G., Kim J.W., Kim J.W., Chae S.K., Kim E., Lee M.S., Yang Y., Lim J.S. Apoptosis-inducing factor (AIF) inhibits protein synthesis by interacting with the eukaryotic translation initiation factor 3 subunit p44 (eIF3g) FEBS Lett. 2006;580:6375–6383. doi: 10.1016/j.febslet.2006.10.049. [DOI] [PubMed] [Google Scholar]
- Klein J.A., Longo-Guess C.M., Rossmann M.P., Seburn K.L., Hurd R.E., Frankel W.N., Bronson R.T., Ackerman S.L. The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature. 2002;419:367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- Koopman W.J., Beyrath J., Fung C.W., Koene S., Rodenburg R.J., Willems P.H., Smeitink J.A. Mitochondrial disorders in children: toward development of small-molecule treatment strategies. EMBO Mol. Med. 2016;8:311–327. doi: 10.15252/emmm.201506131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T.W., Zhang S., Wang Y.T. Excitotoxicity and stroke: identifying novel targets for neuroprotection. Prog. Neurobiol. 2014;115:157–188. doi: 10.1016/j.pneurobio.2013.11.006. [DOI] [PubMed] [Google Scholar]
- Lake N.J., Compton A.G., Rahman S., Thorburn D.R. Leigh syndrome: one disorder, more than 75 monogenic causes. Ann. Neurol. 2016;79:190–203. doi: 10.1002/ana.24551. [DOI] [PubMed] [Google Scholar]
- Li P., Nijhawan D., Budihardjo I., Srinivasula S.M., Ahmad M., Alnemri E.S., Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Li L.Y., Luo X., Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- Lin Y.F., Haynes C.M. Metabolism and the UPR(mt) Mol. Cell. 2016;61:677–682. doi: 10.1016/j.molcel.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.L., Nakagawa A., Skeen-Gaar R., Yang W.Z., Zhao P., Zhang Z., Ge X., Mitani S., Xue D., Yuan H.S. Oxidative stress impairs cell death by repressing the nuclease activity of mitochondrial endonuclease G. Cell. Rep. 2016;16:279–287. doi: 10.1016/j.celrep.2016.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mate M.J., Ortiz-Lombardia M., Boitel B., Haouz A., Tello D., Susin S.A., Penninger J., Kroemer G., Alzari P.M. The crystal structure of the mouse apoptosis-inducing factor AIF. Nat. Struct. Biol. 2002;9:442–446. doi: 10.1038/nsb793. [DOI] [PubMed] [Google Scholar]
- Meyer K., Buettner S., Ghezzi D., Zeviani M., Bano D., Nicotera P. Loss of apoptosis-inducing factor critically affects MIA40 function. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzewska H., Rydzanicz M., Bieganski T., Kosinska J., Mierzewska-Schmidt M., Lugowska A., Pollak A., Stawinski P., Walczak A., Kedra A., Obersztyn E., Szczepanik E., Ploski R. Spondyloepimetaphyseal dysplasia with neurodegeneration associated with AIFM1 mutation - a novel phenotype of the mitochondrial disease. Clin. Genet. 2017;91(1):30–37. doi: 10.1111/cge.12792. [DOI] [PubMed] [Google Scholar]
- Milasta S., Dillon C.P., Sturm O.E., Verbist K.C., Brewer T.L., Quarato G., Brown S.A., Frase S., Janke L.J., Perry S.S., Thomas P.G., Green D.R. Apoptosis-inducing-factor-dependent mitochondrial function is required for T cell but not B cell function. Immunity. 2016;44:88–102. doi: 10.1016/j.immuni.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miramar M.D., Costantini P., Ravagnan L., Saraiva L.M., Haouzi D., Brothers G., Penninger J.M., Peleato M.L., Kroemer G., Susin S.A. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J. Biol. Chem. 2001;276:16391–16398. doi: 10.1074/jbc.M010498200. [DOI] [PubMed] [Google Scholar]
- Miyake N., Wolf N.I., Cayami F.K., Crawford J., Bley A., Bulas D., Conant A., Bent S.J., Gripp K.W., Hahn A., Humphray S., Kimura-Ohba S., Kingsbury Z., Lajoie B.R., Lal D., Micha D., Pizzino A., Sinke R.J., Sival D., Stolte-Dijkstra I., Superti-Furga A., Ulrick N., Taft R.J., Ogata T., Ozono K., Matsumoto N., Neubauer B.A., Simons C., Vanderver A. X-linked hypomyelination with spondylometaphyseal dysplasia (H-SMD) associated with mutations in AIFM1. Neurogenetics. 2017;18(4):185–194. doi: 10.1007/s10048-017-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton S.U., Prabhu S.P., Lidov H.G., Shi J., Anselm I., Brownstein C.A., Bainbridge M.N., Beggs A.H., Vargas S.O., Agrawal P.B. AIFM1 mutation presenting with fatal encephalomyopathy and mitochondrial disease in an infant. Cold Spring Harb. Mol. Case Stud. 2017;3:a001560. doi: 10.1101/mcs.a001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz M.A., Lo E.H., Iadecola C. The science of stroke: mechanisms in search of treatments. Neuron. 2010;67:181–198. doi: 10.1016/j.neuron.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao L.S., Everley R.A., Marino S.M., Lo S.M., De Souza L.E., Gygi S.P., Gladyshev V.N. Mechanism-based proteomic screening identifies targets of thioredoxin-like proteins. J. Biol. Chem. 2015;290:5685–5695. doi: 10.1074/jbc.M114.597245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H., Ohsakaya S., Nagaura Z., Ishihara N., Mihara K. Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 2005;24:1375–1386. doi: 10.1038/sj.emboj.7600614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish J., Li L., Klotz K., Ledwich D., Wang X., Xue D. Mitochondrial endonuclease G is important for apoptosis in C. Elegans. Nature. 2001;412:90–94. doi: 10.1038/35083608. [DOI] [PubMed] [Google Scholar]
- Perocchi F., Gohil V.M., Girgis H.S., Bao X.R., Mccombs J.E., Palmer A.E., Mootha V.K. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010;467:291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perraud A., Akil H., Nouaille M., Petit D., Labrousse F., Jauberteau M.O., Mathonnet M. Expression of p53 and DR5 in normal and malignant tissues of colorectal cancer: correlation with advanced stages. Oncol. Rep. 2011;26:1091–1097. doi: 10.3892/or.2011.1404. [DOI] [PubMed] [Google Scholar]
- Petrungaro C., Zimmermann K.M., Kuttner V., Fischer M., Dengjel J., Bogeski I., Riemer J. The Ca(2+)-dependent release of the Mia40-induced MICU1-MICU2 Dimer from MCU regulates mitochondrial Ca(2+) uptake. Cell Metab. 2015;22:721–733. doi: 10.1016/j.cmet.2015.08.019. [DOI] [PubMed] [Google Scholar]
- Polster B.M., Basanez G., Etxebarria A., Hardwick J.M., Nicholls D.G. Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. J. Biol. Chem. 2005;280:6447–6454. doi: 10.1074/jbc.M413269200. [DOI] [PubMed] [Google Scholar]
- Pospisilik J.A., Knauf C., Joza N., Benit P., Orthofer M., Cani P.D., Ebersberger I., Nakashima T., Sarao R., Neely G., Esterbauer H., Kozlov A., Kahn C.R., Kroemer G., Rustin P., Burcelin R., Penninger J.M. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 2007;131:476–491. doi: 10.1016/j.cell.2007.08.047. [DOI] [PubMed] [Google Scholar]
- Quintas-Cardama A., Qiu Y.H., Post S.M., Zhang Y., Creighton C.J., Cortes J., Kornblau S.M. Reverse phase protein array profiling reveals distinct proteomic signatures associated with chronic myeloid leukemia progression and with chronic phase in the CD34-positive compartment. Cancer. 2012;118:5283–5292. doi: 10.1002/cncr.27568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravagnan L., Gurbuxani S., Susin S.A., Maisse C., Daugas E., Zamzami N., Mak T., Jaattela M., Penninger J.M., Garrido C., Kroemer G. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat. Cell Biol. 2001;3:839–843. doi: 10.1038/ncb0901-839. [DOI] [PubMed] [Google Scholar]
- Rinaldi C., Grunseich C., Sevrioukova I.F., Schindler A., Horkayne-Szakaly I., Lamperti C., Landoure G., Kennerson M.L., Burnett B.G., Bonnemann C., Biesecker L.G., Ghezzi D., Zeviani M., Fischbeck K.H. Cowchock syndrome is associated with a mutation in apoptosis-inducing factor. Am. J. Hum. Genet. 2012;91:1095–1102. doi: 10.1016/j.ajhg.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho P., Sanchez-Monteagudo A., Collado A., Marco-Marin C., Dominguez-Gonzalez C., Camacho A., Knecht E., Espinos C., Lupo V. A newly distal hereditary motor neuropathy caused by a rare AIFM1 mutation. Neurogenetics. 2017;18(4):245–250. doi: 10.1007/s10048-017-0524-6. [DOI] [PubMed] [Google Scholar]
- Sevrioukova I.F. Redox-linked conformational dynamics in apoptosis-inducing factor. J. Mol. Biol. 2009;390:924–938. doi: 10.1016/j.jmb.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova I.F. Apoptosis-inducing factor: structure, function, and redox regulation. Antioxid. Redox Signal. 2011;14:2545–2579. doi: 10.1089/ars.2010.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova I.F. Structure/function relations in AIFM1 variants associated with neurodegenerative disorders. J. Mol. Biol. 2016;428(18):3650–3665. doi: 10.1016/j.jmb.2016.05.004. [DOI] [PubMed] [Google Scholar]
- Shelar S.B., Kaminska K.K., Reddy S.A., Kumar D., Tan C.T., Yu V.C., Lu J., Holmgren A., Hagen T., Chew E.H. Thioredoxin-dependent regulation of AIF-mediated DNA damage. Free Radic. Biol. Med. 2015;87:125–136. doi: 10.1016/j.freeradbiomed.2015.06.029. [DOI] [PubMed] [Google Scholar]
- Shen S.M., Guo M., Xiong Z., Yu Y., Zhao X.Y., Zhang F.F., Chen G.Q. AIF inhibits tumor metastasis by protecting PTEN from oxidation. EMBO Rep. 2015;16:1563–1580. doi: 10.15252/embr.201540536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideris D.P., Tokatlidis K. Oxidative protein folding in the mitochondrial intermembrane space. Antioxid. Redox Signal. 2010;13:1189–1204. doi: 10.1089/ars.2010.3157. [DOI] [PubMed] [Google Scholar]
- Siu P.M., Bae S., Bodyak N., Rigor D.L., Kang P.M. Response of caspase-independent apoptotic factors to high salt diet-induced heart failure. J. Mol. Cell. Cardiol. 2007;42:678–686. doi: 10.1016/j.yjmcc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.J., Long A., Barrow J.H., Macarthur D.C., Coyle B., Grundy R.G., Children's, C. & Leukaemia Group Biological Studies, C Pediatric high-grade glioma: identification of poly(ADP-ribose) polymerase as a potential therapeutic target. Neuro-Oncology. 2011;13:1171–1177. doi: 10.1093/neuonc/nor115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin S.A., Zamzami N., Castedo M., Hirsch T., Marchetti P., Macho A., Daugas E., Geuskens M., Kroemer G. Bcl-2 inhibits the mitochondrial release of an apoptogenic protease. J. Exp. Med. 1996;184:1331–1341. doi: 10.1084/jem.184.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin S.A., Lorenzo H.K., Zamzami N., Marzo I., Snow B.E., Brothers G.M., Mangion J., Jacotot E., Costantini P., Loeffler M., Larochette N., Goodlett D.R., Aebersold R., Siderovski D.P., Penninger J.M., Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- Terziyska N., Lutz T., Kozany C., Mokranjac D., Mesecke N., Neupert W., Herrmann J.M., Hell K. Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions. FEBS Lett. 2005;579:179–184. doi: 10.1016/j.febslet.2004.11.072. [DOI] [PubMed] [Google Scholar]
- Troulinaki K., Bano D. Mitochondrial deficiency: a double-edged sword for aging and neurodegeneration. Front. Genet. 2012;3:244. doi: 10.3389/fgene.2012.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troulinaki K., Büttner S., Marsal Cots A., Maida S., Meyer K., Bertan F., Gioran A., Piazzesi A., Fornarelli A., Nicotera P., Bano D. WAH-1/AIF regulates mitochondrial oxidative phosphorylation in the nematode Caenorhabditis elegans. Cell. Death Disc. 2018 doi: 10.1038/s41420-017-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troutaud D., Petit B., Bellanger C., Marin B., Gourin-Chaury M.P., Petit D., Olivrie A., Feuillard J., Jauberteau M.O., Bordessoule D. Prognostic significance of BAD and AIF apoptotic pathways in diffuse large B-cell lymphoma. Clin. Lymphoma Myeloma Leuk. 2010;10:118–124. doi: 10.3816/CLML.2010.n.016. [DOI] [PubMed] [Google Scholar]
- Turnbull D.M., Rustin P. Genetic and biochemical intricacy shapes mitochondrial cytopathies. Neurobiol. Dis. 2016;92:55–63. doi: 10.1016/j.nbd.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Vahsen N., Cande C., Briere J.J., Benit P., Joza N., Larochette N., Mastroberardino P.G., Pequignot M.O., Casares N., Lazar V., Feraud O., Debili N., Wissing S., Engelhardt S., Madeo F., Piacentini M., Penninger J.M., Schagger H., Rustin P., Kroemer G. AIF deficiency compromises oxidative phosphorylation. EMBO J. 2004;23:4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Empel V.P., Bertrand A.T., Van Der Nagel R., Kostin S., Doevendans P.A., Crijns H.J., De Wit E., Sluiter W., Ackerman S.L., De Windt L.J. Downregulation of apoptosis-inducing factor in harlequin mutant mice sensitizes the myocardium to oxidative stress-related cell death and pressure overload-induced decompensation. Circ. Res. 2005;96:e92–e101. doi: 10.1161/01.RES.0000172081.30327.28. [DOI] [PubMed] [Google Scholar]
- Villanueva R., Ferreira P., Marcuello C., Uson A., Miramar M.D., Peleato M.L., Lostao A., Susin S.A., Medina M. Key residues regulating the reductase activity of the human mitochondrial apoptosis inducing factor. Biochemistry. 2015;54:5175–5184. doi: 10.1021/acs.biochem.5b00696. [DOI] [PubMed] [Google Scholar]
- Viscomi C., Bottani E., Zeviani M. Emerging concepts in the therapy of mitochondrial disease. Biochim. Biophys. Acta. 2015;1847:544–557. doi: 10.1016/j.bbabio.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Wang Y., Hekimi S. Mitochondrial dysfunction and longevity in animals: untangling the knot. Science. 2015;350:1204–1207. doi: 10.1126/science.aac4357. [DOI] [PubMed] [Google Scholar]
- Wang X., Yang C., Chai J., Shi Y., Xue D. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science. 2002;298:1587–1592. doi: 10.1126/science.1076194. [DOI] [PubMed] [Google Scholar]
- Wang Y., Kim N.S., Haince J.F., Kang H.C., David K.K., Andrabi S.A., Poirier G.G., Dawson V.L., Dawson T.M. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci. Signal. 2011;4 doi: 10.1126/scisignal.2000902. ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Karamanlidis G., Tian R. Novel targets for mitochondrial medicine. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aac7410. 326rv3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., An R., Umanah G.K., Park H., Nambiar K., Eacker S.M., Kim B., Bao L., Harraz M.M., Chang C., Chen R., Wang J.E., Kam T.I., Jeong J.S., Xie Z., Neifert S., Qian J., Andrabi S.A., Blackshaw S., Zhu H., Song H., Ming G.L., Dawson V.L., Dawson T.M. A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science. 2016:354. doi: 10.1126/science.aad6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.W., Wang H., Poitras M.F., Coombs C., Bowers W.J., Federoff H.J., Poirier G.G., Dawson T.M., Dawson V.L. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- Yu S.W., Wang Y., Frydenlund D.S., Ottersen O.P., Dawson V.L., Dawson T.M. Outer mitochondrial membrane localization of apoptosis-inducing factor: mechanistic implications for release. ASN Neuro. 2009:1. doi: 10.1042/AN20090046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste V.J., Moubarak R.S., Delettre C., Bras M., Sancho P., Robert N., D'alayer J., Susin S.A. Cysteine protease inhibition prevents mitochondrial apoptosis-inducing factor (AIF) release. Cell Death Differ. 2005;12:1445–1448. doi: 10.1038/sj.cdd.4401687. [DOI] [PubMed] [Google Scholar]
- Zhu C., Wang X., Deinum J., Huang Z., Gao J., Modjtahedi N., Neagu M.R., Nilsson M., Eriksson P.S., Hagberg H., Luban J., Kroemer G., Blomgren K. Cyclophilin a participates in the nuclear translocation of apoptosis-inducing factor in neurons after cerebral hypoxia-ischemia. J. Exp. Med. 2007;204:1741–1748. doi: 10.1084/jem.20070193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Wang X., Huang Z., Qiu L., Xu F., Vahsen N., Nilsson M., Eriksson P.S., Hagberg H., Culmsee C., Plesnila N., Kroemer G., Blomgren K. Apoptosis-inducing factor is a major contributor to neuronal loss induced by neonatal cerebral hypoxia-ischemia. Cell. Death Differ. 2007;14:775–784. doi: 10.1038/sj.cdd.4402053. [DOI] [PubMed] [Google Scholar]
- Zong L., Guan J., Ealy M., Zhang Q., Wang D., Wang H., Zhao Y., Shen Z., Campbell C.A., Wang F., Yang J., Sun W., Lan L., Ding D., Xie L., Qi Y., Lou X., Huang X., Shi Q., Chang S., Xiong W., Yin Z., Yu N., Zhao H., Wang J., Wang J., Salvi R.J., Petit C., Smith R.J., Wang Q. Mutations in apoptosis-inducing factor cause X-linked recessive auditory neuropathy spectrum disorder. J. Med. Genet. 2015;52:523–531. doi: 10.1136/jmedgenet-2014-102961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W.X., Rabinowitz J.D., White E. Mitochondria and Cancer. Mol. Cell. 2016;61:667–676. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]